Abstract
TATA binding protein (TBP) is a key component of the eukaryotic RNA polymerase II (Pol II) transcription machinery that binds to TATA boxes located in the core promoter regions of many genes. Structural and biochemical studies have shown that when TBP binds DNA, it sharply bends the DNA. We used single-molecule FRET (smFRET) to study DNA bending by human TBP on consensus and mutant TATA boxes in the absence and presence of TFIIA. We found that the state of the bent DNA within populations of TBP/DNA complexes is homogeneous; partially bent intermediates were not observed. In contrast to previous ensemble studies, TBP was found to bend a mutant TATA box to the same extent as the consensus TATA box. Moreover, in the presence of TFIIA the extent of DNA bending was not significantly changed, although TFIIA did increase the fraction of DNA molecules bound by TBP. Analysis of the kinetics of DNA bending and unbending revealed that on the consensus TATA box two kinetically distinct populations of TBP/DNA complexes exist, however, the bent state of the DNA is the same in the two populations. Our smFRET studies reveal that human TBP bends DNA in a largely uniform manner under a variety of different conditions, which was unexpected given previous ensemble biochemical studies. Our new observations lead to us to revise the model for the mechanism of DNA binding by TBP and for how DNA bending is affected by TATA sequence and TFIIA.
Transcription of protein-encoding genes is a primary step in gene expression during which mRNA is synthesized from a DNA template. In eukaryotic cells, Pol II synthesizes mRNA, and requires numerous accessory factors to transcribe genes in a regulated manner (1). One such factor is the TFIID (transcription factor IID) complex, which consists of the TATA binding protein (TBP) and TBP associated factors (TAFs). The TFIID complex nucleates preinitiation complex formation at the promoters of genes by binding core promoter elements and recruiting the remaining general transcription factors and Pol II (1). The TBP subunit of TFIID binds to TATA boxes (consensus sequence TATA(A/T)AA(G/A)) that are located approximately 25 basepairs upstream from transcription start sites in a subset of eukaryotic protein-encoding genes (1).
Biochemical experiments and X-ray crystallography have shown that yeast and human TBP are saddle-shaped proteins that bind the minor groove of TATA DNA (2–6). Phenylalanine residues in TBP insert between the bases at positions 1/2 and 7/8 of the TATA box, which disrupts base stacking interactions and bends the DNA away from the protein. TBP-induced DNA bending has previously been described using a two-kink model, which allows the bend angle to be calculated assuming the bent DNA is planar (7, 8). Applying this model to measurements made using fluorescence lifetime and fluorescence resonance energy transfer (FRET) revealed that human and yeast TBP bend the consensus TATA sequence to angles of ~100 and ~80 degrees, respectively (7, 9–11). A more complex model has also been described in which the DNA is both bent and twisted, such that the ends are no longer in the same plane (10); using this model the aforementioned ensemble FRET data would be consistent with a wide range of bend and twist angle combinations.
Several different models have been proposed for the mechanism of TBP binding and bending of TATA DNA using human and yeast TBP. In one model, TBP first interacts with DNA, and then subsequently bends the DNA (12, 13). Using rigorous ensemble lifetime measurements and other biochemical experiments Parkhurst and colleagues concluded that TBP simultaneously binds and bends the DNA (9, 14, 15). In their model, two bound intermediates with unique kinetic profiles exist en route to the final complex; however, each of the intermediates has the same degree of bending as the final complex. Optical tweezer experiments led another group to conclude that a long-lived partially bent intermediate forms between the unbound and fully bent states, suggesting a stepwise bending model. They proposed that this intermediate occurs when one side of the TBP saddle intercalates into the DNA, resulting in a partially bent state (16). Additional studies using other approaches would provide further insight into the mechanism of TBP-induced DNA bending.
The TATA sequence itself affects multiple properties of the TBP/DNA complex. Altering the consensus sequence at one or two positions decreases the affinity of TBP for DNA and the kinetic stability of the TBP/DNA complex (7, 10, 17, 18). In addition, ensemble biochemical studies have concluded that the angle at which human and yeast TBP bend DNA is affected by the TATA sequence; mutant sequences that TBP binds with lower affinity appear less bent compared to the consensus sequence (7, 9, 10, 17, 19). Moreover, for yeast TBP, weaker TATA boxes that appear to be bent to a lesser degree in vitro (7) are less transcriptionally active in cells (7, 20).
The transcription factor IIA (TFIIA) has been shown to regulate the TBP/DNA interaction; it increases the kinetic stability of TBP/DNA complexes at consensus as well as nonconsensus TATA sequences (10, 21–24) and alters the sequence specificity with which TBP binds DNA (22). Moreover, TFIIA has been found to change the extent to which DNA is bent by human TBP. When the planar two-kink model was applied to ensemble FRET data, TFIIA appeared to decrease the bend angle of a consensus TATA sequence and increase the bend angle of a mutant sequence, resulting in both sequences having a similar bend angle (10).
Here, we used single molecule FRET (smFRET) to monitor individual bent and unbent DNA molecules in the presence of human TBP. This system was used to study the extent of heterogeneity in DNA bending by human TBP, and how the bent conformation is affected both by TFIIA and a mutation in the TATA sequence. We found that the DNA molecules within TBP/DNA complexes exist in a homogeneous bent state; we did not observe a population of DNA molecules partially bent by TBP. Moreover, we found that the extent to which the DNA is bent by TBP is the same with consensus and mutant DNA, and in the presence and absence of TFIIA. We also analyzed the kinetics of bending and unbending on consensus and mutant TATA DNA. We found that DNA bending by human TBP displays heterogeneous kinetics on the consensus TATA box, but not a mutant TATA box. Our smFRET results provide unique insights that lead to a re-evaluation of existing models for TBP DNA bending that were derived from ensemble experiments.
Experimental Procedures
Oligonucleotides, proteins, and other reagents
Three oligos labeled at their 5′ ends with Alexa657 (IDT), Alexa555 (Invitrogen), or biotin (IDT) were HPLC purified. The sequences (5′ to 3′) of the acceptor, donor, and biotin linker oligos were Alexa647-CGTCCNNNNNNNNNCCTGCACCACCACCACCACCACCA, Alexa555-CAGGNNNNNNNNNGGACG, and Biotin(dT)-TGGTGGTGGTGGTGGTGGT, respectively, where the specific sequences of the variable regions (indicated with N) are shown in Table 1. Constructs containing a donor oligo, an acceptor oligo, and the biotin-linker oligo were assembled by heating to 95°C for 3 min, then incubating the DNA at 60°C for 45 min and cooling 0.1°C/s to room temperature prior to use. Recombinant human TBP and human TFIIA were expressed and purified from E. coli as previously described (25–27). Fluorescence anisotropy values were measured for DNA constructs containing Alexa555 or Alexa647 in the absence and presence of TBP under the experimental conditions used for smFRET. In all cases the anisotropy values were between 0.21 and 0.24.
Table 1.
Sequences unique to each of the constructs.
| Oligo | Sequence (5′ to 3′) |
|---|---|
| Consensus TATA donor | CTATAAAAG |
| Consensus TATA acceptor | CTTTTATAG |
| TATA(A3) donor | CTAAAAAAG |
| TATA(A3) acceptor | CTTTTTTAG |
| TATA-less donor | TAGAGTCGG |
| TATA-less acceptor | CCGACTCTA |
Catalase (Sigma) and glucose oxidase (Sigma) were resuspended to 6.8 mg/ml and 44 mg/ml, respectively, in storage buffer (50% (v/v) glycerol, 100 mM Tris (pH 7.9), and 50 mM KCl), and stored at −20°C. Streptavidin (1 mg/ml, Sigma) was stored at −20°C in 10 mM Tris (pH 7.9), 10% (v/v) glycerol, 50 mM KCl, and 5 mM MgCl2. D-glucose 10% (w/v) was dissolved in 10 mM Tris (pH 7.9) and 50 mM NaCl, and stored at −20°C. Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid, Sigma) was dissolved in 18 MΩ water to a concentration of 100 mM by adding 4 M NaOH. Trolox was stored at 4°C for no longer than one week.
