Abstract
DNA methylation is a well-characterized epigenetic modification involved in gene regulation and transposon silencing in mammals. It mainly occurs on cytosines at CpG sites but methylation at non-CpG sites is frequently observed in embryonic stem cells, induced pluriotent stem cells, oocytes and the brain. The biological significance of non-CpG methylation is unknown. Here, we show that non-CpG methylation is also present in male germ cells, within and around B1 retrotransposon sequences interspersed in the mouse genome. It accumulates in mitotically arrested fetal prospermatogonia and reaches the highest level by birth in a Dnmt3l-dependent manner. The preferential site of non-CpG methylation is CpA, especially CpApG and CpApC. Although CpApG (and CpTpG) sites contain cytosines at symmetrical positions, hairpin-bisulfite sequencing reveals that they are hemimethylated, suggesting the absence of a template-dependent copying mechanism. Indeed, the level of non-CpG methylation decreases after the resumption of mitosis in the neonatal period, whereas that of CpG methylation does not. The cells eventually lose non-CpG methylation by the time they become spermatogonia. Our results show that non-CpG methylation accumulates in non-replicating, arrested cells but is not maintained in mitotically dividing cells during male germ-cell development.
INTRODUCTION
Cytosine methylation at CpG sites in the genomic DNA is a well-characterized epigenetic modification involved in gene regulation and transposon silencing in mammals (1,2). Upon replication of methylated DNA, cytosines in the newly synthesized strand are initially unmethylated (resulting in a hemimethylated duplex), then become methylated by a maintenance-type DNA methylase, Dnmt1. In addition, the de novo DNA methylases, Dnmt3a and Dnmt3b, also have a role in the maintenance methylation for some genomic regions (3–5). Due to this template-dependent copying mechanism, the pattern of CpG methylation is essentially heritable in somatic cells through cell division.
In contrast, the level of DNA methylation changes dynamically during germ-cell development. First, primordial germ cells (PGCs) undergo genome-wide erasure of methylation and then prospermatogonia, a direct derivative of PGCs in the male gonad, undergo extensive de novo methylation (6,7) (Figure 1A). The prospermatogonia are mitotically arrested, premeiotic cells expressing high levels of the de novo DNA methylase Dnmt3a and its related protein Dnmt3l (8,9). They resume mitotic division for cell expansion and spermatogonial differentiation after birth. Therefore, de novo DNA methylation and its maintenance through cell division can be separately observed in these germ cells. Oocytes are also non-dividing cells that undergo extensive de novo methylation (6,7), but the methylation marks in most genomic regions are not maintained through cell divisions after fertilization (10).
