Abstract
An outbreak of hospital-acquired Acinetobacter baumannii infections, caused by a blaOXA-23-positive carbapenem-resistant strain belonging to international clone II/ST2, was detected in Latvia. The strain was partially equipped with the armA gene and the intI1-aacA4-catB8-aadA1-qacEΔ1 class 1 integron. In addition, the strain carried AbaR25, a novel AbaR4-like resistance island of ∼46,500 bp containing structures similar to the previously described AbaR22 and Tn6167 islands. AbaR25 was characterized by the occurrence of a second copy of Tn6022a interrupted by Tn2006 carrying the blaOXA-23 gene.
TEXT
Acinetobacter baumannii is an aerobic Gram-negative opportunistic pathogen with a remarkable ability to acquire resistance to different classes of antibiotics (1). The increased detection of multidrug-resistant (MDR) and carbapenem-resistant (CR) A. baumannii strains in clinical settings is mainly linked to the global dissemination of a number of highly successful clones, such as international clones I and II and multilocus sequence types (STs) 15 and ST25 (2).
Resistance to carbapenems in A. baumannii is primarily mediated by the production of carbapenem-hydrolyzing β-lactamases (3). Class B metallo-β-lactamases (MBLs) confer high levels of carbapenem resistance as well as resistance to all other β-lactams except for aztreonam, while the substrate profile of class D OXA-type carbapenemases is commonly diverse, with most of these enzymes showing a limited hydrolytic activity against imipenem and meropenem (3). The occurrence of genes encoding aminoglycoside-modifying enzymes (AME) is the main mechanism of resistance to aminoglycosides in A. baumannii (4). However, strains producing the 16S rRNA methylase ArmA have also been identified (5). ArmA has so far been a plasmid-encoded enzyme conferring high levels of resistance to several aminoglycosides (5).
Genomic resistance islands in A. baumannii (AbaR), first detected in 2006, can be sorted into two main models based on their genetic structures (6, 7). The first model, AbaR3-like, consists of Tn6019 as a backbone transposon and has, with the exception of AbaR2, been found only among isolates belonging to international clone I (6–10). The second model, AbaR4-like, consists of Tn6022 as a backbone transposon and has mainly been identified among isolates from international clone II (11–14). Complex structures of AbaR4-like islands, such as Tn6167 and AbaR22, have recently been described (15, 16). Importantly, the AbaR4-like islands have repeatedly been found to be interrupted by Tn2006 carrying the blaOXA-23-like gene (12–15).
The aim of this study was to investigate the molecular epidemiology and antimicrobial resistance characteristics of all the CR A. baumannii blood culture isolates (n = 30) obtained at the P. Stradins University Hospital (SUH) in Riga, Latvia, between May 2008 and December 2009. The study also included five invasive (cerebrospinal fluid and blood culture) CR A. baumannii isolates collected by four hospitals from different cities in Latvia between March and July 2009 (Table 1).
Table 1.
Molecular detection of particular antimicrobial resistance genes and elements in 35 OXA-23-producing A. baumannii isolates collected in Latviaa
| Isolate | Date of isolation (mo/yr) | Hospital | armA | aacA4-catB8-aadA1 | AbaR | Clonal lineage | PFGE | MLST |
|---|---|---|---|---|---|---|---|---|
| K51-65 | 5/2008 | PSCUH | + | + | AbaR25 | Int. II | A1 | ST2 |
| K51-66 | 6/2008 | PSCUH | + | + | AbaR25 | Int. II | A2 | ST2 |
| K51-67 | 7/2008 | PSCUH | − | − | AbaR4 | Int. II | B | ST2 |
| K51-68 | 9/2008 | PSCUH | + | − | AbaR25 | Group 4 | A3 | ST2 |