Preparation of surfaces for smFRET experiments
Glass slides and coverslips (VWR) were cleaned by the following procedure with thorough rinsing in 18 MΩ water between each step. A 1% alconox solution was heated to 90°C and added to a container of glass slides and coverslips and sonicated for 20 min. The slides and coverslips were then sonicated in ethanol for one hour, followed by a 20 min sonication in 1 M KOH. The slides and coverslips were dried by rinsing in ethanol and microwaving for 90 s. Slides and coverslips were flamed over a Bunsen burner and after cooling were silanated by placing them in methanol containing 2% aminosilane (N-(2-Aminoethyl) 3-aminopropyltrimethoxysilane, UCT) for 21 min, with a 1 min sonication after 10 min. Slides and coverslips were rinsed in methanol and baked at 110°C for 5–10 min. A solution of 0.38% (w/v) biotin-PEG-SC (MW 5000, Laysan Bio, Inc.) and 20% (w/v) mPEG-SVA (MW 5000, Laysan Bio, Inc.) in 0.1 M sodium bicarbonate was sandwiched between a coverslip and slide and allowed to react with the silanated glass for 3.5–4 h in a humidified environment (28). The surfaces were rinsed thoroughly with water and dried with nitrogen gas. Reaction chambers were assembled by attaching a coverslip to a slide with an overhang using double-sided tape. Sample chambers were stored in a dark, dry environment until use.
smFRET reaction conditions
A sample chamber was incubated with 0.02 mg/ml streptavidin for 5 min in buffer A (10% (v/v) glycerol, 10 mM Tris (pH 7.9), 50 mM KCl, 1 mM DTT, 0.05 mg/ml BSA, 10 mM HEPES (pH 7.9), 5 mM MgCl2, and 0.1% nonidet P-40). The chamber was washed with excess buffer A prior to flowing in a solution containing 20–40 pM of a 3-oligo construct in buffer A. After 10 min, the chamber was washed with excess buffer A. For all fluorescent measurements with or without proteins, imaging solution was flowed into a chamber, which contained 12% (v/v) glycerol, 14 mM Tris (pH 7.9), 1 mM DTT, 0.05 mg/ml BSA, 10 mM HEPES (pH 7.9), 5 mM MgCl2, 9 mM NaCl, 1 mg/ml glucose oxidase, 11 μg/ml catalase, 0.8% (w/v) D-glucose, 2 mM trolox, 0.1% nonidet P-40, and 150 mM KCl, unless stated otherwise. All smFRET experiments were performed at 22°C. Unless otherwise noted in the text, TBP was at 25 nM and TFIIA was at 15 nM.
Data collection and analysis
FRET data were collected on an objective-based total internal reflection fluorescence microscope (Nikon TE-2000U) equipped with a Cascade II CCD camera, a 1.49 NA immersion objective, and a 532 nm CW laser. Acceptor and donor emission data were collected with MetaMorph software (Molecular Devices) using a 30 ms timescale and a bin of 2.
Movies were analyzed using in-house developed software to identify spot pairs in the donor and acceptor channels. To obtain time traces of donor and acceptor emissions for individual spots, background was subtracted from each frame using an area neighboring the spot. The following equation was used to correct for bleed through of the donor dye into the acceptor channel.
Where A is the background-subtracted signal from the acceptor channel, D is the background-subtracted signal from the donor channel, and α is a factor used to correct for bleedthrough of the donor emission into the acceptor channel; α was 0.14. The ratio of the donor and acceptor detection efficiencies (γ) was one, and the acceptor emmission resulting from direct excitation of the acceptor by the donor laser was approximately 3%; hence, data were not corrected for these two factors. The Alexa555/Alexa647 has a calculated R0 of 51 Å, assuming κ2 = 2/3. Apparent FRET efficiencies (E) were calculated using the equation:
Unsmoothed data were further analyzed using in-house developed software to determine the FRET states and dwell times of each state present throughout individual time traces. Blocks of frames exhibiting photoblinking were removed from the data. FRET states lasting fewer than 3 frames were removed from further analysis. Under our experimental conditions, a minimum change in FRET of 0.05 was required to detect a change between two neighboring states in a time trace. We continuously monitored the sum of the donor and acceptor emissions as a means to assess the effect of TBP and TFIIA on the donor and acceptor fluorophores; this sum remained relatively constant during bending and unbending events.
FRET states were extracted from all data collected under a single condition and binned at 0.02 FRET units to generate the histograms. Gaussian curves were fit to histograms using Prism to obtain mean FRET efficiencies. Dwell times were extracted from the time traces. Time traces that showed multiple state changes on a single DNA molecule were separated into the unbent (lower FRET) and bent (higher FRET) states, and dwell times were extracted for each state. Dwell times were removed for states at the beginning and end of time traces (which included molecules that did not change states throughout the entire time trace), and when a dye blinked during a state. Dwell times for unbent and bent states were binned separately, histogrammed, and fit to monoexponential or biexponential equations using Prism, as noted in the Results section.
Results
Consensus TATA DNA molecules are uniformly bent by TBP
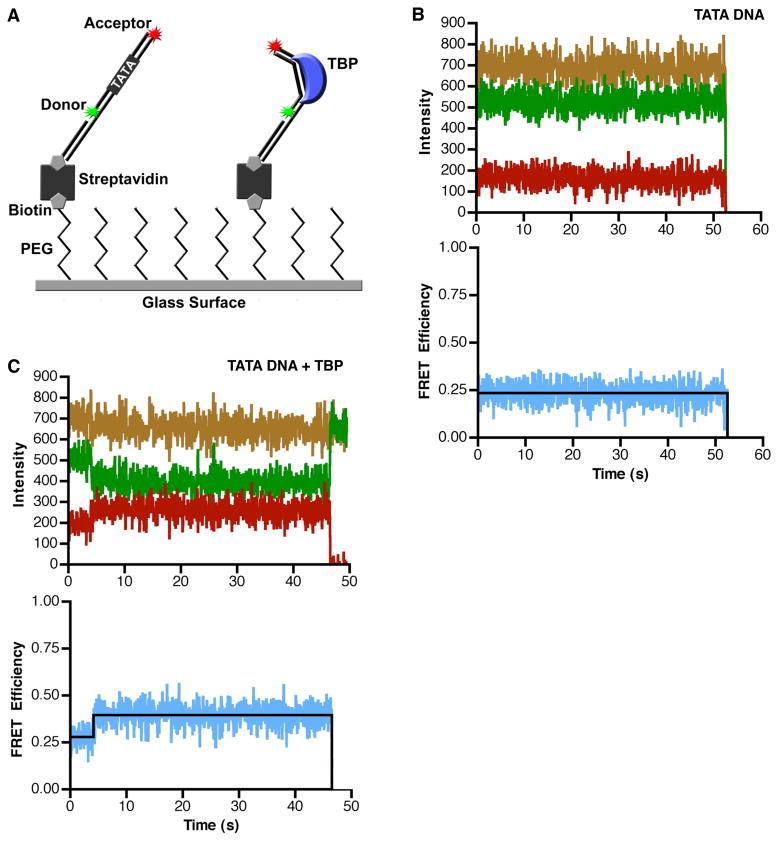
To study DNA bending by human TBP using smFRET, we designed a DNA construct with donor and acceptor fluorophores positioned such that when TBP induced a bend, the distance between the donor and the acceptor fluorophores would decrease, and an increase in FRET efficiency would be observed (Figure 1A). The donor (Alexa 555) and acceptor (Alexa 647) dyes were attached to the 5′-ends of two oligos, that when annealed, formed an 18 bp DNA with a consensus TATA box at its center (sequence from the Adenovirus major late promoter, AdMLP, shown in Table 1) and a 20 nt single stranded 3′ overhang. This single stranded overhang was complementary to the sequence of a third oligo that had a biotin on its 5′-end. After annealing the three-oligo construct, it was attached to a glass surface through a biotin-streptavidin linkage. The sample chamber was placed on a microscope and imaged using total internal reflection fluorescence (TIRF) microscopy. In all experiments the donor was excited with a 532 nm laser and the donor and acceptor emissions were monitored over time with a CCD camera. Apparent FRET efficiencies were calculated as described in the methods.
Figure 1.
