Abstract
In eukaryotic cells a molecular chaperone network associates with translating ribosomes, assisting the maturation of emerging nascent polypeptides. Hsp70 is perhaps the major eukaryotic ribosome-associated chaperone and the first reported to bind cotranslationally to nascent chains. However, little is known about the underlying principles, and function of this interaction. Here, we use a sensitive and global approach to define the cotranslational substrate specificity of the yeast Hsp70 SSB. We find that SSB binds to a subset of nascent polypeptides whose intrinsic properties and slow translation rates hinder efficient cotranslational folding. The SSB-ribosome cycle and substrate recognition is modulated by its ribosome-bound co-chaperone RAC. Deletion of SSB leads to widespread aggregation of newly synthesized polypeptides. Thus, cotranslationally acting Hsp70 meets the challenge of folding the eukaryotic proteome by stabilizing its longer, more slowly translated, and aggregation prone nascent polypeptides.
INTRODUCTION
Generating and maintaining a functional proteome is a major challenge for the cell. Defective protein folding often leads to aggregation, which is deleterious for cell viability (Hartl et al., 2011; Tyedmers et al., 2010). Indeed, a growing number of diseases are associated with impaired protein homeostasis (Luheshi et al., 2008; Powers et al., 2009; Voisine et al., 2010). Accordingly, cells contain an array of molecular chaperones, which facilitate protein folding and promote quality control (Hartl et al., 2011; Kramer et al., 2009; Preissler and Deuerling, 2012; Tyedmers et al., 2010).
Folding during translation is particularly challenging for the cell. Proteins emerge vectorially from the ribosome exit tunnel and cannot fold stably until a domain is fully synthesized (Frydman, 2001). As a result, nascent polypeptide chains are highly susceptible to misfolding and aggregation, particularly given the extreme crowding of the cellular environment (Hartl et al., 2011; Kramer et al., 2009; Preissler and Deuerling, 2012).
Eukaryotic and prokaryotic cells have radically different strategies to assist folding of newly translated proteins (Albanese et al., 2006; Netzer and Hartl, 1997). In prokaryotic cells, which have a simpler proteome and cell architecture, a single ATP-independent ribosome-bound chaperone, called trigger factor associates with many nascent chains (Kramer et al., 2009, Oh et al 2011; Preissler and Deuerling, 2012). Subsequent de novo folding is mostly post-translational and carried out by the same ATP-dependent chaperones that protect the proteome from stress (Agashe et al., 2004; Hartl and Hayer-Hartl, 2009). In contrast, eukaryotes have developed a complex chaperone network that associates with translating ribosomes and facilitates de novo protein folding. These Chaperones Linked to Protein Synthesis (CLIPS) comprise several ATP-dependent and ATP-independent factors that are not induced by stress and are co-regulated with the translational apparatus (Albanese et al., 2006). This is likely due to the fact that eukaryotic proteins are translated with slower kinetics, and comprise far more complex architectures than bacterial proteomes (Koonin et al., 2000; Wang et al., 2011). In addition, the compartmentalization of eukaryotic cells necessitates that many proteins translocate cotranslationally into various organelles.
Hsp70 is the most prominent ribosome-associated chaperone in eukaryotic cells (Frydman, 2001; Kramer et al., 2009). Although it was the first chaperone shown to bind to nascent polypeptides in eukaryotes (Beckmann et al., 1990), its specificity and function in cotranslational protein homeostasis remains undefined. Experiments with model substrates indicate that Hsp70 can bind to nascent chains early in translation and facilitate their folding, translocation, or association with downstream chaperones (Frydman and Hartl, 1996; Frydman et al., 1994; Hartl and Hayer-Hartl, 2009).
Hsp70s are highly conserved and ubiquitous ATP-dependent chaperones implicated in various key aspects of protein homeostasis (Kampinga and Craig, 2010). In yeast, the cytosolic Hsp70s, Ssa1-4 and Ssb1-2 execute distinct and non-overlapping functions (Peisker et al., 2010). The closely related isoforms Ssb1 and Ssb2 (herein termed SSB) associate with translating ribosomes and cotranslationally bind nascent chains, thus making SSB a paradigm to study cotranslationally acting Hsp70s. Like other Hsp70s, the ATPase of SSB is regulated by a J-domain protein, Zuo1, which activates ATP hydrolysis, and by a nucleotide exchange factor, the Hsp110 Sse1. Zuo1 together with the atypical Hsp70 Ssz1 form the ribosome-associated complex RAC (Peisker et al., 2010; Preissler and Deuerling, 2012). Homologs of RAC and Sse1 exist in mammals, indicating a conserved regulation of cotranslational Hsp70s across eukaryotes. Like other cotranslationally acting Hsp70s (Thulasiraman et al., 1999), SSB interacts with many newly synthesized polypeptides in a transient manner (Yam et al., 2005). Cells lacking SSB are viable but exhibit a number of phenotypes such as defects in ribosome biogenesis (Albanese et al., 2010; Koplin et al., 2010) and cellular signaling (von Plehwe et al., 2009). However, its physiological cotranslational substrates are unknown.
An important unanswered question is whether cotranslationally acting Hsp70s fulfill a general role in nascent chain protein homeostasis or a specialized function for a subset of nascent chains. In principle, a general association with most nascent polypeptides is possible, since Hsp70s are thought to recognize linear stretches of hydrophobic amino acids that occur in almost all unfolded protein sequences (Flynn et al., 1991; Rüdiger et al., 1997). Here, we define the function and global specificity of the ribosome-associated Hsp70 SSB and show that it is regulated by its co-factor RAC. A systems analysis of SSB-nascent chain interactions reveals a broad range of physiological substrates. Analysis of the fundamental properties distinguishing cotranslational SSB substrates suggests a preferential association with longer nascent polypeptides enriched in aggregation prone, hydrophobic, and intrinsically disordered regions. Since these properties hinder efficient cotranslational folding, we propose that ribosome-associated Hsp70 evolved to meet the unique challenges of folding the eukaryotic proteome. Indeed, loss of SSB leads to widespread aggregation of newly made polypeptides highlighting the critical role of Hsp70s preventing cotranslational misfolding and downstream aggregation.
RESULTS
The ribosome-associated Hsp70 SSB interacts co- and post-translationally with nascent polypeptides
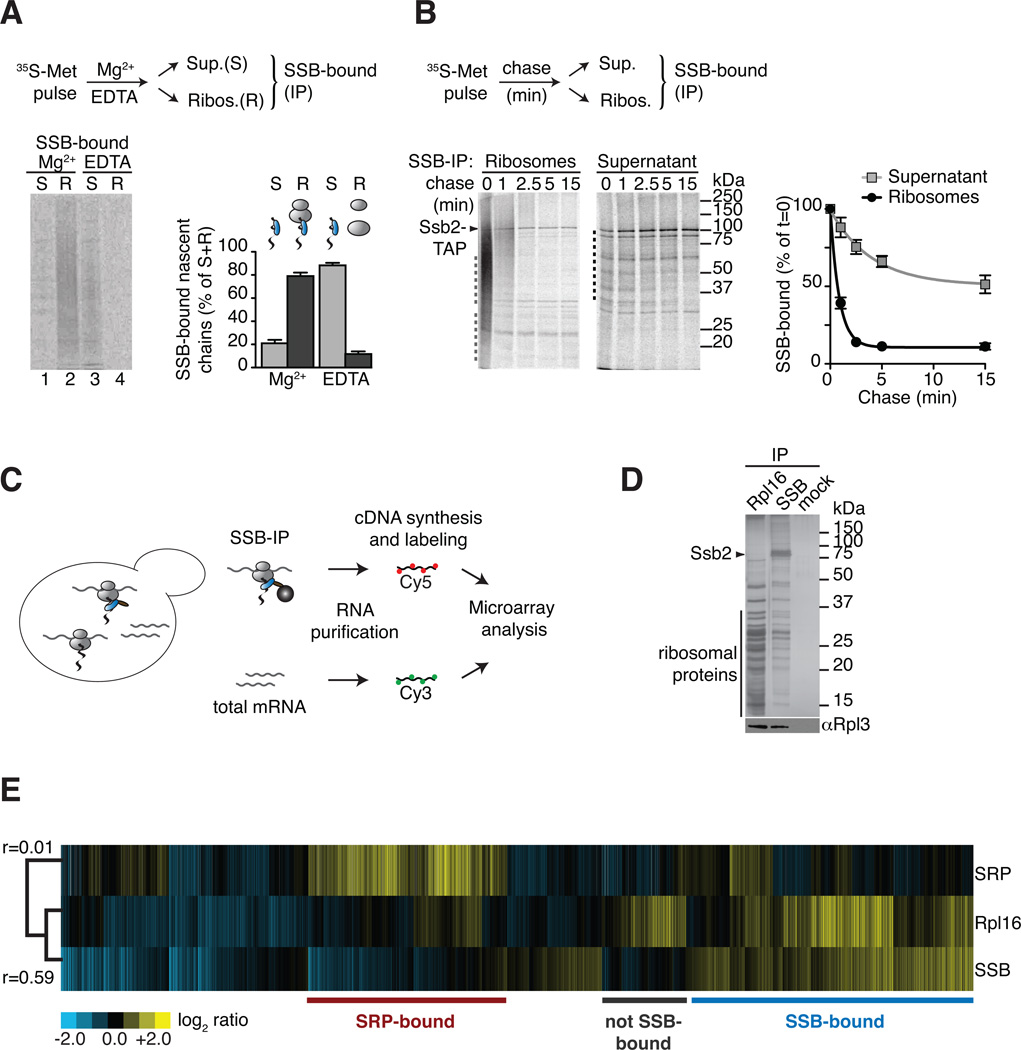
To verify that SSB directly associates with nascent chains, ribosomes with bound 35S-nascent chains (RNC) were generated by pulse-labelling with 35S-methionine (35S-Met) and isolated by sedimentation through a sucrose cushion that separates them from the post-ribosomal supernatant. Alternatively, RNCs were dissociated with EDTA prior to fractionation, releasing the nascent chains into the supernatant fraction. SSB-association to ribosome-bound and to the released nascent chains was assessed by immunoprecipitation (IP)(Figure 1A; Figure S1A). As expected, SSB associated with intact RNCs, as indicated by the smeared distribution of 35S-labeled nascent chains throughout the lane (Figure 1A, lane 2). SSB also remained associated to nascent chains after release from the ribosome into the supernatant (Figure 1A, lane 3). Thus, the interaction between SSB and nascent chains persists even after release from the ribosome.
