SUMMARY
The human gut contains trillions of microorganisms that influence our health by metabolizing xenobiotics, including host-targeted drugs and antibiotics. Recent efforts have characterized the diversity of this host-associated community, but it remains unclear which microorganisms are active and what perturbations influence this activity. Here, we combine flow cytometry, 16S rRNA gene sequencing, and metatranscriptomics to demonstrate that the gut contains a distinctive set of active microorganisms, primarily Firmicutes. Short-term exposure to a panel of xenobiotics significantly affected the physiology, structure, and gene expression of this active gut microbiome. Xenobiotic-responsive genes were found across multiple bacterial phyla, encoding antibiotic resistance, drug metabolism, and stress response pathways. These results demonstrate the power of moving beyond surveys of microbial diversity to better understand metabolic activity, highlight the unintended consequences of xenobiotics, and suggest that attempts at personalized medicine should consider inter-individual variations in the active human gut microbiome.
INTRODUCTION
Within the human gastrointestinal tract lies a diverse and abundant group of microorganisms, the gut microbiota. Metagenomics has enabled the analysis of the aggregate genomes of this microbial community (the gut microbiome) (Consortium, 2012; Qin et al., 2010; Turnbaugh et al., 2009a; Yatsunenko et al., 2012). However, most of these studies are limited to a single phylogenetic marker gene (the 16S rRNA) or to shotgun sequencing community DNA. While powerful to determine community structure and metabolic potential, these tools cannot be used to infer which bacterial members of the community are active, damaged, or responsive to a given compound. Due to these limitations, many fundamental questions linking bacterial activity and identity remain unanswered, despite the importance of the gut microbiota for numerous metabolic processes.
One of these key processes is the resistance to and biotransformation of xenobiotics (compounds foreign to a living organism), including host-targeted drugs and antibiotics (Haiser and Turnbaugh, 2012). Although many clinically relevant drugs are metabolized by the gut microbiome, the specific microorganisms responsible, the molecular mechanisms involved, and the potential impact these compounds have on microbial physiology and gene expression are often unknown. Studies linking bacterial activity, gene expression, and identity are thus essential to identify the active component of the gut microbiota and its response to common and clinically relevant stressors such as xenobiotics.
Single-cell methods, such as flow cytometry and fluorescence-activated cell sorting (FACS), have been used to characterize the physiological structure of environmental and host-associated microbial communities (Ben-Amor et al., 2005; Del Giorgio and Gasol, 2008; Joux and Lebaron, 2000; Shapiro, 2000). From these studies, a general picture of bacterial physiology has emerged in which cells from complex communities can be classified according to their relative nucleic acid content and the state of their membrane (Figure 1A). Relative nucleic acid content is based on differences in the individual cell fluorescence and light scatter signals after nucleic acid staining, and is used as a proxy of cellular activity. Generally, within an ecosystem cells with high nucleic acid content (HNA) are larger but also more active, with higher metabolic activity and growth rates, than cells with low nucleic acid content (LNA) (Gasol et al., 1999; Jellett et al., 1996; Jochem et al., 2004), although the latter are not completely inactive (Bouvier et al., 2007; Wang et al., 2009; Zubkov et al., 2004). The state of the bacterial membrane is another crucial physiological indicator, as cells with a loss of membrane polarity (DiBAC+ cells) or membrane integrity (Pi+ cells) are considered damaged or dying (Del Giorgio and Gasol, 2008). In addition to these single-cell methods, metatranscriptomics (RNA-Seq) allows the direct analysis of the gene expression profiles of the abundant metabolically active microorganisms in the human gut (McNulty et al., 2011; Rey et al., 2010; Turnbaugh et al., 2010; Turnbaugh et al., 2009b). Here, we combine flow cytometry, metatranscriptomics, and metagenomics to identify and characterize the active subset of the gut microbiome, and to determine its short-term responses to xenobiotics.
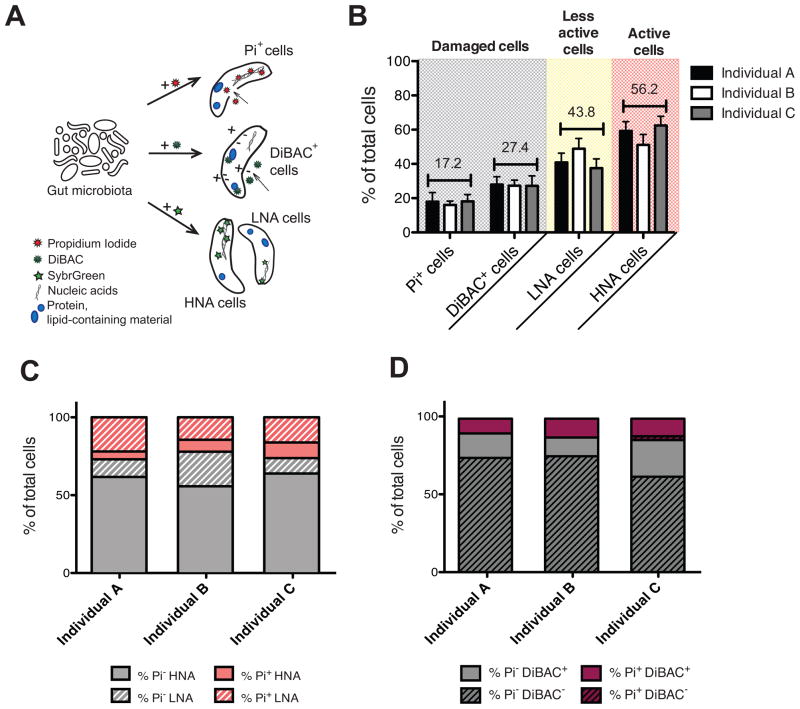
Figure 1. The human gut microbiota is highly active with substantial proportions of damaged cells.
Also see Figures S1, S2 and Table S3. (A) Cellular targets of the fluorescent dyes. The nucleic acid dye Pi enters cells with compromised membranes; DiBAC binds to intracellular lipid-containing material of depolarized cells; and SybrGreen stains the nucleic acids of all bacteria irrespective of their membrane status, identifying two clusters of cells: the low- (LNA) and high- (HNA) nucleic acid containing cells. (B) Average proportions of damaged cells (Pi+ and DiBAC+) and cells with a low (LNA) or high nucleic acid content (HNA) in three unrelated individuals (n=5–10 samples/individual). Values are mean±sem. (C) SybrGreen and Pi dual-staining in 3 unrelated individuals. Pi- cells are in grey, Pi+ cells are in pink, with the HNA (solid) and LNA (stripes) subsets indicated. (D) Pi and DiBAC dual-staining in the same individuals. Pi- cells are in grey, Pi+ cells are in purple, with the DiBAC- (stripes) and DiBAC+ (solid) fractions indicated.