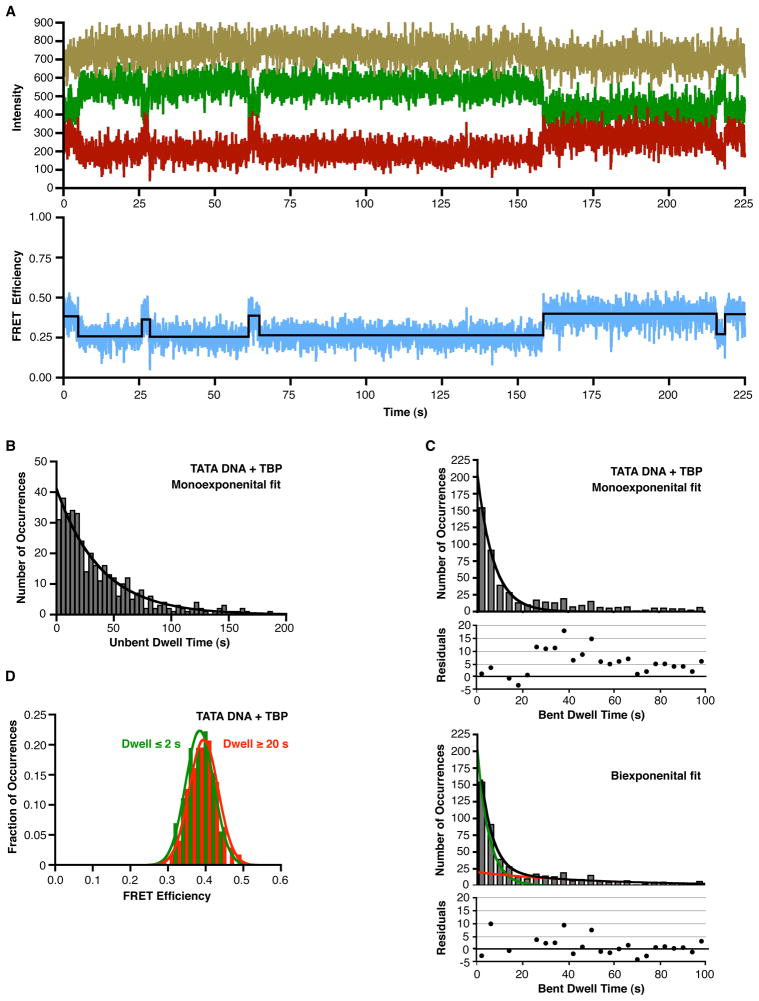
TBP causes an increase in the FRET observed from single immobilized consensus TATA DNA molecules. (A) The single-molecule surface configuration for monitoring TBP-induced DNA bending. When TBP bends the immobilized DNA it causes an increase in FRET efficiency. (B) The consensus TATA construct exhibits constant low FRET in the absence of TBP. The donor dye (green), the acceptor dye (red), and sum of the two dyes (gold) did not change over the course of imaging (upper panel). The calculated FRET at each time point is shown in blue and the derived FRET state of 0.24 is shown in black (lower panel). For the purpose of display, these data were smoothed by averaging the values at 3 time points. (C) TBP causes an increase in FRET consistent with DNA bending. The donor and acceptor dyes show anti-correlated changes in signal intensity (upper panel) and an increase in calculated FRET efficiency (lower panel) at 4 seconds, indicating TBP bent the DNA. At 47 s, the acceptor dye photobleached, causing emission from the donor dye to increase. For the purpose of display, these data were smoothed by averaging the values at 3 time points.
We first imaged DNA in the absence of TBP. An example of an unbound DNA trace is shown in Figure 1B; the donor (green), acceptor (red), and total fluorescence (gold) remained constant over time until the donor dye photobleached at 53 s. The blue trace (lower panel) shows the FRET efficiency calculated from the donor and acceptor emissions, with the black line showing the FRET state fit to the data. In all time courses of DNA imaged in the absence of TBP we observed no noticeable change in FRET efficiency prior to fluorophore photobleaching.
When human TBP was flowed into the sample chamber, we observed changes in the FRET efficiency. A time trace for a single DNA is shown in Figure 1C. Here, the donor and acceptor signals (upper panel) and FRET efficiency (lower panel) remained constant until approximately 4 s, at which time the acceptor intensity increased, and the donor intensity simultaneously decreased. This anti-correlated change in dye intensities resulted in an increase in FRET efficiency from 0.28 to 0.4, indicative of TBP bending the DNA (lower panel of Figure 1C). The FRET efficiency remained steady until the acceptor dye photobleached at 47 s. The sum of the donor and acceptor intensities (gold trace in the upper panel) did not appreciably change throughout the time course, indicating that TBP does not affect the fluorophores themselves.
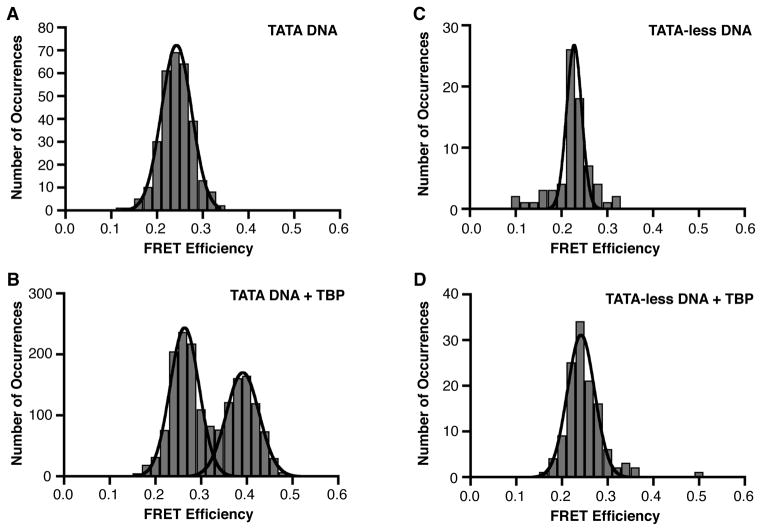
The FRET efficiencies for 301 immobilized consensus TATA DNA molecules in the absence of TBP were plotted on a histogram and fit to a Gaussian, yielding a single peak with a FRET efficiency centered at 0.24 (Figure 2A and Table 2). Analysis of time traces obtained for 477 DNA molecules in the presence of TBP, yielded 1727 distinct FRET occurrences that were histogrammed and fit to Gaussians, yielding two separate peaks with mean FRET efficiencies of 0.26 and 0.39 (Figure 2B). We conclude that the lower FRET state is unbent DNA and the upper FRET state, which depends on the addition of TBP, is the TBP-bound and bent DNA. Table 2 shows the mean FRET values obtained from all Gaussian fits, and Figure S1 shows the standard deviations for the Gaussian fits, as a representation of the widths of the Gaussians.
Figure 2.
TBP/DNA complexes exist in a uniform FRET state. (A) Consensus TATA DNA exists in a homogenous population in a relatively low FRET state. FRET efficiencies for 301 DNA molecules were plotted as a histogram and fit with a Gaussian. The mean FRET efficiency is 0.24. (B) In the presence of TBP, the DNA molecules exist in a two-state population (unbent and bent). Time traces for 477 DNA molecules were analyzed and 1727 distinct FRET occurrences were plotted as a histogram and fit with Gaussians. The low and high FRET states have mean FRET efficiencies of 0.26 and 0.39, respectively. (C) Shown is the histogram and Gaussian fit of FRET efficiencies determined for 72 TATA-less DNA molecules in the absence of TBP. The mean FRET efficiency is 0.23. (D) TBP does not alter the FRET efficiency of a TATA-less DNA construct. The histogram and Gaussian fit of 124 distinct FRET occurrences observed in time traces from 121 TATA-less DNA molecules in the presence of TBP is shown. The mean FRET efficiency is 0.24.
Table 2.
