Abstract
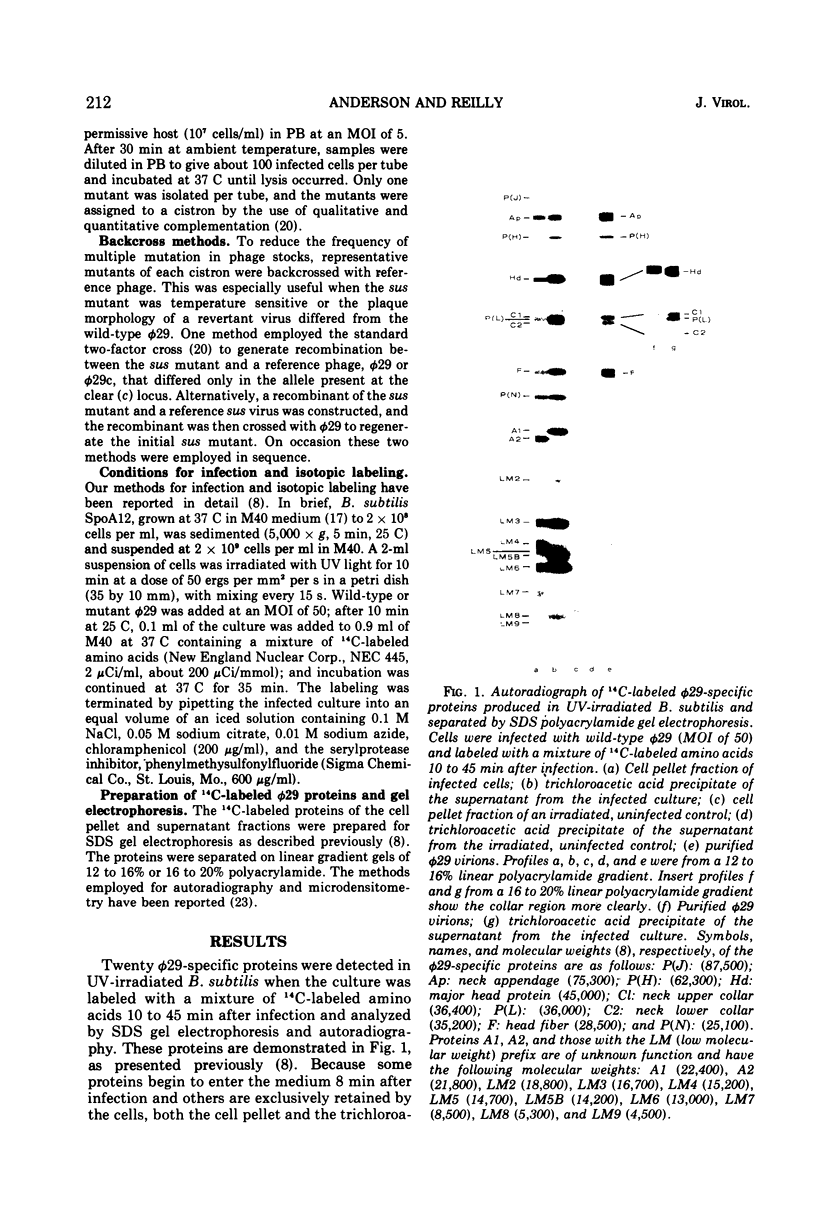
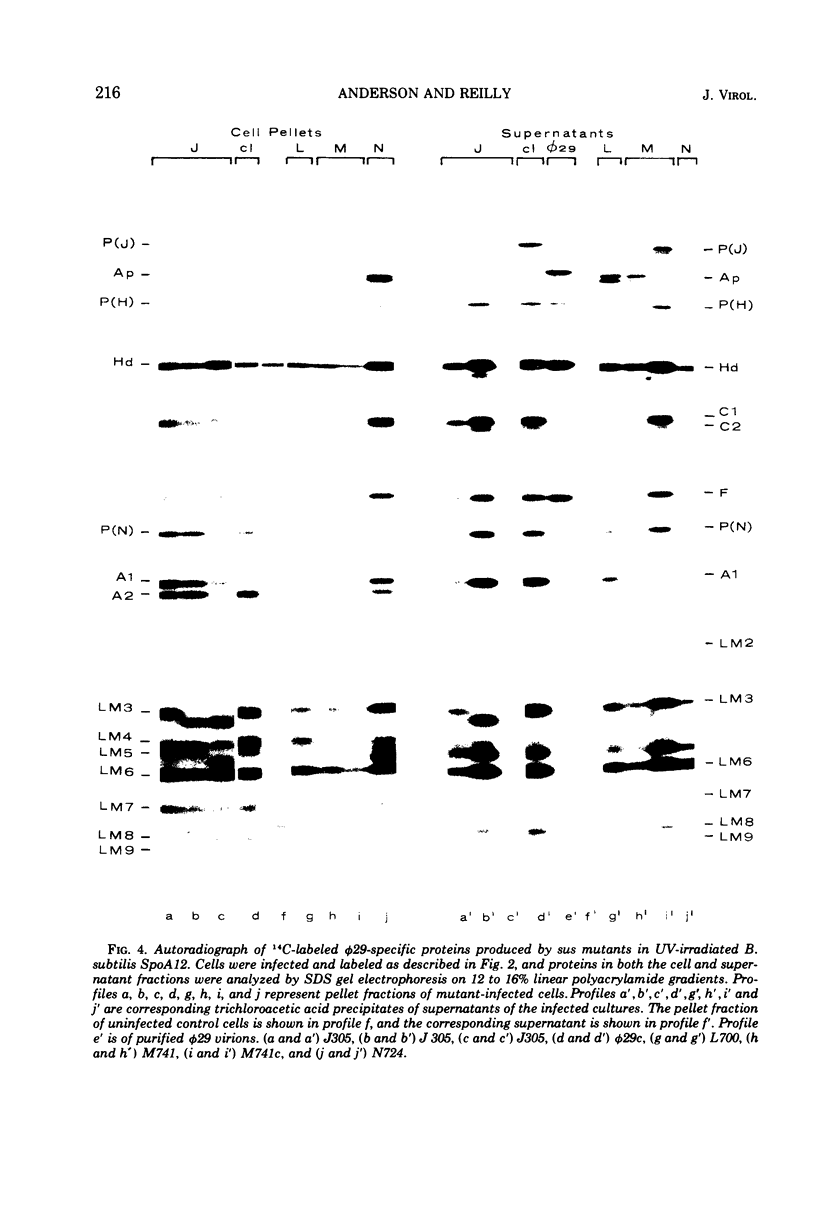
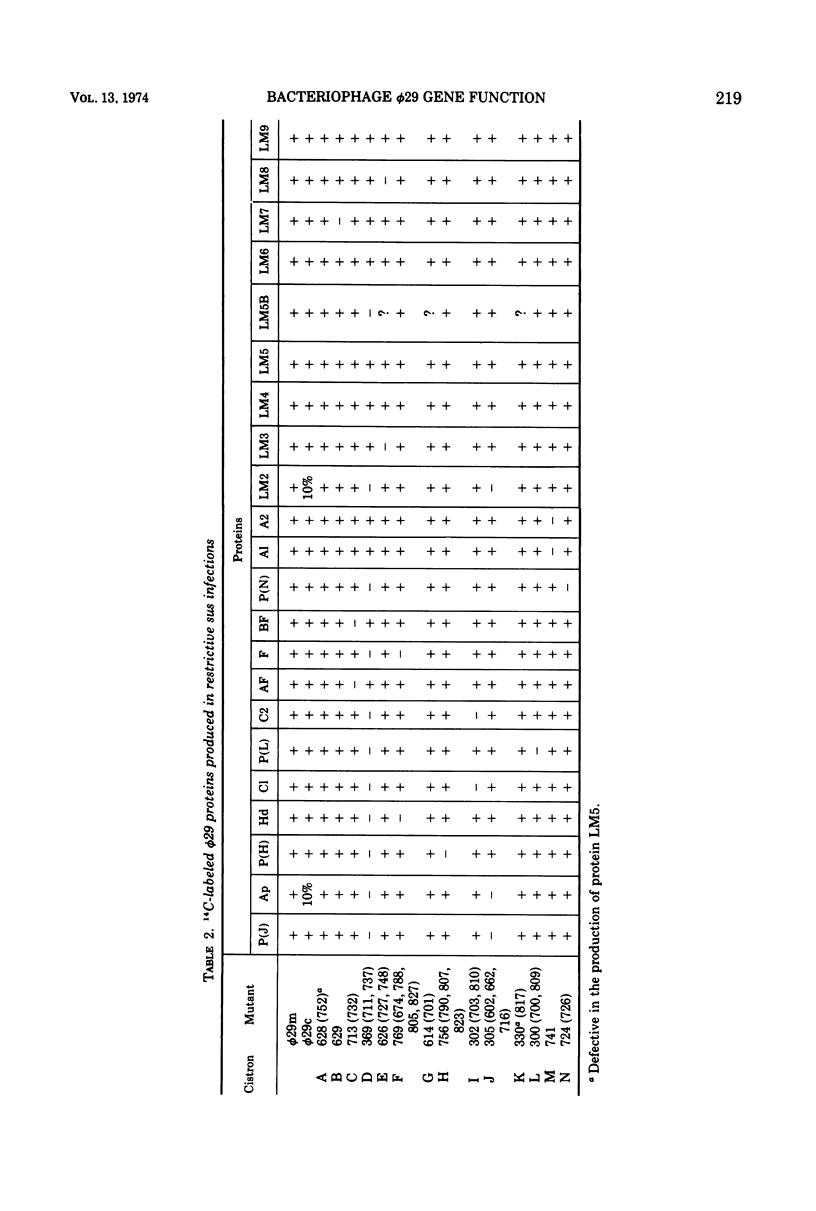
Phage φ29 suppressor-sensitive (sus) mutants of 14 cistrons have been examined for production of 14C-labeled viral-specific proteins in restrictive infections of Bacillus subtilis. Proteins specified by four cistrons (H, J, L, and N) have been resolved and identified by sodium dodecyl sulfate gel electrophoresis and autoradiography, and fragments of the normal polypeptides were detected. Mutants of six cistrons (C, D, E, F, I, and M) demonstrated two or more missing bands in the gel profiles, and thus some of these gene products may have regulatory functions. Mutation was detected in at least five genes coding for low-molecular-weight proteins, but a conditionally lethal mutant in only one of these genes has been isolated. Preliminary evidence that a precursor protein is cleaved to generate the neck appendage structural protein and a low-molecular-weight product has been obtained.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alvarez G., Salas E., Pérez N., Celis J. E. Phi29 bacteriophage structural proteins. J Gen Virol. 1972 Mar;14(3):243–250. doi: 10.1099/0022-1317-14-3-243. [DOI] [PubMed] [Google Scholar]
- Anderson D. L., Hickman D. D., Reilly B. E. Structure of Bacillus subtilis bacteriophage phi 29 and the length of phi 29 deoxyribonucleic acid. J Bacteriol. 1966 May;91(5):2081–2089. doi: 10.1128/jb.91.5.2081-2089.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. L., Mosharrafa E. T. Physical and biological properties of phage phi 29 deoxyribonucleic acid. J Virol. 1968 Oct;2(10):1185–1190. doi: 10.1128/jvi.2.10.1185-1190.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Barnes S. L., Eiserling F. A. Structural proteins of bacteriophage T4. J Mol Biol. 1970 Nov 14;53(3):461–474. doi: 10.1016/0022-2836(70)90077-x. [DOI] [PubMed] [Google Scholar]
