Abstract
Self-incompatibility (SI) is defined as the inability to produce zygotes after self-pollination in a fertile hermaphrodite plant, which has stamens and pistils in the same flower. This structural organization of the hermaphrodite flower increases the risk of self-pollination, leading to low genetic diversity. To avoid this problem plants have established several pollination systems, among which the most elegant system is surely SI. The SI trait can be observed in Brassica crops, including cabbage, broccoli, turnip and radish. To produce hybrid seed of these crops efficiently, the SI trait has been employed in an agricultural context. From another point of view, the recognition reaction of SI during pollen-stigma interaction is an excellent model system for cell-cell communication and signal transduction in higher plants. In this review, we describe the molecular mechanisms of SI in Brassicaceae, which have been dissected by genetic, physiological, and biological approaches, and we discuss the future prospects in relation to associated scientific fields and new technologies.
Keywords: Brassicaceae, cell-cell communication, pollen-stigma interaction, self-incompatibility, signal transduction
1. Introduction
In most species of plants the flowers are hermaphrodite that is both stamen and pistil are present in the same flower. Self-pollination occurs easily in hermaphrodite flowers, and leads to inbreeding depression and decreased genetic variation; therefore plants have evolved several mechanisms to avoid self-pollination. Self-incompatibility (SI) is defined as “the inability of a fertile hermaphrodite seed plant to produce zygotes after self-pollination”, which is surely the most elegant pollination system.1) This trait attracted the attention of Charles Darwin, who published detailed descriptions of pollination and the form of flowers during his studies of evolution.2,3) In the past, the SI phenotype was mostly evaluated by seed formation after open pollination (artificial self-pollination and cross-pollination). However, from the evaluation of seed formation, the phenotype of “incompatibility” and “sterility” cannot be distinguished. However, in relation to biological events, “incompatibility” and “sterility” are clearly different. “Sterility” is caused by non-functional male and/or female components, whereas in “incompatibility” there is a lack of seed formation in a specific male and female combination, both of which are functional. Because the final phenotypes in “incompatibility” and “sterility” are quite similar, this phenomenon was originally termed “self-sterility”. Taking the initial letter of “sterility”, the genetic locus regulating SI was termed the “S locus”, which is still used as the authorized locus name.
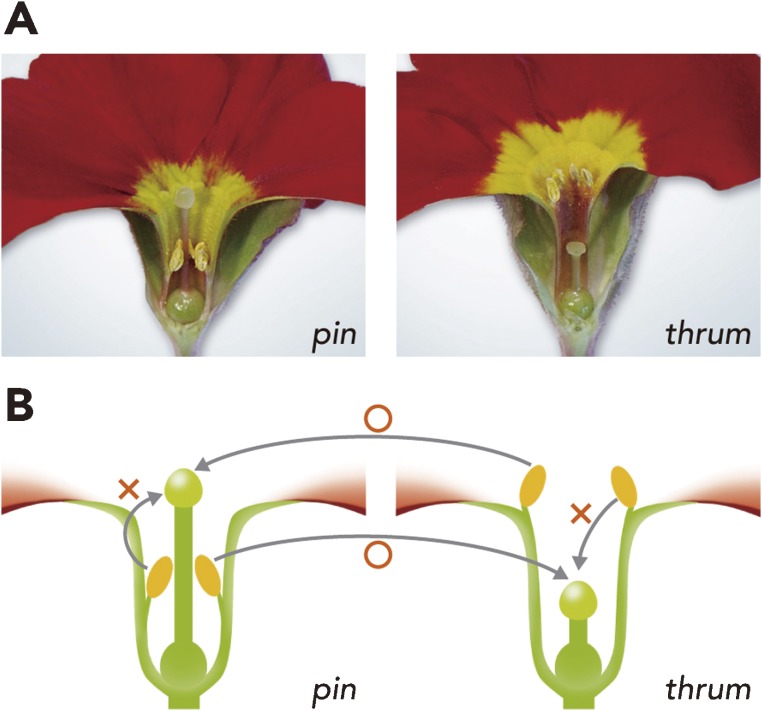
SI is classified morphologically into heteromorphic SI and homomorphic SI. Heteromorphic SI is related to different flower shapes; for example, pin (long pistil and short stamen length) and thrum (short pistil and long stamen length) forms are present in primrose (Primula species), buckwheat (Fagophyrum species) and star fruit (Averrhoa species). Cross pollination between different flower types is compatible and is regulated by a single locus with two alleles, S and s (Fig. 1). The genotype of pin is ss homozygote, and that of thrum is Ss heterozygote. Thus, on cross-pollination, a frequency of 50% each of pin and thrum should be maintained in a wild population under SI regulation. Interestingly, this S gene controls both the incompatibility phenotype and the flower shape (that is length of pistil and stamens). Although several attempts have been made to dissect genes residing at the S locus in plants with heteromorphic SI, no S genes or their products have been identified to date.1)
Figure 1.
Photograph (A) and schematic model (B) of heteromorphic SI in primrose. (A) The pin (long pistil and short stamen length) form is shown on the left, and thrum (short pistil and long stamen length) is on the right. (B) Selfed pollen and pollen derived from the same flower form are incompatible. Crossed pollen and pollen derived from the different flower form are compatible.
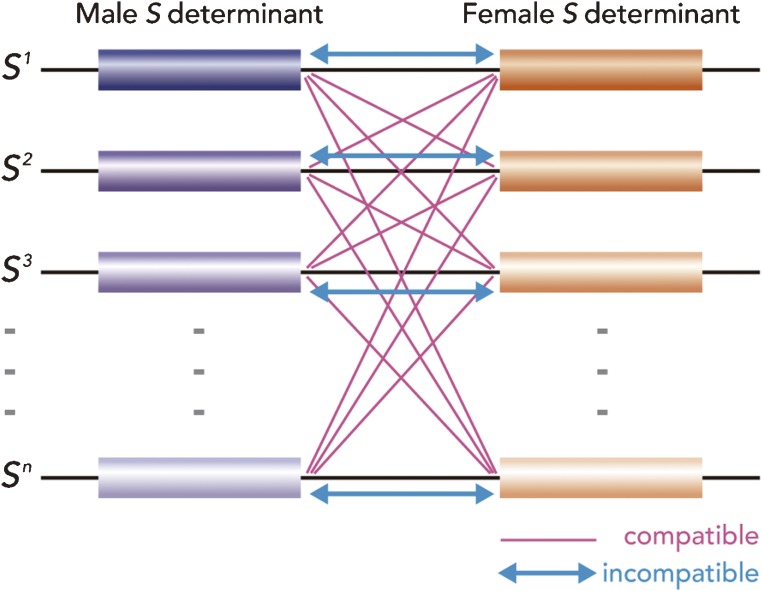
On the other hand, homomorphic SI species have a single flower shape and are classified by the inheritance pattern of the S gene into gametophytic SI and sporophytic SI.1,4) Both SI phenomena are controlled by a single locus, with multiple S alleles (S1, S2, S3, … Sn) in most of the plant species. As described in Fig. 2, the incompatible phenotype is apparent in self-pollination and in cross-pollination between pollen and stigma of two different plants carrying the same S allele. To represent alleles, superscripts are commonly used in general genetic research; however, traditionally, S alleles are denoted by subscripts. In most of our papers and review articles, we have used superscript allele symbols, according to the general genetic convention.4–8) Therefore, in this article, we have again used superscripts for allelic gene representation.
Figure 2.
Schematic model of the S locus. The S locus contains at least two genes, one encoding the male S determinant, which is expressed in anther tapetum surrounding the pollen grains during development, and the other encoding the female S determinant, which is expressed in the pistil (stigma). Both the genes encoding male and female S determinants are inherited as one segregating unit. The number of S alleles is thought to be over 50 to 100 in Brassica species.167–169) When both male and female S determinants are derived from the same S alleles, the incompatible response occurs. In contrast, in the case of a combination of different S alleles from the male and female parents, the compatible response is observed.
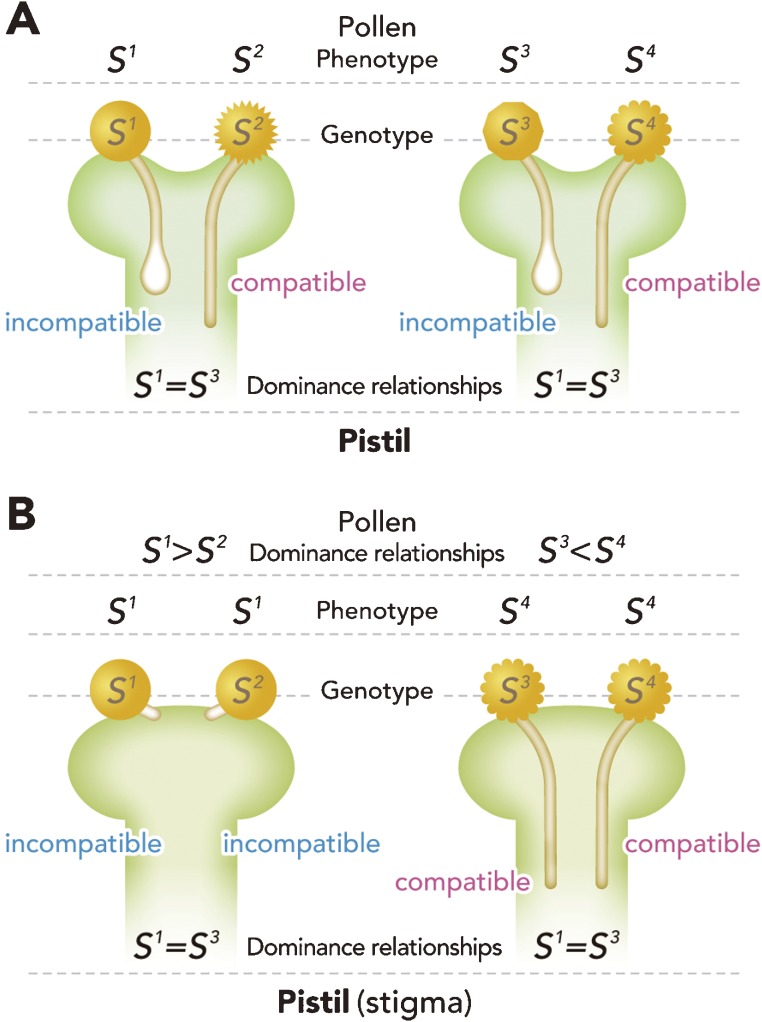
In gametophytic SI (GSI), the S phenotype of pollen is determined by its own haploid S gene. The self-pollen inhibition occurs during pollen tube elongation in the style of the pistil in most GSI. Plant species in families Fabaceae, Onagraceae, Poaceae, Rosaceae, Plantaginaceae and Solanaceae exhibit GSI (Fig. 3A). On the other hand, in sporophytic SI (SSI), the S phenotype of pollen is determined by its parental diploid S gene interaction, so that dominance relationships occur between S alleles (Fig. 3B).9,10) The landmark genetic SSI model, established by Bateman from a series of experiments, is an excellent explanation of S gene behavior, and is still valid today.11–13) The molecular mechanisms of S allele dominance relationships will be described below. In most SSI plant species, from families Asteraceae, Brassicaceae and Convolvulaceae, pollen tube inhibition occurs on the stigma surface (Fig. 4).