Using these complementary techniques, we demonstrate that the gut microbiota contains a distinctive active subset relative to the overall microbial community. The proportion of each physiological fraction is comparable between unrelated healthy individuals, but displays marked temporal variations within each person. Short-term exposure to xenobiotics not only alters bacterial physiology, but also significantly alters the structure of the overall microbial community and each physiological fraction, in addition to gene expression. These responses may provide some clue as to which microorganisms, genes, and pathways are involved in xenobiotic metabolism, and to the general mechanisms for resisting potential damage from these molecules. In a larger sense, we provide a proof-of-principle for future studies combining single cell methods and metatranscriptomics to investigate the response of human and environmental microbiomes to defined perturbations.
RESULTS
Most of the microbial cells in the human gut are active, with substantial proportions of damaged cells
We determined the proportions of four physiological categories in fresh fecal samples immediately after collection: cells with loss of membrane integrity (Pi+), loss of membrane polarity (DiBAC+), and with different relative nucleic acid content (high- HNA, and low- LNA, referred to here as the highly active and less active subsets, respectively) (Figure 1A, S1A, S2A). In total we assayed 21 fecal samples from three unrelated individuals, at 10 different time-points over 9 months (n=5–10 samples/individual). Pi+ cells constituted 17.2±2.1% of the gut microbiota (range across all individuals: 5.5%–41.1%). DiBAC+ cells were 27.4±2.3% of the total community (range: 15.4%–49.6%), and HNA cells were 56.2±3.5% (range: 25.3%–85.2%) (Figure 1B). No significant differences were observed between individuals (p>0.05, Kruskal-Wallis test). However, we observed marked changes within each individual over time, exceeding those found with either biological or staining technical replicates (Figure S2C). These differences could not be attributed to the date or time of sample collection, the time since the last antibiotic use, the time since or the composition of the previous meal, or recent travel history (adjusted p-value>0.05, MINE analysis) (Reshef et al., 2011). We quantified the size distribution of HNA and LNA cells to test if the increased fluorescence observed in HNA cells could be explained by larger cell size, due to division, stochastic variation, or inter-species differences; there was a trend towards increased size but this was not significant (1.216±0.092μm vs. 0.961±0.002μm, respectively; p>0.05, Mann-Whitney test). Thus, on average over half of the gut microbiota had increased metabolic activity not solely attributable to increased cell size, whereas nearly one third could be considered damaged.
Firmicutes dominate the active and damaged subsets of the gut microbiota
We combined fluorescence-activated cell sorting (FACS) with 16S rRNA gene sequencing methods to identify cells within each physiological fraction (referred to here as “FACS-Seq”; Figure S1B, Table S1A). Community membership (presence/absence) was consistent across all physiological categories, with 99/100 of the most abundant species-level bacterial operational taxonomic units (OTUs) and 15/15 of the abundant genera (≥1% 16S rRNA sequences in at least one sample) common to all groups (Figure 2A). We also observed clear inter-individual differences in bacterial community membership, regardless of physiological status (Figure 2B).
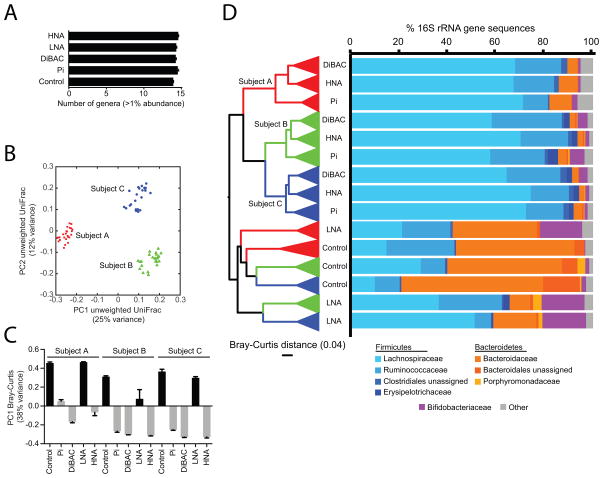
Figure 2. The human gut microbiota contains a distinctive active subset.
Also see Figures S3, S4 and Tables S1, S4, S5. (A) Number of abundant genera (relative abundance ≥1% in at least one sample) found in the Pi+, DiBAC+, LNA, HNA, and control fractions. Values are mean±sem. (B) Unweighted UniFrac clustering of community membership across each subgroup. Samples are labeled by individual in panels B–D: red (individual A), green (individual B), and blue (individual C). (C) Comparisons of microbial community structure, using the Bray-Curtis distance metric. The first principal component is shown across each subgroup and individual. (D) The active and damaged subsets of the gut microbiota are distinctive from the overall unsorted community (Control). UPGMA clustering based on Bray-Curtis distance is shown on the left: replicate samples are collapsed into wedges and three outliers have been removed (see Figure S3A for the full tree). The relative abundance of the major bacterial families in the gut microbiota: Firmicutes (blue), Bacteroidetes (orange), and Actinobacteria (Bifidobacteriaceae; purple). Remaining taxa have been grouped into ‘Other’.
The relative abundance of species-level OTUs in all individuals was altered in highly active cells (HNA), relative to either LNA cells or the overall bacterial community (Figures 2C, D, S3A). HNA cells were significantly enriched for Clostridiales (Firmicutes phylum, 90.8±1.7 vs. 34.8±7.2%) and depleted for Bacteroidales (4.6±1.7 vs. 62.8±7.0%) relative to the overall fecal microbiota (p<0.05, Student’s t-test). By contrast, LNA cells were significantly enriched for Bifidobacteriales relative to the overall community (18.0±0.2 vs. 1.4±0.5%; p<0.001, Student’s t-test). The differences between the HNA and LNA fractions could not be explained by increased genome size or rRNA copy number; as the Clostridiales reference genomes in our database were significantly smaller than the Bacteroidales (3.22±0.15 vs. 4.37±0.18 Mbp; p<10−5, Student’s t-test) and had similar numbers of 16S rRNA genes (2.36±0.25 vs. 2.15±0.40; p>0.05, Student’s t-test). However, Bifidobacteriales did exhibit smaller genomes and less rRNA genes than the Clostridiales, potentially contributing to their enrichment in the LNA fraction (2.27±0.08 vs. 3.22±0.15 Mbp; p<10−5; 1.58±0.29vs. 2.36±0.25 16S rRNA genes; p<0.05, Student’s t-tests). As further validation, cultured gut isolates from the 5 dominant bacterial phyla were analyzed during in vitro growth, confirming that all of the strains could be efficiently stained with SybrGreen and that the proportion of damaged cells was significantly increased for each strain following heat or ethanol treatment (Table S3A).
To further explore these trends, we employed LEfSe, a tool for metagenomic biomarker discovery (Segata et al., 2011). Taxa at multiple phylogenetic depths were found at significantly higher relative abundance within each physiological fraction relative to the others (Table S4A, Figure S3B). Bacteroidetes (Bacteroides), Bifidobacteriales (B. adolescentis and B. longum), and Lachnospiraceae (Roseburia faecis) were enriched in the less active component of the gut microbiota (LNA). Specific groups within the Firmicutes, such as the Ruminococcaceae and certain Clostridiales (Blautia sp., Coprococcus sp.), were associated with the HNA, Pi+, and DiBAC+ fractions, suggesting that members of this diverse phylum vary in their activity levels and level of membrane damage. Overall, the damaged subset of the gut microbiota (Pi+ and DiBAC+ cells) shared a similar community structure to the more active (HNA) fraction (Figures 2C, D, S3A).