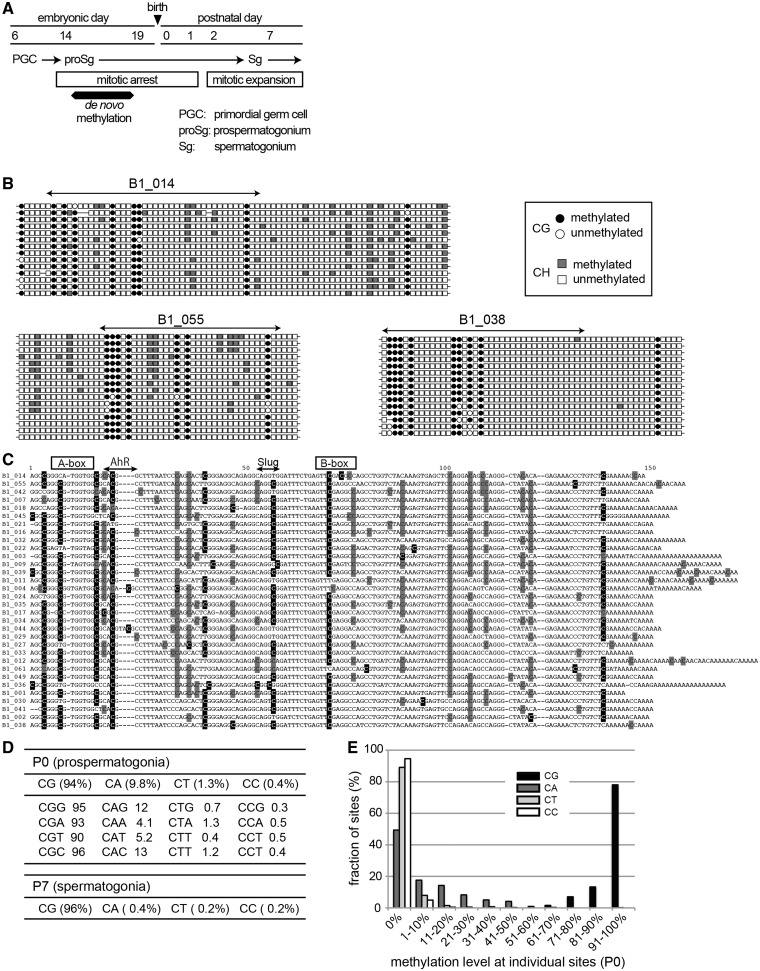
Figure 1.
Non-CpG methylation at B1 retrotransposon loci in neonatal prospermatogonia. (A) Chronology of male germ-cell development from PGCs to spermatogonia. Mitotically arrested prospermatogonia undergo de novo methylation of CpG sites in the fetal testis, and mitosis resumes after birth. (B). Non-CpG methylation at three representative B1 loci detected by bisulfite sequencing in P0 prospermatogonia (see Supplementary Figure S1 for other loci). Circles and squares represent cytosines in CpG and non-CpG (CpH where H is A, T or C) sites, respectively. The methylated cytosines are shown in black for CpG and in gray for CpH. (C) Alignment of the sequences of the 33 B1 loci to visualize the common sites of methylation. Cytosines methylated over 65% at CpG sites are highlighted in black, and those methylated over 5% at non-CpG sites are highlighted in gray. Positions of A- and B-box in the Pol III promoter and the binding sites for the AhR and Slug transcription factors are indicated above. (D) Methylation levels in different sequence contexts. The data for dinucleotides and trinucleotides are shown. (E) Individual CpG and non-CpG sites are categorized according to their methylation levels, and counted.
Short interspersed elements (SINEs) are a class of retrotransposons and are transcribed by RNA plolymerase III (Pol III). Biochemical studies have shown that CpG methylation of the Pol III promoter inhibits transcription of tRNAs and human Alu SINE (11–13). We previously showed that the bulk of CpG sites in B1 retrotransposons, a family of SINEs scattered over the mouse genome, become highly methylated in prospermatogonia and stay methylated throughout the later stages of spermatogenesis (14). We also showed that a mutation in Dnmt3a or Dnmt3l reduces the methylation levels in a locus-specific manner and that the level of B1 methylation is well correlated with the level of B1 expression (14). During the course of that study, we noticed that methylation at non-CpG sites is abundantly present in prospermatogonia. This is interesting because non-CpG methylation has been so far found in oocytes, embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs) and the brain (3,15–21). Although non-CpG methylation dramatically decreases upon differentiation of ESCs and thus could have some relevance to pluripotency (3,19), the biological function of non-CpG methylation remains unknown. In this report, we characterize the non-CpG methylation that we found at B1 retrotransposons in prospermatogonia in detail. We show accumulation of the non-CpG methylation in the non-dividing prospermatogonia and its gradual loss after the resumption of cell division. We also provide evidence suggesting the absence of a template-dependent copying mechanism for non-CpG methylation.