Figure 3.
Relationships between phenotype and genotype in GSI (A) and SSI (B). The pistil (stigma) phenotype presented here is S1S3 codominance, to clearly demonstrate the difference between the GSI and SSI. In both types of SI, pollen is rejected when the SI phenotype of pollen is identical to that of the pistil (stigma). (A) In the case of GSI, the pollen phenotype is identical to the genotype. The pistil phenotype of the GSI is always expressed in codominance. (B) In the case of SSI, the pollen phenotype is determined by interaction of the parental genotype; therefore, there is a dominance relationship at the pollen side. If S1 is dominant over S2, all pollen grains produced from the S1S2 pollen parent show S1 phenotype, and are rejected on the S1S3 (S1 = S3) stigma. In contrast, on the same stigma (S1 = S3), if S3 is recessive to S4 at the pollen side, all pollen grains, which show S4 phenotype, germinate and pollen tube penetration is observed.
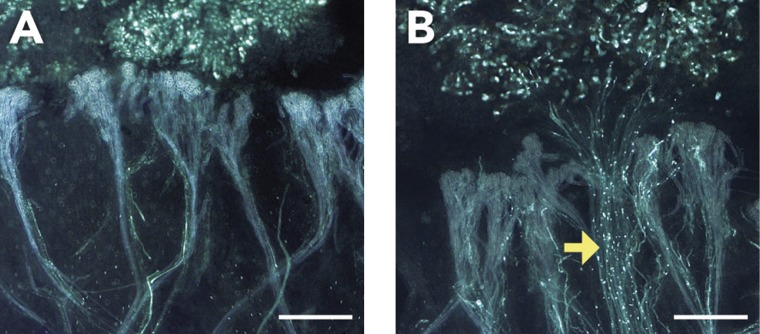
Figure 4.
Micrographs showing (A) the inhibition of self-pollen and the inability of the emerging pollen tubes to invade the papilla cell wall (incompatibility), and (B) the penetration of cross-pollen tubes into the papilla cells (compatibility) in Brassica rapa L. Yellow arrow indicates the penetrated pollen tubes. With aniline blue staining, pollen tubes appear as distinct tubes with prominent dots, because callose plugs in the pollen tubes fluoresce brightly. In contrast, vascular bundles and other tissues do not fluoresce brightly, as shown in (A). Bar = 200 µm. (Courtesy of Masaaki Osaka and Ken-ichiro Hiroi, unpublished data).
SI is interesting in two respects: firstly, SI involves cell-cell communication between pollen/pollen tube and pistil, and signal transduction in the stigma/style; and, secondly, SI is an important trait for F1 hybrid seed production in Brassica rapa (syn. campestris, e.g. Chinese cabbage and turnip), and B. oleracea (e.g. cabbage, cauliflower, broccoli, kohlrabi, kale and Brussels sprouts), which was established in a Japanese seed company before the 2nd World War.6) In other words, we are interested in Brassica SI from the viewpoint of both fundamental biology and agricultural applications. Thus, an understand of the molecular mechanisms of SI in Brassica species is an important research target. In this review article, we will summarize SI research in Brassicaceae species, including molecular cloning of the male and female S determinants, epigenetic regulation of dominance relationships between S alleles, and evolution of the SI system. Finally, we will discuss the future prospect for SI research. Many review articles have been published, and are helpful in understanding this field.1,4–8,14,15)
2. Identification of the S-locus-specific proteins from stigma
In the early SI research, Oenothera (Onagraceae), Malus, Prunus (Rosaceae), Nicotiana, Solanum, Petunia (Solanaceae), Trifolium (Leguminosae), Secale, Pahalaris, Fastuca (Poaceae), Cardamine, Iberis, Brassica (Brassicaceae), Crepis, Cosmos (Asteraceae), Theobroma (Sterculiaceae), Ipomoea (Convolvulaceae) and Antirrhinum (Plantaginaceae), among others, were studied.1) These species were well suited to the establishment of genetic models of SI systems, as described above. However, for the next step, that is identification of S gene products at the protein level, only Brassica and Nicotiana species were used during the 1970s, because the protein products derived from their female S genes are highly expressed in stigma/style tissues. Thus, the SI researchers who selected these species for study had a great advantage for molecular dissection of the S-locus.1)
In the case of the Brassica species, immunological cross-reactivity and isoelectric focusing (IEF) were applied to identify the allelic diversity of the S gene products.16,17) These two methodologies had advantages for the identification of genetic diversity of multi-allelic gene products. For example, in the case of IEF, the molecular masses of allelic gene products are similar. However, they differ in the isoelectric point (pI) because the amino acid composition of their products must be different as a consequence of the allelic diversity and tertiary structure of the proteins. Because the S-locus-specific protein was found to react with concanavalin A (Con A), which is plant lectin,18) it was concluded that this protein has a carbohydrate moiety, and it was named S-locus glycoprotein (SLG).1)
In the 1980s, SLG proteins were extracted and purified from plant material. Furthermore, the cDNA clones encoding SLG genes were obtained from Brassica rapa (syn. campestris) and B. oleracea.19,20) At this time, the SLG was proposed to be the female S gene based on several characteristics, as described below. Firstly, the glycoprotein is directed to be secreted into the cell wall by a hydrophobic signal peptide.19–21) Secondly, twelve cysteine residues were found to be completely conserved in the C-terminal region and variable regions were found in the central and C-terminal regions, from comparisons of over 30 SLGs derived from different S alleles (Fig. 5). When the nucleotide sequences of allelic SLGs were aligned, three highly divergent regions were identified and other regions were found to be highly conserved among S alleles.5) These variable regions were proposed to contribute to S allelic specificity, and could interact with the male S determinant. Thirdly, the glycosyl residues were similar among S alleles, indicating that the S allelic specificity is due to variable regions of SLG itself.1,20) Fourthly, identification of the S allele by PCR methods using primers positioned in the conserved regions has contributed to the breeding of Brassica F1 hybrid varieties.10,22,23) However, this method sometimes resulted in the false amplification of the many other SLG-like genes in the Brassicaceae genome.24–27) To exclude non-S-locus SLG-like sequences, linkage to the S locus had to be confirmed by test pollinations using a Brassica breeding system.5)
Figure 5.
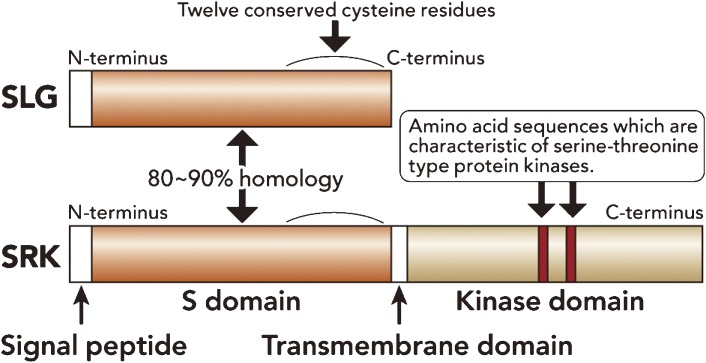
Schematic structure of SLG and SRK. SLG is an S allele-specific secreted glycoprotein, having a hydrophobic signal peptide in the N-terminus and 12 cysteine residues at the C-terminus. SRK is a receptor protein kinase, whose extracellular domain (receptor domain) is similar to SLG. SRK consists of a signal peptide, an SLG-like domain (S-domain), a hydrophobic transmembrane domain and a cytoplasmic catalytic domain (kinase domain) of the serine/threonine protein kinase type.
Furthermore, in the 1980s, to construct cDNA libraries and determine the amino acid sequence of SLG, tens to hundreds of thousands of stigmas, the top part of the pistil, were necessary, because SLG expression is spatiotemporally controlled in mature stigma papilla cells.21,28) Thus, this identification of SLG residing at the S locus gave a great advantage to Brassica SI researchers in the further molecular dissection of the S locus, and subsequently, during the 1990s, in determining the sets of male and female SI genes, as described below. However, at that time, comparisons of SLG to sequences in public databases did not provide any evidence as to the function of SLG in SI recognition.5)
3. Identification and characterization of female S determinants
Following SLG identification, molecular dissection of the S locus was advanced by several important findings. One of the most important milestones was the discovery of ZmPK1, encoding receptor-like protein kinase of maize, whose receptor domain has homology to SLG.29) After this finding, several allelic SRK genes, encoding S-locus receptor-like kinases, located at the S locus, were isolated and characterized from B. rapa, B. oleracea and B. napus. SRK contains three functional domains: an extracellular S domain (receptor domain), a transmembrane domain, and an internal serine/threonine protein kinase domain (Fig. 5).28,30–32) Interestingly, sequence similarity at the amino acid level of SLG and the S domain of SRK within S alleles is over 80%,5) indicating that these two regions arose by duplication within the S locus.28,33)
Thus, at that time, two stigma-specific genes, SLG and SRK, were known to be present at the S locus. However, it was difficult to determine the physical distance between the SLG and SRK genes precisely. In the case of B. oleracea, the size of the S locus was estimated to be 350 kb by PFGE analysis in the early 1990s,34) although this depended on the location of recognition sites of restriction enzymes around the S locus. An S-locus region as large as 350 kb suggested that cloning and sequencing of the S-locus might be quite difficult. However, this was overcome by the second important breakthrough, the successful cloning of the 76-kb genomic fragment containing both SLG and SRK genes in the S9 allele of B. rapa by use of the PAC vector.35) Based on this success, molecular dissection of the whole S locus was possible at the nucleotide sequence level. B. rapa had an advantage over B. oleracea for molecular dissection of the S-locus region because the size of the S locus of B. rapa was smaller than that of B. oleracea.36) Thus, the physical genomic structure of the S-locus region was determined in B. rapa, and also in B. napus (Fig. 6).37–40) Interestingly, in the case of B. napus, the S-locus region was found to have originated from the introgressed B. rapa genome and not from B. oleracea.31,37)
Figure 6.
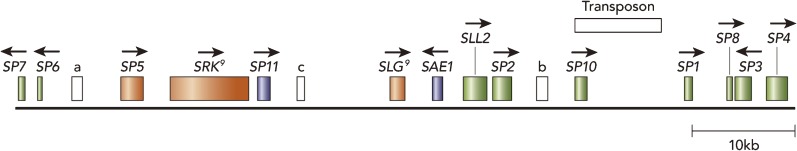
Physical map of the S locus of the S9 line of B. rapa, showing the location of 14 genes, a transposon-like sequence and three open reading frames (ORFs). Filled boxes indicate locations of the 14 genes, and horizontal arrows above the genes indicate directions of their transcription. Three long ORFs (a, b and c) and a transposon-like sequence are represented by open boxes labeled “a,” “b,” “c” and “Transposon.” Three genes represented by orange-colored boxed are specifically expressed in stigma tissues. Two genes represented by purple-colored boxes are specifically expressed in anther tissues. The other nine genes represented by green-boxes are expressed in both vegetative and reproductive tissues.
