Abstract
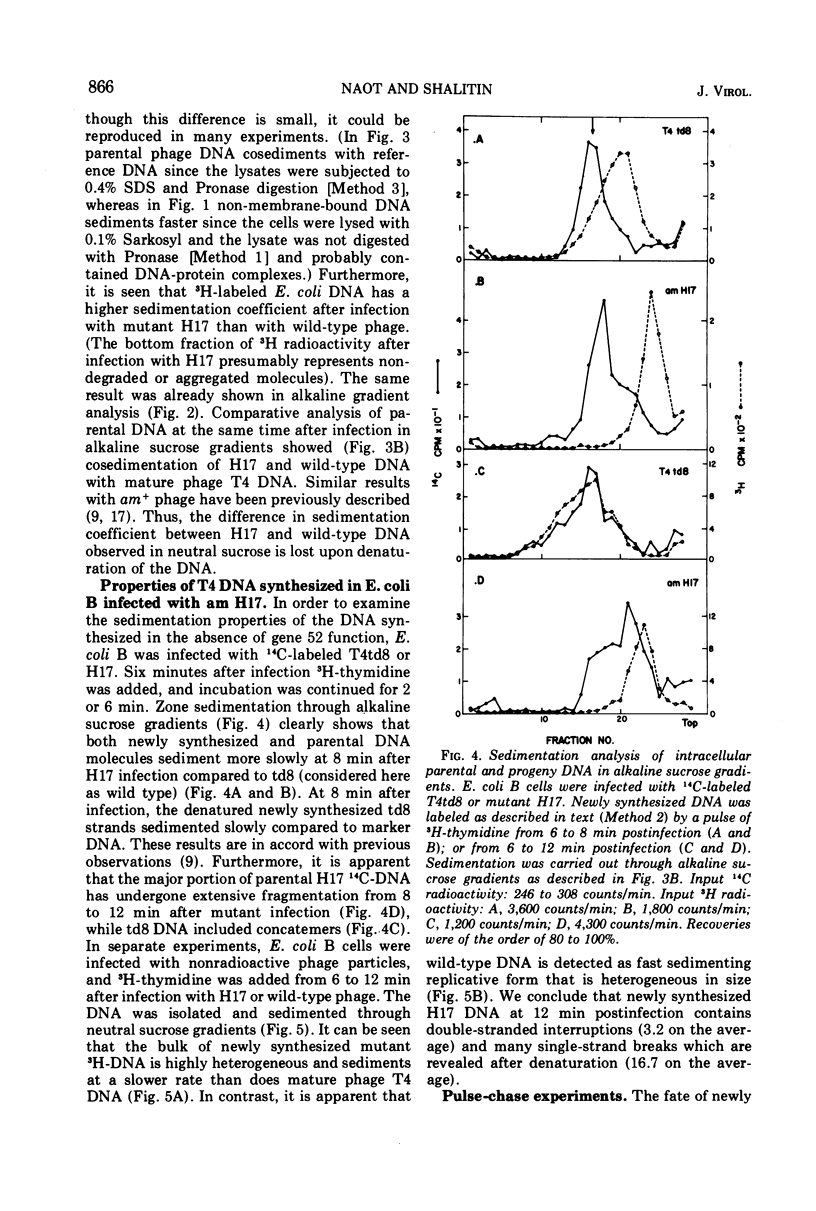
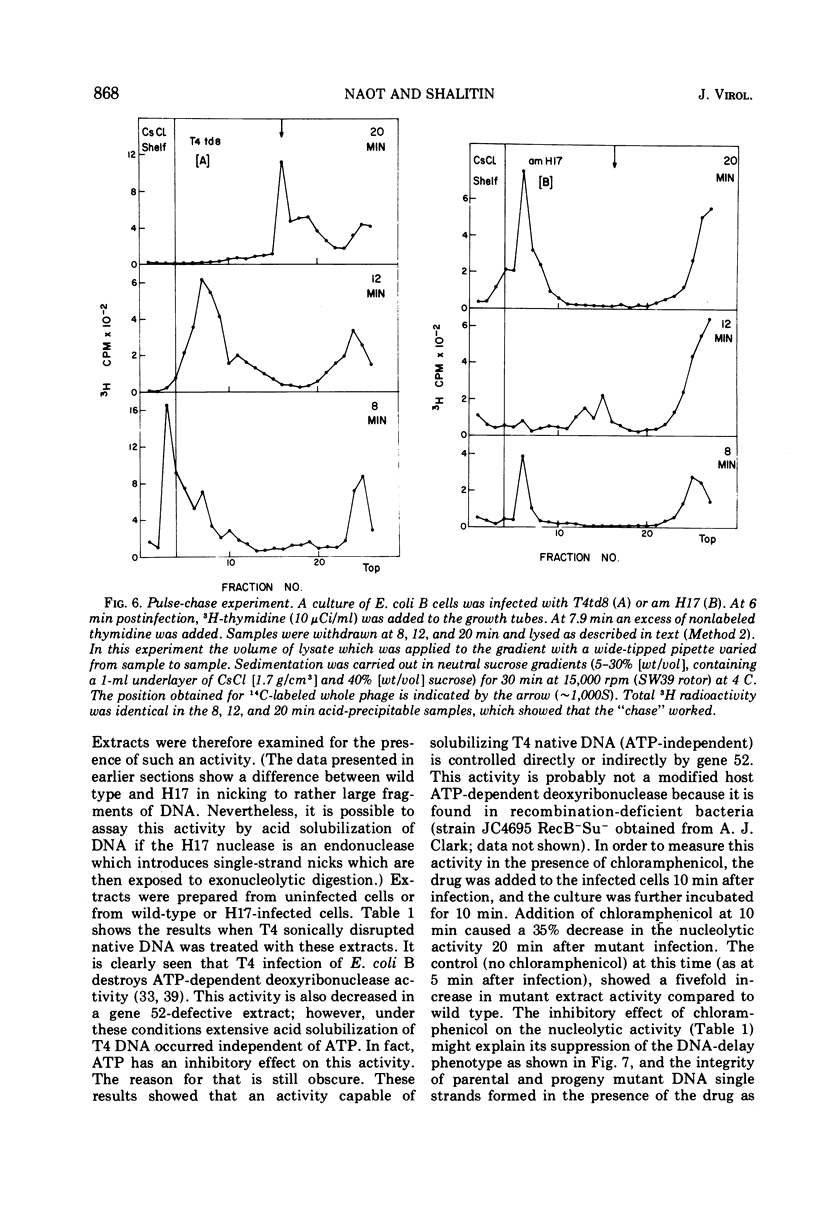
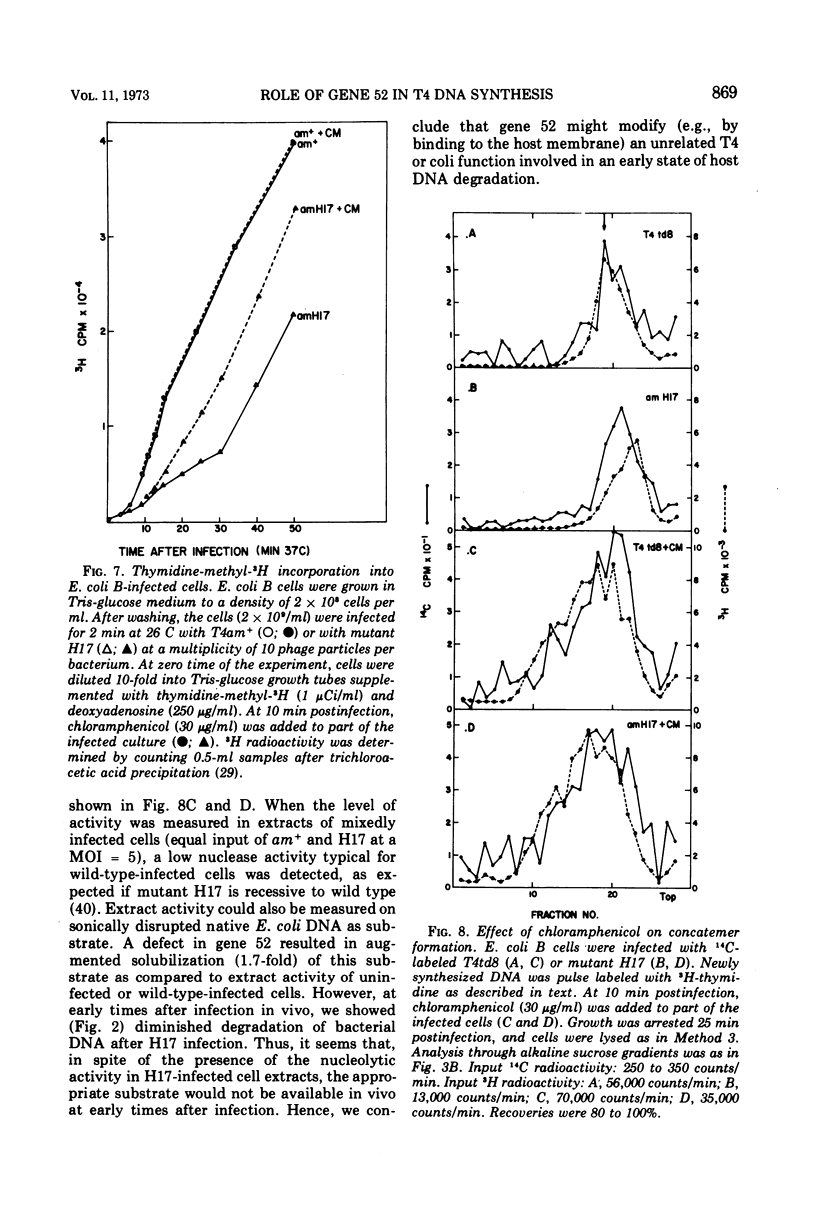
In an attempt to elucidate the mechanism of delayed DNA synthesis in phage T4, Escherichia coli B cells were infected with H17 (an amber mutant defective in gene 52 possessing a “DNA-delay” phenotype). The fate of 14C-labeled H17 parental DNA after infection was followed: we could show that this DNA sediments more slowly in neutral sucrose than wild-type DNA 3 min postinfection. In pulse-chase experiments progeny DNA was found to undergo detachment from the membrane at 12 min postinfection. Reattachment to the membrane was found to be related to an increase in rate of DNA synthesis. A nucleolytic activity that is absent from cells infected by wild-type phage and from uninfected cells could be detected in extracts prepared from mutant-infected cells. In contrast, degradation of host DNA was found to be less extensive in am H17 compared with wild-type infected cells. Addition of chloramphenicol to mutant-infected cells 10 min postinfection inhibited the appearance of a nuclease activity on one hand and suppressed the “DNA-delay” phenotype on the other hand. We conclude that the gene 52 product controls the activity of a nuclease in infected cells whose main function may be specific strand nicking in association with DNA replication. This gene product might directly attack both E. coli and phage T4 DNA, or indirectly determine their sensitivity to degradation by another nuclease.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Lerman L. S. Kinetics and intermediates in the intracellular synthesis of bacteriophage T4 deoxyribonucleic acid. J Mol Biol. 1970 Jun 14;50(2):235–261. doi: 10.1016/0022-2836(70)90190-7. [DOI] [PubMed] [Google Scholar]
