Abstract
Microbial community composition and diversity at a diesel-contaminated railway site were investigated by pyrosequencing of bacterial and archaeal 16S rRNA gene fragments to understand the interrelationships among microbial community composition, pollution level, and soil geochemical and physical properties. To this end, 26 soil samples from four matrix types with various geochemical characteristics and contaminant concentrations were investigated. The presence of diesel contamination significantly impacted microbial community composition and diversity, regardless of the soil matrix type. Clean samples showed higher diversity than contaminated samples (P < 0.001). Bacterial phyla with high relative abundances in all samples included Proteobacteria, Firmicutes, Actinobacteria, Acidobacteria, and Chloroflexi. High relative abundances of Archaea, specifically of the phylum Euryarchaeota, were observed in contaminated samples. Redundancy analysis indicated that increased relative abundances of the phyla Chloroflexi, Firmicutes, and Euryarchaeota correlated with the presence of contamination. Shifts in the chemical composition of diesel constituents across the site and the abundance of specific operational taxonomic units (OTUs; defined using a 97% sequence identity threshold) in contaminated samples together suggest that natural attenuation of contamination has occurred. OTUs with sequence similarity to strictly anaerobic Anaerolineae within the Chloroflexi, as well as to Methanosaeta of the phylum Euryarchaeota, were detected. Anaerolineae and Methanosaeta are known to be associated with anaerobic degradation of oil-related compounds; therefore, their presence suggests that natural attenuation has occurred under anoxic conditions. This research underscores the usefulness of next-generation sequencing techniques both to understand the ecological impact of contamination and to identify potential molecular proxies for detection of natural attenuation.
INTRODUCTION
Anthropogenic contamination with recalcitrant organic compounds has created a large industry for remediation of these polluted sites (1). Among the variety of decontamination tools, in situ biodegradation of contaminants has emerged as a low-cost, less-invasive treatment compared to traditional ex situ techniques. The ubiquity of hydrocarbon-utilizing microorganisms dictates that, with time, degradation will occur to some extent without human intervention—so-called natural attenuation (2). Cultivation-independent molecular approaches have been used to assess the impact of pollution on microbial composition at contaminated sites, and phylogenetic and functional gene-targeted tools have been developed as one of the lines of evidence to monitor microbial populations at natural attenuation sites for degradation potential and activity (3, 4).
To this end, a number of studies have examined how the presence of total petroleum hydrocarbon (TPH) contamination impacts microbial community structure and function. In such studies, chemical analysis of TPH concentration is coupled to molecular analyses. Shifts in bacterial composition have been observed in microcosm experiments due to either the spiking of TPH at specific concentrations and degradation under fully oxic conditions (5, 6) or degradation of TPH coupled to a range of alternative electron acceptors (7). Aerobic degradation of the alkane fraction of TPH specifically has been monitored with real-time PCR targeting phylogenetic and functional gene markers (4, 8–10). The results indicated that microbial community structure and ecological function are clearly impacted by the presence of TPH. However, conclusions from studies on short-term spiking of soils are difficult to translate to long-term in situ natural attenuation conditions.
In addition to changes in microbial populations due to the presence and concentration of contaminants, community composition and diversity are also dependent on the geochemical properties of the subsurface. A number of studies have indicated that various factors dictate the composition and activity of microbes in uncontaminated locations, including geographic region (11), soil type (12), soil pH (11), and land use (13, 14). Furthermore, it has been shown in TPH-contaminated soil that soil type influences the makeup of biodegrading populations (15, 16). This underscores the complexity of factors influencing soil microbial communities and the challenge of elucidating key populations in in situ bioremediation.
In this study, we have investigated microbial diversity and ecological significance in the natural attenuation process through identifying populations present in situ at a diesel-contaminated site. To this end, pyrosequencing of PCR-amplified bacterial and archaeal 16S rRNA gene fragments of both Bacteria and Archaea was utilized as a high-throughput technique to examine the diversity and microbial groups present in contaminated and corresponding clean samples. Whereas previous work examined the community structure in diesel-spiked sediments in microcosms (5, 6), this study investigated field-collected samples representing various soil matrices and contamination levels. The interrelationships among community composition and diversity, contamination, and soil geochemical properties were determined, giving an understanding of the ecological response to changing environmental conditions. Through investigating the impact of long-term in situ diesel contamination, an understanding of natural attenuation characteristics and requirements is developed. This description of microbial community dynamics provides a basis for the development of more-comprehensive theoretical models and molecular proxies for the monitoring of natural attenuation.
MATERIALS AND METHODS
Site description.
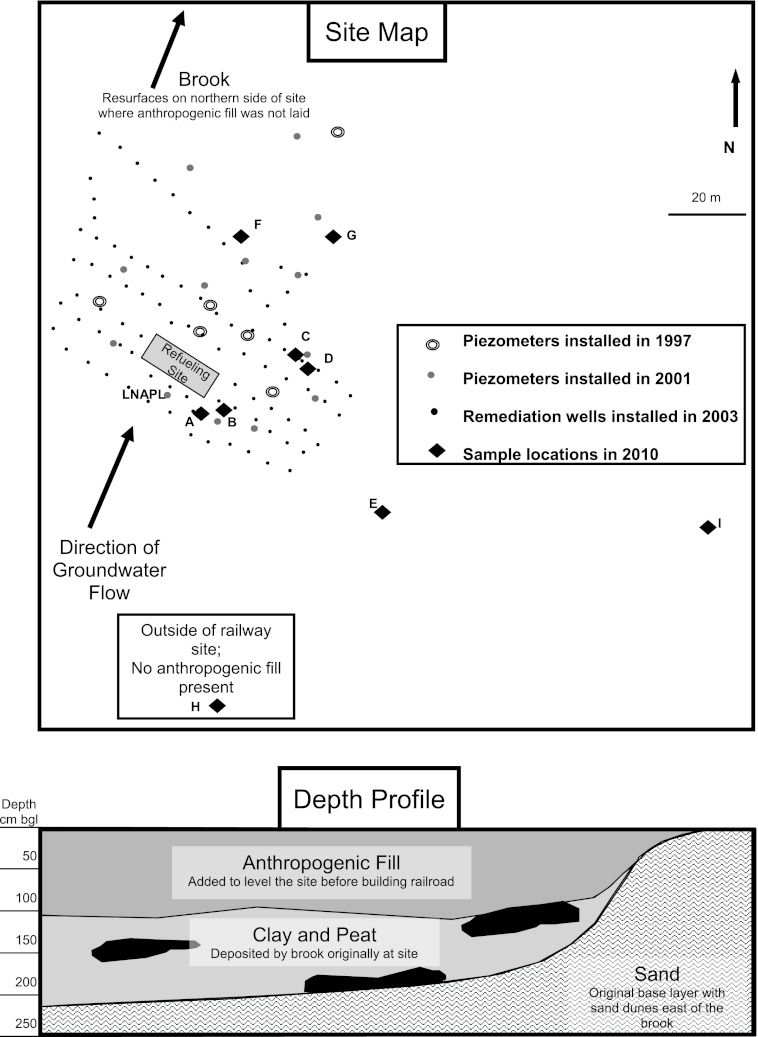
The railway refueling station in Wegliniec, Poland, was contaminated with diesel from 1970 to 2000 (Fig. 1). Eighteen monitoring wells were installed in 1997 and in 2001 to characterize the soil diesel contamination, expressed as TPH concentrations, as well as to monitor contamination in the groundwater. During a previous remediation project in 2003, an additional 89 wells with a depth of 5 m were placed in a tight gridlike pattern for a pump-and-treat style approach. The use of these wells, however, was discontinued within 6 months due to clogging resulting from improper filtering of the wells.
Fig 1.
Site schematic (top) indicating the location of piezometers installed previously (1997 and 2001), inoperable remediation wells (installed in 2003), and sampling locations for the current study (2010), as well as subsurface lithology giving the depth profile corresponding to the site map (bottom). The LNAPL sample was collected in a piezometer from 2001. Sample H was located upstream outside the site in an area that neither encountered diesel contamination nor contains anthropogenic fill. bgl, below ground level.