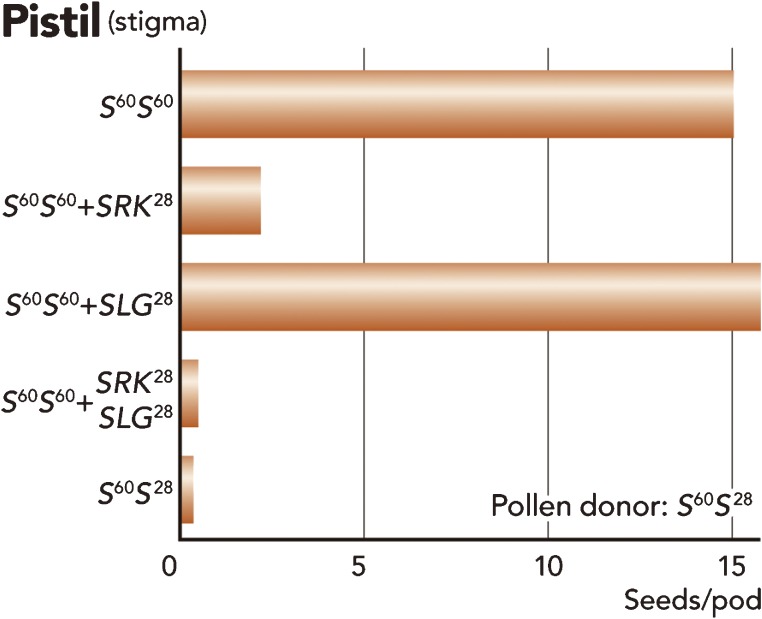
Next, it was important to determine whether SLG and/or SRK are necessary for SI recognition at the stigma side, and for this several gene transformation studies were performed in the 1990s. Loss-of-function experiments showed that SLG and/or SRK are important for the SI recognition reaction.5,6,41–43) In most cases, following introduction of SLG or SRK a change to the self-compatibility (SC) phenotype was observed, which might have been induced by co-suppression as shown by the change in petunia flower color.6,44) In order to overcome co-suppression between endogenous and exogenous SLG and/or SRK genes, Takasaki and co-workers used lower-homology SLG/SRK genes from the two groups of S alleles, class I and class II.45) As described below, S alleles (S haplotypes) of class I (e.g. S28) are dominant over those of class II (e.g. S60) at the pollen side.10,32) Takasaki and co-workers introduced SLG28 and SRK28 into recipient plant lines, S60 homozygote or S28S60 heterozygote of B. rapa. As a result, the transgenic plants harboring SLG28 did not show a change in S phenotype at the stigma side, while the transgenic plants harboring SRK28 showed the S allele specificity at the stigma side, but not in pollen. Interestingly, transgenic plants harboring both SLG28 and SRK28 showed the rigid SI phenotype comparable to non-transgenic S28S60 heterozygotes, indicating that SRK is the female S determinant and SLG is required for full manifestation of the SI response (Fig. 7).45) This additional effect of SLG was not observed in B. napus;46) the difference in SLG effect may be a consequence of the genome organization of the diploid (B. rapa) and amphidiploid (B. napus) species.
Figure 7.
Test pollinations for evaluation of SLG and SRK transgenic plants by seed formation. Line S60 was used as the recipient of the transgene and line S28S60 was used as the pollen tester. In this case, the S phenotype of all pollen was S28 because S28 is dominant over S60. The phenotype of transformants carrying SLG28 was coincident to that of the non-transformant recipient plant, that is S60. In contrast, the seed formation of transformants having SRK28 decreased compared to that of transformants having SLG28 and recipient S60 line. After genetic crossing of the two transformants, which carried SLG28 and SRK28 transgenes, the F1 plant showed a rigid SI phenotype, like the S60S28 heterozygote. SI phenotype was assessed by the number of seeds formed per pod.
4. Identification and characterization of male S determinants
In the course of investigating the identification of the male S determinant, several S-linked genes were found to be unrelated to the male S determinant.47–50) However, another detailed approach, analysis of pollen coat proteins (PCPs), successfully lead to identification of the male S determinant. In Brassica species, PCPs, which are derived from anther tapetum, are essential for pollen development.51) Initially, SLG-interactive PCPs were searched,52) and these interactive molecules were then characterized as cysteine-rich small proteins.53–57) In the course of these experiments, a bioassay system showed that a <10 kDa PCP is the male S determinant.58) Interestingly, a gene encoding a PCP-like protein, SP11, was found to be located at the S locus, near SRK in B. rapa; thus SP11 was the most likely candidate for the male S determinant (Fig. 6).38) Subsequently, it was demonstrated that SP11 is the male S determinant by using bioassay and transgenic experiments in B. rapa.59,60) Around the same time, a gene termed SCR (S-locus cysteine-rich protein) in B. oleracea was also cloned, and identified as the male S determinant.61) Because SP11 and SCR are identical,38,59,61) SP11 will be used as the name of the male S determinant in this review article, in both Brassica and Arabidopsis species.
After identification of SLG, SRK and SP11 genes, many allelic genes were isolated,5,6) which made it possible for us to calculate the allelic diversity. As described above, S alleles were divided into two groups, class I and class II, based on the sequence similarity between allelic genes of the three S-locus genes, SLG, SRK and SP11. In the case of SLG and SRK, within a group, allelic diversity was from a few percent to around 20%. In contrast, between groups, the diversity was over 30%. In contrast, the allelic diversity of SP11 was higher than that of SLG and SRK.5,6) Particularly in the class I alleles of SP11, only certain amino acid residues (glycine, aromatic amino acid and cysteine residues) were conserved in the mature protein.23) From determination of the tertiary structure with 2-D NMR analysis, these amino acids were found to be important for recognition specificity.62) From comparison of SP11 and SRK sequences, these two genes encoding the pollen ligand and stigma receptor, respectively, were found to have co-evolved in a trans-specific mode.6,23,39)
5. Molecular mechanisms of the SI reaction
After identification of both male and female S determinants, the next research targets were the physical interaction between SP11 and SRK, and the SRK-related signaling cascade following this interaction. For the physical interaction between SP11 and SRK, different results have been reported by two laboratories. Shimosato et al.63) showed that both the S-domain and membrane-anchoring domain were necessary for allele-specific interaction by using cross-linking and immunological methods. In contrast, Kachroo et al.64) showed that only the S-domain of SRK could interact with SP11 in the allele-specific manner by using an immunoprecipitation method. In the future, after determination of the tertiary structure of SRK, this inconsistency is likely to be resolved.
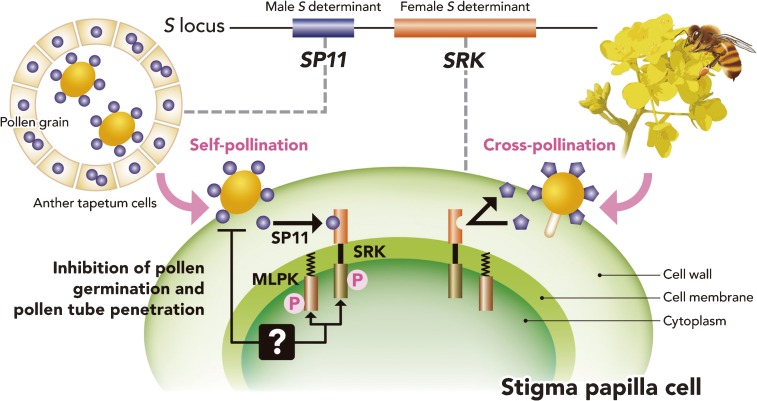
The physical interaction of SP11 and SRK was found to trigger autophosphorylation of SRK in an allele-specific manner.65) The question was then, how does the SI signal transduce in the cytoplasm? In order to clarify the SRK signaling cascade, several SRK-interacting molecules were identified and characterized.66–72) MLPK (M locus protein kinase), identified from the self-compatible line yellow sarson, was found to encode a novel membrane-anchored cytoplasmic protein kinase,71) which interacts with SRK directly.72) ARC1 (arm repeat containing 1) and THL1 (thioredoxin 1) were isolated using a yeast two-hybrid system with the kinase domain of SRK as a bait.66) ARC1 interacts with the kinase domain of SRK in a phosphorylation-dependent manner and has E3 ubiquitin ligase activity, indicating that the proteasome protein degradation system is involved in SI signal transduction. Furthermore, an ARC1 knock-out line showed the self-compatible phenotype.67,69) At present, only MLPK and ARC1 have been identified as positive effectors of the Brassica SI reaction. Other identified factors found to date may be negative effectors of SI.68,71) Recently, novel self-compatible lines of B. rapa have been identified, which will enable progress to be made in understanding the role of other interactive molecules in the complete SI signaling cascade.73) A schematic model of the current understanding of Brassica SI is shown in Fig. 8.
Figure 8.
Schematic model for self-pollen recognition in Brassica species. Male and female S determinant genes, SP11 and SRK, are located at the S locus. SP11 is predominantly expressed in the tapetum cells of anther locules, and accumulates on the pollen surface during pollen maturation. On self-pollination, SP11 molecules penetrate into the papilla cell wall, and interact with SRK in an S-allele-specific manner. Phosphorylated SRK interacts with MLPK. After the subsequent signal transduction, which has not yet been determined, rejection of the self-pollen occurs.
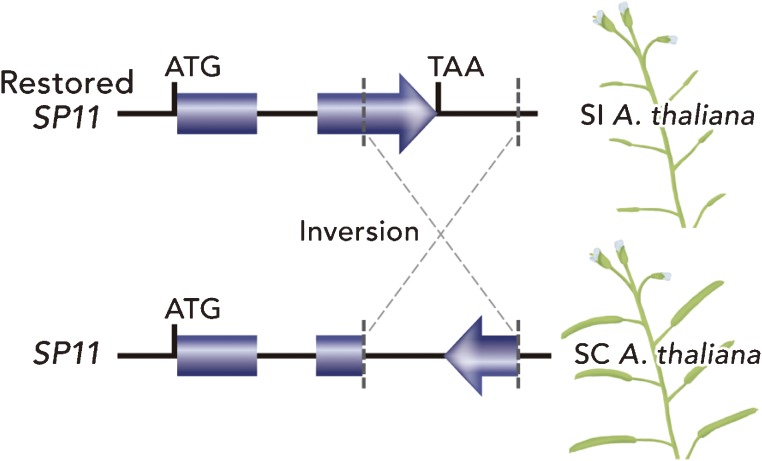
Other approaches to studying the SI signaling cascade have used Arabidopsis species. Because Arabidopsis thaliana is a self-compatible model plant, a very large number of traits of A. thaliana have been studied.74–77) In addition, for research into the evolution of SI and SC, Arabidopsis species are powerful research tools.78–82) As describe above, A. thaliana is a model cruciferous plant, which naturally has an SC phenotype, whereas other species, such as A. halleri and A. lyrata, are self-incompatible. A. thaliana modified to be self-incompatible is a powerful tool for genetic approaches to studying the SI signaling cascade, because the whole genome sequence of A. thaliana has been determined26) and other genetic tools (e.g. T-DNA tag lines, many ecotypes and microarray expression data) are available (http://www.arabidopsis.org/). To produce self-incompatible A. thaliana by transformation, two approaches have been used. Nasrallah and co-workers introduced SP11 and SRK from self-incompatible A. lyrata into self-compatible A. thaliana (C24 ecotype) and the resulting transgenic A. thaliana, with both SP11 and SRK, exhibited the SI trait.83,84) On the other hand, Tsuchimatsu, Suwabe and co-workers introduced only SP11 into the Wei-1 ecotype of A. thaliana, which has a functional SRK but shows SC, to produce a self-incompatible A. thaliana.85) In the course of sequencing analysis of the S-locus genes in self-incompatible A. halleri, the SP11 gene of haplogroup A of A. halleri was found to be quite similar to the disrupted SP11 gene of haplogroup A of A. thaliana; it is suggested that a 213-bp inversion of SP11 might have occurred in A. thaliana. Furthermore, from test-crosses using the pollen of the haplogroup A of A. halleri, it was found that several ecotypes (e.g. Wei-1) were functional on the female side of the SI system, indicating that these ecotypes may have a functional SRK and SI signaling cascade. By using Wei-1 as a representative of these ecotypes, self-incompatible transgenic A. thaliana plants carrying the restored SP11 gene were successfully produced (Fig. 9).85) This transgenic A. thaliana plant with the restored SP11 transgene, in which the recipient and donor are of the same origin, is an impressive achievement because this transformation represents the artificial retrograde evolution of the SI gene.
