Abstract
In order to improve therapeutic outcomes, there is a tremendous need to identify patients who are likely to respond to a given asthma treatment. Pharmacogenomic studies have explained a portion of the variability in drug response and provided an increasing list of candidate genes and SNPs. However, as phenotypic variation arises from a network of complex interactions among genetic and environmental factors, rather than individual genes or SNPs, a multidisciplinary, systems-level approach is required in order to understand the inter-relationships among these factors. Systems biology, which seeks to capture interactions between genetic factors and other variables, offers a promising approach to improved therapeutic outcomes in asthma. This aritcle will review and update progress in the pharmacogenomics of asthma and then discuss the application of systems biology approaches to asthma pharmacogenomics.
Keywords: asthma, genes, GWAS, network medicine, pharmacogenomics, SNP, systems biology
Asthma is a common inflammatory airway disease that is characterized by chronic inflammation, reversible airway obstruction, broncho-spasm and heterogeneous phenotypes. Asthma affects more than 300 million people worldwide and represents a significant public health burden due to its high prevalence (approaching 7–9% in children and adults in developed countries) [201]. Asthma symptoms are treated predominantly through three classes of prescription medications: inhaled short- and long-acting β2 agonists, inhaled and oral corticosteroids and leukotriene antagonists. While these drugs show efficacy in a majority of patients, they are not effective for all patients and also demonstrate significant, reproducible, interindividual variability in therapeutic response [1]. Identifying individuals who are most and least likely to benefit from a given asthma treatment is necessary to improve therapeutic outcomes.
Asthma is a complex disease that is likely the result of multiple gene–gene and gene–environment interactions [2,3]. Genome-wide association studies (GWAS) and candidate-gene studies have attempted to resolve some of this complexity through seeking to identify genes that underlie asthma pathology.
Asthma susceptibility genes identified through GWAS include genes related to both TH2 and non-TH2 immune cellular processes, including T-cell response and differentiation (IL6R [4], DENND1B [5], LRRC32 [6], IL2RB [7], HLA-DQ [7]), recruitment or activation of inflammatory cells (IL1RL1 [8], TSLP [7] and IL33 [7]), cAMP and cell-signaling modulation (PDE4D [9], SMAD3 [7] and ORMDL3 [10]) and apoptosis (GSDMB [7]). Although these loci are important regulators of asthma pathogenesis and may be therapeutic targets, they have not been associated with treatment response in patients. Pharmacogenomic approaches, which investigate the effect of genetic variation on treatment response or treatment-related events, are promising for improving therapeutic targeting in patients. Candidate gene studies and GWAS have identified multiple genes involved in asthma drug response: ADRB2 [11–13], ARG1 [14], ALOX5 [15–18], CYSLTR2 [19], CRHR1 [20], DUSP1 [21], FCER2 [22,23], GLCCI1 [24], GSNOR [25], LTC4S [18,26–29], ABCC1 [17,30], NK2R [31], SLCO2B1 [32], STIP1 [33], TBX21 [20] and others. While these studies have successfully characterized many genes that may explain a proportion of the interindividual variability in patient treatment response, the majority of the heritability of therapeutic response remains unaccounted for [34]. Traditional pharmacogenomic studies have evaluated the effects of single SNPs or genes using genetic models that evaluate individual variables, none of which can individually predict the phenotype. The variability observed in asthma phenotypes is likely to arise from the coordinated effects of multiple genes, pathways and environmental factors. Systems biology seeks to investigate the relationship among these pathways and related factors in order to understand how these relationships impact health and disease [35].
The goal of a systems biology approach is to create a model of the meaningful interactions within a network that best reflects the underlying biology [36–38]. Systems biology is particularly applicable to pharmacogenomic studies, where complex genetic factors contribute to the observed variability in therapeutic response in patients. In addition, this approach can be useful for profiling large, well-characterized asthma cohorts in an effort to improve understanding of asthma phenotypes. Integration of `omics' data with multiple levels of biological, phenotypic and clinical information can then be used to develop predictive models of asthma treatment response. From these models, working hypotheses of the mechanisms of asthma treatment response can be formulated and tested in clinical trials or in cell-based and animal models. The individual components of the model itself, which is represented as a probabilistic graphical network, can also be targeted at nodal points or clusters in order to select potential drug targets or pathways for intervention.
The purpose of this review is to first provide an update of recent progress in the pharmacogenomics GWAS and candidate-gene studies in asthma, and second to discuss the major applications of integrated systems biology approaches to asthma pharmacogenomics.
Progress in the pharmacogenomics of asthma
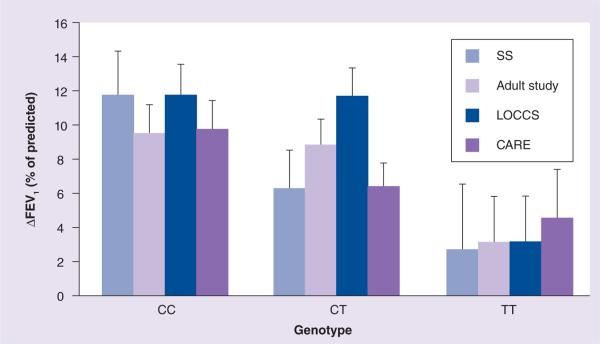
In 2011, the authors completed the first GWAS of asthma treatment response [24]. We genotyped 935 asthmatic participants from one asthma clinical trial: CAMP [39,40], and three replication cohorts, the SOCS [41] and SLIC [42] trials, the Adult Study [43] and the LOCCS [44] trial, with the goal of identifying novel variants associated with response to inhaled glucocorticoids (GCs). To identify markers with the greatest positive association with the primary-outcome phenotype of change in forced expiratory volume in 1 s (ΔFEV1) from baseline during the first 16 months of budesonide therapy, 534,290 SNPs from an initial cohort of 403 CAMP trios (children and their parents) were first screened using a powerful family-based screening algorithm [45], which applies parental genotype information in order to rank the top 100 SNPs with the highest statistical associations [46]. This pharmacogenomic GWAS identified a candidate SNP (rs37972) in a GC pathway gene associated with patient response to inhaled corticosteroids (ICS). This SNP, present in the promoter region of the GLCCI1 gene, was significantly associated with the post-ICS FEV1 change in three of the four replication cohorts (p = 0.0085) (Figure 1). This SNP was in complete linkage disequilibrium (i.e., perfectly correlated) with a functional variant that was experimentally confirmed to reduce both gene expression and pharmacological response to ICS, and which may account for a substantial proportion of poor responders to ICS. A strength of this study was the use of the family-based screening method, which is ideally suited for studies with small sample sizes and limited power; however, the study has important limitations. First, the inclusion of only white subjects limits the generalizability of these findings. Second, the study design required that only the top 100-ranked SNPs be investigated, precluding testing of the majority of SNPs in the GWAS that may also play a role in ICS response. Although the identified SNP had functional consequences on GLCCI1 expression, the presence of unidentified functional variants may also account for these results. Additional mechanistic studies would clarify the role of this (and potentially other) SNPs in relation to ICS response and GLCCI1 function.
Figure 1. GLCCI1 rs37972 SNP genotype predicts inhaled corticosteroid treatment response in asthma patient cohorts.
The rs37972 mutant (T) allele was significantly associated with poorer therapeutic response (measured as the mean ± standard error ΔFEV1, expressed as the percentage of the predicted value), after 4–8 weeks of therapy with inhaled glucocorticoids in four study groups: SOCS and SLIC trials (SS; n = 264), the Adult Study (n = 385), the LOCCS trial (n = 185) and the CARE network trials (n = 101). CC represents the homozygous reference genotype; CT represents the heterozygous genotype; TT represents the variant genotype.
ΔFEV1: Forced expiratory volume in 1 s.
Reproduced with permission from [24] © 2011 Massachusetts Medical Society.
Candidate-gene studies
Several asthma candidate-gene studies have been performed to identify genetic loci associated with therapeutic response to the three major classes of asthma therapy: β2-adrenergic receptor agonists (salmeterol and salbutamol), leukotriene modifiers (LTMs; montelukast and zileuton) and ICS (prednisolone and beclomethasone) [16,20,22,47–49]. For comprehensive reviews of these drug classes, see references [50–54].
The most commonly prescribed drugs for treatment of mild to moderate asthma are β2-agonists, which target ADRB2, a member of the seven-transmembrane, G-protein-coupled receptor family. Bronchodilator response (BDR) to β2-agonists is commonly used as a measure of pharmacological response to β2-agonist medications. The ADRB2 gene consists of a single exon, yet is highly polymorphic and at least 49 SNPs in this gene have been identified and validated [11]. The best studied ADRB2 SNP is a nonsynonymous variant at position 16, encoding for an arginine to glycine amino acid change, Arg16Gly, which decreases ADRB2 gene expression and is clinically and pharmacologically associated with BDR [55]. However, results of these pharmacogenetic studies have been mixed. The earliest investigation of the pharmacogenetics of β2-agonists showed that Arg16 wild-type carriers were more likely to show a positive response to short-acting β2-agonists than subjects who were heterozygous for Arg16Gly, and also had higher predicted values for FEV1 [55]. By contrast, other studies report no significant effect of Arg16 genotype [56–58]. This discrepancy may be attributed to diverse allele frequencies of Arg16Gly in different ethnic groups within the clinical study populations, haplotype effects, as well as differences in environmental and related factors or study design [55–59].