Together, these results indicate that the fecal microbiota harbors a distinctive active subset enriched for Clostridiales. The concordance between the HNA and damaged subsets suggests that these cells are both metabolically active and rapidly turning over, while other members of the community are less active; however, we cannot entirely exclude the possibility that Clostridiales are more sensitive to oxygen exposure during sample collection. To confirm that single cells from human fecal samples can be both active and damaged, we quantified the overlap between the Pi+ and HNA populations using a dual-staining approach; 7.64±1.45% of total cells were both Pi+ and HNA, representing 30.63±4.94% of the total Pi+ cells (Figure 1C). Also, Pi and DiBAC dual-staining revealed that cells with damaged membrane integrity were a subset of the population of cells with altered polarity: 94.0±4.9% of Pi+ cells were also DiBAC+ (Figure 1D).
We validated these findings using metatranscriptomics (RNA-Seq) coupled with metagenomic sequencing of community DNA at two time-points from the same individuals (7.3±1.6 million RNA and 7.0±2.3 million DNA sequencing reads/sample; Table S1B). Most of the gut microbiome was transcriptionally active; the majority of the observed reference genomes were found in the paired RNA-Seq and community DNA from each individual (87.7±3.0%; n=6), whereas all of the 185 most abundant genomes were shared (>121 RPKM on average). However, gene expression profiles from all individuals before and after 4 hours of incubation in rich medium under anaerobic conditions were distinct from gene abundance (Figure S4). Consistent with our FACS-Seq results, genomes from the Firmicutes phylum exhibited significantly higher than expected expression relative to the Bacteroidetes (3.9±0.6 vs. 1.7±0.4 fold-change, p<0.01, Student’s t-test). In total, 15 different Firmicutes taxonomic groups, but no Bacteroidetes, had ≥4-fold higher gene expression relative to abundance (Table S5A), while 2,719 gene clusters had significantly altered expression relative to abundance (Table S5B; p<0.01, >5 fold-change). Rank products analysis identified 985 gene clusters with significantly decreased expression relative to abundance (Table S5C; FDR<0.1, >10 fold-change); these were significantly biased towards Bacteroidetes over Firmicutes (720 versus 162; p<0.0001, χ2 test). Combined, these results are consistent with the hypothesis that a distinctive subset of microorganisms, enriched for Firmicutes, tends to dominate the active fraction of the gut microbiome.
Antibiotics, but not host-targeted drugs, rapidly influence microbial physiology
Having defined the compositional, inter-individual, and temporal variations in the active microbiome, we sought to determine the response of this distinctive subset to short-term perturbations. In particular, exposure to xenobiotics such as antibiotics and host-targeted drugs seemed likely candidates given their long-lasting influence on the composition of the gut microbiota (Dethlefsen and Relman, 2011; Jernberg et al., 2010). We thus conducted incubations with a panel of 14 xenobiotics (6 host-targeted drugs and 8 antibiotics, Figure S1C, Table S2 ) and at lower pH (pH4, pH5, and pH6) to contrast the effects of xenobiotics with those from simple environmental perturbations. Ethanol was the xenobiotics vehicle, and did not result in significant changes of bacterial physiology relative to un-amended samples (p>0.05, Dunn’s test), whereas low pH significantly increased the proportions of damaged cells (Figure 3).
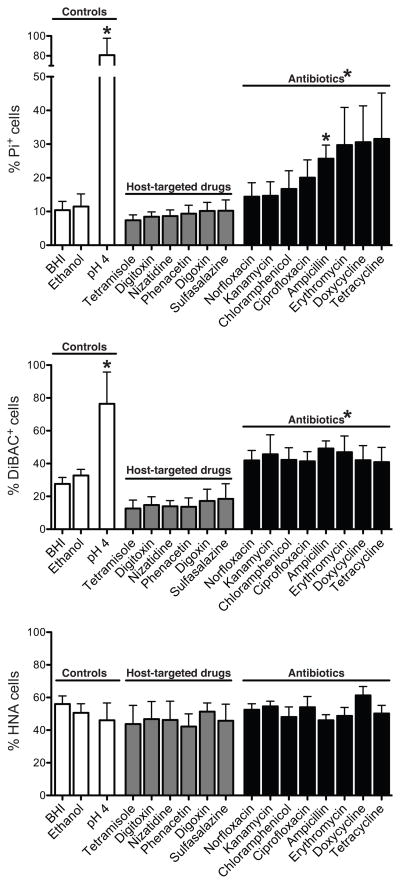
Figure 3. Antibiotics, but not host-targeted drugs, alter microbial physiology.
Also see Figure S5 and Tables S2, S3. Top to bottom: Average proportions of cells with loss of membrane integrity, altered polarity, and high levels of activity across 3 unrelated individuals, determined after incubations with xenobiotics. Control incubations consisted of un-amended BHI medium, BHI medium with the drug vehicle (ethanol), and low pH. Treatments with a significant impact relative to controls are noted (p<0.05, Dunn’s test). Values are mean±sem.
Antibiotics exposure significantly increased the proportions of damaged cells (Figure 3). Averaged across all tested compounds and individuals, DiBAC+ cells increased from 32.7±3.8% to 43.8±2.7%, and Pi+ cells from 11.5±3.7% to 22.9±2.8%, when comparing ethanol controls to antibiotic treatments, respectively (p<0.05, Wilcoxon rank- sum test). The proportion of HNA cells was not significantly altered by antibiotics [51.5±7.4% (control) vs. 51.9±1.8%; p>0.05, Wilcoxon rank-sum test]; this was also the case for total cell abundance [1.26e11±2.34e10 (control) vs. 1.12e11±8.18e9 cells.ml−1], suggesting a rapid increase in membrane damage without complete lysis.
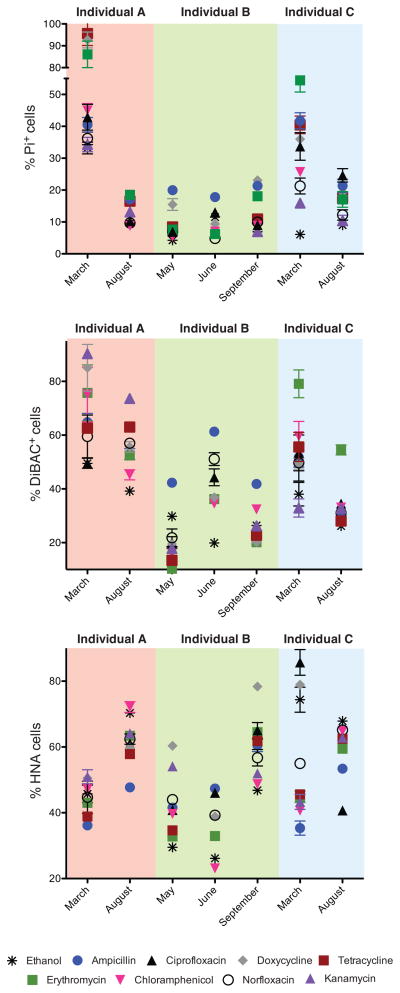
Substantial temporal variation in antibiotic-induced cell damage was observed (Figure 4, Table S3B). These variations could not be attributed to the date or time of sample collection, the time since or the composition of the previous meal, or recent travel history (p>0.05, MINE analysis); but we observed significant associations with the baseline physiology and microbial community structure of each sample. The proportion of depolarized cells (DiBAC+) at the time of sample collection was significantly associated with the average %DiBAC+ cells following antibiotic treatment (p<0.05, Spearman rank correlation). Also, the baseline abundances of the Parabacteroides and Escherichia genera were inversely associated with the mean proportion of Pi+ cells following antibiotic treatment [n=2–3 time-points/individual; MIC>0.98, adjusted p-value<0.05, MINE analysis of all abundant bacterial genera (average relative abundance >1%)].