Figure 1. Global identification of physiological SSB-associated nascent polypeptides.
(A) SSB directly binds nascent polypeptides. Nascent polypeptides were pulse labeled with 35S-methionine (35S-Met), ribosome-nascent chain complexes (RNC) were either stabilized (Mg2+) or dissociated using 25 mM EDTA (EDTA) and fractionated by centrifugation in ribosome-bound (R) and soluble (S). SSB-nascent chain interactions in each fraction were determined after Ssb2-TAP immunopurification (IP), SDS-PAGE and autoradiography, followed by quantification of SSB-bound labeled nascent chains (mean ± SEM, n=3). (B) Kinetics of newly translated polypeptide flux through SSB. Nascent polypeptides were 35S-labeled and chased with cold methionine, samples at indicated time points were processed as in (A). Quantification of SSB-bound polypeptides in ribosome and soluble fractions reflects their co-/post-translational flux through SSB (mean ± SEM, n=4). For totals of (A) and (B) see Figure S1. (C) Scheme: Global identification of cotranslational SSB substrates. SSB-bound RNCs and total mRNAs were isolated, reverse transcribed, and labeled for subsequent microarray analysis. (D) Top: SDS-PAGE and silver staining of IPs of TAP-tagged Rpl16, Ssb2 and untagged cells (mock). Bottom: Immunoblot of ribosomal Rpl3. Ssb2-TAP IP depleted all Ssb2 from the lysate (see Figure S1D). (E) Translation profile of SRP-bound and SSB-bound mRNAs compared to the total Translatome identified by ribosome isolation (Rpl16). Hierarchically clustered heat map showis the average values of three individual experiments in rows; columns represent genes. mRNAs enriched over total RNAs in yellow, mRNAs depleted in blue. Pearson correlation coefficients are shown next to the tree. Comparison with the Translatome shows that some RNCs are preferentially enriched for SSB binding (blue) others are enriched in SRP binding (red). See also Figure S1.
Next, the co- and post-translational flux of newly made nascent polypeptides through SSB was assessed by pulse chase labeling followed by immunoprecipitation of SSB-bound 35S-newly made polypeptides (SSB IP). A large spectrum of ribosome-bound 35S-nascent chains was SSB-bound immediately after biosynthesis (Figure 1B, left panel, t=0) and dissociated with fast kinetics that mirror the rates of elongation; indeed most ribosome-bound nascent chains dissociated from SSB after 2.5 minutes chase (Figure 1B, left panel). Analysis of the post-ribosomal supernatant indicates that a fraction of full-length proteins remains associated post-translationally with SSB (Figure 1B, right panel). Full-length proteins were released from SSB with different kinetics, which may reflect their distinct folding rates (Figure 1B, graph). These results indicate that SSB associates with nascent polypeptides co- and post-translationally; most newly made proteins are released rapidly after synthesis but a set of polypeptides remain guided by SSB over a more prolonged time course.
Global identification of physiological cotranslational substrates of SSB
We next identified cotranslational SSB substrates using a previously described global approach (del Alamo et al., 2011). Briefly, isolation of cotranslationally bound SSB-RNC complexes allows us to identify the chaperone-bound nascent polypeptides through analysis of the mRNAs encoding the substrates (Figure 1C). Direct isolation of ribosomes via the tagged ribosomal protein Rpl16 allowed us to determine the total translational profile in these cells (herein “Translatome”). SDS-PAGE analysis confirmed both SSB and Rpl16 IPs specifically isolated translating ribosomes, as indicated by the characteristic pattern of associated ribosomal proteins, by immunoblotting for the ribosomal protein Rpl3 (Figure 1D) and by the detection of associated mRNA (Figure S1E). Thus, the isolation procedure recovered ribosome-associated nascent chain complexes with SSB, allowing the global identification of its cotranslational nascent substrates. RNC complexes bound to the ER targeting Signal Recognition Particle (SRP) were also analyzed as a specificity control.
SSB-associated nascent chains were identified through their mRNAs using three independent experiments carried out as biological replicates. The experiments were highly reproducible, as underscored by their high correlation coefficient (r=0.92) following hierarchical clustering analysis (Figure S1F). Comparing the SSB-bound dataset to both the Translatome and the cotranslational interactome of SRP revealed clear differences in specificity between SSB and SRP as well as differences between the SSB-interactome and the Translatome (Figure 1E). Our data indicate that SSB does not bind to every nascent chain complex, allowing us to define “SSB-bound” and “not SSB-bound” datasets (Figure 1E; see also Figure 4A). Hierarchical clustering and statistical analyses identified 1,990 mRNAs enriched in the SSB dataset (herein “SSB-bound”)(Table S1), representing 65% of all actively translated proteins.