Mean FRET efficiencies.
| DNA construct | [KCl] mM | TBP | TFIIA | Unbent mean FRET | Bent mean FRET | Unbent 95%CI | Bent 95%CI |
|---|---|---|---|---|---|---|---|
| Cons. TATA | 150 | − | − | 0.24 | N/A | 0.241–0.244 | N/A |
| Cons. TATA | 150 | + | − | 0.26 | 0.39 | 0.261–0.266 | 0.388–0.394 |
| Cons. TATA | 150 | + | + | ND | 0.39 | ND | 0.386–0.398 |
| Cons. TATA | 50 | 0.4x | − | 0.26 | 0.37 | 0.256–0.271 | 0.363–0.380 |
| TATA(A3) | 150 | − | − | 0.25 | N/A | 0.249–0.253 | N/A |
| TATA(A3) | 150 | + | − | 0.26 | ND | 0.259–0.262 | ND |
| TATA(A3) | 150 | + | + | 0.27 | 0.41 | 0.271–0.277 | 0.402–0.411 |
| TATA(A3) | 50 | − | − | 0.25 | N/A | 0.250–0.258 | N/A |
| TATA(A3) | 50 | + | − | 0.27 | 0.36 | 0.262–0.274 | 0.353–0.369 |
| TATA(A3) | 50 | 4x | − | 0.26 | 0.37 | 0.253–0.271 | 0.361–0.379 |
| TATA-less | 150 | − | − | 0.23 | N/A | 0.224–0.231 | N/A |
| TATA-less | 150 | + | − | 0.24 | ND | 0.239–0.244 | ND |
N/A – not applicable
ND – not detected
95% CI – 95% confidence interval for the mean FRET
In order to determine whether the change in FRET efficiency upon addition of TBP was dependent on a TATA box, a TATA-less construct (sequence shown in Table 1) was immobilized and used for smFRET experiments in the absence and presence of TBP. In the absence of TBP, this construct had a FRET efficiency of 0.23 (Figure 2C). A histogram of the TATA-less construct in the presence of TBP resulted in a single peak with a FRET efficiency of 0.24 (Figure 2D). Therefore, we conclude that the higher FRET state observed with the consensus TATA construct (Figure 2B) represents sequence-specific DNA bending by TBP. We further conclude that the TBP/DNA complexes contain TATA DNA in a uniformly bent state.
TFIIA shifts the population of DNA molecules to the bound state but does not change the extent of DNA bending
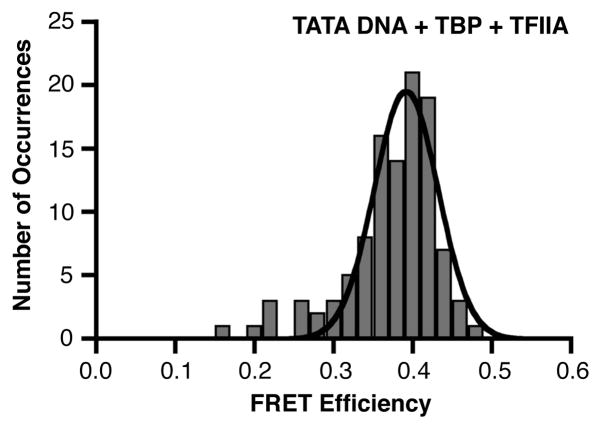
We next asked whether TFIIA could affect either the population distribution or the FRET efficiency of the bent state of TATA DNA molecules in our smFRET system. When TBP and TFIIA were flowed together into a chamber containing the immobilized consensus TATA construct, the DNA population strongly shifted to a higher FRET state (Figure 3). These data fit to a single Gaussian distribution with a peak FRET state of 0.39. We can draw two primary conclusions from these results: 1) TFIIA shifted the population of TATA DNA toward the TBP bound and bent state, consistent with its ability to increase TBP’s affinity for DNA (10, 21, 22), and 2) TFIIA did not change the FRET state of the TBP bound/bent DNA.
Figure 3.
When included with TBP, TFIIA shifts the consensus TATA DNA to a homogenous population in the higher FRET state. Shown is the histogram and Gaussian fit of 107 distinct FRET occurrences observed in time traces from 91 consensus DNA molecules in the presence of TBP and TFIIA; the mean FRET efficiency is 0.39.
TBP bends a mutant TATA DNA similarly to the consensus TATA DNA
We investigated how mutating the TATA sequence changed TBP-induced DNA bending at the single molecule level. We used the TATA(A3) sequence, which contains a single T to A point mutation in the consensus sequence (Table 1) and has previously been shown to bind TBP with significantly lower affinity and to be bent to a lesser extent by TBP in ensemble studies (7, 10, 17). Immobilized mutant TATA(A3) DNA in the absence of TBP exhibited a FRET efficiency similar to that of the consensus TATA DNA (Figure 4A), indicating that, as expected, the point mutation had little, if any, effect on the FRET observed with unbound DNA. After flowing TBP into the sample chamber, we did not observe a higher FRET state in the histogram, indicating that the DNA molecules were unbent (Figure 4B), which is consistent with the weak affinity of TBP for this mutant DNA compared to that of the consensus TATA DNA.
Figure 4.
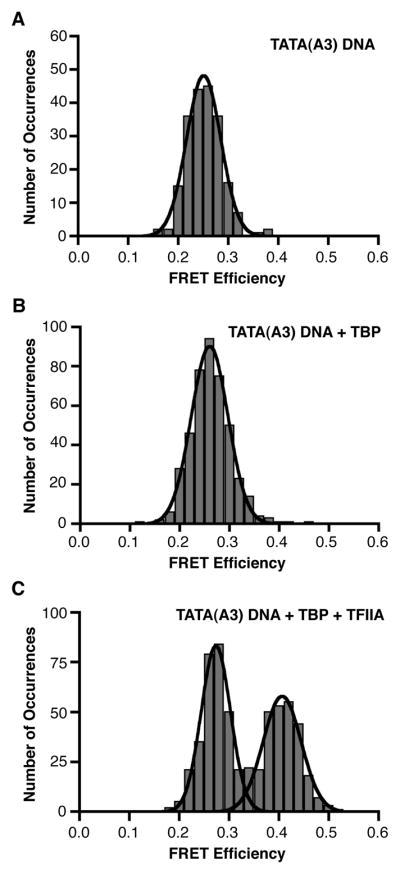
TFIIA facilitates TBP bending the mutant TATA(A3) sequence. (A) In the absence of TBP, the mutant TATA sequence exists in a uniform FRET state similar to that observed with the consensus TATA sequence. The mean FRET efficiency for the Gaussian fit of data from 206 DNA molecules is 0.25. (B) TBP does not shift the TATA(A3) DNA into a higher FRET state. Shown is the histogram and Gaussian fit of 427 distinct FRET occurrences observed in time traces from 414 TATA(A3) DNA molecules in the presence of TBP; the mean FRET efficiency is 0.26. (C) TFIIA facilitates the bending of the TATA(A3) DNA by TBP. The mean FRET efficiencies of the low (319 occurrences) and high (274 occurrences) states obtained from the Gaussian fits of distinct FRET occurrences observed in time traces from 400 TATA(A3) molecules are 0.27 and 0.41, respectively.
We anticipated that TFIIA would increase the extent of TBP binding to the mutant TATA sequence because TFIIA has been shown to stabilize the TBP/DNA complex with both consensus and nonconsensus sequences (10, 21, 22). Indeed, when TFIIA and TBP were flowed together into a chamber containing the immobilized TATA(A3) construct, we observed many molecules that occupied a higher FRET state (Figure 4C) with a mean FRET efficiency of 0.41, which is similar to that observed with TBP/TFIIA on the consensus TATA DNA.
