Summary
α-helical integral membrane proteins critically depend on the correct insertion of their transmembrane α-helices into the lipid bilayer for proper folding, yet a surprisingly large fraction of the transmembrane α-helices in multispanning integral membrane proteins are not sufficiently hydrophobic to insert into the target membrane by themselves. How can such marginally hydrophobic segments nevertheless form transmembrane helices in the folded structure? Here, we show that a transmembrane helix with a strong orientational preference (Ncyt-Clum or Nlum-Ccyt) can both increase and decrease the hydrophobicity threshold for membrane insertion of a neighboring, marginally hydrophobic helix. This effect helps explain the ‘missing hydrophobicity’ in polytopic membrane proteins.
Keywords: membrane protein, positive-inside rule, endoplasmic reticulum
Introduction
Integral membrane proteins abound in all organisms: current estimates range from ~900 in Escherichia coli (Krogh et al., 2001) to ~5,500 in Homo sapiens (Fagerberg et al., 2010). Of these, the majority are multispanning helix-bundle proteins that partake in important cellular functions such as photosynthesis and respiration, active and passive transport of ions, metabolites, and macromolecules, signal transduction, and a multitude of enzymatic reactions. In mammalian cells, proper membrane integration of essentially all but the mitochondrial helix-bundle proteins is mediated by the Sec61 translocon in the endoplasmic reticulum (ER) membrane (White and von Heijne, 2008). Insertion into the ER membrane is co-translational, and the efficiency with which a polypeptide segment partitions into the membrane as a transmembrane α-helix (TMH) depends critically on its hydrophobicity, its overall length, and the precise location of polar and charged residues within the segment (Hessa et al., 2005; Hessa et al., 2007). Failed insertion of a transmembrane helix will result in misfolding of the protein, not only reducing the amount of functional protein in the cell but also placing a burden on the cellular quality control and protein degradation systems.
Predicted apparent free energies of membrane insertion (ΔGapp) of TMHs in proteins of known structure (Hedin et al., 2010; Hessa et al., 2007; Kauko et al., 2010), as well as experimentally determined ΔGapp values (Hedin et al., 2010), have revealed an intriguing pattern. While essentially all TMHs in single-spanning membrane proteins are predicted to insert into the membrane with a high efficiency, a considerable fraction (> 30%) of the TMHs in multispanning membrane proteins are predicted not to be efficiently inserted when taken out of their natural sequence context (Hessa et al., 2007); i.e., they are only marginally hydrophobic. This finding suggests that membrane insertion of TMHs in multispanning membrane proteins in many cases depends on sequence features extrinsic to the hydrophobic segment itself. Such features have indeed been identified and include charged amino acid residues flanking the hydrophobic segment (Hedin et al., 2010; Hessa et al., 2007; Lerch-Bader et al., 2008), interactions between polar residues in adjacent TMHs (Buck et al., 2007; Meindl-Beinker et al., 2006; Zhang et al., 2007), and repositioning of TMHs relative to the membrane during folding and oligomerization (Kauko et al., 2010).
Another extrinsic feature that might influence the membrane-insertion efficiency of a polypeptide segment is the orientational preferences of neighboring TMHs. This possibility was first suggested by Ota and coworkers (Ota et al., 1998), who showed that the strong Nlum-Ccyt orientational preference of TMH8 in the human band 3 protein can force an upstream hydrophilic segment to be inserted across the membrane. Inspired by this lead, we have systematically investigated the effect of the orientational preference of a “controlling” TMH on the membrane insertion efficiency of an upstream, marginally hydrophobic “test” segment. We have manipulated the orientational preference of the controlling TMH by varying its hydrophobicity and by adding positively charged flanking residues at either end. Positively charged residues in membrane proteins are significantly enriched in loops facing the cytosolic side of the membrane – the so-called positive-inside rule (von Heijne, 1986, 1989) – and strategic addition/removal of positively charged residues can be used to manipulate the topological preference of TMHs in a predictable way (Gafvelin et al., 1997; Gafvelin and von Heijne, 1994; Goder and Spiess, 2003; Higy et al., 2004; Seppälä et al., 2010; von Heijne, 1989). Likewise, a high level of hydrophobicity has been reported to promote the Nlum-Ccyt orientation of N-terminal signal-anchor sequences (Goder and Spiess, 2003; Higy et al., 2004; Sakaguchi et al., 1992; Wahlberg and Spiess, 1997).
We now demonstrate that the orientational preference of a TMH can both increase and decrease (depending on the overall topology) the threshold hydrophobicity required for insertion of a neighboring upstream TMH into the ER membrane. We further show that the membrane insertion of the markedly polar and highly conserved TMH8 in the human apical sodium-dependent bile acid transporter depends on the orientational preference of the neighboring TMH9. Our findings highlight a previously unappreciated and likely quite common way by which sequence context can determine the fate of marginally hydrophobic segments in multispanning membrane proteins, and raise intriguing questions regarding the mechanisms whereby translocon-mediated insertion of TMHs can be affected by quite distant topogenic signals in the nascent chain.
Results
Model proteins and membrane-insertion assay
To systematically study the effects exerted by a neighboring TMH on the membrane insertion of a marginally hydrophobic segment, we adapted the Escherichia coli inner membrane protein leader peptidase (Lep) for use as a model protein. Lep has two N-terminal TMHs (TMH1, TMH2) and a large C-terminal catalytic domain (P2). When translated in vitro in reticulocyte lysate in the presence of ER-derived dog pancreas rough microsomes (RMs), Lep adopts an overall Nlum-Clum topology in the microsomal membrane (Johansson et al., 1993). Lep has been used extensively in the past to study the insertion of hydrophobic segments into the ER membrane, e.g. (Hessa et al., 2007).
For the present study, we engineered the P2 domain to create two versions of the model protein, LepIV and LepV, differing in whether TMH2 is present or not, Figure 1. In both constructs, we introduced into the P2 domain three 19-residue hydrophobic Leu/Ala segments (H1-H3) of the general composition XXPX-[nL, (19-n)A]-ZPZZ (abbreviated as 3X/nL/3Z) where n is the number of Leu (L) residues, 19-n the number of Ala (A) residues, and the flanking residues X and Z are either neutral Gly (G) or positively charged Lys (K). To facilitate the interpretation of the results, we replaced all positively charged amino acids between codons 81 and 284 (i.e. from approximately the middle of the loop between TMH1 and H1 until 34 residues downstream of the C-terminal end of H3) in the LepIV construct by alanines (panel A), and did the same for all positively charged residues between codons 80 and 344 (i.e., from the C-terminal end of TMH2 until 42 residues downstream of the C-terminal end of H3) in LepV (panel B). LepIV thus has four hydrophobic segments (TMH1, H1, H2, H3) and LepV has five (TMH1, TMH2, H1, H2, H3); note that H2 is oppositely orientated (Nlum-Ccyt vs. Ncyt-Clum) when forming a TMH in the two model proteins.
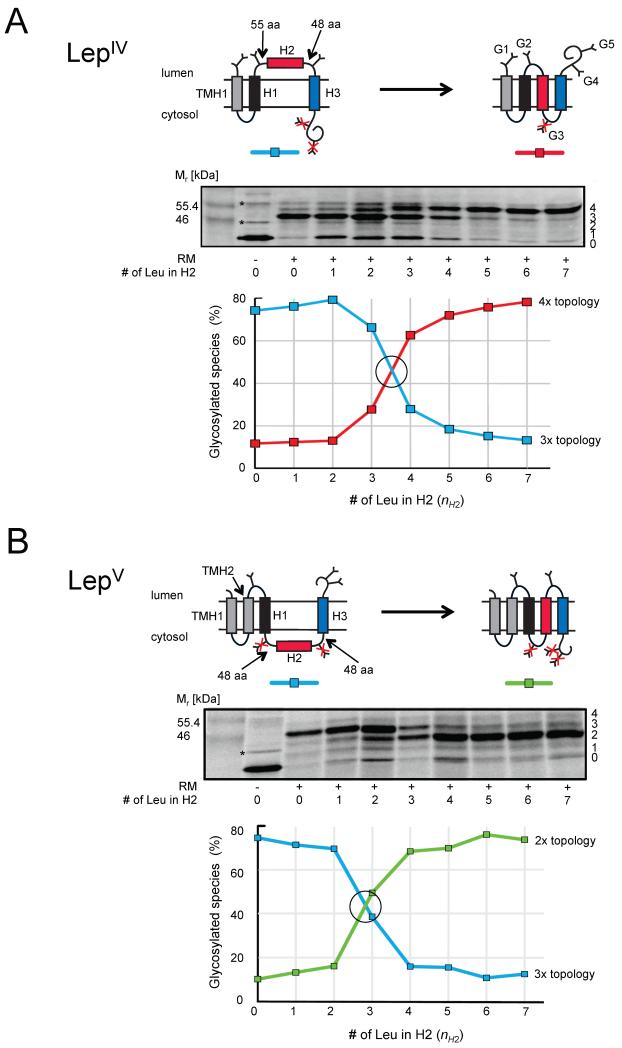
Figure 1.