The β2-adrenergic receptor response is highly complex and involves multiple genes; for a detailed review of this pathway, see references [51,55,60]. For this reason, a major recent focus of more recent candidate-gene studies has been to clarify the role of other genes related to the ADRB2 pathway [61] and how these genes may interact with one another (i.e., gene–gene interactions). Through molecular genetic studies, Tantisira and colleagues first identified AC9 as a potential candidate gene for therapeutic response to β2-agonists [62]. In a large clinical asthmatic cohort, the AC9 Ile772Met SNP was associated with a significant upregulation of ICS-specific response, which was also confirmed in functional assays using AC9-transfected human lung cells [62]. Recently, in a Korean cohort of 86 asthmatics, Kim et al. reported significant associations of the two previously reported AC9 variants, the AC9 Ile772Met and AC9 5′-UTR 150397, with two measures of lung function (ΔFEV1% and change in maximum mid-expiratory flow) following 8 weeks of inhaled combination therapy with budesonide and formoterol [63]. Notably, in patients with one or more variant alleles of AC9 Ile772Met and ADRB2 Arg16Gly polymorphisms, an additive effect on BDR was observed, suggesting that the response to combined treatment of ICS and β2-agonists can be mediated by the ADRB2-AC9 gene–gene interaction in addition to AC9 alone [63].
Other candidate genes for β2-agonist response that have been studied to date include: ARG1 [49], GSNOR [25,64], GCLC [65], CPS1 [25] and THRB [66]. Many of these loci were investigated using gene–gene interaction studies [63]. Various SNPs, including those in linkage disequilibrium with the promoter SNP rs2781659 in ARG1, have been significantly associated with BDR in two separate candidate-gene studies in large asthmatic cohorts [14,49]. In a study of 221 asthmatics, ARG2 SNPs were also associated with BDR, suggesting that both genes are involved in therapeutic response [49]. The improvement of BDR observed in carriers of the ARG1 promoter haplotypes is believed to be due to increased transcription of ARG1, which may promote BDR by reducing bronchial hyper-responsiveness [67,68].
In an African–American cohort, SNPs in several candidate genes – ADRB2, ADCY9, GSNOR, GCLC and CPS1 – were evaluated for their joint association with asthma severity and BDR [25]. In the gene–gene interaction analysis, a promoter SNP in GSNOR (rs1154400) that was associated with acute BDR also interacted with ADRB2 Arg16Gly and ADRB2 Gly27Glu [25]. By multifactor dimensionality reduction prediction method, a multilocus model that included ADRB2 Arg16Gly, ADRB2 Gly27Glu, GSNOR (rs1154400) and CPS1 (rs2230739), was able to predict BDR with an accuracy of 70% [25]. These findings suggest that BDR to β2-adrenergic receptor agonists is dependent upon multiple genes in diverse biological pathways of airway response. In a family-based candidate gene study of 609 Puerto Rican and Mexican asthmatic trios, four SNPs within GSNOR, including a promoter SNP haplotype associated with increased GSNOR transcription, were associated with increased susceptibility to asthma and decreased BDR [64]. A subsequent gene–gene interaction study of the variants within GSNOR and ADRB2 identified significant association of the GSNOR promoter SNP variant allele, ADRB2 Arg16Gly and ADRB2 Gly27Glu genotypes with lower BDR compared with the wild-type carriers in both cohorts [64]. These findings suggest that individuals with the combined genotypes may be at increased risk for poor therapeutic outcomes to β2-agonist treatment. In a recent candidate gene screen, Duan et al. evaluated the association of BDR with 1116 SNPs in 98 genes encoding 59 transcription factors with BDR in an initial asthma cohort (n = 403) and three replication cohorts receiving β2-agonists [66]. The authors identified multiple SNPs that were modestly associated with BDR [66]. An intergenic SNP (rs892940) near the THRB gene was significantly associated with response to β2AR agonists in the initial population and also replicated in the other populations [66]. This locus represents a novel candidate for regulation of β2AR response. Together, these studies demonstrate significant progress in recent efforts to unravel multilocus interactions in the β2-agonist pathway.
5-Lipoxygenase (5-LO) mediates the production of leukotrienes, which are inflammatory mediators generated from arachidonic acid that contribute to a variety of disease states, including asthma. LTMs such as zileuton, montelukast, pranlukast and zafirlukast ameliorate inflammation-induced asthma symptoms by either selectively inhibiting leukotriene production from arachidonic acid (zileuton) or preventing leukotrienes from binding to CYSLTR1, the major leukotriene receptor (montelukast, pranlukast and zafirlukast). For an overview of this molecular pathway, see reviews [17,69,70].
There is significant heterogeneity in response to LTMs, and a genetic basis for this variability was established with the first studies that associated a functional Sp1 tandem repeat promoter variation within the 5-LO-encoding gene (ALOX5) with treatment outcomes [15,16]. Candidate genes involved in the 5-LO pathway include: LTC4S [15,16], CYSLTR1 [18,71], CYSLTR2 [15,16], ABCC1 [15,16], LTA4H [72–74] and SLC02B1 [15,16]. However, in nearly all cases, replication of identified associations has been difficult, and the extent of contribution of the individual genes to LTM response is unclear. For ALOX5, three intronic SNPs with unknown function have demonstrated associations with ΔFEV1 after zileuton (n = 577 asthmatics) [17] and montelukast (n = 252 asthmatics) [18] therapy. Two additional SNPs (rs4987105, Thr120Thr and rs4986832, 5′-UTR) were associated with improved response to montelukast (n = 147 asthmatics) [19]. These associations have not yet been validated, and their functional significance has not been determined. The LTC4S gene promoter polymorphism (A-444C) was associated with improved response to LTMs; however, consistent reproduction of these results has been problematic [19]. Tantisira et al. showed that an intronic LTC4S variant, rs272431, was associated with improvement in FEV1 following zileuton treatment [17]. While no variants of CYSLT1 have been significantly associated with LTM response, two variants in the 3′-UTR of CYSLT2 were associated with an increase in morning peak expiratory flow in 174 asthmatics taking montelukast [18,19].
Recent studies identified genetic determinants of LTM response and plasma levels in transporter genes SLCO2B1 and MRP1 [17,30,32]. Multiple SNPs within MRP1 have been associated with differential responses to montelukast [30] and zileuton [17] treatment; however, the molecular mechanisms of these variants have not yet been determined. The SLCO2B1 gene encodes OAT2B1, which transports montelukast in a concentration-dependent manner and is a probable determinant of montelukast pharmacokinetics [32]. Recently, Mougey et al. identified a nonsynonymous SLCO2B1 SNP, rs12422149 (Arg312Gln), associated with reduced montelukast plasma concentrations during a 1-month or 6-month treatment regimen [32]. In 80 asthma patients who were genotyped for Arg312Gln, heterozygotes had approximately 20% lower montelukast plasma levels at 1 month, and approximately 30% lower concentrations at 6 months, compared with the wild-type carriers [32]. Furthermore, scores for the Asthma Symptom Utility Index (the primary clinical phenotype) showed that Arg312Gln heterozygotes did not show improvement in their symptom scores [32]. These findings suggest that, in carriers of the Arg312Gln genotype, lower systemic exposure to montelukast and a corresponding decrease in pharmacodynamic response are likely to be a result of impaired OAT2B1-mediated intestinal absorption. Screening individuals for the SCLO2B1 Arg312Gln variant (which is present at minor allele frequencies of approximately 20% in the general population) prior to montelukast adminstration may therefore be useful for identifying patients who are unlikely to respond. However, replication of these studies in larger cohorts is necessary in order to make clinical inferences about these results.
GCs initiate their therapeutic effects by forming a complex with the intracellular GC receptor (GR), which then translocates to the nucleus to modulate transcription of inflammatory genes. For a review of this pharmacological pathway in asthma treatment response, see references [50,69]. ICS and oral corticosteroids (e.g., budesonide, prednisolone and beclomethasone) are among the most highly effective drugs for asthma treatment. However, response to these drugs shows significant and repeatable interindividual variability, suggesting that genetic mechanisms may play a role in therapeutic response [50,69].
Efforts to discern the genetics of corticosteroid response in asthma have focused on candidate genes involved in the corticosteroid pathway. In addition to NR3C1, which encodes the GR, other genes were identified including CRHR1 [50,69], TBX21 [50,69], FCER2 [50,69], DUSP1 [50,69], STIP1 [50,69] and NK2R [50,69]. Several studies have reported the NR3C1 gene to be associated with asthma, some of which have demonstrated functional consequences for GC sensitivity [19,75–79]. Recently, Panek and colleagues established that a coding SNP in NR3C1 is associated with moderate/severe bronchial asthma [78]. However, whether this SNP affects GC response via the GR is unknown [78]. An additional SNP encoding a valine to aspartate change at amino acid 641 was shown to alter the affinity of dexamethasone binding to the GR [78]. Molecular genetic studies of GR would benefit asthma pharmacogenetic studies; however, the majority of variants identified in NR3C1 are rare and remain unvalidated.
In addition to the GLCCI1 GWAS, candidate gene SNPs that have been significantly associated with changes in lung function in response to ICS (%ΔFEV1) in larger asthmatic cohorts (n > 400 patients) have been identified in the following genes: CRHR1 (rs242941) and DUSP1 (rs881152) [21] In a candidate gene analysis of a smaller cohort (n = 382 asthma patients), multiple SNPs in STIP1 (rs4980524, rs6591838 and rs2236647) were also associated with variable response to ICS [78]. Additional primary outcome measures investigated in GC candidate-gene studies include airway responsiveness (4-year change) [20], improved asthma control [31], exacerbations [22,23] and plasma IgE levels [22]. In these studies, significant SNP associations with post-ICS airway responsiveness were observed for TBX21 (rs224001) [47]. Single SNP associations with improved asthma control were identified in NK2R (rs7703891) [78] and DUSP1 (rs881152) [21]. In three separate asthma cohorts, an intronic SNP within FCER2 (rs28364072) was associated with increased risk of severe asthma exacerbations while on ICS [78]. The minor SNP allele was also associated with elevated IgE levels, hospital visits and poor asthma control in the replication cohorts [22].