| K51-69 | 10/2008 | PSCUH | + | + | AbaR25 | Int. II | A1 | ND |
| K51-70 | 10/2008 | PSCUH | + | + | AbaR25 | Int. II | A1 | ND |
| K51-71 | 11/2008 | PSCUH | + | + | AbaR25 | Int. II | A1 | ND |
| K51-72 | 12/2008 | PSCUH | + | + | AbaR25 | Int. II | A1 | ST2 |
| K51-73 | 12/2008 | PSCUH | + | + | AbaR25 | Int. II | A1 | ND |
| K51-74 | 1/2009 | PSCUH | + | + | ΔAbaR25 | Int. II | A4 | ND |
| K51-75 | 1/2009 | PSCUH | + | + | AbaR25 | Int. II | A1 | ND |
| K51-76 | 1/2009 | PSCUH | + | + | AbaR25 | Int. II | A5 | ND |
| K51-77 | 1/2009 | PSCUH | + | − | AbaR25 | Int. II | A6 | ND |
| K51-78 | 3/2009 | PSCUH | + | + | AbaR25 | Int. II | A7 | ND |
| K51-79 | 3/2009 | PSCUH | + | + | AbaR25 | Int. II | A6 | ND |
| K51-80 | 3/2009 | PSCUH | + | + | AbaR25 | Int. II | A6 | ND |
| K51-81 | 4/2009 | PSCUH | + | − | AbaR25 | Int. II | A1 | ND |
| K70-64 | 6/2009 | PSCUH | + | + | AbaR25 | Int. II | A8 | ND |
| K70-65 | 6/2009 | PSCUH | + | + | AbaR25 | Int. II | A8 | ND |
| K70-66 | 6/2009 | PSCUH | − | − | AbaR25 | Int. II | A9 | ND |
| K70-67 | 8/2009 | PSCUH | + | + | AbaR25 | Int. II | A10 | ND |
| K70-68 | 9/2009 | PSCUH | − | − | AbaR25 | Int. II | A9 | ND |
| K70-69 | 9/2009 | PSCUH | − | − | AbaR25 | Int. II | A9 | ND |
| K70-70 | 10/2009 | PSCUH | + | + | AbaR25 | Int. II | A10 | ND |
| K70-71 | 10/2009 | PSCUH | − | − | AbaR25 | Int. II | A9 | ND |
| K70-72 | 10/2009 | PSCUH | − | − | AbaR25 | Int. II | A9 | ND |
| K70-73 | 11/2009 | PSCUH | − | − | AbaR25 | Int. II | A9 | ND |
| K70-74 | 12/2009 | PSCUH | − | − | AbaR25 | Int. II | A9 | ST2 |
| K70-75 | 12/2009 | PSCUH | − | − | AbaR25 | Int. II | A4 | ND |
| K70-76 | 12/2009 | PSCUH | − | − | AbaR25 | Int. II | A9 | ND |
| K70-77 | 7/2009 | DRH | + | + | AbaR25 | Int. II | A2 | ND |
| K70-78 | 3/2009 | VzH | + | − | AbaR25 | Int. II | A1 | ND |
| K70-79 | 3/2009 | VH | + | + | AbaR25 | Int. II | A2 | ND |
| K70-80 | 6/2009 | R1H | + | + | AbaR25 | Group 4 | A3 | ST2 |
| K70-81 | 6/2009 | R1H | − | − | AbaR25 | Int. II | A1 | ST2 |
Abbreviations: PSCUH, P. Stradins Clinical University Hospital, Riga; DRH, Daugavpils Regional Hospital, Daugavpils; VzH, Vidzemes Hospital, Valmiera; VH, Venstpils Hospital, Ventspils; R1H, Riga 1st Hospital, Riga; Int., international clone; ST, sequence type; ND, not determined.
Resistance to carbapenems was confirmed in all the isolates (see Table S1 in the supplemental material). Thirty-four isolates showed high levels of resistance to ciprofloxacin. In addition, high levels of resistance to amikacin, gentamicin, and tobramycin were detected in 24 of these isolates. All isolates were susceptible to colistin. Pulsed-field gel electrophoresis (PFGE), using ApaI-digested genomic DNA, assigned all the 34 ciprofloxacin-resistant isolates to indistinguishable or closely related patterns, showing >80% similarity to each other (Table 1) (17). Only the ciprofloxacin-susceptible isolate belonged to a possibly related PFGE pattern showing 70% to 80% similarity to the other patterns. The isolates belonged to international clone II (n = 33) and PCR-based group 4 (n = 2), using two multiplex PCRs targeting the ompA, csuE, and blaOXA-51-like genes (18). Of note, the band pattern of PCR-based group 4 differs from that of international clone II only in the negative result for the csuE allele, which could simply be due to a single polymorphism in the primer annealing regions (19). Multilocus sequence typing (MLST) was performed on eight isolates with different PFGE patterns, including the two isolates from PCR-based group 4 (http://www.pasteur.fr/recherche/genopole/PF8/mlst/). The isolates were all sorted into ST2 (Table 1).