Figure 9.
Restoration to SI phenotype in Arabidopsis thaliana. The Wei-1 ecotype usually has a self-compatible phenotype (formation of large siliques and many seeds). The reason for the SC phenotype Wei-1 is a gene inversion within the 2nd exon of SP11, although SRK is functional. When transformed to restore an active SP11, the transformants showed the self-incompatible phenotype (formation of small siliques and no seeds).
6. Molecular analysis of dominance relationships between S alleles
An important characteristic of Brassica SI is the dominance relationships between S alleles, which are a consequence of the sporophytic behavior of S genes.9–13) Genetic experiments have revealed four characteristics of the dominance relationships of cruciferous plants:9–13) (1) co-dominance is more frequent than dominance/recessiveness; (2) dominance/recessiveness in the pollen is observed more frequently than that in the stigma; (3) dominance relationships act independently in the pollen and stigma; and (4) non-linear dominance relationships are also observed, and are more frequent in the stigma than in the pollen. Discovery of the molecular mechanisms of dominance relationships has been based on these genetic features.
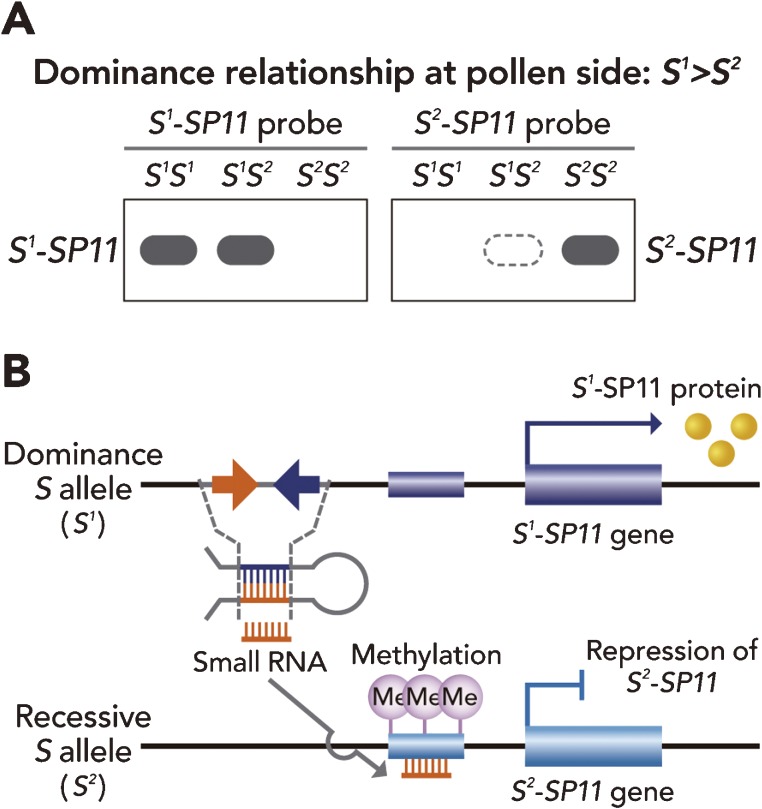
In Brassica pollen, dominance relationships of SI are regulated by transcription of SP11. In sporophytic tapetum cells of anthers of S heterozygous plants, SP11 derived from the dominant allele is normally expressed, whereas expression of the recessive SP11 is significantly suppressed (Fig. 10A).86,87) Interestingly, linear dominance relationships (S9 > S44 > S60 > S40 > S29) are also observed in B. rapa.10,87) In this case, dominance/recessiveness of S44, S60, S40 alleles could be altered, indicating involvement of epigenetic regulation.87) From observation of the methylation level of the promoter region of SP11 in several S heterozygotes, it was shown that the recessive SP11 is specifically methylated in S heterozygotes.88) Recently, it was further demonstrated that the small RNA produced from the dominant allele could activate methylation of the recessive allele. In transgenic experiments, S60 (class II) transformants with a class-I-derived small RNA region (S9) showed SC, and their promoter regions were highly methylated, as in the S9S60 heterozygote, indicating that the small RNA from the dominant allele functions in trans to induce transcriptional silencing of the recessive allele (Fig. 10B).89)
Figure 10.
Schematic model for molecular mechanisms of dominance relationships at the pollen side. (A) In the case where S1 is dominant over S2, dominant transcripts of SP11, S1-SP11, are specifically expressed in the S1S2 heterozygote. However, S2-SP11 transcripts are not detected in the S1S2 heterozygote on RNA gel blot analysis. The results demonstrate that the dominance relationship at the pollen side is regulated at the transcriptional level. (B) In the dominant S allele, small RNA, termed Smi (SP11 methylation inducer), is specifically produced, and its nucleotide sequence is highly similar to the promoter region of the recessive SP11 gene. This small RNA induces the methylation of recessive SP11, and represses the recessive SP11 at the transcriptional level.
In contrast, dominance relationships of SI at the female side appear to be post-transcriptionally regulated by SRK, unlike transcriptional suppression of SP11.90) The different mechanisms operating in the male and female side in the dominance relationships of SI are consistent with the four genetic characteristics described above.9–13)
7. Future prospects
As outlined in this review, the Brassica SI reaction has several interesting biological features, including cell-cell communication, ligand-receptor interaction, signal transduction, phosphorylation cascade, molecular evolution, allelic polymorphism and epigenetic regulation by small RNAs. For an overall understanding of SI events, collaborative research with diverse biological approaches (metabolome, proteome, bioinformatics, phenome, transcriptome, etc.) is necessary (e.g. refs. 91–119).
SI is one of the most interesting phenomena in sexual reproduction. Building on recent progress in sexual plant reproduction research, the molecular processes in male and female gametophyte development have now been extensively dissected in several plant species (e.g. refs. 104, 118–136). Such accumulation of biological knowledge will also contribute to the understand of pollen germination and pollen tube behavior in the SI recognition reaction.
Interestingly, peptide signals similar to SP11 are common in several other related phenomena (fertilization, morphogenesis, plant-microbe interaction, etc.; e.g. refs. 15, 137–141). To elucidate a complete overview of the peptide signaling, further analysis of protein-protein interaction by use of the yeast two-hybrid system, protein cross-linking, etc. is required,142,143) in addition to the genetic analysis described above. To date, binding of SP11 to the SRK receptor, the tertiary structure of SP11 and the variable regions of SP11 have been determined.62) However, the tertiary structure of SRK has not yet been established. When this has been carried out, the precise SP11-SRK allelic-specific interaction will provide new insight into SP11-induced SRK activation, as has been achieved for brassinosteroid and BRI1.144)
As the next important topics for study in pollen-stigma interaction, genetic barriers between different species and interspecific incompatibility are interesting phenomena, and these are also related to the SI reaction. Although genetic analysis of interspecific incompatibility is complicated due to the difficulty or impossibility of obtaining hybrid seeds and their progenies,1) the understanding of molecular mechanisms in the SI recognition reaction will help to identify the molecular players in interspecific incompatibility. In addition, overcoming the barrier of interspecific incompatibility could lead to the establishment of new species. From observations of chromosome hybridization, it was determined that the amphidiploid B. napus originated from crosses between B. rapa and B. oleracea.145) Interestingly, the parental diploids, B. rapa and B. oleracea, show SI phenotype and their amphidiploid, B. napus, has the SC phenotype. In some SC B. napus, the mutated genes conferring SC phenotype have been identified and characterized.146,147) Taken together, these points indicate that further studies of these Brassica SI and SC lines should contribute to our understanding of the molecular mechanism of evolution from SI to SC during polyploidy formation under cultivation.
Very many reports have been published of the early physiological studies.51) In Brassica, breakdown of SI was reported to be caused by several treatments, including CO2 gas, NaCl solution, high temperature, organic solvents and electrical stimulation.148–154) However, the molecular mechanisms involved in overcoming the SI recognition reaction are still unknown. Understanding the mechanisms in overcoming SI will contribute to the discovery of the missing links in our understanding of the physiological aspect of the Brassica SI reaction. Another factor is the cuticle layer of stigma papilla cells, which is important for Brassica SI. Even in an incompatible cross the pollen tube can penetrate into the cuticle layer,155) indicating that there is ‘cutinase’ activity on the surface of pollen grains. Recently, results of studies on cuticle wax in other tissues have been published,156,157) and these are informative for dissecting Brassica SI. Thus, from earlier studies we can obtain many pointers to advance Brassica SI research in the future.
Finally, just as we were starting to write this review article, the huge earthquake (magnitude 9.0) hit the northern part of Japan, around Sendai, on March 11, 2011.158) The Fukushima Daiichi nuclear power station was severely damaged by the huge earthquake and tsunami, and radio-active elements (iodine-131, cesium-137 and other compounds) were released into the environment.159) For the remediation of soil contaminated by radioactive compounds, especially cesium-137, Brassica species are potentially very useful.160–165) Of course, Brassica crops, cabbage, turnip, broccoli, etc., are important as food crops for people in all parts of the world. In addition, the breeding of Brassica crops has the potential to contribute to production of bio-fuel, phytoremediation and phytomining.166) In view of these points, understanding molecular mechanism of the SI recognition reaction in Brassica species will be important in establishing F1 hybrid seed production in the future.
Acknowledgements
The authors thank Soichi Kojima and Masumi Miyano (Tohoku University) for their helpful discussion and critical reading of the manuscript. We thank Masaaki Osaka and Ken-ichiro Hiroi (Tohoku University) for providing the photographs of pollination. We are also thankful to many co-workers in A. Isogai’s and S. Takayama’s groups (Nara Institute of Science and Technology) and our groups involved in this research. We also acknowledge Kokichi Hinata (Tohoku University) for his continuous encouragement to our research. This work was supported in part by Grants-in-Aid for Special Research on Priority Areas (Nos. 18075003 and 18075012) and for Scientific Research on Innovative Areas (Nos. 23113006 and 23113001) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), and a Grant-in-Aid for Young Scientists (S) (No. 20678001) and a Grant-in-Aid for Young Scientists (B) (No. 20780240 and No. 22780002) from the Japan Society for Promotion of Science (JSPS). We apologize to all those whose work in this field could not be cited because of limited space. This review article is dedicated to all the people who were lost or injured in the devastating huge earthquake and tsunami of March 11, 2011, which occurred in the northern part of Japan.