The loss of regulatory control of GC response in asthma creates chronic inflammation, which worsens lung function and leads to exacerbations and poor asthma control despite ICS use. Understanding the functional genotype–phenotype relationships of these clinical symptoms, which are controlled by complex interactions among GC pathway genes, requires additional investigation in efforts to improve quality of life and therapeutic outcomes for patients taking corticosteroids.
In the previous sections of this review, we discussed the most relevant recent findings from asthma pharmacogenomics studies of candidate genes and GWAS, with particular attention to studies with well-powered cohorts and associations of gene–gene and gene–environment interactions. In the subsequent sections, we will discuss how pharmacogenomic associations can be applied toward improved therapeutic outcomes in asthma using systems biology and integrative genomics approaches.
Introduction to systems biology in pharmacogenomics
Despite the existence of numerous pharmacogenomic SNP associations, very few SNPs can explain more than a small percentage of the variability in a pharmacogenomic clinical outcome. Additionally, there is solid evidence of a genetic relationship between treatment outcomes and asthma phenotypes that clearly involves diverse pathways and multiple genes. Recent advances in genomics and other `omics' now provide an unbiased, systems-based approach to investigate both genetic and environmental factors that have direct and indirect effects on gene expression, phenotype and therapeutic outcomes. As a result of these advances, we are now poised to identify relevant biomarkers and design more rational, targeted therapy for inflammatory lung diseases [80].
The role of systems biology in pharmacogenomics is to construct the best model of a biological network from cellular, genomic, proteomic and bioinformatics data to more accurately predict clinical drug response. A systems biology approach generally proceeds in the following manner: data collected from multiple sources are used to generate modeling algorithms that can interpret large data sets and link phenotypic information to genetic, regulatory or protein networks; through iteration and refinement of the original model and testing for accuracy, a final model is obtained, which is used to both predict behavior of the network of interest, and allow the network to be perturbed and manipulated in order to accommodate novel information. For a comprehensive overview of systems biology in pharmacogenomics, see reviews [36,38].
Systems biology in asthma pharmacogenomics
Methodological approaches in systems biology incorporate data mining and network-reconstruction methods with machine-learning algorithms to generate graphical models to infer relationships between the network components. Machine-learning algorithms apply a priori information from known biological pathways to construct representative model networks. For a comprehensive review of machine learning, see reference [81]. Classification-based algorithms, which are valuable in pharmacogenomic analyses for their ability to identify predictive relationships among the variables and generate descriptive models that capture the biological relationships, include decision trees (e.g., Random Forests) [82,83] and representation learning (e.g., cluster analysis and principal components analysis) [84]. Network-based descriptive modeling algorithms, for example, Bayesian networks, allow reverse engineering and graphical visualization of biological networks from data [85–87]. This approach is the most common method used in systems biology data analysis of pharmacogenomic studies [37]. Due to the recent implementation of systems biology toward asthma pharmacogenomic studies, some of the examples to follow relate specifically to asthma rather than asthma treatment response. However, given that asthma is a disease characterized by airways inflammation and reversible airflow obstruction (which the two major categories of asthma therapy target), the findings reported in these studies may be relevant to asthma pharmacogenetics as well.
Bayesian networks
Bayesian networks are useful for descriptive and predictive modeling, and have been used for reconstruction of genetic pathways and cellular networks [87–89]. Bayesian networks are machine-learning algorithms that can be represented by directed acyclic graphs containing nodes (i.e., variables with conditional probability distributions, e.g., genes) and edges (i.e., causal relationships/dependency among the variables, e.g., regulatory relationships among the genes) [85,87]. Bayesian networks and their components are learned directly from the data, and can account for simultaneous associations and interactions among variables. For example, for a known Bayesian network modeling a gene regulatory network, a given node represents a subset of genetic interactions, and inferences about the likely values of other genes in the network can be calculated as a product of relevant conditional probability distributions [87]. Of particular note, the modular nature of Bayesian networks makes them ideal for analysis of large association studies [88,89]. Limitations of Bayesian networks include the choice of variables to include in the model (candidate genes or SNPs), the number of variables to add (e.g., sample size) and the classification or estimation of phenotype, which are often continuous.
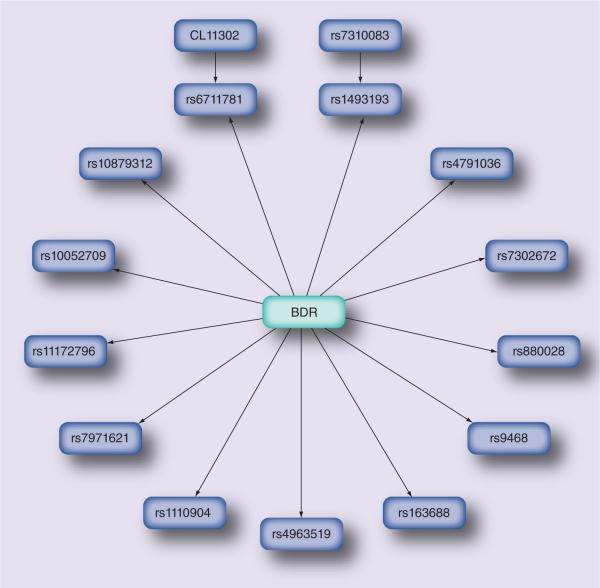
Previously, Bayesian network analysis identified associations of innate immune response genes with asthma and eczema [90], predicted clinical factors that modulate risk of progression to chronic obstructive pulmonary disease based on patient data from electronic medical records [91] and predicted asthma exacerbations [92,93]. Recently, Himes et al. investigated the predictive validity of Bayesian networks in asthma pharmacogenomics using a candidate gene approach to generate a predictive network of SNPs associated with BDR [94]. The Bayesian model was applied to pharmacogenomic data from the CAMP clinical trial to identify a set of genetic predictors of drug response [94]. A set of 254 candidate genes, based on previously published associations with asthma- and therapeutic response-related phenotypes, were genotyped to obtain a subset of 426 associated SNPs, and genetic case–control analysis was conducted to measure the association of these SNPs to BDR [94]. From these data, the authors constructed a Bayesian network of SNPs with the highest likelihood of predicting BDR, which was learned directly from the genetic data (Figure 2) [94]. The Bayesian network model was better at predicting BDR, in comparison with single SNPs or regression models [94]. This approach identified 3.5% of SNPs (15/426) within 5.4% (15/254) of candidate genes that were predictive of BDR [94]. The model showed reasonable predictive accuracy (area under the receiver operating characteristic curve of 0.75) as determined by fivefold cross-validation [94]. When compared with a single-gene approach, the model showed superior predictive accuracy; however, the model was limited by the small sample size (which reduces power to predict genetic effects), the definition of the bronchodilator-response phenotype, the selection of SNPs and candidate genes, and the lack of independent (external) validation [94].
Figure 2. Bayesian network model of SNPs predictive of bronchodilator response.
A Bayesian network, which was learned directly from the data, shows 15 SNPs from 15 candidate genes that are significant predictors of BDR.
BDR: Bronchodilator response.
This study demonstrates both the beneficial applications of Bayesian networks in pharmacogenomics as well as some important limitations. While Bayesian networks automatically learn complex models and can be successfully applied as predictive models of therapeutic response, their success is dependent upon having sufficient power to detect genetic effects and the definition of phenotypes used to build the model [87]. Furthermore, when the number of genes or SNPs greatly outnumbers the sample size, estimation of the model becomes difficult. Nevertheless, Bayesian networks represent a promising approach to pharmacogenomic studies as they provide multivariate networks of SNP–phenotype associations, and are capable of automatically learning complex models and accurately predicting phenotypic response.
Random Forests
Random Forests are randomized decision tree algorithms where each tree makes a decision and votes for the final prediction outcome [83,95–98]. As Random Forests can contain from 500,000 to 1 million variables and can repeatedly evaluate multiple combinations of variables without overfitting, these algorithms are well suited to analyzing GWAS data [95,96]. Importantly, Random Forest models can effectively extract and evaluate information from small numbers of samples, making them a useful option for analyses of pharmacogenomic studies, which are typically constrained by small sample sizes [98].
Random Forests have previously been used to identify susceptibility loci in asthma [99,100]. Xu et al. recently developed a predictive model of asthma exacerbations and clinical attributes including bronchodilator treatment using the Random Forests algorithm [101]. Using GWAS data from 417 Caucasian participants (parent–child trios) in the CAMP study, the authors applied a multistep Random Forests model to rank and select SNPs as predictors of asthma exacerbations [101]. For multiple clinical attributes (e.g., age, prebronchodilator FEV1% and treatment group) and 160 predictor SNPs, which were selected based on Random Forests importance scores, the model predicted severe exacerbations with reasonable success [101]. The authors identified a SNP (rs10496476) in the gene DPP10, which was previously associated with asthma [102], while the remaining SNPs had not previously been reported, suggesting that these SNPs were novel asthma-related associations. The identification of novel SNP–asthma associations by Random Forests may be partially due to the properties of the Random Forests algorithm, which, in contrast to conventional statistical methods for GWAS analysis that only evaluate single-SNP associations, simultaneously evaluates multiple SNPs with additive and interactive effects in the context of multiple clinical factors [101]. The Random Forests approach employed by this investigation underscores the likelihood of complex traits such as asthma exacerbations to be due to a large number of genetic and environmental factors. These results are encouraging examples of how Random Forests methods will be valuable approaches for prediction of complex traits, such as genotype–phenotype relationships in drug response.
Combined methods
Genome-wide gene-expression profiling through microarray analysis and next-generation sequencing provides important information about biological processes by identifying groups of coexpressed genes. Characterization of gene signatures is traditionally inferred through cluster analysis and correlation procedures, which alone are not meant to infer higher-order relationships among genes and gene networks [103]. By combining clustering methods with graphical machine-learning algorithms, subsets of coregulated gene modules that reflect disease states or complex phenotypes can be obtained [104–106]. These modules can then be individually analyzed, for example, to identify novel targets for modulating clinical outcomes, using a variety of integrated statistical and bioinformatic approaches.
