Abstract
A gas chromatography–mass spectrometry approach was employed to evaluate the use of metabolite patterns to differentiate fruit from six commercially grown apple cultivars harvested in 2008. Principal component analysis (PCA) of apple fruit peel and flesh data indicated that individual cultivar replicates clustered together and were separated from all other cultivar samples. An independent metabolomics investigation with fruit harvested in 2003 confirmed the separate clustering of fruit from different cultivars. Further evidence for cultivar separation was obtained using a hierarchical clustering analysis. An evaluation of PCA component loadings revealed specific metabolite classes that contributed the most to each principal component, whereas a correlation analysis demonstrated that specific metabolites correlate directly with quality traits such as antioxidant activity, total phenolics, and total anthocyanins, which are important parameters in the selection of breeding germplasm. These data sets lay the foundation for elucidating the metabolic basis of commercially important fruit quality traits.
Keywords: apple, correlation analysis, hierarchical clustering analysis, metabolomics, principal component analysis, quality trait
INTRODUCTION
Apples (Malus × domestica Borkh.) are one of the most important tree fruit crops grown in midlatitude climate zones. Attractiveness to consumers is determined by numerous factors, including appearance, firmness, taste, and perceived health benefits.1 These quality traits are ultimately based on the metabolic makeup of a given fruit. For example, sweetness depends primarily on the amounts and composition of various sugars and sugar alcohols, acidity is determined mostly by the amounts and composition of organic acids, firmness is controlled by the properties of chemically complex cell wall polymers, color is determined by the quantity and composition of carotenoids and anthocyanins in the peel, and browning of fruit tissue after cutting is associated with the oxidation of hydroxycinnamic acids.2–7 Potential health benefits of apple fruit consumption are thought to result from the biological activities of numerous phytochemicals, which are particularly enriched in the peel and include, among others, polyphenolic metabolites and triter-penes.8
There are various targeted assays to quantitate specific classes of phytochemicals. The field of metabolomics, in contrast, aims at quantitating all metabolites in a given cell, tissue, organ, or organism.9 Although it is currently impossible to cover the entire metabolome of apple fruit in a single analysis, there has been significant progress in the development of technologies for the nontargeted analysis of relevant subsets of the metabolome. Whereas nuclear magnetic resonance spectroscopy is an excellent choice for a first-pass analysis of the more abundant metabolites, the majority of metabolomics studies with fruit employ a combination of a chromatographic separation coupled to a mass spectrometric (MS) detector.10,11 GC-MS-based metabolomics has been used successfully to assess phenotypic differences in apple and pear fruit subjected to postharvest treatments, such as cold or low-oxygen storage or irradiation with UV–vis light.12–16 Metabolomics using an LC-MS platform was employed recently to assess the authenticity of fruit juices, including those from apple.17 Metabolic changes during ripening have been studied by a metabolomic analysis of strawberry fruit.18,19 Attempts have also been made to correlate metabolomics data for apple fruit volatiles with quality traits and fungal infection.20–24 Füzfai et al.25 evaluated cultivar differences in sour cherry, apple, and ber fruits using GC-MS. Metabolomic imaging techniques have been used to determine the spatial gradients of metabolite accumulation in apple fruit.26
In this study we obtained nontargeted GC-MS-based metabolomics data from fruit (peel and flesh) of six commercially grown apple cultivars (‘Red Delicious’, ‘Golden Delicious’, ‘Cox’s Orange Pippin’, ‘Fuji’, ‘Gala’, and ‘Granny Smith’). Phenotypic variation (including metabolite diversity) is ultimately dependent on interactions between genotype (in this case the cultivar), the developmental stage of the fruit, and environmental factors (e.g., soil type, weather, and management practices).27 To ensure that conclusions drawn from the statistical analyses of metabolomics data obtained with various apple cultivars would be robust, we collected fruit from two harvest years (2003 and 2008) and different commercial orchards in Washington State. Using various statistical methods we demonstrate the utility of our metabolomics approach to differentiate apple cultivars and correlate specific metabolites with commercially relevant quality traits such as antioxidant activity, phenolic content, and anthocyanin amounts.
MATERIALS AND METHODS
Chemicals
All authentic standards were purchased from Sigma-Aldrich (St. Louis, MO, USA) except for maslinic acid, which was purchased from Cayman Chemical (Ann Arbor, MI, USA), and α-tocopherol, which was purchased from Matreya (Pleasant Gap, PA, USA).
Fruit Provenance and Harvesting
Cultivars ‘Golden Delicious’, ‘Red Delicious’, ‘Gala’, ‘Fuji’, and ‘Granny Smith’ were obtained from six commercial orchards in central Washington State (Yakima Valley and Columbia Basin) between August and October 2008. Also in 2008, ‘Cox’s Orange Pippin’ apples were obtained from three orchards located in western Washington and at Washington State University’s R. B. Tukey Horticulture Farm in Pullman, WA, USA. Fruit from the same cultivars (with the exception of ‘Cox’s Orange Pippin’) were also processed from a 2003 harvest (as a year-to-year control). In general, semiarid, mesoclimatic conditions prevailed among all orchards. Trees were mature (>5 years) and on standard Malling rootstocks. Similar crop loads were maintained to ensure marketable fruit size. Most soils in the region have a sandy-loam texture of similar fertility, and therefore, fertilizer practices do not vary appreciably for a given cultivar. Similar pest and pathogen pressures exist throughout the region, with codling moth and powdery mildew being the principal insect pest and pathogen, respectively. Thus, pest and disease management practices are similar across the region.
Fruit was harvested at commercial maturity based on standard measurements (see the Supporting Information) of flesh firmness, soluble solids concentration, starch–iodine index, and the nonred, surface background color. Peel tissue from undamaged fruits of similar size and color (separate biological replicates) was taken from the sun-exposed equator of the fruit with a razor blade, finely chopped, and immediately frozen in liquid nitrogen. Flesh tissue, without peel, was taken with a cork borer (15 mm diameter) at the sun-exposed equator of the fruit (separate biological replicates). We limited our tissue sampling to a cylinder of cortical tissue taken perpendicularly from the outer 10 mm of tissue. Given that this was <20% of the distance from the surface to the center of the fruit, our sampling procedure would be unlikely to have traversed the most conspicuous physiological boundary that separates the cortical from carpel tissue (i.e., core) of the fruit. Flesh and peel tissue samples (six biological and three technical replicates) were separately ground with a mortar and pestle in the presence of liquid nitrogen and the tissue homogenates stored at −80 °C until further analysis.