The site was originally the location of a brook running from south to north across the site. This created a peat and clay layer on top of the base sand, flanked by higher sandy dunes on the eastern side of the site. In order to level the site for the construction of the railroad, the brook was filled with a heterogeneous layer containing coarse sand, gravel, and fine brown sand. Thus, within the contaminated area, 3 to 4 lithological layers were encountered: anthropogenic fill, followed by peat and/or clay, and finally, natural sand. To the east of the site, essentially two layers were present, an upper more-organic rich brown sand followed by very light sand.
The initial location of refueling was at the southwest corner of the site and was the source of contamination. In this area, up to 70 cm of light nonaqueous-phase liquid (LNAPL) of TPH was observed in piezometers in 2010, and solid-phase concentrations were higher than 20,000 mg/kg of dry matter. LNAPL has spread across the site in the same direction as the groundwater, which was in a north-northeast direction. The thicker layer of anthropogenic fill deposited on the northern area of the site was contaminated due to the groundwater transportation of dissolved diesel. The extent of weathering varied across the site, as observed from the color and texture of the contamination in piezometers.
Soil sampling.
Sampling was performed in 2010 at nine locations across the site, including near the refueling station (sites A and B), just downstream therefrom (sites C and D), and farther removed from the station (sites E, F, and G). Additionally, samples were taken where no anthropogenic fill was present, either because the location was upstream and just outside the rail yard (site H) or due to the natural lithology of the subsurface (site I). Hand augers were used to collect layers of sediment at various depths for each sampling location. A full description of the number of samples collected at each location and their depth is given in Table 1. The samples were collected and stored in large high-density polyethylene (HDPE) containers for further characterization. Additionally, the pure-product LNAPL sample was collected and stored in a glass tube. All samples were stored at 4°C.
Table 1.
Physical, geochemical, and contaminant characteristics of soil samplesa
| Sample | Depth (cm bgl) | Groundwater level (cm bgl) | Soil matrix | Organic matter (%) | pH | TPH concn (g TPH/kg)b |
|---|---|---|---|---|---|---|
| A1 | 0–100 | 230 | Fill | 10 | 7.10 | 4.26 |
| A2 | 130–230 | Fill | 11 | 6.88 | 22.21 | |
| B1 | 0–70 | 170 | Fill | 9 | 7.41 | — |
| B2 | 80–150 | Clay | 5 | 5.15 | 2.88 | |
| B3 | 150–170 | Peat | 12 | 6.50 | 17.82 | |
| B4 | 150–250 | Peat | 10 | 6.80 | 16.31 | |
| C1 | 20–90 | 200 | Fill | 9 | 7.55 | — |
| C2 | 90–180 | Peat | 6 | 7.37 | — | |
| C3 | 200–220 | Peat | 7 | 7.02 | 6.38 | |
| D1 | 0–130 | 200 | Fill | 13 | 7.35 | — |
| D2 | 130–160 | Clay | 8 | 7.17 | — | |
| D3 | 170–200 | Clay | 7 | 6.98 | 9.99 | |
| D4 | 200–230 | Peat | 54 | 6.18 | 11.44 | |
| D5 | 230–250 | Sand | 3 | 6.22 | 13.65 | |
| E1 | 40–120 | 120 | Fill | 5 | 6.51 | — |
| E2 | 120–130 | Fill | 35 | 7.11 | 8.47 | |
| F1 | 80–150 | 190 | Sand | 2 | 7.37 | — |
| F2 | 190–220 | Sand | 2 | 7.01 | 1.28 | |
| G1 | 50–120 | Unknown; >180 | Fill | 4 | 7.17 | — |
| G2 | 120–160 | Fill | 5 | 7.33 | — | |
| G3 | 160–180 | Fill | 4 | 7.31 | — | |
| H1 | 10–40 | 40 | Peat | 29 | 6.55 | — |
| H2 | 40–70 | Peat | 56 | 5.48 | — | |
| H3 | 70–100 | Sand | 5 | 5.82 | — | |
| I1 | 10–40 | 240 | Sand | 2 | 4.42 | — |
| I2 | 70–240 | Sand | 1 | 4.23 | — |
bgl, below ground level.
—, clean sample without diesel contamination.
Subsamples were taken directly from the sediment layers in the field using sterile metal spatulas and placed in sterile 50-ml tubes for molecular analysis. These samples were immediately frozen in liquid nitrogen, stored on dry ice during transport (1 day), and stored at −80°C prior to further processing.
Chemical analyses.
Soil samples were sieved at 2 mm and analyzed for a variety of parameters. Organic matter content was determined as the weight difference due to ignition at 550°C. pH was measured in the supernatant of a 1:2.5 solid-to-liquid (g/ml) ratio suspension of soil sample in 0.01 M CaCl2 (17). Diesel concentrations, measured as TPH, were determined by extraction and quantification with gas chromatography. Extraction was performed according to the NEN 5733 solvent shake extraction protocol with acetone and hexane (18). For pure-product samples, 0.2 g LNAPL was dissolved in 25 ml hexane. Hexane was dried with Na2SO4, and TPH was measured on an HP 5890 gas chromatograph (GC) with a simulated distillation (Sim Dist) column and flame ionization detector with nitrogen as the carrier gas. After 5 min at an initial temperature of 40°C, the temperature was increased at 10°C per minute to a final temperature of 300°C. TPH was determined for samples portraying a diesel chromatogram pattern as the area between C10 and C40. Carbon number fractions were determined by using the retention times of a boiling point standard. Samples lacking a chromatogram indicative of diesel but, rather, typical for trace amounts of organic matter extracted with hexane were considered clean for the purposes of this analysis.
DNA extraction, amplification, and pyrosequencing of the 16S rRNA gene.
Total microbial community DNA was extracted from 0.5 g of each soil sample using the FastDNA spin kit for soil (MP Biomedicals, Solon, OH) according to the manufacturer's instructions. The amount and quality of the extracted DNA (average molecular size and purity) were estimated on 1% agarose gels and using NanoDrop spectrophotometer readings (Thermo Scientific, Wilmington, DE). Extracted DNA was stored at −20°C for downstream applications.
In this study, we used a two-step PCR protocol as previously described (19). With this approach, tags and adapters were added in a second round of PCR amplification. This method facilitates DNA amplification from complex biological samples (20), increases reproducibility, and recovers higher genetic diversity, avoiding the amplification bias that is introduced by using long fusion primers in one-step PCR amplification (21).
The first PCR amplification was performed using the modified primers 341F (CCTAYGGGRBGCASCAG) and 806R (GGACTACNNGGGTATCTAAT) (20) to amplify a 466-bp fragment of the 16S rRNA gene flanking the V3 and V4 regions. The PCR mix (40 μl) contained 1× Phusion HF buffer containing 2.5 mM MgCl2 (Finnzymes, Vantaa, Finland), 0.2 mM deoxynucleoside triphosphate (dNTP) mixture, 0.8 U Phusion hot start DNA polymerase, 2 μM each primer, 25 μg bovine serum albumin (BSA; molecular grade, 10 mg ml−1 stock), and 1 μl template. The PCR conditions were an initial activation of the hot start polymerase at 98°C for 30 s, followed by 30 cycles of 98°C for 5 s, 56°C for 20 s, and 72°C for 20 s, and a final extension at 72°C for 5 min. Analysis of PCR products was done on a 1% agarose gel stained with ethidium bromide, and bands of the correct size were cut and purified using the Montage gel extraction kit (Millipore, Billerica, MA). The quality and concentration of the purified DNA was determined through analysis on a 1% agarose gel stained with ethidium bromide (Gel Doc; Bio-Rad, Hercules, CA) and using NanoDrop. A second round of PCR was performed as described above, except that primers with an adapter and barcodes of 10 nucleotides in length were used (see Table S1 in the supplemental material). Furthermore, the number of PCR cycles was reduced to 15. The PCR products were analyzed on 1% (wt/vol) agarose gel, and DNA was extracted from the agarose gel as described above, quantified using the Quant-iTdsDNA high-sensitivity assay kit and the Qubit fluorometer (Invitrogen, Grand Island, NY), and mixed in approximately equal concentrations (4 × 106 copies μl−1) to ensure equal representation of each sample. These samples were pooled with other samples (total of 72 samples) and were sequenced on one of the two regions of a 70_75 GS PicoTiterPlate (PTP) by using a Titanium kit and GS FLX system according to the manufacturer's instructions (Roche, Branford, CT).