Classification algorithms such as weighted voting classifiers have been applied to predictive modeling of asthma drug response phenotypes [107]. To search for genes that accurately predict responders and nonresponders to GC treatment, microarray gene-expression profiles from peripheral blood mononuclear cells from GC-sensitive and GC-resistant asthma patients were examined [106]. The analysis identified 923 genes that were significantly up- or down-regulated in response to treatment with cytokines in vitro and that had reversed expression in the GC responders after treatment with GC [106]. To identify genes predictive of GC response, the most extreme subsets of GC responders and GC-resistant cohorts were split into equally sized training and test sets, and a weighted voting algorithm was applied [106]. At baseline, the GC-response phenotype could be predicted with 60% accuracy. When profiles from the most extreme phenotypes were evaluated, a resulting subset of 15 genes predicted patient phenotypes with an accuracy of 84%. By reverse transcription PCR validation, 11 of the 15 genes could predict a GC response phenotype from an independent asthma cohort with 84% accuracy [106]. Among these genes, NF-κB could independently predict GC response with an accuracy of 81.25% [106]. These findings suggest that a few GC-response genes can predict response to GC in asthmatics with high accuracy. However, as the authors trained their algorithm on extreme phenotypes, the GC-resistant study population consisted of individuals who were truly steroid resistant, a situation which affects only a small minority of asthmatics (0.5%).
Module networks analysis is a machine-learning algorithm that incorporates graphical models with clustering and regression methods [104–106]. This method was developed to identify regulatory gene modules and coregulated genes, using gene expression data and a priori biological information [106]. The algorithm requires an input gene-expression data set (e.g., DNA microarray data set) and a list of candidate regulatory genes (e.g., transcription factors). The procedure applies an expectation maximization algorithm to iteratively search and partition genes into modules with associated regression trees specifying a set of genes that regulate the other genes or processes within a module. Given a set of modules and regression trees, the algorithm first learns the best regression tree for each module and then determines the module whose associated regression tree best predicts the behavior for each gene, repeating this procedure until convergence is reached. The resulting output, a list of modules and their associated regulation programs, may then be evaluated for biological significance by database annotation. These annotated modules can then be further analyzed to infer functionally related genes and gene networks, and to derive testable hypotheses, thereby providing a powerful means to generate new insights into complex regulatory processes.
Using publicly available microarray expression data, Novershtern et al. performed module networks analysis to identify significantly coregulated gene modules and potential regulatory networks within these modules, by projecting the discovered modules to annotated protein interaction networks using the Ingenuity Knowledge Base [108]. A complete map of all network interactions is publicly available online at the interactive AsthmaMap website [108,202]. The authors identified four distinct responses to asthma treatment, defined by early response, general induction, repression and dependence on IL13 induction [108]. Modeling of IL13-dependent response using protein interaction networks identified a key regulatory gene for IL13-dependent therapeutic responses in allergic lung inflammation, TGF-β1 (Figure 3) [108]. This effort underscores the utility of using complementary analytic approaches to identify novel regulatory pathways that are only apparent when multiple levels of information are simultaneously analyzed.
Figure 3. Module network analysis of a treatment-dependent protein interaction network.
Ingenuity pathway analysis of a treatment response-induced, IL13-dependent cluster identified key regulatory genes TGF-β1 and JUNB. Eight of the network proteins were regulated by TGF-β1, which in turn is activated or induced by THBS1, MMP14 and IL13 in a positive-feedback loop. Proteins are represented by nodes, and edges indicate all interactions other than coexpression. Nodes of dark blue indicate IL13-dependent expression (i.e., genes that are induced by treatment only in the presence of IL13).
Reproduced with permission from [108] © 2008, American Thoracic Society.
A systems biology perspective of asthma phenotypes
A goal of systems biology is to integrate the relevant clinical and pharmacogenomic phenotypes with genomic and expression information to explore the relationships among genes and to identify disease susceptibility loci. Because pharmacogenomic studies are designed to relate drug response (i.e., phenotype) to genotype, the definition of accurate and repeatable phenotypes is crucial in order to avoid bias. While tremendous effort has been exerted toward identifying the genotypic basis for variation in asthma phenotypes, comparably little effort has focused on accurately defining phenotypic features and integrating these efforts toward discerning environmental influences on phenotypes. In genetic studies of complex traits such as asthma, misclassification of phenotypes through measurement errors and poor phenotypic definition can bias the observed relative risk or relative odds of response. These errors have contributed to the lack of reproducibility of results obtained through large-scale genomic studies. Due to the differing levels of asthma severity, diversity and complexity of emerging phenotypic definitions of asthma, researchers have called for the re-evaluation of asthma, not as a single disease, but as an entity of multiple diverse phenotypes [6,109].
Phenotypic definitions in asthma pharmacogenomics have traditionally been categorized into two functional response classes. The first class comprises diverse factors that directly determine drug pharmacokinetics and pharmacodynamics; that is, `drug response', and the second class reflects factors that influence disease outcome, symptoms, therapeutic effects and disease pathogenesis; that is, `target response' (reviewed in [110]). The two functional-response classes can be further divided into phenotypic subcategories based on the effects of genetic variability and environmental perturbation on the pharmacological properties of a drug (Figure 4). These phenotypic categories include pharmacokinetic and pharmacodynamic response, idiosyncratic drug reactions and the natural history of the disease (Figure 4). The majority of pharmacogenomic studies to date have focused on pharmacokinetic phenotypes. This is primarily due to the direct relationship between drug levels in vivo and genetic variation in the metabolic enzymes and transporters that govern the absorption, disposition and elimination of a specific drug. By contrast, fewer associations with pharmacodynamic effects and idiosyncratic drug reaction associations have been identified, which reflects the complexity of most drug target and hypersensitivity-response pathways and the reduced power to detect true differences in response pathways versus single-drug targets.
Figure 4. Pharmacogenetic categories of asthma phenotypes.
Combined effects of genetic and environmental variation categorically affect drug and target response in four distinct ways: pharmacokinetics (therapeutic concentrations at the target site), pharmacodynamics (drug–target interactions), idiosyncratic drug reactions (immune hypersensitivity and drug–drug interactions) and genetic variation related to the natural history of the disease (pathogenesis and symptomatic response).
IDR: Idiosyncratic drug reaction; NHD: Natural history of the disease; PD: Pharmacodynamic; PK: Pharmacokinetic.
As previously discussed in this review, many recent large-scale genomic studies have identified candidate genes and pathways involved in symptomatic response and disease progression of asthma. Most of these studies have focused on early-onset or childhood clinical asthma phenotypes, such as asthma severity, quantitative measures of lung function and exacerbations (e.g., hospitalizations and emergency room visits; reviewed in [111]). Childhood asthma is believed to be a result of TH2-type immune responses linked to numerous additional covariate phenotypes, many of which have not been fully characterized (reviewed in [6]). However, recent progress in the characterization of asthma phenotypes suggests up to half of mild-to-moderate adult-onset asthmatics show little evidence of TH2 processes (reviewed in [6]). From a natural history of disease perspective, non-TH2 asthma may represent a subgroup of patients who are more likely to be poor responders to corticosteroid therapy, probably due to the lack or suppression of TH2 pathway response (reviewed in [6]).
Due to the overlap of genetic pathways involved in asthma pathogenesis and pharmacological response to treatment (e.g., corticosteroid-targeting of TH2 pathways), discerning true pharmacogenomic associations from those associated with the natural history of the disease has been difficult. In particular, clarification of pharmacogenetic associations jointly with disease-related phenotypes such as symptomatic response (which incorporates asthmatic immune response pathways as well as pharmacokinetic and pharmacodynamic phenotypes) is challenging. One solution to the problem of asthma phenotype complexity may be found in moving away from traditional epidemiological approaches for disease classification and hypothesis development, and embracing systems-level methods that can readily integrate the complexity of multidimensional asthmatic phenotypes. Recently, several investigators have utilized clustering and network-based approaches to evaluate asthma, atopy and wheezing [6,112–115]. These methods have been able to differentiate severity of phenotype [114], but cannot always provide reproducible results among different studies. Network methods that cluster phenotypes based on similarity and/or severity prior to genetic analysis would also be useful for identifying functional or disease modules that more closely reflect the true relationships between genotype and drug or target responses. The resulting networks could also be combined with metabolome or transcriptome profiles to improve phenotypic characterization of a disease or biological process. For example, Inouye et al. applied network analysis of genome-wide genetic and transcriptional variation with blood lipid measurements from 500 unrelated individuals to identify a tissue-specific immune response network module associated with blood lipid levels, inflammatory and allergic processes and a single SNP that regulates serum IgE levels [116].
Integrative pharmacogenomics
Genomic technologies including expression microarrays, high-throughput genotyping and next-generation sequencing platforms have significantly advanced the identification of causal variants. By integrating expression data with genotype data through functional genomic screens, investigators can identify genetic variants contributing to the expression and activity of genes and pathways involved in drug response (i.e., functional SNPs), thereby enabling the rapid discovery of pharmacogenetic biomarkers that alter gene expression and function.
An integrative pharmacogenomic study might involve first obtaining genotype data (i.e., SNPs) through pharmacogenomic GWAS or candidate-gene studies in an appropriate asthma patient cohort. One historical method of obtaining gene-expression data (e.g., by microarray) is from immortalized B-lymphocytes, which can potentially be derived from the original patient population, and which have been pharmacologically treated or left untreated with the study drug. The expression quantitative trait loci (eQTL; reviewed in reference [117]), which are defined as genome-wide expression SNPs with the greatest association to pharmacologically induced expression differences in the asthmatic cell lines, are identified through statistical analysis of the expression data. The most salient of eQTLs are then tested for clinical validation using genomic information from the previously completed clinical trial of asthmatics taking drug therapy. The eQTL can be used to weight or otherwise be statistically linked with the genomic information, and then modeled through various statistical or systems biology approaches, including pathway modeling and multivariate analysis [117]. Through integrating genome-wide expression data with population genomics, pharmacogenetic eQTLs can be rapidly identified.