Membrane insertion of H2 test segments of varying hydrophobicity (H2:3G/(1-7)L/3G) into dog pancreas rough microsomes (RM). H1 and H3 both have the composition 3G/7L/3G. (A) LepIV constructs. Molecules with 0-4 added glycans are indicated on the right-hand side of the gel and two background bands present also when translation is carried out in the absence of added RMs are indicated by *. The graph shows the fractions of the two dominating species with, respectively, three (blue) and four (red) glycans (corresponding to the two topologies shown at the top). The crossover point (I50) between the 3x and 4x topologies is at nH2 = 3.5 (black circle); at this point H2 is inserted into the membrane in 50% of the molecules. (B) LepV constructs. The graph shows the fractions of the two dominating species with, respectively, three (blue) and two (green) glycans. In this case, I50 = 2.8 (black circle). See also Figure S1.
For H1 we used the sequence GGPGALAALALAALAALALAALAGPGG (i.e., nH1 = 7 and X, Z = G; shorthand H1:3G/7L/3G), for H2 we used X, Z = G and varied nH2 in the interval 0-7 (H2:3G/(0-7)L/3G), and for H3 we varied both X, Z, and nH3 and also used longer flanks of the composition XXXXPX- and -ZPZZZZ (abbreviated as H3:5X/nL/3Z and H3:3X/nL/5Z) in some constructs (see Table S1 for sequences of all constructs). Apparent free energies of membrane insertion for single segments of this kind have previously been measured using in vitro translation in the presence of RMs (Hessa et al., 2005; Hessa et al., 2007; Lundin et al., 2008); the threshold hydrophobocity for membrane insertion (i.e., ΔGapp = 0 kcal/mol) of a single Nlum-Ccyt-orientated [nL,(19-n)Ala] segment is n = 3.2. Our main objective in the present study was to use the LepIV and LepV model proteins to determine if the charge of the flanking residues (X, Z) or the hydrophobicity dictated by the number of Leu residues (nH3) of the controlling H3 segment influences the threshold hydrophobicity required for membrane insertion of the H2 test segment.
H1, H2, and H3 are separated by loops that are either 48 or 55 residues in length and each loop harbors a single acceptor site (Asn-X-Thr) for N-linked glycosylation. To ensure unhindered access to the oligosaccharyl transferase, the acceptor sites are placed ≥ 15 amino acids away from the nearest hydrophobic segment (Nilsson and von Heijne, 1993; Popov et al., 1997). In addition, we placed two additional glycan acceptor sites downstream of H3 in LepIV and LepV, and one acceptor site in a short extension added upstream of TMH1 in LepIV. All five acceptor sites (G1-G5) can be efficiently glycosylated in the reticulocyte-RM in vitro translation system, Figure S1. Since N-linked glycosylation, carried out by the ER-bound oligosaccharyl transferase complex, can only take place in the lumen of the ER (Imperiali and Hendrickson, 1995), assaying the modification state of the glycan acceptor sites makes it possible to determine which topology any particular LepIV or LepV construct adopts in the ER membrane. Two examples are shown in Figure 1. In both examples, H1 and H3 have the composition 3G/7L/3G, while the composition of the H2 segments is 3G/(0-7)L/3G, i.e., the number of Leu residues in H2, nH2, is varied between 0 and 7. For the LepIV constructs (panel A), the dominating topology changes from one with three glycosylated acceptor sites (3x topology; H2 not inserted) to one with four glycosylated sites (4x topology; H2 inserted) as nH2 increases from 0 to 7; the crossover point (I50) between the two topologies is at nH2 = 3.5 (see Table S2 for I50 values for all series of constructs). Because TMH1, H1, and H3 have hydrophobicities that are well above the threshold of n = 3.2 determined previously for insertion of single Nlum-Ccyt orientated TMHs into the ER membrane (Hessa et al., 2005), we can confidently assign the two dominating topologies as shown in the Figure. Evidently, the I50 for membrane insertion of the Nlum-Ccyt orientated H2 segment in this context is not far from the single-TMH threshold.
The H2 segment behaves similarly when present in the LepV construct (panel B). Even though its membrane orientation is reversed to Ncyt-Clum in this case, the crossover point is not much changed (I50 = 2.8). The slight drop in I50 is consistent with earlier results obtained for single TMHs where the threshold for membrane insertion was found to be somewhat lower for a Ncyt-Clum orientated segment compared to Nlum-Ccyt orientated segment (Lundin et al., 2008).
Positively charged residues flanking H3 can alter the threshold hydrophobicity of H2
With the LepIV and LepV model proteins in hand, we can now ask how the I50 threshold for membrane insertion of H2 varies as a function of its membrane orientation (Nlum-C in the LepIV context, Ncyt-Clum in the LepV context) and as a function of the composition of H3 (X, Z, n). In order to keep the number of constructs within a manageable range, we have focused on the extremes in terms of flanking charges and hydrophobicity of H3, but have also analyzed some intermediary combinations in order to obtain a more complete picture. The results for the LepIV constructs are shown in Figure 2, and for the LepV constructs in Figure 3. We will first discuss the effects of adding positively charged Lys residues to the ends of H3 (X, Z = K vs. X, Z = G), and then turn our attention to the role of H3 hydrophobicity (nH3).
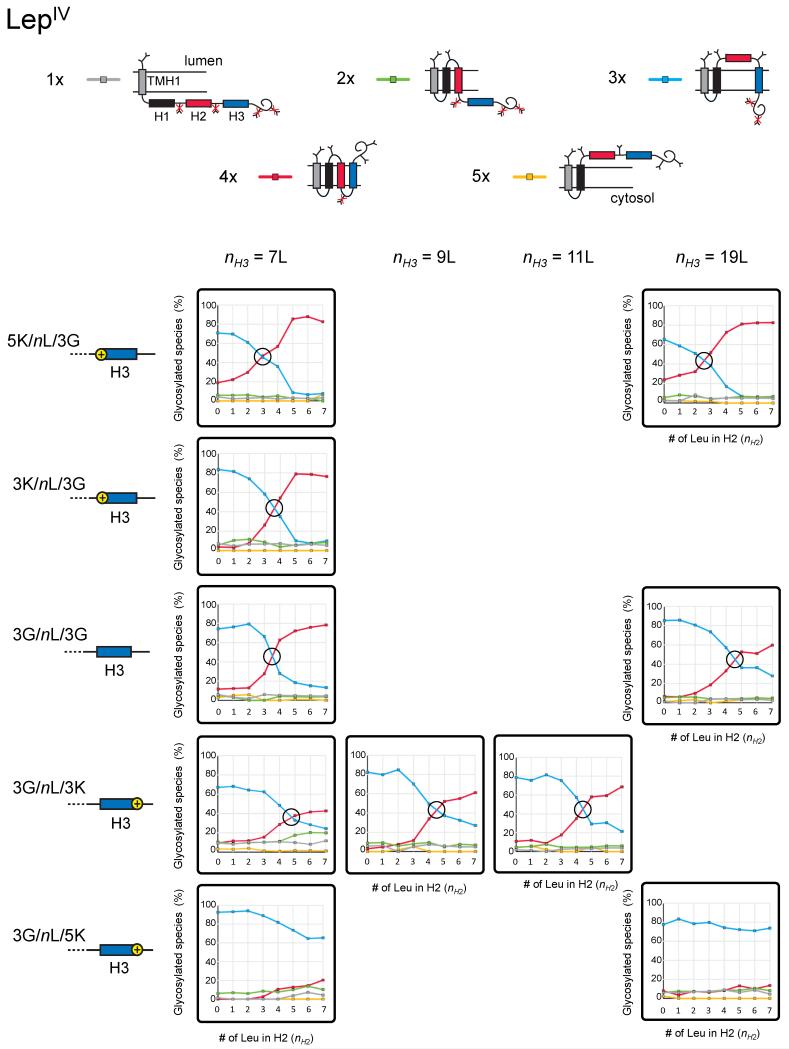
Figure 2.