- Dube S. K., Rudland P. S. Control of translation by T4 phage: altered binding of disfavoured messengers. Nature. 1970 May 30;226(5248):820–823. doi: 10.1038/226820a0. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Suppressor system in Bacillus subtilis 168. J Bacteriol. 1969 Mar;97(3):1397–1402. doi: 10.1128/jb.97.3.1397-1402.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley L. A., Reilly B. E., Hagen E. W., Anderson D. L. Viral protein synthesis in bacteriophage phi 29-infected Bacillus subtilis. J Virol. 1973 Nov;12(5):1149–1159. doi: 10.1128/jvi.12.5.1149-1159.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda J., Cone R. Analysis of T4 phage proteins. I. Conversion of precursor proteins into lower molecular weight peptides during normal capsid formation. Proc Natl Acad Sci U S A. 1970 Aug;66(4):1275–1281. doi: 10.1073/pnas.66.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Kornberg A., Alberts B. M. Stimulation of T4 bacteriophage DNA polymerase by the protein product of T4 gene 32. J Mol Biol. 1971 Nov 28;62(1):39–52. doi: 10.1016/0022-2836(71)90129-x. [DOI] [PubMed] [Google Scholar]
- Kellenberger E., Der Kamp C. K.-V. On a modification of the gene product P23 according to its use as subunit of either normal capsids of phage T4 or of polyheads. FEBS Lett. 1970 Jun 1;8(3):140–144. doi: 10.1016/0014-5793(70)80247-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Loskutoff D. J., Pène J. J., Andrews D. P. Gene expression during the development of Bacillus subtilis bacteriophage phi 29. I. Analysis of viral-specific transcription by deoxyribonucleic acid-ribonucleic acid competition hybridization. J Virol. 