Figure 4. The physiological response of microbial communities to antibiotics varies over time within each individual.
Also see Figure S5 and Tables S2, S3. Top to bottom: Proportions of cells with loss of membrane integrity, altered polarity, and high levels of activity after antibiotics exposure. Values for vehicle controls are indicated with asterisks. When visible, error bars represent staining replicates. Values are mean±sem.
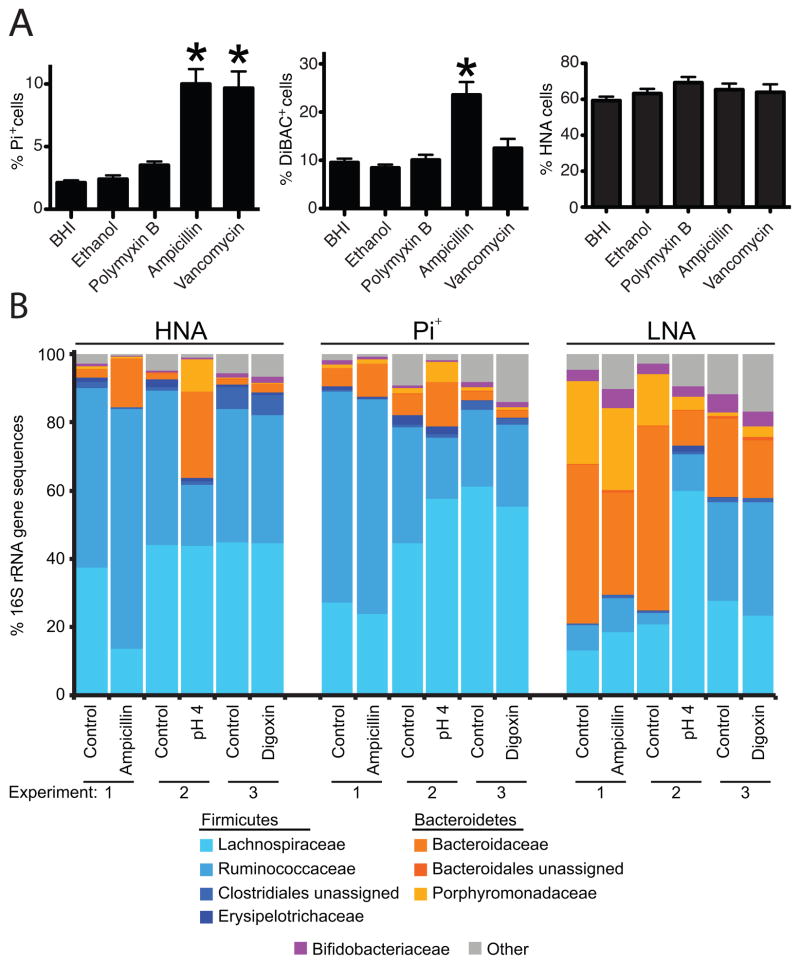
Altogether, 75% of the antibiotics tested had significant impacts on cell damage (Table S3B). However, ampicillin was the only compound that consistently increased the proportion of Pi+ cells when averaged across all individuals and all time-points (Figure 3; p<0.05, Dunn’s test); suggesting that antibiotics targeting cell wall synthesis in Gram-positive bacteria may consistently induce membrane damage. This was further tested with another cell wall synthesis inhibitor specific to Gram-positive bacteria (vancomycin) and a cell membrane disrupter specific to Gram-negative bacteria (polymyxin B; n=3 individuals at 2 time-points; Table S2). Both cell wall synthesis inhibitors significantly induced cell damage, whereas no significant effects were observed following exposure to polymyxin B (Figure 5A).
Figure 5. Antibiotics targeting cell wall biosynthesis consistently increase cell damage, and change the structure of the active and damaged gut microbiota.
Also see Figure S5 and Tables S1, S2, S4. (A) Average proportions of cells with loss of membrane integrity, altered polarity, and high levels of activity following exposure to antibiotics targeting the bacterial cell wall and membrane (2 time-points/individual). Values are mean±sem. (B) Changes to the highly active (HNA), less active (LNA), and damaged (Pi+) gut microbiota following exposure to ampicillin, digoxin, and low pH. Each experiment represents FACS-Seq analysis of samples from a single individual with or without treatment. Bars represent the average across all technical replicates (n=2–3 replicates).
Analysis of microbial community structure before and after exposure to antibiotics revealed significant differences, although they were minor relative to inter-individual and temporal variations (Figure S5A). ANOVA and LefSe-based comparisons of species-level OTUs between all 8 treatments revealed multiple OTUs and taxonomic groups with significantly altered relative abundance following antibiotic treatment (Tables S4B, C); 90% of the affected OTUs were Firmicutes. These antibiotic-responsive OTUs were affected in unique ways by different classes of compounds; for example, an OTU representing Faecalibacterium prausnitzii increased 4.5-fold after ampicillin treatment, but decreased after exposure to the other compounds. Repeated administrations of ampicillin to fecal samples collected from one individual revealed that it significantly reduced the expansion of Enterobacteriales during the 4-hour incubation [13.7±4.3% (vehicle control) vs. 0.6±0.2% (ampicillin), p<0.05, Kruskal-Wallis test; Figure S5B].
In contrast, the host-targeted drugs did not exhibit a pronounced impact on microbial physiology, even when tested at high concentrations (100-fold concentration increase, p>0.05, Wilcoxon rank-sum test). Subtle changes in community structure were observed when performing LefSe analysis across all 6 treatments (Table S4C), but samples mainly still grouped by individual and not by treatment (Figure S5C). Incubations at low pH significantly modified community structure, increasing the relative abundance of Clostridiales [56.4±1.2% (pH4)], relative to the control (37.2±10.0 (pH7); p<0.01, Student’s t-test of pH4 vs. pH5–7].
Finally, we sought to determine if exposure to xenobiotics or low pH could alter the structure of the active or damaged gut microbiota. Fecal samples were collected and exposed to ampicillin, digoxin, or low pH for 4 hours prior to FACS-Seq. The results confirmed our original characterization of the active and damaged gut microbiota, with an increased abundance of Clostridiales in the active and damaged subset, in addition to a higher level of Bacteroidales and Bifidobacteriales in the less active fraction (Figure 5B). Furthermore, LefSe analysis revealed that low pH and ampicillin exposure resulted in significant changes to the active and damaged gut microbiota (Table S4D), namely an increased relative abundance of members of the Bacteroidetes phylum. Digoxin also led to a significantly higher relative abundance of the Faecalibacterium genus (Firmicutes phylum) in the Pi+ subset (15.0±0.9% after digoxin treatment versus 10.5±1.2% in controls; Table S4D). Together, these results emphasize that despite shared features of the active and damaged gut microbiota between healthy individuals, it can be rapidly altered following exposure to xenobiotics.