Biographies
Profile
Masao Watanabe was born in Ehime Prefecture in 1965, and received his Ph.D. in 1994 from Tohoku University under the direction of Prof. Kokichi Hinata. He was an Assistant Professor at Tohoku University (1991–1997), Associate Professor at Iwate University (1997–2005). Since 2005, he has been a Professor of Plant Reproductive Genetics, Tohoku University. His research interest is the molecular dissection of self-incompatibility in Brassica species. He received JSPS PRIZE in 2011.
Keita Suwabe was born in Kanagawa Prefecture in 1974, and received his Ph.D. in 2004 from Mie University under the direction of Prof. Yasuo Kowyama. Since 2009, he has been an Associate Professor, Mie University. His research interest is the molecular dissection of pollen-stigma interaction in higher plants. He received Japanese Society of Breeding Award for Young Scientist in 2012.
Go Suzuki was born in Shizuoka Prefecture in 1971, and received his Ph.D. in 1999 from Tohoku University under the direction of Prof. Kokichi Hinata. He was an Assistant Professor at Osaka Kyoiku University (1999–2003). Since 2003, he has been an Associate Professor, Osaka Kyoiku University. His research interest is the functional analysis of genomic organization in higher plant genes. He received Japanese Society of Breeding Award for Best Paper in 2005.
References
- 1).De Nettancourt, D. (2001) Incompatibility and Incongruity in Wild and Cultivated Plants, 2nd ed., Springer, Berlin, Heidelberg, New York, pp. 1–322. [Google Scholar]
- 2).Darwin, C. (1876) The Effects of Cross- and Self-fertilisation in the Vegetable Kingdom. John Murray, London, pp. 1–482. [Google Scholar]
- 3).Darwin, C. (1877) The Different Forms of Flowers on Plants of the Same Species. John Murray, London, pp. 1–352. [Google Scholar]
- 4).Hinata K., Watanabe M., Toriyama K., Isogai A. (1993) A review of recent studies on homomorphic self-incompatibility. Int. Rev. Cytol. 143, 257–296 [Google Scholar]
- 5).Watanabe, M. and Hinata, K. (1999) Self-incompatibility. In Biology of Brassica Coenospecies (ed. Gomez-Campo, C.). Elsevier, Amsterdam, pp. 149–183. [Google Scholar]
- 6).Watanabe, M., Suzuki, G. and Takayama, S. (2008) Chapter 7: Milestones identifying self-incompatibility genes in Brassica species: From old stories to new findings. In Self-incompatibility in Flowering Plants—Evolution, Diversity, and Mechanisms (ed. Franklin-Tong, V.E.), Springer, Berlin, Heidelberg, New York, pp. 151–172. [Google Scholar]
- 7).Suzuki G. (2009) Recent progress in plant reproduction research: the story of the male gametophyte through to successful fertilization. Plant Cell Physiol. 50, 1857–1864 [DOI] [PubMed] [Google Scholar]
- 8).Suwabe K., Suzuki G., Watanabe M. (2010) Achievement of genetics in plant reproduction research: past decade for coming decade. Genes Genet. Syst. 85, 297–310 [DOI] [PubMed] [Google Scholar]
- 9).Thompson K.F., Taylor J.P. (1966) Non-linear dominance relationships between S alleles. Heredity 21, 345–362 [Google Scholar]
- 10).Hatakeyama K., Watanabe M., Takasaki T., Ojima K., Hinata K. (1998) Dominance relationships between S-alleles in self-incompatible Brassica campestris L. Heredity 79, 241–247 [Google Scholar]
- 11).Bateman A.J. (1952) Self-incompatibility systems in angiosperms. I. Theory. Heredity 6, 285–310 [Google Scholar]
- 12).Bateman A.J. (1954) Self-incompatibility systems in angiosperms. II. Iberis amara. Heredity 8, 305–332 [Google Scholar]
- 13).Bateman A.J. (1955) Self-incompatibility systems in angiosperms. III. Cruciferae. Heredity 9, 53–68 [Google Scholar]
- 14).Franklin-Tong, V.E. (2008) Self-incompatibility in Flowering Plants-evolution, Diversity, and Mechanisms. Springer, Berlin, pp. 1–313. [Google Scholar]
- 15).Higashiyama T. (2010) Peptide signaling in pollen-pistil interactions. Plant Cell Physiol. 51, 177–189 [DOI] [PubMed] [Google Scholar]
- 16).Nasrallah M.E., Wallace D.H. (1967) Immunogenetics of self-incompatibility in Brassica oleracea. Heredity 22, 519–527 [Google Scholar]
- 17).Nishio T., Hinata K. (1977) Analysis of S-specific proteins in stigmas of Brassica oleracea L. by isoelectric focusing. Heredity 38, 391–396 [Google Scholar]
- 18).Nishio T., Hinata K. (1978) Stigma proteins in self-incompatible Brassica campestris L. and self-compatible relatives, with special reference to S-allele specificity. Jpn. J. Genet. 53, 27–33 [Google Scholar]
- 19).Nasrallah J.B., Kao T.-h., Goldberg M.L., Nasrallah M.E. (1985) A cDNA clone encoding an S-locus-specific glycoprotein from Brassica oleracea. Nature 318, 263–267 [Google Scholar]
- 20).Takayama S., Isogai A., Tsukamoto C., Ueda Y., Hinata K., Okazaki K., Suzuki A. (1987) Sequences of S-glycoproteins, products of the Brassica campestris self-incompatibility locus. Nature 326, 102–104 [Google Scholar]
- 21).Kishi-Nishizawa N., Isogai A., Watanabe M., Hinata K., Yamakawa S., Shojima S., Suzuki A. (1990) Ultrastructure of papillar cells in Brassica campestris revealed by liquid helium rapid-freezing and substitution-fixation method. Plant Cell Physiol. 31, 1207–1219 [Google Scholar]
- 22).Brace J., Ockendon D.J., King G.J. (1993) Development of a method for the identification of S-alleles in Brassica oleracea based on digestion of PCR-amplified DNA with restriction endonucleases. Sex. Plant Reprod. 6, 133–138 [Google Scholar]
- 23).Watanabe M., Ito A., Takada Y., Ninomiya C., Kakizaki T., Takahata Y., Hatakeyama K., Hinata K., Suzuki G., Takasaki T., Satta Y., Shiba H., Takayama S., Isogai A. (2000) Highly divergent sequences of the pollen self-incompatibility (S) gene in class-I S haplotypes of Brassica campestris (syn. rapa) L. FEBS Lett. 473, 139–144 [DOI] [PubMed] [Google Scholar]
- 24).Suzuki G., Watanabe M., Toriyama K., Isogai A., Hinata K. (1995) Molecular cloning of members of the S-multigene family in self-incompatible Brassica campestris L. Plant Cell Physiol. 36, 1273–1280 [PubMed] [Google Scholar]
- 25).Suzuki G., Watanabe M., Kai N., Matsuda N., Toriyama K., Takayama S., Isogai A., Hinata K. (1997) Three members of the S-multigene family are linked to the S locus of Brassica. Mol. Gen. Genet. 256, 257–264 [DOI] [PubMed] [Google Scholar]
- 26).The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 [DOI] [PubMed] [Google Scholar]
- 27).Kai N., Suzuki G., Watanabe M., Isogai A., Hinata K. (2001) Sequence comparisons among dispersed members of the Brassica S multigene family in an S9 genome. Mol. Genet. Genomics 265, 526–534 [DOI] [PubMed] [Google Scholar]
- 28).Watanabe M., Takasaki T., Toriyama K., Yamakawa S., Isogai A., Suzuki A., Hinata K. (1994) A high degree of homology exists between the protein encoded by SLG and the S receptor domain encoded by SRK in self-incompatible Brassica campestris L. Plant Cell Physiol. 35, 1221–1229 [DOI] [PubMed] [Google Scholar]
- 29).Walker J.C., Zhang R. (1990) Relationship of a putative receptor protein kinase from maize to the S-locus glycoprotein of Brassica. Nature 345, 743–746 [DOI] [PubMed] [Google Scholar]
- 30).Stein J.C., Howlett B., Boyes D.C., Nasrallah M.E., Nasrallah J.B. (1991) Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. U.S.A. 88, 8816–8820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Goring D.R., Rothstein S.J. (1992) The S-locus receptor kinase gene in a self-incompatible Brassica napus line encodes a functional serine/threonine kinase. Plant Cell 4, 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Hatakeyama K., Takasaki T., Watanabe M., Hinata K. (1998) Molecular characterization of S locus genes, SLG and SRK, in a pollen-recessive self-incompatibility haplotype of Brassica rapa L. Genetics 149, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Suzuki G., Watanabe M., Isogai A., Hinata K. (1997) Highly conserved 5′-flanking regions of two self-incompatibility genes, SLG9 and SRK9. Gene 191, 123–126 [DOI] [PubMed] [Google Scholar]
- 34).Boyes D.C., Nasrallah J.B. (1993) Physical linkage of the SLG and SRK genes at the self-incompatibility locus of Brassica oleracea. Mol. Gen. Genet. 236, 369–373 [DOI] [PubMed] [Google Scholar]
- 35).Suzuki G., Watanabe M., Toriyama K., Isogai A., Hinata K. (1997) Direct cloning of the Brassica S locus by using a P1-derived artificial chromosome. Gene 199, 133–137 [DOI] [PubMed] [Google Scholar]
- 36).Suzuki G., Watanabe M., Nishio T. (2000) Physical distances between S-locus genes in various S haplotypes of Brassica rapa and B. oleracea. Theor. Appl. Genet. 101, 80–85 [Google Scholar]
- 37).Cui Y., Brugiere N., Jackman L., Bi Y.-M., Rothstein S.J. (1999) Structural and transcriptional comparative analysis of the S locus regions in two self-incompatible Brassica napus lines. Plant Cell 11, 2217–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Suzuki G., Kai N., Hirose T., Fukui K., Nishio T., Takayama S., Isogai A., Watanabe M., Hinata K. (1999) Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 153, 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Kimura R., Sato K., Fujimoto R., Nishio T. (2002) Recognition specificity of self-incompatibility maintained after the divergence of Brassica oleracea and Brassica rapa. Plant J. 29, 215–223 [DOI] [PubMed] [Google Scholar]
- 40).Shiba H., Kenmochi M., Sugihara M., Iwano M., Kawasaki S., Suzuki G., Watanabe M., Isogai A., Takayama S. (2003) Genomic organization of the S-locus region of Brassica. Biosci. Biotechnol. Biochem. 67, 622–626 [DOI] [PubMed] [Google Scholar]
- 41).Shiba H., Hinata K., Suzuki A., Isogai A. (1995) Breakdown of self-incompatibility in Brassica by the antisense RNA of the SLG gene. Proc. Jpn. Acad., Ser. B 71, 81–83 [Google Scholar]
- 42).Takasaki T., Hatakeyama K., Watanabe M., Toriyama K., Isogai A., Hinata K. (1999) Introduction of SLG (S locus glycoprotein) alters the phenotype of endogenous S haplotype, but confers no new S haplotype specificity in Brassica rapa L. Plant Mol. Biol. 40, 659–668 [DOI] [PubMed] [Google Scholar]
- 43).Takasaki T., Hatakeyama K., Watanabe M., Toriyama K., Hinata K. (2001) Homology-dependent suppression of stigma phenotype by an antisense S-locus glycoprotein (SLG) gene in Brassica rapa L. Breed. Sci. 51, 89–94 [Google Scholar]