PCR assays were used to detect antimicrobial resistance genes encoding the OXA carbapenemases (blaOXA-51-like, blaOXA-23-like, blaOXA-24-like, and blaOXA-58-like), metallo-β-lactamases (blaVIM, blaGIM, blaIMP, and blaSPM), and 16S rRNA methylases (armA, rmtA, rmtB, rmtC, rmtD, and npmA) (20). All the isolates were positive for blaOXA-51-like and blaOXA-23-like, while none of them carried the blaOXA-24-like, blaOXA-58-like, or metallo-β-lactamase genes. The armA gene was detected in all the isolates (n = 24) showing high levels of resistance to amikacin, gentamicin, and tobramycin (Table 1). Furthermore, sequence analysis detected the occurrence of the intI1-aacA4-catB8-aadA1-qacEΔ1 class 1 integron in isolate K51-65. Subsequent PCR assays (see Tables S2 to S4 in the supplemental material) confirmed the occurrence of this integron in 20 isolates (Table 1). Interestingly, all the class 1 integron-positive isolates carried the armA gene, indicating the succeeding acquisition of armA first and the intI1-aacA4-catB8-aadA1-qacEΔ1 integron second.
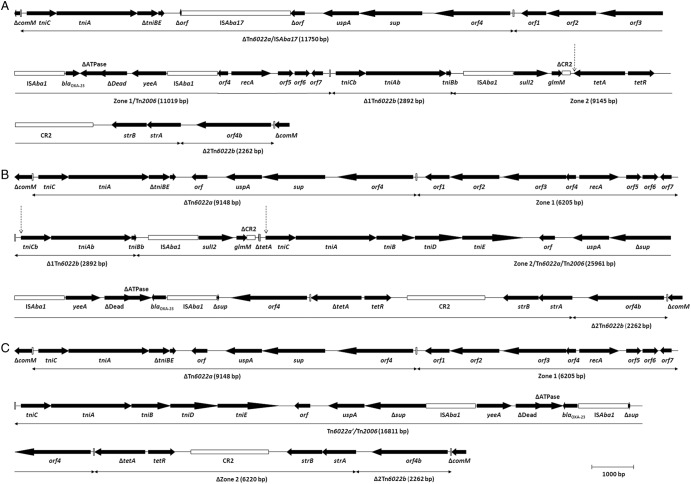
The comM gene was interrupted in all the isolates, indicating the occurrence of AbaR islands. AbaR25, a novel AbaR4-like island of 46,469 bp, was detected and fully sequenced in isolate K51-65 (Fig. 1; see also Table S2 in the supplemental material). AbaR25 was most similar to Tn6167 and successively consisted of (i) ΔTn6022a (9,148 bp) at the left-hand end; (ii) zone 1 (6,205 bp), including seven conserved open reading frames of unknown function and a proposed tyrosine integrase gene; (iii) Δ1Tn6022b (2,892 bp); (iv) zone 2 (9,049 bp), including the ISAba1-sul2-ΔCR2-tetA-tetR-CR2-strB-strA configuration; and (v) Δ2Tn6022b (2,262 bp), containing orf4b, at the right-hand end (15). Interestingly, the tetA gene of zone 2 was interrupted by a sequence of 11,998 bp, representing a complete second copy of Tn6022a. In addition, the sup gene of this Tn6022a was interrupted by a sequence of 4,805 bp (Tn2006). Transposition of Tn6022a/Tn2006 into tetA was associated with the standard 5-bp target duplication (11). PCR assays (see Tables S3 and S4 in the supplemental material) confirmed the occurrence of AbaR25 in 33 isolates and detected the occurrence of ΔAbaR25, a variant form of AbaR25, in one additional isolate (Table 1). ΔAbaR25 was fully sequenced in isolate K51-74 and found to be identical to AbaR25 except for the occurrence of an internal deletion of 5,822 bp (Fig. 1). The deletion included ∼2,750 bp of Δ1Tn6022b, ISAba1-sul2-ΔCR2-ΔtetA of zone 2, and ∼150 bp of Tn6022a/Tn2006. The deletion was most likely due to a single intramolecular recombination event between Δ1Tn6022b and the corresponding region of Tn6022a/Tn2006. This was indicated by the occurrence of Tn6022a′/Tn2006, a novel structure characterized by a mosaic sequence derived from the two recombined segments. On the other hand, sequence analysis of the AbaR island in the ciprofloxacin-susceptible isolate (K51-67) detected an island of 16,808 bp showing 99.9% similarity with AbaR4 (GenBank accession numbers JN107991 and CP001182) (7, 12).
Fig 1.
Structures of three different Acinetobacter baumannii resistance islands: Tn6167 (A), AbaR25 (B), and ΔAbaR25 (C) (GenBank accession numbers JN968483, JX481978, and JX481979, respectively). The genes and open reading frames (orf) are shown by labeled arrows, with the arrowhead indicating the direction of transcription. The mobile elements ISAba17, ISAba1, and CR2 are shown as labeled open boxes. Inverted repeats are shown as vertical bars. The vertical dashed arrow in panel A defines the transposition site of the second copy of Tn6022a in AbaR25. The vertical dashed arrows in panel B define the deletion in ΔAbaR25. The genes and genetic structures are drawn to scale.