- Altman S., Meselson M. A T4-induced endonuclease which attacks T4 DNA. Proc Natl Acad Sci U S A. 1970 Jul;66(3):716–721. doi: 10.1073/pnas.66.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S. K., Warren R. J. Bacteriophage-induced inhibition of host functions. II. Evidence for multiple, sequential bacteriophage-induced deoxyribonucleases responsible for degradation of cellular deoxyribonucleic acid. J Virol. 1969 Jun;3(6):549–556. doi: 10.1128/jvi.3.6.549-556.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Howe C., Kozinski A. W. Structure of the replicating DNA from bacteriophage T4. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3049–3053. doi: 10.1073/pnas.68.12.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earhart C. F. The association of host and phage DNA with the membrane of Escherichia coli. Virology. 1970 Oct;42(2):420–436. [PubMed] [Google Scholar]
- Earhart C. F., Tremblay G. Y., Daniels M. J., Schaechter M. DNA replication studied by a new method for the isolation of cell membrane-DNA complexes. Cold Spring Harb Symp Quant Biol. 1968;33:707–710. doi: 10.1101/sqb.1968.033.01.079. [DOI] [PubMed] [Google Scholar]
- Frankel F. R., Batcheler M. L., Clark C. K. The role of gene 49 in DNA replication and head morphogenesis in bacteriophage T4. J Mol Biol. 1971 Dec 28;62(3):439–463. doi: 10.1016/0022-2836(71)90147-1. [DOI] [PubMed] [Google Scholar]
- Frankel F. R. DNA replication after T4 infection. Cold Spring Harb Symp Quant Biol. 1968;33:485–493. doi: 10.1101/sqb.1968.033.01.056. [DOI] [PubMed] [Google Scholar]
- Frankel F. R. Evidence for long DNA strands in the replicating pool after T4 infection. Proc Natl Acad Sci U S A. 1968 Jan;59(1):131–138. doi: 10.1073/pnas.59.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Haskell E. H., Daverin C. I. Pre-fork synthesis: a model for DNA replication. Proc Natl Acad Sci U S A. 1969 Nov;64(3):1065–1071. doi: 10.1073/pnas.64.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercules K., Munro J. L., Mendelsohn S., Wiberg J. S. Mutants in a nonessential gene of bacteriophage T4 which are defective in the degradation of Escherichia coli deoxyribonucleic acid. J Virol. 1971 Jan;7(1):95–105. doi: 10.1128/jvi.7.1.95-105.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda J., Mathews E. DNA replication in vivo by polynucleotide-ligase defective mutants of T4. II. Effect of chloramphenicol and mutations in other genes. J Mol Biol. 1971 Jan 28;55(2):155–179. doi: 10.1016/0022-2836(71)90189-6. [DOI] [PubMed] [Google Scholar]