Analysis of the pyrosequencing data set.
Pyrosequencing data were analyzed using the QIIME pipeline (http://qiime.org/) (22). Sequences were deemed of low quality and were removed if they did not comply with the following default quality parameters: (i) include a perfect match to the sequence tag and the 16S rRNA gene primer, (ii) be at least 200 bp in length, (iii) have no ambiguous bases, and (iv) have no homopolymers longer than 6 nucleotides. Once trimmed and assigned to samples, data were processed using the QIIME's UCLUST method in order to cluster the sequences into operational taxonomic units (OTUs) at the 97% similarity level. A set of representative sequences (the most abundant sequence in each OTU) were aligned using PyNAST (23) against the Greengenes core set (24). Possible chimeric sequences were identified using QIIME's ChimeraSlayer and subtracted from the previously generated OTU list, producing a nonchimeric nonredundant OTU list. The taxonomic affiliation of each OTU was determined using the RDP Classifier at a confidence threshold of 80% (25). Aligned sequences (OTU representatives) were used to generate a phylogenetic tree with FastTree (26). The complete pyrosequencing data set is available at the European Bioinformatics Institute (http://www.ebi.ac.uk/ena/data/view/ERP001958) under accession number ERP001958.
Statistical analyses.
In order to relate the changes in microbial community composition to environmental variables, redundancy analysis (RDA) was used as implemented in the CANOCO 4.5 software package (Biometris, Wageningen, The Netherlands). The presence/absence and relative abundances of OTUs obtained from the pyrosequencing data set were used. The environmental variables tested were presence/absence of contamination, TPH concentration, pH, organic matter, and ground water level. All of the environmental data were transformed as log(1 + x). A Monte Carlo permutation test based on 999 random permutations was used to determine whether or not and which of the experimental variables significantly contributed to explaining the observed variance in composition of microbial communities. The community structure was visualized via ordination triplots with scaling focused on intersample differences. For all statistical analyses, correlations were considered highly significant at a P value of <0.001 and significant at a P value of <0.05. The significance of the observed differences for carbon fraction distribution and Shannon and Chao1 diversity indexes were verified using the Student t test.
RESULTS AND DISCUSSION
General characteristics of the soil samples.
Due to both the lithological and contamination patterns at the site, characterization of the 26 samples indicated considerable variation in the physical and geochemical properties of the soils collected (Table 1). The organic matter content correlated well with the soil type, with fill and sand samples generally below 10% and 2%, respectively, and peat samples above 10% organic matter in most cases. pH varied across samples from 4.2 to 7.5; sandy soils showed a slightly lower pH. The TPH concentrations of the 11 contaminated samples varied from 1.3 g/kg to 22.2 g/kg. The highest levels of contamination were encountered at sampling sites near the refueling location and at or below the groundwater level.
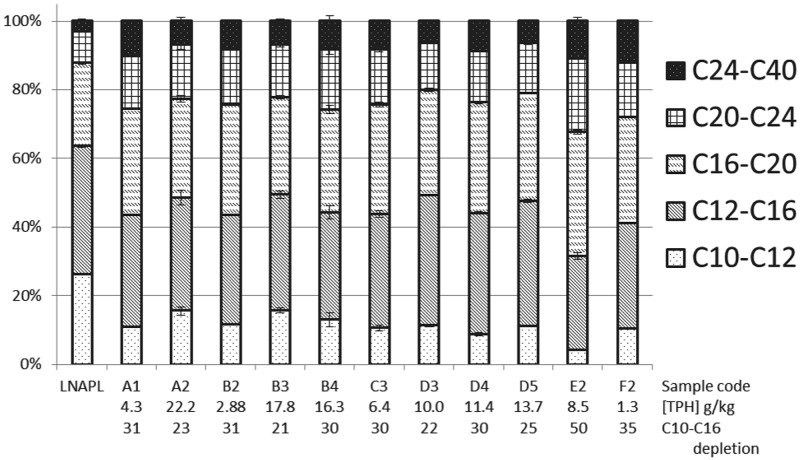
The distribution and composition of the TPH contamination were indicative of natural in situ degradation processes. Diesel concentrations were notably reduced in samples obtained above the groundwater level in the source area where the contamination originated; this indicates that degradation may occur in the vadose zone. The TPH carbon number fraction distribution also points toward degradation. An LNAPL sample collected in the source zone at the location of refueling shows that the original diesel contamination contained mainly low-molecular-weight (LMW) hydrocarbons, with 63% of the mass from compounds in the C10-to-C16 range (Fig. 2). Soil samples were overall depleted in LMW compounds compared to the LNAPL. Samples well above the groundwater level (site A1) or farther downstream (sites E2 and F2) showed notably less LMW, with up to 50% of the C10-to-C16 fraction depleted relative to the LNAPL signature. Reductions in the amount of LMW compounds were also observed across the site; the difference in relative carbon fraction abundance was significant for fractions C10 to C12 and C12 to C16 when comparing source samples A2 and B3 to plume samples E2 and F2 (P = 0.02).
Fig 2.
TPH carbon fraction distribution in LNAPL and contaminated samples. Carbon fraction percentages are the relative distribution within each sample to allow comparison irrespective of TPH concentration. C10-to-C16 depletion is calculated using the composition of the LNAPL as the starting material. Absolute TPH concentrations are given below each sample name. Error bars on soil samples are the standard deviation of the average of each fraction for triplicate TPH extractions.
This shift in contaminant composition can most likely be attributed to biological degradation. As the LNAPL source spreads across the site, microbial degradation occurs, thereby degrading LMW compounds. Microbial preference for low-molecular-weight compounds has been observed previously (27, 28). These results suggest that natural attenuation potential is present at this location and that biodegradation processes are ongoing, which is further corroborated by observed changes in microbial community composition, as described below.
General analysis of the pyrosequencing data set.
The phylogenetic composition and diversity of the microbial communities present in the soil samples collected throughout the diesel-contaminated railway site were profiled using pyrosequencing of PCR-amplified bacterial and archaeal 16S rRNA gene fragments. After trimming and quality filtering of the raw reads, the sequences were clustered into operational taxonomic units (OTUs) using a similarity threshold of 97%, yielding a total of 40,912 high-quality, nonchimeric OTUs. These OTUs represented a total of 201,585 reads, distributed in 7,753 ± 1,032 (mean ± standard deviation) sequences per sample with an average length of 401.8 bp. Representative sequences of each OTU were classified into the domains Bacteria (91.7% of the total data set) and Archaea (7.7%), using the RDP Classifier with a confidence threshold of 80%. A small proportion of sequences (0.6%) could not be classified at the domain level.
Relatively high proportions of sequences, ranging from 2% to 46% (average, 21%), could not be classified below the phylum level. These considerable proportions of unclassified bacterial and archaeal sequences fall in the range of values obtained in other recent studies applying tag-pyrosequencing to different soil and sediment ecosystems, where, in principle, a high microbial diversity is expected. For example, Inceoglu et al. found 15 to 45% unclassified bacteria in a pyrosequencing data set obtained from potato field soil samples (20), and a similar range of unclassified bacterial reads (11 to 24%) was found in a collection of forest and grassland soils (13). A possible interpretation of these high proportions of unclassified sequences could be that they belong to as-yet-uncultured, unrecognized, or novel bacterial or archaeal species.