Fruit Maturity Measurements and Determination of Quality Trait Parameters
Apples (10–20) from each sample were weighed and their cortex firmness and soluble solids concentration measured. Fruit cortex firmness after skin removal was measured with a 11 mm diameter, stainless steel cylinder on an automated Güss Fruit Texture Analyzer (model GS-20, software version 5.0, Güss Manufacturing Ltd., Strand, South Africa) set at 16 mm/s forward speed, 1.11 N trigger threshold, 10 mm/s measuring speed, 8 mm measure distance, and 35 mm/s reverse speed. Soluble solids concentration was measured in juice expressed from a 10 mm thick × 15 mm diameter cylinder of fruit cortical tissue taken just under the skin with a digital refractometer (Atago model PR-101 Palette Refractometer, Atago Co., Ltd., Tokyo, Japan). Two measurements each of firmness and soluble solids were made at the equator on opposite sides of each fruit. The starch–iodine index for individual fruit from each sample orchard was evaluated using a previously reported method.28 Briefly, each fruit was cut in half across the equator, sprayed with a potassium iodide solution, and incubated for 5 min, and then its pattern of staining was compared to the Cornell starch–iodine index chart for apples.
Antioxidant activity of hydrophilic and lipophilic fractions was measured by the end-point 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS)/hydrogen peroxide/peroxidase (horseradish peroxidase, HRP, type VI-A) method with modifications.29,30 Specifically, 100 mg of powdered, frozen apple tissue was extracted in 700 μL of 50 mM MES (pH 6.0) and 700 μL of ethyl acetate, mixed for 30 s, and centrifuged at 13000g for 10 min at 4 °C. The organic (top) and aqueous (bottom) phases were separately transferred to new vials for measurements of lipophilic and hydrophilic antioxidant activities (LAA and HAA, respectively). For both fractions, 40 μL of 1 mM H2O2, 100 μL of 15 mM ABTS, and 10 μL of 3.3 U HRP were placed in 1 mL quartz cuvettes and gently shaken for 10 s, after which 830 μL of 50 mM phosphate buffer (pH 7.4) was added and mixed with a stir paddle. Absorbance was monitored at 734 nm on a UV–visible spectrophotometer until stable (<10 s), and then 20 μL (for HAA) or 40 μL (for LAA) extract was added, mixed with a stir paddle, and monitored at 734 nm until absorbance reached a minimum. HAA and LAA were calculated from the absorbance difference and expressed on the basis of Trolox equivalents from standard curves of 5 mM Trolox diluted in 50 mM MES buffer (pH 6.0) or 100% (v/v) ethyl acetate, respectively, and measured as described for the samples. HAA and LAA were summed to estimate total antioxidant activity (TAA) as Trolox equivalent antioxidant capacity (TEAC).
Total phenolic (TP) compounds were measured with the Folin–Ciocalteu phenol (F–C) reagent with modifications.31 Specifically, 1 mL of 80% (v/v) methanol was added to powdered, frozen apple tissue (100 mg of peel or 250 mg of flesh) in microcentrifuge tubes. Samples were mixed and allowed to extract for 1 h at room temperature and then overnight at −20 °C, followed by centrifugation at 14000g for 20 min at 4 °C. The supernatant was removed and extraction of the pellet was repeated twice as described above, with supernatants being combined after each extraction and then made up to 4 mL with 80% (v/v) methanol after the final extraction. TP compounds were assayed by adding 400 μL of sample extract into two 15 mL tubes containing 600 μL of 80% (v/v) methanol, 5 mL of 10% (v/v) F–C reagent, and either 4 mL of saturated Na2CO3 (75 g/L) or 4 mL of water. Tubes were thoroughly mixed and incubated at room temperature for 2 h. One milliliter aliquots from the sample tubes containing Na2CO3 or water were added to 1.5 mL plastic cuvettes, and the absorbance of each was measured at 760 nm in a UV–visible spectrophotometer. The concentration of phenolic compounds was determined by subtracting the absorbance of samples containing Na2CO3 from those not containing this chemical, and was quantified as gallic acid (3,4,5-trihydroxybenzoic acid) equivalents based on standard curves.
For the determination of total anthocyanin (TA) levels, powdered, frozen apple tissue (100 mg of peel and 250 mg of flesh) was extracted in 1 mL of 1% (v/v) HCl–methanol.32 After incubation for 24 h at −20 °C, sample tubes were centrifuged at 14000g for 10 min at 4°C. Extraction with HCl–methanol as above was repeated twice. Following centrifugation on day 4, supernatants were decanted into 15 mL plastic tubes and made up to 3 mL volumes with HCl–methanol. Anthocyanin concentrations were determined by measuring the absorbance at 530 nm, expressed as cyanidin-3-O-galactoside equivalents based on standard curves.
Tissue Processing and Extraction
Aliquots of 50–55 mg of frozen flesh or peel tissue were placed in a 1.5 mL Eppendorf vial, assigned a randomized numerical identifier, and cryogenically milled to a fine powder for 2 min (MM301, Retsch, Haan, Germany). To each sample was added 1 mL of 70% aqueous MeOH, and the mixture was vortexed for 1 min. Suspensions were flash-frozen in liquid nitrogen, thawed in a 34 °C water bath for 2 min, and incubated for 30 min at 23°C. Samples were centrifuged at 16000g for 5 min, and the resulting extract was removed and filtered through a 0.45 μm PTFE cartridge filter. Extracts were dried under reduced pressure (EZ-Bio, GeneVac, Gardiner, NY, USA), and the residue was suspended in 500 μL of H2O (containing 0.1 mg/mL hydroxyproline as an internal standard) and 500 μL of ethyl acetate (containing 0.1 mg/mL nonadecane as an internal standard) by vigorous mixing for 5 min. For improved phase separation samples were centrifuged at 3220g. Aliquots of the nonpolar ethyl acetate (300 μL) and polar aqueous (250 μL) phases were transferred to 2 mL borosilicate glass vials and the extracts dried separately. The derivatization method for metabolites was modified from that of Roessner et al.33 Briefly, 50 μL of pyridine containing 20 mg/mL methoxylamine and 0.5 mg/mL each of pentadecane and docosane (retention time standards) was added to each dried residue and the mixture incubated for 30 min at 47 °C. After the addition of 50 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA), samples were incubated for another 60 min at 47 °C. Each reaction mix was transferred to a deactivated glass insert, which was then placed back in the original 2 mL borosilicate glass vial. When samples were not analyzed directly, the dried residue was stored at −80 °C before derivatization.
