Abstract
Hydroxyl radicals predominantly react with the C8 of purines forming 7,8-dihydro-8-oxoguanine (8oxoG) and 7,8-dihydro-8-oxoadenine (8oxoA) adducts, which are highly mutagenic in mammalian cells. The majority of oxidized DNA bases are removed by DNA glycosylases in the base excision repair pathway. Here, we report for the first time that human thymine-DNA glycosylase (hTDG) and Escherichia coli mismatch-specific uracil-DNA glycosylase (MUG) can remove 8oxoA from 8oxoA•T, 8oxoA•G and 8oxoA•C pairs. Comparison of the kinetic parameters of the reaction indicates that full-length hTDG excises 8oxoA, 3,N4-ethenocytosine (εC) and T with similar efficiency (kmax = 0.35, 0.36 and 0.16 min−1, respectively) and is more proficient as compared with its bacterial homologue MUG. The N-terminal domain of the hTDG protein is essential for 8oxoA–DNA glycosylase activity, but not for εC repair. Interestingly, the TDG status had little or no effect on the proliferation rate of mouse embryonic fibroblasts after exposure to γ-irradiation. Nevertheless, using whole cell-free extracts from the DNA glycosylase-deficient murine embryonic fibroblasts and E. coli, we demonstrate that the excision of 8oxoA from 8oxoA•T and 8oxoA•G has an absolute requirement for TDG and MUG, respectively. The data establish that MUG and TDG can counteract the genotoxic effects of 8oxoA residues in vivo.
INTRODUCTION
Accumulation of oxidative damage to DNA has been implicated in diverse human diseases, including cancer, neurological disorders and premature aging. Hydroxyl radicals (OH•) preferentially react with C8 atom of purines to generate 7,8-dihydro-8-oxoguanine (8oxoG), 7,8-dihydro-8-oxoadenine (8oxoA) and formamidopyrimidines (Fapy) residues in DNA (1–2). Although 8oxoA is present at approximately one-tenth level of 8oxoG (1), 8oxoA and FapyA are the most abundant oxidized adenine adducts in irradiated DNA samples (3). The presence of 8oxoA in duplex DNA does not perturb its B-helix conformation and DNA synthesis (4). Structural studies have shown that both oxidized purines are present predominantly in keto form and can adopt ‘syn’ conformation (5). In vitro, DNA polymerases almost exclusively incorporate dTMP opposite to 8oxoA in the presence of all four dNTPs (6). In Escherichia coli, no mutagenesis was observed on transfection of the single-stranded M13 phage DNA containing 8oxoA (7). The studies on mutagenesis in mammalian cells, however, showed that 8oxoA can induce mutations at a significant rate (8–9). Kamiya et al. (8) reported that in 3T3 mouse fibroblasts, 8oxoA induces A→C and A→G mutations when incorporated in the sequence context 5′-CCXAG-3′ of an exogenous double-stranded c-Ha-ras gene and that the mutagenic potency of 8oxoA was similar to the one of 8oxoG. However, Tan et al. (9) compared the mutagenic property of 8oxoA in two different sequence contexts in single-stranded DNA vectors in simian kidney cells. At variance with the previous study, they detected mainly A→C transversion with frequencies at least four times lower than for 8oxoG.
The majority of oxidative DNA base damage is removed by the base excision repair (BER) pathway. BER is initiated by a DNA glycosylase that cleaves the N-glycosylic bond between the abnormal base and the deoxyribose, generating either an abasic site or single-stranded break in DNA. The E. coli Fpg and Nei DNA glycosylases excise 8oxoA poorly, whereas the eukaryotic 8oxoG–DNA glycosylases (OGG1) and mammalian endonuclease VII-like protein 1 (NEIL1) can efficiently remove 8oxoA, but only when it is paired with cytosine (10–11). Based on these observations, it was hypothesized that in eukaryotes, 8oxoA•C base pairs may occur by misincorporation of the oxidized DNA precursor 8-oxo-2′-deoxyadenosine 5′-triphosphate (8oxodATP) opposite to cytosine during DNA replication. Indeed, the human (2′-deoxy)ribonucleoside 5′-triphosphate pyrophosphohydrolase (MTH1) protein, a dNTPs pool sanitizing enzyme, hydrolyses with good efficiency 8oxodATP in contrast to its E. coli counterpart MutT protein, which lacks activity on this oxidized precursor (12). Interestingly, an unidentified mono-functional mammalian DNA glycosylase, distinct from OGG1, which can remove 8oxoA when paired with guanine, has been also described (13). However, so far, no DNA glycosylase excising 8oxoA from its naturally occurring pair with thymine has been reported.