Analysis of LepIV constructs in which both the hydrophobicity (nH2) of H2 and the flanking residues (Gly, Lys) and hydrophobicity (nH3) of H3 are varied. Topologies characterized by different numbers of added glycans are shown on top, with their corresponding color codes as used in the small panels showing quantitations. The composition of the H3 segment in terms of number of Leu (nH3) and flanking residues are indicated next to the cartoon representations of the H3 segments on the left. The crossover points (I50) between the dominating topologies are indicated (black circles). See also Table S1, Table S2, and Figure S2.
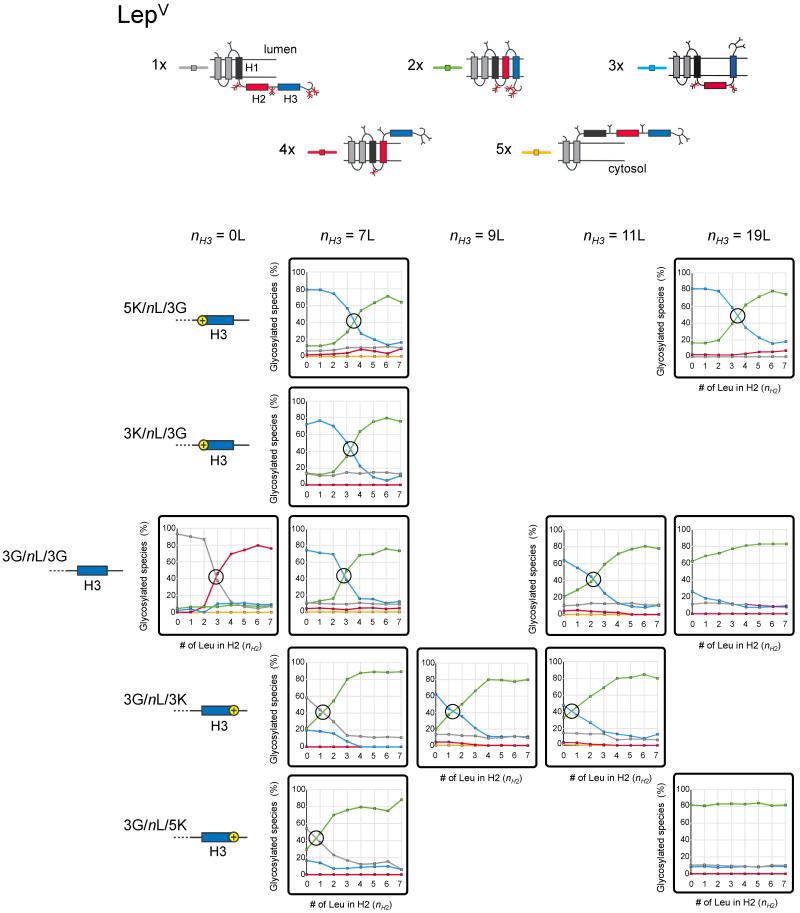
Figure 3.
Analysis of LepV constructs in which both the hydrophobicity (nH2) of H2 and the flanking residues (Gly, Lys) and hydrophobicity (nH3) of H3 are varied. Topologies characterized by different numbers of added glycans are shown on top, with their corresponding color codes as used in the small panels showing quantitations. The composition of the H3 segment in terms of number of Leu (nH3) and flanking residues are indicated next to the cartoon representations of the H3 segments on the left. The crossover points (I50) between the dominating topologies are indicated (black circles). See also Table S1 and Table S2, and Figure S3.
Based on previous findings demonstrating that positively charged residues flanking TMHs can have non-local effects on membrane-protein topology (Gafvelin and von Heijne, 1994; Seppälä et al., 2010), we surmised that adding Lys residues either to the N terminus (X = K) or to the C terminus (Z = K) of H3 might affect the I50 threshold for H2, and that these effects should be in opposite directions in the LepIV and LepV constructs. This is indeed the case, as can best be seen by comparing Figures 2 and 3. Starting with the LepIV data in Figure 2, for an H3 segment with 7 Leu (nH3 = 7) there is little effect on the H2 I50 value upon changing the N-terminal flanking residues from Gly to Lys, even when five Lys residues are added (I50 is reduced from 3.5 to 3.0 when going from H3:3G/7L/3G to H3:5K/7L/3G). The effect is more pronounced for a very hydrophobic H3 with 19 Leu (nH3 = 19; I50 is reduced from 4.6 to 2.6 in going from H3:3G/19L/3G to H3:5K/19L/3G). In both cases, the change in I50 is in the direction expected from the positive-inside rule, i.e., the transition from the 3x to the 4x topology (in which a positively charged N-terminal end of H3 is located on the “correct”, cytoplasmic side of the ER membrane) happens at a lower I50 threshold when Lys residues are added to the N terminus of H3. A possible caveat is that a weakly hydrophobic H2 segment that is initially translocated across the membrane may be prevented from being pulled back into a transmembrane configuration by early glycosylation of the G3 site between H2 and H3 (c.f., Figure 6A below), and hence that the change in I50 is underestimated in these experiments. To rule this out, we repeated the determination of I50 with H3:3K/7L/3G in a construct lacking the G3 site. As seen in Figure S2, there is no significant change in I50 when the G3 site is absent.
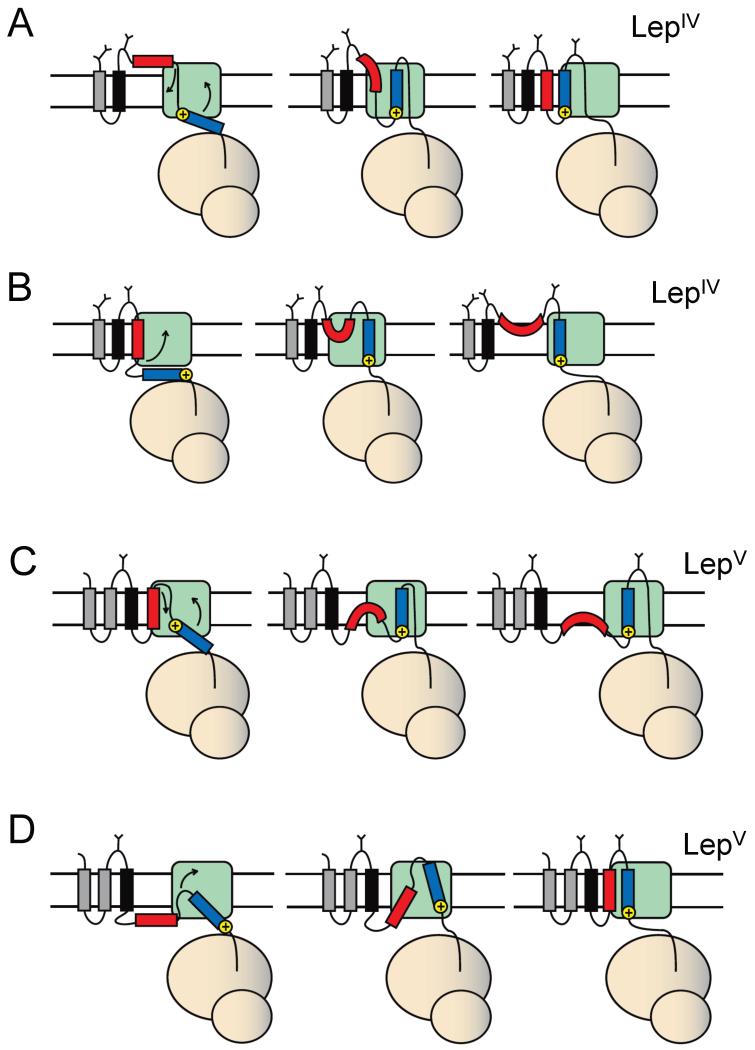
Figure 6.
