Abstract
In Bacillus subtilis, the response regulator DegU and its cognate kinase, DegS, constitute a two-component system that regulates many cellular processes, including exoprotease production and genetic competence. Phosphorylated DegU (DegU-P) activates its own promoter and is degraded by the ClpCP protease. We observed induction of degU by glucose in sporulation medium. This was abolished in two mutants: the ccpA (catabolite control protein A) and clpC disruptants. Transcription of the promoter of the operon containing clpC (PclpC) decreased in the presence of glucose, and the disruption of ccpA resulted in derepression of PclpC. However, this was not directly mediated by CcpA, because we failed to detect binding of CcpA to PclpC. Glucose decreased the expression of clpC, leading to low cellular concentrations of the ClpCP protease. Thus, degU is induced through activation of autoregulation by a decrease in ClpCP-dependent proteolysis of DegU-P. An electrophoretic mobility shift assay showed that CcpA bound directly to the degU upstream region, indicating that CcpA activates degU through binding. The bound region was narrowed down to 27 bases, which contained a cre (catabolite-responsive element) sequence with a low match to the cre consensus sequence. In a footprint analysis, CcpA specifically protected a region containing the cre sequence from DNase I digestion. The induction of degU by glucose showed complex regulation of the degU gene.
INTRODUCTION
Glucose is a very easily metabolized carbon source and thus is one of the preferred carbon sources for bacteria. Addition of glucose to growth medium has profound effects on bacterial cell physiology (1). The transcription factor CcpA plays a central role in catabolite regulation in low-GC Gram-positive bacteria (2, 3). CcpA is activated by association with P-Ser-Hpr, which is Ser-phosphorylated Hpr produced by HprK. Hpr is a histidine-containing phosphotransfer protein involved in sugar transport. Activation of HprK is triggered by accumulation of fructose-1,6-biphosphate, whose cellular concentrations are increased by the incorporation of a preferred sugar such as glucose. CcpA binds to a degenerate pseudopalindromic sequence known as the cre (catabolite-responsive element) sequence (4). CcpA functions negatively and positively for gene transcription, and its function is determined by the location of the cre sequence in many cases (2). Upstream binding of CcpA from the core promoter results in transcriptional activation, but it represses transcription when it binds to regions in the core promoter or at downstream locations, including within the gene open reading frame (ORF) (2). Many of the genes regulated by CcpA are related to steps in carbon metabolism, such as glycolysis, gluconeogenesis, and the tricarboxylic acid (TCA) cycle, and to nitrogen metabolism. Several global analyses, such as DNA microarray analyses, chromatin immunoprecipitation (ChIP) analyses, and in silico searching for cre sequences in the genome revealed that more than 300 genes are directly or indirectly regulated by CcpA (5–11). In DNA microarray analyses and chromatin immunoprecipitation analyses, many previously unidentified CcpA-dependent genes were detected, leading to expansion of the functional roles of CcpA. We note that degU was not identified as a CcpA-dependent gene in these analyses.
DegU is a response regulator that belongs to the NarL subfamily (12). It is activated by phosphorylation of a single Asp site on its receiver domain by the cognate kinase DegS (13). The degU gene resides within an operon with the upstream degS gene (14, 15). DegU, a DNA-binding protein that recognizes AT-rich octamers in a variety of arrangements (12, 16), controls many genes and biological processes (17, 18). In its low-level phosphorylation state, including its nonphosphorylated state, DegU activates comK (a master regulator of genetic competence) and the fla/che operon (12, 14, 19, 20). In its high-level phosphorylation state, DegU activates degU itself and induces many target genes encoding extracellular degrading enzymes (20–25). Thus, degU is activated by an autoregulatory loop through the P3 promoter (26, 27). In addition, high levels of DegU-P result in repression of motility (25, 28). The P2 promoter, which is located upstream of the degU P3 promoter, has been known to be activated by the transcription factor TnrA either under nitrogen starvation conditions or in a disruptant of glnA, encoding glutamine synthetase (27, 29). These observations indicate that there is some nutrient-dependent regulation of degU. Furthermore, the AAA+ protease ClpCP specifically degrades the phosphorylated form of DegU (DegU-P), leading to modulation of the DegU autoactivation loop (26, 30). DegU is also regulated by protein-protein interactions, including the RapG-PhrG system. RapG inhibits the binding of DegU to DNA, and the extracellular pentapeptide PhrG inhibits RapG activity (31). The other protein-protein interaction includes the small protein SwrA, which is a swarming motility regulator (32). SwrA interacts with the N-terminal region of DegU, leading to stabilization of the DegU dimer (16).
In this paper, we report that the addition of glucose induced expression of the degU gene, leading to increased expression of two DegU-P-driven genes. The utilization of the promoter of the operon containing clpC (PclpC) was subject to CcpA-dependent glucose repression. Glucose decreased the expression of clpC, leading to low cellular concentrations of the ClpCP protease. Thus, degU was induced through activation of autoregulation. On the other hand, CcpA bound directly to the degU upstream region in electrophoretic mobility shift assay (EMSA) and footprint analyses, indicating that CcpA activated degU by binding to it. These analyses revealed a cre sequence with a low match to the consensus sequence in degU. The cre sequence itself in degU showed a low affinity for CcpA, while in the more natural context (using a longer DNA fragment containing cre as a probe), the cre sequence in degU showed a stronger affinity for CcpA.
MATERIALS AND METHODS
Bacterial strains, culture media, and general methods.
All of the Bacillus subtilis strains used for this study are listed in Table 1. Schaeffer's nutrient broth-based sporulation medium and Luria-Bertani medium (LB medium; Lennox, Difco, MI) were used for the determination of β-galactosidase activity (1). For B. subtilis transformation, modified competence medium was used (13). Escherichia coli JM103 cells for DNA manipulation were grown in LB medium. The concentrations of antibiotics used were described previously (35). Synthetic oligonucleotides were prepared commercially by Tsukuba Oligo Service (Ibaraki, Japan) and are listed in Table S1. KOD+-neo DNA polymerase I (Toyobo, Osaka, Japan) with high fidelity was used for PCR.
Table 1.