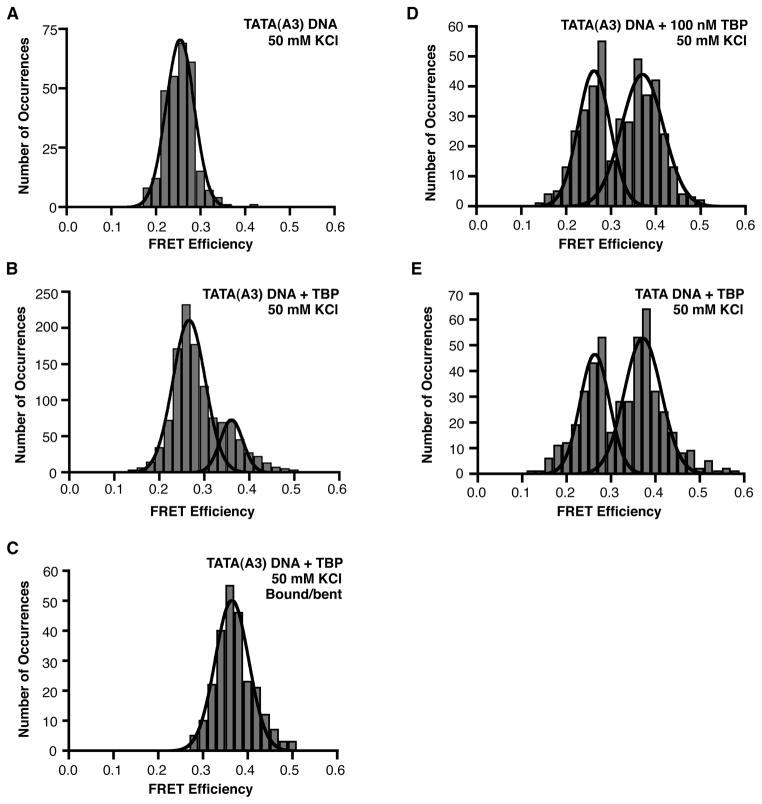
To study TBP bending the TATA(A3) construct in the absence of TFIIA, we reduced the KCl concentration from 150 mM to 50 mM; the lower salt concentration did not affect the FRET of the TATA(A3) construct in the absence of TBP (Figure 5A). In the presence of TBP, a higher FRET state indicative of bending emerged on the TATA(A3) DNA with a mean efficiency of 0.36 (Figure 5B). To ensure that the unbent states in the histogram did not affect the Gaussian fit of the bound/bent states, we histogrammed just the bent states. These bound/bent states were obtained only from those traces in which a distinct transition from a low FRET state to high FRET state, and back to a low FRET state was observed. The Gaussian fit of the histogram yielded a mean FRET efficiency of 0.36 (Figure 5C), identical to that observed when the unbent and bent states were histogrammed together (Figure 5B). We also performed the experiment at a 4-fold higher concentration of TBP, which we expected would increase the fraction of TATA(A3) DNA molecules occupied by TBP. As shown in Figure 5D, this resulted in a shift of the population toward the bound/bent state, which exhibited a mean FRET efficiency of 0.37. To enable a more direct comparison to the consensus TATA box, we monitored TBP bending the consensus TATA construct at 50 mM KCl, which yielded a mean FRET efficiency for the bound/bent states of 0.37 (Figure 5E). At 50 mM KCl, the mean FRET efficiency values for the bent state on the TATA(A3) construct (0.36 and 0.37) and the consensus TATA DNA (0.37) are indistinguishable. Taken together, our results indicate that the extent of bending of the TATA(A3) construct is the same as that of the consensus TATA element, although it remains possible that the two constructs exist in different bent conformations that result in similar FRET values.
Figure 5.
TBP bends the TATA(A3) sequence to a similar extent as the consensus TATA box. (A) Shown is the histogram of FRET efficiencies for 282 TATA(A3) molecules in the absence of TBP at 50 mM KCl; the mean FRET is 0.25. (B) TBP shifts a portion of the TATA(A3) DNA into a higher FRET state at 50 mM KCl. 1165 distinct FRET occurrences observed in time traces from 642 TATA(A3) molecules were histogrammed; the low and high FRET states have mean FRET efficiencies of 0.27 and 0.36, respectively. (C) Bent states on the TATA(A3) DNA were histogrammed and fit to a single Gaussian; the mean FRET efficiency is 0.36. (D) Raising the concentration of TBP to 100 nM increases the number of bound/bent TATA(A3) DNA molecules. 421 distinct FRET occurrences observed in time traces from 141 TATA(A3) molecules were histogrammed; the low and high FRET states have mean FRET efficiencies of 0.26 and 0.37, respectively. (E) TBP bending the consensus TATA box at 50 mM KCl. The experiment was performed with 10 nM TBP. 467 distinct FRET occurrences observed in time traces from 244 TATA molecules were histogrammed; the low and high FRET states have mean FRET efficiencies of 0.26 and 0.37, respectively.
TBP bends consensus TATA DNA, but not the TATA(A3) mutant DNA, in two kinetically distinct complexes bent to the same extent
We observed that many DNA molecules that switched between unbent and bent FRET states, which enabled us to investigate the kinetics with which TBP bends TATA DNA as well as the kinetics with which bent complexes revert back to unbent DNA. Figure 6A shows a representative time trace of a DNA molecule swiching between low FRET (unbent) and higher FRET (bent) states; the donor (green), acceptor (red), and total (gold) fluorescence are shown on the top plot and the FRET efficiency (blue) and FRET state fit (black line) are shown in the bottom plot. To evaluate the kinetics of TBP-DNA bending, we histogrammed the dwell times for unbent states that were flanked by bent states on the consensus TATA box in the presence of TBP at 150 mM KCl (for example, the four low FRET states in Figure 6A). A monoexponential fit of the data yielded an observed rate constant of 0.026 s−1 (Figure 6B). We compare this value, as well as the values of the other rate constants we measure by smFRET, to published data in the Discussion.
Figure 6.
Unbending consensus TATA DNA is biphasic. (A) Representative time trace of a single DNA molecule swiching between low FRET (unbent) and higher FRET (bent) states. The top plot shows the donor emission in green, the acceptor in red, and the sum of the donor and acceptor in gold. The bottom plot shows the corresponding FRET in blue, with the black line indicating the FRET state fit of the data. For the purpose of display, these data were smoothed by averaging the values at 3 time points. (B) Analysis of the rate at which TBP bends the consensus TATA box. Dwell times for the unbent occurrences observed at 150 mM KCl were histogrammed and fit to a monoexponential. The observed rate constant is 0.026 ± 0.003 s−1, where the error represents the 95% confidence interval. (C) Unbending of the consensus TATA box occurs in two distinct kinetic phases. Dwell times for the bent occurrences observed at 150 mM KCl were histogrammed and fit to a monoexponential (upper panel) or a biexponential (lower panel). Residuals are plotted for each of the fits. The two rate constants obtained from the biexponential fit are 0.19 ± 0.03 s−1 and 0.019 ± 0.008 s−1, where the error represents the 95% confidence interval (D) TATA DNA molecules in complexes with different kinetic stabilities are bent to the same extent. FRET efficiencies were separately histogrammed for bent occurrences having dwell times ≤ 2 s (green) or ≥ 20 s (red). Gaussian fits showed mean FRET efficiencies of 0.38 and 0.39, respectively.
To investigate the kinetics which which TBP/DNA complexes revert from the bent state to unbent DNA, we histogrammed the dwell times for bent states that were flanked by unbent states. The histogram was fit to a monoexponential, however all of the bins with dwell times longer than 25 s were above the curve (Figure 6C). This is apparent in the plot of residuals below the histogram. For this reason we fit the histogram of bent state dwell times to a biexponential (Figure 6C). The black curve shows the biexponential fit, the green and red curves show the respective single exponential decay curves for the fast and slow populations generated using the rate constants obtained from the biexponential fit (0.19 s−1 and 0.019 s−1, respectively). The biexponential fit of the data was much better as indicated by the lack of a directional trend in the residuals (plotted below the histogram). This indicates at least two kinetically distinct bent TBP/TATA complexes exist that revert back to unbent DNA with different rates. Moreover, single DNA molecules exhibited both the faster and the slower rates of un-bending (see Figure 6A). To investigate the extent of bending in these kinetically distinct populations, we independently histogrammed the FRET efficiencies of less stable bent complexes (dwell time for the bent state ≤ 2 s) and more stable bent complexes (dwell time for the bent state ≥ 20 s). As shown in Figure 6D, the two histograms have very similar mean FRET efficiences, 0.38 for the less stable complexes and 0.39 for the more stable complexes. Therefore, the DNA in these kinetically different complexes is bent to the same extent.