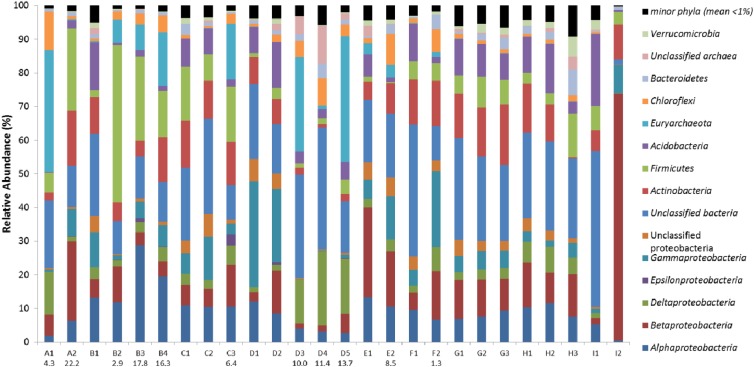
The OTUs were classified into the domains Bacteria (27 phyla) and Archaea (2 phyla). On average for the complete data set, the major microbial phyla were Proteobacteria (36.6% ± 14.5% of the reads), Firmicutes (9.0% ± 10.2%), Actinobacteria (9.3% ± 5.1%), Acidobacteria (6.6% ± 5.5%), Euryarchaeota (6.1% ± 11.3%), Chloroflexi (2.8% ± 3.0%), Bacteroidetes (1.8% ± 1.7%), and Verrucomicrobia (1.1% ± 1.3%) (Fig. 3; also see Table S2 in the supplemental material).
Fig 3.
Relative abundance of dominant microbial phyla and Proteobacteria classes (>1% average abundance for the total pyrosequencing data set). Unclassified bacteria and archaea correspond to sequences that could only be classified to the domain level. Unclassified Proteobacteria correspond to sequences that could only be classified to the phylum level. Groups that were observed at less than 1% average abundance were grouped in minor phyla (see Fig. S1 in the supplemental material). Values below sample names indicate contamination as g TPH/kg soil.
Initial examination of the phylogenetic distribution identified sample I2 as an outlier. Whereas the relative abundance of individual phyla does not surpass 54.4% in any other sample, 83.6% of sequences were classified as Proteobacteria in sample I2 (Fig. 3). Additionally, 88% of Proteobacteria were from the Betaproteobacteria (Fig. 3; also see Table S3 in the supplemental material). Sampling was performed at location I to provide a clean sand sample as a control for comparison to contaminated sand samples. However, visual comparison of I2, a coarse light-colored sand, to other clean sand samples (F1, H3, and I1), which were notably darker and finer, showed that I2 was dissimilar. This was further confirmed by geochemical parameters (Table 1), phylogenetic distribution (Fig. 3), and alpha-diversity (Table 2; see below). Redundancy analysis (see Fig. 5) indicated that, along with I2, I1 is also not representative of the clean samples.
Table 2.
Microbial richness and diversity as determined by Chao1 estimator and Shannon index
| Sample | Contamination | Chao1 estimate of OTU richness | 95% CIa | Shannon index (H′) | No. of high-quality sequences |
|---|---|---|---|---|---|
| A1 | Contaminated | 3,127 | 2,804; 3,520 | 6.49 | 8,462 |
| A2 | Contaminated | 3,259 | 2,994; 3,575 | 8.59 | 5,259 |
| B1 | Clean | 13,024 | 11,963; 14,222 | 10.69 | 8,285 |
| B2 | Contaminated | 4,493 | 4,183; 4,853 | 9.02 | 7,876 |
| B3 | Contaminated | 8,348 | 7,599; 9,211 | 8.77 | 8,054 |
| B4 | Contaminated | 6,431 | 5,847; 7,112 | 8.81 | 7,636 |
| C1 | Clean | 11,783 | 10,891; 12,785 | 10.8 | 7,781 |
| C2 | Clean | 9,226 | 8,499; 10,053 | 10.25 | 7,458 |
| C3 | Contaminated | 5,315 | 4,848; 5,860 | 8.25 | 7,527 |
| D1 | Clean | 6,794 | 6,235; 7,437 | 8.86 | 8,200 |
| D2 | Clean | 7,811 | 7,184; 8,528 | 9.44 | 8,158 |
| D3 | Contaminated | 4,227 | 3,783; 4,761 | 7.18 | 9,474 |
| D4 | Contaminated | 3,849 | 3,472; 4,302 | 7.43 | 8,258 |
| D5 | Contaminated | 4,587 | 4,155; 5,099 | 7.52 | 8,192 |
| E1 | Clean | 6,948 | 6,346; 7,646 | 9.17 | 7,691 |
| E2 | Contaminated | 6,529 | 5,983; 7,159 | 9.54 | 7,639 |
| F1 | Clean | 14,683 | 13,506; 16,005 | 10.94 | 7,798 |
| F2 | Contaminated | 5,439 | 4,932; 6,032 | 9.08 | 5,619 |
| G1 | Clean | 9,820 | 8,941; 10,826 | 10.44 | 5,663 |
| G2 | Clean | 11,929 | 11,075; 12,884 | 11.06 | 7,711 |
| G3 | Clean | 14,447 | 13,512; 15,483 | 11.16 | 10,332 |
| H1 | Clean | 12,824 | 11,878; 13,885 | 11.19 | 7,637 |
| H2 | Clean | 12,637 | 11,767; 13,609 | 11.27 | 8,294 |
| H3 | Clean | 7,716 | 7,227; 8,269 | 10.69 | 7,821 |
| I1 | Clean | 4,382 | 3,987; 4,851 | 8.54 | 7,867 |
| I2 | Clean | 521 | 463; 612 | 3.75 | 6,893 |
CI, confidence interval.
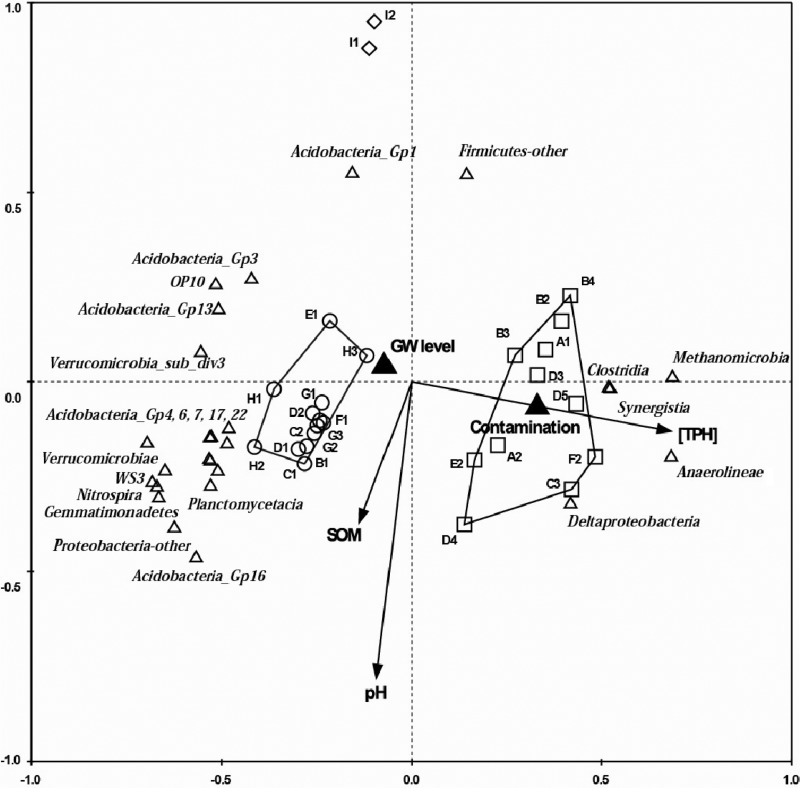
Fig 5.
Redundancy analysis triplot showing relationship between microbial composition at class level and soil geochemical and physical properties as described in Table 1. Clean samples, shown by open circles, cluster on the left quadrant; contaminated samples, shown by open squares, cluster on the right quadrant. Outliers I1 and I2 are shown by open diamonds. Classes are indicated with triangles, environmental parameters with arrows, and nominal environmental variables with filled triangles. The eigenvalues of the first (x) and second (y) canonical axes are 0.265 and 0.073, respectively. The plot can be interpreted qualitatively by following the direction of arrows for environmental parameters. The arrow length corresponds to variance that can be explained by the environmental variable. The direction of an arrow indicates an increasing magnitude of the environmental variable. The perpendicular distance between classes and environmental variable axes in the plot reflects their correlations. The smaller the distance, the stronger the correlation. SOM, soil organic matter; GW level, above groundwater level; [TPH], concentration of TPH.