- Koerner J. F. Enzymes of nucleic acid metabolism. Annu Rev Biochem. 1970;39:291–322. doi: 10.1146/annurev.bi.39.070170.001451. [DOI] [PubMed] [Google Scholar]
- Kozinski A. W. Molecular recombination in the ligase negative T4 amber mutant. Cold Spring Harb Symp Quant Biol. 1968;33:375–391. doi: 10.1101/sqb.1968.033.01.044. [DOI] [PubMed] [Google Scholar]
- Kozinski A., Lorkiewicz Z. K. Early intracellular events in the replication of T4 phage DNA, IV. Host-mediated single-stranded breaks and repair in ultraviolet-damaged T4 DNA. Proc Natl Acad Sci U S A. 1967 Nov;58(5):2109–2116. doi: 10.1073/pnas.58.5.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter E. M., Wiberg J. S. Degradation of cytosin-containing bacterial and bacteriophage DNA after infection of Escherichia coli B with bacteriophage T4D wild type and with mutants defective in genes 46, 47 and 56. J Mol Biol. 1968 Dec;38(3):395–411. doi: 10.1016/0022-2836(68)90394-x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lang D., Mitani M. Simplified quantitative electron microscopy of biopolymers. Biopolymers. 1970;9(3):373–379. doi: 10.1002/bip.1970.360090310. [DOI] [PubMed] [Google Scholar]
- Litwin S., Shahn E., Kozinski A. W. Interpretation of sucrose gradient sedimentation pattern of deoxyribonucleic acid fragments resulting from random breaks. J Virol. 1969 Jul;4(1):24–30. doi: 10.1128/jvi.4.1.24-30.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh R. C., Breschkin A. M., Mosig G. Origin and direction of bacteriophage T4 DNA replication. II. A gradient of marker frequencies in partially replicated T4 DNA as assayed by transformation. J Mol Biol. 1971 Sep 14;60(2):213–233. doi: 10.1016/0022-2836(71)90289-0. [DOI] [PubMed] [Google Scholar]