GC-MS Data Acquisition and Peak Identification
Using a CombiPAL autosampler (LEAP Technologies, Carrboro, NC, USA), 1 μL sample aliquots were injected onto a fused silica column installed in a 6890N GC coupled to a 5973 mass selective detector (Agilent Technologies, Santa Clara, CA, USA). The GC was equipped with a split/splitless liner with deactivated glass wool packing. Samples were analyzed in the random order originally assigned for processing, and care was taken to ensure that samples were on the autosampler for no longer than 12 h before injection. The derivatized polar and nonpolar extracts of each sample were first injected in splitless mode (GC-MS-3 method). The polar extract was also injected with a 50:1 split to analyze the more abundant sugars and organic acids (GC-MS-4 method). The GC inlet temperature was set to 280 °C and the inlet pressure to 0.8 bar. The column used was a 30 m × 250 μm i.d., 0.25 μm, HP-5MS (J&W Scientific, Folsom, CA, USA) with a carrier gas flow of 1.3 mL/min. The oven temperature program for the GC-MS-3 method (in splitless mode) involved an initial temperature of 70 °C, a ramp of 2 °C/min to 220 °C, and a fast ramp of 4 °C/min to 300 °C, followed by a 14 min hold. The GC-MS-4 method (50:1 split injection) used an oven temperature program with an initial temperature of 70 °C, a ramp of 2 °C/min to 220 °C, a ramp of 4 °C/min to 275 °C, and a fast ramp of 30 °C/min to 300 °C, which was held for 5 min. All other conditions were the same as for the GC-MS-3 method. The MSD thermal transfer line was set to 280 °C for both methods, and the MSD settings were as follows: MS source at 230 °C, MS quadrupole at 150 °C, and scan range at m/z 50–550. Data acquisition, instrument control, and peak quantitation were performed using the ChemStation software package (rev E.01.00; Agilent Technologies, Santa Clara, CA, USA). Analytes were tentatively identified on the basis of a comparison of retention indices and mass spectra with those of authentic standards. For this purpose the NIST05 (National Institute of Standards and Technology, Gaithersburg, MD, USA) and the Fiehn GC-MS Metabolomics RTL Library (Agilent Technologies), as well as our own custom-built library, were employed. For quantitation purposes a unique target ion was selected for each peak. A custom report macro was generated to export raw data into a Microsoft EXCEL-compatible format.
Data Processing and Statistical Analysis
Raw signal abundance values were log10-transformed and adjusted for internal standard amount and original sample weight. Results for the three chromatographic runs per sample (nonpolar extract, splitless; polar extract, splitless; and polar extract, split) were collapsed into a single data set. Apple peel and flesh data sets were processed separately. Using the EXCEL add-on StatistiXL (StatistiXL, Nedlands, WA, Australia), a factorial analysis of variance (ANOVA) with Tukey post hoc tests was used to differentiate between the different apple varieties. Components with an ANOVA P value >0.05 were eliminated and the preprocessed data subjected to a principal component analysis (PCA) using StatistiXL. The case-wise PCA scores for the first three principal components were uploaded into the Systat 13 software package (Systat Software Inc., Chicago, IL, USA) for three-dimensional plotting. In addition, the ANOVA preprocessed data were range-scaled in Systat 13 before being analyzed by hierarchical clustering algorithms (complete linkage and Pearson’s correlation distance methods). To evaluate the correlation of fruit quality traits and apple fruit metabolite patterns, the values from the fruit quality measurements were log10-scaled and integrated into the peel and flesh metabolite data matrices. A Pearson correlation analysis was performed in Microsoft EXCEL using the StatistiXL add-in. Only correlations that passed a Sidak multiple testing correction (P value ≤ 2.16 × 10–4) were considered to be significant.34
RESULTS AND DISCUSSION
Analyte Extraction and Analysis
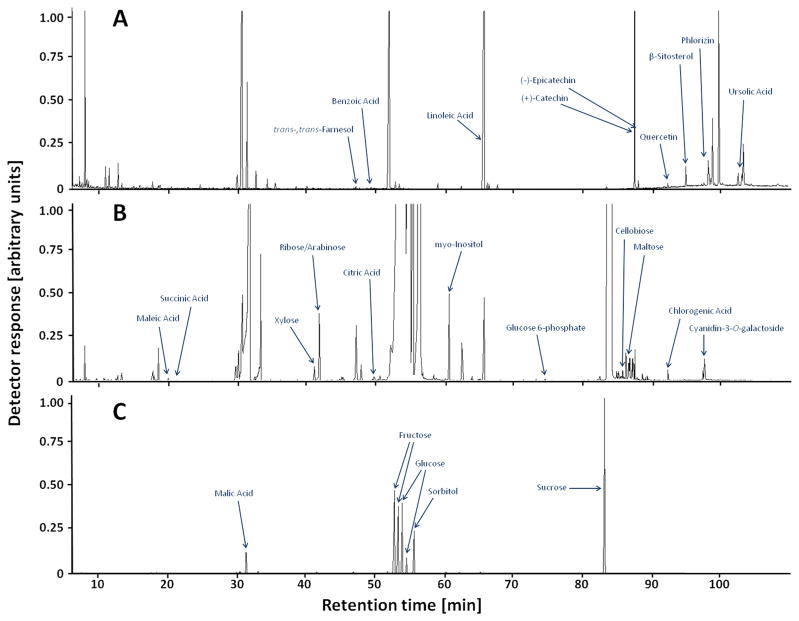
The GC-MS-based broad-spectrum profiling of plant metabolites involved in central metabolism was first reported for the analysis of Arabidopsis thaliana leaves and potato tubers.33,35 These pioneering methods were then adapted to interrogate the analogous subset of the metabolome in various other plants, organs, tissues, and specialized cell types.36 Metabolomics has been used to evaluate metabolic changes associated with postharvest storage conditions in apple.12–16 Our analysis deviated from these published tree fruit extraction protocols in several important aspects:37 (1) we used a protocol that involves the partitioning of analytes into a nonpolar organic phase and a polar aqueous phase; (2) a shallow oven ramp (2–4 °C/min) was employed to ensure adequate separation of analytes; (3) separate chromatographic runs were performed with a splitless injection to capture low abundance metabolites in both the nonpolar (Figure 1A) and polar (Figure 1B) extracts; and (4) high-abundance metabolites in the polar fraction were quantified after a 50:1 split injection (Figure 1C). We tracked a total of 248 mass tags, of which 75 were quantified from nonpolar extracts, 163 from polar extracts in splitless injection mode, and 10 from polar extracts in 50:1 split injection mode.
Figure 1.
Representative GC-MS total ion current chromatograms of apple peel extracts: (A) nonpolar fraction (splitless injection); (B) polar fraction (splitless); (C) polar fraction (split 50:1).
Our GC-MS analysis covered a broad range of different metabolite classes, some of which, for example, sugars (mono-and disaccharides), sugar alcohols, sugar phosphates, organic acids, fatty acids, amino acids, and sterols, are routinely detected in GC-MS. However, because of the relevance of polyphenolic metabolites in apple fruit, it is important to note that we also detected flavan-3-ols (e.g., (+)-catechin and (−)-epicatechin), dihydrochalcone glycosides (e.g., phloridzin), flavonols (e.g., quercetin), anthocyanidins (e.g., cyanidin), anthocyanins (e.g., cyanidin-3-O-glactoside), and phenolic acids (e.g., chlorogenic acid). Interestingly, although flavonoids and phenolic acids are most commonly analyzed by LC-MS, their TMS derivatives were readily detectable in our analysis, thus greatly enhancing the utility of the data, as these metabolites have been associated with antioxidant capacity and other potentially health-related traits.39 Various additional unknown metabolites with retention times and mass fragmentation patterns consistent with polyphenols or their glycosides were detected, which is further evidence for a relatively broad coverage of the metabolome.