The majority of the most-abundant bacterial OTUs were affiliated with Proteobacteria, which were observed at relative abundances of 10.4 to 82.9%, and all five major classes were represented (Fig. 3; also see Tables S3 and S4 in the supplemental material). Proteobacteria have been identified in many studies as the predominant phylum in soil samples (5, 13, 14, 29), playing an integral role in nutrient cycling (30). For example, the two most-abundant bacterial OTUs were classified within the genera Variovorax (OTU 7163, 4.1% of the total number of sequences, family Comamonadaceae, Betaproteobacteria), and Methylocystis (OTU 64060, 1.4%, family Methylocystaceae, Alphaproteobacteria). The ability of Alpha-, Beta-, and Gammaproteobacteria to utilize aliphatic and aromatic compounds has previously been established (31–33), and shifts in their abundance are often noted upon contamination with TPH or during bioremediation (34–36).
OTUs with sequences similar to Methanosaeta in the Methanosaetaceae family in the archaeal phylum Euryarchaeota showed remarkably high abundance. These OTUs were observed in many contaminated samples and represented more than 30% of all 16S rRNA gene sequences in samples A1 and D5 (Fig. 3; also see Table S2 in the supplemental material). A thorough analysis of archaeal abundance and diversity is given below.
The phyla Firmicutes, Actinobacteria, and Acidobacteria, found in this study in high abundances, have been noted previously in work examining uncontaminated soil samples (13, 14, 37). Furthermore, these phyla have been observed in soils contaminated with aliphatic or aromatic compounds, both in the polluted samples and the clean reference soil (7, 29, 38, 39). Bacteroidetes, which were not dominant in this study, have been found in clean and contaminated samples at similar relative abundances (1.9% and 1.8%, respectively). In a previous work using pyrosequencing to study uncontaminated soils, Bacteriodetes was identified as quite prevalent, with relative abundances of 15 to 25% (14); a different study, where pyrosequencing was also utilized to compare community structure in forest versus grasslands, placed Bacteriodetes into the rare-phylum group with <1% overall relative abundance (13).
Microbial diversity in pristine and contaminated samples.
Alpha-diversity estimations, based on OTUs defined at a genetic distance of 3% (i.e., sequence identity of 97%), showed that there was a significant reduction in microbial richness and diversity in the contaminated samples. Differences in the Chao1 richness estimator and Shannon diversity indexes were highly significant when comparing clean (uncontaminated) and contaminated samples (P = 0.00003 and P = 0.0002, respectively) (Table 2). When each soil matrix type (fill, peat, clay, and sand) was considered separately, only fill and peat samples showed a significant difference in microbial diversity between clean and contaminated samples (P < 0.05). The clay group was too small to assess the significance of observed differences in diversity. Within the sand group, diversity indices in clean and contaminated samples were not significantly different, most likely due to the low TPH concentration in sample F2, leading to a nonsignificant microbial response to the contamination. The peat and fill groups contained samples with much higher TPH concentrations in the polluted samples.
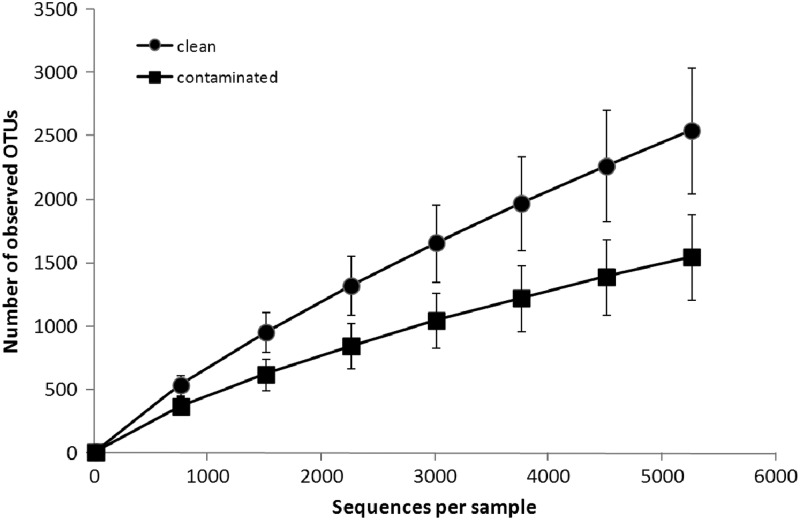
Finally, rarefaction curves estimating OTU richness confirmed the difference between clean and contaminated samples (Fig. 4), where clean samples were predicted to harbor higher microbial species richness than contaminated samples when excluding the outlier sample I2. Since no similar correlations between the other soil properties and richness were observed, the noted change in richness could be attributed to the presence of TPH contamination. No notable correlation between TPH concentration and diversity was observed, indicating that the presence of contamination, rather than its concentration, dictates changes in diversity. Apparently, a threshold concentration of TPH is required to elicit changes in diversity, above which no correlations of concentration and diversity would be observed, at least for the range of concentrations observed at this field site.
Fig 4.
Rarefaction curves indicating the average observed number of operational taxonomic units (OTUs) for clean samples and contaminated samples without the outlier I2 of the clean group. Error bars indicate ±standard deviation.
Correlations between soil properties, including contamination and microbial community composition.
Differences in microbial community composition have been attributed to soil properties, including vegetation and land use (13), soil matrix type (12, 16), and soil pH (37). We performed multivariate analysis in order to determine the impact of environmental parameters on the community structure. Here, geochemical soil characteristics, including organic matter content, pH, sample depth relative to the groundwater level, and soil matrix type, were considered. There was no significant correlation between overall community composition with either organic matter content or sample depth. However, redundancy analysis (RDA) indicated that the relative abundance of some microbial phyla, such as Acidobacteria subgroup 16 and unclassified Proteobacteria, was correlated with organic matter content (Fig. 5).
pH was found to contribute significantly to explaining the observed variation in community composition (P = 0.008). Acidobacteria subgroup 1 was correlated with low pH, while Acidobacteria subgroup 16 was somewhat correlated with increasing pH values (Fig. 5). High abundances of Acidobacteria subgroup 1 have previously been observed in samples with pH below 5.5, and high abundances of Acidobacteria subgroup 16 in samples above pH 6 (40). Additionally, all Acidobacteria subgroups were negatively correlated with the presence of TPH (Fig. 5), which has been observed previously (41). A similarly deleterious effect on the abundance and diversity of Acidobacteria was observed due to both the short-term and long-term presence of 2,4,6-trinitrotoluene (42), further supporting the conclusion that Acidobacteria are particularly sensitive to changes in soil conditions, including the presence of organic contamination.
RDA was also performed in order to analyze the impact on community composition of diesel contamination in general and, more specifically, the TPH concentration (Fig. 5). The presence of diesel in soil samples contributed significantly to explaining the variation in microbial community structure (P = 0.002), indicating that contamination, above all other geochemical characteristics, dictates microbial diversity. Contaminated samples were positively correlated with TPH contamination (Fig. 5; right cluster), whereas the clean samples were negatively correlated with TPH contamination (Fig. 5; left cluster). However, clean samples I1 and I2 show a divergent pattern. The concentration of TPH present was found to correlate with the relative abundance of a number of phyla (Fig. 5 and 6), including Chloroflexi, Firmicutes, and Euryarchaeota, as described in the following sections.