Overall, our results detected the occurrence of a strain, representing 34 out of 35 isolates, responsible for a prolonged ongoing outbreak/endemic status of hospital-acquired infections in Latvia. The strain was linked to international clone II/ST2 and carried the blaOXA-23-like carbapenemase gene within a novel AbaR4-like island. The strain was partially equipped with the armA gene, and the armA-positive subdivision of this strain has subsequently acquired the aacA4-catB8-aadA1 class 1 integron. The occurrence of minor differences among the PFGE patterns and limited variations in the phenotypic and genotypic resistance characteristics was anticipated since the isolates were collected over a prolonged period of more than 1 year (17). As previously described, a linkage was detected between international clone II/ST2 and the armA, aacA4-catB8-aadA1, and AbaR4-like antimicrobial resistance elements (2, 14). Further studies are required in order to determine the evolution and geographical and clonal distribution of the different AbaR islands in A. baumannii.
Nucleotide sequence accession numbers.
The nucleotide sequences of AbaR25 and ΔAbaR25 were deposited in the GenBank nucleotide database under accession numbers JX481978 and JX481979, respectively.
Supplementary Material
ACKNOWLEDGMENTS
Bjørg Haldorsen and Bettina Aasnæs are acknowledged for excellent technical assistance. We thank the Genotyping of Pathogens and Public Health platform (Institut Pasteur) for coding MLST alleles and profiles available at www.pasteur.fr/recherche/genopole/PF8/mlst.
Part of this project was funded by a research grant from the Northern-Norway Regional Health Authority, Latvian Research Council, and National Research Programme.
Footnotes
Published ahead of print 10 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01783-12.
REFERENCES
- 1. Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951 [DOI] [PubMed] [Google Scholar]
- 2. Karah N, Sundsfjord A, Towner K, Samuelsen O. 2012. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15:237–247 [DOI] [PubMed] [Google Scholar]
- 3. Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836 [DOI] [PubMed] [Google Scholar]
- 4. Nemec A, Dolzani L, Brisse S, van den Broek P, Dijkshoorn L. 2004. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 53:1233–1240 [DOI] [PubMed] [Google Scholar]
- 5. Cho YJ, Moon DC, Jin JS, Choi CH, Lee YC, Lee JC. 2009. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn. Microbiol. Infect. Dis. 64:185–190 [DOI] [PubMed] [Google Scholar]
- 6. Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7 doi:10.1371/journal.pgen.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJ, Sicheritz-Ponten T, De Bellis G, Visca P, Cassone A, Carattoli A. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Post V, White PA, Hall RM. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:1162–1170 [DOI] [PubMed] [Google Scholar]
- 10. Krizova L, Dijkshoorn L, Nemec A. 2011. Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrob. Agents Chemother. 55:3201–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nigro SJ, Hall RM. 2012. Antibiotic resistance islands in A320 (RUH134), the reference strain for Acinetobacter baumannii global clone 2. J. Antimicrob. Chemother. 67:335–338 [DOI] [PubMed] [Google Scholar]
- 12. Hamidian M, Hall RM. 2011. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J. Antimicrob. Chemother. 66:2484–2491 [DOI] [PubMed] [Google Scholar]
- 13. Hornsey M, Loman N, Wareham DW, Ellington MJ, Pallen MJ, Turton JF, Underwood A, Gaulton T, Thomas CP, Doumith M, Livermore DM, Woodford N. 2011. Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J. Antimicrob. Chemother. 66:1499–1503 [DOI] [PubMed] [Google Scholar]
- 14. Seputiene V, Povilonis J, Suziedeliene E. 2012. Novel variants of AbaR resistance islands with a common backbone in Acinetobacter baumannii isolates of European clone II. Antimicrob. Agents Chemother. 56:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nigro SJ, Hall RM. 2012. Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J. Antimicrob. Chemother. 67:1342–1346 [DOI] [PubMed] [Google Scholar]
- 16. Zhou H, Zhang T, Yu D, Pi B, Yang Q, Zhou J, Hu S, Yu Y. 2011. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob. Agents Chemother. 55:4506–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807–815 [DOI] [PubMed] [Google Scholar]
- 19. Towner KJ, Levi K, Vlassiadi M. 2008. Genetic diversity of carbapenem-resistant isolates of Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 14:161–167 [DOI] [PubMed] [Google Scholar]
- 20. Karah N, Haldorsen B, Hermansen NO, Tveten Y, Ragnhildstveit E, Skutlaberg DH, Tofteland S, Sundsfjord A, Samuelsen O. 2011. Emergence of OXA-carbapenemase- and 16S rRNA methylase-producing international clones of Acinetobacter baumannii in Norway. J. Med. Microbiol. 60:515–521 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.