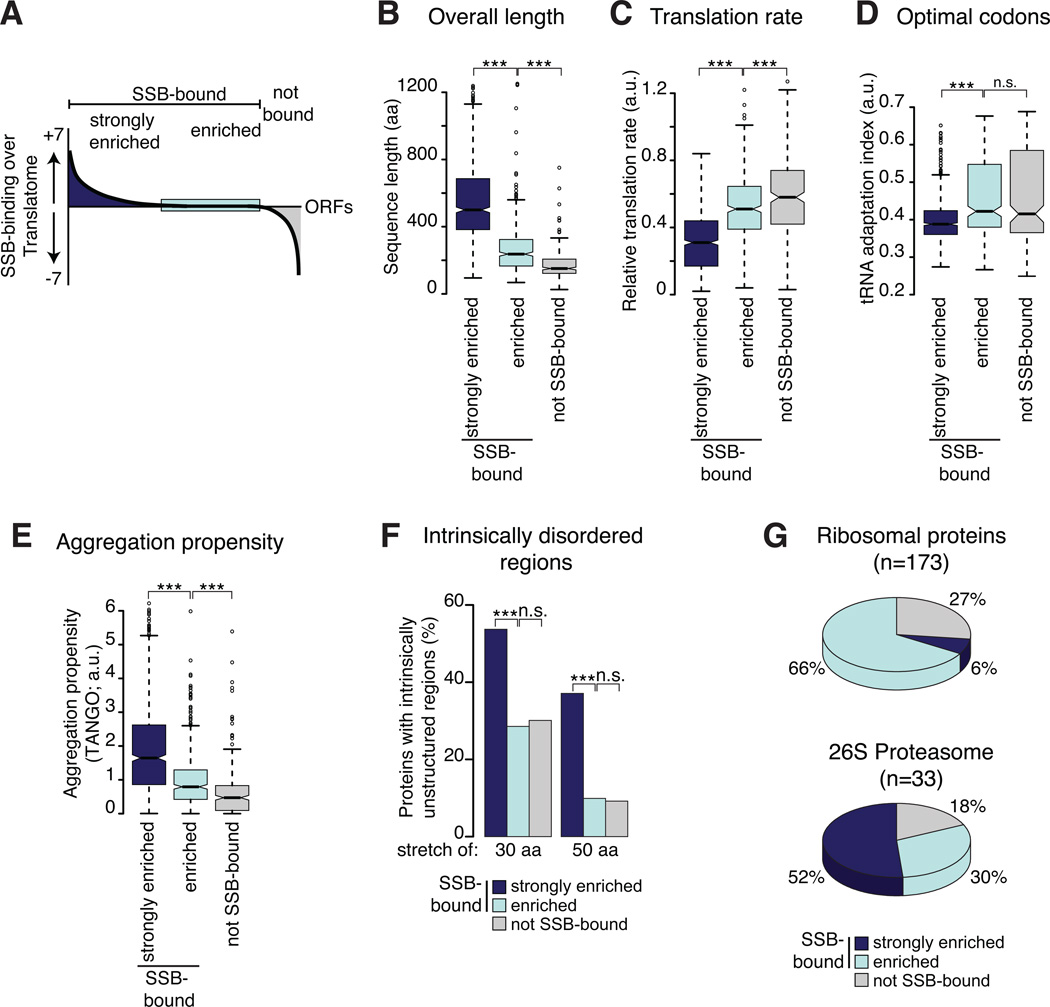
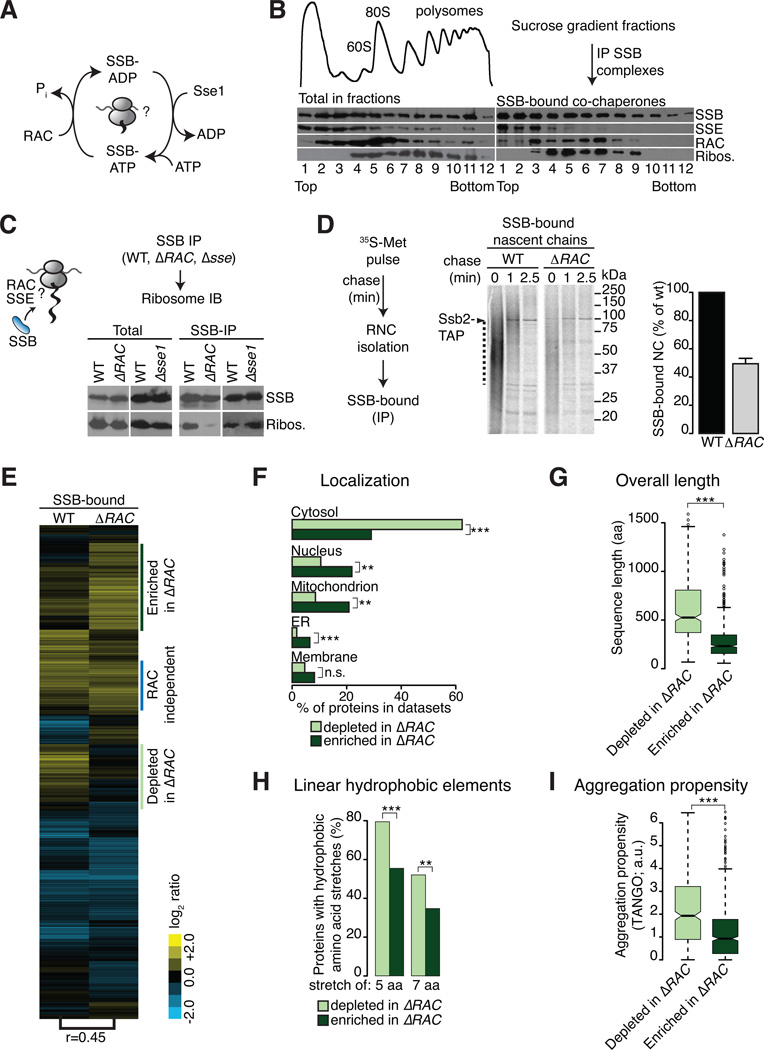
Figure 4. Nascent chain properties modulate the strength of SSB association.
(A) The degree of enrichment of SSB substrates over the Translatome was determined by statistical analysis comparing the enrichment of each mRNA in the RNC of SSB-bound to the Translatome. mRNAs with positive enrichment scores were termed “strongly enriched” (dark blue), mRNAs with negative scores were termed “not SSB-bound” (grey), and equally enriched mRNAs in both dataset were classified “enriched” (light blue). (B)– (F) SSB substrate properties were compared to those of the “not SSB-bound” set. The Translatome serves as reference. As in Figure 3, only protein properties of cytosolic and nuclear proteins are shown. (G) Small and rapidly translated subunits of abundant complexes contain SSB-bound, enriched and not bound subunits. *p ≤ 0.01; **p≤ 10−4; ***p ≤10−10, n.s. not significant. See also Figure S4.
The selective association of SSB with newly made nascent polypeptides
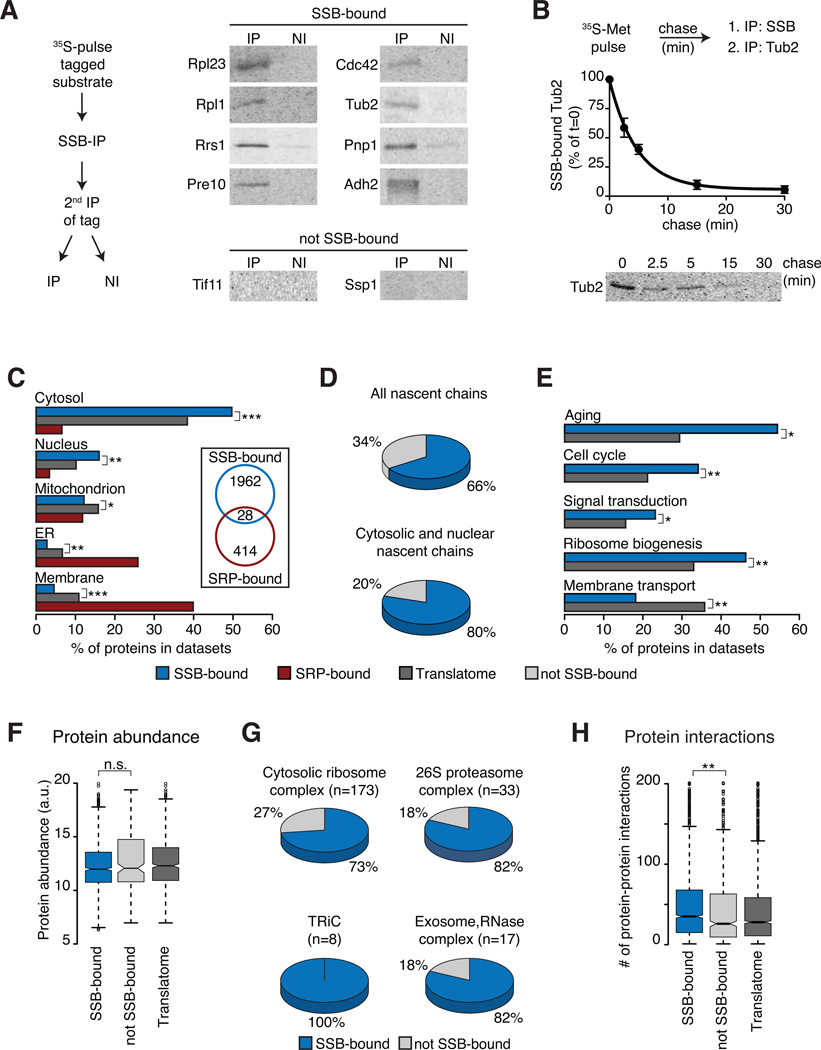
To directly confirm the specific interaction of SSB with its identified substrates, both highly and moderately translated candidates were selected from the “SSB-bound“ and “not SSB-bound“ categories (Figure S2A). Proteins were expressed carrying N-terminal epitope tags and their de novo interaction with SSB was determined by IP following a 2 min 35S-Met pulse (Figure 2A and Figure S2A). An initial non-denaturing SSB IP revealed many labeled nascent polypeptides associated with SSB (Figure S2A). A second IP against the N-terminal tag examined the binding of the specific 35S-labeled candidate protein to SSB (Figure 2A). While all the substrates selected from the “SSB-bound” set were indeed associated with SSB, polypeptides corresponding to “not SSB-bound” mRNAs did not associate with SSB (Figure 2A and Figure S2A). Pulse-chase analysis together with sequential IP further demonstrated that individual substrates flux through SSB following translation (Figure 2B for tubulin, see also below Figure 3H) as expected for transient nature of chaperone-substrates complexes. These analyses indicate that SSB directly binds the substrates identified in our global approach.
Figure 2. Selectivity of SSB for specific nascent polypeptides.
(A) Direct biochemical interaction of SSB with candidate substrates from Figure 1. N-terminally tagged substrates were briefly 35S-labeled, SSB-bound, labeled substrates were isolated by SSB-IP, and enrichment through a 2nd IP for the tag (IP). Non-immune controls were done in parallel (NI). Samples were analyzed by SDS-PAGE and autoradiography. (B) Flux of identified substrates through SSB. N-terminally tagged Tub2 was 35S-labeled and chased for the indicated times. Samples were processes as in (A) and quantified (mean ± SEM, n=3). (C) Subcellular localization of SSB and SRP substrates versus the Translatome is plotted as fraction of the datasets (%). Insert highlights low overlap between SSB and SRP substrates. (D) SSB substrates within the Translatome (top) and fractions of cytosolic and nuclear proteins. (E) SSB substrates are enriched in key cytoplasmic regulatory functions, but not in membrane proteins (Aging n=68; Cell-Cycle n=533; Sign. transd. n=224; Rib. biog. n=406; Mem. transp. n=187), plotted as fraction of the datasets (%). (F) Protein abundance of SSB-bound and not SSB-bound proteins. (G) Many subunits of large oligomeric complexes are substrates of SSB. (H) Enrichment of protein-protein interactions among SSB substrates. *p ≤ 0.01; **p≤ 10−4; ***p ≤10−10, n.s. not significant. See also Figure S2.