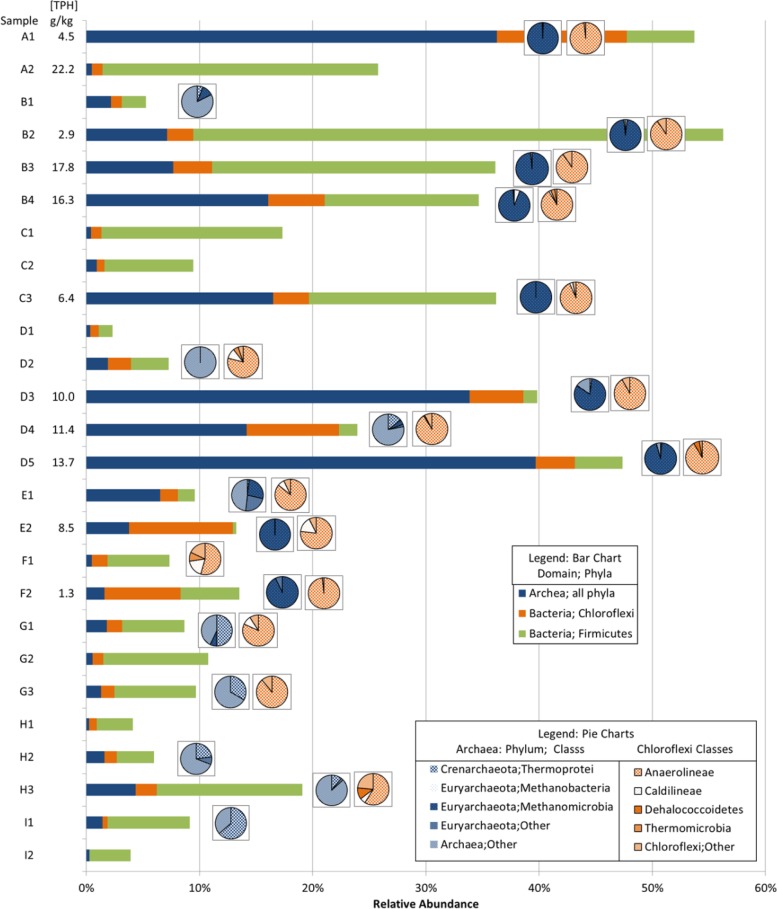
Fig 6.
Comparison of relative abundances within total data set of archaeal and bacterial phyla and classes associated with contamination. TPH concentrations are given next to sample names. Abundances of Archaea, Chloroflexi, and Firmicutes are given in the bar graph using the x axis. For samples in which the relative abundance of total Archaea or Chloroflexi exceeds 1%, pie charts are included for class level distribution.
Impact of TPH contamination on soil microbial communities.
A highly significant reduction in OTU richness and microbial diversity was observed to be correlated with TPH contamination, as described above. Previous works investigating microbial diversity have found that contamination may yield an increase (43–45) or a decrease (36, 46) in diversity. The reduction in diversity observed in our data set lies in stark contrast to the results of the only other pyrosequencing-based study investigating the impact of diesel contamination on microbial community structure (5). In that study, an increase in diversity and richness was observed 23 days after spiking and was attributed to the addition of hydrocarbon substrates with diverse structures. Whereas similar short-term effects are possible in the field, our study investigated microbial diversity under natural in situ conditions in soil contaminated for more than 40 years with diesel. Reduced diversity is most probably caused by the selective ecological pressures of TPH contamination, including (i) the introduction of toxic TPH concentrations and biodegradation products (47–49), (ii) the dominance of a pool of less-diverse, easily degradable diesel-originating hydrocarbon substrates compared to complex soil organic matter compounds (27, 28, 50), (iii) the presence of imbalanced C:N:P ratios due to the influx of carbon-rich diesel, leading to competition for nutrients and reduced nutrient availability for the microbial community originating from the pristine system (6, 51, 52), and (iv) possible electron acceptor limitation for the native microbial community in the subsurface due to microbial TPH mineralization (48, 51).
We further explored the specific microbial groups within the phyla Chloroflexi, Firmicutes, and Euryarchaeota identified to be associated with contamination. Although these phyla were found in nearly all samples, the combined relative abundances in contaminated samples were on average 33% (compared to 7% in clean samples) and reached more than 50% in contaminated samples A1 and B3 (Fig. 6; bar graph). Additionally, the high relative abundance in contaminated samples of certain classes in the aforementioned phyla explains the reduced richness noted above (Fig. 6; pie graphs). This difference was reflected in the apparent class composition of Chloroflexi and Euryarchaeota in contaminated samples. Whereas a number of classes were represented in clean samples, in the presence of TPH, OTUs classified as Euryarchaeota were almost exclusively from the class Methanomicrobia and Chloroflexi from the class Anaerolineae. The increased abundance of the phyla Euryarchaeota, Chloroflexi, and Firmicutes in association with hydrocarbon-impacted samples, as found here, is in line with observations made in previous studies (5, 53–56).
Euryarchaeota have been observed to be the dominant archaeal phylum in heavily oil-contaminated environments (53). The most-dominant OTU classified in the class Methanomicrobia showed 98% identity to the 16S rRNA genes of members of the Methanosaeta, microorganisms that have previously been observed in oil sands (55) and have been observed in consortiums able to perform anaerobic mineralization of long alkane chains (57, 58). This genus is known to solely perform acetate fermentation (59), under methanogenic conditions, indicating that methanogenic processes could prevail at the contaminated locations.
The presence and high abundance of Euryarchaeota in contaminated samples in the current study lies in contrast to previous pyrosequencing studies that reported on archaeal abundance and diversity in clean soil samples. A study on archaeal diversity from 146 uncontaminated soil samples collected across six continents showed that the relative abundance of Archaea averaged 2% and was less than 16% in all samples; the main phylum observed was Crenarchaeota (60). The observed differences in archaeal phylum diversity and abundance between the current study on TPH-contaminated samples and clean soils investigated previously point toward a role of contamination in archaeal community composition.
A major OTU in contaminated samples showed 99% sequence identity to members of the family Anaerolineae within the Chloroflexi. Anaerolineae comprise obligate anaerobes, often found under sulfate-reducing conditions (56). This Anaerolineae OTU showed 100% sequence identity to a 16S rRNA gene sequence of an uncultured Anaerolineae phylotype that was also observed at a tar oil-contaminated site with toluene degradation (56). Although present at higher relative abundances in contaminated samples, OTUs related to this class were also observed in a variety of clean samples (E1, G1, and H3), indicating that the subsurface was most likely naturally anoxic. Finally, members of the Firmicutes have been identified in diesel-contaminated samples either within a bacterial consortium (5, 54) or as an isolate selected for biosurfactant production to improve hydrocarbon bioavailability (61).
Current knowledge regarding the physiology of characterized phylotypes showing sequence similarity to members of the microbial phyla identified to be predominantly associated with contaminated soils in this study indicates the presence of anoxic conditions. Anoxic subsurface conditions have implications for the potential for and especially the rate of natural attenuation. Although biodegradation of aromatics and alkanes can occur at low redox potentials, the degradation rates are severely reduced compared to those in oxic environments (62).
Implications of community structure in contaminated samples for natural attenuation.
In this study, we explored the long-term effect of TPH contamination on the microbial community at a site with a diesel contamination history of 40 years, giving microorganisms the ability to adapt and eventually utilize the present aliphatic and aromatic hydrocarbons. As a result, the impact of diesel on the microbial community structure is illuminated beyond the initial shifts in composition associated with the addition of carbon substrates and/or toxicants in the short term.
The increasing prevalence of natural attenuation projects due to risk-based remediation regulations requires improved understanding of the microbial populations present and active in contaminated soils. Regulations generally require multiple lines of evidence, including (i) historical data showing a stabilization or reduction in plume size, (ii) hydrogeological or geochemical indications of the type of natural attenuation processes, and (iii) proof of biological degradation potential through the use of microcosm studies or molecular diagnostics (63, 64). The current study is a step toward the further development of molecular tools to determine microbial degradation potential in support of natural attenuation of TPH-contaminated locations.
We identified a number of OTUs showing high similarity to microbes known to be involved in TPH degradation. These were mainly anaerobic microbes, including members of the phyla Euryarchaeota, Chloroflexi, and Firmicutes. This finding correlates with the degradation potential that was found at the site, based on the shifts in diesel carbon number distribution (Fig. 2). The soil samples, especially those from the vadose zone or in the plume, had notably fewer LMW compounds, with up to 50% of the C10-to-C16 fraction having been depleted, compared to the composition of the LNAPL source. Microbial preference for degradation of the more mobile and thus high-risk LMW compounds fits well into risk-based natural attenuation approaches. Such microbial degradation potential is another line of evidence, in addition to traditional chemical analyses of soil and groundwater, indicating the usefulness of a full understanding of the microbial community in order to substantiate the choice for natural attenuation of a site.