Models for how H3 segments (blue) with different orientational preferences imparted by flanking positively charged residues may affect the membrane insertion of H2 (red) in LepIV (panels A, B) and in LepV (panels C, D). The positively charged residues are on the N terminus of H3 in A and C, and on the C terminus in B and D. See text for discussion.
The effects on the H2 I50 value are much stronger when changing the C-terminal flanking residues of H3 in LepIV from Gly to Lys. For the H3:3G/7L/3K constructs I50 is 4.8 (compared to 3.5 for the H3:3G/7L/3G construct), and for the H3:3G/7L/5K constructs the 3x topology, in which the H2 segment is translocated across the membrane, persists even for an H2:3G/7L/3G segment (i.e., I50 >> 7). The direction of the change in I50 is as predicted by the positive-inside rule, since the positively charged C-terminal end of H3 is now on the “incorrect”, lumenal side of the membrane in the 4x topology.
Turning to the LepV constructs in Figure 3, the effects on H2 are again in the direction predicted by the positive-inside rule. For the H3 constructs with 7 Leu (nH3 = 7) the effect seen when changing the N-terminal Gly residues to Lys is small (I50 increases from 2.8 for H3:3G/7L/3G to 3.5 for H3:5K/7L/3G). The effect is more marked for the nH3 = 19 constructs: I50 < 0 for H3:3G/19L/3G and increases to 3.4 for H3:5K/19L/3G. In the H3:5K/19L/3G series of constructs we see a lower molecular weight, singly glycosylated product when nH2 ≤ 3 (shown in Figure S3 for the H2:3G/0L/3G construct). This product co-migrates on the gel with a construct truncated immediately after H3 (but not with a construct truncated immediately after H2 or a construct starting at the internal Met92 in Lep located 15 residues downstream of TMH2). It is unclear how this form is generated, but given its size and the fact that it carries one glycan it should have the same topology as the full-length protein with 3x topology. We have therefore included it in the quantitation of the fraction of 3x topology for the H3:5K/19L/3G constructs in Figure 3; the change in the I50 value is small (from 3.4 to 3.0) if this form is not included.
As seen for LepIV, for H3 segments with nH3 = 7, C-terminal Gly to Lys changes have a stronger effect on I50, which is reduced from 2.8 for H3:3G/7L/3G to 0.7 for H3:3G/7L/5K. Interestingly, for the LepV H3:3G/7L/3K and H3:3G/7L/5K constructs, the 1x topology predominates when H2 is marginally hydrophobic (nH2 = 0), i.e., neither the H2 nor the H3 segment spans the membrane in these cases.
We conclude that the threshold hydrophobicity (I50) required for membrane insertion of the H2 segment can be systematically tuned both up and down by manipulation of the charge bias across H3.
The hydrophobicity of H3 can affect the threshold hydrophobicity of H2
As noted above, the hydrophobicity of H3 can influence the I50 threshold for H2. To study this more systematically, we varied the hydrophobicity of H3 (nH3) in the absence of any flanking charges (X, Z = G) for both LepIV and LepV. For LepIV, an increase in nH3 from 7 to 19 Leu leads to a modest increase in the H2 I50 value from 3.5 for H3:3G/7L/3G to 4.6 for H3:3G/19L/3G, Figure 2. The direction of the change is consistent with the previous finding that an increase in the hydrophobicity of an N-terminal signal-anchor sequence favors the Nlum-Ccyt orientation (Goder and Spiess, 2003), i.e., in the LepIV context an increase in nH3 would favor the 3x topology. The picture is the same for most of the LepV constructs. There is not much effect on I50 when changing nH3 from 0 to 11 Leu when the flanking residues are Gly, Figure 3. Going from H3:3G/11L/3G to H3:3G/19L/3G, however, we see a substantial reduction in I50 (from 2.2 to < 0), again consistent with the expectation that an increase in nH3 should lead to a stronger preference for H3 having the Nlum-Ccyt orientation, favoring the 2x topology for LepV.
Combining flanking charged residues and hydrophobicity
Both the hydrophobicity of H3 and its flanking positively charged residues can influence the H2 I50 threshold, and the two properties can obviously be combined. The extreme cases in Figures 2 and 3 are the H3:5K/19L/3G and H3:3G/19L/5K segments. For LepIV, changing the N-terminal flanking residues of H3 from Gly to Lys has a modest effect on I50, which is reduced from 4.6 for H3:3G/19L/3G to 2.6 for H3:5K/19L/3G, but the same change at the C terminus of H3 makes a huge difference, essentially abolishing the membrane insertion of the H2 segment (I50 >> 7 for H3:3G/19L/5K).
The situation for LepV is a little different in that N-terminal Lys residues have a major effect: I50 < 0 for the H3:3G/19L/3G segment and increases to 3.4 for H3:5K/19L/3G. C-terminal Lys residues in H3 also appear to have a strong effect (compare H3:3G/19L/3G and H3:3G/19L/5K), although we cannot determine the precise I50 values in this case since they are both < 0. Clearly, when the orientational preferences imparted on H3 by positively charged residues and hydrophobicity work in concert, very strong effects on the H2 I50 threshold can be elicited.
Topological frustration
“Frustrated” topologies are topologies in which segments that are sufficiently hydrophobic to form transmembrane helices when on their own are prevented from doing so by neighboring helices with strong orientational preferences (Gafvelin and von Heijne, 1994). In the present data set, there are a few examples of topological frustration. For LepV, frustration of H3 is seen in the form of the 1x topology for the H3:3G/7L/3K and H3:3G/7L/5K constructs, Figure 3. In these cases, the C-terminal Lys residues prevent H3 from inserting with its C-terminal end towards the lumen, while at the same time H2 is not sufficiently hydrophobic to allow formation of the 2x topology. However, by increasing the hydrophobicity of H3 to nH3 = 9L (H3:3G/9L/5K), the orientational effect of the C-terminal Lys residues can be overcome and H3 now inserts in the “incorrect” Ncyt-Clum orientation. A more intriguing case of topological frustration is the 3x topology observed for LepIV H3:3G/7L/5K and H3:3G/19L/5K constructs with hydrophobic H2 segments (H2:3G/(5-7)L/3G), Figure 2. In these constructs, the strong Nlum-Ccyt orientational preference of the H3 segment apparently forces the H2 segment to translocate across the membrane even though the hydrophobicity of the H2 segment is well above the previously determined threshold for its membrane insertion, nH2 = 3.2 (Hessa et al., 2005).
Kinetics of membrane insertion
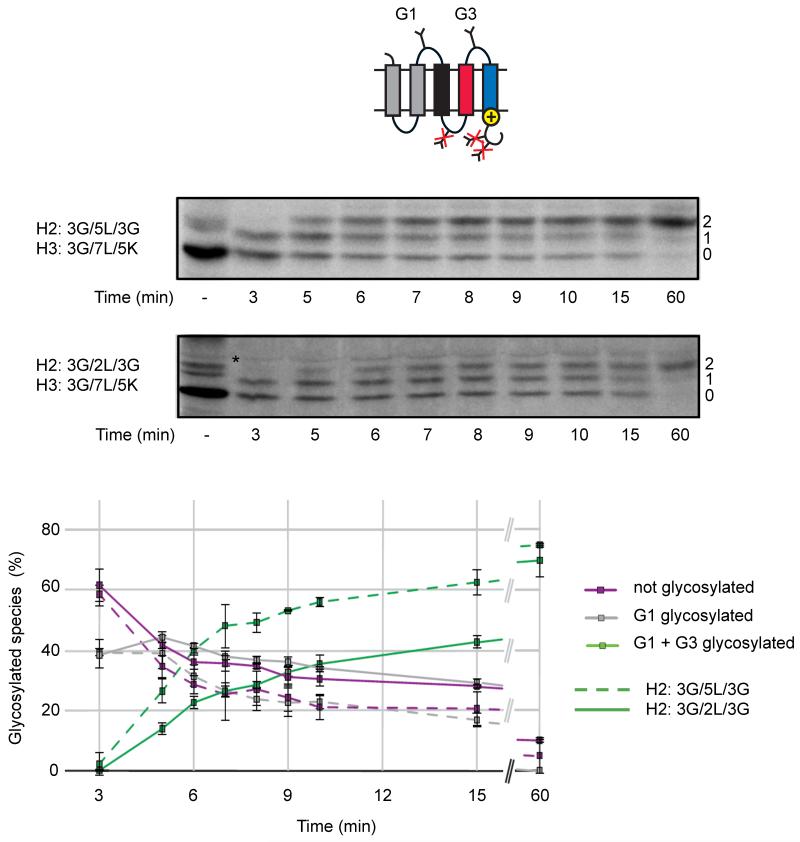
As shown above, positively charged residues on the C terminus of H3 can exert a long-range effect on the hydrophobicity threshold for insertion of H2. In particular, in the LepV H3:3G/7L/5K construct the I50 threshold is at nH2 = 0.7, compared to nH2 = 2.8 in the absence of C-terminal Lys residues (construct H3:3G/7L/3G), Figure 3. This observation raises the question whether the membrane insertion of an H2 segment with nH2 < 2.8 is delayed until the H3 segment reaches the translocon in the LepV H3:3G/7L/5K construct, compared to the kinetics of insertion of an H2 segment that is sufficiently hydrophobic to insert on its own?
