Abstract
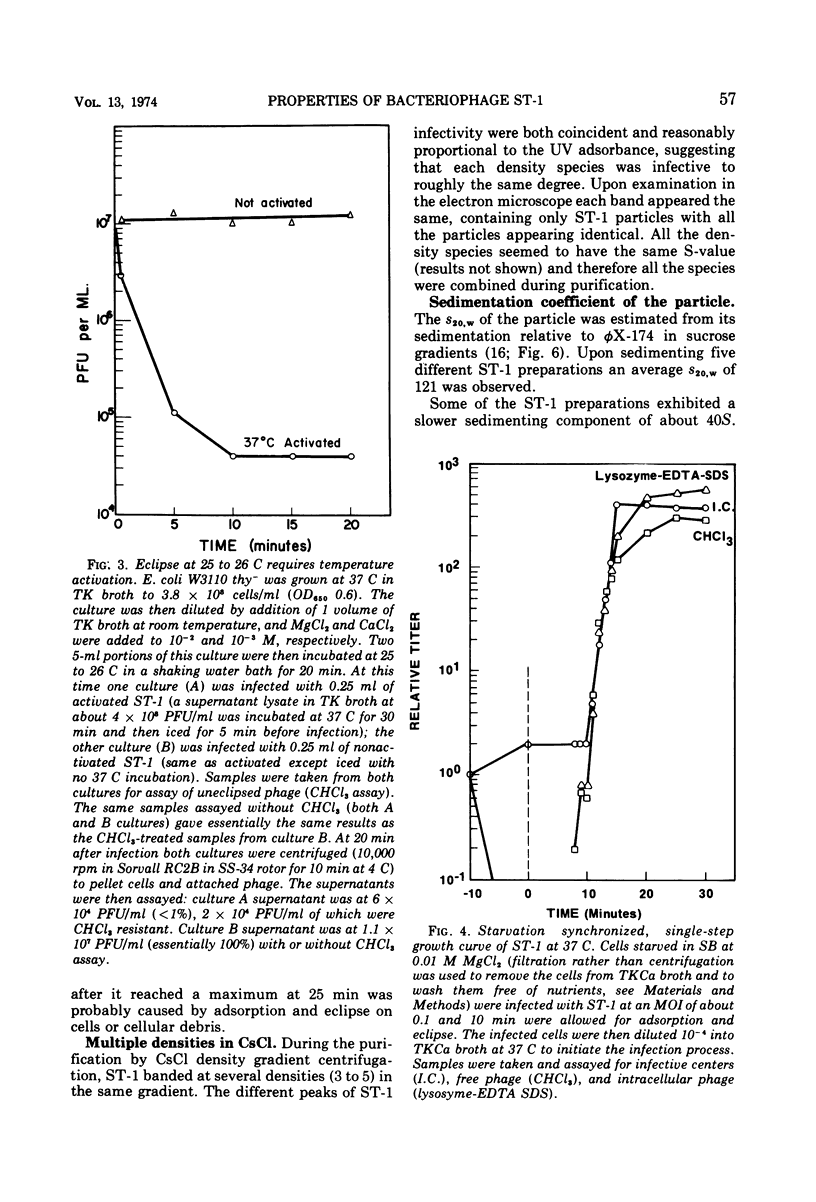
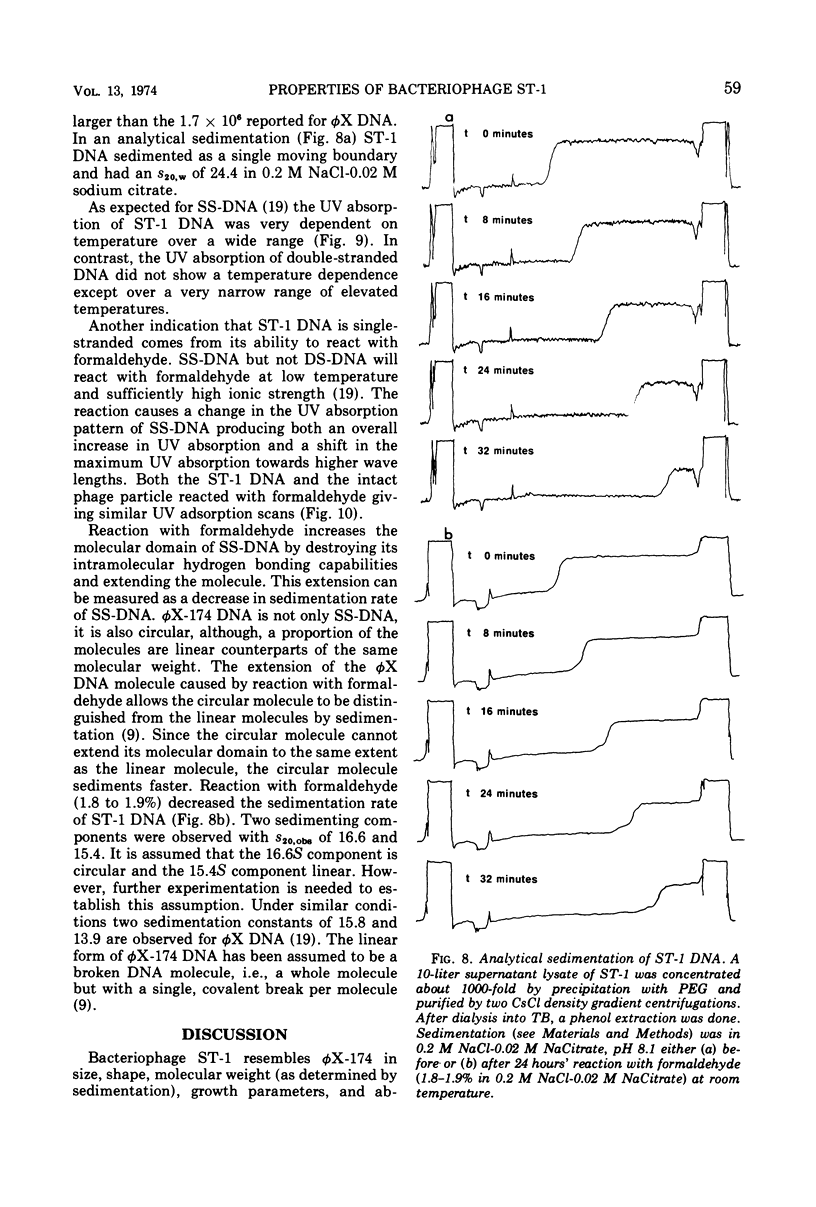
Bacteriophage ST-1 is shown to be a small, isometric, single-stranded deoxyribonucleic acid (SS-DNA) virus with a diameter of about 260 nm. Standard methods for growth, assay, preparation of high-titer lysates, and purification of the phage are suggested. ST-1 infects K-12 and not C strains of Escherichia coli and requires a divalent cation to adsorb to susceptible bacteria. Adsorption also requires an activation of the particle brought on by incubation at 37 C. The latent and eclipse periods are essentially identical (9 to 11 min) in ST-1 infections, with an average burst size about 250 phages per cell. Multiple densities of ST-1 infectivity are observed during purification in CsCl gradients. The virus recovered from different densities has the same sedimentation coefficient and, therefore, all phage containing fractions are pooled during purification. The purified ST-1 particle has a sedimentation coefficient of 121S relative to φX-174 (114S) in a sucrose gradient and a molecular weight of 6.8 × 106 (as estimated from its relative sedimentation). The nucleic acid is assumed to be SS-DNA on the basis of (i) the specific incorporation of 3H-thymine, (ii) the dependence of its UV absorption on temperature, and (iii) its reaction with formaldehyde. ST-1 SS-DNA sediments at 24.4S relative to φX-174 SS-DNA (23.8S).
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRADLEY D. E. THE STRUCTURE OF SOME BACTERIOPHAGES ASSOCIATED WITH MALE STRAINS OF ESCHERICHIA COLI. J Gen Microbiol. 1964 Jun;35:471–482. doi: 10.1099/00221287-35-3-471. [DOI] [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichrodt J. F., Berends-Van Abkoude E. R. Bacteriophages related to phi X174 showing a transition between two forms with different heat sensitivity and adsorp- tion behavior. Virology. 1968 Feb;34(2):366–370. doi: 10.1016/0042-6822(68)90251-1. [DOI] [PubMed] [Google Scholar]
- Bleichrodt J. F., van Abkoude E. R. The transition between two forms of bacteriophage phi-X174 differing in heat sensitivity and adsorption characteristics. Virology. 1967 May;32(1):93–102. doi: 10.1016/0042-6822(67)90256-5. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. A comparative study of some properties of the phi-X174 type bacteriophages. Can J Microbiol. 1970 Oct;16(10):965–971. doi: 10.1139/m70-165. [DOI] [PubMed] [Google Scholar]