The redox conditions noted here also allow more-specific direct molecular analyses of bioremediation under such natural attenuation conditions. Typical investigations into TPH degradation include utilization of quantitative PCR assays targeting genes encoding enzymes involved in aerobic TPH degradation, such as the alkane monooxygenases AlkB and P450 (33, 65). In the case of this site, any response to such alkane monooxygenase primers with a DNA template would indicate whether or not there is a potential for aerobic degradation. However, the potential for continued natural attenuation under the prevailing anoxic conditions would not be illuminated, and the exploitation of any existing aerobic degradation potential would require extensive manipulation of in situ circumstances.
This work highlights a number of potential molecular proxies which, at this site, were indicative of long-term diesel contamination and natural attenuation. The reduced diversity noted in samples with TPH, in addition to being ecologically relevant, means that a large portion of the microbial community could be targeted with relatively few molecular assays. Thus, microbial characterization of such a site could be simplified, as fewer microbial species need to be detected to identify the potential for degradation. OTUs related to Archaea, specifically of the class Methanomicrobia in the phylum Euryarchaeota, are particularly associated with TPH contamination in this study and have been observed in previous work to be present in microbial consortiums performing alkane degradation (53, 55, 57). This class dominates archaeal sequences in most contaminated samples, where Archaea relative abundance reaches up to 35%. Additionally, sequences identified as belonging to the phyla Chloroflexi (specifically Anaerolineae) and Firmicutes show higher abundances in diesel-contaminated samples than in clean samples. Previous work identifying these phyla in TPH-contaminated locations and describing their role in hydrocarbon degradation supports the choice of these phyla for consideration as potential targets for molecular-based techniques (5, 54, 56, 61). Additional studies investigating the microbial community composition at TPH-contaminated locations where natural attenuation has been observed are required to further develop and validate this approach.
The results presented here indicate that a number of taxa that are correlated with TPH contamination can be identified at a contaminated site, providing a first insight into the potential for degradation of the TPH contamination present. This knowledge can be further used to examine the microbial population in terms of ecological relevance in order to understand the underlying conditions that are conducive to degradation processes. Finally, with the increasing prevalence of natural attenuation projects due to risk-based remediation regulations, an understanding of the microbial populations present in contaminated soils is needed for the acceptance of monitored natural attenuation as a remediation strategy.
Supplementary Material
ACKNOWLEDGMENTS
This research is funded by the European Union Consortium Upsoil, a Seventh Framework Programme within Theme 6, number 226956 (www.upsoil.eu), and by the Ecogenomics Innovation Centre (Ecolinc; www.ecogenomics.nl).
We thank Pauline van Gaans (Deltares, The Netherlands), Janusz Krupanek and Mariusz Kalisz (Instytut Ekologii Terenow Uprzemyslowionych, Poland), and Jan Marek (Przedsiebiorstwo Oczyszczania Wodi Ziemi, Poland) for logistical support during field sampling. Additionally, Detmer Sipkema (Wageningen University, The Netherlands) is thanked for his support throughout the laboratory and analytical phases.
Footnotes
Published ahead of print 9 November 2012
Supplemental material for this article may be found at 10.1128/AEM.02747-12.
REFERENCES
- 1. Singh A. 2009. Biological remediation of soil: an overview of global market and available technologies, p 1–19 In Singh A, Ward OP. (ed), Advances in applied bioremediation, vol 17 Springer-Verlag, Berlin, Germany [Google Scholar]
- 2. Wiedemeier TH, Swanson MA, Moutoux DE, Gordon EK, Wilson JT, Wilson BH, Kampbell DH, Haas PE, Miller RN, Hansen JE, Chapelle FH. 1998. Technical protocol for evaluating natural attenuation of chlorinated solvents in ground water. EPA/600/R-98/128. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 3. Kloos K, Munch JC, Schloter M. 2006. A new method for the detection of alkane-monooxygenase homologous genes (alkB) in soils based on PCR-hybridization. J. Microbiol. Methods 66:486–496 [DOI] [PubMed] [Google Scholar]
- 4. Powell SM, Ferguson SH, Bowman JP, Snape I. 2006. Using real-time PCR to assess changes in the hydrocarbon-degrading microbial community in Antarctic soil during bioremediation. Microb. Ecol. 52:523–532 [DOI] [PubMed] [Google Scholar]
- 5. dos Santos HF, Cury JC, do Carmo FL, dos SantosAL, Tiedje J, van Elsas JD, Rosado AS, Peixoto RS. 2011. Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS One 6:e16943 doi:10.1371/journal.pone.0016943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Margesin R, Hammerle M, Tscherko D. 2007. Microbial activity and community composition during bioremediation of diesel-oil-contaminated soil: effects of hydrocarbon concentration, fertilizers, and incubation time. Microb. Ecol. 53:259–269 [DOI] [PubMed] [Google Scholar]
- 7. Kasai Y, Takahata Y, Hoaki T, Watanabe K. 2005. Physiological and molecular characterization of a microbial community established in unsaturated, petroleum-contaminated soil. Environ. Microbiol. 7:806–818 [DOI] [PubMed] [Google Scholar]
- 8. Alvarez VM, Santos SC, Casella Rda C, Vital RL, Sebastin GV, Seldin L. 2008. Bioremediation potential of a tropical soil contaminated with a mixture of crude oil and production water. J. Microbiol. Biotechnol. 18:1966–1974 [PubMed] [Google Scholar]
- 9. Salminen JM, Tuomi PM, Jorgensen KS. 2008. Functional gene abundances (nahAc, alkB, xylE) in the assessment of the efficacy of bioremediation. Appl. Biochem. Biotechnol. 151:638–652 [DOI] [PubMed] [Google Scholar]
- 10. Yergeau E, Arbour M, Brousseau R, Juck D, Lawrence JR, Masson L, Whyte LG, Greer CW. 2009. Microarray and real-time PCR analyses of the responses of high-Arctic soil bacteria to hydrocarbon pollution and bioremediation treatments. Appl. Environ. Microbiol. 75:6258–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS. 2003. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 69:1800–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, Herold N, Schöning I, Schrumpf M, Daniel R. 2011. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 6:e17000 doi:10.1371/journal.pone.0017000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bundy JG, Paton GI, Campbell CD. 2002. Microbial communities in different soil types do not converge after diesel contamination. J. Appl. Microbiol. 92:276–288 [DOI] [PubMed] [Google Scholar]
- 16. Powell SM, Bowman JP, Ferguson SH, Snape I. 2010. The importance of soil characteristics to the structure of alkane-degrading bacterial communities on sub-Antarctic Macquarie Island. Soil Biol. Biochem. 42:2012–2021 [Google Scholar]
- 17. Pansu M, Gautheyrou J. 2006. Handbook of soil analysis: mineralogical, organic and inorganic methods. Springer, Berlin, Germany [Google Scholar]
- 18. Nederlands Normalisatie Instituut 1997. Soil-determination of mineral oil content in soil and sediments with gas chromatography, NEN 5733. Nederlands Normalisatie Instituut, Delft, The Netherlands [Google Scholar]
- 19. Acosta-Martinez V, Dowd S, Sun Y, Allen V. 2008. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 40:2762–2770 [Google Scholar]
- 20. Inceoglu O, Abu Al-Soud W, Salles JF, Semenov AV, van Elsas JD. 2011. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS One 6:e23321 doi:10.1371/journal.pone.0023321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berry D, Ben Mahfoudh K, Wagner M, Loy A. 2011. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol. 77:7846–7849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang WJ, Dyen M, Spagnuolo L, Simon P, Whyte L, Ghoshal S. 2010. Biodegradation of semi- and non-volatile petroleum hydrocarbons in aged, contaminated soils from a sub-Arctic site: laboratory pilot-scale experiments at site temperatures. Chemosphere 80:319–326 [DOI] [PubMed] [Google Scholar]