Differentiating Tree Fruit Cultivars on the Basis of Principal Component Analysis
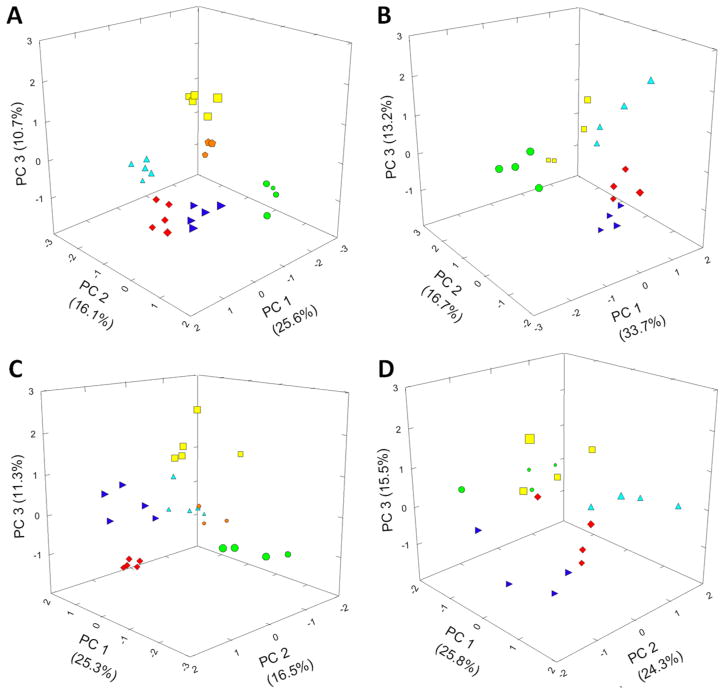
To reduce the complexity of our data set obtained with apple fruit harvested in 2008 (248 mass tags, 6 apple cultivars, 3–6 replicates per cultivar), normalized raw data (adjusted for sample weight and internal standard loading) were log10-transformed, ANOVA-filtered (P < 0.05), and then subjected to a PCA. In the PCA model for apple peel extracts the case-wise principal component (PC) scores for PC1, PC2, and PC3 were 25.6, 16.1, and 10.7% (total of 52.4%) of the variance, respectively (Figure 2A). Highly positive loadings on the PC1 axis were obtained for ‘Red Delicious’ and, to a lesser extent, the ‘Fuji’ cultivar, whereas the loading for ‘Granny Smith’ was strongly negative. The loadings on PC2 were highly positive for ‘Fuji’ and negative for ‘Gala’. ‘Golden Delicious’ had the most positive loading on PC3, followed by ‘Cox’s Orange Pippin’, whereas other cultivars had neutral to slightly negative values. The replicates of each individual apple cultivar had similar scores for all three PCs, although the scores differed significantly from cultivar to cultivar, thus leading to a separation of cultivars in a three-dimensional plot (Figure 2A). The standard deviation and standard error of PC scores for different apple cultivars were smaller than the difference between the means of PC scores between cultivars, indicating that the separation of cultivars by PCA was statistically significant. The same analysis was performed with data acquired from an apple fruit harvest in 2003 (with the exception that ‘Cox’s Orange Pippin’ was not included). Once again, a separation of all apple cultivars was observed in a three-dimensional PCA plot (Figure 2B). The loadings for each individual cultivar were different from those obtained with fruit harvested in 2008, which could be a reflection of environmental effects on metabolite abundances. In this context, it is important to note that average maximum temperatures during the growing season (May–September) were markedly different between 2003 and 2008, with maximum temperatures in 2003 being well above average (i.e., +2.5–4.0 °C, except for May) and temperatures closer to average in 2008 (±1.5 °C).40 The fact that a statistical analysis enabled us to still capture cultivar-to-cultivar variation demonstrates the robustness of our GC-MS metabolomics approach for cultivar differentiation.
Figure 2.
Principal component analysis of metabolomics data from extracts of (A) apple fruit peel (harvest 2008), (B) apple fruit peel (harvest 2003), (C) apple fruit flesh (harvest 2008), and (D) apple fruit flesh (harvest 2003). PC, principal component. Symbols are used to indicate loadings for different apple cultivars: ‘Golden Delicious’, yellow squares; ‘Gala’, light blue triangles; ‘Red Delicious’, red diamonds; ‘Fuji’, dark blue triangles; ‘Granny Smith’, green circles; ‘Cox’s Orange Pippin’, orange pentagons.
The PCA model for metabolites in apple flesh extracts gave case-wise PC scores of 25.3% (PC1), 16.5% (PC2), and 11.3% (PC3) (total of 53.1%) of the total variance (Figure 2C). Four cultivars were positively loaded on PC1 (in order of loading scores: ‘Gala’, ‘Cox’s Orange Pippin’, ‘Fuji’, and ‘Red Delicious’), and ‘Golden Delicious’ had a slightly negative score, whereas the loading of ‘Granny Smith’ was strongly negative. On PC2 ‘Red Delicious’ and ‘Fuji’ had positive loadings, whereas the loadings of ‘Golden Delicious’, ‘Gala’, and ‘Cox’s Orange Pippin’ were negative. ‘Golden Delicious’ was the only cultivar with a highly positive loading on PC3 and ‘Red Delicious’ the only one with a significantly negative loading. A three-dimensional PCA plot (Figure 2C) based on apple flesh extract data indicated that the six cultivars were generally distinguishable (lower variance between replicates than cultivar-to-cultivar variance), but the separation was not as clear-cut as with peel data. In particular, ‘Gala’ and ‘Cox’s Orange Pippin’ occupied partially overlapping positions in a three-dimensional plot. A general separation of cultivars in a PCA plot was also observed with fruit flesh from a 2003 harvest (Figure 2D), which again validates the robustness of our analytical platform. One of the reasons the flesh samples did not cluster as tightly as those obtained from peel might be that the flesh profile is dominated by sugars and sugar alcohols (as discussed in more detail below), and these types of metabolites are fairly similar in the flesh of fruit from different cultivars. In contrast, many of the natural products of peel accumulate differentially in the cultivars investigated here (more details below), and this natural variation is reflected in our data sets and statistical analyses (see the Supporting Information).