MATERIALS AND METHODS
Cell preparation and bisulfite-PCR sequencing
EpCAM-positive germ cells were collected from testes of C57BL/6 J mice at P0, P2, P5 and P7 as described previously (14). To collect fetal germ cells at E16.5, green fluorescent protein (GFP)-positive cells were sorted from the testes of Oct4-GFP transgenic embryos. Genomic DNA preparation, bisulfite conversion and PCR were done as described previously (14). Typically, 100–200 ng of genomic DNA was treated with bisulfite, and an aliquot equivalent to 5–10 ng of genomic DNA was used for PCR with ExTaq HS (TAKARA BIO, Ohtsu, Japan). To monitor the efficiency of C-to-T conversion, λ phage DNA that had no methylated cytosine (Promega, Madison, WI) was included in the bisulfite reaction. All sequenced clones were used for the analysis. To validate the purity of germ-cell preparations, methylation of the imprinted Lit1 differentially methylated region (DMR) was also analysed using the same bisulfite-treated DNA. Among at least 40 sequenced clones, none showed methylation in the Lit1 DMR, suggesting a germ-cell purity of >95%, given that only the maternal Lit1 DMR allele is methylated in somatic cells and no allele is methylated in male germ cells. Bisulfite PCR primers for the 33 B1 loci were previously described (14). The sequences of the primers for the DMRs, CpG islands and control λ phage DNA are shown in Supplementary Table S1.
Hairpin-bisulfite PCR analysis
Genomic DNA (100 ng) was digested with TaqαI (New England Biolab, Ipswich, MA) and ligated with a synthetic oligonucleotide (5'-CGTGAACTGAGGTCGGAAGACCTCAGTTCA-3') that forms a hairpin structure. The ligation product was denatured at 98°C for 10 min in 0.3 M NaOH, treated with 9.17 M bisulfite (22) and used for PCR as described (14). The hairpin-bisulfite PCR primers for the three B1 loci are listed in Supplementary Table S1.
RESULTS AND DISCUSSION
We collected prospermatogonia at postnatal day 0 (P0) and studied DNA methylation states of 33 B1 loci by bisulfite sequencing. The bisulfite sequencing method is unable to distinguish between 5-methylcytosine (5mC) and 5-hydroxylmethylcytosine (5hmC), but hereinafter we regard bisulfite-resistant cytosines as 5mC, given that 5hmC is present much less than 5mC in cells/tissues examined so far, and almost exclusively at CpG sites in ESCs (23). As shown in Figure 1B, C and Supplementary Figure S1, significant methylation was found at non-CpG sites, in addition to CpG sites, both within and around the B1 sequences. This was not due to imcomplete bisulfite conversion, because the failure rate was only 0.2–0.5%, as determined using the control unmethylated DNA (λ phage DNA) mixed to the samples. On average, the methylation level at non-CpG sites (total of ∼2700) in P0 prospermatogonia was 4.8%, which was higher than the levels in ESCs (2.6%), iPSCs (3.2%), oocytes (3.4–3.8%) and the brain (1.5–2.2%) (16,20,21). Of all methylated cytosines, 77% were at CpG sites and 23% were at non-CpG sites. When the methylation level was determined for each dinucleotide sequence, it was 94%, 9.8%, 1.3% and 0.4% at CpG, CpA, CpT and CpC sites, respectively (Figure 1D). About half of CpA sites showed methylation levels of higher than 5%, whereas the vast majority of CpT and CpC sites showed no methylation (Figure 1E). Thus, CpA is the most preferred non-CpG sites. Among the CpA sites, the CpApC (15%) and CpApG sites (12%) were more frequently methylated than the CpApA and CpApT sites (Figure 1D). Interestingly, despite that the 33 B1 loci that we analysed were from various genomic locations, some non-CpG sites at equivalent positions in the B1 sequences were more preferentially methylated (Figure 1C). A subfamily of B1 sequences, designated as B1-X35S, has been shown to bind the AhR and Slug transcription factors, and the binding was involved in the formation of a chromatin boundary (24,25). However, the preferentially methylated non-CpG sites did not overlap with these binding sites or the RNA Polymerase III promoter (A-box and B-box) (Figure 1C).