To address this issue, we measured the kinetics of glycosylation of the G3 site that is located in the loop between H2 and H3 using an assay in which chain elongation is synchronized by addition of the translation-initiation inhibitor aurintricarboxylic acid (ATA) 4 min after mixing of the mRNA into the in vitro translation system and then removing aliquots of the reaction at regular intervals, mixing each aliquot with Triton X-100 to disrupt the microsomal membranes and thereby prevent further glycan addition, and continuing the incubation up to 60 min to ensure that all chains are fully elongated (Monné et al., 1999; Rothman and Lodish, 1977).
As seen in Figure 4, in a LepV H3:3G/7L/5K construct with H2:3G/5L/3G, i.e., with an H2 segment that is sufficiently hydrophobic to insert by itself, the half-maximal glycosylation of the G3 site (i.e., the G3 site is glycosylated in 35-40% of the chains) is reached ~6 min after translation initiation. The rate of glycosylation of the G3 site in the corresponding LepV H3:3G/7L/5K construct with H2:3G/2L/3G (i.e., an H2 segment that cannot insert by itself) is significantly slower with half-maximal glycosylation of the G3 site reached at ~11 min, yet the final level of glycosylation of the G3 site after 60 min incubation is roughly the same for both constructs (70-75%). This is the expected outcome if insertion of the H3:3G/7L/5K segment into the translocon is required to force the insertion of H2 in the H2:3G/2L/3G construct (c.f., Figure 6D below). As seen by the rather slow increase in the glycosylation of G3 at later chase times, a significant population of molecules seems to be glycosylated post-translationally for the H2:3G/2L/3G construct (on average, the 363 residues long chain should be fully synthesized by ~12 min since the average translation rate in our RM system is ~0.5 residues/sec (Monné et al., 1999))
Figure 4.
Kinetics of glycan addition to two LepV constructs where the H3 segment has the composition H3:3G/7L/5K and the H2 segment has the composition H2:3G/5L/3G (dashed lines) or H2:3G/2L/3G (full lines). Both constructs adopt the same final topology shown in the cartoon. Molecules with 0-2 added glycans are indicated on the right-hand side of the gels and a background band present also when translation is carried out in the absence of added RMs is indicated by *. The graph shows the fractions of the different species with, respectively, zero (purple), one (gray), and two (green) glycans as a function of the time after the addition of mRNA to the translation mix. Error bars indicate ±1 SD.
Effects on membrane insertion exerted by a neighboring TMH in a natural protein
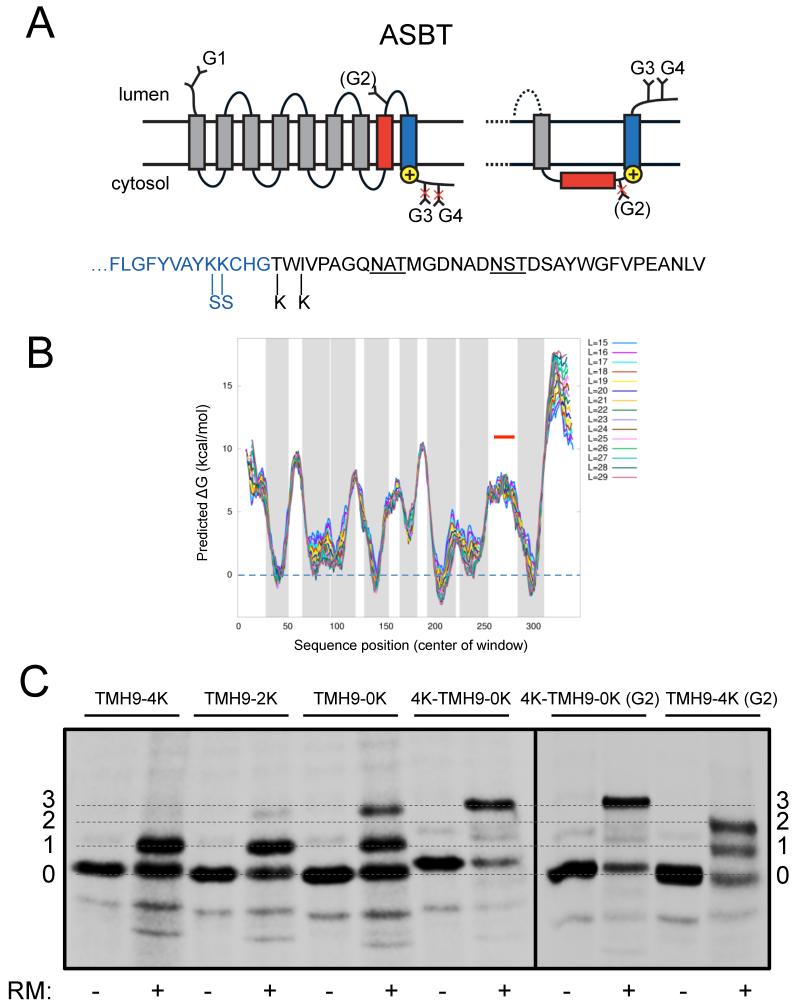
In order to validate the effects of a neighboring TMH on the membrane insertion propensity of marginally hydrophobic TMHs demonstrated above in a natural protein, we perused the data base of multispanning membrane proteins of known three-dimensional structure, looking for cases where a TMH predicted not to insert into the membrane by itself was immediately followed by a hydrophobic TMH with a strong orientational preference. We focused on a suitable candidate protein, the human apical sodium-dependent bile acid transporter (ASBT), in which the very polar and highly conserved Ncyt-Clum orientated TMH8 is followed by TMH9 which has a high hydrophobicity and is flanked by a strong orientational determinant in the form of a cluster of C-terminal positively charged residues (Hu et al., 2011), Figure 5A, B. Our attention was drawn to the ASBT protein by a previous study showing that TMH8 only inserts into the ER membrane in the presence of TMH9, but not when TMH9 is deleted (Hallén et al., 1999).
Figure 5.
Membrane insertion of TMH8 in the human ASBT protein depends on the orientational preference of TMH9. (A) Topology diagram. THM8 is in red and TMH9 in blue. The positions of the glycan acceptor sites G1-G4 and the locations before and after TMH9 where positively charged lysine residues are removed or added are indicated. The alternative topology of the C-terminal TMH8-TMH9 region seen in the 4K-TMH9-0K construct is also shown. The sequence of the C-terminal tail is shown below with the positions of the Lys mutations indicated and the G3 and G4 glycan acceptor sites underlined; the LepV-derived segment is in black. (B) Hydrophobicity plot produced by the ΔG prediction server (http://dgpred.cbr.su.se/); gray shading indicates predicted TMHs. TMH8 (indicated by the red line) is too polar to score as a TMH. L is the length of the moving window used for averaging. (C) SDS-PAGE analysis of ASBT constructs. Molecules with 0-3 added glycans are indicated. See Table S1 for sequences.