- Brown L. R., Dowell C. E. Replication of coliphage M-13. II. Intracellular deoxyribonucleic acid forms associated with M-13 infection of mitomycin C-treated cells. J Virol. 1968 Nov;2(11):1296–1307. doi: 10.1128/jvi.2.11.1296-1307.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIERS W., SINSHEIMER R. L. The structure of the DNA of bacteriophage phi-X174. III. Ultracentrifugal evidence for a ring structure. J Mol Biol. 1962 Oct;5:424–434. doi: 10.1016/s0022-2836(62)80031-x. [DOI] [PubMed] [Google Scholar]
- Gefter M. L., Hirota Y., Kornberg T., Wechsler J. A., Barnoux C. Analysis of DNA polymerases II and 3 in mutants of Escherichia coli thermosensitive for DNA synthesis. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3150–3153. doi: 10.1073/pnas.68.12.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann-Berling H., Kaerner H. C., Knippers R. Small bacteriophages. Adv Virus Res. 1966;12:329–370. doi: 10.1016/s0065-3527(08)60852-0. [DOI] [PubMed] [Google Scholar]
- Holt S. C., Trüper H. G., Takács B. J. Fine structure of Ectothiorhodospira mobilis strain 8113 thylakoids: chemical fixation and freeze-etching studies. Arch Mikrobiol. 1968;62(2):111–128. doi: 10.1007/BF00410398. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Sinsheimer R. L. The process of infection with bacteriophage phi-X174. X. Mutations in a phi-X Lysis gene. J Mol Biol. 1966 Jul;18(3):429–447. doi: 10.1016/s0022-2836(66)80035-9. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Sinsheimer R. L. Process of infection with bacteriophage phi-X174. XIV. Studies on macromolecular synthesis during infection with a lysis-defective mutant. J Mol Biol. 1967 Aug 28;28(1):87–94. doi: 10.1016/s0022-2836(67)80079-2. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Schekman R., Wickner W., Westergaard O., Brutlag D., Geider K., Bertsch L. L., Kornberg A. Initiation of DNA synthesis: synthesis of phiX174 replicative form requires RNA synthesis resistant to rifampicin. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2691–2695. doi: 10.1073/pnas.69.9.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J. A., Gross J. D. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol Gen Genet. 1971;113(3):273–284. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- Wickner R. B., Wright M., Wickner S., Hurwitz J. Conversion of phiX174 and fd single-stranded DNA to replicative forms in extracts of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3233–3237. doi: 10.1073/pnas.69.11.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]