Strains and plasmids used for this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| 168 | trpC2 | Laboratory stock |
| TF10 | trpC2 ccpA (Cmr) | 33 |
| OAM632 | trpC2 hprK (Pmr) | This study |
| OAM588 | trpC2 clpC (Tcr) | 26 |
| CU741T | trpC2 leuC7 tnrA (Cmr) | 27 |
| 168 glnA14 | trpC2 glnA (Spr) | 29 |
| OAM415 | trpC2 amyE::degU-lacZ U-WT (Cmr) | 26 |
| OAM633 | trpC2 amyE::degU-lacZ U-del11 (Cmr) | This study |
| OAM634 | trpC2 amyE::degU-lacZ U-M15 (Cmr) | This study |
| OAM516 | trpC2 amyE::degU-lacZ U-del3 (Cmr) | 26 |
| OAM636 | trpC2 amyE::degU-lacZ U-P2 (Cmr) | This study |
| OAM637 | trpC2 amyE::degU-lacZ U-P2 (Cmr) glnA(Spr) | This study |
| OAM638 | trpC2 amyE::degU-lacZ U-P2 (Cmr) glnA(Spr) tnrA(Cmr::Kmr) | This study |
| OAM639 | trpC2 amyE::degU-lacZ U-P2 (Cmr::Tcr) glnA(Spr) ccpA(Cmr) | This study |
| OAM420 | trpC2 amyE::degU-lacZ U-WT (Cmr) clpC(Tcr) | 26 |
| OAM416 | trpC2 amyE::degU-lacZ U-WT (Cmr) degU(Kmr) | 26 |
| OAM640 | trpC2 thrC::degU-lacZ U-WT (Spr) | This study |
| OAM641 | trpC2 thrC::degU-lacZ U-WT (Spr) ccpA (Cmr) | This study |
| OAM642 | trpC2 thrC::degU-lacZ U-WT (Spr) hprK (Pmr) | This study |
| OAM643 | trpC2 thrC::degU-lacZ U-WT (Spr) tnrA (Cmr) | This study |
| OAM371 | trpC2 amyE::sacB-lacZ S-WT (Cmr) | 20 |
| OAM372 | trpC2 amyE::sacB-lacZ S-WT (Cmr) degU(Kmr) | 20 |
| OAM367 | trpC2 amyE::bpr-lacZ B-WT (Cmr) | 23 |
| OAM368 | trpC2 amyE::bpr-lacZ B-WT (Cmr) degU(Kmr) | 23 |
| OAM644 | trpC2 amyE::PclpC-bgaB (Cmr::Tcr) | This study |
| OAM645 | trpC2 amyE::PclpC-bgaB (Cmr::Tcr) ccpA (Cmr) | This study |
| OAM646 | trpC2 amyE::PclpC-bgaB (Cmr::Tcr) hprK(Pmr) | This study |
| Plasmids | ||
| pDG1729 | Insertion vector for thrC, spectinomycin and erythromycin resistance genes, and lacZ | 34 |
| pDG1729-degU | pDG1729 carrying a promoter region of degU (positions −184 to +56) | 26 |
| pIS284 | Insertion vector for amyE, chloramphenicol resistance gene, and lacZ | 20 |
| pIS284-degU-del11 | pIS284 carrying a promoter region of degU (positions −151 to +56) | This study |
| pIS284-degU-M15 | pIS284 carrying a promoter region of degU (positions −151 to +56), with CG-to-TT mutation (positions −135 and −136) | This study |
| pIS284-degU-P2 | pIS284 carrying a promoter region of degU (positions −184 to −34) | This study |
| pPhl2 | Phleomycin resistance | 35 |
| pPhl2-hprK | pPhl2 carrying part of hprK | This study |
| ECE75 | Ampicillin resistance; Cmr::Tcr | BGSC |
| ECE73 | Ampicillin resistance; Cmr::Kmr | M. Steinmetz |
| His-CcpA | pQE30-based vector for production of CcpA-His | 36 |
| His-MecA | pQE8-based vector for production of MecA-His | This study |
Plasmid construction.
The plasmids used in this study are listed in Table 1. To construct pPhl2-hprK, a PCR product was prepared using the relevant primer pair (see Table S1 in the supplemental material), and the DNA fragment was digested with BamHI and BglII and ligated into the BamHI-digested pPhl-2 plasmid (35). To construct pIS284-degUdel11, -degUM15, and -degU-P2, PCR products were prepared using the relevant primer pairs (see Table S1). The DNA fragments were digested with BamHI and HindIII and then ligated into the similarly digested pIS284 plasmid (20). To construct His-MecA, a PCR product was prepared using the relevant primer pair (see Table S1), and the DNA fragment was digested with BamHI and then ligated into the BamHI-digested pQE8 plasmid (Qiagen, Hilden, Germany). The sequences cloned into all plasmids were confirmed.
β-Galactosidase assays.
Samples were collected at hourly intervals, and the level of β-galactosidase activity was determined as described previously (35).
Protein purification.
The procedures used to induce protein production and purify proteins from E. coli were described previously (31). His-tagged CcpA was expressed as a soluble protein in E. coli M15 cells carrying pRep4 and was purified by stepwise elution with imidazole from a Ni affinity column (36). After SDS-PAGE analysis of the fractions, the purified protein was dialyzed against TEDG buffer (37).
Western blot assay.
Cells were harvested and lysed using a French press in Tris-EDTA (TE) buffer containing 1 mM phenylmethylsulfonyl fluoride. The whole-cell lysate was separated into supernatant and pellet by centrifugation. Soluble proteins were separated by 12.5% SDS-PAGE. An anti-MecA polyclonal antibody was raised against purified His-MecA in rabbits and prepared by JBioS Inc. (Saitama, Japan). The Western blot assay was conducted using a BM chemiluminescence Western blotting kit (Roche, Basel, Switzerland). We used polyclonal anti-DegU (38), anti-ClpP (39), and anti-ClpC (40) antibodies for analyses. Quantification of band intensities was described previously (26).
EMSA and DNase I footprint assay.
Probes were prepared by PCR amplification using an appropriate primer and the biotinylated primer shown in Table S1 in the supplemental material. For EMSA, the indicated amounts of His-CcpA were incubated with 5 nM DNA probe in 12 μl of 0.5× TEDG buffer containing 20 mM KCl, 1 mM MgCl2, and an appropriate amount of poly(dI-dC) as a nonspecific competitor (GE Healthcare, United Kingdom) for 15 min at 37°C. After addition of 2 μl of loading buffer (26), the samples were separated in polyacrylamide gels containing the indicated concentrations of acrylamide. Electrophoresis was performed in 0.25× Tris-borate-EDTA (TBE) buffer at 4°C. For the footprint assay, the reaction mixture was similar to that used for the EMSA, except that 30 nM probe DNA was included in the mixture. After incubation for 15 min at 37°C, 0.05 unit of DNase I (TaKaRa, Kyoto, Japan) was added, and the mixture was incubated for 3 min at 28°C. After addition of 25 μl TE buffer, phenol-chloroform-isoamyl alcohol (25:25:1) extraction was performed, followed by ethanol precipitation with carrier tRNA. The samples were separated in a 6% polyacrylamide gel containing 7 M urea. Electrophoresis was performed in a TBE buffer. Biotin-labeled DNA was detected as described previously (35).
RESULTS
Glucose-induced degU expression.
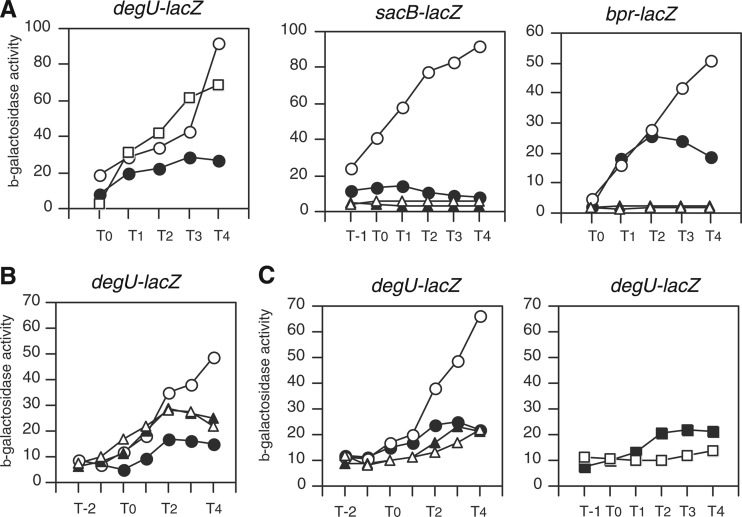
During the course of studying the expression of degU driven by its own promoter, we observed that degU-lacZ was induced by addition of glucose to the sporulation medium (Fig. 1A, left panel). Thereafter, we always incorporated 2% (wt/vol) glucose into the medium. This effect was dependent on the composition of the medium, as the expression of degU-lacZ in LB medium was slightly reduced by glucose (see Fig. S1A in the supplemental material). Next, we examined the expression of DegU-driven genes in the presence of glucose to confirm whether the positive effect was observed ubiquitously among genes regulated by DegU. As reported previously, there are more than 120 DegU regulon genes, and many of them are under complex regulation by multiple transcription factors, such as that encoded by aprE (18, 41). Thus, we chose the following two genes for analyses: the bpr gene, which is regulated only by DegU (23), and the sacB gene, whose cis element (which is responsible for sucrose induction) was deleted, resulting in activation of the sacB-lacZ fusion solely by DegU (20). As shown in Fig. 1A, glucose induced expression of both fusions. In addition, the induction of both genes was abolished by disruption of degU (Fig. 1A). These observations confirmed that the sacB and bpr genes were induced in a DegU-dependent manner by glucose.
