Abstract
The detection of anaerobic hydrocarbon degrader populations via catabolic gene markers is important for the understanding of processes at contaminated sites. Fumarate-adding enzymes (FAEs; i.e., benzylsuccinate and alkylsuccinate synthases) have already been established as specific functional marker genes for anaerobic hydrocarbon degraders. Several recent studies based on pure cultures and laboratory enrichments have shown the existence of new and deeply branching FAE gene lineages, such as clostridial benzylsuccinate synthases and homologues, as well as naphthylmethylsuccinate synthases. However, established FAE gene detection assays were not designed to target these novel lineages, and consequently, their detectability in different environments remains obscure. Here, we present a new suite of parallel primer sets for detecting the comprehensive range of FAE markers known to date, including clostridial benzylsuccinate, naphthylmethylsuccinate, and alkylsuccinate synthases. It was not possible to develop one single assay spanning the complete diversity of FAE genes alone. The enhanced assays were tested with a range of hydrocarbon-degrading pure cultures, enrichments, and environmental samples of marine and terrestrial origin. They revealed the presence of several, partially unexpected FAE gene lineages not detected in these environments before: distinct deltaproteobacterial and also clostridial bssA homologues as well as environmental nmsA homologues. These findings were backed up by dual-digest terminal restriction fragment length polymorphism diagnostics to identify FAE gene populations independently of sequencing. This allows rapid insights into intrinsic degrader populations and degradation potentials established in aromatic and aliphatic hydrocarbon-impacted environmental systems.
INTRODUCTION
Hydrocarbon contaminants are harmful to organisms and the environment, but they are also substrates for microbial degradation under both oxic and anoxic conditions. The aerobic degradation of hydrocarbons occurs readily but is constrained by oxygen limitation, especially in subsurface habitats. Therefore, degradation at many sites either naturally or anthropogenically impacted by hydrocarbons relies on anaerobic pathways. Investigating the structure and function of the involved anaerobic hydrocarbon-degrading microbial communities is thus crucial for the understanding of environmental hydrocarbon breakdown (e.g., see reference 1).
During the last few decades, several pathways for anaerobic hydrocarbon catabolism have been unraveled (2). Consequently, researchers have begun to trace the respective microbes in environmental systems based on the detection of the involved catabolic genes. Currently, the genes of enzymes for different peripheral and central catabolic reactions have been established as so-called functional markers for anaerobic hydrocarbon degradation (summarized by Kuntze et al. [3]). Among these, glycyl radical enzymes responsible for the initial activation of hydrocarbons with methyl- and methylene groups via addition to fumarate (i.e., fumarate addition) are currently the most widely used (see references 4 to 6 and see Table S1 in the supplemental material). This approach was initially introduced to target benzylsuccinate synthase (BSS) genes involved in toluene degradation by Betaproteobacteria (4). The utility of targeting fumarate-adding enzymes (FAEs) is limited not only to the detection of organisms involved in the oxidation of toluene, xylenes, and ethylbenzene (2). The same substrate activation principle is also used in alkylsuccinate synthases (ASS [7]; also named methylalkylsuccinate synthases [MAS] [8]) for long-chain as well as short-chain alka(e)nes (9) and in naphthylmethylsuccinate synthases (NMS) for 2-methylnaphthalene activation (10).
A number of primers targeting the α subunit of BSS and ASS genes for the analysis of environmental degrader populations have been developed (see Table S1 in the supplemental material). The first PCR and quantitative PCR (qPCR) primers targeting bssA genes of nitrate-reducing Betaproteobacteria were introduced by Beller et al. (4). The assay of Winderl et al. (6) extended the range of detectable hydrocarbon-degrading microbes to iron- and sulfate-reducing Deltaproteobacteria and revealed partially novel, site-specific degrader populations at three different tar oil-impacted aquifers in Germany. Staats et al. (11) applied an altered primer targeting bssA of iron-reducing degraders first developed by Botton et al. (12) at an aquifer contaminated by landfill leachate. Here, retrieved bssA sequences were related to the betaproteobacterial bssA sequence type of Georgfuchsia toluolica (13) rather than the Geobacter populations expected from 16S rRNA gene studies. Recently, Callaghan et al. (5) also introduced assays for ASS genes, evolved from existing bssA primers, on the basis of the small number of pure-culture assA sequences available. These optimized primer sets were applied to DNA extracted from propane- and paraffin-degrading enrichments as well as several aquifers and freshwater and estuarine habitats contaminated with alkanes, revealing an environmental diversity of assA genes similar to that known for bssA genes.
The study conducted by Winderl et al. (6) revealed several unassigned, deeply branching bssA lineages, the so-called T and F clusters. F1 bssA was later identified to belong to the dominating Desulfobulbaceae degrader population (14) on-site, while F2 was identified as a novel bssA sequence type of the Peptococcaceae (15). Other studies on hydrocarbon-contaminated aquifers by Callaghan et al. (5) and Yagi et al. (16) and on xylene-degrading enrichments by Herrmann et al. (17) also corroborated the existence of new and deeply branching FAE lineages, in addition to the described BSS, NMS, and ASS lineages. Furthermore, some new anaerobic hydrocarbon degraders belonging to the Clostridia were recently discovered: Desulfitobacterium aromaticivorans UKTL, using fumarate addition for toluene activation (18), and strain BF, possessing a bss-homologous gene (19), despite oxidizing benzene and not toluene. We observed that bssA or homologous gene amplicons are not readily obtained from these strains with the available bssA primers. Moreover, the NMS genes recently described in naphthalene-degrading marine strains NaphS2, NaphS3, and NaphS6 (20) and aquifer sediment enrichment strain N47 (21) are not targeted by available primers at all. Consequently, the potential of FAE genes as markers for the degradation of various hydrocarbons by putatively novel species may not yet be fully exploited by existing assays.
FAE gene lineages dominating at given sites indicate relevant degradation potentials and should not be missed, if optimum bioremediation strategies are to be designed. Thus, there is a need for the development and application of more comprehensive primers for FAE gene detection in general. Qualitative gene detection is also a prerequisite for the design of quantitative (qPCR) assays, which may be useful for the inference of in situ degradation rates (4, 22).
The aim of this work was to assess FAE gene pools in different terrestrial and marine environments impacted by hydrocarbons via more universal detection assays. We hypothesize that the existing FAE detection systems do not yet optimally recover the full diversity of catabolic lineages, such as clostridial bssA and also nmsA homologues. We test whether one universal primer pair can be developed or whether several primer sets targeting overlapping amplicons in the FAE gene alignment are needed to recover most lineages. Moreover, for the routine and possibly high-throughput detection of FAE genes in the field and microcosms, rapid diagnostics via fingerprinting may be helpful to trace hydrocarbon degrader communities in space and time. Thus, we propose a novel, dual-digest terminal restriction fragment length polymorphism (T-RFLP) fingerprinting method for sequencing-independent diagnostics of major FAE gene lineages.
MATERIALS AND METHODS
Primer design.
Primers were designed using an updated version of the bssA ARB database originally set up by Winderl et al. (6). Several new bssA, nmsA, and assA (also known as masD) sequences were added (see Fig. 1). The primer sets FAE-B, FAE-N, and FAE-KM, presented in Table 1, were designed to match the same target region as the original primers of Winderl et al. (6), which were also applied in this study. The binding sites for the new primers were found by searching the FAE alignment for conserved, distinctive protein motifs common for major gene clusters and using the DNA sequences of those protein motifs for primer design.