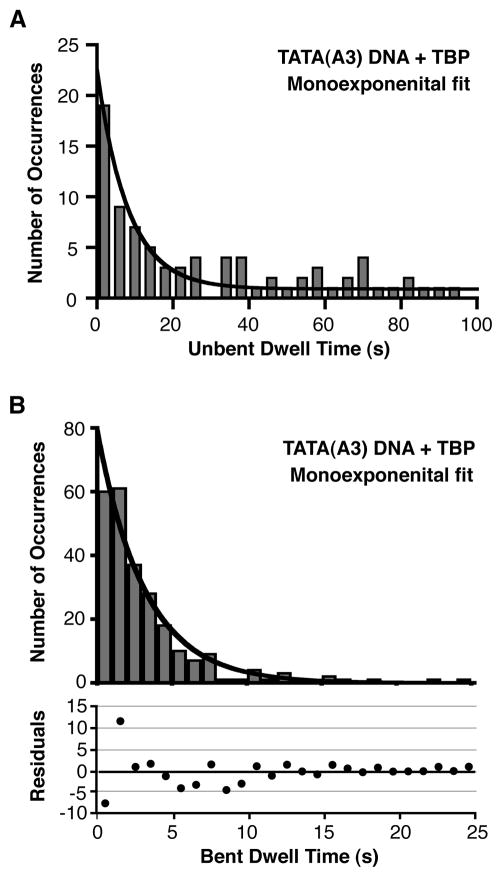
We also analyzed the kinetics of bending and unbending for complexs formed on the mutant TATA(A3) DNA. The rate of DNA bending was assessed by histogramming the unbent state dwell times and fitting to a monoexponential, which yielded an observed rate constant of 0.12 s−1 (Figure 7A). To assess the kinetics with which the bent state decays on the TATA(A3) construct, the bent state dwell times were histogrammed. Surprisingly, the bent state dwell times for the TBP/TATA(A3) DNA complex fit well to a monoexponential (Figure 7B), yielding a rate constant of 0.31 s−1. Therefore, bent complexes on the mutant TATA DNA decay with a different kinetic profile compared to bent complexes on the consensus TATA DNA.
Figure 7.
Unbending on the TATA(A3) DNA in monophasic. (A) Analysis of the rate at which TBP bends the TATA(A3) construct. Dwell times for the unbent occurrences observed at 50 mM KCl were histogrammed and fit to a monoexponential. The observed rate constant is 0.12 ± 0.03 s−1, where the error represents the 95% confidence interval (B) Unbending of the TATA(A3) construct. Dwell times for the bent occurrences observed at 50 mM KCl were histogrammed and fit to a monoexponential. Residuals are plotted below the rate plot. The rate constant obtained is 0.31 ± 0.04 s−1, where the error represents the 95% confidence interval
Discussion
Here we used smFRET to study the heterogeneity and kinetics of DNA bending by human TBP. We found that with either a consensus or mutant TATA box the populations of DNA molecules bound by TBP were bent to a uniform FRET state in both the absence and presence of TFIIA. Throughout our studies, we did not observe populations of TBP/DNA complexes with intermediate bent states, consistent with a mechanism for TBP-DNA binding that lacks partially bent intermediates. Our kinetic analyses suggest that the mechanism of TBP-DNA bending differs on the consensus and mutant TATA boxes. Together our single molecule studies led to several unexpected observations: 1) mutating the TATA box did not significantly decrease the extent of bending, as was previously concluded from ensemble experiments, 2) although TFIIA profoundy increased the affinity of TBP for DNA, it did not change the extent of DNA bending, as was previously concluded from ensemble experiments, 3) for the consensus TATA box, the transition from bent DNA to unbent DNA was biphasic, whereas that from the mutant TATA(A3) construct was monophasic. Together our smFRET studies provide new insight into the mechanism of DNA bending by TBP that is distinct from earlier ensemble experiments and lead us to a revised model for TBP-induced DNA bending. Hence, our work contributes to an ever-growing list of systems for which single-molecule technqiues have resolved heterogeneous kinetics and conformational states (29–31).
Populations of TBP/DNA complexes exist in a uniform bent state
Within the time and FRET resolution of our experiments, our data show that TBP bends consensus and non-consensus TATA DNA uniformly in the presence and absence of TFIIA. Under each of the experimental conditions containing TBP, we observed only one subpopulation with a FRET state consistent with bent DNA. This suggests a high degree of homogeneity in the conformation of TBP/TATA complexes regardless of DNA sequence or the presence of TFIIA. It is possible that different DNA conformations within TBP-bound complexes give rise to the same FRET state if the twist in the DNA is considered. We have modeled and discussed this concept in a previous study that used ensemble FRET to monitor TBP-induced DNA bending, and found that many combinations of bend and twist angles can give rise to a given FRET value (10). We also used a crystal structure of the human TBP/TATA DNA complex to create a model of an 18 bp TBP-bent DNA with a centered TATA box (5). This showed that the distance between the 5′ carbons on the ends of the bent DNA is 54.7 Å. The distance between the fluorphores in the bent conformation cacluated from our smFRET data is 55.3 Å. Hence, our data suggest that the bend we observe in smFRET is similar to that observed in the crystal structure; however, the distance calculated from our smFRET values is only an estimate due to underlying assumptions in the orientation and environment of the dyes. It is also possible that TBP can bind DNA prior to bending the TATA box (12, 13), which would be resolved in future smFRET experiments involving 3 fluorphores, two labeling the DNA (to monitor bending by FRET) and one on TBP (to monitor DNA binding).
In a general sense, our results are consistent with a model developed by Parkhurst, Brenowitz, and colleagues in which TBP simultaneously binds and bends TATA DNA. In their experiments using yeast TBP, complexes containing partially bent DNA were not observed (14, 15). By contrast, single molecule optical tweezer experiments of DNA bending by yeast TBP led to the conclusion that a partially bent, long-lived intermediate forms en route to the fully bent state (16). We do not observe a distinct third FRET state between the unbent and bent states in the histograms of our smFRET data with human TBP as would be predicted from the optical tweezer study. Hence, our data best support a model in which TBP uniformly bends DNA via a mechanism that does not involve a partially bent intermediate.
Mutating the TATA box strongly reduces the affinity of TBP DNA binding, but does not affect the extent of DNA bending
Our smFRET data show that TBP binds and bends TATA(A3) DNA with a much reduced affinity compared to the consensus sequence, which corroborates data from ensemble biochemical experiments (7, 10). Indeed, we did not observe TATA(A3) DNA molecules in the higher FRET state (i.e. bent) in the presence of TBP under our standard conditions (Figure 4B). We did, however, observe TATA(A3) molecules in the higher FRET state when either TFIIA was included with TBP (Figure 4C) or the KCl concentration was reduced (Figures 5B, 5C, and 5D). Both of these observations are consistent with previous ensemble experiments showing that TBP/DNA complexes are more stable at lower salt concentrations and/or in the presence of TFIIA (10).
Our smFRET results lead us to conclude that TBP bends the TATA(A3) mutant DNA to the same extent as the consensus sequence (smFRET states of 0.36–0.37 and 0.37 at 50 mM KCl, respectively). This similarity was not expected based on previous ensemble studies that compared DNA bending by human and yeast TBP on these two sequences (7, 10, 17). Given the change in bending reported in the ensemble studies and our measured smFRET efficiency of 0.37 for the bent consensus TATA box, we expected to observe an smFRET state of approximately 0.29 for the TATA(A3) mutant. Instead we clearly observed a bent population with a FRET state of 0.36–0.37, therefore, the mutant TATA(A3) DNA can be far more bent than would be predicted from ensemble studies. We cannot, however, rule out that a weakly populated 0.29 FRET state exists since this would be difficult to resolve from the unbent DNA. The inconsistency between smFRET and ensemble studies with human TBP is likely due to the weak binding affinity of TBP for the TATA(A3) sequence. In the previous ensemble FRET studies with human TBP (10), if the TATA(A3) DNA was not fully occupied, then the measured FRET would have been an average of the FRET from free DNA and bent DNA in the population, resulting in a lower calculated bend angle for the population. Ensemble studies with yeast TBP have also concluded that TBP bends TATA(A3) DNA significantly less than consensus TATA DNA (7, 17). It will be interesting to further investigate DNA binding by yeast TBP on consensus and mutant TATA boxes using single molecule approaches.
Many studies have shown that mutating the TATA sequence away from consensus results in decreased affinity for binding TBP as well as a reduction in transcriptional activity both in vitro and in cells (7, 17, 20). Some ensemble studies have concluded that decreased DNA bending by TBP correlates with decreased affinity and decreased transcriptional activity (7, 9). Our smFRET results show that there in no discernable difference in bending between the consensus and TATA(A3) constructs although there is a substantial difference in affinity, hence the degree of bending does not strongly correlate with binding affinity. This is consistent with crystal structures in which TBP induced the same conformational change in 10 different naturally occurring TATA box variants, most of which reduced transcriptional activity (32).