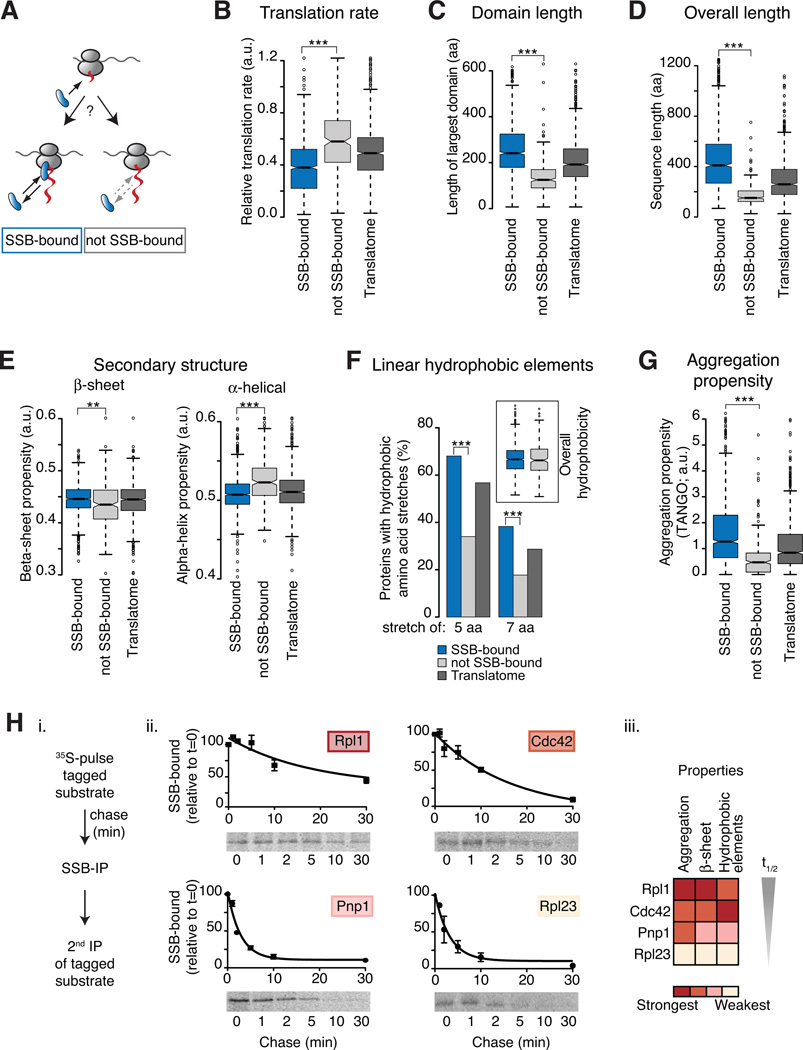
Figure 3. Underlying properties characterize SSB association with nascent polypeptides.
(A) Specific nascent chain properties determine cotranslational SSB binding or lack thereof. (B–G) Analysis of intrinsic properties of the SSB substrates compared to those in the “not SSB-bound” dataset. The Translatome serves as reference. Only protein properties of cytosolic and nuclear localized proteins are shown since they represent the majority of SSB-bound substrates and they undergo maturation in the cellular compartment where SSB is localized. However, the conclusions were generally applicable for all SSB substrates. SSB substrates were found to differ significantly from non-SSB-bound nascent polypeptides for the indicated properties (B–G). (H) Association kinetics of SSB with substrates correlates with intensities of substrate properties i. N-terminally tagged substrates were 35S-Met pulse labeled and chased for the indicated times. SSB-bound, labeled substrates were isolated by SSB-IP, and enrichment through a 2nd IP for the tag. ii. SDS-PAGE and autoradiography shows SSB-bound 35S-labeled substrates; flux through SSB was measured by quantification (mean ± SEM, n=3). iii. Heat map represents the intensities of intrinsic sequence features. *p ≤ 0.01; **p≤ 10−4; ***p ≤10−10. See also Figure S3.
Systems level analysis of SSB specificity
We next examined the overall characteristics of SSB-associated nascent polypeptides. In contrast to SRP, which binds secretory and membrane proteins, SSB preferentially binds to cytosolic and nuclear proteins (Figure 2C). The overlap between SSB-bound and SRP-bound nascent chains was negligible (Figure 2C, inset; Figure 1E). These results suggest that SSB and SRP binding to nascent polypeptides is mutually exclusive and that SSB binds to a large subset of approximately 80% of nascent chains encoding cytosolic and nuclear proteins (Figure 2D).
Gene Ontology (GO) categories revealed a selectivity of SSB for certain substrates. Cellular processes such as aging, signal transduction and ribosome biogenesis were enriched among SSB substrates, while proteins involved in membrane transport or mitochondria organization were depleted in the SSB dataset (Figure 2E and Figure S2B). SSB interactors comprised many different protein folds, although some domains were enriched as indicated by GO annotations such as ATPase activity and DNA-binding (Figure S2C).
SSB specificity was not determined by protein abundance. While abundant proteins such as metabolic enzymes (Figure S2B) do bind cotranslationally to SSB, we did not detect a strong link between SSB binding and overall protein abundance (Figure 2F). Importantly, this finding indicates that our approach was not biased towards enrichment of abundant, highly translated proteins (see also Figure 3B).
SSB substrates are enriched for subunits of oligomeric complexes and are engaged in a significantly higher number of protein-protein interactions than the Translatome (Figure 2G, 2H). For instance, all subunits of the chaperonin TRiC/CCT, the elongator complex, and most subunits of the large 26S proteasome and 40S and 60S ribosomal particles were SSB-associated (Figure 2G). This suggests a potential role of SSB in stabilizing free subunits of oligomeric complexes, which may display a higher number of exposed contact interfaces.
Cotranslational SSB binding is determined by intrinsic nascent chain properties
While every polypeptide contains at least one potential Hsp70-binding site (Flynn et al., 1991), our translation-wide analysis clearly indicates that not every polypeptide emerging from the ribosome binds to SSB. This raises the question of what determines cotranslational association with Hsp70. To test whether intrinsic properties of the translated polypeptide determine SSB-association, we compared features distinguishing the “SSB-bound” from the “not SSB-bound” dataset (Figure 3A).
Strikingly, one of the major features distinguishing SSB-bound and not SSB-bound nascent chains was a characteristic of their mRNA. Analysis of relative translation rates revealed that the majority of SSB substrates were translated with moderate or slow translation rates compared to non-SSB-bound chains or the Translatome (Figure 3B). SSB-bound nascent chains also had a lower tRNA adaptation index (tAI; Figure S3A) an orthogonal metric of translation efficiency (dos Reis et al., 2004). Analysis of the nascent chain sequence themselves also revealed intrinsic polypeptide features that promote SSB binding. SSB substrates contain longer individual domains (Figure S3B). For instance, the longest domain of each individual SSB-bound protein was on average significantly larger than the domains of nascent chains not bound by SSB (Figure 3C). Because long domains can be considered as bottlenecks for structure formation, we estimated folding rates for individual domains using parameters optimized to predict folding rates of small proteins (Ouyang and Liang, 2008). Indeed, we found that the predicted folding rates for domains of SSB substrates were lower than for domains of non-SSB substrates (Figure S3C). SSB substrates also tend to be longer (Figure 3D), and enriched in multi-domain polypeptides (Figure S3D). However, length alone is clearly not the sole determinant since most ribosomal proteins and 20S proteasome subunits also bind to SSB, despite the fact that many are below 30 kDa. Rather, our data suggest that complexity for cotranslational structural formation determines binding to SSB. Slowly translated regions, which may spend more time unfolded on the ribosome, or domains with low cotranslational folding require SSB-association for stabilization while emerging from the ribosomal tunnel.
We next examined if physicochemical properties linked to slow cotranslational folding correlate with SSB binding. Nascent chains cannot complete tertiary folding until a domain is synthesized, but can cotranslationally adopt some secondary structures. Notably, SSB-bound nascent polypeptides had a higher content of beta-sheets than the non-SSB-bound set; conversely alpha-helical propensity was reduced in the SSB substrates (Figure 3E). Intrinsically disordered regions (IDR) of 30 and 50 amino acids in length were significantly enriched among SSB-bound nascent chains (Figure S3E). Perhaps beta-sheet rich regions and IDRs delay the cotranslational formation of folded structures and thus promote SSB association. Indeed, alpha-helices can form very early during protein synthesis (Lu and Deutsch, 2005; Woolhead et al., 2004), while beta-domains are discontinuous in sequence and thus less favored to fold cotranslationally, and likely to produce aggregation-prone intermediates. These results support the idea that SSB serves to protect folding-challenged polypeptides as they emerge from the ribosome.