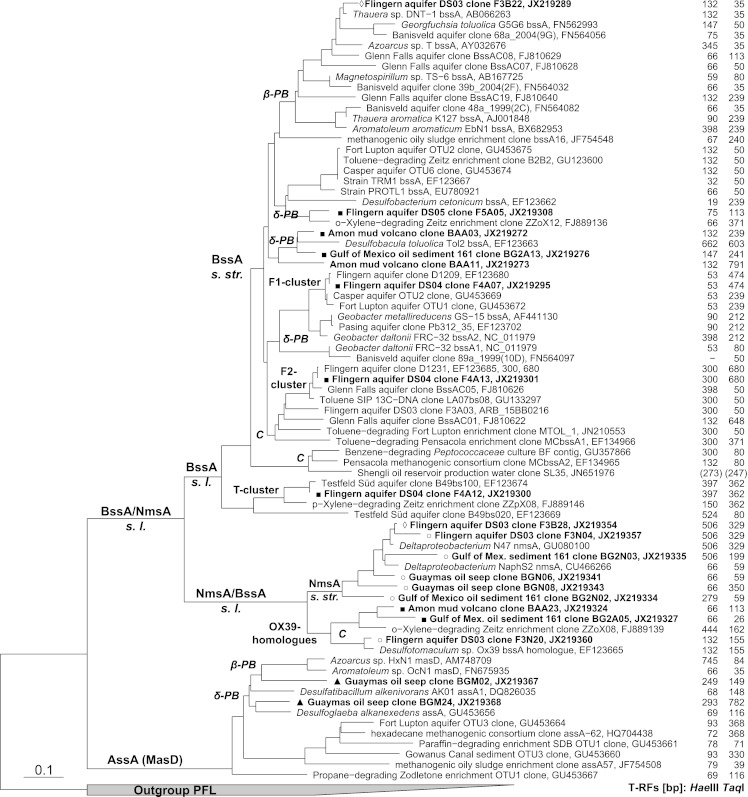
Fig 1.
Current overview of FAE gene phylogeny and affiliation of clones detected in this study. Sequences retrieved in this study are in bold and marked with different symbols according to the primer sets used: ◊, set FAE-B; ■, bssA at 52°C; ○, set FAE-N; ▲, set FAE-KM. Abbreviations of FAE lineages (as identified via enrichments or pure cultures): C, Clostridia; β-P, Betaproteobacteria; δ-P Deltaproteobacteria. The numbers to the right give the T-RFs predicted in silico for amplicons generated with the 8543r primer and a digest with HaeIII or TaqI. The T-RF sizes in parentheses are tentatively extrapolated, as some sequence entries end before the primer site. s. str. sensu stricto; s. l., sensu lato; PFL pyruvate formate lyase.
Table 1.
FAE-gene targeted primer sets designed and employed in this study
| Primer set | Annealing temp (°C) | Primer, positiona | Sequence (5′–3′) | Targeted FAE lineagesb |
|---|---|---|---|---|
| bssAc | 58/52 | 7772f | GAC ATG ACC GAC GCS ATY CT | bssA sensu lato |
| 8546r | TC GTC GTC RTT GCC CCA YTT | |||
| FAE-B | 58 | 7768f | C AAY GAT TTA ACC RAC GCC AT | Clostridial bssA, bssA sensu lato, nmsA |
| 8543r | TC GTC RTT GCC CCA YTT NGG | |||
| FAE-N | 58 | 7363f | TC GCC GAG AAT TTC GAY TTG | nmsA sensu stricto |
| 7374f | TTC GAY TTG AGC GAC AGC GT | |||
| 8543r | TC GTC RTT GCC CCA YTT NGG | |||
| FAE-KM | 58 | FAE-Kf, 7757f-1 | TCG GAC GCG TGC AAC GAT CTG A | assA |
| FAE-Kf, 7757f-2 | TCG GAC GCG TGC AAC GCC CTG A | |||
| FAE-Mf, 7766f | TGT AAC GGC ATG ACC ATT GCG CT | |||
| 8543r | TC GTC RTT GCC CCA YTT NGG |
Strains, samples, and DNA extraction.
Desulfotignum toluenicum DSMZ 18732, Desulfatibacillum alkenivorans DSMZ 16219, Desulfatibacillum aliphaticivorans DSMZ 15576, and Geobacter metallireducens DSMZ 7210 were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; Braunschweig, Germany). The strains were grown anaerobically in 120-ml serum bottles with butyl stoppers and the media specified by DSMZ. A toluene-degrading Desulfosporosinus sp. enrichment (15), Desulfitobacterium aromaticivorans UKTL, Azoarcus sp. strain T, strain BF, and strain N47 originated from our institute strain collection (Institute of Groundwater Ecology). The Desulfosporosinus sp. enrichment and D. aromaticivorans UKTL were cultivated anaerobically as previously described (15, 18). DNA was extracted from collected biomass by bead beating and a phenol-chloroform extraction (23). Azoarcus sp. T was cultivated and extracted as described by Winderl et al. (6). Strain BF was cultivated as described by Kunapuli et al. (24), and the DNA was extracted as previously described (19). Strain N47 was cultivated and extracted as described by Selesi et al. (21).
Flingern tar oil-contaminated aquifer sediment was sampled in June 2009 and extracted as described by Pilloni et al. (25) with a total of four replicate sediment DNA extracts per chosen depth. Three depths (6.85 m, 7.15 m, and 7.25 m below the surface) spanning the lower plume fringe were chosen, as those depths were previously identified to be benzene, toluene, ethylbenzene, and o-, m-, and p-xylene (BTEX) degradation hot spots (26, 27). Furthermore, DNA from different terrestrial hydrocarbon-degrading enrichments (Paclele Mici mud volcano and Gölzau refinery-contaminated aquifer) was extracted with a FastDNA Spin kit for soil (MP Biomedicals, Irvine, CA). Further information on the sampling sites and enrichments are provided by Alain et al. (28) and Feisthauer et al. (29). Gulf of Mexico hydrocarbon seep sediment was sampled from station 161 as described by Orcutt et al. (30). DNA was extracted according to the protocol of Zhou et al. (31), and cleanup was performed with a Wizard DNA cleanup system (Promega, Fitchburg, WI). Marine samples from the Nyegga area methane seeps were taken at pockmarks located on the edge of the Norwegian continental slope; the sampling is described by Van Gaever et al. (32). The DNA was extracted with a Power soil RNA extraction kit and DNA elution accessory kit (MO BIO Laboratories, Carlsbad, CA). Sediment samples from the Amon mud volcano in the eastern Nile deep-sea fan (station 929, dive 240 [33]) and Guaymas Basin hydrocarbon seeps in the Gulf of California (dive 5473 [34]) were collected from below a microbial mat. The DNA was extracted as described by Kleindienst et al. (S. Kleindienst, F.-A. Herbst, F. von Netzer, R. Amann, J. Peplies, M. von Bergen, T. Lueders, J. Seifert, F. Musat, and K. Knittel, unpublished data) from a 2- to 20-cm sediment depth (Amon mud volcano) and from a 0- to 10-cm sediment depth (Guaymas, fine-grained sediments) according to the protocol described by Zhou et al. (31) and based on mechanical, chemical, and enzymatic lyses.
PCR amplification.
FAE gene fragments were amplified via PCR using available and newly developed primer sets (Table 1) in a Mastercycler ep gradient (Eppendorf, Hamburg, Germany) with the following cycling conditions: a 3-min initial degradation, 30 to 40 cycles of amplification (30 s at 94°C, 30 s at 52°C or 58°C, 60 s at 72°C), and 5 min at 72°C of terminal extension. The annealing temperature was 58°C for all primers, as initially published (6), but also lowered comparatively to 52°C (15) for the bssA primer set. The 50-μl PCR mixture contained nuclease-free water (Promega, Fitchburg, WI), 1× PCR buffer, 1.5 mM MgCl2 (Fermentas, Thermo Fisher Scientific, Waltham, MA), 10 μg bovine serum albumin (Roche, Basel, Switzerland) 0.1 mM deoxynucleoside triphosphates, 0.3 μM primer (biomers.net, Ulm, Germany), 1.25 U Taq polymerase, and DNA in the range of ∼0.2 to 20 ng. Amplicon quality was checked with gel electrophoresis in a 1.5% agarose gel. Cleanup of the amplicons was done with a PCRextract kit (5Prime, Hamburg, Germany) according to the manufacturer's protocol before further processing.