Dissecting the Contribution of Individual Mass Tags to Principal Components
A further examination of the component loadings of apple PCA models by extracting eigenvectors enabled an assessment of which individual mass tags were associated with cultivar differences. The positive loadings of peel extracts from ‘Red Delicious’ and ‘Fuji’ on PC1 correlated with positive component loadings of triterpene/sterols, flavonoids, phenolic acids, stearic acid, anthocyanins, and several carbohydrates. The strongly negative loading of ‘Granny Smith’ on PC1 corresponded to negative component loadings for three organic acids (malic, maleic, and citric acid) and several carbohydrates. The positive PC2 loading of ‘Fuji’ peel extracts correlated with positive component loadings of various carbohydrates (including glucose and sorbitol). The strong positive loading of ‘Golden Delicious’ peel extracts on PC3 corresponded to positive component loadings of myo-inositol, several carbohydrates (some with unknown structure), a putative sugar phosphate, and succinic acid. Consistently negative loadings on PC3 were detected for ‘Red Delicious’ and ‘Granny Smith’, which correlated with the relative abundances of unsaturated fatty acids (oleic and linoleic acid) and several carbohydrates (see Supporting Information).
The positive PC1 loadings of flesh extract data for several cultivars (‘Gala’, ‘Cox’s Orange Pippin’, ‘Fuji’, and ‘Red Delicious’) correlated mostly with positive component loadings of various mono- and disaccharides, whereas organic acids (malic and maleic acid) and several carbohydrates contributed to the negative loading of ‘Granny Smith’. Positive PC2 loadings of flesh (‘Red Delicious’ and ‘Fuji’) were associated primarily with positive component loadings of several mono-and disaccharides as well as (−)-epicatechin. The moderately negative PC2 loadings of all other cultivars were in accordance with negative component loadings of myo-inositol, various organic acids (malic, succinic, and maleic acid), and two amino acids (valine and pyroglutamic acid, which is formed from glutamate and glutamine during derivatization). As in peel, the highly positive PC3 loading of ‘Golden Delicious’ was associated with high levels of myo-inositol and several carbohydrates. The component loadings contributing to the negative PC3 loading of ‘Red Delicious’ were enriched in flavan-3-ols ((+)-catechin and (−)-epicatechin) and various carbohydrates. The fairly strong correlation of specific metabolite classes with cultivar loadings indicated that our metabolomics data should be tested for correlations between metabolites and fruit traits (see Supporting Information).
Correlating Metabolomics Data with Genetic Relationships among Apple Cultivars
To further interrogate our apple metabolomics data, we used, in addition to PCA, hierarchical clustering analysis (HCA) as an complementary multivariate analysis approach (Figure 3). A heat map (see the Supporting Information) was used as a graphical representation of metabolomics data, where the individual values contained in a matrix of cultivar replicates (x-axis) and metabolite abundances (y-axis) are represented with a color code. For the peel data set, the replicates for each cultivar were placed in one clade, indicating that the biological variation among fruits of one cultivar harvested at different locations was less than the cultivar-to-cultivar variance. The clustering of peel samples based on metabolite patterns sorted apple cultivars into two major clades (clade 1, ‘Fuji’, ‘Red Delicious’, ‘Gala’, ‘Golden Delicious’, and ‘Cox’s Orange Pippin’; clade 2, ‘Granny Smith’; Figure 3A). Clade 1 split into two primary subclades: the first subclade contained ‘Fuji’, ‘Red Delicious’, and ‘Gala’, whereas the second subclade consisted of ‘Golden Delicious’ and ‘Cox’s Orange Pippin’ (Figure 3A).
Figure 3.
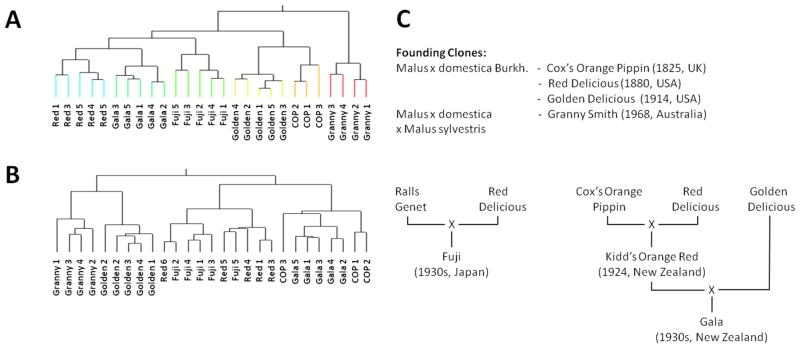
Hierarchical clustering of metabolomics data from extracts of (A) apple fruit peel and (B) apple fruit flesh. (C) Outline of the genetic relationships between apple cultivars.
The clustering of flesh metabolomics data was not as clear-cut. A full separation of cultivars was not achieved in all cases (i.e., one ‘Red Delicious’ sample clustered with ‘Fuji’ and one ‘Cox’s Orange pippin’ sample clustered with ‘Gala’). Another difference was that flesh metabolite accumulation patterns of ‘Golden Delicious’ were more closely related to those of ‘Granny Smith’ than those of other cultivars (Figure 3B). The chromatograms of flesh samples were very much dominated by common mono- and disaccharides, which may be one of the reasons for an insufficient separation of genotypes in this analysis, as discussed before in the context of PCA.
Interestingly, the clustering of peel generally reflected the genetic relationships among apple cultivars (Figure 3C). ‘Red Delicious’, ‘Golden Delicious’, and ‘Cox’s Orange Pippin’ belong to the founding clones.41 ‘Fuji’ was generated by crossing ‘Red Delicious’ with ‘Ralls Genet’ (not included in this analysis), and one would expect ‘Fuji’ to be positioned near ‘Red Delicious’ (true for both peel and flesh clustering). The ancestry of ‘Gala’ is more complicated. A cross of ‘Red Delicious’ and ‘Cox’s Orange Pippin’ generated ‘Kidd’s Orange Red’. ‘Gala’ emerged from a further cross of ‘Kidd’s Orange Red’ with ‘Golden Delicious’.41 In both the peel and flesh clusters, ‘Gala’ grouped with ‘Red Delicious’. In our HCA of peel samples ‘Granny Smith’ formed an outgroup (Figure 3C). This could be due to differences in origins between apple cultivars. In terms of its genetics, ‘Granny Smith’ was propagated from a chance seedling of unknown identity, but it has been suggested that Malus sylvestris may have been a significant contributor to its gene pool.42
Correlating Metabolomics Data with Fruit Quality Traits
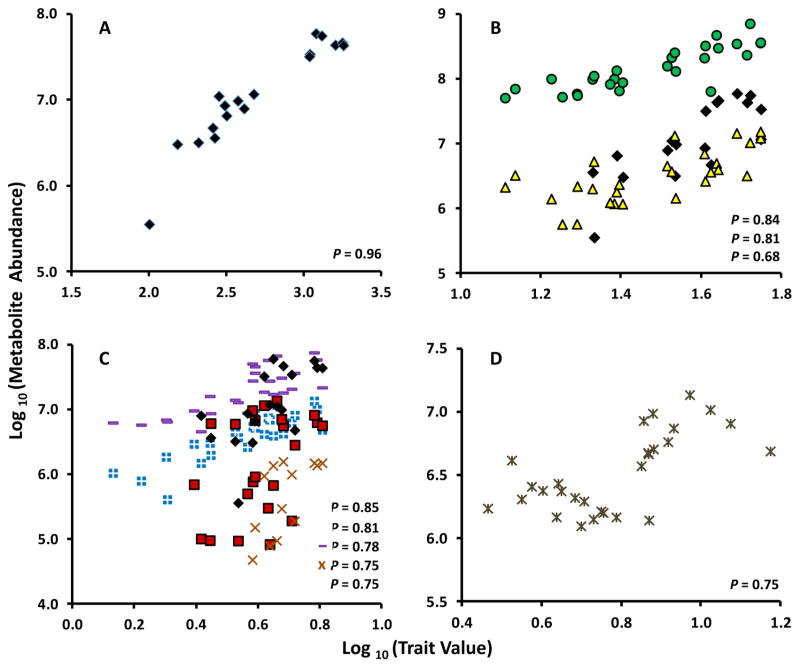
To obtain further insights into the metabolic basis of fruit quality traits, we correlated metabolomic data with traditional fruit quality measurements for peel and flesh samples. An almost perfect correction (R (Pearson coefficient) = 0.96, P value = 2.7 × 10−15) was observed between total anthocyanin (TA) amounts and a peak at 96.7 min in the hydrophilic extracts. The main anthocyanins in apple cultivars with reddish fruit skin had been described as cyanidin glycosides.43,44 Because volatile derivatives of anthocyanins and other polyphenols are amenable to GC-MS analyses,45 we purchased authentic standards of candidate metabolites and, on the basis of retention time and mass fragmentation patterns, were able to annotate several additional mass tags. Importantly, the above-mentioned mass tag was identified as the trimethylsilylated derivative of cyanidin-3-O-galactoside (Figure 4A).