Fig 1.
Glucose induction of expression of DegU-dependent genes and abolishment of induction in ccpA and clpC mutants. Cells were grown in sporulation medium without (closed symbols) or with (open symbols) 2% (wt/vol) glucose. Cells were sampled hourly. β-Galactosidase activities are shown in Miller units. The x axis represents the growth time (h) relative to the end of vegetative growth (T0). (A) Glucose induction of DegU-dependent gene expression. Each gene fusion is indicated above the relevant panel. (Left) Squares, 1% glucose; open circles, 2% glucose. (Middle and right panels) Circles, wild-type cells; triangles, degU cells. (B) Expression of wild-type degU-lacZ fusion in the clpC strain (triangles). Data for wild-type cells are shown in circles. (C) Expression of wild-type degU-lacZ fusion in ccpA (left) and hprK (right) strains. Circles, wild type; triangles, ccpA strain; squares, hprK strain.
Disruption of clpC and ccpA abolishes glucose induction of degU.
Previously, we reported that ClpCP degrades DegU-P. Thus, the expression of degU is enhanced in the clpC strain because DegU-P is not degraded in that strain (26). Based on these regulatory mechanisms, it is possible that addition of glucose somehow affects ClpCP-dependent regulation of degU. To test this possibility, we examined the expression of degU in the clpC strain in medium containing glucose. Addition of glucose did not further enhance the increased expression of degU in the clpC strain (Fig. 1B). The level of degU expression in the clpC strain did not reach that in the wild-type strain in the presence of glucose. This suggested that another mechanism(s) besides the ClpCP system affected expression.
The transcription factor CcpA is one of the primary regulators of glucose induction, and HprK produces active CcpA through production of P-Ser-Hpr (2). Thus, we examined the expression of degU in the presence of glucose in the ccpA strain. As expected, the induction of degU by glucose was abolished by disruption of ccpA (Fig. 1C, left panel), indicating that CcpA was involved in this phenomenon. Without glucose, disruption of ccpA resulted in a slight decrease in degU expression. We obtained a similar result with the hprK strain (Fig. 1C, right panel), confirming the involvement of HprK-activated CcpA in the glucose induction of degU.
To further confirm the glucose effect on degU expression, we used the synthetic competence medium MC with the addition of either glucose or glycerol. Compared to the expression of degU-lacZ in glucose-based competence medium, the levels of degU expression were lower in glycerol-based competence medium (see Fig. S1A in the supplemental material). This is because in the glycerol-based medium, CcpA is inactivated to derepress glycerol incorporation (42). This result again confirmed the CcpA-dependent control of degU expression.
Since glucose had no effect on degU expression in the clpC mutant, the simplest explanation is that the effects of the CcpA and ClpC proteins are interdependent. To test this idea, we constructed a ccpA and clpC double mutant and examined its β-galactosidase activity. As shown in Fig. S1B in the supplemental material, the clpC mutation did not enhance the expression of wild-type degU-lacZ compared to that observed in the ccpA strain. This suggested that CcpA and ClpC are somehow interdependent through an unknown mechanism.
Western analysis of DegU.
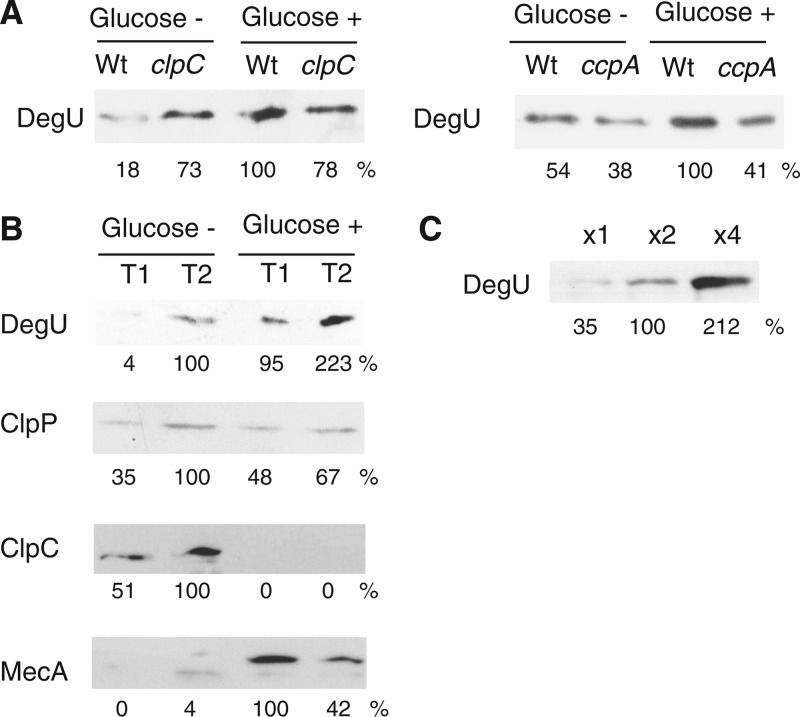
Next, we confirmed the glucose induction of degU at the protein level. The relationship between band intensities and protein amounts (Fig. 2C) showed a rough linearity, which justified quantification of the observed bands. As shown in Fig. 2A and B, addition of glucose increased the amount of the DegU protein to various extents, consistent with the results of degU-lacZ expression. In both the clpC and ccpA strains, addition of glucose did not increase the amount of DegU (Fig. 2A). Using the same soluble extracts of whole-cell lysates, we investigated the protein levels of the ClpCP degradation machinery associated with MecA. Surprisingly, the ClpC protein was not detected in extracts from the culture medium containing glucose, while similar levels of the ClpP protein were detected in extracts both with and without glucose (Fig. 2B). Moreover, the amount of MecA, a substrate for ClpCP (43), was clearly enhanced. In addition, there was only a slight increase in mecA transcription (see Fig. S2 in the supplemental material). Thus, the increased amount of MecA was consistent with the disappearance of ClpC. Consequently, when glucose was added to the sporulation medium, ClpCP was not functional because of the lack of ClpC, leading to enhanced degU expression. Like MecA, ClpE is a substrate for ClpCP (44). Thus, there was abundant ClpE in the ClpC-depleted cells. We note that the anti-ClpC antibody cross-reacted with ClpE under strong detection conditions, including when high concentrations of secondary antibody were used (data not shown).
Fig 2.
Western blot analysis of DegU and ClpCP, with and without glucose. Cells were grown in sporulation medium. Times of sample collection and addition of 2% (wt/vol) glucose are indicated above the panels. In each panel, relative band intensities are indicated under the panel. In the sets of experiments, equal amounts of proteins were applied to the lanes. (A) DegU in clpC and ccpA strains. (B) Effects of glucose on DegU, ClpP, ClpC, and MecA. (C) Relationship between protein amounts and band intensities.
CcpA-dependent catabolite repression of clpC.