Functional eQTL studies, where gene transcripts are evaluated as a quantitative phenotype in genetic association studies, are becoming increasingly valuable for identifying and validating pharmacogenomic loci. These studies are particularly useful for identifying treatment-specific genetic determinants of regulation of gene expression, and are well-suited to pathway-based modeling approaches. Through an extensive global eQTL mapping study, Grundberg et al. queried the genome expression profiles of cultured human osteoblast-like cells in response to treatment with dexamethasone, a potent GC steroid hormone, and identified dexamethasone-specific eQTLs in which approximately 60 genes per sample showed significant evidence of dexamethasone-specific allele-specific expression differences [118]. A top dexamethasone eQTL locus encoded TNC, an extracellular matrix protein involved in inflammation that is increased in the lung tissue of asthmatics [119] and is known to be downregulated by dexamethasone [118,119]. After determining that dexamethasone-specific downregulation of TNC occurs in a genotype-dependent manner, the authors sought to determine whether dexamethasone-dependent, heritable cis-regulation of TNC expression was responsible for the observed variability in therapeutic response to ICS in asthma patients. Six expression SNPs within the TNC locus were tested for association with response (FEV1 measured before and after 2 months of corticosteroid treatment) to daily inhaled budesonide treatment in 170 children with mild-to-moderate persistent asthma, who had been randomized to receive either daily ICS treatment or placebo. Of these, four SNPs (rs955387-A [β = −6.99; p = 0.005], rs10982634-C, [β = −6.01; p = 0.01], rs10817727-G [β = −5.78; p = 0.02] and rs12380804-A [β = −8.09, p = 0.02]) demonstrated significant associations with treatment response. These results demonstrate the utility of functional genomics in identifying specific pharmacogenomic eQTLs for asthma treatment response.
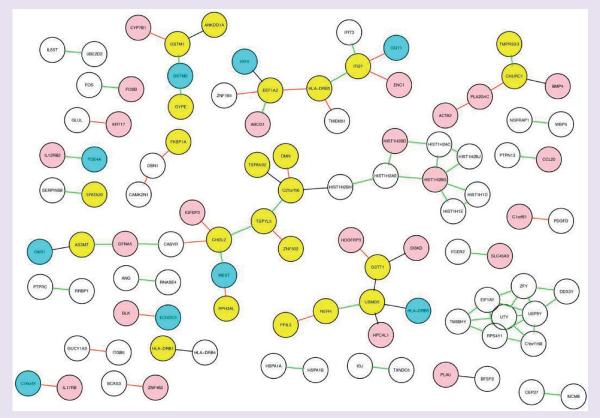
Many eQTL studies have identified regulatory variants located near target transcripts (e.g., cis-acting variants), of which only a small number are heritably expressed [120–122]. Identifying the epistatic effects that are likely to explain the remaining heritable variation is a difficult problem owing to the large number of tests in proportion to the number of samples analyzed. Bayesian networks and other graphical network models that can estimate millions of combinations of variables, such as GGMs, have been applied in the context of integrative genomics in an effort to circumvent this problem [124–125]. Recently, Chu et al. employed this approach to investigate gene–SNP associations [126]. A GGM was developed to infer SNP–gene networks from an asthma integrated genomic data set. The model was constructed using a multistep approach based on conditional independence of SNPs, using partial correlation to define the gene–SNP interaction matrix, and false-discovery rate multiple testing and posterior probability threshold to estimate the significance of the interactions. To validate the model, the authors created an asthma integrative genomics data set, using the genome-wide genotype data from the CAMP GWAS of patients on ICS (n = 229) and whole-genome gene expression data from CD4+ T lymphocytes isolated from a subset of these CAMP participants (n = 154) [126]. GGM was applied to 534,290 SNP markers identified in the GWAS and 3203 RefSeq-annotated transcript profiles from the CD4+ lymphocyte samples, from which 513,203 probable gene–gene associations were identified [126]. The model was independently validated using a publicly available gene expression set in the Gene Expression Omnibus [127] from CD4+ lymphocytes in asthmatic and normal subjects [126]. Using a stringent threshold (posterior probability > 0.9) a significant proportion of gene–gene interactions (1913 interactions) and 40 hub genes with >100 significant connections to other genes were found, suggesting that these genes are of biological importance in CD4+ lymphocytes (Figure 5) [126]. To demonstrate the utility of the model, one of the significant hub networks, containing the β2 subunit of the IL12RB2, which has important immunomodulatory roles in CD4+ lymphocytes and airway disease, was evaluated. The GGM found 306 IL12Rβ2-interacting genes with 5611 cis-acting SNPs; following false-discovery rate adjustment, the model selected 225 SNP–gene pairs with significant association, including two asthma-related genes, RAP1A and TBKBP1 [127]. GGM was also applied to nonhub genes in order to identify regulatory variants for genes associated with a second asthma candidate gene, IL1B, for which it identified 353 significant SNP–ene pairs [127]. These results demonstrate a novel application of graphical network models for inferring gene–SNP associations that reflect the underlying biological network between genes and phenotypes. The integration of clinical phenotypes with expression and genotype information to identify novel candidate genes represents an exciting frontier in asthma pharmacogenomics.
Figure 5. Integrative gene network of 86 common edges from the CAMP and HapMap consortium (GeneVar) cohorts.
From a subset of 608 genes, a subnetwork of 86 edges appeared in both CAMP and GeneVar networks using a threshold of posterior probability of 0.9. Hub nodes are indicated in pink for CAMP, cyan for GeneVar and yellow for both. The direction of the edges is shown by the edge color (green: both positive; black: both negative; red: opposite).
Reproduced with permission from [126] © 2009 BioMed Central.
Conclusion
Significant recent progress has been made in the pharmacogenomics of asthma treatment response. Through candidate gene studies and GWAS, novel pharmacogenetic loci and SNPs associated with therapeutic response to B2-agonists and GCs have been identified. An important finding in these studies is that treatment response is significantly influenced by the interaction of mutiple genes (i.e., gene–gene interactions), SNPs and haplotypes. These data provide strong evidence of involvement of genetic networks in asthma therapeutic response phenotypes and demonstrate a need for prospective clinical studies of gene–gene interactions as well as genome-wide approaches that interrogate all available SNPs in order to identify true markers of asthma treatment response.
Several recent studies have applied network-based, integrative methods to identify genotype–phenotype relationships in asthma pharmacogenomics. SNP networks generated through Bayesian modeling and Random Forests classifiers have successfully predicted BDR and asthma exacerbations. Module networks analysis, which generates probabilistic graphical models of gene expression and function, was used to create a global map of asthma that contains network modules associated with treatment response. Importantly, these findings can infer significant genotype–phenotype relationships that are otherwise difficult to discern.
Integrative genomic approaches that combine genome-wide genotype data with gene-expression profiles have been used to identify novel candidate genes and infer biological networks associated with therapeutic response. The results of these studies illustrate the utility of systems biology approaches in asthma. Using data from an omics data set, researchers can develop descriptive and predictive models of drug-response phenotypes that incorporate multiple genetic and environmental factors.
While the systems biology methods offer promising solutions, their direct applicability in clinical practice and drug development is hindered by some significant limitations. The greatest limitation is that many of these methods are still under development and have not been widely validated. In addition, there has been little independent replication of findings resulting from these studies. The clinical translation and predictability of these methods therefore remains to be seen; however, increased efforts to validate methodologies and a focus on replicating initial results will expedite this process. Given its relevance and utility, we anticipate that the systems biology paradigm will soon become standard practice in pharmacogenomic research in asthma.
Future perspective
The initial asthma pharmacogenomics GWAS represents the first of many pharmacogenomic GWAS to come. These future GWAS will drive the development of larger well-characterized cohorts, new and improved study design and statistical analysis strategies that take into account the unique challenges presented by genome-wide pharmacogenomic studies. In addition, recent progress in next-generation sequencing technologies such as whole-genome sequencing, exome sequencing, RNA-sequencing and chromatin immunoprecipitation sequencing represent an exciting frontier in asthma pharmacogenomics. These technologies will lead to an abundance of genetic information, and integrating these data with other relevant clinical and biological information will provide informative data sets for predictive modeling. However, these efforts are expected to introduce significant methodological and bioinformatic challenges. As data analysis becomes more complex, the development and refinement of software tools that provide improved and more rapid knowledge discovery, data mining and network engineering activities is anticipated to overcome some of these challenges.
Executive summary.
Progress in asthma pharmacogenomics
-
■
The first pharmacogenomic genome-wide association study of inhaled corticosteroids response in an asthma cohort identified a functional variant in GLCCI1, a glucocorticoid pathway gene.
-
■
Gene–gene interactions of various candidate loci are associated with treatment response.
Systems biology
-
■
Systems biology approaches seek to generate the best model of a biological network from large-scale `omics' data.
Systems biology approaches in asthma
-
■
Bayesian networks and Random Forests are capable of modeling and accurately predicting phenotypic response from genomic and expression data.
-
■
Integration of modeling with expression data provides a system-level view of asthma.
-
■
Incorporating epistasis and directionality effects will improve the accurate prediction of pharmacogenomic phenotypes through utilization of clustering and network-based approaches.
Integrative pharmacogenomics
-
■
Integrating genomic data with expression quantitative trait loci can identify candidate genes and infer biological networks associated with therapeutic response.