To address whether the non-CpG methylation is specific to B1 sequences or not, we examined the methylation status of three paternally methylated DMRs (the H19, Dlk1/Meg3 and Rasgrf1 DMRs) in P0 prospermatogonia. Non-CpG methylation was detected in the DMRs with the levels comparable to those in B1 loci (Supplementary Figure S2A). We also examined seven CpG islands that are methylated in spermatozoa (26). Whereas three of them were not methylated even at CpG sites (data not shown), the other four had methylation at non-CpG sites in P0 prospermatogonia (Supplementary Figure S2B). These results suggest that non-CpG methylation is widespread in the prospermatogonia.
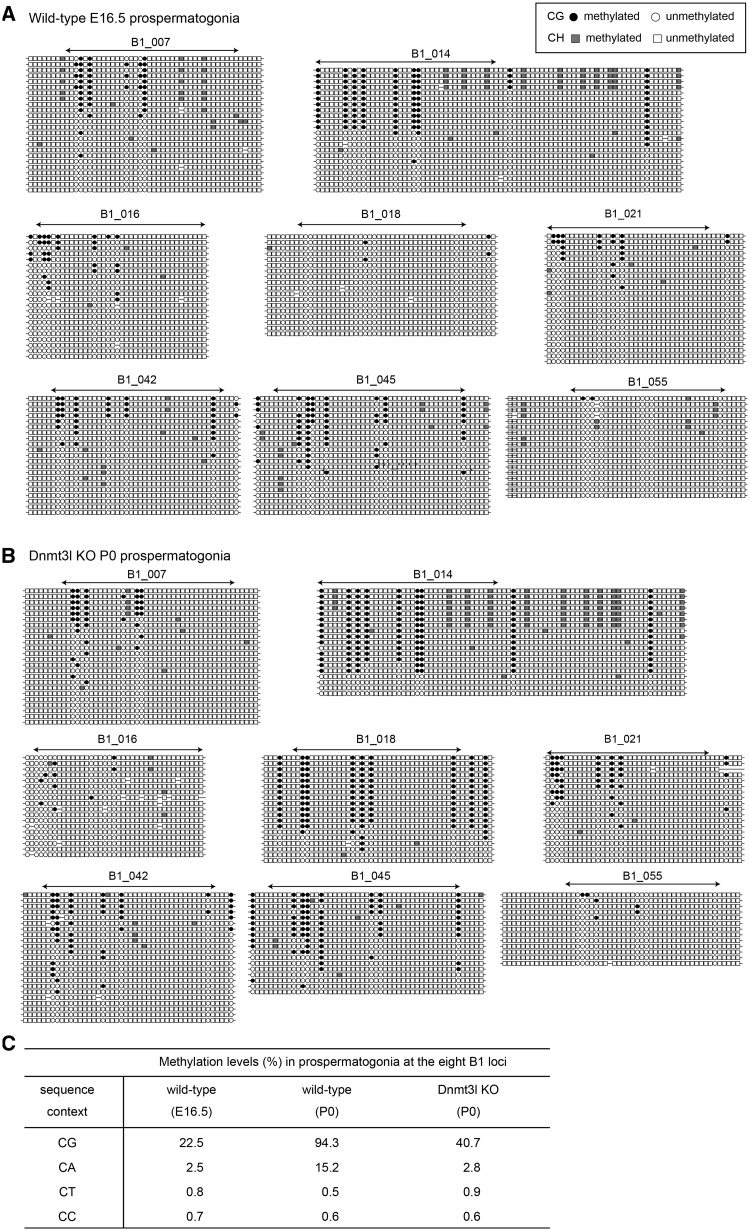
It is known that de novo CpG methylation of the DMRs and retrotransposons occurs in prospermatogonia between embryonic day 14.5 (E14.5) and P0 (27). The presence of non-CpG methylation in P0 prospermatogonia suggested their co-occurence with CpG methylation in fetal prospermatogonia. We selected eight B1 loci that showed high levels of non-CpG methylation in P0 prospermtogonia (B1_007, B1_014, B1_016, B1_018, B1_021, B1_042, B1_045 and B1_055) (Supplementary Figure S1) and examined their methylation status in prospermatogonia at embryonic day 16.5 (E16.5) (Figure 2A and C). The average CpA methylation level at E16.5 was six times less than that at P0, indicating that these non-CpG sites become de novo methylated in prospermatogonia during late gestation. In the same period, the methylation level at CpG sites increased four times. We previously showed that mutation in Dnmt3a or Dnmt3l affects CpG methylation at B1 loci (14). We therefore analysed P0 prospermatogonia from Dnmt3l knockout mice (28) for the eight loci (Figure 2B). The CpA methylation level was significantly reduced (>5-fold, P < 10−13) by the Dnmt3l mutation (Figure 2B and C), suggesting that the non-CpG methylation in prospermatogonia is dependent on Dnmt3l.