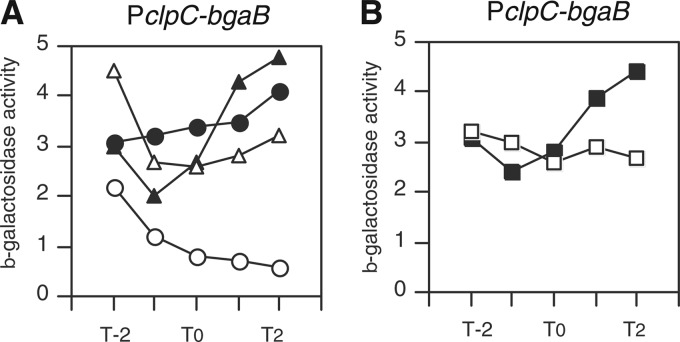
According to the Western blot analysis of ClpC, we speculated that CcpA may downregulate the expression of clpC driven by the promoter of the four-gene ctsR-mcsA-mcsB-clpC operon (45, 46). We analyzed a PclpC-bgaB fusion to explore this possibility. As shown in Fig. 3A, PclpC-bgaB was repressed by addition of glucose, and disruption of ccpA at least partially derepressed the expression of the fusion in the presence of glucose. The disruption of hprK also derepressed PclpC-bgaB from glucose repression (Fig. 3B). These results clearly demonstrated that the clpC operon is repressed by glucose in a CcpA-dependent manner. It was reported that CtsR represses expression of its own operon (45). Therefore, we examined the effects of glucose and disruption of ccpA in a ctsR disruptant. We observed essentially the same results. In the ctsR background, the expression of clpC-bgaB was enhanced to about 25 Miller units without glucose and decreased to about 5 Miller units with glucose (data not shown). The repression was not observed in the ccpA strain (data not shown). These results again demonstrated the CcpA-dependent repression of the operon and showed that this regulation was independent of CtsR.
Fig 3.
CcpA-dependent glucose repression of PclpC. Cells were grown in sporulation medium without (closed symbols) or with (open symbols) 2% (wt/vol) glucose. Cells were sampled hourly. β-Galactosidase activities are shown in Miller units. The x axis represents the growth time (h) relative to the end of vegetative growth (T0). (A) Circles, wild type; triangles, ccpA strain. (B) Squares, hprK strain.
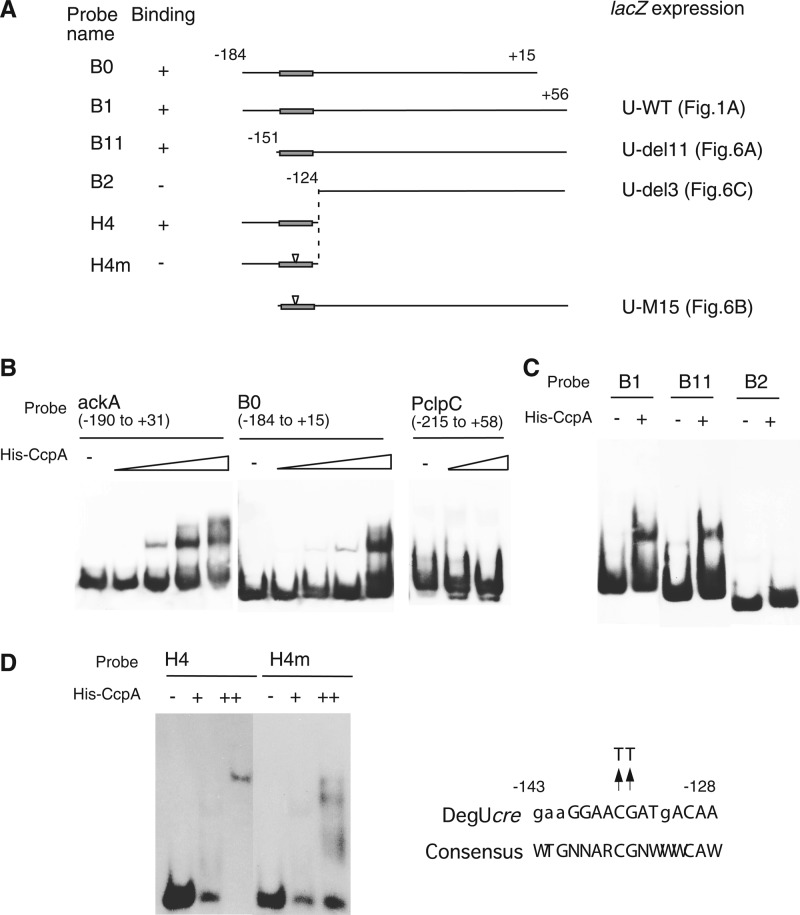
These observations raised the possibility that CcpA might bind to the regulatory region of PclpC. Thus, we used the same region of PclpC cloned into the PclpC-bgaB fusion (positions −215 to +58 relative to the transcription start site) as a probe for EMSA. Unfortunately, we did not observe any binding under the conditions used (Fig. 4B), suggesting an indirect regulation of PclpC by CcpA.
Fig 4.
Binding of His-CcpA to degU. Various probes were prepared by PCR amplification using the primers shown in Table S1 in the supplemental material. (A) Structures of probes. Solid rectangles on lines show cre sites. Numbers indicate nucleotide positions relative to the transcription start site. Relationships between probes and fusions with corresponding regions fused to lacZ are indicated. (B to D) Each reaction mixture contained poly(dI-dC) (1 μg [B], 0.7 μg [C], or 0 μg [D]) as a nonspecific competitor and probe. Samples were analyzed by nondenaturing polyacrylamide gel electrophoresis on 4.5% gels (B and C) or 11% gels (D). In each panel, all lanes shared the same gel electrophoresis conditions. For panel B, probes were incubated with increasing amounts of His-tagged CcpA (0, 12.5, 25, 50, and 100 nM for ackA and B0 and 0, 50, and 100 nM for PclpC). For panels C and D, probes were incubated with His-tagged CcpA (+, 75 nM; ++, 150 nM).
Direct binding of CcpA to the degU promoter region.
The results of the epistatic analyses using the degU-lacZ fusion (Fig. 1C) suggested that CcpA might be involved in glucose induction of degU through its binding to degU. To investigate this possibility, we examined the direct binding of CcpA to the degU promoter. In most cases, CcpA does not bind specifically to its target if it is not associated with P-Ser-Hpr (2, 47). In several experiments, however, His-tagged CcpA bound specifically to its target without P-Ser-Hpr, probably because of the positive charge of histidines and/or structural changes in the protein (36, 48–50). Thus, we used His-CcpA for the analysis. Using the ackA promoter and PclpC as positive and negative controls, respectively, we conducted an EMSA using His-CcpA and the degU promoter region (positions −184 to +15 relative to the transcription start site of the P3 promoter). As shown in Fig. 4B, CcpA bound to the degU probe in the presence of the nonspecific competitor poly(dI-dC). To narrow down the binding region, we used several 5′- and 3′-deleted probes (Fig. 4A). CcpA bound to the B1 and B11 probes but not to the B2 probe, indicating that the region from positions −151 to −124 was required for binding (Fig. 4C). This was further confirmed by the binding of CcpA to the H4 probe (positions −184 to −124). The region spanning positions −151 to −124 contained a cre-like sequence, although this sequence showed a low match to the consensus cre sequence (Fig. 4A). We introduced mutations into the core CG nucleotides in this cre sequence in the H4 probe, creating the H4m probe. In EMSAs, the H4m probe did not show a discrete band but instead showed a smeared pattern, suggesting dissociation of CcpA from the DNA-protein complex during electrophoresis. This observation demonstrated the low affinity of the H4m probe for CcpA compared with that of the H4 probe (Fig. 4D). This further confirmed the specific binding of CcpA to the cre sequence in degU. Next, we performed an EMSA using this sequence itself as a probe. CcpA bound efficiently to the cre sequence of ackA, while it bound rather poorly to the degU-derived probe (see Fig. S3 in the supplemental material). However, the binding to the cre sequence in degU was considered specific, because the probe spanning positions −172 to −152 in degU did not bind (see Fig. S3). In summary, CcpA bound efficiently to the cre sequence in degU when nonspecific DNA was present beside the cre sequence, and binding occurred irrespective of the sequence on the 5′ or 3′ side. This is based on the appearance of discrete bands in EMSAs with the H4 and B11 probes.