1973 Jan;11(1):78–86. doi: 10.1128/jvi.11.1.78-86.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loskutoff D. J., Pène J. J. Gene expression during the development of Bacillus subtilis bacteriophage phi29. II. Resolution of viral-specific ribonucleic acid molecules. J Virol. 1973 Jan;11(1):87–97. doi: 10.1128/jvi.11.1.87-97.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosharrafa E. T., Schachtele C. F., Reilly B. E., Anderson D. L. Complementary Strands of Bacteriophage phi29 Deoxyribonucleic Acid: Preparative Separation and Transcription Studies. J Virol. 1970 Dec;6(6):855–864. doi: 10.1128/jvi.6.6.855-864.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez E., Ramírez G., Salas M., Viñuela E. Structural proteins of bacteriophage phi 29. Virology. 1971 Sep;45(3):567–576. doi: 10.1016/0042-6822(71)90172-3. [DOI] [PubMed] [Google Scholar]
- Polsinelli M., Beretta M. Genetic Recombination in Crosses Between Streptomyces aureofaciens and Streptomyces rimosus. J Bacteriol. 1966 Jan;91(1):63–68. doi: 10.1128/jb.91.1.63-68.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REILLY B. E., SPIZIZEN J. BACTERIOPHAGE DEOXYRIBONUCLEATE INFECTION OF COMPETENT BACILLUS SUBTILIS. J Bacteriol. 1965 Mar;89:782–790. doi: 10.1128/jb.89.3.782-790.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez G., Méndez E., Salas M., Viñuela E. Head-neck connecting protein in phage phi29. Virology. 1972 Apr;48(1):263–265. doi: 10.1016/0042-6822(72)90134-1. [DOI] [PubMed] [Google Scholar]
- Reilly B. E., Zeece V. M., Anderson D. L. Genetic study of suppressor-sensitive mutants of the Bacillus subtilis bacteriophage phi 29. J Virol. 1973 May;11(5):756–760. doi: 10.1128/jvi.11.5.756-760.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARABHAI A. S., STRETTON A. O., BRENNER S., BOLLE A. CO-LINEARITY OF THE GENE WITH THE POLYPEPTIDE CHAIN. Nature. 1964 Jan 4;201:13–17. doi: 10.1038/201013a0. [DOI] [PubMed] [Google Scholar]