Short-term exposure to xenobiotics affects community-wide gene expression
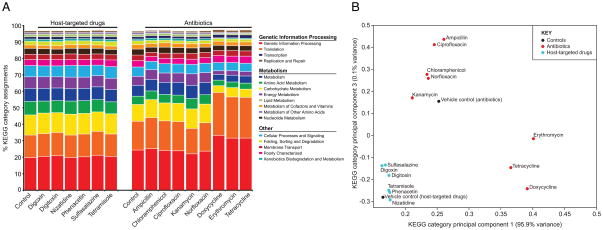
In order to more comprehensively characterize the influence of xenobiotics on the active gut microbiome, we tested for changes in community-wide gene expression following xenobiotics exposure. RNA-Seq profiles were obtained after the same 4-hour incubations with host-targeted drugs, antibiotics, vehicle controls, and low pH used for the analysis of microbial physiology and community structure (Figures S1 C, D and Table S1B; 565±142 thousand mRNA sequencing reads/sample). To obtain a high-level view of the transcriptional response to xenobiotics, we tallied the number of normalized counts assigned to each KEGG metabolic pathway and higher-level category. In general, the treatment and control samples shared a similar distribution of pathways (Figure S6A; note, all correlations were >0.9) and categories (Figure 6A). Transcriptional profiles were most similar when from the same individual and the same time-point. However, clear deviations from this pattern occurred after exposure to tetracycline and macrolide antibiotics that inhibit translation (Figure 6B). All three treatments resulted in an increased expression of genetic information processing genes (e.g. transcription and translation; 24.8±0.4 vs. 32.8±0.7% of assignments in controls compared to treatments, p<10−5, Student’s t-test). Analysis of KEGG module and pathway abundance with HUMAnN and LEfSe confirmed and extended these trends; antibiotics induced the expression of modules for tRNA biosynthesis, translation, vitamin biosynthesis, and phosphate transport, whereas host-targeted drugs induced modules for membrane transport, the pentose phosphate pathway and xenobiotic metabolism/biodegradation (Tables S4 E, F).
Figure 6. Impact of xenobiotics on the expression of KEGG categories.
Also see Figure S6 and Table S2, S4, S7. Expression levels of genes assigned to high-level KEGG categories following short-term exposure to xenobiotics. (A) The % of normalized counts assigned to each category is shown for the 14 treatments and their vehicle controls. (B) Principal component analysis of KEGG category expression levels for the 14 treatments and controls. The mean expression profile for each group was clustered using the “princomp” function in Matlab.
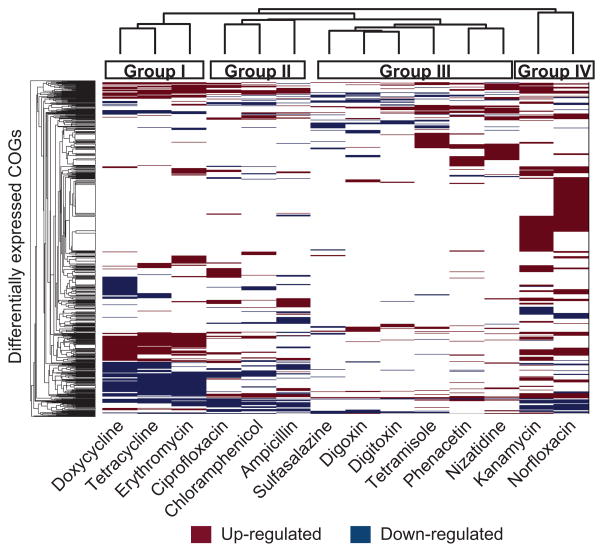
A complementary analysis using the COG ontology (Tatusov et al., 2000) and the ShotgunFunctionalizeR package (Kristiansson et al., 2009) revealed qualitatively similar clustering within each individual (Figure S6B), which was quantitatively confirmed by comparing Spearman distance matrices constructed for each individual (p<0.05, Mantel test). A total of 832 orthologous groups were identified with significantly different expression levels between the treatments and controls (Table S6). We then clustered the transcriptional responses to the 14 xenobiotics, revealing 4 major response groups (Figure 7). Drugs from the same class were nearest neighbors, including the cardiac glycosides (digoxin and digitoxin) and the tetracyclines (doxycycline and tetracycline); but the fluoroquinolones (ciprofloxacin and norfloxacin) resulted in divergent responses. Exposure to antibiotics resulted in a more dramatic transcriptional response relative to host-targeted drugs (146±29 vs. 67±17 up-regulated COGs and 97±13 vs. 38±8 down-regulated COGs; p<0.05, Student’s t-test). A subset of these groups was consistently differentially expressed across the majority of compounds, potentially representing conserved pathways for responding to xenobiotics or environmental stress (Supplemental Experimental Procedures).
Figure 7. Short-term exposure to antibiotics and host-targeted drugs results in altered community-wide gene expression.
Also see Figures S6 and Tables S2, S4, S6, S7. COGs up- or down-regulated after exposure to xenobiotics are labeled red and blue, respectively (adjusted p-value<0.01, AIC<300). COGs with significantly different expression between controls collected on separate days were excluded. Treatments and COGs were clustered using the “clusterGram” function in Matlab. The four major treatment groups are noted by colored boxes on top of the heatmap.
Xenobiotics induce the expression of genes for drug resistance and metabolism
We then searched for gene clusters affected by exposure to xenobiotics (Figure S1D). In total, we identified 1,728 gene clusters differentially expressed after antibiotics exposure (p-value<0.05, >2 fold-change; Tables S7A–H) and 328 after exposure to host-targeted drugs (Tables S7I–N). A more stringent rank products analysis revealed 350 gene clusters with significantly decreased expression after the addition of antibiotics (FDR<0.1; Table S7Q): 64.3% (225) were in response to ampicillin and 88.9% of those (200) were attributed to the Bifidobacterium and Collinsella genera, isolates of which are sensitive to β-lactam antibiotics (Le Blay et al., 2007).
These gene-centric results, combined with our metabolic pathway, module, and functional group analyses, provide insight into the microbial genes and genomes involved in drug resistance and metabolism in the human gut. For instance, recent studies of cultured gut bacteria have demonstrated that in addition to inhibiting their intended target, antibiotics induce metabolic stress (Kohanski et al., 2007), a process related to proton motive force (Lee and Collins, 2012). Our results suggest that these processes also occur within the intact human gut microbiota: exposure to antibiotics induced the expression of key genes affecting stress response (e.g., cell death regulator MazG, heat shock regulator protein HspR), proton motive force, antibiotic resistance (e.g., tetracycline resistance protein, MATE efflux family protein), and phage induction (Tables S7A–H and Supplemental Experimental Procedures).