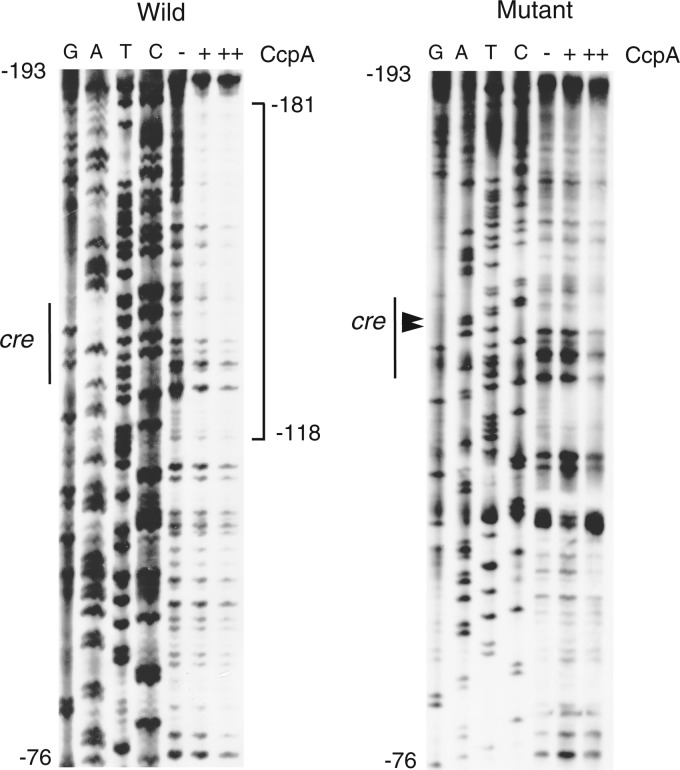
Footprint analysis of CcpA binding to cre sequence in degU.
To further analyze CcpA binding to degU, we performed a footprint experiment. As shown in Fig. 5, CcpA clearly protected the region corresponding to the H4 probe from DNase I attack. This result was consistent with the results of EMSA using the H4 probe. In previous binding studies (48–51), CcpA protected the relatively narrow regions containing the cre sequence, except for the cre sequence in the lev gene, for which a long footprint pattern was observed (about 65 bases). Thus, our results showed some differences from those of previous studies. We do not know the stoichiometry that underlies these results, that is, whether one dimer binds nonspecifically upstream of the cre sequence and the other binds specifically to the cre sequence or one dimer binds to the cre sequence while concomitantly interacting with the upstream region, protecting it from DNase I attack. We introduced the same mutations for H4m (replacement of the core CG nucleotides) into the probe and conducted another footprint experiment. We observed much more reduced binding, again demonstrating the importance of the cre sequence in degU in CcpA binding (Fig. 5).
Fig 5.
Footprint analysis of His-CcpA binding to degU. To analyze the wild-type template strand, oligonucleotide pairs DegU-Pro-Biotin3/DegU-pIS-B1 and DegU-pIS-H3/DegU-pIS-B1 were used for PCR amplification to generate the probe and sequencing ladder template, respectively. To analyze the mutant template strand, the oligonucleotide pair DegU-Pro-Biotin3/DegU-pIS-B1 in addition to mCre-degU-F/mCre-degU-R was used to create a mutant probe for PCR-mediated mutagenesis (35). DegU-pIS-H3 and DegU-pIS-B1 were used for PCR amplification of the resultant mutant probe to generate the sequencing ladder template. The probe spanning positions −184 to +15 relative to the transcription start site, which carries the additional nine bases derived from the PCR primer, was incubated with His-CcpA (+, 75 nM; ++, 150 nM) in buffer containing poly(dI-dC) (0.5 μg) as a nonspecific competitor (the buffer had the same composition as the buffer used for EMSA) and subjected to DNase I cleavage. A sequencing ladder is shown, with G, A, T, and C lanes. The square bracket shows the protected region. The two mutated nucleotides are indicated by arrowheads.
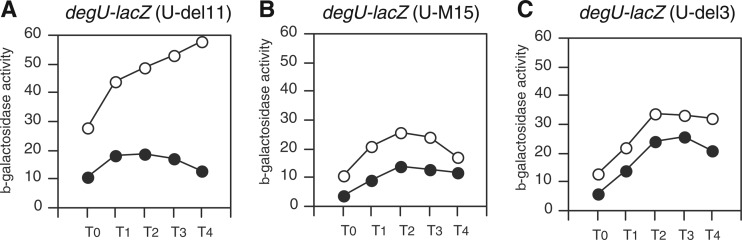
Mutational analysis of cre sequence in degU, using degU-lacZ.
To confirm the in vitro results in vivo, we used degU-lacZ fusions with deletions or mutations. As expected, the expression of B11, whose 5′ end was position −151, was induced in the presence of glucose (Fig. 6A). This was expected because the fusion contained the cre sequence. However, the expression of the other two fusions, B11m and B2, was not fully induced in the presence of glucose (Fig. 6B and C), because B11m carried a mutated cre sequence and B2 had no cre sequence. These results were consistent with those of the in vitro binding study and demonstrated the in vivo functionality of the cre sequence in glucose induction of degU. The partial induction of the B11m and B2 fusions in the presence of glucose can be attributed to the lack of ClpCP degradation machinery resulting from glucose addition (Fig. 2B). When the ccpA gene was disrupted, ClpCP no longer affected the expression of degU-lacZ (see Fig. S1B in the supplemental material). Thus, these two observations seem to contradict each other. Perhaps the ccpA disruption and deletion/mutation of cre in degU result in different regulatory effects.
Fig 6.
Effects of deletion/mutation of cre on expression of degU-lacZ. Cells were grown in sporulation medium without (closed symbols) or with (open symbols) 2% (wt/vol) glucose. Cells were sampled hourly. β-Galactosidase activities are shown in Miller units. The x axis represents the growth time (h) relative to the end of vegetative growth (T0).
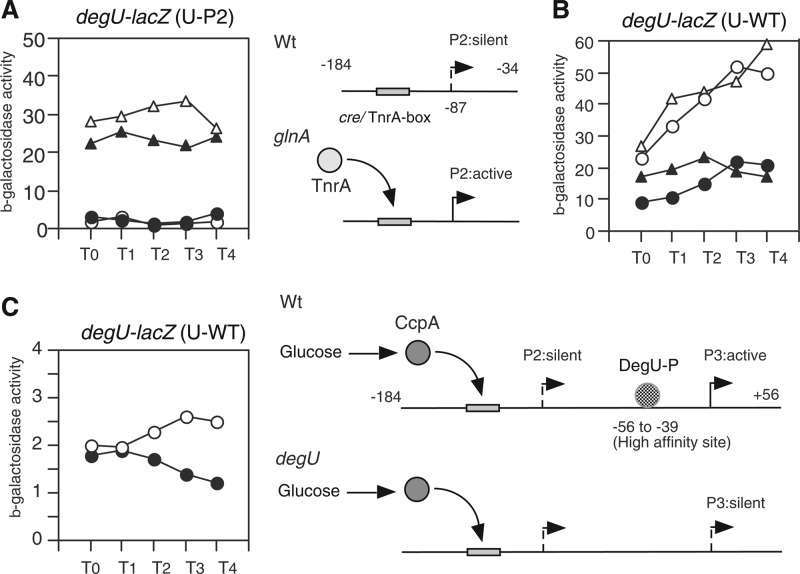
The P2 promoter is silent and is not induced by glucose.