- 44).Napoli C., Lemieux C., Jorgensen R. (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous gene in trans. Plant Cell 2, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Takasaki T., Hatakeyama K., Suzuki G., Watanabe M., Isogai A., Hinata K. (2000) The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403, 913–916 [DOI] [PubMed] [Google Scholar]
- 46).Cui Y., Bai Y.-M., Brugiere N., Arnold M.A., Rothstein S.J. (2000) The S-locus glycoprotein and the S receptor kinase are sufficient for self-pollen rejection in Brassica. Proc. Natl. Acad. Sci. U.S.A. 97, 3713–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Watanabe M., Shiozawa H., Isogai A., Suzuki A., Takeuchi T., Hinata K. (1991) Existence of S-glycoprotein-like proteins in anthers of self-incompatible species of Brassica. Plant Cell Physiol. 32, 1039–1047 [Google Scholar]
- 48).Boyes D.C., Nasrallah J.B. (1995) An anther-specific gene encoded by an S locus haplotype of Brassica produces complementary and differentially regulated transcripts. Plant Cell 7, 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Yu K., Schafer U., Glavin T.L., Goring D.R., Rothstein S.J. (1996) Molecular characterization of the S locus in two self-incompatible Brassica napus lines. Plant Cell 8, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Watanabe M., Suzuki G., Toriyama K., Takayama S., Isogai A., Hinata K. (1999) Two anther-expressed genes downstream of SLG9: identification of a novel S-linked gene specifically expressed in anthers at the uninucleate stage of Brassica campestris (syn. rapa) L. Sex. Plant Reprod. 12, 127–134 [Google Scholar]
- 51).Heslop-Harrison J. (1975) Incompatibility and the pollen-stigma interaction. Annu. Rev. Plant Physiol. 26, 403–425 [Google Scholar]
- 52).Doughty J., Hedderson F., McCubbin A., Dickinson H.G. (1993) Interaction between a coating-borne peptide of the Brassica pollen grain and stigmatic S (self-incompatibility)-locus-specific glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 90, 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Hiscock S.J., Doughty J., Willis A.C., Dickinson H.G. (1995) A 7-kDa pollen coating-borne peptide from Brassica napus ineracts with S-locus glycoprotein and S-locus related glycoprotein. Planta 196, 367–374 [DOI] [PubMed] [Google Scholar]
- 54).Stanchev B.S., Doughty J., Scutt C.P., Dickinson H.G., Croy R.R.D. (1996) Cloning of PCP1, a member of a family of pollen coat protein (PCP) genes from Brassica oleracea encoding novel cysteine-rich proteins involved in pollen-stigma interactions. Plant J. 10, 303–313 [DOI] [PubMed] [Google Scholar]
- 55).Toriyama K., Hanaoka K., Okada T., Watanabe M. (1998) Molecular cloning of a cDNA encoding a pollen extracellular protein as a potential source of a pollen allergen in Brassica rapa. FEBS Lett. 424, 234–238 [DOI] [PubMed] [Google Scholar]
- 56).Doughty J., Dixon S., Hiscock S.J., Willis A.C., Parkin I.A.P., Dickinson H.G. (1998) PCP-A1, a difensin-like Brassica pollen coat protein that binds the S locus glycoprotein, is the product of gametophytic gene expression. Plant Cell 10, 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Takayama S., Shiba H., Iwano M., Asano K., Hara M., Che F.-S., Watanabe M., Hinata K., Isogai A. (2000) Isolation and characterization of pollen coat proteins of Brassica campestris that interact with S locus-related glycoprotein 1 involved in pollen-stigma adhesion. Proc. Natl. Acad. Sci. U.S.A. 97, 3765–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Stephenson A.G., Doughty J., Dixon S., Elleman C., Hiscock S., Dickinson H.G. (1997) The male determinant of self-incompatibility in Brassica oleracea is located in the pollen coating. Plant J. 12, 1351–1359 [Google Scholar]
- 59).Takayama S., Shiba H., Iwano M., Shimosato H., Che F.-S., Kai N., Watanabe M., Suzuki G., Hinata K., Isogai A. (2000) The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. U.S.A. 97, 1920–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Shiba H., Takayama S., Iwano M., Shimosato H., Funato M., Nakagawa T., Che F.-S., Suzuki G., Watanabe M., Hinata K., Isogai A. (2001) A pollen coat protein, SP11/SCR, determines the pollen S-specificity in the self-incompatibility of Brassica species. Plant Physiol. 125, 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Schopher C.R., Nasrallah M.E., Nasrallah J.B. (1999) The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700 [DOI] [PubMed] [Google Scholar]
- 62).Mishima M., Takayama S., Sasaki K.-i., Jee J.-g., Kojima C., Isogai A., Shirakawa M. (2003) Structure of the male determinant factor for Brassica self-incompatibility. J. Biol. Chem. 278, 36389–36395 [DOI] [PubMed] [Google Scholar]
- 63).Shimosato H., Yokota N., Shiba H., Iwano M., Entani T., Che F.-S., Watanabe M., Isogai A., Takayama S. (2007) Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in Brassica self-incompatibility. Plant Cell 19, 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Kachroo A., Schopher C.R., Nasrallah J.B. (2001) Allelic-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293, 1824–1826 [DOI] [PubMed] [Google Scholar]
- 65).Takayama S., Shimosato H., Shiba H., Funato M., Che F.-S., Watanabe M., Iwano M., Isogai A. (2001) Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413, 534–538 [DOI] [PubMed] [Google Scholar]
- 66).Gu T., Mazzurco M., Sulaman W., Matias D.D., Goring D.R. (1998) Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. U.S.A. 95, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Stone S.L., Arnold M., Goring D.R. (1999) A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286, 1729–1731 [DOI] [PubMed] [Google Scholar]
- 68).Cabrillac D., Cock J.M., Dumas C., Gaude T. (2001) The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat protein. Nature 410, 220–223 [DOI] [PubMed] [Google Scholar]
- 69).Stone S.L., Anderson E.M., Mullen R.T., Goring D.R. (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15, 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Vanoosthuyse V., Tichtinsky G., Dumas C., Gaude T., Cock J.M. (2003) Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphatase with the Brassica oleracea S-locus receptor kinase. Plant Physiol. 133, 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Murase K., Shiba H., Iwano M., Che F.-S., Watanabe M., Isogai A., Takayama S. (2004) A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303, 1516–1519 [DOI] [PubMed] [Google Scholar]
- 72).Kakita M., Murase K., Iwano M., Matsumoto T., Watanabe M., Shiba H., Isogai A., Takayama S. (2007) Two distinct forms of M-locus protein kinase localize to the plasmamembrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19, 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73).Isokawa S., Osaka M., Shirasawa A., Kikuta R., Komatsu S., Horisaki A., Niikura S., Takada Y., Shiba H., Isogai A., Takayama S., Suzuki G., Suwabe K., Watanabe M. (2010) Novel self-compatible lines of Brassica rapa L. isolated from the Japanese bulk-populations. Genes Genet. Syst. 85, 87–96 [DOI] [PubMed] [Google Scholar]
- 74).Bergelson J., Roux F. (2010) Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat. Rev. Genet. 11, 867–879 [DOI] [PubMed] [Google Scholar]
- 75).Ge X., Chang F., Ma H. (2010) Signaling and transcriptional control of reproductive development in Arabidopsis. Curr. Biol. 20, R988–R997 [DOI] [PubMed] [Google Scholar]
- 76).Liepman A.H., Wightman R., Geshi N., Turner S.R., Scheller H.V. (2010) Arabidopsis—a powerful model system for plant cell wall research. Plant J. 61, 1107–1121 [DOI] [PubMed] [Google Scholar]
- 77).Walley J.W., Dehesh K. (2010) Molecular mechanisms regulating rapid stress signaling networks in Arabidopsis. J. Interg. Plant Biol. 52, 354–359 [DOI] [PubMed] [Google Scholar]
- 78).Shimizu K.K., Cork J.M., Caicedo A.L., Mays C.A., Moore R.C., Olsen K.M., Ruzsa S., Coop G., Bustamante C.G., Awadalla P., Purugganan M.D. (2004) Darwinian selection on a selfing locus. Science 306, 2081–2084 [DOI] [PubMed] [Google Scholar]
- 79).Bechsgaard J.S., Castric V., Charlesworth D., Vekemans X., Schierup M.H. (2006) The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23, 1741–1750 [DOI] [PubMed] [Google Scholar]
- 80).Sherman-Broyles S., Boggs N., Farkas A., Liu P., Vrebalv J., Nasrallah M.E., Nasrallah J.B. (2007) S-locus and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19, 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81).Tang C., Toomajian C., Sherman-Broyles S., Plagnol V., Guo Y.-L., Hu T.T., Clark R.M., Nasrallah J.B., Weigel D., Nordborg M. (2007) The evolution of selfing in Arabidopsis thaliana. Science 317, 1070–1072 [DOI] [PubMed] [Google Scholar]
- 82).Shimizu K.K., Shimizu-Inatsugi R., Tsuchimatsu T., Purugganan M.D. (2008) Muliple orgins of self-compatibility in Arabidopsis thaliana. Mol. Ecol. 17, 704–714 [DOI] [PubMed] [Google Scholar]
- 83).Nasrallah M.E., Liu P., Nasrallah J.B. (2002) Generation of self-incompatible Arabidopsis thaliana by transfer of two S-locus genes from A. lyrata. Science 297, 247–249 [DOI] [PubMed] [Google Scholar]
- 84).Nasrallah M.E., Liu P., Sherman-Broyles S., Broggs N.A., Nasrallah J.B. (2004) Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc. Natl. Acad. Sci. U.S.A. 101, 16070–16074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85).Tsuchimatsu T., Suwabe K., Shimizu-Inatsugi R., Isokawa S., Pavlidis P., Städler T., Suzuki G., Takayama S., Watanabe M., Shimizu K.K. (2010) Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464, 1342–1346 [DOI] [PubMed] [Google Scholar]
- 86).Shiba H., Iwano M., Entani T., Ishimoto K., Shimosato H., Funato M., Che F.-S., Satta Y., Ito A., Takada Y., Watanabe M., Isogai A., Takayama S. (2002) The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell 14, 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Kakizaki T., Takada Y., Ito A., Suzuki G., Shiba H., Takayama S., Isogai A., Watanabe M. (2003) Linear dominance relationship among four class-II S-haplotypes in pollen side determined by the expression of SP11 in Brassica self-incompatibility. Plant Cell Physiol. 44, 70–75 [DOI] [PubMed] [Google Scholar]