T-RFLP.
The reverse primers 8546r and 8543r were labeled for T-RFLP with 5′-6-carboxyfluorescein (FAM). After amplicon purification, these were digested in two separate preparations with TaqI (Fermentas, Thermo Fisher Scientific, Waltham, MA) and HaeIII (New England BioLabs, Ipswich, MA) for 2 h in a thermocycler at 65 and 37°C, respectively. The digests were purified with DyeEx columns (Qiagen, Hilden, Germany). The T-RFLP run was conducted on an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA) as described by Pilloni et al. (14). The electropherograms were processed with the PeakScanner (version 1.0) and GeneMapper (version 4.0) programs (Applied Biosystems, Foster City, CA). Further analysis was performed with the T-REX online software (35) using the default settings. Noise filtering was done on the basis of the peak height with the standard deviation multiplier set to 1. Terminal restriction fragments (T-RFs) were defined by aligning peaks with a clustering threshold of 1 bp. T-RF affiliation in dual fingerprints was assigned via searching TaqI digests first for database entries with a corresponding in silico T-RF. If multiple entries with potentially matching T-RFs were present, the corresponding data from the HaeIII digests were used for unambiguous T-RF affiliation to a defined FAE gene lineage.
Cloning and sequencing.
Selected purified, nonlabeled FAE gene amplicons were cloned with a TOPO-XL kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The sequencing of the Flingern samples (primer bssA, 52°C, all three depths; primer sets FAE-B and FAE-N, only the 6.85-m depth; 30 clones were picked per five samples) was performed on an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA) as described by Winderl et al. (6). Samples of Amon mud volcano (bssA primers, 58°C), Gulf of Mexico hydrocarbon seeps (bssA and FAE-N primers, 58°C), and Guaymas hydrocarbon seeps (with primer sets FAE-N and FAE-KM) were sequenced by GATC (Konstanz, Germany). FAE gene tree reconstruction was done in ARB on the basis of an amino acid alignment via a Phylip distance matrix with the Fitch algorithm as described by Winderl et al. (6).
Nucleotide sequence accession numbers.
The sequences were deposited with GenBank under accession numbers JX219271 to JX219368.
RESULTS
Establishment of optimized primer sets.
No universally conserved protein motif that would allow the development of a single, universal primer pair was found across the different FAE gene lineages (shown in Fig. 1). Therefore, several primer sets called FAE-B, FAE-N, and FAE-KM, shown in Table 1, were designed in silico to optimally target an overlapping FAE gene region for robust phylogenetic comparison. All newly developed forward primers target sequence motifs not targeted by available assays prior to this study. The new FAE-B forward primer was predicted to target betaproteobacterial bssA, the recently discovered clostridial bssA homologues (F2 cluster, strain BF), as well as nmsA homologues (strain N47, NaphS strains). The two new FAE-N forward primers specifically target a sequence motif found only in nmsA of strain N47 and NaphS strains 400 bp upstream of the other forward primer sites. This primer set was aimed to distinguish nmsA sensu stricto amplicons from the homologous bssA-nmsA sensu lato amplicons retrieved by set FAE-B. The new FAE-KM set contained two different forward primer combinations: forward primer combination FAE-K to target sequences related to the assA genes of Desulfatibacillum alkenivorans and forward primer FAE-M to target relatives of strain HxN1 and OcN1 masD. FAE-K forward primers consisted of two different primers with two positions, instead of one with two ambiguity codes, to avoid four possible homopolymers and a subsequent loss of specificity. The primer sets FAE-B, FAE-N, and FAE-KM shared the same modified reverse primer 8543r, as the targeted sequence motif is conserved in all known FAE genes. This is documented for other, similar primers targeting this region (5, 6, 36) (see Table S1 in the supplemental material).
Primer performance in pure cultures and environmental samples.
The lineage-specific FAE gene performance of the different primer sets predicted in silico was verified, wherever possible, using pure-culture DNA, as shown in Table 2. Our previous bssA primer set (6) did not amplify FAE genes of Desulfitobacterium aromaticivorans UKTL, clostridial strain BF, strain N47, Desulfitobacterium alkenivorans PF2803, or D. aliphaticivorans CV2803. Apart from the bssA gene of Azoarcus sp. T, the new primer set FAE-B did indeed amplify the FAE genes of these selected clostridial strains and also nmsA of strain N47. As intended, set FAE-N amplified only nmsA of strain N47. Primer set FAE-KM amplified only assA of D. alkenivorans PF2803 and D. aliphaticivorans CV2803. A combination of all possible forward primers with one reverse primer in a single PCR failed to produce amplicons.
Table 2.
Experimentally tested performance of different FAE gene-targeted primers in selected strains and enrichments degrading hydrocarbons via fumarate addition
| Gene | FAE lineage | Culture | Substrate | Reference | Amplicon production with primer seta: |
|||
|---|---|---|---|---|---|---|---|---|
| bssA | FAE-B | FAE-N | FAE-KM | |||||
| bssA sensu lato | Proteobacteria | Azoarcus sp. T | Toluene | 37 | + | + | − | − |
| Geobacter metallireducens GS-15 | Toluene | 38 | + | − | − | − | ||
| Desulfotignum toluenicum H3 | Toluene | 39 | + | − | − | − | ||
| Clostridia | Desulfosporosinus sp. enrichment | Toluene | 15 | + | + | − | − | |
| Desulfitobacterium aromaticivorans UKTL | Toluene | 18 | − | + | − | − | ||
| Strain BF | Benzene | 19 | − | + | − | − | ||
| nmsA | Proteobacteria | Strain N47 | 2-Methyl-naphthalene | 21 | − | + | + | − |
| assA | Desulfatibacillum alkenivorans PF2803 | Alkanes | 40 | − | − | − | + | |
| Desulfatibacillum aliphaticivorans CV2803 | Alkanes | 41 | − | − | − | + | ||
+, a clear amplicon with the correct size was observed; −, no amplicon was obtained.
Subsequently, the performance of the primer sets was tested with samples originating from different hydrocarbon-degrading enrichments or impacted environmental systems. The samples in Table 3 were chosen as they were exposed to and actively degrading different aliphatic or aromatic hydrocarbons: single compounds (enrichments), confined mixtures (e.g., natural gas at marine seeps), or complex mixtures of hydrocarbons (oil, asphalt). According to the known hydrocarbon substrates of those samples and the potential FAEs supposedly involved in their degradation, we predicted expected FAE gene lineages to be recovered by our primer sets: (i) bssA-nmsA sensu lato amplified by primers bssA and FAE-B, (ii) nmsA sensu stricto amplified by primer set FAE-N, and (iii) assA amplified by primer set FAE-KM (Fig. 1). The results are presented in Table 3.
Table 3.
Verification of FAE assay performance with DNA from selected hydrocarbon-impacted enrichments and environmental samples
| Sample | Region | Reference(s) | Environment, sample type | Substrate(s) | Expected FAE lineage | Resultsa |
|||
|---|---|---|---|---|---|---|---|---|---|
|
bssA-nmsA sensu lato |
nmsA sensu stricto with FAE-N | assA with FAE-KM | |||||||
| bssA | FAE-B | ||||||||
| Paclele Mici mud volcano | Romania | 28 | Terrestrial, enrichment | Hexadecane | assA | − | − | − | + |
| Amon mud volcano | Mediterranean Sea, Nile deep-sea fan | 42 | Marine, sediment | Methane, ethane, propane, butane | assA-like? | +b | − | − | − |
| Nyegga methane seeps | Norwegian continental ridge | 32 | Marine, sediment | Methane, natural gas | assA-like? | − | − | − | + |
| Chapopote asphalt volcano station GeoB106-17-6 | Gulf of Mexico | 42, 43 | Marine, sediment | Asphalt, alkanes, aromatics, methane | FAE | + | + | − | + |
| Hydrocarbon seep station 161 | Gulf of Mexico | 30, 42 | Marine, sediment | Oil, asphalt, alkanes, aromatics | FAE | +b | +b | +b | + |
| Guaymas Basin hydrocarbon seep | Gulf of California | 42 | Marine, sediment | Methane, alkanes, aromatics | FAE, especially assA | − | − | +b | +b |
| Gölzau aquifer, former refinery | Germany | 29 | Terrestrial, enrichment | Toluene | bssA | + | − | + | − |
| Butane | assA | + | + | + | + | ||||
| Methylnaphthalene | nmsA | + | − | + | − | ||||
| Flingern aquifer, former gas work site | Germany | 25 | Terrestrial, sediment | BTEX, PAHc | bssA-nmsA sensu lato | +b | +b | +b | +d |
+, a clear amplicon with the expected size was observed; −, no amplicon.