As reported previously (27), the degU gene is preceded by the transcription factor TnrA-dependent promoter P2, which is activated by the disruption of glnA (27). To examine the involvement of the P2 promoter in glucose induction of degU, we constructed a fusion bearing the P2 promoter alone (U-P2) (Fig. 7A). U-P2 showed no β-galactosidase activity in the presence of 2% glucose, indicating that glucose did not activate the P2 promoter (Fig. 7A). As a control, we introduced the glnA disruption into the strain bearing this fusion, and as expected, the resultant strain showed β-galactosidase activity. This expression was dependent on TnrA, as the further introduction of a tnrA disruption into the strain abolished the P2 expression (data not shown). This is consistent with former observations (27). The addition of glucose slightly enhanced the expression of TnrA-dependent expression of P2 (Fig. 7A). This enhancement is thought to also be CcpA dependent, since this slight enhancement of the expression of P2 was not observed in the glnA and ccpA double mutant (data not shown). It should be noted that the putative TnrA box (TGGAAGGAACGATGACA; positions −145 to −129 relative to the transcription start site of P3) and the cre site overlap at the same region. It is not known at present, however, whether CcpA and TnrA might bind to the same sequence concomitantly. Moreover, we examined the expression of the degU fusion bearing the P2 and P3 promoters in a tnrA disruptant. As shown in Fig. 7B, the disruption of tnrA did not affect the glucose induction ratio or the expression levels of the fusion. This result further demonstrated that P2 is not related to the glucose induction of degU. The P3 promoter is activated by DegU-P, and thus the fusion bearing only P3 and the DegU-binding site (U-del5) showed no β-galactosidase activity in the degU disruptant (26). This indicated that P3 was solely dependent on DegU-P. In addition, we note that P2 is not influenced by degU disruption (27). To confirm that P2 is silent under wild-type conditions, we examined the β-galactosidase activity of the U-WT fusion in the degU background. Only marginal activities were observed without glucose, and a minor enhancement of activities was seen in the presence of glucose (Fig. 7C). This finally demonstrated that P2 is silent and that P3 but not P2 contributes to glucose induction. The slight enhancement of expression of the fusion is due to CcpA's action on the residual P3 promoter activity.
Fig 7.
Effects of glucose addition on P2 promoter. Cells were grown in sporulation medium without (closed symbols) or with (open symbols) 2% (wt/vol) glucose. Each fusion is indicated above the relevant panel. (A) Circles, wild-type cells; triangles, glnA cells. Schematic representations of the structure of U-P2 and the regulation of P2 are shown alongside the panel. Numbers indicate nucleotide positions relative to the transcription start site for P3. (B) Circles, wild-type cells; triangles, tnrA cells. (C) Circles, degU cells. Schematic representations of the regulation of the U-WT fusion bearing the P2 and P3 promoters are shown alongside the panel. Numbers indicate nucleotide positions relative to the transcription start site for P3.
DISCUSSION
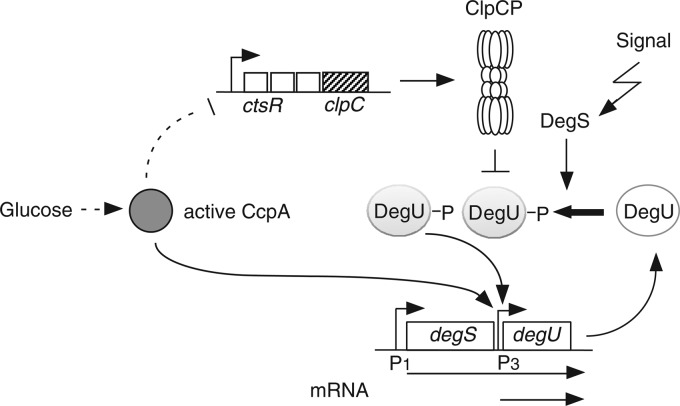
In this report, we present the direct and indirect regulatory mechanisms of glucose induction of degU (Fig. 8). Evidence is presented that CcpA activates degU transcription by binding to its upstream region. Moreover, catabolite repression of clpC reduces the amount of ClpC, leading to activation of the autoregulatory loop of degU. The effects of ccpA mutation and/or addition of glucose to various media on bacterial growth have been analyzed in several genomewide global analyses (5–8, 10, 11). None of the previous studies reported induction of the expression of DegU-driven genes, including the degU gene itself, by glucose and CcpA binding to degU. There are several possible reasons for this. The previous studies did not use the same conditions as those in our study, that is, growth in sporulation medium supplemented with glucose and serial sampling from log phase to early stationary phase. Thus, induction of the expression of DegU-driven genes is dependent on the medium conditions and sampling points. For example, in LB medium, the expression of degU-lacZ was slightly repressed by the addition of glucose, while in competence medium the induction by glucose was clearly observed in early stationary phase (see Fig. S1A in the supplemental material). In the ChIP analysis, CcpA did not bind to the control region of degU or to those of the genes for which CcpA-dependent direct regulation has been verified (2, 6). This might be due to the fact that the ChIP analysis provided a snapshot of the CcpA-binding pattern of cells in the exponential growth phase in M9 medium containing glucose and malate (6).
Fig 8.
Schematic representation of glucose induction mechanism of degU. Arrows and the T bar indicate activation and inhibition, respectively. Bent arrows indicate promoters. The dotted line indicates an indirect effect.
Many biosynthetic enzymes are targets for Clp-dependent degradation in the glucose-starved stationary phase (9). Thus, it is possible that ClpCP protease activity is enhanced after glucose starvation. Although this hypothesis remains to be verified, our finding that the transcription of the clpC gene is repressed by glucose is consistent with the pattern of regulation under glucose starvation conditions. Moreover, a global transcriptome analysis reported that transcription of the clpC gene was enhanced in a CcpA-dependent manner in the absence of glucose (39), which is consistent with our findings. In the current study, our results showed that repression of clpC is indirectly regulated by CcpA, because CcpA did not bind to the PclpC probe. Since partial derepression of clpC in the ccpA strain was observed, we investigated the effects of other trans-acting factors, such as CggR, CcpB, CcpC, and CcpN, which might be responsible for glucose-dependent regulation of PclpC expression (2, 6). Disruption of these genes did not affect reduced expression from PclpC in the presence of glucose (M. Ogura and H. Ishii, unpublished results). These results indicated that there might be an as yet unknown direct trans-acting factor for regulation of clpC. Otherwise, PclpC might be regulated indirectly, for example, by a change in metabolic state.
There are a few genes, such as ackA, pta, and the ilv-leu operon, that are directly activated by CcpA (36, 47, 52, 53). Their gene products are involved in consumption of acetyl-coenzyme A (acetyl-CoA) produced by accumulated pyruvate (2). Genes encoding polyketide synthase for bacillaene synthesis have been detected among the DegU-activated genes (14, 18, 54). This might be one reason for glucose activation of degU, because polyketide synthesis consumes acetyl-CoA (54). The cre sequences in these genes are located upstream of the core promoter, which enables these cre sequences to function as positive cis sequences. Thus, it is reasonable that the cre sequence in degU is located at positions −143 to −128 relative to the transcription start site, assuming DNA looping mediated by some DNA-binding proteins, e.g., DegU-P and/or CcpA, which would enable an interaction of CcpA with RNA polymerase over a long distance. In our previous study using various 5′-deleted degU-lacZ fusions, we observed a 3-fold decrease in degU-lacZ expression after deletion of the region from positions −184 to −124 when cells were grown in a glucose-based competence medium (26). Thus, we speculated that an as yet unknown transcription factor is involved in degU regulation. This unknown factor could be CcpA.