Hydrophobicity also exposes non-native polypeptides to misfolding and aggregation. Notable, we did not detect any correlation between the overall hydrophobicity of the full-length protein and cotranslational binding to SSB (Figure 3F inset). However, short linear hydrophobic elements were significantly enriched in SSB-bound proteins. Most (~70%) of SSB-bound substrates carried at least one, but often more, stretches of 5 or more consecutive hydrophobic amino acids. In contrast, less than 35% of the not SSB-bound proteins had such hydrophobic stretches (Figure 3F). A similar trend was also observed for longer hydrophobic linear elements of at least 7 residues. These short linear stretches of hydrophobic amino acids may provide binding sites for SSB on the nascent chains. SSB substrates are also enriched in short linear sequences with high cross-beta sheet aggregation propensity, as determined by the TANGO algorithm (Fernandez-Escamilla et al., 2004). The fact that SSB-bound nascent chains were significantly enriched in linear aggregation-prone sequences, suggests that these elements are important factors for SSB association (Figure 3G).
We next chose a representative panel of substrates with different properties and examined their de novo flux through SSB using pulse-chase experiments and sequential IP (Figure 3Hi). The SSB association kinetics diverged significantly for these substrates (Figure 3Hii), and correlated with above-determined parameters such as the propensity to aggregate, beta-sheet propensity, and number of hydrophobic elements of the substrate (Figure 3Hiii). For instance, Rpl1 and Cdc42, substrates enriched in such features, had extended SSB interaction kinetics, while Rpl23 and Pnp1, which contain weaker features, interacted more transiently with SSB (Figure 3Hii). These findings point to a direct correlation between sequence properties that characterize the susceptibility to misfolding and aggregation during translation and the length of association with SSB.
Intrinsic nascent polypeptide properties modulate the strength of SSB association
We exploited the quantitative nature of our data to distinguish between nascent polypeptides that are strongly enriched in SSB binding over the Translatome (“strongly enriched”), those unfavored for SSB binding (“not SSB-bound”), and those with similar enrichment within the SSB and the Translatome sets (“enriched”)(Figure 4A). This analysis provided further insight into the determinants conferring cotranslational Hsp70 binding. Properties that either slow cotranslational folding or render nascent chains susceptible to inappropriate interactions and misfolded states, e.g. domain length, translation rate or aggregation propensity, directly correlated with recruitment of SSB (Figure 4B–4E), suggesting that Hsp70 functions to protect vulnerable cotranslational intermediates. Similar correlations were observed for secondary structure and the presence of linear hydrophobic stretches (Figure S4B–S4D). SSB-enriched nascent chains were also distinguished by the presence of intrinsically disordered regions (Figure 4F). The strongest SSB interactors correspond to lower abundance proteins (Figure S4E), consistent with the previously noted evolutionary pressure to limit the abundance of more aggregation prone proteins (Calloni et al., 2012; Tartaglia et al., 2010). However, abundant complexes such as the ribosome and the 26S proteasome contain subunits in each of these categories (Figure 4G). Thus, the enrichment for SSB binding is not strictly determined by size or abundance but rather by the severity of the challenges hindering cotranslational folding.
Co-chaperones regulate the specificity of SSB
We analyzed whether the co-chaperones RAC and Sse1 regulate the ribosome cycle and cotranslational specificity of SSB. We first examined the association of RAC and Sse1 with SSB and ribosomes using sucrose density gradients. As reported, SSB was evenly distributed between soluble (Figure 5B, lanes 1–3) and ribosomal fractions (Figure 5B, lanes 4–12) (Albanese et al., 2006; Nelson et al., 1992). RAC and Sse1 co-migrated with polysomes, albeit with different association patterns; most of RAC was ribosome-associated while most of Sse1 was in the soluble fraction (Figure 5B). The SSB-co-chaperone interactions on and off the ribosome were examined in each fraction of the gradient by IP followed by immunoblot analysis (IB) (Figure 5B, right panel). RAC only associated with SSB in the ribosome-containing fractions suggesting that this complex regulates SSB at the ribosome (Figure 5B, right panel, lanes 3–8). In contrast, Sse1 was predominantly associated with SSB in the soluble, non-ribosome associated fractions (Figure 5B, right panel, lanes 1–3), indicating a post-translational context for the Sse1-SSB interaction.
Figure 5. A co-chaperone network regulates the cotranslational substrate cycle of SSB.
(A) Postulated function of RAC and Sse1 in SSBs nucleotide cycle. (B) Association of RAC and Sse1 with SSB on/off ribosomes. Top left: OD254 reading of polysome profiles after sucrose gradient fractionation. Bottom left: Immunoblot of ribosomes and distribution of chaperones in fractions. Right panel: SSB-IP, SDS-PAGE and immunoblot analysis from each gradient fraction examining the association of RAC and Sse1 with soluble and ribosome-bound SSB. As described, larger polysomal complexes are less stable during IP due to the very high molecular weight of the complex (Inada et al., 2002). (C) RAC but not Sse1 stimulate SSB-ribosome-association. SSB-IPs from wild type (WT), ΔRAC (Δzuo1/Δssz1) and Δsse1 cells and immunoblot for SSB-bound ribosomal protein Rpl3. Left: Totals; right: Immunoblot of IPs. For controls see Figure S5A. (D) Loss of RAC decreases the cotranslational flux of nascent polypeptides through SSB. SSB binding to RNC complexes was assessed by 35S-pulse-chase analysis as in Figure 1B (scheme left), autoradiography (middle) and quantification of SSB-bound radiolabeled nascent chains as indicated by the dotted line (right panel). Time=0 values were adjusted over Totals (Figure S5B) and plotted relative to WT (mean ± SEM, n=3). (E) RAC modulates the cotranslational specificity of SSB. Hierarchically clustered heat map of SSB-bound mRNAs in WT and ΔRAC cells (column=average of three experiments; row=single genes). Enriched SSB-bound mRNAs are in yellow, blue displays depleted mRNAs. Pearson correlation coefficients between experiments are indicated at the bottom of the tree. (F)–(I) Comparison of cotranslational substrate properties that are depleted or enriched for SSB binding in ΔRAC. Statistical analysis determined SSB-substrates less enriched or lost in ΔRAC cells (“depleted in ΔRAC”, light green) and those enriched in SSB binding in ΔRAC cells (“enriched in ΔRAC”, dark green). Importantly, enrichment or depletion for SSB-binding in ΔRAC cells was not due to up or down regulation of mRNAs on a transcriptional level. *p ≤ 0.01; **p≤ 10−4; ***p ≤10−10, n.s. not significant. See also Figure S5.
We next compared the SSB-ribosome association in wild type (WT), ΔRAC (Δssz1Δzuo1) and Δsse1 cells. SSB complexes were isolated by IP and the presence of ribosomes detected by IB. Deletion of RAC, but not Sse1, impaired SSB association with ribosomes (Figure 5C). Pulse-chase analysis compared SSB-nascent polypeptide binding in WT and ΔRAC cells (Figure 5D, schematic). The overall translation rate was not impaired in ΔRAC cells (Figure S5B) but RAC deletion significantly impaired SSB binding to nascent chains (Figure 5D t=0, and graph). Thus, the co-chaperones of SSB modulate its ribosome cycle: RAC acts on ribosome-associated SSB to enhance cotranslational binding to nascent chains while Sse1 binds SSB post-translationally following release from ribosomes.
To examine whether RAC also affects the cotranslational specificity of SSB, SSB-associated nascent polypeptides in ΔRAC cells were identified through their mRNAs. Consistent with decreased ribosome and nascent chain association of SSB, the IPs of SSB from ΔRAC cells yielded less RNA than for WT cells (not shown). Analysis of SSB-associated mRNAs in WT and ΔRAC cells revealed distinct patterns of enrichment, indicating that RAC does modulate SSB specificity (Figure 5E). While a subset of SSB-associated nascent chains was unaffected in ΔRAC cells, loss of RAC reduced binding to some nascent chains while enhancing SSB-binding to others. Statistical comparison identified the mRNA of those SSB-bound nascent chains depleted in ΔRAC cells (20% of all SSB-bound, “depleted in ΔRAC”, light green) and those enriched in ΔRAC cells (33% of all SSB-bound, “enriched in ΔRAC”, dark green). Analysis of the properties of these subsets indicated that loss of RAC relaxes the specificity of SSB binding. For instance, cytosolic proteins were lost from SSB binding in ΔRAC cells, while nuclear, mitochondrial and membrane proteins were SSB-enriched (see Figure 5F for SSB depleted and enriched subpopulations and Figure S5C for all SSB-bound messages). Loss of RAC reduced the fraction of SSB-bound nascent polypeptides with longer sequences (Figure 5G and Figure S5D), decreased the enrichment in hydrophobic elements (Figure 5H and Figure S5E) and in regions with higher aggregation propensity (Figure 5I and Figure S5F). These experiments uncover the complex modulation of cotranslational Hsp70 specificity by its co-chaperone RAC.