Our data also provide multiple candidate genes for the microbial biotransformation of xenobiotics (Tables S6, S7). For example, the cardiac glycosides digoxin and digitoxin undergo microbial reduction (Lindenbaum et al., 1981; Magnusson et al., 1982), and both compounds induced reductases, transcriptional regulators, and small molecule/sugar transporters (Tables S7I–N and Supplemental Experimental Procedures). Nizatidine, known to undergo N-oxide bond cleavage (Sousa et al., 2008), induced the expression of drug/metabolite transporters and enzymes acting on nitrogen bonds. Phenacetin, an aromatic compound that undergoes N-deacetylation (Smith and Griffiths, 1974), uniquely induced the expression of genes annotated as aromatic ring-cleaving dioxygenases and acylaminoacyl-peptidases. Sulfasalazine, known to undergo azo reduction (Sousa et al., 2008), induced the expression of gene clusters annotated as encoding thioredoxins and nitrate reductases. Finally, tetramisole, which undergoes a thiazole ring opening and disulfide bond formation (Sousa et al., 2008), induced the expression of sulfoxide reductases, genes involved in disulfide bond formation, and sulfate transport systems.
DISCUSSION
Moving beyond DNA-based measurements of microbial communities
Over the last few years, the wealth of information describing the organismal and genetic diversity found in the complex microbial communities colonizing different body habitats has allowed us to better assess the key factors influencing community assembly and metabolic potential. However, these largely DNA-based studies have provided limited insight into the metabolic activity of their microbial affiliates and their molecular responses to common perturbations, such as xenobiotics exposure. Through a combination of flow cytometry, 16S rRNA gene sequencing, and metatranscriptomics, we characterized the active subset of the human gut microbiome, and its response to a common and clinically relevant perturbation: exposure to host-targeted drugs and antibiotics.
Single-cell methods have allowed the description of bacterial physiology in several ecosystems, and linked physiological categories to different levels of activity and biomass (Davey and Winson, 2003; Del Giorgio and Gasol, 2008; Wang et al., 2009). We successfully applied this approach to determine the physiology of cells in the gut microbiota, presenting evidence for four distinct physiological categories ranging from highly active to severely damaged cells. More than half of the gut microbiota can be considered as highly active, with HNA proportions comparable to values from very productive aquatic systems, such as estuaries and marshes (Bouvier et al., 2007). This high level of activity is likely sustained by our regular dietary input of nutrients and the ability of the gut microbiota to metabolize glycoproteins from saliva, gastric juice, and mucus (Blaut, 2011).
FACS-Seq analysis of cells from each physiological category revealed that the active subset was enriched for Clostridiales, and the less active subset was enriched for Bifidobacteriales, consistent with recent reports using complementary methods (Gosalbes et al., 2011; Peris-Bondia et al., 2011), and supported by our comparisons of gene expression and abundance. Our results suggest that the composition of the active gut microbiota cannot simply be explained by methodological biases in staining, an increased rate of cell division, larger cell or genome size, or more copies of the 16S rRNA gene. In aquatic ecosystems, numerous studies have demonstrated that the HNA cell population has an increased rate of metabolic activity [e.g., labeled leucine or thymidine uptake, observation of dividing cells (Bouvier and Maurice, 2011; Jellett et al., 1996; Wang et al., 2009)]. Similar studies on the gut microbiota are needed to determine the extent to which these trends are conserved in host-associated microbial communities.
In addition to high proportions of metabolically active cells, we also report that one third of the gut microbiota is composed of damaged cells with impaired membrane polarity (DiBAC+). More severely damaged cells with compromised membrane integrity (Pi+) constitute less than one quarter of the total community, consistent with previous reports (Ben-Amor et al., 2005). These findings support a view of the human gut as an ecosystem harboring productive bacterial communities, with a surprising level of temporal variation in activity and cell damage given the overall stability of strong drivers of bacterial physiology, such as temperature, pH, and nutrient availability (Blaut, 2011; Walter and Ley, 2011). More work is needed to identify the factors responsible for these temporal dynamics, e.g. dietary changes, phage exposure, bile acids, host immunity, or other ill-defined factors.
Ecological tradeoffs between activity level and cell damage
Our FACS-Seq analysis demonstrates that both the active and damaged subsets are enriched for Clostridiales, suggesting that environmental stressors may more readily influence these groups. Multiple observations support this hypothesis: the majority of the species-level OTUs significantly impacted by antibiotic treatment were Clostridiales; antibiotics exposure increased the overall level of gene expression of the Clostridiales (185,500±41,386 vs. 489,362±59,430 RPKM in controls relative to antibiotic treatments; p<0.001, Student’s t-test); the most consistent physiological responses were after exposure to antibiotics targeting Gram-positive bacteria (of which Clostridiales are the dominant member); and dual-staining revealed that up to one-third of Pi+ cells could also be HNA cells.
Together, these findings prompt the hypothesis that members of the gut microbiota may inhabit distinct ecological niches, defined not only by physical location and resource utilization, but also by their level of metabolic activity. A similar hypothesis has been proposed for pathogens, wherein metabolically active cells exhibit increased susceptibility to antibiotics (Levin and Rozen, 2006; Martinez and Rojo, 2011). Studies of genetically tractable commensal gut microorganisms in vitro and in mouse models will be necessary to test to what degree differing levels of metabolic activity influences bacterial fitness when controlling for differences in diet, host genotype, and microbial community structure.
Implications for personalized medicine
The magnitude of the physiological response to each xenobiotic varied substantially between individuals and over time, although antibiotics generally had a greater impact than host-targeted drugs. Our results are consistent with multiple, not necessarily mutually exclusive, mechanisms that could be responsible for these variations. First, we detected associations between the composition of each individual’s gut microbiota and the extent of cell damage, consistent with previous studies demonstrating that bacterial susceptibility to antibiotics can be highly species- or even strain-specific (Mortimer et al., 2000; Novo et al., 2000; Wickens et al., 2000). Second, the degree of response could be driven by variations in the prevalence and/or expression of xenobiotic-induced genes impacting drug metabolism, small molecule transport, antibiotic resistance, stress response, and/or the production of protective molecules.
Elucidating the underlying mechanisms for xenobiotic resistance and metabolism in the active human gut microbiome will not only provide novel insight into host-microbial interactions and biochemistry, but may also provide indications for unexplained patient-to-patient variations in drug efficacy and toxicity. Our results provide a view of the degree and consistency of the microbial response to a panel of xenobiotics. As expected, antibiotics had the strongest effects, whether assessing physiology, community structure, or gene expression, the latter being significantly affected by all of the tested xenobiotics. In particular, cell wall synthesis inhibitors targeting Gram-positive bacteria consistently increased cell damage, whereas compounds inhibiting translation dramatically altered gene expression profiles. Surprisingly, we observed a divergent transcriptional response to the fluoroquinolones, despite the same mechanism of action, prompting more studies to determine whether these closely related compounds differ in their ability to elicit off-target effects or in their spectrum of susceptible gut microorganisms.