Experimental analyses of the relationship between cre sequences and CcpA-dependent regulation of corresponding genes have shown the intrinsic degenerative nature of the cre sequence (2) (Table 2). The cre sequences in Table 2 are examples of those with this degenerative characteristic, and all have had their CcpA-binding properties validated experimentally. Although the cre sequence in degU showed a low-level match to the consensus cre sequence, our results show that it can function as a CcpA-binding site and a positive cis sequence if some “scaffold” sequence is available. The “scaffold” might strengthen the searching efficiency of the binding site by CcpA through nonspecific binding of CcpA. A recent idea on how a transcription factor achieves specific binding includes its sliding on DNA adjacent to a specific binding site through nonspecific binding (56). This observation might increase the number of possible CcpA-binding sites in the B. subtilis genome, which has been estimated to be approximately 150 sites (2). Thus, these findings are important for further research on the function and binding modes of CcpA. Moreno et al. pointed out that in their transcriptome analyses, recognizable cre sites were virtually absent from the operons that are activated by glucose in a CcpA-dependent manner, although a large majority of the genes subject to catabolite repression were found in operons bearing one or more cre sites (10). Our results may partly explain this observation.
Table 2.
Comparison of cre sequences with low-level matching
| Gene name or consensus | cre sequencea | No. of matching nt | In vivo bindingb | Reference |
|---|---|---|---|---|
| Consensus | WTGNNARCGNWWWCAW | 4 | ||
| cydA | TTGAAAtgaATccttg | 8 | + | 50 |
| resA | gTaAAAACGCTTTCtA | 13 | − | 55 |
| citZ | ATGTAAGCaTTTTCtT | 14 | + | 48 |
| degU | gaaGGAACGATgACAA | 12 | − | This study |
Underlining shows the most conservative nucleotides. Lowercase letters show mismatched nucleotides.
Data from ChIp-chip experiments in reference 6.
We used His-CcpA instead of intact CcpA with P-Ser-Hpr. The two proteins have similar DNA affinities, with Kd (dissociation constant) values of approximately 5 to 10 nM for the cre sequence in the ackA gene (4, 36). Both types of CcpA protein at similar concentrations were also reported to achieve protection from DNase I digestion in the ilvB promoter (52, 53). In addition, there are several reports of specific DNA binding of His-CcpA to target sequences (36, 48–50), indicating the full functionality of in vitro DNA binding of His-CcpA. In fact, the His tag in His-CcpA increases DNA binding about 6-fold over that of CcpA in the absence of P-Ser-Hpr, while there is no stimulatory effect of P-Ser-Hpr addition (57). Thus, our result that His-CcpA bound to the cre sequence in degU is reasonable, because His-CcpA seems to have authentic sequence specificity.
CcpA indirectly regulates many genes through its effect on regulators. CcpA directly regulates three response regulator genes, citT, phoP, and resD, in two-component regulatory systems (49, 55, 58). These three regulators are subject to catabolite repression, while the degU gene is activated by glucose. The rational explanation for the difference is an issue to be elucidated in the future.
The CcpA-binding site is located upstream of the low-affinity DegU-P-binding site (positions −104 to −87), which raises the possibility that CcpA may interact with DegU-P. Recent reports discussed possible interactions between CcpA and other transcription factors, for example, CodY on the ilv-leu operon and ackA promoters (52, 53, 59). Further research is required to clarify the possible interaction between CcpA and DegU-P.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. Kawamura (clpC-bgaB fusion), S. H. Fisher (168 glnA14), U. Gerth (anti-ClpC antibody), K. Turgay (anti-ClpP antibody), and T. M. Henkin and A. L. Sonenshein (His-CcpA strain) for kindly supplying the bacterial strains, plasmids, and antibodies used in this study. We thank S. Ishikawa for fruitful discussions about CcpA and for a critical reading of the paper.
This work was supported by JSPS KAKENHI grant 24580123.
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01881-12.
REFERENCES
- 1. Schaeffer P, Millet J, Aubert J. 1965. Catabolite repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54: 704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fujita Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73: 245–259 [DOI] [PubMed] [Google Scholar]
- 3. Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5: 917–927 [DOI] [PubMed] [Google Scholar]
- 4. Schumacher MA, Sprehe M, Bartholomae M, Hillen W, Brennan RG. 2011. Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators. Nucleic Acids Res. 39: 2931–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blencke HM, Homuth G, Ludwig H, Mäder U, Hecker M, Stülke J. 2003. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5: 133–149 [DOI] [PubMed] [Google Scholar]
- 6. Buescher JM, Liebermeister W, Jules M, Uhr M, Muntel J, Botella E, Hessling B, Kleijn RJ, Le Chat L, Lecointe F, Mäder U, Nicolas P, Piersma S, Rügheimer F, Becher D, Bessieres P, Bidnenko E, Denham EL, Dervyn E, Devine KM, Doherty G, Drulhe S, Felicori L, Fogg MJ, Goelzer A, Hansen A, Harwood CR, Hecker M, Hubner S, Hultschig C, Jarmer H, Klipp E, Leduc A, Lewis P, Molina F, Noirot P, Peres S, Pigeonneau N, Pohl S, Rasmussen S, Rinn B, Schaffer M, Schnidder J, Schwikowski B, Van Dijl JM, Veiga P, Walsh S, Wilkinson AJ, Stelling J, Aymerich S, Sauer U. 2012. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 335: 1099–1103 [DOI] [PubMed] [Google Scholar]
- 7. Lorca GL, Chung YJ, Barabote RD, Weyler W, Schilling CH, Saier MH., Jr 2005. Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, HPr, and HprK. J. Bacteriol. 187: 7826–7839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lulko AT, Buist G, Kok J, Kuipers OP. 2007. Transcriptome analysis of temporal regulation of carbon metabolism by CcpA in Bacillus subtilis reveals additional target genes. J. Mol. Microbiol. Biotechnol. 12: 82–95 [DOI] [PubMed] [Google Scholar]
- 9. Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 28: 1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moreno MS, Schneider BL, Maile RR, Weyler W, Saier MH., Jr 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39: 1366–1381 [DOI] [PubMed] [Google Scholar]
- 11. Yoshida K, Kobayashi K, Miwa Y, Kang CM, Matsunaga M, Yamaguchi H, Tojo S, Yamamoto M, Nishi R, Ogasawara N, Nakayama T, Fujita Y. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimane K, Ogura M. 2004. Mutational analysis of the helix-turn-helix region of Bacillus subtilis response regulator DegU, and identification of cis-acting sequences for DegU in the aprE and comK promoters. J. Biochem. 136: 387–397 [DOI] [PubMed] [Google Scholar]
- 13. Kunst F, Msadek T, Rapoport G. 1994. Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis, p 1–20 In Piggot PJ, Moran CP, Jr, Youngman P. (ed), Regulation of bacterial differentiation. American Society for Microbiology, Washington, DC [Google Scholar]
- 14. Kobayashi K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66: 395–409 [DOI] [PubMed] [Google Scholar]
- 15. Msadek T, Kunst F, Henner D, Klier A, Rapoport G, Dedonder R. 1990. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J. Bacteriol. 172: 824–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ogura M, Tsukahara K. 2012. SwrA regulates assembly of Bacillus subtilis DegU via its interaction with N-terminal domain of DegU. J. Biochem. 151: 643–655 [DOI] [PubMed] [Google Scholar]
- 17. Mader U, Antelmann H, Buder T, Dahl MH, Hecker M, Homuth G. 2002. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet. Genomics 268: 455–467 [DOI] [PubMed] [Google Scholar]