Amplicons were actually cloned and sequenced as displayed in Fig. 1.
PAH, polycyclic aromatic hydrocarbon.
Forty PCR cycles were necessary to obtain an amplicon.
For samples exposed to confined mixtures of hydrocarbons, like those from the mud volcanoes and the Nyegga methane seeps, amplicons were obtained only for single FAE gene lineages. In contrast, several major FAE gene lineages were obtained for samples exposed to a broader mixture of hydrocarbons. For example, all expected FAE lineages were detected for the Gölzau enrichments and respective substrates, but also additional catabolic marker lineages were always present. Multiple FAE gene lineages were also detected at the Flingern aquifer and the Gulf of Mexico hydrocarbon seep, while only both nmsA sensu stricto and assA could be detected in the Guaymas Basin hydrocarbon seep sediments.
In order to substantiate the amplicon-based results, selected samples were chosen for sequencing in small clone libraries: Amon mud volcano (6 FAE sequences out of 25 clones with bssA primers, 58°C), the Gulf of Mexico hydrocarbon seep (9 FAE sequences out of 25 clones with bssA primers, 58°C; 1 FAE sequence out of 25 clones with FAE-B primers; and 6 FAE sequences out of 25 clones with FAE-N primers), and the Guaymas Basin hydrocarbon seep (8 FAE sequences out of 25 clones with FAE-N primers and 2 FAE sequences out of 25 clones with FAE-KM primers). The clones (Fig. 1) were clearly affiliated with the expected FAE lineages; the rest of the clone libraries were sequences that resulted from unspecific amplification. Amon mud volcano clones were mostly related to Desulfobacula toluolica bssA, with one assigned to more deeply branching nmsA homologues. Guaymas Basin hydrocarbon seep clones generated with set FAE-KM were related to betaproteobacterial masD and Desulfoglaeba alkanexedens assA sequences. From all three sites, clones related to nmsA sensu stricto of NaphS strains (all from set FAE-N) or bssA homologues were retrieved with primer sets FAE-N, FAE-B, and bssA at 52°C.
Hidden FAE gene lineages revealed by dual-digest T-RFLP analysis in the tar oil-contaminated Flingern aquifer.
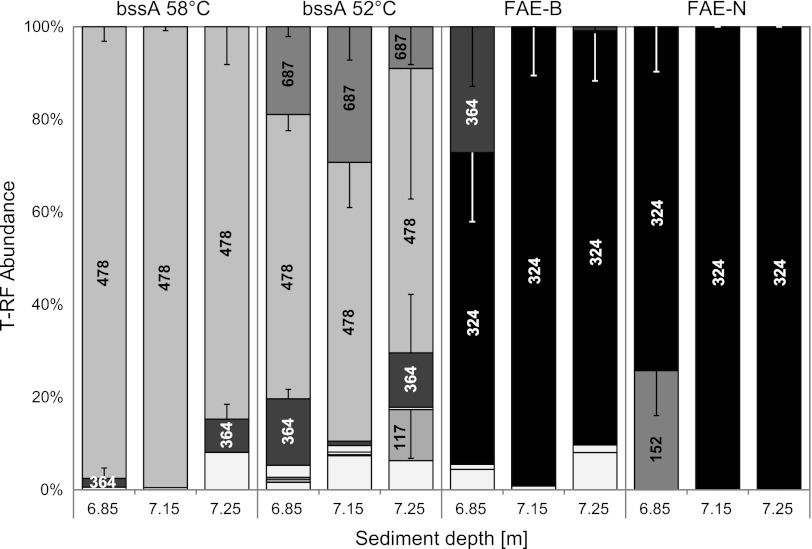
A more extensive assay performance test and sequencing-independent T-RFLP-based FAE gene lineage identification were conducted with sediment from the well-investigated tar oil-contaminated aquifer in Flingern (6, 44). This aquifer is known to host two specific bssA populations at the lower plume fringe, the hot spot of sulfidogenic BTEX degradation (26, 27): dominating Desulfobulbaceae F1-cluster bssA (6, 14) and the less abundant clostridial F2-cluster bssA (6, 15). As the sediment studied here is from a later sampling period (2009), we could not be sure whether to expect the same bssA populations. We have previously introduced the use of a bssA-based T-RFLP fingerprinting system allowing sequencing-independent lineage identification at reasonable confidence from samples whose sequences are known from prior clone library analysis (15). However, several different bssA lineages were observed to display identical T-RFs with TaqI restriction (Fig. 1; e.g., a 50-bp T-RF for certain Clostridia, Betaproteobacteria, and Deltaproteobacteria). For better diagnostic confidence, we now introduce a second, parallel digest with HaeIII, in addition to TaqI, to unambiguously identify T-RF.
The application of the different primer sets and TaqI-digested T-RFLP for the three chosen depths from the Flingern aquifer revealed the presence of a number of tentative bssA T-RFs, as shown in Fig. 2. The identity of those bssA lineages was assigned via the T-RF sizes predicted for sequences in our ARB database and further confirmed by parallel HaeIII digest as well as, for proof of principle, by cloning and sequencing (Table 4 and Fig. 1; HaeIII digest raw data not shown). With the standard bssA primers and settings, the Desulfobulbaceae F1-cluster bssA T-RF (478 bp, TaqI) clearly dominated over the three chosen depths, as was expected from previous work (6, 15). However, already at a lowered annealing temperature for reduced PCR selectivity (15), a T-RF representing the clostridial F2-cluster bssA (687 bp, TaqI) and the unidentified Testfeld Süd T1-cluster (6) bssA homologues (364 bp, TaqI) became detectable. By applying the new FAE-B primer set, strain N47 nmsA-like sequences (324 bp, TaqI) became detectable at all three investigated depths, while the F1 and F2 clusters disappeared. With the FAE-N primers, it was, furthermore, possible to detect a T-RF (152 bp, TaqI) at the 6.85-m depth tentatively assigned to the nmsA-associated bssA homologue previously detected for Desulfotomaculum sp. strain OX39.
Fig 2.
Comparative FAE gene T-RF retrieval from distinct depths of the tar oil-contaminated Flingern aquifer with different assays. Distinct T-RF sizes (bp) are indicated by gray scales and labeling within bars and correspond to the FAE gene lineages identified in Table 4. T-RF abundance is averaged over results from four independent DNA extractions; error bars represent standard deviations and are partially shown only as negative or only as positive standard deviations, when the graphical layout allows.
Table 4.