A metagenomic perspective of drug resistance and metabolism
Our results also emphasize the many impacts of antibiotics on the gut microbiome, including the induction of antibiotic resistance genes, stress response pathways, and bacteriophage elements. It will be important to perform time series experiments to determine how the short-term alterations to microbial physiology and gene expression described here translate into long-term changes to microbial community structure following exposure to antibiotics (Dethlefsen and Relman, 2011; Jernberg et al., 2010).
Although >40 host-targeted drugs are modified by members of the gut microbiota, with important implications for drug efficacy and toxicity, the microorganisms and the genetic machinery involved often remain unknown (Haiser and Turnbaugh, 2012). These results show that despite minimal short-term effects on microbial physiology or community structure, all six host-targeted drugs tested resulted in significant changes to the genes expressed by the human gut microbiome. This catalogue of xenobiotic-induced microbial genes includes candidate genes for drug import, metabolism, and transcriptional regulation, providing the basis for follow-up studies aimed at confirming their predicted activities and determining their distribution among members of the human gut microbiota. It will be important to measure the concentration of these host-targeted drugs along the length of the gastrointestinal tract in humans or animal models, coupled to a detailed profile of the concentration and activity of the microbial enzymes of interest. Finally, these results raise the question as to whether exposure to xenobiotics has provided enough selective pressure to lead to microbial systems fine-tuned for specific drugs, or if the observed responses represent crosstalk between xenobiotics and systems evolved to respond to other natural targets. Detailed structure-function analyses, coupled to an in-depth characterization of the endogenous compounds produced by the host and the gut microbiota will be necessary to answer these questions.
The integrated characterization of the host, microbial, and environmental factors governing the response of the gut microbiome to xenobiotics could ultimately be used for the design of novel diagnostic tests predicting drug pharmacokinetics or novel therapeutic interventions. At a minimum, these results emphasize that in order to understand the ultimate fate of xenobiotics and their impact on human health, it is important to consider their intended, or often unintended, effects on the active gut microbiome.
EXPERIMENTAL PROCEDURES
Sample collection
All human studies were done under the guidance of the Harvard Committee on the Use of Human Subjects in Research (Protocol #F18510). The three recruited unrelated healthy subjects were free of gastrointestinal disorders and did not use antibiotics during the course of the experiment. Additional subject information is provided in the Supplemental Experimental Procedures. Fresh fecal samples (n=5–10 samples/individual) were immediately diluted 1:10 in PBS, centrifuged at 700g for 1min, and transferred to an anaerobic chamber.
Bacterial physiology
Bacterial physiology was assessed with 3 fluorescent stains (Figure 1A): propidium iodide (Pi), bis-(1,3-dibutylbarbituric acid)trimethine oxonol [DiBAC4(3)], and SybrGreen I, allowing the observation of cells with loss of membrane integrity, membrane polarity and with distinct nucleic acid content, respectively (Del Giorgio and Gasol, 2008). Cells were stained in the dark under anaerobic conditions after testing sample preparation and staining conditions (Figure S2B). The percentages of Pi+, DiBAC+, LNA and HNA cells were calculated relative to the total bacterial counts obtained by SybrGreen I staining.
Fluorescence-activated cell sorting of bacterial physiological categories
Fecal samples from the same individuals were sorted on a MoFlo cytometer equipped with a 488nm excitation and 530nm band pass filter for detection (Becton Dickinson). For each individual, we sorted 50,000 bacterial events of each physiological category (Figure S1B), and processed them according to published protocols, with 3 consecutive freeze-heating (100°C) cycles in liquid nitrogen and a thermocycler (Guillebault et al., 2010), before PCR amplification. We also determined the average size of sorted HNA and LNA bacteria in all individuals using a “Multisizer 3” Coulter Counter (20μm aperture) according to the manufacturer’s guidelines (Beckman Coulter).
16S rRNA gene sequencing and analysis
Samples were kept at −80°C until DNA extraction with the PowerSoil bacterial DNA extraction kit (MoBio, Carlsbad CA). The V4 region of the 16S rRNA gene was PCR amplified in triplicate, and the resulting amplicons were cleaned, quantified, and sequenced on the Illumina HiSeq platform according to published protocols (Caporaso et al., 2012; Caporaso et al., 2011), with custom barcoded primers (Table S1A). Sequences were analyzed with the QIIME software package (Quantitative Insights Into Microbial Ecology) (Caporaso et al., 2010) in addition to custom Perl scripts to analyze alpha (within sample) and beta (between sample) diversity. LEfSe analysis (Segata et al., 2011) and OTU significant tests by ANOVA were performed on filtered datasets; rare OTUs with <1,000 sequence counts or found in <10 samples were excluded.
Ex vivo incubations with xenobiotics
All incubations took place for 4 hours under anaerobic conditions, in the dark, and at 37°C. Fresh fecal samples from the same individuals were diluted 1:10 in freshly prepared liquid BHI (pH7; Becton Dickinson). We tested 6 host-targeted drugs and 8 antibiotics (Figure S1C, Table S2). Control incubations consisted of BHI with or without the drug vehicle (ethanol). Bacterial physiology, community structure, and gene content were assessed as previously described, prior to the incubations. Following the incubations, we determined bacterial physiology, community structure, and gene expression patterns. We set the amount of each xenobiotic (10μg.ml−1) to be near to or lower than the estimated level remaining in the small intestine and colon (Baselt, 2008; Raaflaub and Dubach, 1975; Somogyi et al., 1995).
RNA-seq sample preparation and analysis pipeline
Methods for microbial mRNA sequencing and analysis were as previously described (McNulty et al., 2011; Rey et al., 2010; Turnbaugh et al., 2010; Turnbaugh et al., 2009b) (see Figure S1D and Supplemental Experimental Procedures).
Statistical Analysis
Data are expressed as mean±sem, unless otherwise noted. Statistics were performed using GraphPad Prism (v.5), Microsoft Excel (v. 14), and the MINE software package (Reshef et al., 2011). Significance was accepted at p<0.05, unless otherwise noted.
Supplementary Material
HIGHLIGHTS.
The gut microbiome is highly active, similar to nutrient-rich productive systems.
Firmicutes dominate the active and damaged subsets of the gut microbiome.
Both antibiotics and host-targeted drugs rapidly alter the active gut microbiome.
Xenobiotics induce genes for drug metabolism, drug resistance, and stress response.
Acknowledgments
We thank Rob Knight and Donna Berg-Lyons for contributing barcoded 16S rRNA primers; Claire Reardon, Jiangwen Zhang, and Christian Daly for sequencing support; Brian Tilton and Thierry Bouvier for flow cytometry assistance; and Curtis Huttenhower, Nicola Segata, and Jovian Yu for bioinformatics support. We are indebted to our colleagues Andrew Murray, Erin O’Shea, Bodo Stern, Lawrence David, Julie Button, Rachel Dutton, and Benjamin Wolfe for their helpful suggestions. This work was supported by the NIH (P50 GM068763).
Footnotes
ACCESSION NUMBERS
RNA-Seq data is deposited in the Gene Expression Omnibus (GEO) database (accession number GSE38151); 16S rRNA gene sequencing reads are deposited in MG-RAST (accession number 1319).