Conclusion
-
■
Novel pharmacogenetic loci and gene pathways can be inferred using a systems biology approach.
Acknowledgments
This work was supported by grants from the NIH (NIH T32 HL007427, U01 HL065899, R01 HL092197 and R01 NR013391). No writing assistance was utilized in the production of this manuscript.
Footnotes
Financial & competing interests disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br. Med. Bull. 2000;56(4):1054–1070. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 2.Holloway JW, Arshad SH, Holgate ST. Using genetics to predict the natural history of asthma? J. Allergy Clin. Immunol. 2010;126(2):200–209. doi: 10.1016/j.jaci.2010.06.006. quiz 210-211. [DOI] [PubMed] [Google Scholar]
- 3.Ober C, Vercelli D. Gene–environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27(3):107–115. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med. 2012;18(5):716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira MA, McRae AF, Medland SE, et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur. J. Hum. Genet. 2011;19(4):458–464. doi: 10.1038/ejhg.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sleiman PM, Flory J, Imielinski M, et al. Variants of DENND1B associated with asthma in children. N. Engl. J. Med. 2010;362(1):36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 7.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Bjornsdottir US, Halapi E, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009;41(3):342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 9.Himes BE, Hunninghake GM, Baurley JW, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am. J. Hum. Genet. 2009;84(5):581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moffatt MF, abesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 11.Hawkins GA, Tantisira K, Meyers DA, et al. Sequence, haplotype, and association analysis of ADRbeta2 in a multiethnic asthma case–control study. Am. J. Respir. Crit. Care Med. 2006;174(10):1101–1109. doi: 10.1164/rccm.200509-1405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reihsaus E, Innis M, MacIntyre N, Liggett SB. Mutations in the gene encoding for the beta 2-adrenergic receptor in normal and asthmatic subjects. Am. J. Respir. Cell Mol. Biol. 1993;8(3):334–339. doi: 10.1165/ajrcmb/8.3.334. [DOI] [PubMed] [Google Scholar]
- 13.Drysdale CM, McGraw DW, Stack CB, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl Acad. Sci. USA. 2000;97(19):10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vonk JM, Postma DS, Maarsingh H, Bruinenberg M, Koppelman GH, Meurs H. Arginase 1 and arginase 2 variations associate with asthma, asthma severity and beta2 agonist and steroid response. Pharmacogenet. Genomics. 2010;20(3):179–186. doi: 10.1097/FPC.0b013e328336c7fd. [DOI] [PubMed] [Google Scholar]
- 15.Duroudier NP, Tulah AS, Sayers I. Leukotriene pathway genetics and pharmacogenetics in allergy. Allergy. 2009;64(6):823–839. doi: 10.1111/j.1398-9995.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- 16.Drazen JM, Yandava CN, Dube L. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat. Genet. 1999;22(2):168–170. doi: 10.1038/9680. [DOI] [PubMed] [Google Scholar]
- 17.Tantisira KG, Lima J, Sylvia J, Klanderman B, Weiss ST. 5-lipoxygenase pharmacogenetics in asthma: overlap with Cys-leukotriene receptor antagonist loci Pharmacogenet. Genomics. 2009;19(3):244–247. doi: 10.1097/FPC.0b013e328326e0b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lima JJ, Zhang S, Grant A, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am. J. Respir. Crit. Care Med. 2006;173(4):379–385. doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klotsman M, York TP, Pillai SG, et al. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet. Genomics. 2007;17(3):189–196. doi: 10.1097/FPC.0b013e3280120043. [DOI] [PubMed] [Google Scholar]
- 20.Tantisira KG, Lake S, Silverman ES, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum. Mol. Genet. 2004;13(13):1353–1359. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 21.Jin Y, Hu D, Peterson EL, et al. Dual-specificity phosphatase 1 as a pharmacogenetic modifier of inhaled steroid response among asthmatic patients. J. Allergy Clin. Immunol. 2010;126(3):618–625. e1–2. doi: 10.1016/j.jaci.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tantisira KG, Silverman ES, Mariani TJ, et al. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J. Allergy Clin. Immunol. 2007;120(6):1285–1291. doi: 10.1016/j.jaci.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Koster ES, Maitland-van der Zee AH, Tavendale R, et al. FCER2 T2206C variant associated with chronic symptoms and exacerbations in steroid-treated asthmatic children. Allergy. 2011;66(12):1546–1552. doi: 10.1111/j.1398-9995.2011.02701.x. [DOI] [PubMed] [Google Scholar]
- 24.Tantisira KG, Lasky-Su J, Harada M, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N. Engl. J. Med. 2011;365(13):1173–1183. doi: 10.1056/NEJMoa0911353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore PE, Ryckman KK, Williams SM, Patel N, Summar ML, Sheller JR. Genetic variants of GSNOR and ADRB2 influence response to albuterol in African–American children with severe asthma. Pediatr. Pulmonol. 2009;44(7):649–654. doi: 10.1002/ppul.21033. [DOI] [PubMed] [Google Scholar]
- 26.Sampson AP, Siddiqui S, Buchanan D, et al. Variant LTC(4) synthase allele modifies cysteinyl leukotriene synthesis in eosinophils and predicts clinical response to zafirlukast. Thorax. 2000;55(Suppl. 2):S28–S31. doi: 10.1136/thorax.55.suppl_2.S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asano K, Shiomi T, Hasegawa N, et al. Leukotriene C4 synthase gene A(-444)C polymorphism and clinical response to a CYS-LT(1) antagonist, pranlukast, in Japanese patients with moderate asthma. Pharmacogenetics. 2002;12(7):565–570. doi: 10.1097/00008571-200210000-00009. [DOI] [PubMed] [Google Scholar]
- 28.Mastalerz L, Nizankowska E, Sanak M, et al. Clinical and genetic features underlying the response of patients with bronchial asthma to treatment with a leukotriene receptor antagonist. Eur. J. Clin. Invest. 2002;32(12):949–955. doi: 10.1046/j.1365-2362.2002.01088.x. [DOI] [PubMed] [Google Scholar]
- 29.Whelan GJ, Blake K, Kissoon N, et al. Effect of montelukast on time-course of exhaled nitric oxide in asthma: influence of LTC4 synthase A(-444)C polymorphism. Pediatr. Pulmonol. 2003;36(5):413–420. doi: 10.1002/ppul.10385. [DOI] [PubMed] [Google Scholar]
- 30.Lima JJ. Treatment heterogeneity in asthma: genetics of response to leukotriene modifiers. Mol. Diagn. Ther. 2007;11(2):97–104. doi: 10.1007/BF03256228. [DOI] [PubMed] [Google Scholar]
- 31.Ye YM, Lee HY, Kim SH, et al. Pharmacogenetic study of the effects of NK2R G231E G>A and TBX21 H33Q C>G polymorphisms on asthma control with inhaled corticosteroid treatment. J. Clin. Pharm. Ther. 2009;34(6):693–701. doi: 10.1111/j.1365-2710.2009.01054.x. [DOI] [PubMed] [Google Scholar]
- 32.Mougey EB, Feng H, Castro M, Irvin CG, Lima JJ. Absorption of montelukast is transporter mediated: a common variant of OATP2B1 is associated with reduced plasma concentrations and poor response. Pharmacogenet. Genomics. 2009;19(2):129–138. doi: 10.1097/FPC.0b013e32831bd98c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawkins GA, Lazarus R, Smith RS, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J. Allergy Clin. Immunol. 2009;123(6):1376–1383.e7. doi: 10.1016/j.jaci.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010;11(6):446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nadeau JH, Dudley AM. Genetics. Systems genetics. Science. 2011;331(6020):1015–1016. doi: 10.1126/science.1203869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koster ES, Rodin AS, Raaijmakers JA, Maitland-van der Zee AH. Systems biology in pharmacogenomic research: the way to personalized prescribing? Pharmacogenomics. 2009;10(6):971–981. doi: 10.2217/pgs.09.38. [DOI] [PubMed] [Google Scholar]
- 37.Rodin AS, Gogoshin G, Boerwinkle E. Systems biology data analysis methodology in pharmacogenomics. Pharmacogenomics. 2011;12(9):1349–1360. doi: 10.2217/pgs.11.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L. Pharmacogenomics: a systems approach. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010;2(1):3–22. doi: 10.1002/wsbm.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control. Clin. Trials. 1999;20(1):91–120. [PubMed] [Google Scholar]
- 40.Long-term effects of budesonide or nedocromil in children with asthma. The Childhood Asthma Management Program Research Group. N. Engl. J. Med. 2000;343(15):1054–1063. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 41.Lazarus SC, Boushey HA, Fahy JV, et al. Long-acting beta2-agonist monotherapy vs continued therapy with inhaled corticosteroids in patients with persistent asthma: a randomized controlled trial. JAMA. 2001;285(20):2583–2593. doi: 10.1001/jama.285.20.2583. [DOI] [PubMed] [Google Scholar]
- 42.Lemanske RF, Jr, Sorkness CA, Mauger EA, et al. Inhaled corticosteroid reduction and elimination in patients with persistent asthma receiving salmeterol: a randomized controlled trial. JAMA. 2001;285(20):2594–2603. doi: 10.1001/jama.285.20.2594. [DOI] [PubMed] [Google Scholar]