SSB maintains solubility of aggregation-prone nascent polypeptides
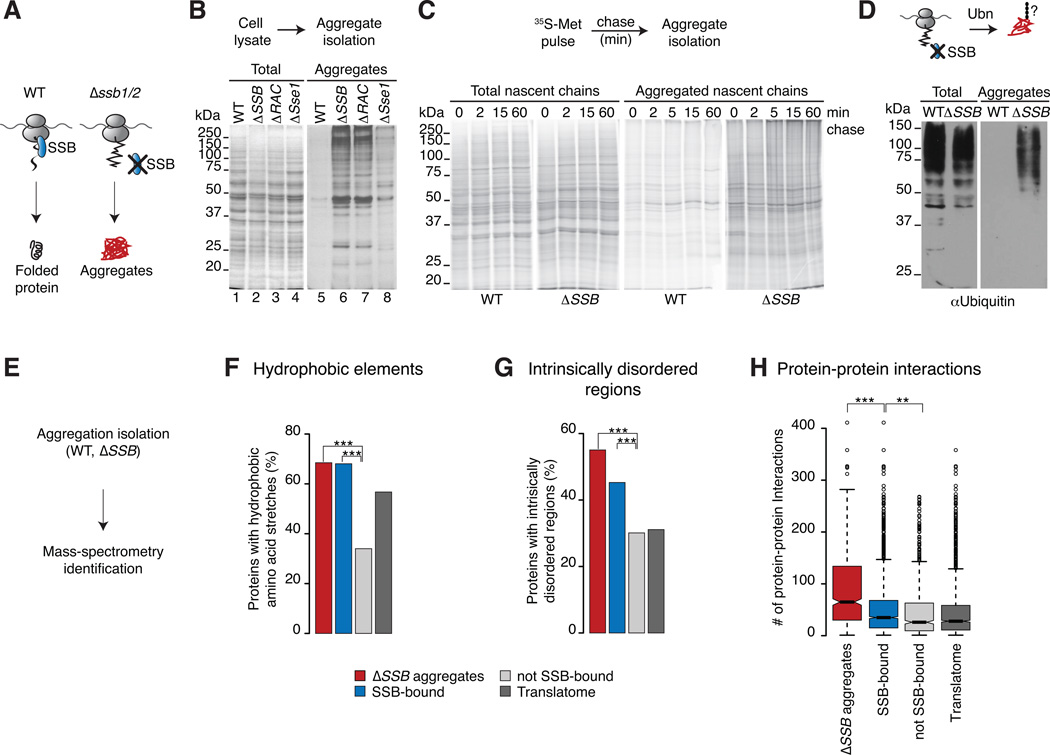
We next tested whether SSB prevents misfolding and aggregation of polypeptides as they emerge from the ribosome by comparing the presence of protein aggregates in WT and ΔSSB (Δssb1/2) cells. Loss of SSB led to widespread protein aggregation (Figure 6B, lane 6) absent from WT cells (Figure 6B, lane 5). Expression of only Ssb1 or Ssb2 alone sufficed to prevent formation of insoluble aggregates, indicating their close functional overlap (Figure S1A). Deletion of RAC and Sse1 had distinct effects on protein aggregation. Loss of RAC led to similar intensities and patterns of aggregated proteins as loss of SSB, consistent with the reduced binding of aggregation prone nascent chains to SSB in ΔRAC cells. The absence of Sse1 caused insolubility of proteins of mostly higher molecular weight (Figure 6B, lanes 6 to 8),
Figure 6. SSB maintains solubility of aggregation-prone nascent polypeptides.
(A) We hypothesize that SSB prevents aggregation of newly synthesized proteins. (B) Loss of SSB or RAC leads to widespread and loss of Sse1 leads to partial aggregation. The presence of insoluble proteins in WT, mutant cells was examined by SDS-PAGE and silver-staining (left panel: Totals; right panel: aggregates). (C) Loss of SSB leads to rapid aggregation of newly synthesized proteins. Newly made proteins of WT and ΔSSB cells were 35S-Met pulse labeled followed by a chase with cold methionine. Aggregates were isolated and analyzed by SDS-PAGE and autoradiography. (D) Proteins aggregated in ΔSSB cells are ubiquitylated as shown by SDS-PAGE and immunoblotting for Ub. (E) Global identification of aggregates in WT and ΔSSB cells and mass spectrometry. (F–G) Comparison of intrinsic properties between aggregates in ΔSSB cells, SSB substrates and non-SSB-bound proteins. The Translatome serves as reference. Only protein properties of cytosolic and nuclear localized proteins are shown. Proteins that aggregate in ΔSSB cells have similar intrinsic properties as cotranslational substrates of SSB. (H) Distinct enrichment of protein-protein interactions in protein aggregates of ΔSSB cells. *p ≤ 0.01; **p≤ 10−4; ***p ≤10−10. See also Figure S6.
Among the aggregates in ΔSSB were many ribosomal proteins (Figure S6B) as previously described (Koplin et al., 2010), consistent with our finding that many ribosomal proteins are cotranslational substrates of SSB. Pulse-chase analysis indicated that loss of SSB causes early aggregation of de novo translated polypeptides. Nascent chains were labeled for 2 min with 35S-Met in WT or ΔSSB cells and aggregates were isolated at various time points during the chase (Figure 6C). Strikingly, a large fraction of nascent polypeptides aggregated in ΔSSB cells at the earliest time points examined, indicating that the lack of SSB leads to very rapid and early aggregation of polypeptides emerging from the ribosome. The production of insoluble misfolded polypeptides in ΔSSB cells was further supported by the high degree of ubiquitylation of the aggregates (Figure 6D), which indicates that these misfolded proteins were targeted by the ubiquitin-proteasome pathway.
Mass spectrometry identification of aggregates in ΔSSB cells revealed significant overlap between proteins aggregated in ΔSSB cells and cotranslational SSB substrates (Figure S6C). It appears that a subset of SSB substrates aggregates in the absence of this chaperone, since aggregates in ΔSSB showed the same characteristics as cotranslational SSB substrates (Figure S6D), including enrichment in short hydrophobic elements (Figure 6F), regions of local disorder (Figure 6G) as well as increased length (Figure S6E) and slower translation rates (Figure S6F). Polypeptides aggregated in ΔSSB cells are also enriched in predicted aggregation-prone sequences (Figure S6G). Thus, the absence of SSB severely affects the solubility of SSB substrates.
We noted that proteins aggregated in ΔSSB cells have more interaction partners than the SSB-bound and Translatome datasets (Figure 6H). This provides a plausible rationale for their enhanced insolubility and suggests that a combination of aggregation propensity, length and the exposure of protein interaction interfaces results in enhanced aggregation of newly made proteins in the absence of SSB.
The ΔSSB aggregates contained a higher fraction of essential proteins than cotranslational SSB substrates (Figure S6H). We estimated that ~5% of newly translated polypeptides in ΔSSB cells are found in aggregates. Given the stringency of our aggregate purification procedure, this is likely an underestimate of the fraction of misfolded or aggregated polypeptides in ΔSSB cells. We propose that loss of biosynthetic capacity, reduction in the levels of essential proteins, and accumulation of misfolded proteins contributes to the slow growth phenotype ΔSSB cells (Figure S6H).
The proteins aggregated in the absence of RAC and Sse1 were also analyzed. Consistent with the patterns of insoluble proteins between ΔSSB, ΔRAC and Δsse1 cells (Figure 6B), most ΔRAC aggregates overlapped with the ΔSSB dataset (94% overlap; Figure S6I, upper panel), while aggregates in Δsse1 overlapped to a smaller extent (58% overlap, Figure S6I lower panel). These data support the idea that RAC closely regulates the cotranslational action of SSB while Sse1 acts on SSB posttranslationally with additional functions regulating other cytosolic Hsp70s.
DISCUSSION
Although eukaryotic Hsp70 was first shown to associate with translating ribosomes over 20 years ago (Beckmann et al., 1990; Nelson et al., 1992), the function and specificity of this interaction remained undefined. Here we establish the underlying principles governing the cotranslational role of Hsp70 maintaining the eukaryotic protein homeostasis. Our analysis finds a remarkable correspondence between the cotranslational requirement for Hsp70 and biophysical and chemical polypeptide properties associated with reduced folding and enhanced aggregation (Tartaglia and Vendruscolo, 2010; O'Brien et al., 2012).