Figure 2.
De novo methylation of non-CpG sites occurs concomitantly with de novo CpG methylation. (A) Site-by-site representation of the bisulfite sequencing data for the eight selected B1 loci in E16.5 prospermatogonia. (B) Methylation status of the same B1 loci in Dnmt3l knockout (KO) P0 prospermatogonia. (C) Methylation levels of cytosines at B1 loci in each sequence context.
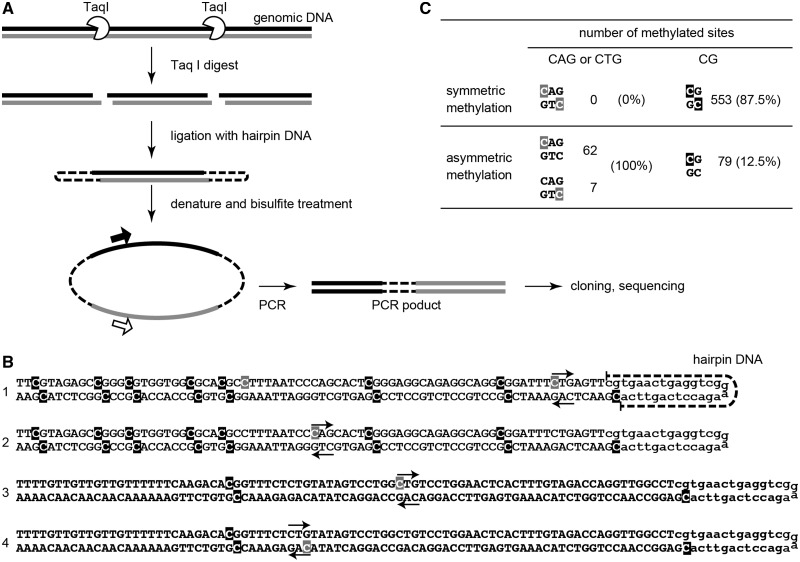
It is known that higher plants have methylation at both CpHpG and CpHpH sites (where H = A, C or T) (2). At CpHpG sites, the two cytosines on the different DNA strands are symmetrically methylated (29,30), and the chromo-methylase CMT3 is responsible for this methylation (31,32). A previous study in human ESCs and iPSCs showed that the levels of methylation at given CpHpG sites differ between the two strands (19), consistent with the fact that no CMT3-like methylase is found in mammals. However, in that study, the bisulfite reads were from different DNA duplexes and the read numbers were small. Therefore, whether the CpHpG sites are indeed hemimethylated in the same duplex has been an open question. To address this issue, we used the hairpin-bisulfite sequencing method (33) with some modifications (Figure 3A). Briefly, genomic DNA was digested with TaqI, ligated to synthetic hairpin forming DNA, and subjected to bisulfite reaction followed by PCR. The PCR primers were designed so that the amplicon consisted of the top- and bottom-strand sequences from a single duplex separated by the hairpin DNA sequence. The results obtained from three B1 loci showed that 87.5% of CpG sites were symmetrically methylated in P0 prospermatogonia (Figure 3B and C). In contrast, we detected no symmetric methylation at any CAG or CTG sites (the cumulative number of tested sites was 69) (Figure 3B and C). This suggests that a template-dependent copying mechanism does not operate in methylation at CpHpG sites. Thus, most or all of non-CpG methylation is ascribed to de novo methylation. The dinucleotide preference observed at the non-CpG sites (CpA>CpT>CpC) in prospermatogonia is well correlated with the target preference of the recombinant Dnmt3a protein (34,35). Indeed, in human and mouse ESCs and mouse oocytes, a depletion of the de novo methylases resulted in greatly reduced non-CpG methylation (3,17,18,20, K. Shirane et al. submitted for publication).