- 43.Bielory L, Picone F, Rabinowitz P, et al. Multicentre, randomised, parallel-group study of the efficacy and tolerability of flunisolide administered once daily via AeroChamber in the treatment of mild to moderate asthma. Clin. Drug Investig. 2000;19:93–101. [Google Scholar]
- 44.Peters S, Anthonisen N, Castro M, et al. Randomized comparison of strategies for reducing treatment in mild persistent asthma. N. Engl. J. Med. 2007;356:2027–2039. doi: 10.1056/NEJMoa070013. [DOI] [PubMed] [Google Scholar]
- 45.Van Steen K, McQueen MB, Herbert A, et al. Genomic screening and replication using the same data set in family-based association testing. Nat. Genet. 2005;37(7):683–691. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- 46.Lange C, Lyon H, DeMeo D, Raby B, Silverman EK, Weiss ST. A new powerful non-parametric two-stage approach for testing multiple phenotypes in family-based association studies. Hum. Hered. 2003;56(1–3):10–17. doi: 10.1159/000073728. [DOI] [PubMed] [Google Scholar]
- 47.Israel E, Chinchilli VM, Ford JG, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004;364(9444):1505–1512. doi: 10.1016/S0140-6736(04)17273-5. [DOI] [PubMed] [Google Scholar]
- 48.Tantisira KG, Hwang ES, Raby BA, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc. Natl Acad. Sci. USA. 2004;101(52):18099–18104. doi: 10.1073/pnas.0408532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duan QL, Gaume BR, Hawkins GA, et al. Regulatory haplotypes in ARG1 are associated with altered bronchodilator response. Am. J. Respir. Crit. Care Med. 2011;183(4):449–454. doi: 10.1164/rccm.201005-0758OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tse SM, Tantisira K, Weiss ST. The pharmacogenetics and pharmacogenomics of asthma therapy. Pharmacogenomics J. 2011;11(6):383–392. doi: 10.1038/tpj.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tantisira K, Weiss S. The pharmacogenetics of asthma treatment. Curr. Allergy Asthma Rep. 2009;9(1):10–17. doi: 10.1007/s11882-009-0002-9. [DOI] [PubMed] [Google Scholar]
- 52.Lima JJ, Blake KV, Tantisira KG, Weiss ST. Pharmacogenetics of asthma. Curr. Opin. Pulm. Med. 2009;15(1):57–62. doi: 10.1097/MCP.0b013e32831da8be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss ST, Litonjua AA, Lange C, et al. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics J. 2006;6(5):311–326. doi: 10.1038/sj.tpj.6500387. [DOI] [PubMed] [Google Scholar]
- 54.Tantisira KG, Weiss ST. The pharmacogenetics of asthma: an update. Curr. Opin. Mol. Ther. 2005;7(3):209–217. [PubMed] [Google Scholar]
- 55.Chung LP, Waterer G, Thompson PJ. Pharmacogenetics of beta2 adrenergic receptor gene polymorphisms, long-acting beta-agonists and asthma. Clin. Exp. Allergy. 2011;41(3):312–326. doi: 10.1111/j.1365-2222.2011.03696.x. [DOI] [PubMed] [Google Scholar]
- 56.Taylor DR, Epton MJ, Kennedy MA, et al. Bronchodilator response in relation to beta2-adrenoceptor haplotype in patients with asthma. Am. J. Respir. Crit. Care Med. 2005;172(6):700–703. doi: 10.1164/rccm.200501-092OC. [DOI] [PubMed] [Google Scholar]
- 57.Carter-Pokras OD, Gergen PJ. Reported asthma among Puerto Rican, Mexican–American, and Cuban children, 1982 through 1984. Am. J. Public Health. 1993;83(4):580–582. doi: 10.2105/ajph.83.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Homa DM, Mannino DM, Lara M. Asthma mortality in U.S. Hispanics of Mexican, Puerto Rican, and Cuban heritage, 1990–1995. Am. J. Respir. Crit. Care Med. 2000;161(2 Pt 1):504–509. doi: 10.1164/ajrccm.161.2.9906025. [DOI] [PubMed] [Google Scholar]
- 59.Choudhry S, Ung N, Avila PC, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am. J. Respir. Crit. Care Med. 2005;171(6):563–570. doi: 10.1164/rccm.200409-1286OC. [DOI] [PubMed] [Google Scholar]
- 60.von Homeyer P, Schwinn DA. Pharmacogenomics of beta-adrenergic receptor physiology and response to beta-blockade. Anesth. Analg. 2011;113(6):1305–1318. doi: 10.1213/ANE.0b013e31822b887e. [DOI] [PubMed] [Google Scholar]
- 61.Moore PE. Influence of gene–gene interactions on response to albuterol therapy. Pharmacogenomics. 2011;12(1):1–3. doi: 10.2217/pgs.10.192. [DOI] [PubMed] [Google Scholar]
- 62.Tantisira KG, Small KM, Litonjua AA, Weiss ST, Liggett SB. Molecular properties and pharmacogenetics of a polymorphism of adenylyl cyclase type 9 in asthma: interaction between beta-agonist and corticosteroid pathways. Hum. Mol. Genet. 2005;14(12):1671–1677. doi: 10.1093/hmg/ddi175. [DOI] [PubMed] [Google Scholar]
- 63.Kim SH, Ye YM, Lee HY, Sin HJ, Park HS. Combined pharmacogenetic effect of ADCY9 and ADRB2 gene polymorphisms on the bronchodilator response to inhaled combination therapy. J. Clin. Pharm. Ther. 2011;36(3):399–405. doi: 10.1111/j.1365-2710.2010.01196.x. [DOI] [PubMed] [Google Scholar]
- 64.Choudhry S, Que LG, Yang Z, et al. GSNO reductase and beta2-adrenergic receptor gene–gene interaction: bronchodilator responsiveness to albuterol. Pharmacogenet. Genomics. 2010;20(6):351–358. doi: 10.1097/FPC.0b013e328337f992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polonikov AV, Ivanov VP, Solodilova MA, Khoroshaya IV, Kozhuhov MA, Panfilov VI. The relationship between polymorphisms in the glutamate cysteine ligase gene and asthma susceptibility. Respir. Med. 2007;101(11):2422–2424. doi: 10.1016/j.rmed.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 66.Duan QL, Du R, Lasky-Su J, et al. A polymorphism in the thyroid hormone receptor gene is associated with bronchodilator response in asthmatics. Pharmacogenomics J. 2012 doi: 10.1038/tpj.2011.56. doi:10.1038/tpj.2011.56. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maarsingh H, Zuidhof AB, Bos IS, et al. Arginase inhibition protects against allergen-induced airway obstruction, hyperresponsiveness, and inflammation. Am. J. Respir. Crit. Care Med. 2008;178(6):565–573. doi: 10.1164/rccm.200710-1588OC. [DOI] [PubMed] [Google Scholar]
- 68.Meurs H, McKay S, Maarsingh H, et al. Increased arginase activity underlies allergen-induced deficiency of cNOS-derived nitric oxide and airway hyperresponsiveness. Br. J. Pharmacol. 2002;136(3):391–398. doi: 10.1038/sj.bjp.0704725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duan QL, Tantisira KG. Pharmacogenetics of asthma therapy. Curr. Pharm. Des. 2009;15(32):3742–3753. doi: 10.2174/138161209789649510. [DOI] [PubMed] [Google Scholar]
- 70.Tantisira KG, Drazen JM. Genetics and pharmacogenetics of the leukotriene pathway. J. Allergy Clin. Immunol. 2009;124(3):422–427. doi: 10.1016/j.jaci.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.York TP, Vargas-Irwin C, Anderson WH, van den Oord EJ. Asthma pharmacogenetic study using finite mixture models to handle drug-response heterogeneity. Pharmacogenomics. 2009;10(5):753–767. doi: 10.2217/pgs.09.19. [DOI] [PubMed] [Google Scholar]
- 72.Kotani H, Kishi R, Mouri A, et al. Influence of leukotriene pathway polymorphisms on clinical responses to montelukast in Japanese patients with asthma. J. Clin. Pharm. Ther. 2012;37(1):112–116. doi: 10.1111/j.1365-2710.2011.01248.x. [DOI] [PubMed] [Google Scholar]
- 73.Via M, De Giacomo A, Corvol H, et al. The role of LTA4H and ALOX5AP genes in the risk for asthma in Latinos. Clin. Exp. Allergy. 2010;40(4):582–589. doi: 10.1111/j.1365-2222.2009.03438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tcheurekdjian H, Via M, De Giacomo A, et al. ALOX5AP and LTA4H polymorphisms modify augmentation of bronchodilator responsiveness by leukotriene modifiers in Latinos. J. Allergy Clin. Immunol. 2010;126(4):853–858. doi: 10.1016/j.jaci.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bray PJ, Cotton RG. Variations of the human glucocorticoid receptor gene (NR3C1): pathological and in vitro mutations and polymorphisms. Hum. Mutat. 2003;21(6):557–568. doi: 10.1002/humu.10213. [DOI] [PubMed] [Google Scholar]
- 76.Hawkins GA, Amelung PJ, Smith RS, et al. Identification of polymorphisms in the human glucocorticoid receptor gene (NR3C1) in a multi-racial asthma case and control screening panel. DNA Seq. 2004;15(3):167–173. doi: 10.1080/10425170410001704517. [DOI] [PubMed] [Google Scholar]
- 77.Pietras T, Panek M, Tworek D, et al. The Bcl I single nucleotide polymorphism of the human glucocorticoid receptor gene h-GR/NR3C1 promoter in patients with bronchial asthma: pilot study. Mol. Biol. Rep. 2011;38(6):3953–3958. doi: 10.1007/s11033-010-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Panek M, Pietras T, Antczak A, Gorski P, Kuna P, Szemraj J. The role of functional single nucleotide polymorphisms of the human glucocorticoid receptor gene NR3C1 in Polish patients with bronchial asthma. Mol. Biol. Rep. 2011;39(4):4749–4757. doi: 10.1007/s11033-011-1267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sayers I, Hall IP. Pharmacogenetic approaches in the treatment of asthma. Curr. Allergy Asthma Rep. 2005;5(2):101–108. doi: 10.1007/s11882-005-0082-0. [DOI] [PubMed] [Google Scholar]
- 80.Auffray C, Adcock IM, Chung KF, Djukanovic R, Pison C, Sterk PJ. An integrative systems biology approach to understanding pulmonary diseases. Chest. 2010;137(6):1410–1416. doi: 10.1378/chest.09-1850. [DOI] [PubMed] [Google Scholar]
- 81.Mitchell T. Machine Learning. McGraw Hill; Columbus, OH, USA: 1997. [Google Scholar]
- 82.Russel S, Norvig P. Artificial Intelligence: A Modern Approach. 2nd Edition Prentice Hall; NJ, USA: 2003. [Google Scholar]
- 83.Brieman L. Random Forests. Mach. Learn. 2001;45(1):5–32. [Google Scholar]