In order to dissect the role of the C-terminal charge cluster flanking TMH9, we replaced the last 32 residues in the C-terminal tail downstream of the charge cluster with a similar-length segment lacking positively charged residues from the C terminus of LepV. In order to probe the topology of the TMH8-TMH9 region of ASBT we introduced two glycan acceptor sites in the C-terminal tail (G3, G4), and, in two constructs, one additional site in TMH8 (G2) that had already been shown to become partially glycosylated when present in the wild type protein (Zhang et al., 2004). A naturally glycosylated acceptor site in the N-terminal extracellular tail of ASBT (G1) served as a convenient marker for proper targeting of the protein to the microsomal membrane.
As seen in Figure 5C, a construct with a total of four positively charged lysines in the C-terminal charge cluster (construct TMH9-4K) is only glycosylated on the single N-terminal acceptor site G1 (G2 is lacking in this construct). With only two Lys residues in the C-terminal charge cluster (construct TMH9-2K), a small fraction of triply glycosylated molecules modified on G1, G3, and G4 is apparent; this fraction increases further when there are no Lys in the C-terminal tail (construct TMH9-0K) and becomes predominant when four Lys are added at the N terminus of TMH9 (construct 4K-TMH9-0K). Thus, the orientation of TMH9 is almost completely inverted in the latter construct compared to its orientation in wild type ASBT and in construct TMH9-4K. Consistent with the inversion of TMH9, the G2 acceptor site added near the lumenal end of TMH8 is partially modified when introduced in construct TMH9-4K (as it is in the wild type protein (Zhang et al., 2004)) but is not glycosylated when introduced in construct 4K-TMH9-0K.
We conclude that membrane insertion of the markedly polar TMH8 in ASBT is completely dependent on the orientational preference of its more hydrophobic neighbor TMH9, similar to the observations made above using engineered model proteins. As TMH8 contains four of the nine charged or polar residues in ASBT that form the two Na+-binding sites and one polar residue important for substrate binding, and also has an unusual break in the α-helical conformation near the middle of the membrane that is involved in a critical cross-over interaction with the likewise well-conserved TMH3 (Hu et al., 2011), its polar character is clearly the result of a strong selection pressure. The competing requirements of polar character and efficient membrane integration of TMH8 explain the need for TMH9 to be designed with an orientational preference that ‘forces’ TMH8 into the membrane. Since polar residues in TMHs are generally well conserved and often functionally important (Illergård et al., 2011), we expect that the hydrophobicity of many or most marginally hydrophobic TMHs is similarly constrained by functional requirements.
Discussion
A few earlier studies (Enquist et al., 2009; Hedin et al., 2010; Ota et al., 1998) have suggested the possibility that the membrane-insertion efficiency of a TMH may not always be a function only of local sequence properties (e.g., hydrophobicity, flanking charged residues) or specific interactions with a neighboring TMH. Here, we have asked to what extent the orientational preference of a TMH can affect the threshold for membrane insertion of a neighboring TMH. The answer is: To a surprisingly large degree, at least when extreme levels of hydrophobicity and positive-charge bias across a TMH are brought into play. Thus, the H2 test segment in the LepIV H3:3G/19L/5K construct does not form a TMH even when being very hydrophobic (H2:3G/7L/3G), Figure 2, while the H2 segment in the LepV H3:3G/19L/5K construct forms a TMH even when being of low hydrophobicity (H2:3G/0L/3G), Figure 3. The effect can be reasonably strong also under more moderate conditions. In the case of the LepIV model protein, the thresholds for membrane insertion of the Nlum-Ccyt orientated H2 segment are I50 = 3.6 for the H3:3K/7L/3G construct and I50 = 4.8 for the H3:3G/7L/3K construct; for the Ncyt-Clum orientated H2 segment in the LepV model protein the corresponding values are I50 = 3.3 and I50 = 1.2. Expressed as a difference in apparent free energy of membrane insertion (Hessa et al., 2007), two Ala-to-Leu replacements corresponds to ΔΔGapp = −1.2 kcal/mol, which goes a long way towards eliminating the discrepancy between the ΔGapp-distributions found for single-spanning and multispanning membrane proteins alluded to in the Introduction (Hessa et al., 2007).
The effect exerted by H3 on the H2 segment is long-range: Lys residues in the C-terminal flank of H3 are 67 residues away from the C-terminal end of the H2 segment in both the LepIV and LepV constructs. Given that it only takes a ~40 residues long nascent chain to span the distance from the ribosomal P-site to the cytoplasmic end of the translocon channel (Becker et al., 2009; Bhushan et al., 2010), the flanking Lys residues have not been incorporated into the nascent chain at the point when the H2 segment has reached the translocon. Yet, in LepIV constructs such as H3:3G/7L/5K and H3:3G/19L/5K, H2 segments containing at least 7 Leu are forced to the lumenal side of the membrane. How could this come about? A possible model is depicted in Figure 6B. In this model, the H2 segment initially adopts a transmembrane Nlum-Ccyt orientation, either being wholly integrated into the membrane or being kept in the lateral gate region of the Sec61 translocon. The strong orientational preference of the H3 segment makes it adopt the same Nlum-Ccyt orientation, forcing the C-terminal end of H2 across the membrane. The final topology adopted by the protein would be one where H2 forms a re-entrant loop from the lumenal side of the membrane and H3 is inserted across the membrane in its preferred Nlum-Ccyt orientation.
Another example of long-range action is provided by the LepV H3:3G/7L/3K and H3:3G/7L/5K constructs, in which the H2 segment inserts as a TMH at considerably lower hydrophobicity thresholds than in the corresponding H3:3G/7L/3G construct (I50 = 1.2 and 0.7 vs. 2.8), Figure 3. A model similar to the one above may be advanced in this case, Figure 6D: when the H2 segment is not in itself sufficiently hydrophobic to insert as a TMH it remains on the cytoplasmic side until the H3 segment starts interacting with the translocon. At this point, the strong Nlum-Ccyt orientational preference of H3 pulls H2 into the membrane. The slow modification kinetics of the G3 glycosylation site in the LepV H3:3G/7L/5K (H2:3G/2L/3G) construct, Figure 4, is consistent with such a model. If H2 is of very low hydrophobicity (H2:3G/0L/3G), the cost of inserting it into the membrane may be too high and H3 also remains on the cytoplasmic side of the membrane, possibly forming a re-entrant loop.
It is notable that Lys residues added to the C terminus of the H3 segment in general have somewhat stronger effects on the I50 for H2 than have Lys residues added to the N terminus of H3; this is true for both the LepIV and LepV model proteins. In the case of LepIV, it thus seems difficult to “pull in” an H2 segment that has already been translocated to the lumenal side, Figure 6A, and in the case of LepV it seems difficult to “pull out” an H2 segment that has already been inserted, Figure 6C.
How common is this way of promoting the membrane insertion of a marginally hydrophobic TMH in natural proteins? While no comprehensive study has been carried out so far, some clear examples have already been found. In the human band 3 protein (a chloride-bicarbonate exchanger), the Ncyt-Clum orientated TMH7 requires the downstream TMH8 for efficient membrane insertion; notably, the Nlum-Ccyt orientational preference of TMH8 is strong enough even to pull an upstream, non-hydrophobic segment unrelated to TMH7 into the membrane (Ota et al., 1998). Another example has been found in the human P-glycoprotein (a drug efflux protein) where the marginally hydrophobic, Ncyt-Clum orientated TMH9 depends on the more hydrophobic, Nlum-Ccyt orientated TMH10 for membrane insertion (Enquist et al., 2009). As shown above, a similar situation exists in the human apical sodium-dependent bile acid transporter ASBT in which the very polar and functionally important Ncyt-Clum orientated TMH8 only inserts into the ER membrane in the presence of TMH9, which has a high hydrophobicity and is flanked by a strong orientational determinant in the form of a cluster of C-terminal positively charged residues, Figure 5. Band 3, P-glycoprotein, and ASBT represent large protein families and provide examples of the situation shown in Figure 6D.