Figure 3.
Asymmetric methylation at CpHpG sites. (A) Procedure for hairpin-bisulfite sequencing. (B) Examples of the reads obtained by hairpin-bisulfite sequencing. (C) Results of hairpin-bisulfite sequencing of three B1 loci. The numbers of sites with symmetric or asymmetric methylation are shown for CAG (and CTG) and CG sites. Methylated cytosines are highlighted.
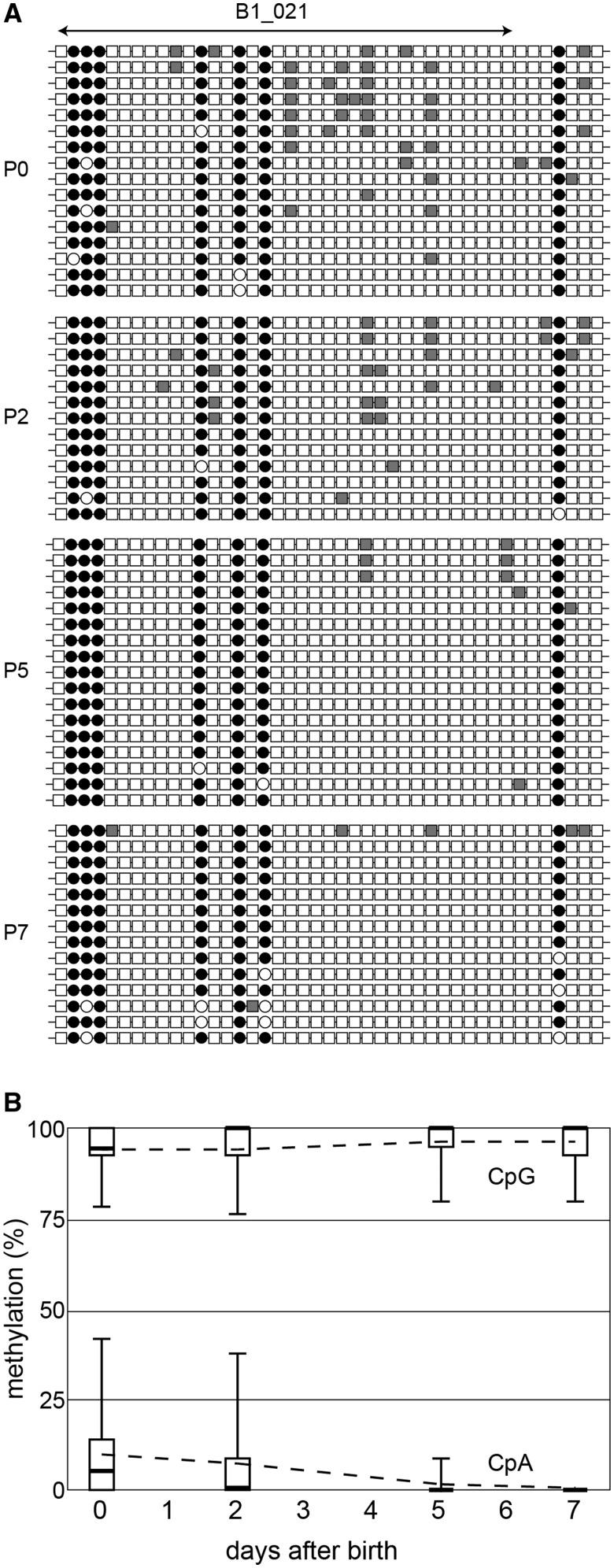
Next, we studied the fate of non-CpG methylation after birth, during the transition from the prospermatogonia to spermatogonia. The mitotically arrested prospermatogonia resume mitotic cell division at this stage. Proliferating spermatogonia at P7 showed very low levels of methylation at all three non-CpG dinucleotides (0.2–0.4%) (Figure 1D), which were comparable to the error rate of bisulfite conversion (0.2–0.5%). This suggests that non-CpG methylation decreases along with mitotic divisions. We then studied the time course of the loss of non-CpG methylation between P0 and P7. Whereas the CpG methylation levels in the 33 B1 loci stayed around 95% at all time points, the non-CpG methylation levels declined gradually (Figure 4A and Supplementary Figure S1). For example, the average CpA methylation levels were 9.8, 7.4, 1.5 and 0.4% at P0, P2, P5 and P7, respectively (Figure 4B). These results suggest that the de novo methylation activity is not high in postnatal germ cells and that the lack of a copying mechanism for non-CpG methylation results in a gradual loss of this methylation after every DNA replication. The Dnmt3a and Dnmt3l expression levels are high in prospermatogonia but become low after birth, and the Dnmt3b expression level is relatively low in prospermatogonia (8,9).
Figure 4.
Loss of non-CpG methylation after the resumption of mitosis in postnatal development. (A) Gradual loss of methylation at a representative B1 locus in the transition period from prospermatogonia to spermatogonia (P0, P2, P5 and P7). CpG sites are shown in circles and non-CpG sites are shown in squares, with open and filled (black or gray) symbols representing unmethylated and methylated cytosines, respectively. (B) Methylation levels of the 33 B1 loci at CpG sites and CpA sites at P0, P2, P5 and P7. The boxes indicate 75 and 25 percentiles, with the thick line inside being the median. The ends of the wiskers represent 95 and 5 percentiles. The dashed line indicates the average.