Sequencing-independent identification of T-RF lineages from Flingern sediment via dual-digest T-RFLPa
| Observed fragment size (bp) |
In silico fragment sizeb (bp) |
Candidate lineage | No. of FAE clones | Primer sets | ||
|---|---|---|---|---|---|---|
| TaqI | HaeIII | TaqI | HaeIII | |||
| 364 | 398 | 362 | 397 | Unidentified bssA homologues, unaffiliated (Testfeld Süd T1) | 8 | bssA, 52°C; bssA, 58°C; FAE-B |
| 478 | 51 | 474 | 52 | Deltaproteobacteria bssA, Desulfobulbaceae (F1 cluster) | 14 | bssA, 52°C; bssA, 58°C |
| 678 | 304 | 670 | 300 | Clostridial bssA Desulfosporosinus sp. related (F2 cluster) | 22 | bssA, 52°C |
| 324 | 505 | 329 | 506 | Deltaproteobacteria nmsA, Desulfobacteraceae (N47 related) | 18 | FAE-B, FAE-N |
| 152 | 130 | 155 | 132 | Clostridial bssA homologues, Desulfotomaculum related | 1 | FAE-N |
| 132 | 35 | 132 | Betaproteobacteria bssA, Thauera sp. strain DNT-1 | 2 | FAE-B | |
| 117 | 76 | 113 | 75 | Deltaproteobacteria bssA, Desulfobacterium cetonicum bssA | 1 | bssA, 52°C |
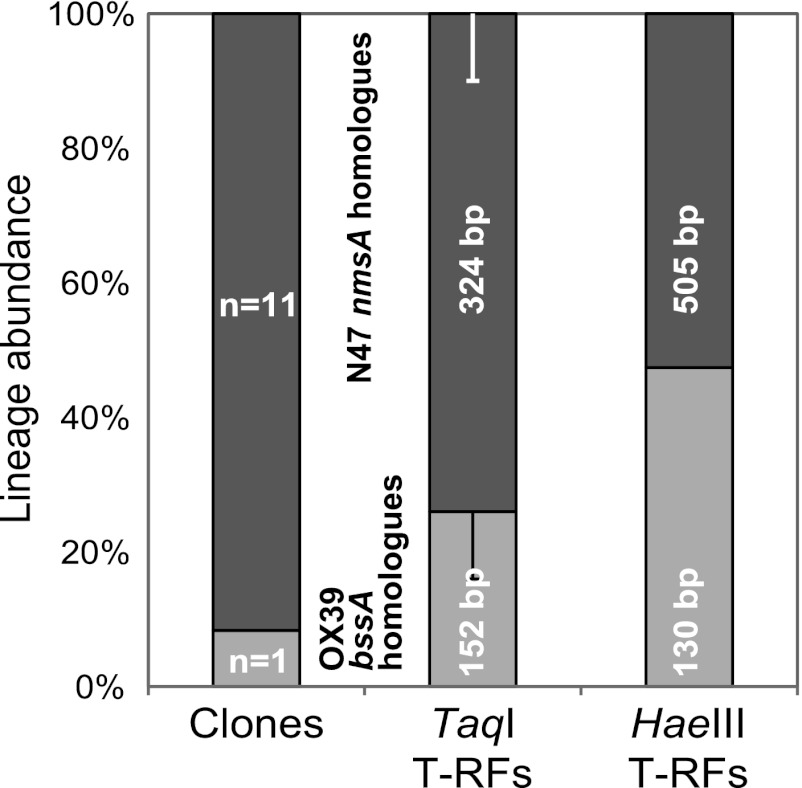
On the basis of previous FAE gene sequence data from the site (6), one minor T-RF (117/76 bp, TaqI/HaeIII), found with FAE-B primers at 7.25 m, was not assignable in silico and was thus identified by cloning and sequencing to represent a new relative of Desulfobacterium cetonicum bssA (67% amino acid sequence identity, clone F5A05; Fig. 1). Another FAE population became detectable at 6.85 m with primer set FAE-B (35-bp T-RF), which was revealed by HaeIII digestion and cloning to represent betaproteobacterial bssA (Table 4; clone F3B22 in Fig. 1). This population was missed in standard TaqI-digested fingerprints, as such short fragments (<50 bp) are usually not considered since they cannot be effectively differentiated from primer-dimers and other short artifacts usually observed in T-RFLP raw data. All expected HaeIII T-RFs (summarized in Table 4), corresponding to the primary TaqI T-RFs in Fig. 2 and as predicted via clones from the same samples (Fig. 1), were readily detectable (HaeIII data not shown). However, recovery of comparative T-RF abundance in both restriction assays was not semiquantitative, as illustrated for one Flingern sample in Fig. 3.
Fig 3.
Exemplary comparison of semiquantitative FAE gene lineage recovery in Flingern sediments (depth, 6.85 m) retrieved with the FAE-N primer set via different methods: cloning and sequencing (a total of 12 clones) as well as T-RFLP analyses with the different restriction enzymes. Dark gray bars, abundance of FAE gene sequence types affiliated with N47 nmsA; light gray bars, the abundance of those affiliated with OX39 bssA homologues.
DISCUSSION
The present study focuses on functional gene analyses for degrader detection at contaminated sites. Our new primer sets are able to survey directly and rapidly (in fingerprinting) the structure of natural FAE gene pools in hydrocarbon-degrading systems. Among all described anaerobic hydrocarbon degradation pathways, activation by fumarate addition is presently one of the most intensively investigated. Other pathways, such as oxygen-independent hydroxylation or carboxylation, either are known for only a small selection of substances (2) or are just about to be understood biochemically (45, 46). Thus, at present, FAE genes can be regarded as the most comprehensive genetic marker for investigating anaerobic hydrocarbon degraders and degradation in the environment.
Assay performance in pure cultures and environmental samples.
Our tests for FAE lineage detectability at the amplicon level (Tables 2 and 3) clearly show that major FAE gene populations are not detectable with the established bssA primer set alone. Thus, not all catabolic potentials for fumarate addition can be detected with this assay, as we demonstrate for terrestrial and marine samples. The clones retrieved in this study prove that the simultaneous application of our new suite of primers is able to recover a far wider range of proteobacterial and clostridial FAE gene lineages than previously described. Furthermore, our primer suite now allows a robust phylogenetic comparison of all generated amplicons. This was our main motivation to introduce the new FAE-KM primer set for assA detection, since the primary assA primers published by Callaghan et al. (5) generate FAE amplicons only partially overlapping with our other bssA and homologous genes.
In part, our novel primer sets are based on a combination of different forward primers, which may potentially complicate the interpretation of amplification results. Yet, mixing of different primers is commonly used in multiplex PCR (47). The application of such methods is not only common to the analysis of nutritional or clinical samples but also established for the analysis of complex environmental samples (e.g., see references 48 and 49). For example, Knaebel and Crawford (50) employed multiplex primers for the detection of aerobic functional genes. There, amplification was shown to be very sensitive but not linear for all template ratios. Still, the influence of sample origin, the targeted gene, and inhibitors from the sampled material was shown to introduce considerably greater bias to PCR results (e.g., on template ratios and detectability) than the influence of the use of multiple primers. Because of this, we consider the introduction of multiple forward primers in some sets to be connected to an acceptable bias, especially since catabolic gene analyses and fingerprinting are already known to be much less robust in a semiquantitative manner than analyses with ribosomal markers (51).
Another parameter of concern was the low frequency of actual FAE gene clones observed, especially for the libraries from the marine sites. Nonintended inserts were mostly shorter primer concatemers or other PCR artifacts. With Flingern aquifer DNA, the yield of good clones was always substantially higher (between 40 and 60%). This is why we confined the verification of our T-RFLP diagnostics to the Flingern samples, as PCR artifacts certainly complicate the interpretation of T-RFLP fingerprints. However, other studies have also reported low FAE sequence yields and cloning efficiencies for certain sample types (5, 6), which may indicate general problems in applying these degenerate primers to PCRs with complex environmental DNA. Here, a possible remedy would be the introduction of cloning-independent, high-throughput next-generation pyrotag sequencing of FAE amplicons, which would still produce sufficient amounts of good reads, even if short reads from biases in pyrotag amplification were abundant. Nevertheless, it was already shown by Winderl et al. (6) that the use of degenerate primers is important for finding previously undetected novel FAE gene lineages. This is a prerequisite for any subsequent design of lineage-specific qPCR assays (44), potentially even allowing a more quantitative inference of in situ biodegradation rates based on the degraders themselves (e.g., see reference 22).