- 18. Ogura M, Yamaguchi H, Yoshida K, Fujita Y, Tanaka T. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29: 3804–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamoen LW, van Werkhoven AF, Venema G, Dubnau D. 2000. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 97: 9246–9251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsukahara K, Ogura M. 2008. Promoter selectivity of the Bacillus subtilis response regulator DegU, a positive regulator of the fla/che operon and sacB. BMC Microbiol. 8: 8 doi:10.1186/1471-2180-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ohsawa T, Tsukahara K, Ogura M. 2009. Bacillus subtilis response regulator DegU is a direct activator of pgsB transcription involved in γ-poly-glutamic acid synthesis. Biosci. Biotechnol. Biochem. 73: 2096–2102 [DOI] [PubMed] [Google Scholar]
- 22. Stanley NR, Lazazzera BA. 2005. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-gamma-dl-glutamic acid production and biofilm formation. Mol. Microbiol. 57: 1143–1158 [DOI] [PubMed] [Google Scholar]
- 23. Tsukahara K, Ogura M. 2008. Characterization of DegU-dependent expression of bpr in Bacillus subtilis. FEMS Microbiol. Lett. 290: 8–13 [DOI] [PubMed] [Google Scholar]
- 24. Veening JW, Igoshin OA, Eijlander RT, Nijland R, Hamoen LW, Kuipers OP. 2008. Transient heterogeneity in extracellular protease production by Bacillus subtilis. Mol. Syst. Biol. 4: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verhamme DT, Kiley TB, Stanley-Wall NR. 2007. DegU co-ordinates multicellular behavior exhibited by Bacillus subtilis. Mol. Microbiol. 65: 554–568 [DOI] [PubMed] [Google Scholar]
- 26. Ogura M, Tsukahara K. 2010. Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP. Mol. Microbiol. 75: 1244–1259 [DOI] [PubMed] [Google Scholar]
- 27. Yasumura A, Abe S, Tanaka T. 2008. Involvement of nitrogen regulation in Bacillus subtilis degU expression. J. Bacteriol. 190: 5162–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amati G, Bisicchia P, Galizzi A. 2004. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J. Bacteriol. 186: 6003–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wray LV, Jr, Ferson AE, Rohrer K, Fisher SH. 1996. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 93: 8841–8845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frees D, Savijoki K, Varmanen P, Ingmer H. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63: 1285–1295 [DOI] [PubMed] [Google Scholar]
- 31. Ogura M, Shimane KK, Asai K, Ogasawara N, Tanaka T. 2003. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol. Microbiol. 49: 1685–1697 [DOI] [PubMed] [Google Scholar]
- 32. Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19: 3083–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayashi K, Kensuke T, Kobayashi K, Ogasawara N, Ogura M. 2006. Bacillus subtilis RghR (YvaN) represses rapG and rapH, which encode inhibitors of expression of the srfA operon. Mol. Microbiol. 59: 1714–1729 [DOI] [PubMed] [Google Scholar]
- 34. Guerout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180: 57–61 [DOI] [PubMed] [Google Scholar]
- 35. Ogura M, Ohshiro Y, Hirao S, Tanaka T. 1997. A new Bacillus subtilis gene, med, encodes a positive regulator of comK. J. Bacteriol. 179: 6244–6253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moir-Blais TR, Grundy FJ, Henkin TM. 2001. Transcriptional activation of the Bacillus subtilis ackA promoter requires sequences upstream of the CcpA binding site. J. Bacteriol. 83: 2389–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mukai K, Kawata-Mukai M, Tanaka T. 1992. Stabilization of phosphorylated Bacillus subtilis DegU by DegR. J. Bacteriol. 174: 7954–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hata M, Ogura M, Tanaka T. 2001. Involvement of stringent factor RelA in expression of the alkaline protease gene aprE in Bacillus subtilis. J. Bacteriol. 183: 4648–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gerth U, Kock H, Kusters I, Michalik S, Switzer RL, Hecker M. 2008. Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J. Bacteriol. 190: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kirstein J, Schlothauer T, Dougan DA, Lilie H, Tischendorf G, Mogk A, Bukau B, Turgay K. 2006. Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 25: 1481–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogura M, Matsuzawa A, Yoshikawa H, Tanaka T. 2004. Bacillus subtilis SalA (YbaL) negatively regulates expression of scoC, which encodes the repressor for the alkaline exoprotease gene, aprE. J. Bacteriol. 186: 3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kleijn RJ, Buescher JM, Le Chat L, Jules M, Aymerich S, Sauer U. 2010. Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. J. Biol. Chem. 285: 1587–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Turgay K, Hahn J, Burghoorn J, Dubnau D. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17: 6730–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gerth U, Kirstein J, Mostertz J, Waldminghaus T, Miethke M, Kock H, Hecker M. 2004. Fine-tuning in regulation of Clp protein content in Bacillus subtilis. J. Bacteriol. 186: 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krüger E, Hecker M. 1998. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J. Bacteriol. 180: 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nanamiya H, Ohashi Y, Asai K, Moriya S, Ogasawara N, Fujita M, Sadaie Y, Kawamura F. 1998. ClpC regulates the fate of a sporulation initiation sigma factor, sigmaH protein, in Bacillus subtilis at elevated temperatures. Mol. Microbiol. 29: 505–513 [DOI] [PubMed] [Google Scholar]
- 47. Presecan-Siedel E, Galinier A, Longin R, Deutscher J, Danchin A, Glaser P, Martin-Verstraete I. 1999. Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J. Bacteriol. 181: 6889–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim HJ, Roux A, Sonenshein AL. 2002. Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes. Mol. Microbiol. 45: 179–190 [DOI] [PubMed] [Google Scholar]
- 49. Puri-Taneja A, Paul S, Chen Y, Hulett FM. 2006. CcpA causes repression of the phoPR promoter through a novel transcription start site, P(A6). J. Bacteriol. 188: 1266–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Puri-Taneja A, Schau M, Chen Y, Hulett FM. 2007. Regulators of the Bacillus subtilis cydABCD operon: identification of a negative regulator, CcpA, and a positive regulator, ResD. J. Bacteriol. 189: 3348–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martin-Verstraete I, Deutscher J, Galinier A. 1999. Phosphorylation of HPr and Crh by HprK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J. Bacteriol. 118: 2966–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shivers RP, Sonenshein AL. 2005. Bacillus subtilis ilvB operon: an intersection of global regulons. Mol. Microbiol. 56: 1549–1559 [DOI] [PubMed] [Google Scholar]
- 53. Tojo S, Satomura T, Morisaki K, Deutscher J, Hirooka K, Fujita Y. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 56: 1560–1573 [DOI] [PubMed] [Google Scholar]
- 54. Butcher RA, Schroeder FC, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. 2007. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 104: 1506–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Choi SK, Saier MH., Jr 2006. Mechanism of CcpA-mediated glucose repression of the resABCDE operon of Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 11: 104–110 [DOI] [PubMed] [Google Scholar]
- 56. Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. 2012. The lac repressor displays facilitated diffusion in living cells. Science 336: 1595–1598 [DOI] [PubMed] [Google Scholar]
- 57. Aung-Hilbrich LM, Seidel G, Wagner A, Hillen W. 2002. Quantification of the influence of HPrSer46P on CcpA-cre interaction. J. Mol. Biol. 319: 77–85 [DOI] [PubMed] [Google Scholar]
- 58. Yamamoto H, Murata M, Sekiguchi J. 2000. The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol. Microbiol. 37: 898–912 [DOI] [PubMed] [Google Scholar]
- 59. Wünsche A, Hammer E, Bartholomae M, Völker U, Burkovski A, Seidel G, Hillen W. 2012. CcpA forms complexes with CodY and RpoA in Bacillus subtilis. FEBS J. 279: 2201–2214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.