In summary, our results show that a hydrophobic TMH with a strong orientational preference can to a significant extent control whether or not a neighboring, marginally hydrophobic segment forms a transmembrane helix. This is a clear demonstration of how distant sequence features may impact the overall topology of a membrane protein. In a different sequence context, clusters of positively charged Lys residues have been shown to be able to increase the membrane insertion efficiency of an upstream marginally hydrophobic transmembrane segment from quite far away (Fujita et al., 2010; Lerch-Bader et al., 2008), underscoring the importance of the intricate and so far poorly understood dynamics underlying translocon-mediated assembly of multispanning membrane proteins (Skach, 2009). Such effects are not yet explicitly incorporated into current topology-prediction programs (Elofsson and von Heijne, 2007), but are to some degree implicit in the use of the positive-inside rule in these algorithms.
We now know of at least four ways that nature can ensure efficient membrane insertion of marginally hydrophobic TMHs: by the addition of positively charged flanking residues to the cytoplasmic end of the TMH (Hedin et al., 2010; Hessa et al., 2007; Lerch-Bader et al., 2008), by repositioning of the TMH relative to the membrane during folding or oligomerization (Kauko et al., 2010), by specific interactions with a neighboring TMH (Buck et al., 2007; Meindl-Beinker et al., 2006; Zhang et al., 2007), and, as shown here, by reduction of the threshold hydrophobicity required for insertion imparted by a neighboring TMH with a strong orientational preference. Future work will need to address which of these are more commonly used in nature and how they may act in combination.
Experimental procedures
Enzymes and chemicals
All chemicals were from Sigma-Aldrich (St. Louis, MO, US) unless stated otherwise. [35S]-Met was from PerkinElmer (Boston, MA, US). All enzymes were from Fermentas (Burlington, Ontario, CA) except Phusion DNA polymerase that was from Finnzymes (Espoo, FI) and SP6 polymerase that was from Promega (Madison, WI, US). The QuickChange™ Site-directed Mutagenesis kit was from Stratagene (La Jolla, CA, US) and oligonucleotides were from Cybergene (Stockholm, SE) and Eurofins MWG Operon (Ebersberg, DE). Plasmid pGEM1, RNasin, nuclease treated reticulocyte lysate, TnT® Quick transcription/translation system, and deoxynucleotides were from Promega (Madison, WI, US). Gene fragments #1 and #2 (see Table S1) were from GenScript USA Inc. (Piscataway, NJ, US).
DNA manipulations
To create the basic LepV construct starting from the lepB gene in the pGEM1 vector (Hessa et al., 2005), the PCR-amplified gene fragment #1 (for complete amino acid sequences, see Table S1) was introduced replacing the corresponding lepB gene part between a newly created SmaI restriction site in codons 88-89 and a SacI restriction site immediately downstream of the lepB gene. The SacI restriction site was then removed and a new SacI site generated in LepV codons 159-160. Other restriction sites within LepV that were used are a Xho1 site in codons 130-131 flanking H1 together with the SacI site, an Apa1 site at codons 195-196 and a Mfe1 site at codons 227-228 flanking H2, a Spe1 site at codons 264-265 and a Kpn1 site at codons 293-294 flanking H3. The five glycan acceptor sites for N-linked glycosylation were located as follows: G1 in codons 96-98, G2 in codons 172-174, G3 in codons 241-243, G4 in codons 308-310, and G5 in codons 317-319 (all codon positions refer to the LepV sequence).
The same strategy was used to create the basic LepIV construct, starting from a Lep construct with a, by 24 residues extended, N-terminus carrying a 24-residue N-terminal extension that includes an Asn-Ser-Thr glycan acceptor site (Lundin et al., 2008). The PCR-amplified gene fragment #2 (for amino acid sequence see Table S1) was introduced between a SpeI restriction site in Lep codons 81-82 and a SacI restriction site immediately downstream of the Lep gene. The SacI site was then removed. Other restriction sites within LepIV that were used are a Kpn1 site in codons 110-111 flanking H1 together with the SpeI site described above, a Xho1 site in codons 194-195 flanking H2, and an Apa1 site at codons 232-233 and a Mfe1 site at codons 261-262 flanking H3. The five glycosylation sites for N-linked glycosylation were located as follows: G1 in codons 3-5, G2 in codons 122-124, G3 in codons 207-209, G4 in codons 276 -278, and G5 in codons 336-338 (all codon positions refer to the LepIV sequence).
For series 9, 10, 19, and 20 (Table S1), constructs encoding H3 segments 3X/nL/3Z with n = 9 or 11 and either (X = G, Z = K) or (X = K, Z = G), all flanked by 3′ Spe1 and 5′ Kpn1 sites in case of LepV or flanked by 3′ Apa1 and 5′ Mfe1 sites in case of LepIV, were generated by annealing two pairs of complementary oligonucleotides (18-48 nucleotides long) with overlapping overhangs followed by annealing of these pairs and ligation with the cut vector (Hessa et al., 2005; Lundin et al., 2008). Double-stranded oligonucleotides encoding all other test segment with X, Z = G were PCR-amplified from Lep-constructs described previously (Hessa et al., 2005; Hessa et al., 2007) with primers containing appropriate restriction sites for subsequent cloning and introduction into LepIV or LepV replacing H2 or H3. Conventional site-directed mutagenesis was used to replace X, Z = G flanks with X, Z = K flanks and to increase the number of lysines in the flanks from 3 to 5 by replacing the two amino acids immediately adjacent to the 3K flanks by lysine.
To generate the basic ASBT construct with the C-terminal LepV tail, the ASBT cDNA from residue 1 to 316 was PCR-amplified using a forward primer with a 5′ overhang containing a XbaI restriction site and a Kozak sequence and a reverse primer prolonged with a SmaI restriction site and subsequently introduced into empty pGEM1. The LepV tail was PCR-amplified from LepV (construct #1, residues 300 - 334) with primers containing SmaI restriction sites and ligated into SmaI digested ASBT-pGEM1. The removal of the SmaI site between the ASBT and the LepV gene parts, the replacement of the two lysines by serines in the ASBT part directly after TMH9, the replacement of the threonine and the isoleucine by lysines in the LepV tail, and the introduction of a glycan acceptor site in amino acids 266 - 268 in the ASBT part was done by conventional site-directed mutagenesis.
The identity of all constructs was confirmed by sequencing of plasmid DNA at Eurofins MWG Operon (Ebersberg, DE).
Expression in vitro
Constructs cloned in pGEM1 were transcribed and translated in the TnT® Quick coupled transcription/translation system. 150-200 ng DNA template, 1 μl [35S]-methionine (5 μCi) and 1 μl dog pancreas rough microsomes were added at the start of the reaction, and samples were incubated for 90 min at 30 °C (in some duplicate experiments the reaction volume was decreased by 50%). Translation products were analyzed by SDS-PAGE and proteins bands were visualized in a Fuji FLA-3000 phosphoimager (Fujifilm, Tokyo, JP) using the Image Reader V1.8J/Image Gauge V 3.45 software (Fujifilm). The MultiGauge (Fujifilm) software was used to generate a 2D intensity profile of each gel lane and the multi-Gaussian fit program from the Qtiplot software package (www.qtiplot.ro) was used to calculate the peak areas of the protein bands.
The fraction of proteins with n added glycans was calculated as the peak area of the n-times glycosylated protein band divided by the summed peak areas of all bands from glycosylated protein (non-glycosylated protein was not included in the calculation, as it represents chains not successfully targeted to the microsomal membrane). The glycosylation levels vary by no more than ±5% between repeated experiments (96% of all repeat measurements fall within this interval).
Measurement of glycosylation kinetics
To generate mRNA, the constructs with ID 131 and 134 (see Table S1) were transcribed using a standard SP6 polymerase transcription protocol (Monné et al., 1999) for 60 min at 37 °C. For translations, a reaction mix composed of 40.5 μl reticulocyte lysate, 4.5 μl amino acid mix minus methionine (1 mM), 4.5 μl [35S]-methionine (5 μCi), 4.5 μl dog pancreas rough microsomes, and 2 μl RNasin (40 U/μl) was incubated for 90 sec at 30 °C, whereupon translation was initiated by addition of 2.8 μg mRNA. After 4 min, 1.8 μl aurintricarboxylic acid (ATA, 3.5 mM) was added to inhibit further translation initiation. 5 μl samples were transferred to tubes containing 0.5 μl 10% Triton X100 at specific time points after ATA addition as indicated in Figure 4 and incubation was continued at 30 °C until 60 min after addition of ATA. Reference samples without dog pancreas rough microsomes were run in parallel, and all translation products were analyzed as described under Expression in vitro.