Degrader diversity and on-site hydrocarbon degradation processes.
As the present study was entirely based on gene detection, assumptions on actually ongoing hydrocarbon-degrading processes in the examined samples are not straightforward. Functional implications may have been stronger for transcript detection, but respective attempts were unsuccessful and indicate general problems with extraction of mRNA from these anaerobic systems.
Several distinct FAE gene lineages were detected in the Gölzau enrichments, even though they had been cultivated on monosubstrates for over a year. This either suggests a surprising diversity of catabolic potentials involved in substrate degradation in these enrichments or indicates a memory effect from initially diverse degrader populations in situ. At present, a more detailed investigation of the catabolic genes and potentials in the Gölzau samples is still under way.
Assumptions on ongoing hydrocarbon-degrading processes are also not facile for the gas seepage sites. The occurrence of hydrocarbons (>C4) other than natural gas is known for the Paclele Mici mud volcano (28), but for the Amon mud volcano and Nyegga methane seeps, there have been no assessments for the presence of hydrocarbons higher than C4 to date (32, 33, 52). While the detection of putative assA homologues is expected for natural gas seepages due to the degradation of short-chain alkanes via fumarate addition (5, 53), the discovery of only bssA genes related to Desulfobacula toluolica in Amon mud volcano sediments was unexpected. This could imply two different scenarios: (i) the microbes carrying this bssA sequence type may actually be involved in the degradation of short-chain alkanes, despite the phylogenetic placement of their marker genes with toluene-degrading bssA sensu stricto, or (ii) the detection of these FAE gene types could be attributed to the actual presence of aromatic hydrocarbons via carryover from nearby oil deposits, i.e., by gas ebullition, as is known for the Amon mud volcano's neighbor mud volcano Isis (54). The latter would imply that the true alkane-catabolizing gene markers at the site remained undetected, despite our comprehensive parallel PCR screening. Although FAE substrate specificity for methylated aromatics and nongaseous alkanes does not usually overlap among degrader isolates (55), this may still be possible for short-chain alkanes like butane or propane, which would favor the first scenario.
Finally, although our FAE-B primer set does detect a selection of several clostridial bssA genes and homologues in silico as well in pure-culture screenings, we were able to find the clostridial F2-cluster bssA sequences only in Flingern sediments using our classical bssA primer pair at a lowered annealing temperature. It seems that the FAE-B primer set has, despite its proven ability to amplify clostridial bssA in pure-culture DNA, a bias for nmsA in environmental samples, as observed with T-RFLP data retrieved from Flingern (Fig. 2). A lowered annealing temperature, as also successfully employed for extending the coverage of the bssA primer set, may help to overcome this limitation and is currently being tested.
Novel nmsA sequences found in different environments.
With our new primer sets FAE-B and FAE-N, we provide the first evidence that nmsA and homologous sequences can be detected in the environment, regardless of sample origin. This raises a question about the relevance of the respective catabolic pathway: addition of fumarate to methylnaphthalene. Recent work by Oka et al. (56) showed metabolites of 2-methylnaphthalene carboxylation to be present at a contaminated aquifer. Additionally, methylation and subsequent fumarate addition were first presumed to be the degradation pathway for naphthalene (57). However, recent results suggest that carboxylation (46) is the principal degradation mechanism in nmsA-containing strain N47 (21), abating the potential for nmsA to be a marker for naphthalene degradation. DiDonato et al. (58) found that fumarate addition to 2-methylnaphthalene and toluene is possible with NMS alone for strain NaphS2. Therefore, the nmsA sequences retrieved in this study could potentially play a role not only in 2-methylnaphthalene catabolism but also in toluene degradation. This concept may also help to better explain the phylogenetic placement of the bssA homologue previously retrieved from the toluene-degrading clostridial strain OX39 (6), which does not degrade 2-methylnaphthalene (59). Yet, its bssA-homologous sequence is untypically placed with respect to the now widely documented typical clostridial bssA sequence type (F2 cluster; Fig. 1). Thus, the new sequences related to OX39 that were found (Amon mud volcano clone BAA23, Gulf of Mexico sediment clone BG2A05, and Flingern aquifer clone F3N20) may also be also monoaromatics-utilizing degraders of clostridial affiliation.
Hidden lineages revealed by sequencing-independent T-RFLP screening in Flingern sediment.
Our results for the Flingern samples demonstrate, in contrast to previous findings (6), that there are indeed different intrinsic FAE gene populations to be found in this contaminated aquifer. The apparent dominance of the Desulfobulbaceae F1-cluster bssA reported previously may thus be an effect of PCR selection. We show that the clostridial F2-cluster bssA sequences are not consistently detected over the three selected depths by our standard bssA primer set (58°C), while putative toluene degraders related to Desulfosporosinus spp. have been identified in sediments from the same site sampled in 2006 (14) and are abundant in 16S rRNA pyrotag libraries of sediments taken in 2009 (25).
The identification of degrader lineages on the basis of T-RFLP results was straightforward at the Flingern site, as there are already ample bssA sequence data available from that site (6, 44). We cannot claim that our new approach has an absolute diagnostic capability; sequencing will still be necessary to identify FAE lineages for sites with no a priori sequence information available. Nevertheless, the double digest can add diagnostic security in case of overlapping T-RF sizes for unrelated FAE lineages (e.g., an HaeIII T-RF of 132 bp or a TaqI T-RF of 50 bp could both represent distinct beta- and deltaproteobacterial as well as clostridial bssA lineages, as illustrated in Fig. 1), where dual-digest results can mutually exclude options. Thus, the double-digest T-RFLP described here can be used as a tool for monitoring FAE gene populations at sites with minimized sequencing effort.
Conclusions.
In essence, we demonstrate that the simultaneous use of parallel FAE primer sets is necessary to recover a wide diversity of FAE gene lineages in hydrocarbon-impacted environmental systems. Via amplicon screening as well as sequencing-independent T-RFLP diagnostics, we reveal that distinct FAE gene lineages may be missed with available bssA primer sets, especially clostridial bssA and proteobacterial nmsA homologues. The detection of these additional degrader populations in distinct hydrocarbon-impacted systems allows extending our grasp of the biodiversity and ecology of bacteria involved in anaerobic hydrocarbon degradation in the environment, which is relevant for a better understanding of natural attenuation processes.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Wilfried Röling (University of Amsterdam) for giving us access to Banisveld bssA sequences before publication; Drazenka Selesi and Nidal Abu Laban (Helmholtz Zentrum München) for DNA of strains N47 and BF and for giving us access to sequence information before publication; and the officers, crews, and shipboard scientific parties of the cruises M67-2 (RV Meteror), Vicking (RV Pourquoi Pas?), SO174 (RV Sonne), AT 15-56 (RV Atlantis), and MSMB-3 (RV Maria S. Merian).
This work was financed by grants from the Deutsche Forschungsgemeinschaft within Priority Programme 1319. Further support came from the Max Planck Society.
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at 10.1128/AEM.02362-12.