TFIIA does not substantially change the extent to which TATA DNA is bent
Our smFRET experiments showed that the fraction of DNA molecules occupying the TBP-induced higher FRET state increased in the presence of TFIIA for both the consensus and mutant TATA(A3) constructs, indicating that TFIIA facilitates TBP binding to and bending DNA. This is consistent with previous data showing that TFIIA increases the affinity and kinetic stablity with which TBP interacts with DNA (10, 21–23). Although TFIIA increased the portion of DNA molecules in the higher FRET state, it did not appreciably change the mean FRET efficiency of the TBP-bent state on either the consensus or TATA(A3) constructs. Therefore, in this system TFIIA does not alter the extent, or angle, at which DNA is bent by TBP, which is consistent with crystal structures of the human and yeast TFIIA/TBP/TATA and TBP/TATA complexes (33–35). Our previous ensemble FRET studies indicated that TFIIA decreased the FRET efficiency of TBP-bent consensus TATA DNA, which reflected either a change in the angle at which TBP bent the DNA within the TATA box, the angle at which the DNA emerging from the TBP/DNA complex twisted, or a combination of changes in both bend and twist angles (10). We performed ensemble experiments using smFRET conditions, and observed no effect of TFIIA on the FRET efficiency of TBP-bent DNA. It is possible that contacts between TFIIA and upstream DNA in the 38 bp smFRET construct restrict changes in the bend or twist angle that could occur with the 14 bp DNA construct used in the original study.
smFRET studies of the kinetics of bending and unbending reveal new aspects of the mechanism of DNA binding by human TBP
Our smFRET experiments allowed us to investigate the kinetics with which TBP bends DNA as well as the kinetics with which bent DNA in complexes reverts back to unbent DNA. For the consensus TATA box in the presence of 25 nM TBP, a histogram of the dwell times of unbent DNA occurrences fit nicely with a monoexpoential, yielding a pseudo first order rate constant of 0.026 s−1. Taking into account the TBP concentation provides a calculated second order rate constant of 1 × 106 M−1s−1, which is consistent with previous studies that have measured rates of TBP/DNA association that are well below the diffusion limit (11, 13, 18, 36, 37). A histogram of the dwell times of bent DNA occurrences did not fit nicely with a monoexpoential, but instead was fit with a biexponential yielding rate constants of 0.19 s−1 and 0.019 s−1. The second of these two rate constants compares well with our previous measurement of the rate of unbending using ensemble FRET (0.013 s−1, (10)). Observing biphasic decay in our smFRET data shows that two populations of TBP/consensus TATA complexes formed, having distinct kinetic stabilites. The DNA in these complexes was bent to the same extent. We used the bent and unbent dwell times to calculate an apparent KD for the TBP/DNA interaction, presuming binding and bending are concerted. For the data collected at 50 mM KCl (Figure 5E) we obtain an appKD of 6 nM, and for the data collected at 150 mM KCl (Figure 2B) we obtain an appKD of 78 nM. Both the appKD values and the 13-fold difference in affinity due to salt concentration are consistent with previous measurements for yeast TBP (18).
When we analyzed the kinetics on the TATA(A3) sequence we found that histograms of both the dwell times of unbent occurrences and bent occurrences were nicley fit with monoexponentials, yielding rate constants of 0.12 s−1 and 0.31 s−1, respectively. The affinity of TBP binding to the TATA(A3) DNA is much lower than binding to the consensus TATA box (7, 17). Our data suggest that this is largely due to the mutation causing a dramatic decrease in the kinetic stability of the bent DNA complexes, as opposed to an effect on the rate of bending. The observed rate of bending on the TATA(A3) DNA was faster than what we observed on the consensus TATA DNA. We believe that the difference in the rates of bending likely reflects salt-dependent differences in the rates of binding; the unbent dwell times on the mutant DNA were measured at 50 mM KCl and on the consensus DNA at 150 mM KCl. Previous kinetic studies of the TBP/DNA interaction have shown that the rate of association increases as the salt concentration decreases (18).
We were unable to determine rate constants from data obtained in the presence of TFIIA on both the consensus and TATA(A3) constructs due to the fact that: 1) we did not observe a population with a low FRET state, hence we were unable to obtain dwell times for unbent molecules in the presence of TBP, and 2) high FRET occurrences photobleached prior to transitioning back to a low FRET state, hence we could not obtain dwell times for bent molecules. The rate constant for photobleaching in our system is approximately 0.004 s−1, which provides an upper limit for the rate constant for unbending in the presence of TFIIA on the consensus and TATA(A3) constructs, which is consistent with previous measurements (10).
Our kinetic analyses lead us to reconsider models for DNA binding and bending by TBP. Previous studies concluded from global analysis of ensemble biochemical data that human TBP binds and bends the AdMLP TATA box via a three step linear mechanism (9). In this model, the two intermediates and the final bent complex all contain DNA bent to the same extent. We observe two kinetically distinct populations of complexes containing similarly bent DNA; however, a third kinetically distinct population was not observed in our smFRET studies. In the published model, the first bent intermediate is predicted to form within seconds and dissociate 7 times more rapidly than the rate at which it moves forward to the second intermediate. In a smFRET experiment this would predict that a given DNA molecule would undergo mutliple relatively rapid bending/unbending transitions (dwell time of the bent state would be less than 1 s) prior to transitioning into a stable bent state. We do not observe this in our smFRET traces with the AdMLP consensus TATA box, even though our time resolution (30 ms) would allow us to observe these rapid transitions if they occurred.
Our kinetic data with the AdMLP conensus TATA box can be approximated by either of the following two bent state models.
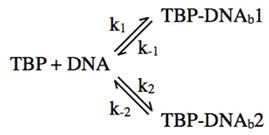 |
In both the linear model and branched model the two DNA bound complexes (TBP-DNAb1 and TBP-DNAb2) contain DNA bent to the same extent, but the formation and dissociation of the two complexes are governed by distinct rate constants. It remains possible that other steps exist that occur too rapidly to observe in our system (e.g. states with dwell times less than 30 ms). Because the FRET states for both kinetically distinct complexes are the same, our studies do not allow us to distinguish between these two models. The measurement of two rate constants for unbending does not preclude one model over the other, and we cannot say with certainty that either of the apparent rate constants we measure is k−1 or k−2. Previous studies allow for speculation on the structural or mechanistic basis for observing two kinetic populations. For example, TBP binds the consensus TATA box in two orientations (38); it is possible that the two kinetic populations represent TBP bound in two different orientations with the bend induced in each orientation being the same. It is also possible multiple populations allow for increased specificity in the binding reaction, with one reflecting a less-stable “check-point” and the other reflecting a more-stable final complex. Although in this scenario, the less-stable complex would be bent to the same extent as the final complex.
With the TATA(A3) construct we did not observe biphasic decay of bent complexes. It is possible that k2 is sharply decreased on the mutant construct, therefore in the linear model the transition from the first bent complex to the second would not occur, and in the branched model the second complex would not form to a significant extent. Alternatively, a second population of complexes might be present on the TATA(A3) construct, but the formation and dissociation of these complexes occurs too rapidly to resolve in our system. Finally, for both the consensus and TATA(A3) constructs in the presence of TFIIA the complexes we observe were kinetically very stable. If two kinetically distinct populations exist in the presence of TFIIA they both must have average dwell times that are much longer than the rate of photobleaching in our system (half time for decay of over 180 s). Alternatively, TFIIA could force complexes into a single kinetic state by pushing the reaction to the second complex in the linear pathway (e.g. decreasing k−1 or k−2) or to the more stable complex in the branched pathway.
Supplementary Material
Acknowledgments
The authors would like to thank Anne Marie Blair and Matthew Guerra for their input, Art Pardi, Haemi Lee, and Vasiliy Fomenko for establishing the TIRF microscope and providing advice on its use, Amanda Carpenter for advice on surface preparation, and Brian Ziemba for guidence on the fluorescence anisotropy measurements. We would also like to thank Art Pardi for helpful discussions on smFRET, data analysis, and programming.