- Schachtele C. F., De Sain C. V., Anderson D. L. Transcription during the development of bacteriophage phi29: definition of "early" and "late" phi29 ribonucleic acid. J Virol. 1973 Jan;11(1):9–16. doi: 10.1128/jvi.11.1.9-16.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtele C. F., De Sain C. V., Hawley L. A., Anderson D. L. Transcription during the development of bacteriophage phi 29: production of host- and phi 29-specific ribonucleic acid. J Virol. 1972 Dec;10(6):1170–1178. doi: 10.1128/jvi.10.6.1170-1178.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtele C. F., Reilly B. E., De Sain C. V., Whittington M. O., Anderson D. L. Selective replication of bacteriophage phi29 deoxyribonucleic acid in 6-(p-hydroxyphenylazo)-uracil-treated Bacillus subtilis. J Virol. 1973 Jan;11(1):153–155. doi: 10.1128/jvi.11.1.153-155.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A. New small polypeptides associated with DNA-dependent RNA polymerase of Escherichia coli after infection with bacteriophage T4. Proc Natl Acad Sci U S A. 1972 Mar;69(3):603–607. doi: 10.1073/pnas.69.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera A., Salas M., Viñuela E. Temperature-sensitive mutants affected in DNA synthesis in phage phi29 of Bacillus subtilis. Eur J Biochem. 1972 Dec 4;31(2):367–371. doi: 10.1111/j.1432-1033.1972.tb02542.x. [DOI] [PubMed] [Google Scholar]
- Tosi M., Anderson D. L. Antigenic properties of bacteriophage phi 29 structural proteins. J Virol. 1973 Dec;12(6):1548–1559. doi: 10.1128/jvi.12.6.1548-1559.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]