Since a high level of non-CpG methylation was first detected in pluripotent cells such as ESCs and iPSCs, a correlation of non-CpG methylation with pluripotency has been discussed (19). However, the detection of non-CpG methylation in oocytes, brain cells and prospermatogonia suggests that the above may not be the case. Rather, the feature common to these cell types seems relatively high expression levels of the de novo methylase(s) (6,7,36,37). Indeed, in human ESCs and iPSCs, the level of non-CpG methylation is better correlated with the expression levels of the de novo methylases than those of pluripotency factors (20).
In summary, we showed that non-CpG methylation accumulates within and around B1 SINE sequences, as well as the paternally methylated DMRs and some CpG islands, in the mitotically arrested prospermatogonia during fetal development. The observed non-CpG methylation level was relatively high compared with other cells, but this does not necessarily mean that prospermatogonia have high levels of non-CpG methylation throughout the genome, as non-CpG methylation is relatively high in the Alu SINE sequences in human ESCs (20). Our hairpin-bisulfite sequencing revealed that non-CpG sites including symmetrical CpHpG sites are hemimethylated. Therefore, it appears that no template-dependent copying mechanism exsists for non-CpG methylation in prospermatogonia. Rather, almost all non-CpG methylation is ascribed to de novo methylation. Perhaps, the presence and the level of non-CpG methylation are determined by the balance between the de novo methylation activity and the rate of cell proliferation. In the case of B1 retrotransposons, the non-CpG methylation may not have a role in regulation of transcription or boundary function as shown above, but further studies are needed to know whether non-CpG methylation in other genomic regions has a function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1 and 2.