Supplementary Material
Highlights.
-
–
ER-insertion propensity of a TMH depends on the orientational bias of nearby TMHs
-
–
This helps explain the ‘missing hydrophobicity’ in polytopic membrane proteins
-
–
Membrane insertion of TMH8 in the human ASBT protein depends on TMH9
Acknowledgments
This work was supported by grants from Magnus Bergvalls Stiftelse, Henrik Granholms Stiftelse, Stiftelsen för Gamla Tjänarinnor, and the Swedish Foundation for International Cooperation in Research and Higher Education (STINT) to IMN, from the Swedish Cancer Foundation, the Swedish Research Council, the Swedish Foundation for Strategic Research, and the European Research Council (ERC-2008-AdG 232648) to GvH, and from the Sigrid Juselius Foundation and the Magnus Ehrnrooths stiftelse to KKH. We gratefully thank Prof. Dr. B. Dobberstein, Heidelberg University, for providing dog pancreas rough microsomes and Dr. David Drew, Imperial College, London, for the ASBT cDNA and for insightful comments.
References
- Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Buck TM, Wagner J, Grund S, Skach WR. A novel tripartite motif involved in aquaporin topogenesis, monomer folding and tetramerization. Nat Struct Mol Biol. 2007;14:762–769. doi: 10.1038/nsmb1275. [DOI] [PubMed] [Google Scholar]
- Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- Enquist K, Fransson M, Boekel C, Bengtsson I, Geiger K, Lang L, Pettersson A, Johansson S, von Heijne G, Nilsson I. Membrane-integration characteristics of two ABC transporters, CFTR and P-glycoprotein. J Mol Biol. 2009;387:1153–1164. doi: 10.1016/j.jmb.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Fagerberg L, Jonasson K, von Heijne G, Uhlén M, Berglund L. Prediction of the human membrane proteome. Proteomics. 2010;10:1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- Fujita H, Kida Y, Hagiwara M, Morimoto F, Sakaguchi M. Positive charges of translocating polypeptide chain retrieve an upstream marginal hydrophobic segment from the endoplasmic reticulum lumen to the translocon. Molecular biology of the cell. 2010;21:2045–2056. doi: 10.1091/mbc.E09-12-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafvelin G, Sakaguchi M, Andersson H, von Heijne G. Topological rules for membrane protein assembly in eukaryotic cells. J Biol Chem. 1997;272:6119–6127. doi: 10.1074/jbc.272.10.6119. [DOI] [PubMed] [Google Scholar]
- Gafvelin G, von Heijne G. Topological “frustration” in multi-spanning E. coli inner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- Goder V, Spiess M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 2003;22:3645–3653. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallén S, Brändén M, Dawson PA, Sachs G. Membrane insertion scanning of the human ileal sodium/bile acid co-transporter. Biochemistry. 1999;38:11379–11388. doi: 10.1021/bi990554i. [DOI] [PubMed] [Google Scholar]
- Hedin LE, Öjemalm K, Bernsel A, Hennerdal A, Illergard K, Enquist K, Kauko A, Cristobal S, von Heijne G, Lerch-Bader M, et al. Membrane insertion of marginally hydrophobic transmembrane helices depends on sequence context. J Mol Biol. 2010;396:221–229. doi: 10.1016/j.jmb.2009.11.036. [DOI] [PubMed] [Google Scholar]
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Higy M, Junne T, Spiess M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry. 2004;43:12716–12722. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- Hu NJ, Iwata S, Cameron AD, Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter, ASBT. Nature. 2011 doi: 10.1038/nature10450. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illergård K, Kauko A, Elofsson A. Why are polar residues within the membrane core evolutionary conserved? Proteins. 2011;79:79–91. doi: 10.1002/prot.22859. [DOI] [PubMed] [Google Scholar]
- Imperiali B, Hendrickson T. Asparagine-linked glycosylation: Specificity and function of oligosaccharyl transferase. Bioorg Medicinal Chem. 1995;3:1565–1578. doi: 10.1016/0968-0896(95)00142-5. [DOI] [PubMed] [Google Scholar]
- Johansson M, Nilsson I, von Heijne G. Positively charged amino acids placed next to a signal sequence block protein translocation more efficiently in Escherichia coli than in mammalian microsomes. Mol Gen Genet. 1993;239:251–256. doi: 10.1007/BF00281625. [DOI] [PubMed] [Google Scholar]
- Kauko A, Hedin LE, Thebaud E, Cristobal S, Elofsson A, von Heijne G. Repositioning of transmembrane alpha-helices during membrane protein folding. J Mol Biol. 2010;397:190–201. doi: 10.1016/j.jmb.2010.01.042. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer E. Predicting transmembrane protein topology with a hidden Markov model. Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lerch-Bader M, Lundin C, Kim H, Nilsson I, von Heijne G. Contribution of positively charged flanking residues to the insertion of transmembrane helices into the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2008;105:4127–4132. doi: 10.1073/pnas.0711580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C, Kim H, Nilsson I, White SH, von Heijne G. Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for N(in)-C(out) and N(out)-C(in) transmembrane helices. Proc Natl Acad Sci U S A. 2008;105:15702–15707. doi: 10.1073/pnas.0804842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl-Beinker NM, Lundin C, Nilsson I, White SH, von Heijne G. Asn- and Asp-mediated interactions between transmembrane helices during translocon-mediated membrane protein assembly. EMBO Rep. 2006;7:1111–1116. doi: 10.1038/sj.embor.7400818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monné M, Gafvelin G, Nilsson R, von Heijne G. N-tail translocation in a eukaryotic polytopic membrane protein - Synergy between neighboring transmembrane segments. Eur J Biochem. 1999;263:264–269. doi: 10.1046/j.1432-1327.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- Nilsson I, von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J Biol Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- Ota K, Sakaguchi M, von Heijne G, Hamasaki N, Mihara K. Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol Cell. 1998;2:495–503. doi: 10.1016/s1097-2765(00)80149-5. [DOI] [PubMed] [Google Scholar]
- Popov M, Tam LY, Li J, Reithmeier RAF. Mapping the ends of transmembrane segments in a polytopic membrane protein - Scanning N-glycosylation mutagenesis of extracytosolic loops in the anion exchanger, Band 3. J Biol Chem. 1997;272:18325–18332. doi: 10.1074/jbc.272.29.18325. [DOI] [PubMed] [Google Scholar]
- Rothman J, Lodish H. Synchronised transmembrane insertion and glycosylation of a nascent membrane protein. Nature. 1977;269:775–780. doi: 10.1038/269775a0. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M, Tomiyoshi R, Kuroiwa T, Mihara K, Omura T. Functions of signal and signal-anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc Natl Acad Sci USA. 1992;89:16–19. doi: 10.1073/pnas.89.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä S, Slusky JS, Lloris-Garcera P, Rapp M, von Heijne G. Control of membrane protein topology by a single C-terminal residue. Science. 2010;328:1698–1700. doi: 10.1126/science.1188950. [DOI] [PubMed] [Google Scholar]
- Skach WR. Cellular mechanisms of membrane protein folding. Nat Struct Mol Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- Wahlberg JM, Spiess M. Multiple determinants direct the orientation of signal-anchor proteins: the topogenic role of the hydrophobic signal domain. J Cell Biol. 1997;137:555–562. doi: 10.1083/jcb.137.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- Zhang EY, Phelps MA, Banerjee A, Khantwal CM, Chang C, Helsper F, Swaan PW. Topology scanning and putative three-dimensional structure of the extracellular binding domains of the apical sodium-dependent bile acid transporter (SLC10A2) Biochemistry. 2004;43:11380–11392. doi: 10.1021/bi049270a. [DOI] [PubMed] [Google Scholar]
- Zhang L, Sato Y, Hessa T, von Heijne G, Lee JK, Kodama I, Sakaguchi M, Uozumi N. Contribution of hydrophobic and electrostatic interactions to the membrane integration of the Shaker K+ channel voltage sensor domain. Proc Natl Acad Sci U S A. 2007;104:8263–8268. doi: 10.1073/pnas.0611007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.