- 88).Shiba H., Kakizaki T., Iwano M., Tarutani Y., Watanabe M., Isogai A., Takayama S. (2006) Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat. Genet. 38, 297–299 [DOI] [PubMed] [Google Scholar]
- 89).Tarutani Y., Shiba H., Iwano M., Kakizaki T., Suzuki G., Watanabe M., Isogai A., Takayama S. (2010) Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466, 983–986 [DOI] [PubMed] [Google Scholar]
- 90).Hatakeyama K., Takasaki T., Suzuki G., Nishio T., Watanabe M., Isogai A., Hinata K. (2001) The S receptor kinase gene determines dominance relationships in stigma expression of self-incompatibility in Brassica. Plant J. 26, 69–76 [DOI] [PubMed] [Google Scholar]
- 91).Endo M., Matsubara H., Kokubun T., Masuko H., Takahata Y., Tsuchiya T., Fukuda H., Demura T., Watanabe M. (2002) DNA microarray is useful for identification of anther-specific genes in model legume, Lotus japonicus. FEBS Lett. 514, 229–237 [DOI] [PubMed] [Google Scholar]
- 92).Amagai M., Ariizumi T., Endo M., Hatakeyama K., Kuwata C., Shibata D., Toriyama K., Watanabe M. (2003) Identification of reproductive organ-specific genes of cruciferous model plant, Arabidopsis thaliana, by using a combination of Arabidopsis macroarray and mRNA derived from Brassica oleracea. Sex. Plant Reprod. 15, 213–220 [Google Scholar]
- 93).Endo M., Tsuchiya T., Saito H., Matsubara H., Hakozaki H., Masuko H., Kamada M., Higashitani A., Takahashi H., Fukuda H., Demura T., Watanabe M. (2004) Identification and molecular characterization of novel anther-specific genes in Oryza sativa L. by using cDNA microarray. Genes Genet. Syst. 79, 213–226 [DOI] [PubMed] [Google Scholar]
- 94).Yoshida K.T., Endo M., Nakazono M., Fukuda H., Demura T., Tsuchiya T., Watanabe M. (2005) cDNA microarray analysis of gene expression changes during pollination, pollen-tube elongation, fertilization, and early embryogenesis in rice pistils. Sex. Plant Reprod. 17, 269–275 [Google Scholar]
- 95).Suwabe K., Suzuki G., Takahashi H., Shiono K., Endo M., Yano K., Fujita M., Masuko H., Saito H., Fujioka T., Kaneko F., Kazama T., Mizuta Y., Kawagishi-Kobayashi M., Tsutsumi N., Kurata N., Nakazono M., Watanabe M. (2008) Separated transcriptomes of male gametophyte and tapetum in rice: validity of a laser microdissection (LM) microarray. Plant Cell Physiol. 49, 1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Hobo T., Suwabe K., Aya K., Suzuki G., Yano K., Ishimizu T., Fujita M., Kikuchi S., Hamada K., Miyano M., Fujioka T., Kaneko F., Kazama T., Mizuta Y., Takahashi H., Shiono K., Nakazono M., Tsutsumi N., Nagamura Y., Kurata N., Watanabe M., Matsuoka M. (2008) Various spatiotemporal expression profiles of anther-expressed genes in rice. Plant Cell Physiol. 49, 1417–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97).Endo M., Tsuchiya T., Hamada K., Kawamura S., Yano K., Ohshima M., Higashitani A., Watanabe M., Kawagishi-Kobayashi M. (2009) High temperatures cause male sterility in rice plants with transcriptional alterations during pollen development. Plant Cell Physiol. 50, 1911–1922 [DOI] [PubMed] [Google Scholar]
- 98).Fujita M., Horiuchi Y., Ueda Y., Mizuta Y., Kubo T., Yano K., Yamaki S., Tsuda K., Nagata T., Niihama M., Kato H., Kikuchi S., Hamada K., Mochizuki T., Ishimizu T., Iwai H., Tsutsumi N., Kurata N. (2010) Rice expression atlas in reproductive development. Plant Cell Physiol. 51, 2060–2081 [DOI] [PubMed] [Google Scholar]
- 99).Mochida K., Shinozaki K. (2010) Genomics and bioinformatics resources for crop improvement. Plant Cell Physiol. 51, 497–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100).Oda S., Kaneko F., Yano K., Fujioka T., Masuko H., Park J.-I., Kikuchi S., Hamada K., Endo M., Nagano K., Nagamura Y., Kawagishi-Kobayashi M., Suwabe K., Suzuki G., Watanabe M. (2010) Morphological and gene expression analysis under cool temperature conditions in rice anther development. Genes Genet. Syst. 85, 107–120 [DOI] [PubMed] [Google Scholar]
- 101).Sun W., Xu X., Zhu H., Liu A., Liu L., Li J., Hua X. (2010) Comparative transcriptomic profiling of a salt-tolerant wild tomato species and a salt-sensitive tomato cultivar. Plant Cell Physiol. 51, 997–1006 [DOI] [PubMed] [Google Scholar]
- 102).Yamakawa H., Hakata M. (2010) Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 51, 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103).Aya K., Suzuki G., Suwabe K., Hobo T., Takahashi H., Shiono K., Yano K., Tsutsumi N., Nakazono M., Nagamura Y., Matsuoka M., Watanabe M. (2011) Comprehensive network analysis of anther-expressed genes in rice by the combination of 33 laser microdissection and 143 spatiotemporal microarrays. PLoS ONE 6, e26162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104).Boavida L., Borges F., Becker J.D., Feijo J. (2011) Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathway during pollen-stigma interactions in Arabidopsis. Plant Physiol. 155, 2066–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105).Du H., Liang Y., Pei K., Ma K. (2011) UV radiation-responsive proteins in rice leaves: a proteomic analysis. Plant Cell Physiol. 52, 306–316 [DOI] [PubMed] [Google Scholar]
- 106).Hamada K., Hongo K., Suwabe K., Shimizu A., Nagayama T., Abe R., Kikuchi S., Yamamoto N., Fujii T., Yokoyama K., Tsuchida H., Sano K., Mochizuki T., Oki N., Horiuchi Y., Fujita M., Watanabe M., Matsuoka M., Kurata N., Yano K. (2011) OryzaExpress: an integrated database of gene expression networks and omics annotations in rice. Plant Cell Physiol. 52, 220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107).Ito-Inaba Y., Hida Y., Matsumura H., Masuko H., Yazu F., Terauchi R., Watanabe M., Inaba T. (2012) The gene expression landscape of thermogenic skunk cabbage suggests critical roles for mitochondorial and vacuolar metabolic pathways in the regulation of thermogenesis. Plant Cell Environ. 35, 554–566 [DOI] [PubMed] [Google Scholar]
- 108).Matsuo M., Hachisu R., Tabata S., Fukuzawa H., Obokata J. (2011) Transcriptome analysis of respiration-responsive genes in Chlamydomonas reinharditii: mitochondorial retrograde signaling coordinates the genes for cell proliferation with energy-producing metabolism. Plant Cell Physiol. 52, 333–343 [DOI] [PubMed] [Google Scholar]
- 109).Mochida K., Uehara-Yamaguchi Y., Yoshida T., Sakurai T., Shinozaki K. (2011) Global landscape of a co-expressed gene network in barley and its application to gene discovery in Triticeae crops. Plant Cell Physiol. 52, 785–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110).Mochida K., Shinozaki K. (2011) Advances in omics and bioinformatics tools for systems analyses of plant function. Plant Cell Physiol. 52, 2017–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111).Nagamura Y., Antonio B.A., Sato Y., Miyao A., Namiki N., Yonemaru J.-i., Minami H., Kamatsuki K., Shimura K., Shimizu Y., Hirochika H. (2011) Rice TOGO browser: a platform to retrieve integrated information on rice functional and applied genomics. Plant Cell Physiol. 52, 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112).Obayashi T., Nishida K., Kasahara K., Kinoshita K. (2011) ATTED-II update: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 52, 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113).Okabe Y., Asamizu E., Saito T., Matsukura C., Ariizumi T., Brès C., Rothan C., Mizoguchi T., Ezura H. (2011) Tomato TILLING technology: development of reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 52, 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114).Saito T., Ariizumi T., Okabe Y., Asamizu E., Hiwasa-Tanase K., Fukuda N., Mizoguchi T., Yamazaki Y., Aoki K., Ezura H. (2011) TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 52, 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115).Su C.-l., Chao Y.-T., Chang Y.-C.A., Chen W.-C., Chen C.-Y., Lee A.-Y., Hwa K.T., Shih M.-C. (2011) De novo assembly of expressed transcripts and global analysis of the Phalaonopsis aphrodite transriptome. Plant Cell Physiol. 52, 1501–1514 [DOI] [PubMed] [Google Scholar]
- 116).Swarbreck S.M., Lindquist E.A., Ackerly D.D., Anderson G.L. (2011) Analysis of leaf and root transcriptomes of soil-grown Avena barbata. Plant Cell Physiol. 52, 317–332 [DOI] [PubMed] [Google Scholar]
- 117).Uchida N., Sakamoto T., Kurata T., Tasaka M. (2011) Identification of EMS-induced casual mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 52, 716–722 [DOI] [PubMed] [Google Scholar]
- 118).Hafidh S., Breznenová K., Růžička P., Feciková J., Čapková V., Honys D. (2012) Comprehensive analysis of tobacco pollen transcriptome unveils common pathways in polar cell expansion and underlying heterochronic shift during spermatogenesis. BMC Genomics 12, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119).Xu X.H., Chen H., Sang Y.L., Wang F., Ma J.P., Gao X.-Q., Zhang X.S. (2012) Identification of genes specifically or preferentially expressed in maize silk reveals similarity and diversity in transcript abundance of different dry stigmas. BMC Genomics 13, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120).Takada Y., Nakanowatari T., Sato J., Hatakeyama K., Kakizaki T., Ito A., Suzuki G., Shiba H., Takayama S., Isogai A., Watanabe M. (2005) Genetic analysis of intra-species unilateral incompatibility between cultiver Osome and Turkish line in Brassica rapa (syn. campestris) L. Sex. Plant Reprod. 17, 211–217 [Google Scholar]
- 121).Masuko H., Endo M., Saito H., Hakozaki H., Park J.I., Kawagishi-Kobayashi M., Takada Y., Okabe T., Kamada M., Takahashi H., Higashitani A., Watanabe M. (2006) Anther-specific genes, which expressed through microsporogenesis, are temporally and spatially regulated in model legume, Lotus japonicus. Genes Genet. Syst. 81, 57–62 [DOI] [PubMed] [Google Scholar]
- 122).Kazama T., Nakamura T., Watanabe M., Sugita M., Toriyama K. (2008) Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 55, 619–628 [DOI] [PubMed] [Google Scholar]