REFERENCES
- 1. Widdel F, Boetius A, Rabus R. 2006. Anaerobic biodegradation of hydrocarbons including methane, p 1028–1049 In The prokaryotes, vol 2 Ecophysiological and biochemical aspects. Springer Verlag, New York, NY [Google Scholar]
- 2. Heider J. 2007. Adding handles to unhandy substrates: anaerobic hydrocarbon activation mechanisms. Curr. Opin. Chem. Biol. 11:188–194 [DOI] [PubMed] [Google Scholar]
- 3. Kuntze K, Vogt C, Richnow H-H, Boll M. 2011. Combined application of PCR-based functional assays for the detection of aromatic-compound-degrading anaerobes. Appl. Environ. Microbiol. 77:5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beller HR, Kane SR, Legler TC, Alvarez PJ. 2002. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 36:3977–3984 [DOI] [PubMed] [Google Scholar]
- 5. Callaghan AV, Davidova IA, Savage-Ashlock K, Parisi VA, Gieg LM, Suflita JM, Kukor JJ, Wawrik B. 2010. Diversity of benzyl- and alkylsuccinate synthase genes in hydrocarbon-impacted environments and enrichment cultures. Environ. Sci. Technol. 44:7287–7294 [DOI] [PubMed] [Google Scholar]
- 6. Winderl C, Schaefer S, Lueders T. 2007. Detection of anaerobic toluene and hydrocarbon degraders in contaminated aquifers using benzylsuccinate synthase (bssA) genes as a functional marker. Environ. Microbiol. 9:1035–1046 [DOI] [PubMed] [Google Scholar]
- 7. Callaghan AV, Wawrik B, Ni Chadhain SM, Young LY, Zylstra GJ. 2008. Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes. Biochem. Biophys. Res. Commun. 366:142–148 [DOI] [PubMed] [Google Scholar]
- 8. Grundmann O, Behrends A, Rabus R, Amann J, Halder T, Heider J, Widdel F. 2008. Genes encoding the candidate enzyme for anaerobic activation of n-alkanes in the denitrifying bacterium, strain HxN1. Environ. Microbiol. 10:376–385 [DOI] [PubMed] [Google Scholar]
- 9. Widdel F, Grundmann O. 2010. Biochemistry of the anaerobic degradation of non-methane alkanes, p 909–924 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany [Google Scholar]
- 10. Annweiler E, Materna A, Safinowski M, Kappler A, Richnow HH, Michaelis W, Meckenstock RU. 2000. Anaerobic degradation of 2-methylnaphthalene by a sulfate-reducing enrichment culture. Appl. Environ. Microbiol. 66:5329–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Staats M, Braster M, Röling WFM. 2011. Molecular diversity and distribution of aromatic hydrocarbon-degrading anaerobes across a landfill leachate plume. Environ. Microbiol. 13:1216–1227 [DOI] [PubMed] [Google Scholar]
- 12. Botton S, van Harmelen M, Braster M, Parsons JR, Röling WFM. 2007. Dominance of Geobacteraceae in BTX-degrading enrichments from an iron-reducing aquifer. FEMS Microbiol. Ecol. 62:118–130 [DOI] [PubMed] [Google Scholar]
- 13. Weelink SA, van Doesburg W, Saia FT, Rijpstra WI, Röling WF, Smidt H, Stams AJM. 2009. A strictly anaerobic betaproteobacterium Georgfuchsia toluolica gen. nov., sp. nov. degrades aromatic compounds with Fe(III), Mn(IV) or nitrate as an electron acceptor. FEMS Microbiol. Ecol. 70:575–585 [DOI] [PubMed] [Google Scholar]
- 14. Pilloni G, von Netzer F, Engel M, Lueders T. 2011. Electron acceptor-dependent identification of key anaerobic toluene degraders at a tar-oil-contaminated aquifer by Pyro-SIP. FEMS Microbiol. Ecol. 78:165–175 [DOI] [PubMed] [Google Scholar]
- 15. Winderl C, Penning H, von Netzer F, Meckenstock RU, Lueders T. 2010. DNA-SIP identifies sulfate-reducing Clostridia as important toluene degraders in tar-oil-contaminated aquifer sediment. ISME J. 4:1314–1325 [DOI] [PubMed] [Google Scholar]
- 16. Yagi JM, Suflita JM, Gieg LM, DeRito CM, Jeon C-O, Madsen EL. 2010. Subsurface cycling of nitrogen and anaerobic aromatic hydrocarbon biodegradation revealed by nucleic acid and metabolic biomarkers. Appl. Environ. Microbiol. 76:3124–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herrmann S, Vogt C, Fischer A, Kuppardt A, Richnow H-H. 2009. Characterization of anaerobic xylene biodegradation by two-dimensional isotope fractionation analysis. Environ. Microbiol. Rep. 1:535–544 [DOI] [PubMed] [Google Scholar]
- 18. Kunapuli U, Jahn MK, Lueders T, Geyer R, Heipieper HJ, Meckenstock RU. 2010. Desulfitobacterium aromaticivorans sp. nov. and Geobacter toluenoxydans sp. nov., iron-reducing bacteria capable of anaerobic degradation of monoaromatic hydrocarbons. Int. J. Syst. Evol. Microbiol. 60:686–695 [DOI] [PubMed] [Google Scholar]
- 19. Abu Laban N, Selesi D, Rattei T, Tischler P, Meckenstock RU. 2010. Identification of enzymes involved in anaerobic benzene degradation by a strictly anaerobic iron-reducing enrichment culture. Environ. Microbiol. 12:2783–2796 [DOI] [PubMed] [Google Scholar]
- 20. Musat F, Galushko A, Jacob J, Widdel F, Kube M, Reinhardt R, Wilkes H, Schink B, Rabus R. 2009. Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria. Environ. Microbiol. 11:209–219 [DOI] [PubMed] [Google Scholar]
- 21. Selesi D, Jehmlich N, von Bergen M, Schmidt F, Rattei T, Tischler P, Lueders T, Meckenstock RU. 2010. Combined genomic and proteomic approaches identify gene clusters involved in anaerobic 2-methylnaphthalene degradation in the sulfate-reducing enrichment culture N47. J. Bacteriol. 192:295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kazy S, Monier A, Alvarez P. 2010. Assessing the correlation between anaerobic toluene degradation activity and bssA concentrations in hydrocarbon-contaminated aquifer material. Biodegradation 21:793–800 [DOI] [PubMed] [Google Scholar]
- 23. Lueders T, Manefield M, Friedrich MW. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73–78 [DOI] [PubMed] [Google Scholar]
- 24. Kunapuli U, Lueders T, Meckenstock RU. 2007. The use of stable isotope probing to identify key iron-reducing microorganisms involved in anaerobic benzene degradation. ISME J. 1:643–653 [DOI] [PubMed] [Google Scholar]
- 25. Pilloni G, Granitsiotis MS, Engel M, Lueders T. 2012. Testing the limits of 454 pyrotag sequencing: reproducibility, quantitative assessment and comparison to T-RFLP fingerprinting of aquifer microbes. PLoS One 7:e40467 doi:10.1371/journal.pone.0040467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anneser B, Pilloni G, Bayer A, Lueders T, Griebler C, Einsiedl F, Richters L. 2010. High resolution analysis of contaminated aquifer sediments and groundwater—what can be learned in terms of natural attenuation? Geomicrobiol. J. 27:130–142 [Google Scholar]
- 27. Jobelius C, Ruth B, Griebler C, Meckenstock RU, Hollender J, Reineke A, Frimmel FH, Zwiener C. 2011. Metabolites indicate hot spots of biodegradation and biogeochemical gradients in a high-resolution monitoring well. Environ. Sci. Technol. 45:474–481 [DOI] [PubMed] [Google Scholar]
- 28. Alain K, Holler T, Musat F, Elvert M, Treude T, Krüger M. 2006. Microbiological investigation of methane- and hydrocarbon-discharging mud volcanoes in the Carpathian Mountains, Romania. Environ. Microbiol. 8:574–590 [DOI] [PubMed] [Google Scholar]
- 29. Feisthauer S, Siegert M, Seidel M, Richnow HH, Zengler K, Gründger F, Krüger M. 2010. Isotopic fingerprinting of methane and CO2 formation from aliphatic and aromatic hydrocarbons. Org. Geochem. 41:482–490 [Google Scholar]