- 84.Junker B, Schreiber F. Analysis of Biological Networks. John Wiley & Sons; NJ, USA: 2008. [Google Scholar]
- 85.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. Morgan Kaufmann Publishers; CA, USA: 1988. [Google Scholar]
- 86.Rodin AS, Boerwinkle E. Mining genetic epidemiology data with Bayesian networks I: Bayesian networks and example application (plasma apoE levels) Bioinformatics. 2005;21(15):3273–3278. doi: 10.1093/bioinformatics/bti505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR. A primer on learning in Bayesian networks for computational biology. PLoS Comput. Biol. 2007;3(8):e129. doi: 10.1371/journal.pcbi.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sebastiani P, Ramoni M. On the use of Bayesian networks to analyse survey data. Res Off. Statist. 2001;41(1):53–64. [Google Scholar]
- 89.Ramoni M, Sebastiani P. Bayesian methods: intelligent data analysis. In: Hand DJ, Berthold MR, editors. Intelligent Data Analysis: an Introduction. Springer; NY, USA: 2003. pp. 128–166. [Google Scholar]
- 90.Sharma S, Poon A, Himes BE, et al. Association of variants in innate immune genes with asthma and eczema. Pediatr. Allergy Immunol. 2011;23(4):315–323. doi: 10.1111/j.1399-3038.2011.01243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Himes BE, Dai Y, Kohane IS, Weiss ST, Ramoni MF. Prediction of chronic obstructive pulmonary disease (COPD) in asthma patients using electronic medical records. J. Am. Med. Inform. Assoc. 2009;16(3):371–379. doi: 10.1197/jamia.M2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanders DL, Aronsky D. Prospective evaluation of a Bayesian Network for detecting asthma exacerbations in a Pediatric Emergency Department. AMIA. Annu. Symp. Proc. 2006;1085 [PMC free article] [PubMed] [Google Scholar]
- 93.Sanders DL, Aronsky D. Detecting asthma exacerbations in a pediatric emergency department using a Bayesian network. AMIA. Annu. Symp. Proc. 2006:684–688. [PMC free article] [PubMed] [Google Scholar]
- 94.Himes BE, Wu AC, Duan QL, et al. Predicting response to short-acting bronchodilator medication using Bayesian networks. Pharmacogenomics. 2009;10(9):1393–1412. doi: 10.2217/pgs.09.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rodin AS, Litvinenko A, Klos K, et al. Use of wrapper algorithms coupled with a random forests classifier for variable selection in large-scale genomic association studies. J. Comput. Biol. 2009;16(12):1705–1718. doi: 10.1089/cmb.2008.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goldstein BA, Hubbard AE, Cutler A, Barcellos LF. An application of Random Forests to a genome-wide association dataset: methodological considerations and new findings. BMC Genet. 2010;11:49. doi: 10.1186/1471-2156-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nonyane BA, Foulkes AS. Application of two machine learning algorithms to genetic association studies in the presence of covariates. BMC Genet. 2008;9:71. doi: 10.1186/1471-2156-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun YV. Multigenic modeling of complex disease by random forests. Adv. Genet. 2010;72:73–99. doi: 10.1016/B978-0-12-380862-2.00004-7. [DOI] [PubMed] [Google Scholar]
- 99.Bureau A, Dupuis J, Falls K, et al. Identifying SNPs predictive of phenotype using random forests. Genet. Epidemiol. 2005;28(2):171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- 100.De Lobel L, Geurts P, Baele G, Castro-Giner F, Kogevinas M, Van Steen K. A screening methodology based on Random Forests to improve the detection of gene–gene interactions. Eur. J. Hum. Genet. 2010;18(10):1127–1132. doi: 10.1038/ejhg.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu M, Tantisira KG, Wu A, et al. Genome Wide Association Study to predict severe asthma exacerbations in children using random forests classifiers. BMC Med. Genet. 2011;12:90. doi: 10.1186/1471-2350-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weiss ST, Raby BA, Rogers A. Asthma genetics and genomics 2009. Curr. Opin. Genet. Dev. 2009;19(3):279–282. doi: 10.1016/j.gde.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 103.Sun N, Zhao H. Genomic approaches in dissecting complex biological pathways. Pharmacogenomics. 2004;5(2):163–179. doi: 10.1517/phgs.5.2.163.27488. [DOI] [PubMed] [Google Scholar]
- 104.Xu X, Wang L, Ding D. Learning module networks from genome-wide location and expression data. FEBS Lett. 2004;578(3):297–304. doi: 10.1016/j.febslet.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 105.Joshi A, De Smet R, Marchal K, Van de Peer Y, Michoel T. Module networks revisited: computational assessment and prioritization of model predictions. Bioinformatics. 2009;25(4):490–496. doi: 10.1093/bioinformatics/btn658. [DOI] [PubMed] [Google Scholar]
- 106.Segal E, Shapira M, Regev A, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003;34(2):166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 107.Hakonarson H, Bjornsdottir US, Halapi E, et al. Profiling of genes expressed in peripheral blood mononuclear cells predicts glucocorticoid sensitivity in asthma patients. Proc. Natl Acad. Sci. USA. 2005;102(41):14789–14794. doi: 10.1073/pnas.0409904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Novershtern N, Itzhaki Z, Manor O, Friedman N, Kaminski N. A functional and regulatory map of asthma. Am. J. Respir. Cell Mol. Biol. 2008;38(3):324–336. doi: 10.1165/rcmb.2007-0151OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372(9643):1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 110.Tantisira K, Drazen JM. Pharmacogenetics. In: Shapiro SJ, Lomas D, Weiss ST, Silverman E, editors. Respiratory Genetics (Volume 1) Hodder Education; London, UK: 2005. pp. 191–216. [Google Scholar]
- 111.Tamari M, Tomita K, Hirota T. Genome-wide association studies of asthma. Allergol. Int. 2011;60(3):247–252. doi: 10.2332/allergolint.11-RAI-0320. [DOI] [PubMed] [Google Scholar]
- 112.Haldar P, Pavord ID, Shaw DE, et al. Cluster analysis and clinical asthma phenotypes. Am. J. Respir. Crit. Care Med. 2008;178(3):218–224. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Isozaki A, Shoda T, Mimura S, et al. A trial of hierarchical cluster analysis of hospitalized wheezy infants and young children based on clinical factors. Arerugi. 2011;60(5):586–592. [PubMed] [Google Scholar]
- 114.Fitzpatrick AM, Teague WG, Meyers DA, et al. Heterogeneity of severe asthma in childhood: confirmation by cluster analysis of children in the National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program. J. Allergy Clin. Immunol. 2011;127(2):382–389. e1–13. doi: 10.1016/j.jaci.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weatherall M, Travers J, Shirtcliffe PM, et al. Distinct clinical phenotypes of airways disease defined by cluster analysis. Eur. Respir. J. 2009;34(4):812–818. doi: 10.1183/09031936.00174408. [DOI] [PubMed] [Google Scholar]
- 116.Inouye M, Silander K, Hamalainen E, et al. An immune response network associated with blood lipid levels. PLoS Genet. 2010;6(9):e1001113. doi: 10.1371/journal.pgen.1001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Franke L, Jansen RC. eQTL analysis in humans. Methods Mol. Biol. 2009;573:311–328. doi: 10.1007/978-1-60761-247-6_17. [DOI] [PubMed] [Google Scholar]
- 118.Grundberg E, Adoue V, Kwan T, et al. Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet. 2011;7(1):e1001279. doi: 10.1371/journal.pgen.1001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Laitinen A, Altraja A, Kampe M, Linden M, Virtanen I, Laitinen LA. Tenascin is increased in airway basement membrane of asthmatics and decreased by an inhaled steroid. Am. J. Respir. Crit. Care Med. 1997;156(3 Pt 1):951–958. doi: 10.1164/ajrccm.156.3.9610084. [DOI] [PubMed] [Google Scholar]
- 120.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39(10):1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 121.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat. Genet. 2007;39(10):1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Butte AJ, Tamayo P, Slonim D, Golub TR, Kohane IS. Discovering functional relationships between RNA expression and chemotherapeutic susceptibility using relevance networks. Proc. Natl Acad. Sci. USA. 2000;97(22):12182–12186. doi: 10.1073/pnas.220392197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schadt EE. Novel integrative genomics strategies to identify genes for complex traits. Anim. Genet. 2006;37(Suppl. 1):18–23. doi: 10.1111/j.1365-2052.2006.01473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schadt EE, Lamb J, Yang X, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005;37(7):710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schafer J, Strimmer K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics. 2005;21(6):754–764. doi: 10.1093/bioinformatics/bti062. [DOI] [PubMed] [Google Scholar]
- 126.Chu JH, Weiss ST, Carey VJ, Raby BA. A graphical model approach for inferring large-scale networks integrating gene expression and genetic polymorphism. BMC Syst. Biol. 2009;3:55. doi: 10.1186/1752-0509-3-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 201.American Lung Association Epidemiology and Statisticus Unit. Trends in Asthma Morbidity and Mortality: American Lung Association 2011 www.lung.org/finding-cures/our-research/trend-reports/asthma-trend-report.pdf.
- 202.AsthmaMap http://compbio.cs.huji.ac.il/AsthmaMap.