Role of Hsp70 SSB within the cotranslational chaperone network
The Hsp70 SSB associates with approximately 70% of newly translated polypeptides in the yeast cell, with a strong enrichment observed for approximately 45% of nascent polypeptides. The cotranslational SSB interactome included many polypeptides encoding subunits of oligomeric complexes, such as TRiC, the 26S proteasome, the ribosome, and the exosome (Figure 2G). In addition, many ribosome biogenesis factors, kinases, such as the glucose-sensing kinase Snf1, and carbohydrate metabolism proteins cotranslationally associate with SSB. Our findings may thus explain the plurality of previously unrelated phenotypes observed for ΔSSB cells which ranged from ribosome biogenesis defects (Albanese et al., 2010; Koplin et al., 2010) to altered metabolic sensing and signaling (von Plehwe et al., 2009).
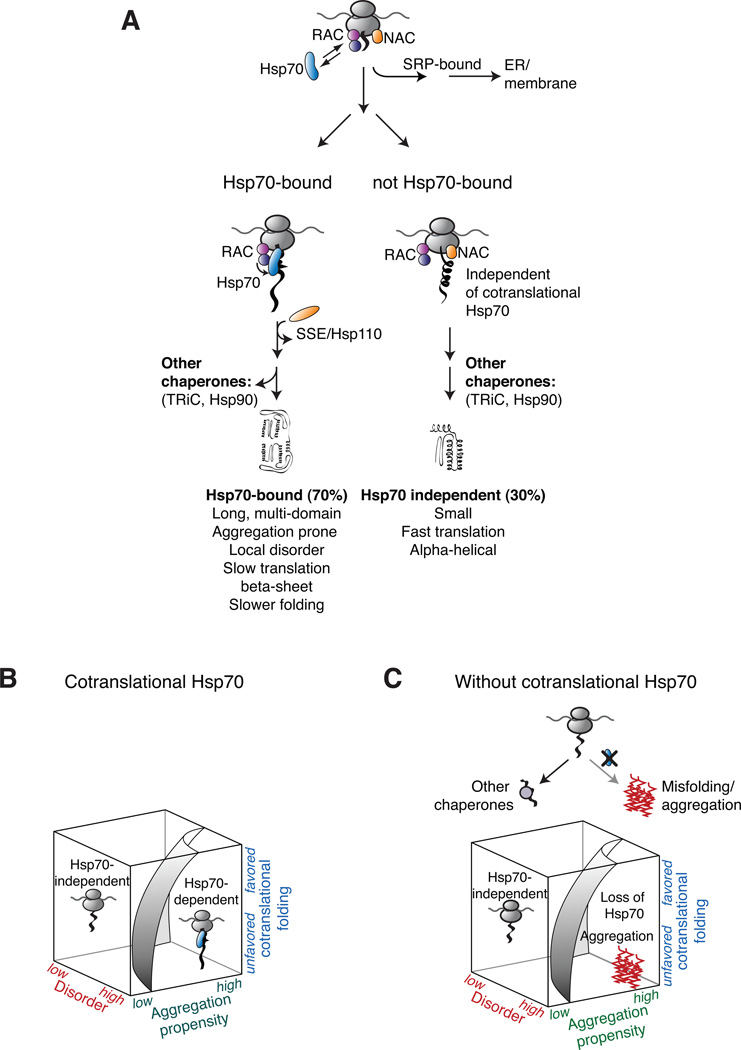
Our data provide novel insights into the division of labor and the selectivity amongst the cotranslationally acting chaperones and factors (Preissler and Deuerling, 2012). SSB binds preferentially to nascent chains of nuclear and cytosolic proteins; while the different isoforms of the nascent polypeptide-associated complex NAC bind to virtually all nascent chains, including mitochondrial and secretory proteins (del Alamo et al., 2011). Of note, the cotranslational binding to SSB and the ER-delivery factor SRP appear to be mutually exclusive, unlike what is observed for NAC, suggesting an early sorting mechanism of these factors for nascent chains at the ribosomes (Figure 7A). A few SSB-bound nascent chains localize to the ER or the membrane; these may correspond to SRP-independent substrates (Plath and Rapoport, 2000) or to membrane proteins with cytosolic domains. Interestingly, many SSB-bound nascent chains encode proteins known to also require the assistance of additional chaperones, such as TRiC for tubulin and Hsp90 for kinases, for completion of folding. Perhaps SSB provides a more general, early acting chaperone function upstream of these more specialized chaperone systems (Figure 7A).
Figure 7. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis.
(A) The role of cotranslational acting Hsp70s in protecting nascent polypeptides. Hsp70 associates with approximately 70% of newly translated polypeptides with a strong enrichment of cytosolic and nuclear proteins. The cotranslational specificity of Hsp70 for its substrates is modulated by the co-chaperone RAC. Early sorting of SSB and SRP results in mutually exclusive binding to nascent chains at the ribosomes. Maturation of not Hsp70-bound proteins is likely facilitated by other chaperone like NAC. (B) Co-translationally acting Hsp70 meets the challenge of folding the eukaryotic proteome by protecting newly translated polypeptides challenged in co-translational folding. (C) Loss of co-translational acting Hsp70 leads to widespread aggregation of newly made polypeptides with properties hindered in efficient co-translational folding.
A co-chaperone network regulates the SSB ribosome cycle and substrate specificity. The interaction of SSB with its co-chaperone network is spatially segregated with respect to the ribosome (Figure 5). RAC appears to stabilize the SSB interaction with ribosomes while Sse1 interacts mostly post-translationally with SSB (Figure 5B). RAC contributes to, but is not essential for, substrate binding to SSB. However, deletion of RAC relaxes the specificity of SSB. The enhanced association of SSB with a different set of polypeptides in ΔRAC cells could be due to their slower cotranslational maturation in the absence of RAC. RAC and Sse1 are conserved in mammalian cells, suggesting their role is conserved across eukaryotes (Peisker et al., 2010).
Underlying principles of cotranslational Hsp70 recruitment
Analysis of the properties of SSB-bound nascent polypeptides gave insight into the underlying principles determining cotranslational Hsp70 association. SSB-specificity appears strongly modulated by the cotranslational context, and enhanced among polypeptides with slower translation rates (Figure 3B) as well as those enriched in a set of intrinsic features in the nascent chains themselves. SSB binding correlated with both the domain length and the number of domains in the translating polypeptide, in good agreement with theoretical considerations highlighting domain length as a limit and constraint for foldability of proteins in the cell (Lin and Zewail, 2012; O'Brien et al., 2012).
SSB substrates were also enriched in beta-sheet regions and long intrinsically disordered stretches. These sequence elements hinder the cotranslational formation of folded structures and may thus require Hsp70 stabilization. We also found a striking enrichment in short hydrophobic sequence stretches that promote aggregation (Figure 3F). Based on the know specificity of Hsp70 substrate binding domains for linear hydrophobic stretches (Flynn et al., 1991; Rüdiger et al., 1997), it is likely that these motifs are directly recognized by Hsp70 in the context of a ribosome-bound unstructured or partially folded domain. Taken together, SSB association appears to be tailored to protect polypeptides whose length or complex architecture challenges cotranslational folding causing prolonged exposure of partially unfolded domains during synthesis (Figure 7B).
Cotranslational function of Hsp70s in the cellular folding landscape
The coordinated cotranslational folding and assembly of large architectures is a highly challenging process (Duncan and Mata, 2011). The presence of exposed hydrophobic protein interfaces make the unassembled subunits very sensitive to aggregation and thus require continued protection by a chaperone until full complex assembly (Figure 7B and Figure 7C). We find that SSB plays a key role preventing aggregation during translation (Figure 7C). Our data suggest that newly made proteins with extensive protein-protein interactions may be particularly prone to aggregation. The extensive misfolding and insolubility of newly translated polypeptides in ΔSSB cells likely contributes to their slow growth phenotype, even though other cytosolic Hsp70s and/or NAC can partially substitute for SSB (Koplin et al., 2010; Yam et al., 2005).
The avoidance of aggregation has been proposed as major driving force in the evolutionary design of naturally occurring proteins (Dobson, 2003). The remarkable enrichment in highly aggregation prone linear stretches within the cotranslational SSB-interactome suggests that Hsp70s evolved to recognize these short linear aggregation prone sequence stretches as they emerge from the ribosome. Hsp70 specificity may be the cellular response to the inevitable presence of aggregation prone regions in complex proteins, which may be buried within the folded structure, but are exposed during translation.
Our study uncovers the contribution of cotranslational Hsp70s to overall folding of the eukaryotic proteome. Eukaryotes support extensive cotranslational domain folding and even assembly (Duncan and Mata, 2011; Frydman et al., 1994; Netzer and Hartl, 1997), unlike bacteria, which shift the folding process toward a post-translational route (Agashe et al., 2004). This likely reflects the increased complexity of the eukaryotic proteome, which consists of longer, more complex proteins with a higher incidence of intrinsically unstructured regions (Koonin et al., 2000; Wang et al., 2011). The cotranslational association of eukaryotic Hsp70 evolved to meet these challenges by protecting nascent chains encoding proteins of a complex structural nature thereby preventing unfavorable intra- and inter-chain contacts leading to aggregation. The specificity and cotranslational action of Hsp70s appears fundamentally suited to the evolution of long and complex proteins in the eukaryotic proteome.