Abbreviations
- PEG
polyethylene glycol
- mPEG
monomethoxypolyethylene glycol
- smFRET
single molecule fluorescence resonance energy transfer
- TBP
TATA binding protein
- Pol II
RNA polymerase II
- TAFs
TBP-associated factors
- TFIIA
transcription factor IIA
- TFIID
transcription factor IID
Footnotes
Acknowledgements: This work was funded by grant MCB-0919935 from the National Science Foundation. R.H.B. was supported in part by NIH Training Grant T32 GM07135.
Supporting Information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 2.Starr DB, Hawley DK. TFIID binds the minor groove of the TATA box. Cell. 1991;67:1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- 3.Lee DK, Horikoshi M, Roeder RG. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991;67:1241–1250. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 5.Nikolov DB, Chen H, Halay ED, Hoffman A, Roeder RG, Burley SK. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc Natl Acad Sci USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Parkhurst KM, Powell RM, Brenowitz M, Parkhurst LJ. DNA bends in TATA-binding protein-TATA complexes in solution are DNA sequence-dependent. J Biol Chem. 2001;276:14614–14622. doi: 10.1074/jbc.M004402200. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Parkhurst KM, Powell RM, Parkhurst LJ. DNA sequence-dependent differences in TATA-binding protein-induced DNA bending in solution are highly sensitive to osmolytes. J Biol Chem. 2001;276:14623–14627. doi: 10.1074/jbc.M004401200. [DOI] [PubMed] [Google Scholar]
- 9.Whittington JE, Delgadillo RF, Attebury TJ, Parkhurst LK, Daugherty MA, Parkhurst LJ. TATA-binding protein recognition and bending of a consensus promoter are protein species dependent. Biochemistry. 2008;47:7264–7273. doi: 10.1021/bi800139w. [DOI] [PubMed] [Google Scholar]
- 10.Hieb AR, Halsey WA, Betterton MD, Perkins TT, Kugel JF, Goodrich JA. TFIIA changes the conformation of the DNA in TBP/TATA complexes and increases their kinetic stability. J Mol Biol. 2007;372:619–632. doi: 10.1016/j.jmb.2007.06.061. [DOI] [PubMed] [Google Scholar]
- 11.Masters KM, Parkhurst KM, Daugherty MA, Parkhurst LJ. Native human TATA-binding protein simultaneously binds and bends promoter DNA without a slow isomerization step or TFIIB requirement. J Biol Chem. 2003;278:31685–31690. doi: 10.1074/jbc.M305201200. [DOI] [PubMed] [Google Scholar]
- 12.Zhao X, Herr W. A regulated two-step mechanism of TBP binding to DNA: a solvent-exposed surface of TBP inhibits TATA box recognition. Cell. 2002;108:615–627. doi: 10.1016/s0092-8674(02)00648-7. [DOI] [PubMed] [Google Scholar]
- 13.Hoopes BC, LeBlanc JF, Hawley DK. Kinetic analysis of yeast TFIID-TATA box complex formation suggests a multi-step pathway. J Biol Chem. 1992;267:11539–11547. [PubMed] [Google Scholar]
- 14.Parkhurst KM, Brenowitz M, Parkhurst LJ. Simultaneous binding and bending of promoter DNA by the TATA binding protein: real time kinetic measurements. Biochemistry. 1996;35:7459–7465. doi: 10.1021/bi9530301. [DOI] [PubMed] [Google Scholar]
- 15.Parkhurst KM, Richards RM, Brenowitz M, Parkhurst LJ. Intermediate species possessing bent DNA are present along the pathway to formation of a final TBP-TATA complex. J Mol Biol. 1999;289:1327–1341. doi: 10.1006/jmbi.1999.2835. [DOI] [PubMed] [Google Scholar]
- 16.Tolic-Norrelykke SF, Rasmussen MB, Pavone FS, Berg-Sorensen K, Oddershede LB. Stepwise bending of DNA by a single TATA-box binding protein. Biophys J. 2006;90:3694–3703. doi: 10.1529/biophysj.105.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starr DB, Hoopes BC, Hawley DK. DNA bending is an important component of site-specific recognition by the TATA binding protein. J Mol Biol. 1995;250:434–446. doi: 10.1006/jmbi.1995.0388. [DOI] [PubMed] [Google Scholar]
- 18.Petri V, Hsieh M, Jamison E, Brenowitz M. DNA sequence-specific recognition by the Saccharomyces cerevisiae “TATA” binding protein: promoter-dependent differences in the thermodynamics and kinetics of binding. Biochemistry. 1998;37:15842–15849. doi: 10.1021/bi981072u. [DOI] [PubMed] [Google Scholar]
- 19.Bareket-Samish A, Cohen I, Haran TE. Signals for TBP/TATA box recognition. J Mol Biol. 2000;299:965–977. doi: 10.1006/jmbi.2000.3797. [DOI] [PubMed] [Google Scholar]
- 20.Wobbe CR, Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imbalzano AN, Zaret KS, Kingston RE. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J Biol Chem. 1994;269:8280–8286. [PubMed] [Google Scholar]
- 22.Bonham AJ, Neumann T, Tirrell M, Reich NO. Tracking transcription factor complexes on DNA using total internal reflectance fluorescence protein binding microarrays. Nucleic Acids Res. 2009;37:e94. doi: 10.1093/nar/gkp424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weideman CA, Netter RC, Benjamin LR, McAllister JJ, Schmiedekamp LA, Coleman RA, Pugh BF. Dynamic interplay of TFIIA, TBP and TATA DNA. J Mol Biol. 1997;271:61–75. doi: 10.1006/jmbi.1997.1152. [DOI] [PubMed] [Google Scholar]
- 24.Pugh BF. Control of gene expression through regulation of the TATA-binding protein. Gene. 2000;255:1–14. doi: 10.1016/s0378-1119(00)00288-2. [DOI] [PubMed] [Google Scholar]
- 25.Weaver JR, Kugel JF, Goodrich JA. The sequence at specific positions in the early transcribed region sets the rate of transcript synthesis by RNA polymerase II in vitro. J Biol Chem. 2005;280:39860–39869. doi: 10.1074/jbc.M509376200. [DOI] [PubMed] [Google Scholar]
- 26.Ozer J, Moore PA, Bolden AH, Lee A, Rosen CA, Lieberman PM. Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 27.Sun X, Ma D, Sheldon M, Yeung K, Reinberg D. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev. 1994;8:2336–2348. doi: 10.1101/gad.8.19.2336. [DOI] [PubMed] [Google Scholar]
- 28.Joo C, Ha T. Single-molecule FRET with total internal reflection microscopy. Single-Molecule Techniques: A Laboratory Manual. 2008:3–36. [Google Scholar]
- 29.Tinoco IJ, Gonzalez RLJ. Biological mechanisms, one molecule at a time. Genes Dev. 2011;25:1205–1231. doi: 10.1101/gad.2050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 31.Shi J, Dertouzos J, Gafni A, Steel D. Application of single-molecule spectroscopy in studying enzyme kinetics and mechanism. Methods Enzymol. 2008;450:129–157. doi: 10.1016/S0076-6879(08)03407-1. [DOI] [PubMed] [Google Scholar]
- 32.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan S, Hunziker Y, Sargent DF, Richmond TJ. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 34.Geiger JH, Hahn S, Lee S, Sigler PB. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 35.Bleichenbacher M, Tan S, Richmond TJ. Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J Mol Biol. 2003;332:783–793. doi: 10.1016/s0022-2836(03)00887-8. [DOI] [PubMed] [Google Scholar]
- 36.Grove A, Galeone A, Yu E, Mayol L, Geiduschek EP. Affinity, stability and polarity of binding of the TATA binding protein governed by flexure at the TATA Box. J Mol Biol. 1998;282:731–739. doi: 10.1006/jmbi.1998.2058. [DOI] [PubMed] [Google Scholar]
- 37.Hoopes BC, LeBlanc JF, Hawley DK. Contributions of the TATA box sequence to rate-limiting steps in transcription initiation by RNA polymerase II. J Mol Biol. 1998;277:1015–1031. doi: 10.1006/jmbi.1998.1651. [DOI] [PubMed] [Google Scholar]
- 38.Cox JM, Hayward MM, Sanchez JF, Gegnas LD, van der Zee S, Dennis JH, Sigler PB, Schepartz A. Bidirectional binding of the TATA box binding protein to the TATA box. Proc Natl Acad Sci U S A. 1997;94:13475–13480. doi: 10.1073/pnas.94.25.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.