Figure 4.
Correlation analysis of quality traits and the relative amounts of specific metabolites in apple fruit: (A) total anthocyanins (TA) in peel; (B) total antioxidant activity (TAA) in peel; (C) total phenolics (TP) in peel; and (D) TAA in flesh. Cyanidin-3-O-galactoside, black diamonds; (−)-epicatechin, green circles; (+)-catechin, yellow triangles; mass tag BML-GCMS3-E1-34.10-348.1, light blue squares with cross; mass tag BML-GCMS3-H1-87.68-525.3, red squares; mass tag BML-GCMS3-E1-32.46-348.1, purple hyphen; mass tag BML-GCMS3-H1-100.34-394.1, orange ×; and myo-inositol in flesh (D).
The total antioxidant activity (TAA) measurements for peel tissue correlated with two flavan-3-ols, (−)-epicatechin (R = 0.84, P = 4.28 × 10−8) and (+)-catechin (R = 0.68, P = 9.9 × 10−5) (Figure 4A), cyanidin-3-O-galactoside (R = 0.83, P = 6.62 × 10−8), and several as yet unidentified metabolites with mass spectra characteristic for polyphenols. (−)-Epicatechin and (+)-catechin contributed most prominently to the antioxidant activity of lipophilic metabolites, whereas cyani-din-3-O-galactoside was the most highly correlated hydrophilic metabolite. The correlation of TAA with flavan-3-ols is in agreement with prior work.46 The contribution of cyanidin-3-O-galactoside to TAA is particularly relevant in apple varieties with reddish fruit skin (most prominently in ‘Red Delicious’). We calculated a weaker correlation between TAA and the flavonoids quercetin (P = 0.005) and phloridzin (P = 0.02) than reported previously.46 It is conceivable that these discrepancies are due to the fact that many of the phenolic glycosides in our analysis have not yet been identified. The fact that the majority of peaks remain unidentified is a general challenge with metabolomics approaches and limits their utility with regard to linking metabolites with traits. To project the field beyond the proof-of-concept stage, it will thus be important to increase the coverage and identification of analytes in metabolomics analyses. Common data repositories and searchable databases are a first step in this direction.47,48
The values for peel total phenolic (TP) content correlated with a larger number of metabolites, some of which do not correspond to known metabolites in our in-house database. The list of true polyphenolic metabolites with high correlation coefficients contained cyanidin-3-O-galactoside (R = 0.75, P = 6.35 × 10−6) and (−)-epicatechin (R = 0.67, P = 1.5 × 10−4) (Figure 4B). However, several other highly correlated metabolites had retention times and mass spectra indicative of sugars (mono-and disaccharides). The very high concentrations of sugars in apple fruit obviously interfered with the Folin–Ciocalteu assay for phenolic content, which is a well-known problem with this method.31
In the apple fruit flesh data set, there was a much weaker correlation between TA, TAA, and TP with any of the detected metabolites, which is probably the result of lower flesh antho-cyanin levels in the selected cultivars (compared to peel) and the generally low antioxidant activity of flesh.49,50 The only metabolite that correlated significantly with TAA was myo-inositol (R = 0.64, P = 1.9 × 10−4) (Figure 4D). Although myo-inositol has not been demonstrated to exert strong antioxidant activities, it is the biosynthetic precursor for ascorbic acid,51 a well-established antioxidant of apple flesh.49 In this context it is important to note that ascorbic acid was not reported in this study as the quantitation was unreliable due to a coelution with the much more abundant monosaccharide fructose.
The LC-MS analysis of fruit extracts from a segregating F1 population of a cross between apple cultivars ‘Prima’ and ‘Fiesta’ was used previously to integrate metabolite data with the existing reference genetic linkage map, thus allowing the mapping of metabolite quantitative trait loci (mQTL).52,53 The same approach was used successfully to map mQTLs for apple fruit volatiles.54 Apple linkage group 16 was found to contain an mQTL hotspot for various phenylpropanoid metabolites, which correlated with the location of genes putatively involved in the phenylpropanoid pathway.53 This knowledge can now be used in marker-assisted breeding, which should be greatly aided by the available apple genome sequence.55 The here presented GC-MS approach could be used in an analogous fashion to discover markers for breeding, with a particular emphasis on quality traits. It is conceivable that potentially health-related traits in tree fruit could become marketable in the future, and a good understanding of the metabolic basis of these traits would thus be desirable. We focused our current study on TA, TAA, and TP, but it is obvious that metabolite composition also underlies other important fruit quality traits such as firmness, soluble solids (Brix), starch–iodine index, and postharvest stability,56 and approaches such as the one described here have the potential to contribute to unraveling the metabolic networks regulating these traits.
Supplementary Material
Acknowledgments
Funding
This study was supported by the Emerging Research Issues for Washington Agriculture Initiative of the Agricultural Research Center at Washington State University.
We acknowledge the valuable assistance of Dr. Ines Hanrahan of the Washington Tree Fruit Research Commission for obtaining the commercial apple cultivars and Leonard Fuller of the Seattle Tree Fruit Society for obtaining the ‘Cox’s Orange Pippin’ apples. We also acknowledge the laboratory assistance of Margaret Collier and John Nguyen.
Footnotes
The authors declare no competing financial interest.