- 30. Orcutt BN, Joye SB, Kleindienst S, Knittel K, Ramette A, Reitz A, Samarkin V, Treude T, Boetius A. 2010. Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments. Deep Sea Res. Part II Top. Stud. Oceanogr. 57:2008–2021 [Google Scholar]
- 31. Zhou J, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van Gaever S, Raes M, Pasotti F, Vanreusel A. 2010. Spatial scale and habitat-dependent diversity patterns in nematode communities in three seepage related sites along the Norwegian Sea margin. Mar. Ecol. 31:66–77 [Google Scholar]
- 33. Mastalerz V, de Lange GJ, Dählmann A. 2009. Differential aerobic and anaerobic oxidation of hydrocarbon gases discharged at mud volcanoes in the Nile deep-sea fan. Geochim. Cosmochim. Acta 73:3849–3863 [Google Scholar]
- 34. Didyk BM, Simoneit BRT. 1989. Hydrothermal oil of Guaymas Basin and implications for petroleum formation mechanisms. Nature 342:65–69 [Google Scholar]
- 35. Culman S, Bukowski R, Gauch H, Cadillo-Quiroz H, Buckley D. 2009. T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics 10:171 doi:10.1186/1471-2105-10-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Washer CE, Edwards EA. 2007. Identification and expression of benzylsuccinate synthase genes in a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 73:1367–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Achong GR, Rodriguez AM, Spormann AM. 2001. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J. Bacteriol. 183:6763–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lovley DR, Giovannoni SJ, White DC, Champine JE, Phillips EJP, Gorby YA, Goodwin S. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336–344 [DOI] [PubMed] [Google Scholar]
- 39. Ommedal H, Torsvik T. 2007. Desulfotignum toluenicum sp. nov., a novel toluene-degrading, sulphate-reducing bacterium isolated from an oil-reservoir model column. Int. J. Syst. Evol. Microbiol. 57:2865–2869 [DOI] [PubMed] [Google Scholar]
- 40. Cravo-Laureau C, Matheron R, Joulian C, Cayol J-L, Hirschler-Rea A. 2004. Desulfatibacillum alkenivorans sp. nov., a novel n-alkene-degrading, sulfate-reducing bacterium, and emended description of the genus Desulfatibacillum. Int. J. Syst. Evol. Microbiol. 54:1639–1642 [DOI] [PubMed] [Google Scholar]
- 41. Cravo-Laureau C, Matheron R, Cayol J-L, Joulian C, Hirschler-Réa A. 2004. Desulfatibacillum aliphaticivorans gen. nov., sp. nov., an n-alkane- and n-alkene-degrading, sulfate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 54:77–83 [DOI] [PubMed] [Google Scholar]
- 42. Kleindienst S, Ramette A, Amann R, Knittel K. 2012. Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ. Microbiol. 14:2689–2710 [DOI] [PubMed] [Google Scholar]
- 43. Bohrmann G, Spiess V, Böckel B, Boetius A, Boles M, Brüning M, Buhmann S, Cruz Melo C, Dalthorp-Moorhouse M, Dehning K, Ding F, Escobar-Briones E, Enneking K, Felden J, Fekete N, Freidank T, Gassner A, Gaytan A, Geersen J, Hinrichs K-U, Hohnberg J, Kasten S, Keil H, Klar S, Klaucke I, Kuhlmann J, MacDonald I, Meinecke G, Mortera C, Naehr T, Nowald N, Ott C, Pacheco Muñoz J, Pelaez JR, Ratmeyer V, Renken J, Reuter M, Sackmann V, Sahling H, Schubotz F, Schewe F, Stephan S, Thal J, Trampe A, Truscheit T, Viehweger M, Wilhelm TT, Wegner G, Wenzhöfer F, Zabel M. 2008. Fluid seepage in the Gulf of Mexico. In Report and preliminary results of R/V Meteor cruise M67/2a and 2b, Balboa-Tampico-Bridgetown, 15 March-24 April 2006 Berichte aus dem Fachbereich Geowissenschaften, Universität Bremen, Bremen, Germany [Google Scholar]
- 44. Winderl C, Anneser B, Griebler C, Meckenstock RU, Lueders T. 2008. Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar oil contaminant plume. Appl. Environ. Microbiol. 74:792–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meckenstock RU, Mouttaki H. 2011. Anaerobic degradation of non-substituted aromatic hydrocarbons. Curr. Opin. Biotechnol. 22:406–414 [DOI] [PubMed] [Google Scholar]
- 46. Mouttaki H, Johannes J, Meckenstock RU. 2012. Identification of naphthalene carboxylase as a prototype for the anaerobic activation of non-substituted aromatic hydrocarbons. Environ. Microbiol. 14:2770–2774 [DOI] [PubMed] [Google Scholar]
- 47. Settanni L, Corsetti A. 2007. The use of multiplex PCR to detect and differentiate food- and beverage-associated microorganisms: a review. J. Microbiol. Methods 69:1–22 [DOI] [PubMed] [Google Scholar]
- 48. Volkmann H, Schwartz T, Kirchen S, Stofer C, Obst U. 2007. Evaluation of inhibition and cross-reaction effects on real-time PCR applied to the total DNA of wastewater samples for the quantification of bacterial antibiotic resistance genes and taxon-specific targets. Mol. Cell. Probes 21:125–133 [DOI] [PubMed] [Google Scholar]
- 49. Xiong X, Yin X, Pei X, Jin P, Zhang A, Li Y, Gong W, Wang Q. 2012. Retrieval of glycoside hydrolase family 9 cellulase genes from environmental DNA by metagenomic gene specific multi-primer PCR. Biotechnol. Lett. 34:875–882 [DOI] [PubMed] [Google Scholar]
- 50. Knaebel DB, Crawford RL. 1995. Extraction and purification of microbial DNA from petroleum-contaminated soils and detection of low numbers of toluene, octane and pesticide degraders by multiplex polymerase chain reaction and Southern analysis. Mol. Ecol. 4:579–592 [DOI] [PubMed] [Google Scholar]
- 51. Lueders T, Friedrich MW. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vanreusel A, Andersen AC, Boetius A, Connelly D, Cunha MR, Decker C, Hilario A, Kormas KA, Maignien LL, Olu K, Pachiadaki M, Ritt B, Rodrigues C, Sarrazin J, Tyler T, Van Gaever S, Vanneste H. 2009. Biodiversity of cold seep ecosystems along the European margins. Oceanography 22:110–127 [Google Scholar]
- 53. Kniemeyer O, Musat F, Sievert SM, Knittel K, Wilkes H, Blumenberg M, Michaelis W, Classen A, Bolm C, Joye SB, Widdel F. 2007. Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria. Nature 449:898–901 [DOI] [PubMed] [Google Scholar]
- 54. Mastalerz V, de Lange GJ, Dählmann A, Feseker T. 2007. Active venting at the Isis mud volcano, offshore Egypt: origin and migration of hydrocarbons. Chem. Geol. 246:87–106 [Google Scholar]
- 55. Rabus R, Jarling R, Lahme S, Kühner S, Heider J, Widdel F, Wilkes H. 2011. Co-metabolic conversion of toluene in anaerobic n-alkane-degrading bacteria. Environ. Microbiol. 13:2576–2586 [DOI] [PubMed] [Google Scholar]
- 56. Oka AR, Phelps CD, Zhu X, Saber DL, Young LY. 2011. Dual biomarkers of anaerobic hydrocarbon degradation in historically contaminated groundwater. Environ. Sci. Technol. 45:3407–3414 [DOI] [PubMed] [Google Scholar]
- 57. Safinowski M, Meckenstock RU. 2006. Methylation is the initial reaction in anaerobic naphthalene degradation by a sulfate-reducing enrichment culture. Environ. Microbiol. 8:347–352 [DOI] [PubMed] [Google Scholar]
- 58. DiDonato RJ, Jr, Young ND, Butler JE, Chin K-J, Hixson KK, Mouser P, Lipton MS, DeBoy R, Methé BA. 2010. Genome sequence of the deltaproteobacterial strain NaphS2 and analysis of differential gene expression during anaerobic growth on naphthalene. PLoS One 5:e14072 doi:10.1371/journal.pone.0014072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morasch B, Schink B, Tebbe CC, Meckenstock RU. 2004. Degradation of o-xylene and m-xylene by a novel sulfate-reducer belonging to the genus Desulfotomaculum. Arch. Microbiol. 181:407–417 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.