- Miller R. C., Jr, Kozinski A. W. Early intracellular events in the replication of bacteriophage T4 deoxyribonucleic acid. V. Further studies on the T4 protein-deoxyribonucleic acid complex. J Virol. 1970 Apr;5(4):490–501. doi: 10.1128/jvi.5.4.490-501.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. C., Kozinski A. W., Litwin S. Molecular Recombination in T4 Bacteriophage Deoxyribonucleic Acid: III. Formation of Long Single Strands During Recombination. J Virol. 1970 Mar;5(3):368–380. doi: 10.1128/jvi.5.3.368-380.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray R. E., Mathews C. K. Biochemistry of DNA-defective amber mutants of bacteriophage T4. II. Intracellular DNA forms in infection by gene 44 mutants. J Mol Biol. 1969 Sep 14;44(2):249–262. doi: 10.1016/0022-2836(69)90173-9. [DOI] [PubMed] [Google Scholar]
- Naot Y., Shalitin C. Defective concatemer formation in cells infected with deoxyribonucleic acid-delay mutants of bacteriophage T4. J Virol. 1972 Oct;10(4):858–862. doi: 10.1128/jvi.10.4.858-862.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashad N., Hosoda J. Role of genes 46 and 47 in bacteriophage T4 reproduction. II. Formation of gaps on parental DNA of polynucleotide ligase defective mutants. J Mol Biol. 1972 Oct 14;70(3):617–635. doi: 10.1016/0022-2836(72)90562-1. [DOI] [PubMed] [Google Scholar]
- Richardson C. C., Masamune Y., Live T. R., Jacquemin-Sablon A., Weiss B., Fareed G. C. Studies on the joining of DNA by polynucleotide ligase of phage T4. Cold Spring Harb Symp Quant Biol. 1968;33:151–164. doi: 10.1101/sqb.1968.033.01.019. [DOI] [PubMed] [Google Scholar]
- Shalitin C., Kahana S. Conversion of T4 gene 46 mutant deoxyribonucleic acid into nonviable bacteriophage particles. J Virol. 1970 Sep;6(3):353–362. doi: 10.1128/jvi.6.3.353-362.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin C., Naot Y. Role of gene 46 in bacteriophage T4 deoxyribonucleic acid synthesis. J Virol. 1971 Aug;8(2):142–153. doi: 10.1128/jvi.8.2.142-153.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin C., Sarid S. Differential effect of polyamines on T4 morphogenesis. J Virol. 1967 Jun;1(3):559–568. doi: 10.1128/jvi.1.3.559-568.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Emrich J., Stahl M. M. Chromosome structure in phage t4, iii. Terminal redundancy and length determination. Proc Natl Acad Sci U S A. 1967 Feb;57(2):292–295. doi: 10.1073/pnas.57.