Figure of hierarchical clustering analysis and tables of processed metabolomics data and statistical analyses. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Harker FR, Gunson FA, Jaeger SR. The case for fruit quality: an interpretive review of consumer attitudes, and preferences for apples. Postharvest Biol Technol. 2003;28:333–347. [Google Scholar]
- 2.Harker FR, Marsh KB, Young H, Murray SH, Gunson FA, Walker SB. Sensory interpretation of instrumental measurements 2: sweet and acid taste of apple fruit. Postharvest Biol Technol. 2002;24:241–250. [Google Scholar]
- 3.Merzlyak MN, Melø TB, Naqvi KR. Effect of anthocyanins, carotenoids, and flavonols on chlorophyll fluorescence excitation spetra in apple fruit: signature analysis, assessment, modeling, and relevance to photoprotection. J Exp Bot. 2008;59:349–359. doi: 10.1093/jxb/erm316. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Gao H, Zhao L, Liao X, Chen F, Wang Z, Hu X. Chemical compositional characterization of some apple cultivars. Food Chem. 2007;103:88–93. [Google Scholar]
- 5.Oraguzie N, Alspach P, Volz R, Whitworth C, Ranatunga C, Weskett R, Harker R. Postharvest assessment of fruit quality parameters in apple using both instruments and an expert panel. Postharvest Biol Technol. 2009;52:279–287. [Google Scholar]
- 6.Costa F, Peace CP, Stella S, Serra S, Musacchi S, Bazzani M, Sansavini S, van der Weg E. QTL dynamics for fruit firmness and softening around an ethylene-dependent polygalacturonase gene in apple (Malus × domestica Borkh.) J Exp Bot. 2010;61:3029–3039. doi: 10.1093/jxb/erq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaki K, Saito E, Taniguchi K, Joshita K, Murata M. Role of chlorogenic acid quinone and interaction of chlorogenic acid quinone and catechins in the enzymatic browning of apple. Biosci, Biotechnol Biochem. 2001;75:829–832. doi: 10.1271/bbb.100444. [DOI] [PubMed] [Google Scholar]
- 8.Boyer J, Liu RH. Apple phytochemicals and their health benefits. Nutr J. 2004;3:5. doi: 10.1186/1475-2891-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tohge T, Mettler T, Arrivault S, Carroll AJ, Stitt M, Fernie AR. From models to crop species: caveats and solutions for translational metabolomics. Front Plant Physiol. 2011;2:61. doi: 10.3389/fpls.2011.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward JL, Baker JM, Beale MH. Recent applications of NMR spectroscopy in plant metabolomics. FEBS J. 2007;274:1126–1131. doi: 10.1111/j.1742-4658.2007.05675.x. [DOI] [PubMed] [Google Scholar]
- 11.Lei Z, Huhman D, Sumner LW. Mass spectrometry strategies in metabolomics. J Biol Chem. 2011;286:25435–25442. doi: 10.1074/jbc.R111.238691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudell DR, Mattheis JP, Curry EA. Prestorage ultraviolet-white light irradiation alters apple peel metabolome. J Agric Food Chem. 2008;56:1138–1147. doi: 10.1021/jf072540m. [DOI] [PubMed] [Google Scholar]
- 13.Rudell DR, Mattheis JP, Hertog MLATM. Metabolomic change precedes apple superficial scald symptoms. J Agric Food Chem. 2009;57:8459–8466. doi: 10.1021/jf901571g. [DOI] [PubMed] [Google Scholar]
- 14.Rudell DR, Mattheis JP. Superficial scald development and related metabolism is modified by postharvest light irradiation. Postharvest Biol Technol. 2009;51:174–182. [Google Scholar]
- 15.Lee J, Mattheis JP, Rudell DR. Antioxidant treatment alters metabolism associated with internal browning in ‘Braeburn’ apples during controlled atmosphere storage. Postharvest Biol Technol. 2012;68:32–42. [Google Scholar]
- 16.Pedreschi R, Franck C, Lammertyn J, Erban A, Kopka J, Hertog M, Verlinden B, Nicolaï B. Metabolic profiling of ‘Conference’ pears under low oxygen stress. Postharvest Biol Technol. 2009;51:123–130. [Google Scholar]
- 17.Vaclavic L, Schreiber A, Lacina O, Cajka T, Hajslova J. Liquid chromatography-mass spectrometry-based metabolomics for authenticity assessment of fruit juices. Metabolomics. 2012 doi: 10.1007/s11306–011–0371–7. in press [Epub ahead of print] [DOI] [Google Scholar]
- 18.Fait A, Hanhineva K, Beleggia R, Dai N, Rogachev I, Nikiforova VJ, Fernie AR, Aharoni A. Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol. 2008;148:730–750. doi: 10.1104/pp.108.120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Wang X, Yu O, Tang J, Gu X. Metabolic profiling of strawberry (Fragaria × ananassa Duch.) during fruit development and maturation. J Exp Bot. 2010;62:1103–1118. doi: 10.1093/jxb/erq343. [DOI] [PubMed] [Google Scholar]
- 20.Song J, Gardner BD, Holland JF, Beaudry RM. Rapid analysis of volatile flavor compounds in apple fruit using SPME and GC/time-of-flight mass spectrometry. J Agric Food Chem. 1997;45:1801–1807. [Google Scholar]
- 21.Zini E, Biasioli F, Gasperi F, Mott D, Aprea E, Märk TD, Patocchi A, Gessler C, Komjanc M. QTL mapping of volatile compounds in ripe apples detected by proton transfer reaction-mass spectrometry. Euphytica. 2005;145:269–279. [Google Scholar]
- 22.Rowan DD, Hunt MB, Alspach PA, Whitworth CJ, Oraguzie NC. Heritability and genetic and phenotypic correlations of apple (Malus × domestica) fruit volatiles in a genetically diverse breeding population. J Agric Food Chem. 2009;57:7944–7952. doi: 10.1021/jf901359r. [DOI] [PubMed] [Google Scholar]
- 23.Schmarr HG, Bernhardt J. Profiling analysis of volatile compounds from fruits using comprehensive two-dimensional gas chromatography and image processing techniques. J Chromatogr, A. 2010;1217:565–574. doi: 10.1016/j.chroma.2009.11.063. [DOI] [PubMed] [Google Scholar]
- 24.Vikram A, Prithiviraj B, Hamzehzarghani H, Kushalappa AC. Volatile metabolite profiling to discriminate diseases of McIntosh apple inoculated with fungal pathogens. J Sci Food Agric. 2004;84:1333–1340. [Google Scholar]
- 25.Füzfai Z, Katona ZF, Kovács E, Molnár-Perl I. Simultaneous identification and quantitation of sugar, sugar alcohol, and carboxylic acid contents of sour cherry, apple, and ber fruits, as their trimethylsilyl derivatives, by gas chromatography-mass spectrometry. J Agric Food Chem. 2004;52:7444–7452. doi: 10.1021/jf040118p. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Cha S, Yeung ES. Colloidal graphite-assisted laser desorption/ionization MS and MSn of small molecules. 2. Direct profiling and MS imaging of small metabolites from fruits. Anal Chem. 2007;79:6575–6584. doi: 10.1021/ac0706170. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Bink MCAM, Volz RK, Bus VGM, Chagné D. Towards genomic selection in apple (Malus × domestica Borkh.) breeding programmes: prospects, challenges and strategies. Tree Genet Genomes. 2012;8:1–14. [Google Scholar]