- 28. Olson JJ, Mills GL, Herbert BE, Morris PJ. 1999. Biodegradation rates of separated diesel components. Environ. Toxicol. Chem. 18:2448–2453 [Google Scholar]
- 29. Militon C, Boucher D, Vachelard C, Perchet G, Barra V, Troquet J, Peyretaillade E, Peyret P. 2010. Bacterial community changes during bioremediation of aliphatic hydrocarbon-contaminated soil. FEMS Microbiol. Ecol. 74:669–681 [DOI] [PubMed] [Google Scholar]
- 30. Kersters K, Vos Gillis M, Swings J, Vandamme P, Stackebrandt E. 2006. Introduction to proteobacteria, p 3–37 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The Prokaryotes: a handbook on the biology of bacteria, 3rd ed, vol 5 Springer, New York, NY [Google Scholar]
- 31. Greer CW, Whyte LG, Niederberger TD. 2010. Microbial communities in hydrocarbon-contaminated temperate, tropical, alpine, and polar soils, p 2313–2328 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer-Verlag, Berlin, Germany [Google Scholar]
- 32. Parales RE. 2010. Hydrocarbon degradation by betaproteobacteria, p 1715–1724 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer-Verlag, Berlin, Germany [Google Scholar]
- 33. van Beilen JB, Funhoff EG. 2007. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 74:13–21 [DOI] [PubMed] [Google Scholar]
- 34. Baek KH, Yoon BD, Kim BH, Cho DH, Lee IS, Oh HM, Kim HS. 2007. Monitoring of microbial diversity and activity during bioremediation of crude oil-contaminated soil with different treatments. J. Microbiol. Biotechnol. 17:67–73 [PubMed] [Google Scholar]
- 35. Popp N, Schlomann M, Mau M. 2006. Bacterial diversity in the active stage of a bioremediation system for mineral oil hydrocarbon-contaminated soils. Microbiology 152:3291–3304 [DOI] [PubMed] [Google Scholar]
- 36. Vinas M, Sabate J, Espuny MJ, Solanas AM. 2005. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl. Environ. Microbiol. 71:7008–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3:442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Allen JP, Atekwana EA, Duris JW, Werkema DD, Rossbach S. 2007. The microbial community structure in petroleum-contaminated sediments corresponds to geophysical signatures. Appl. Environ. Microbiol. 73:2860–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saul DJ, Aislabie JM, Brown CE, Harris L, Foght JM. 2005. Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol. Ecol. 53:141–155 [DOI] [PubMed] [Google Scholar]
- 40. Jones A, Escobar M, Serlin C, Sercu B, Holden P, Stollar R, Murphy P. 2009. Microbial community composition assessment during in situ chemical oxidation with permanganate, p. K-14 Proc. 10th Int. In Situ On-Site Bioremediat. Symp Batelle Press, Baltimore, Maryland [Google Scholar]
- 41. Yergeau E, Sanschagrin S, Beaumier D, Greer CW. 2012. Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high Arctic soils. PLoS One 7:e30058 doi:10.1371/journal.pone.0030058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. George IF, Liles MR, Hartmann M, Ludwig W, Goodman RM, Agathos SN. 2009. Changes in soil Acidobacteria communities after 2,4,6-trinitrotoluene contamination. FEMS Microbiol. Lett. 296:159–166 [DOI] [PubMed] [Google Scholar]
- 43. Juck D, Charles T, Whyte L, Greer C. 2000. Polyphasic microbial community analysis of petroleum hydrocarbon contaminated soils originating from 2 northern Canadian communities. FEMS Microbiol. Ecol. 33:241–249 [DOI] [PubMed] [Google Scholar]
- 44. Kaplan CW, Kitts CL. 2004. Bacterial succession in a petroleum land treatment unit. Appl. Environ. Microbiol. 70:1777–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zucchi M, Angiolini L, Borin S, Brusetti L, Dietrich N, Gigliotti C, Barbieri P, Sorlini C, Daffonchio D. 2003. Response of bacterial community during bioremediation of an oil-polluted soil. J. Appl. Microbiol. 94:248–257 [DOI] [PubMed] [Google Scholar]
- 46. Bordenave S, Goni-Urriza MS, Caumette P, Duran R. 2007. Effects of heavy fuel oil on the bacterial community structure of a pristine microbial mat. Appl. Environ. Microbiol. 73:6089–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Labud V, Garcia C, Hernandez T. 2007. Effect of hydrocarbon pollution on the microbial properties of a sandy and a clay soil. Chemosphere 66:1863–1871 [DOI] [PubMed] [Google Scholar]
- 48. Leahy JG, Colwell RR. 1990. Microbial-degradation of hydrocarbons in the environment. Microbiol. Rev. 54:305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stroud JL, Paton GI, Semple KT. 2007. Microbe-aliphatic hydrocarbon interactions in soil: implications for biodegradation and bioremediation. J. Appl. Microbiol. 102:1239–1253 [DOI] [PubMed] [Google Scholar]
- 50. Marchal R, Penet S, Solano-Serena F, Vandecasteele JP. 2003. Gasoline and diesel oil biodegradation. Oil Gas Sci. Technol. 58:441–448 [Google Scholar]
- 51. Zhou E, Crawford RL. 1995. Effects of oxygen, nitrogen, and temperature on gasoline biodegradation in soil. Biodegradation 6:127–140 [DOI] [PubMed] [Google Scholar]
- 52. Zytner RG, Salb AC, Stiver WH. 2006. Bioremediation of diesel fuel contaminated soil: comparison of individual compounds to complex mixtures. Soil Sediment Contam. 15:277–297 [Google Scholar]
- 53. Liu RY, Zhang Y, Ding R, Li D, Gao YX, Yang M. 2009. Comparison of archaeal and bacterial community structures in heavily oil-contaminated and pristine soils. J. Biosci. Bioeng. 108:400–407 [DOI] [PubMed] [Google Scholar]
- 54. Peixoto R, Chaer GM, Carmo FL, Araujo FV, Paes JE, Volpon A, Santiago GA, Rosado AS. 2011. Bacterial communities reflect the spatial variation in pollutant levels in Brazilian mangrove sediment. Antonie Van Leeuwenhoek 99:341–354 [DOI] [PubMed] [Google Scholar]
- 55. Penner TJ, Foght JM. 2010. Mature fine tailings from oil sands processing harbour diverse methanogenic communities. Can. J. Microbiol. 56:459–470 [DOI] [PubMed] [Google Scholar]
- 56. Winderl C, Anneser B, Griebler C, Meckenstock RU, Lueders T. 2008. Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar oil contaminant plume. Appl. Environ. Microbiol. 74:792–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lanoil BD, Sassen R, La Duc MT, Sweet ST, Nealson KH. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zengler K, Richnow HH, Rossello-Mora R, Michaelis W, Widdel F. 1999. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401:266–269 [DOI] [PubMed] [Google Scholar]
- 59. Kleikemper J, Pombo SA, Schroth MH, Sigler WV, Pesaro M, Zeyer J. 2005. Activity and diversity of methanogens in a petroleum hydrocarbon-contaminated aquifer. Appl. Environ. Microbiol. 71:149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Menezes Bento FM, Camargo FAD, Okeke BC, Frankenberger WT. 2005. Diversity of biosurfactant producing microorganisms isolated from soils contaminated with diesel oil. Microbiol. Res. 160:249–255 [DOI] [PubMed] [Google Scholar]
- 62. Suarez MP, Rifai HS. 1999. Biodegradation rates for fuel hydrocarbons and chlorinated solvents in groundwater. Bioremediat. J. 3:337–362 [Google Scholar]
- 63. Beck P, Mann B. 2010. A technical guide for demonstrating monitored natural attenuation of petroleum hydrocarbons in groundwater, CRC CARE Technical Report no. 15. CRC for Contamination Assessment and Remediation of the Environment, Adelaide, Australia [Google Scholar]
- 64. Wisconsin Department of Natural Resources 2004. Naturally occurring biodegradation as a remedial action option for soil contamination. Interim guidance (revised). PUBL-SW-515-95. Bureau for Remediation and Redevelopment, Wisconsin Department of Natural Resources, Madison, WI: http://dnr.wi.gov/files/pdf/pubs/rr/rr515.pdf [Google Scholar]
- 65. Vilchez-Vargas R, Geffers R, Suárez-Diez M, Conte I, Waliczek A, Kaser VS, Kralova M, Junca H, Pieper DH. 20 April 2012, posting date Analysis of the microbial gene landscape and transcriptome for aromatic pollutants and alkane degradation using a novel internally calibrated microarray system. Environ. Microbiol. [Epub ahead of print.] doi:10.1111/j.1462-2920.2012.02752.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.