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner D., Oishi M. The effect of bacteriophage T4 infection on an ATP-dependent deoxyribonuclease in Escherichia coli. Biochim Biophys Acta. 1971 Feb 11;228(3):767–769. doi: 10.1016/0005-2787(71)90747-7. [DOI] [PubMed] [Google Scholar]
- WIBERG J. S., DIRKSEN M. L., EPSTEIN R. H., LURIA S. E., BUCHANAN J. M. Early enzyme synthesis and its control in E. coli infected with some amber mutants of bacteriophage T4. Proc Natl Acad Sci U S A. 1962 Feb;48:293–302. doi: 10.1073/pnas.48.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. J., Bose S. K. Bacteriophage-induced inhibition of host functions. I. Degradation of Escherichia coli deoxyribonucleic acid after T4 infection. J Virol. 1968 Apr;2(4):327–334. doi: 10.1128/jvi.2.4.327-334.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner R. Initiation and propagation of growing points in the DNA of phage T4. Cold Spring Harb Symp Quant Biol. 1968;33:501–507. doi: 10.1101/sqb.1968.033.01.058. [DOI] [PubMed] [Google Scholar]
- Wiberg J. S. Mutants of bacteriophage T4 unable to cause breakdown of host DNA. Proc Natl Acad Sci U S A. 1966 Mar;55(3):614–621. doi: 10.1073/pnas.55.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson J., Dressler D., Magazin M. Bacteriophage T7 DNA replication: a linear replicating intermediate (gradient centrifugation-electron microscopy-E. coli-DNA partial denaturation). Proc Natl Acad Sci U S A. 1972 Feb;69(2):499–504. doi: 10.1073/pnas.69.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y. A reduced activity of a deoxyribonuclease requiring ATP in Escherichia coli infected by bacteriophage T4. Biochim Biophys Acta. 1971 Nov 19;247(4):535–541. doi: 10.1016/0005-2787(71)90690-3. [DOI] [PubMed] [Google Scholar]
- Yegian C. D., Mueller M., Selzer G., Russo V., Stahl F. W. Properties of the DNA-delay mutants of bacteriophage T4. Virology. 1971 Dec;46(3):900–919. doi: 10.1016/0042-6822(71)90090-0. [DOI] [PubMed] [Google Scholar]