Supplemental Information includes Supplemental Experimental Procedures, 6 figures, and 7 tables and can be found with this article online at XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baselt RC. Disposition of toxic drugs and chemicals in man. 8. Foster City, California: Biomedical Publications; 2008. [Google Scholar]
- Ben-Amor K, Heilig H, Smidt H, Vaughan EE, Abee T, de Vos WM. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl Environ Microbiol. 2005;71:4679–4689. doi: 10.1128/AEM.71.8.4679-4689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaut M. Ecology and physiology of the intestinal tract. In: Compans RW, Cooper MD, Honjo T, Koprowski H, Melchers F, Oldstone MBA, Vogt PK, Gleba YY, Malissen B, Aktories K, editors. Curr Top Microbiol Immunol. Berlin Heidelberg: Springer-Verlag; 2011. [Google Scholar]
- Bouvier T, del Giorgio PA, Gasol JM. A comparative study of the cytometric characteristics of High and Low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ Microbiol. 2007;9:2050–2066. doi: 10.1111/j.1462-2920.2007.01321.x. [DOI] [PubMed] [Google Scholar]
- Bouvier T, Maurice CF. A single-cell analysis of virioplankton adsorption, infection, and intracellular abundance in different bacterioplankton physiologic categories. Microb Ecol. 2011;62:669–678. doi: 10.1007/s00248-011-9862-3. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey HM, Winson MK. Using flow cytometry to quantify microbial heterogeneity. Curr Issues Mol Biol. 2003;5:9–15. [PubMed] [Google Scholar]
- Del Giorgio PA, Gasol JM. Physiological structure and single-cell activity in marine bacterioplankton. In: Kirchman DL, editor. Microbial ecology of the oceans. Wiley-Blackwell; 2008. pp. 243–285. [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasol JM, Zweifel UL, Peters F, Fuhrman JA, Hagstrom A. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl Environ Microbiol. 1999;65:4475–4483. doi: 10.1128/aem.65.10.4475-4483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes MJ, Durban A, Pignatelli M, Abellan JJ, Jimenez-Hernandez N, Perez-Cobas AE, Latorre A, Moya A. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillebault D, Laghdass M, Catala P, Obernosterer I, Lebaron P. Improved method for bacterial cell capture after flow cytometry cell sorting. Appl Environ Microbiol. 2010;76:7352–7355. doi: 10.1128/AEM.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Turnbaugh PJ. Is it time for a metagenomic basis of therapeutics? Science. 2012;336:1253–1255. doi: 10.1126/science.1224396. [DOI] [PubMed] [Google Scholar]
- Jellett JF, Li WKW, Dickie PM, Boraie A, Kepkay PE. Metabolic activity of bacterioplankton communities assessed by flow cytometry and single carbon substrate utilization. Mar Ecol-Prog Ser. 1996;136:213–225. [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- Jochem FJ, Lavrentyev PJ, First MR. Growth and grazing rates of bacteria groups with different apparent DNA content in the Gulf of Mexico. Mar Biol. 2004;145:1213–1225. [Google Scholar]
- Joux F, Lebaron P. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microb Infect. 2000;2:1523–1535. doi: 10.1016/s1286-4579(00)01307-1. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Kristiansson E, Hugenholtz P, Dalevi D. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics. 2009;25:2737–2738. doi: 10.1093/bioinformatics/btp508. [DOI] [PubMed] [Google Scholar]
- Le Blay G, Lacroix C, Zihler A, Fliss I. In vitro inhibition activity of nisin A, nisin Z, pediocin PA-1 and antibiotics against common intestinal bacteria. Lett Appl Microbiol. 2007;45:252–257. doi: 10.1111/j.1472-765X.2007.02178.x. [DOI] [PubMed] [Google Scholar]
- Lee HH, Collins JJ. Microbial environments confound antibiotic efficacy. Nat Chem Biol. 2012;8:6–9. doi: 10.1038/nchembio.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nat Rev Microbiol. 2006;4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- Lindenbaum J, Rund DG, Butler VPJ, Tse-Eng D, Saha JR. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. New Engl J Med. 1981;305:789–794. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- Magnusson JO, Bergdahl B, Bogentoft C, Johnsson UE. Metabolism of digoxin and absorption site. Br J Clin Pharmacol. 1982;14:284–285. doi: 10.1111/j.1365-2125.1982.tb01976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JL, Rojo F. Metabolic regulation of antibiotic resistance. FEMS Microbiol Rev. 2011;35:768–789. doi: 10.1111/j.1574-6976.2011.00282.x. [DOI] [PubMed] [Google Scholar]
- McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer FC, Mason DJ, Gant VA. Flow cytometric monitoring of antibiotic-induced injury in Escherichia coli using cell-impermeant fluorescent probes. Antimicrob Agents Chemother. 2000;44:676–681. doi: 10.1128/aac.44.3.676-681.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo DJ, Perlmutter NG, Hunt RH, Shapiro HM. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob Agents Chemother. 2000;44:827–834. doi: 10.1128/aac.44.4.827-834.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris-Bondia F, Latorre A, Artacho A, Moya A, GDA The active human gut microbiota differs from the total microbiota. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0022448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaflaub J, Dubach UC. On the pharmacokinetics of phenacetin in man. Eur J Clin Pharmacol. 1975;8:261–265. doi: 10.1007/BF00567125. [DOI] [PubMed] [Google Scholar]
- Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC. Detecting novel associations in large data sets. Science. 2011;334:1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem. 2010;285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro HH. Microbial analysis at the single-cell level: tasks and techniques. J Microbiol Methods. 2000;42:3–16. doi: 10.1016/s0167-7012(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Smith GE, Griffiths LA. Metabolism of n-acetylated and o-alkylated drugs by the intestinal microflora during anaerobic incubation in vitro. Xenobiotica. 1974;4:477–487. doi: 10.3109/00498257409052100. [DOI] [PubMed] [Google Scholar]
- Somogyi AA, Bochner F, Hetzel D, Williams DB. Evaluation of the intestinal absorption of erythromycin in man: absolute bioavailability and comparison with enteric coated erythromycin. Pharm Res. 1995;12:149–154. doi: 10.1023/a:1016215510223. [DOI] [PubMed] [Google Scholar]
- Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008;363:1–25. doi: 10.1016/j.ijpharm.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009a;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009b;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hammes F, Boon N, Chami M, Egli T. Isolation and characterization of low nucleic acid (LNA)-content bacteria. ISME J. 2009:889–902. doi: 10.1038/ismej.2009.46. [DOI] [PubMed] [Google Scholar]
- Wickens HJ, Pinney RJ, Mason DJ, Gant VA. Flow cytometric investigation of filamentation, membrane patency, and membrane potential in Escherichia coli following ciprofloxacin exposure. Antimicrob Agents Chemother. 2000;44:682–687. doi: 10.1128/aac.44.3.682-687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubkov MV, Allen JI, Fuchs BM. Coexistence of dominant groups in marine bacterioplankton community - a combination of experimental and modelling approaches. J Mar Biol Assoc UK. 2004;84:519–529. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.