FUNDING
Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan [22770009, 21200037 to K.I. and 20062010 to H.S.]; Career Center for Women Researchers’ Hand-in-Hand Program at Kyushu University (Research Assistants Dispatching Program to T.I.). Funding for open access charge: Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan [20062010 to H.S.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Kenichiro Hata [National Research Institute for Child Health and Development, Japan] for Dnmt3l knockout mice. The authors also thank Ms Ayako Ogawa and Dr Tomoko Tahira for technical assistance and Dr Motoko Unoki for her helpful comments on the manuscript.
REFERENCES
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012;8:e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 7.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 8.La Salle S, Mertineit C, Taketo T, Moens PB, Bestor TH, Trasler JM. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev. Biol. 2004;268:403–415. doi: 10.1016/j.ydbio.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Sakai Y, Suetake I, Shinozaki F, Yamashina S, Tajima S. Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr. Patterns. 2004;5:231–237. doi: 10.1016/j.modgep.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu WM, Schmid CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993;21:1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kochanek S, Renz D, Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 1993;12:1141–1151. doi: 10.1002/j.1460-2075.1993.tb05755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Besser D, Gotz F, Schulze-Forster K, Wagner H, Kroger H, Simon D. DNA methylation inhibits transcription by RNA polymerase III of a tRNA gene, but not of a 5S rRNA gene. FEBS Lett. 1990;269:358–362. doi: 10.1016/0014-5793(90)81193-r. [DOI] [PubMed] [Google Scholar]
- 14.Ichiyanagi K, Li Y, Watanabe T, Ichiyanagi T, Fukuda K, Kitayama J, Yamamoto Y, Kuramochi-Miyagawa S, Nakano T, Yabuta Y, et al. Locus- and domain-dependent control of DNA methylation at mouse B1 retrotransposons during male germ cell development. Genome Res. 2011;21:2058–2066. doi: 10.1101/gr.123679.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289:41–48. doi: 10.1016/s0378-1119(02)00469-9. [DOI] [PubMed] [Google Scholar]
- 19.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiraishi M, Hayatsu H. High-speed conversion of cytosine to uracil in bisulfite genomic sequencing analysis of DNA methylation. DNA Res. 2004;11:409–415. doi: 10.1093/dnares/11.6.409. [DOI] [PubMed] [Google Scholar]
- 23.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roman AC, Benitez DA, Carvajal-Gonzalez JM, Fernandez-Salguero PM. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc. Natl Acad. Sci. USA. 2008;105:1632–1637. doi: 10.1073/pnas.0708366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roman AC, Gonzalez-Rico FJ, Molto E, Hernando H, Neto A, Vicente-Garcia C, Ballestar E, Gomez-Skarmeta JL, Vavrova-Anderson J, White RJ, et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21:422–432. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- 28.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 29.Meyer P, Niedenhof I, ten Lohuis M. Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrida. EMBO J. 1994;13:2084–2088. doi: 10.1002/j.1460-2075.1994.tb06483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 31.Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 33.Laird CD, Pleasant ND, Clark AD, Sneeden JL, Hassan KM, Manley NC, Vary JC, Jr, Morgan T, Hansen RS, Stoger R. Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc. Natl Acad. Sci. USA. 2004;101:204–209. doi: 10.1073/pnas.2536758100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki A, Suetake I, Miyagawa J, Fujio T, Chijiwa T, Sasaki H, Tajima S. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 2001;29:3506–3512. doi: 10.1093/nar/29.17.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J. Mol. Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe D, Suetake I, Tada T, Tajima S. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech. Develop. 2002;118:187–190. doi: 10.1016/s0925-4773(02)00242-3. [DOI] [PubMed] [Google Scholar]
- 37.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.