- 28.Blanpied GD, Silsby KJ. Information Bulletin. Cornell Cooperative Extension; Ithaca, NY: 1992. Predicting the harvest date window for apples; p. 221. [Google Scholar]
- 29.Arnao MB, Cano A, Acosta M. The hydrophilic and lipophilic contribution to total antioxidant activity. Food Chem. 2001;73:239–244. [Google Scholar]
- 30.Cano A, Hernández-Ruíz J, García-Cánovas F, Acosta M, Arnao MB. An end-point method for estimation of the total antioxidant activity in plant material. Phytochem Anal. 1998;9:196–202. [Google Scholar]
- 31.Singleton VL, Orthofer R, Lamuela-Raventós RM. Analysis of total phenols and other oxidation substrates antioxidants by means of Folin-Ciocalteu reagent. Methods Enzymol. 1999;299:152–178. [Google Scholar]
- 32.Siegelman HW, Hendricks SB. Photo-control of alcohol, aldehyde and anthocyanin production in apple skin. Plant Physiol. 1958;33:409–413. doi: 10.1104/pp.33.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Simultaneous analysis of metabolites in potato tuber by chromatography-mass spectrometry. Plant J. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- 34.Abdi H. Bonferroni and Sidak corrections for multiple comparisons. In: Salkind NJ, editor. Encyclopedia of Measurement and Statistics. Sage; Thousand Oaks, CA: 2007. pp. 103–107. [Google Scholar]
- 35.Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 36.Saito K, Matsuda F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol. 2010;61:463–489. doi: 10.1146/annurev.arplant.043008.092035. [DOI] [PubMed] [Google Scholar]
- 37.Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 2003;133:84–99. doi: 10.1104/pp.103.023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.André C, Castanheira I, Cruz JM, Paseiro P, Sanches-Silva A. Analytical strategies to evaluate antioxidants in food: a review. Trends Food Sci Technol. 2010;21:229–246. [Google Scholar]
- 39.Hyson DA. A comprehensive review of apples and apple components and their relationship to human health. Adv Nutr. 2011;2:408–420. doi: 10.3945/an.111.000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.PRISM Climate Group. [accessed Aug 5, 2012]; http://www.prism.oregonstate.edu.
- 41.Noiton DAM, Alspach PA. Founding clones, inbreeding, coancestry, and status of modern apple cultivars. J Am Soc Hortic Sci. 1996;121:773–782. [Google Scholar]
- 42.Harrison N, Harrison R. On the evolutionary history of the domesticated apple. Nat Genet. 2011;43:1043–1044. doi: 10.1038/ng.935. [DOI] [PubMed] [Google Scholar]
- 43.Duncan IJ, Dustman RB. The anthocyanin pigment of the Winesap apple. J Am Chem Soc. 1936;58:1511–1514. [Google Scholar]
- 44.Lu Y, Foo LY. Identification and quantitation of major polyphenols in apple pomace. Food Chem. 1997;59:187–194. [Google Scholar]
- 45.Füzfei Z, Molnár-Perl I. Gas chromatographic-mass spectro-metric fragmentation study of flavonoids as their trimethylsilyl derivatives: analysis of flavonoids, sugars, carboxylic and amino acids in model systems and in Citrus fruits. J Chromatogr A. 2007;1149:88–101. doi: 10.1016/j.chroma.2007.01.060. [DOI] [PubMed] [Google Scholar]
- 46.Lee KW, Kim YJ, Kim DO, Lee HJ, Lee CY. Major phenolics in apple and their contribution to the total antioxidant capacity. J Agric Food Chem. 2003;51:6516–6520. doi: 10.1021/jf034475w. [DOI] [PubMed] [Google Scholar]
- 47.Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, Fernie AR, Kopka J. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005;579:1332–1337. doi: 10.1016/j.febslet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 48.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 49.Wolfe K, Wu X, Liu RH. Antioxidant activity of apple peels. J Agric Food Chem. 2003;51:609–614. doi: 10.1021/jf020782a. [DOI] [PubMed] [Google Scholar]
- 50.Tsao R, Yang R, Young JC, Zhu H. Polyphenolic profiles in eight apple cultivars using high-performance liquid chromatography (HPLC) J Agric Food Chem. 2003;51:6347–6353. doi: 10.1021/jf0346298. [DOI] [PubMed] [Google Scholar]
- 51.Torabinejad J, Donahue JL, Gunesekera BN, Allen-Daniels MJ, Gillaspy GE. VTC4 is a bifunctional enzyme that affects myo-inositol and ascorbate biosynthesis. Plant Physiol. 2009;150:951–961. doi: 10.1104/pp.108.135129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maliepaard C, Alston FH, van Arkel G, Brown LM, Chevreau E, Dunemann F, Evans KM, Gardiner S, Guilford P, van Heusden AW, Laurens JJF, Lynn JR, Manganaris AG, den Nijs APM, Periam N, Rikkerink E, Roche P, Ryder C, Sansavini S, Schmidt H, Tartarini S, Verhaegh JJ, Vrielink-van Ginkel M, King GJ. Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theor Appl Genet. 1998;97:60–73. [Google Scholar]
- 53.Khan SA, Chibon PY, de Vos RC, Schipper BA, Walraven E, Beekwilder J, van Dijk T, Finkers R, Visser RG, van de Weg EW, Bovy A, Cestaro A, Velasco R, Jacobsen E, Schouten HJ. Genetic analysis of metabolites in apple fruits indicates an mQTL hotspot for phenolic compounds on linkage group 16. J Exp Bot. 2012;63:2895–2908. doi: 10.1093/jxb/err464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunemann F, Ulrich D, Boudichevaskaia A, Grafe C, Weber WE, Dunemann F, Ulrich D, Boudichevaskaia A, Grafe C, Weber WE. QTL mapping of aroma compounds analysed by headspace solid-phase microextraction gas chromatography in the apple progeny ‘Discovery’ × ‘Prima’. Mol Breed. 2009;23:501–521. [Google Scholar]
- 55.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagné D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouzé P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner R, Gardiner E, Skolnick SE, Egholm M, Van de Peer M, Salamini Y, Viola FR. The genome of the domesticated apple (Malus × domestica Borkh.) Nat Genet. 2010;42:833–841. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 56.Hanhineva K, Aharoni A. Metabolomics in fruit development. In: Jain SM, Brar DS, editors. Molecular Techniques in Crop Improvement. Springer Science; Berlin, Germany: 2010. pp. 675–693. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.