EXPERIMENTAL PROCEDURES
Yeast strains and Plasmids
Yeast strains were from (Winzeler et al., 1999) or created by mating and sporulation. Chromosomally integrations of Rpl16-TAP and Ssb2-TAP were obtained from Open Biosystems or by integrating Ssb2-TAP at the endogenous locus. N-terminally tagged Ssb2 was from (Albanese et al., 2006). ORFs of candidate substrates were expressed from GPD promoter except HA-Tub2, which was expressed from galactose-inducible promoter. For a detailed description see Supplemental Experimental Procedures.
Biochemical Procedures
Affinity purifications and microarray analysis were as described in (del Alamo et al., 2011); sucrose density fractionation was as in (Albanese et al., 2010) and 35S-methionine pulse labeling was as in (Yam et al., 2005). For IPs from RNCs and supernatant fractions, cells were lysed in buffer A (50 mM Hepes-KOH [pH 7.5], 140 mM KCl, 10 mM MgCl2, 0.1 % NP-40, 0.1 mg/ml CHX, 0.5 mM DTT and protease inhibitor). Pre-cleared lysates were fractionated in supernatant and ribosomal pellets by centrifugation through a 25 % sucrose cushion in buffer A for 20 min at 200,000 g. SSB-associated nascent chains were immunopurified as in (del Alamo et al., 2011).
Aggregates were isolated as in (Koplin et al., 2010). For pulse chase experiments followed by aggregate isolation, cells were starved for 30 min in medium without methionine. Cells were pulse labeled for 2 min with 100 µCi/ml 35S-methionine and chased with 20 mM cold methionine. At indicated time-points, aggregates were isolated as in (Koplin et al., 2010). Mass-spectrometry analysis was as described in (Sephton et al., 2011).
Bioinformatic Analysis
Microarray data were analyzed using the SAM algorithm (Tusher et al., 2001) that statistically tests for differences in gene expression by gene-specific t-tests. SSB-bound substrates were defined as polypeptides whose encoding mRNAs were significantly enriched in the SSB-IP over the total cellular mRNAs at a false discovery rate (FDR) of 0.05. Similar criteria were applied to the Translatome (Rpl16-IP) and SRP-substrates (Srp54-IP). The degree of enrichment of SSB substrates over the Translatome was determined by a “two-class unpaired” SAM with an FDR=1,relative enrichment of SSB-substrates between datasets obtained in WT and ΔRAC cells was done accordingly.
GO annotations, as well as the composition of macromolecular complexes, protein sequence length and numbers of protein-protein interactions were retrieved from the Saccharomyces Genome Database (www.yeastgenome.org). Relative translation rates were from (Arava et al., 2003) and the tRNA adaptation index from (dos Reis et al., 2004). Sequence hydrophobicity profiles were computed from the Kyte & Doolittle scale (Kyte and Doolittle, 1982). Protein disorder was predicted with the Disopred program (Ward et al., 2004). Secondary structure propensity scales were taken from (Deleage and Roux, 1987), the organization of protein domains from (Malmstrom et al., 2007) and protein aggregation propensities with the TANGO algorithm (Fernandez-Escamilla et al., 2004). All statistical analyses were performed in R (www.r-project.org). Bar plots represent the relative fraction of a dataset, box plots graph the median (solid line), 25% and 75% quartiles and 1.5× the interquartile range (dashed lines). Statistical significance for categorical variables is based on a Fisher test (column graphs) and Wilcoxon test (box plots).
Supplementary Material
HIGHLIGHTS.
Yeast Hsp70 SSB binds cotranslationally to a defined set of nascent polypeptides
SSB shows specificity for substrates highly challenged in cotranslational folding
The heterodimeric co-chaperone RAC modulates cotranslational SSB specificity
Loss of SSB function leads to widespread misfolding and aggregation
ACKNOWLEDGMENTS
We thank Dr. Patrick Brown and Dr. Dan Hogan for help and advice with the microarray experiments. We also thank Dr. Raul Andino, Karolin Dorn and members of the JF laboratory for critical reading of the manuscript; Dr. Elke Deuerling for the kind gift of anti-Sse antibody and the protocol for aggregate isolation, Dr. Scott W. Moye-Rowley for the kind gift of the anti-Ssz1 antibody. This work was supported by the Grant NIH1RO1 GM-56433 to JF and NIH R21RR025822 to JP as well as NIHF32AG038259 to EBD. FW and SP were supported by the EMBO Long-term Fellowship. MdA was partially supported by the Spanish Ministry of Education/FECYT postdoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interest.
Supplemental Information
Supplemental Information includes 1 Supplemental Table and 6 Supplemental Figures and can be found online at XXXXX.
REFERENCES
- Agashe VR, Guha S, Chang H-C, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell. 2004;117:199–209. doi: 10.1016/s0092-8674(04)00299-5. [DOI] [PubMed] [Google Scholar]
- Albanese V, Reissmann S, Frydman J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. The Journal of cell biology. 2010;189:69–81. doi: 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese V, Yam AY-W, Baughman J, Parnot C, Frydman J. Systems Analyses Reveal Two Chaperone Networks with Distinct Functions in Eukaryotic Cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann RP, Mizzen LE, Welch WJ. Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, Chang H-c, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl UF. DnaK Functions as a Central Hub in the E. coli Chaperone Network. Cell Reports. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biology. 2011;9 doi: 10.1371/journal.pbio.1001100. e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Research. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CDS, Mata J. Widespread cotranslational formation of protein complexes. PLoS genetics. 2011;7 doi: 10.1371/journal.pgen.1002398. e1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla A-M, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nature biotechnology. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annual review of biochemistry. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nature Structural & Molecular Biology. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Inada T, Winstall E, Tarun SZ, Jr, Yates JR, 3rd, Schieltz D, Sachs AB. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. Rna. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–576. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. The Journal of cell biology. 2010;189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nature Structural & Molecular Biology. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lin MM, Zewail AH. Hydrophobic forces and the length limit of foldable protein domains. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9851–9856. doi: 10.1073/pnas.1207382109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Deutsch C. Folding zones inside the ribosomal exit tunnel. Nature Structural & Molecular Biology. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- Luheshi LM, Crowther DC, Dobson CM. Protein misfolding and disease: from the test tube to the organism. Curr Opin Chem Biol. 2008;12:25–31. doi: 10.1016/j.cbpa.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Malmstrom L, Riffle M, Strauss CE, Chivian D, Davis TN, Bonneau R, Baker D. Superfamily assignments for the yeast proteome through integration of structure prediction with the gene ontology. PLoS Biology. 2007;5:e76. doi: 10.1371/journal.pbio.0050076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- O'Brien EP, Vendruscolo M, Dobson CM. Prediction of variable translation rate effects on cotranslational protein folding. Nat Commun. 2012;3:868. doi: 10.1038/ncomms1850. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Liang J. Predicting protein folding rates from geometric contact and amino acid sequence. Protein Sci. 2008;17:1256–1263. doi: 10.1110/ps.034660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisker K, Chiabudini M, Rospert S. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochimica et Biophysica Acta. 2010;1803:662–672. doi: 10.1016/j.bbamcr.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Plath K, Rapoport TA. Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. The Journal of cell biology. 2000;151:167–178. doi: 10.1083/jcb.151.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annual review of biochemistry. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends in Biochemical Sciences. 2012 doi: 10.1016/j.tibs.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. The EMBO journal. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. The Journal of biological chemistry. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia GG, Dobson CM, Hartl FU, Vendruscolo M. Physicochemical determinants of chaperone requirements. Journal of Molecular Biology. 2010;400:579–588. doi: 10.1016/j.jmb.2010.03.066. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Vendruscolo M. Proteome-level interplay between folding and aggregation propensities of proteins. Journal of Molecular Biology. 2010;402:919–928. doi: 10.1016/j.jmb.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- Voisine C, Pedersen JS, Morimoto RI. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis. 2010;40:12–20. doi: 10.1016/j.nbd.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Plehwe U, Berndt U, Conz C, Chiabudini M, Fitzke E, Sickmann A, Petersen A, Pfeifer D, Rospert S. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev. 2009;23:2102–2115. doi: 10.1101/gad.529409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Kurland CG, Caetano-Anolles G. Reductive evolution of proteomes and protein structures. Proc Natl Acad Sci U S A. 2011;108:11954–11958. doi: 10.1073/pnas.1017361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. Journal of Molecular Biology. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- Yam AY-W, Albanese V, Lin H-TJ, Frydman J. Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. The Journal of biological chemistry. 2005;280:41252–41261. doi: 10.1074/jbc.M503615200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.