- 123).Fujioka T., Kaneko F., Kazama T., Suwabe K., Suzuki G., Makino A., Mae T., Endo M., Kawagishi-Kobayashi M., Watanabe M. (2008) Identification of small RNAs in late developmental stage of rice anthers. Genes Genet. Syst. 83, 281–284 [DOI] [PubMed] [Google Scholar]
- 124).Kurasawa K., Matsui A., Yokoyama R., Kuriyama T., Yoshizumi T., Matsui M., Suwabe K., Watanabe M., Nishitani K. (2009) The AtXTH28 gene, a xyloglucan endotransglucosylase/hydrolase, is involved in automatic self-pollination in Arabidopsis thaliana. Plant Cell Physiol. 50, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125).Fujii S., Yamada M., Fujita M., Itabashi E., Hamada K., Yano K., Kurata N., Toriyama K. (2010) Cytoplasmic-nuclear genomic barriers in rice pollen development revealed by comparison of global gene expression profiles among five independent cytoplasmic male sterile lines. Plant Cell Physiol. 51, 610–620 [DOI] [PubMed] [Google Scholar]
- 126).Ishiguro S., Nishimori Y., Yamada M., Saito H., Suzuki T., Nakagawa T., Miyake H., Okada K., Nakamura K. (2010) The Arabidopsis FLAKY POLLEN1 gene encodes a 3-hydroxy-3-methylglutaryl-coenzyme A synthase required for development of tapetum-specific organelles and fertility of pollen grains. Plant Cell Physiol. 51, 896–911 [DOI] [PubMed] [Google Scholar]
- 127).Ishimizu T., Kodama H., Ando T., Watanabe M. (2010) Molecular evidence that most RNAs required for germination and pollen tube growth are stored in the mature pollen grain in petunia. Genes Genet. Syst. 85, 259–263 [DOI] [PubMed] [Google Scholar]
- 128).Li W.-Q., Zhang X.-Q., Xia C., Deng Y., Ye D. (2010) MALE GAMETOPHYTE DEFECTIVE 1, encoding the FAd subunit of mitochondrial F1F0-ATP synthase, is essential for pollen formation in Arabidopsis thaliana. Plant Cell Physiol. 51, 923–935 [DOI] [PubMed] [Google Scholar]
- 129).Park J.-I., Ishimizu T., Suwabe K., Sudo K., Masuko H., Hakozaki H., Nou I.-S., Suzuki G., Watanabe M. (2010) UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 51, 981–996 [DOI] [PubMed] [Google Scholar]
- 130).Aki S., Nakai H., Aoyama T., Oka A., Tsuge T. (2011) AtSAP130/AtSF3b-3 function is required for reproduction in Arabidopsis thaliana. Plant Cell Physiol. 52, 1330–1339 [DOI] [PubMed] [Google Scholar]
- 131).Huang J.-C., Chang L.-C., Wang M.-L., Guo C.-L., Chung M.-C., Jauh G.-Y. (2011) Identification and exploration of pollen tube small proteins encoded by pollination-induced protein. Plant Cell Physiol. 52, 1546–1559 [DOI] [PubMed] [Google Scholar]
- 132).Huang M.-D., Caroline-Hsing Y.-I., Huang H.C. (2011) Transcriptomes of the anther sporophyte: availability and uses. Plant Cell Physiol. 52, 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133).Kobayashi M., Kouzu N., Inami A., Toyooka K., Konishi Y., Matsuoka K., Matoh T. (2011) Characterization of Arabidopsis CTP: 3-deoxy-D-manno-2-octulosonate cytidylyltransferase (CMP-KDO synthethase), the enzyme that activates KDO during rhamnogalacturonan II biosynthesis. Plant Cell Physiol. 52, 1832–1843 [DOI] [PubMed] [Google Scholar]
- 134).Liu M.-C., Wang B.-J., Huang J.-K., Wang C.-S. (2011) Expression, localization and function of a cis-prenyltransferase in the tapetum and microspores of lily anthers. Plant Cell Physiol. 52, 1487–1500 [DOI] [PubMed] [Google Scholar]
- 135).Wang C.-W., Chen W.-C., Lin L.-J., Lee C.-T., Tseng T.-H., Leu W.-M. (2011) OIP30, a RuvB-like DNA helicase 2, is a potential substrate for the pollen-predominant OsCPK25/26 in rice. Plant Cell Physiol. 52, 1641–1656 [DOI] [PubMed] [Google Scholar]
- 136).Yang H., Liu P., Wang Y., Ma H. (2012) The transcriptome landscape of Arabidopsis male meiocytes from high-throughput sequencing: the complexity and evolution of the meiotic process. Plant J. 65, 503–516 [DOI] [PubMed] [Google Scholar]
- 137).Matsubayashi Y., Sakagami Y. (2006) Peptide hormones in plants. Annu. Rev. Plant Biol. 57, 649–674 [DOI] [PubMed] [Google Scholar]
- 138).Kawaguchi M., Minamisawa K. (2010) Plant-microbe communications for symbiosis. Plant Cell Physiol. 51, 1377–1380 [DOI] [PubMed] [Google Scholar]
- 139).Kondo Y., Hirakawa Y., Kieber J.J., Fukuda H. (2011) CLE peptides can negatively regulated protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol. 52, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140).Goto H., Okuda S., Mizukami A., Mori H., Sasaki N., Kurihara D., Higashiyama T. (2011) Chemical visualization of an attractant peptide, LURE. Plant Cell Physiol. 52, 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141).Matsubayashi Y. (2011) Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 52, 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142).Hattori Y., Nagai K., Furukawa S., Song X.-J., Kawano R., Sakakibara H., Wu J., Matsumoto T., Yoshimura A., Kitano H., Matsuoka M., Mori H., Ashikari M. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030 [DOI] [PubMed] [Google Scholar]
- 143).Shinya T., Osada T., Desaki Y., Hatamoto M., Yamanaka Y., Hirano H., Takai R., Che F.-S., Kaku H., Shibuya N. (2010) Characterization of receptor proteins using affinity cross-linking with biotinylated ligands. Plant Cell Physiol. 51, 262–270 [DOI] [PubMed] [Google Scholar]
- 144).She J., Han Z., Kim T.-W., Wang J., Cheng W., Chang J., Shi S., Wang J., Yang M., Wang Z.-Y., Chai J. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145).U N. (1935) Genome-analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7, 389–452 [Google Scholar]
- 146).Goring D.R., Glavin T.L., Schafer U., Rothstein S.J. (1993) An S receptor kinase gene in self-incompatible Brassica napus has 1-bp deletion. Plant Cell 5, 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147).Okamoto S., Odashima M., Fujimoto R., Sato Y., Kitashiba H., Nishio T. (2007) Self-compatibility in Brassica napus is caused by independent mutations in S-locus genes. Plant J. 50, 391–400 [DOI] [PubMed] [Google Scholar]
- 148).Tatebe T. (1964) Studies on the physiological mechanism of self-incompatibility in Japanese radish II. Breakdown of self-incompatibility by chemical treatment. J. Hort. Assoc. Jpn. 37, 227–230 [Google Scholar]
- 149).Gonai H., Hinata K. (1969) Studies on self-incompatibility in Brassica: influence of organic solvents on seed fertility. Jpn. J. Breed. 19 (Suppl. 1), 153–154(in Japanese) [Google Scholar]
- 150).Nakanishi T., Esashi Y., Hinata K. (1969) Control of self-incompatibility by CO2 gas in Brassica. Plant Cell Physiol. 10, 925–927 [Google Scholar]
- 151).Roggen H.P.J.R., van Dijk A.J., Dorsman C. (1972) ‘Electric aided’ pollination: a method of breaking incompatibility in Brassica oleracea L. Euphytica 21, 181–184 [Google Scholar]
- 152).Nakanishi T., Hinata K. (1973) An effective time for CO2 gas treatment in overcoming self-incompatibility in Brassica. Plant Cell Physiol. 14, 873–879 [Google Scholar]
- 153).Okazaki K., Hinata K. (1987) Repressing the expression of self-incompatibility in crucifers by short-term high temperature treatment. Theor. Appl. Genet. 73, 496–500 [DOI] [PubMed] [Google Scholar]
- 154).Monterio A.A., Gabelman W.H. (1988) Use of sodium chloride solution to overcome self-incompatibility in Brassica campestris. Hortic. Sci. 23, 876–877 [Google Scholar]
- 155).Kanno T., Hinata K. (1969) An electron microscopic study of the barrier against pollen-tube growth in self-incompatible Cruciferae. Plant Cell Physiol. 10, 213–216 [Google Scholar]
- 156).Kamigaki A., Kondo M., Mano S., Hayashi M., Nishimura M. (2009) Suppression of peroxisome biogenesis factor 10 reduces cuticular wax accumulation by disrupting the ER network in Arabidopsis thaliana. Plant Cell Physiol. 50, 2034–2046 [DOI] [PubMed] [Google Scholar]
- 157).Takahashi K., Shimada T., Kondo M., Tamai A., Mori M., Nishimura M., Hara-Nishimura I. (2010) Ectopic expression of an esterase, which is a candidate for the unidentified plant cutinase, causes cuticular defects in Arabidopsis thaliana. Plant Cell Physiol. 51, 123–131 [DOI] [PubMed] [Google Scholar]
- 158).Monastersky R. (2011) Giant shock rattles ideas about quake behaviour. Nature 471, 274. [DOI] [PubMed] [Google Scholar]
- 159).Brumfiel G. (2011) The meltdown that wasn’t. Nature 471, 417–418 [DOI] [PubMed] [Google Scholar]
- 160).Buysse J., Van den Brande K., Merckx R. (1996) Genotypic differences in the uptake and distribution of radiocaesium in plants. Plant Soil 178, 265–271 [Google Scholar]
- 161).Lasat M.M., Norvel W.A., Kochian L.V. (1997) Potential phytoextraction of 137Cs from a contaminated soil. Plant Soil 195, 99–106 [Google Scholar]
- 162).Lasat M.M., Fuhrmann M., Ebbs S.D., Cornish J.E., Kochian L.V. (1998) Phytoremediation of a radiocesium-contaminated soil: evaluation of cesium-137 bioaccumulation in the shoots of three plant species. J. Environ. Qual. 27, 165–169 [Google Scholar]
- 163).Dushenkov S., Mikheev A., Rrokhnevsky A., Ruchko M., Sorochinsky B. (1999) Phytoremediation of radiocesium-contaminated soil in the vicinity of Chernobyl, Ukraine. Environ. Sci. Technol. 33, 469–475 [Google Scholar]
- 164).Zhu Y.-G., Smolders E. (2000) Plant uptake of radiocaecium: a review of mechanisms, regulation and application. J. Exp. Bot. 51, 1635–1645 [DOI] [PubMed] [Google Scholar]
- 165).Staunton S., Hinsinger P., Guivarch A., Brechignac F. (2003) Root uptake and translocation of radiocaesium from agricultural soils by various plant species. Plant Soil 254, 443–455 [Google Scholar]
- 166).Brooks R.R., Chambers M.F., Nicks L.J., Robinson B.H. (1999) Phytomining. Trends Plant Sci. 3, 359–362 [Google Scholar]
- 167).Nou I.S., Watanabe M., Isogai A., Shiozawa H., Suzuki A., Hinata K. (1991) Variation of S-alleles and S-glycoproteins in a naturalized population of selfincompatible Brassica campestris L. Jpn. J. Genet. 66, 227–239 [Google Scholar]
- 168).Nou I.S., Watanabe M., Isogai A., Hinata K. (1993) Comparison of S-alleles and S-glycoproteins between two wild populations of Brassica campestris in Turkey and Japan. Sex. Plant Reprod. 6, 79–86 [Google Scholar]
- 169).Nou I.S., Watanabe M., Isuzugawa K., Isogai A., Hinata K. (1993) Isolation of S-allele from a wild population of Brassica campestris L. at Balcesme, Turkey and their characterization by S-glycoprotein. Sex. Plant Reprod. 6, 71–78 [Google Scholar]