The human thymine-DNA glycosylase (hTDG) was first biochemically characterized for its ability to remove T mispaired with G (14). More detailed characterization showed that hTDG exhibits a wide substrate specificity; it excises 3,N4-ethenocytosine (εC) (15,16), thymine glycol (17), 5-hydroxycytosine (18), mismatched uracil (19) and its derivatives with modifications at the C5 position (20). The hTDG protein exhibits an extremely low turnover rate that relies on a high affinity for the generated apurinic/apyrimidinic (AP) site (21). Human major AP endonuclease 1 (APE1) can activate hTDG by increasing the dissociation rate of hTDG from AP site (22). A second factor facilitating hTDG enzymatic turnover is the SUMOylation (Small Ubiquitin-like Modifier protein = SUMO) of hTDG by SUMO-1 and SUMO-2/3, which reduces drastically its affinity for AP sites and increases its enzymatic turnover towards DNA substrates (23). The hTDG protein interacts with the transcriptional co-activator CBP/p300 (CREB-binding protein/p300), and the resulting hTDG/CBP/p300 complex is competent for both BER and histone acetylation (24). The hTDG protein enhances the CBP/p300 transcriptional activity and, in turn, CBP/p300 acetylates hTDG. Acetylation of hTDG regulates the recruitment of APE1. Thus, TDG functions are tightly regulated by post-translational modifications (SUMOylation and acetylation) and by protein–protein interactions. Interestingly, TDG knockout mice are embryonically lethal, possibly because of aberrant de novo DNA methylation of CpG-rich promoters of developmental genes, which results in failure to establish and/or maintain cell type-specific gene expression programs during embryonic development (25,26). Recently, it has been demonstrated that oxidized derivatives of 5-methylcytosine (5mC) in CpG sequence contexts are efficiently processed by human TDG, suggesting a direct involvement of TDG-initiated BER in protection of gene promoters from aberrant DNA methylation (27,28). Escherichia coli mismatch-specific DNA glycosylase MUG was identified by sequence homology to mammalian TDG and was characterized as enzyme excising uracil from G•U mismatch with a possible role as a back up enzyme for the classical uracil–DNA glycosylase (UDG) (29). MUG is a small 18.6 kDa protein, able to remove various ε-bases, including εC (16,30) and 1,N2-ethenoguanine (31) and also with low efficiency hypoxanthine and 5-hydroxycytosine residues when present in DNA (32).
At present, the mechanisms of 8oxoA repair when paired with thymine are only poorly investigated. Here, we report that E. coli MUG and human TDG are able to excise 8oxoA in 8oxoA•T pairs. To address the physiological relevance of the MUG and hTDG-catalysed excision of 8oxoA, we measured kinetic parameters using various DNA substrates and characterized 8oxoA–DNA glycosylase activities in whole cell-free extracts from BER deficient cells. The role of MUG/TDG-initiated BER in counteracting oxidized adenine residues in duplex DNA is discussed.
MATERIALS AND METHODS
Chemicals, reagents and proteins
Cell culture media were purchased from Invitrogen (Life Technologies SAS, Saint Aubin, France). Restriction enzymes and T4 DNA ligase were from New England Biolabs France (Evry, France). The E. coli BL21 (DE3) cells were from Novagen-EMD4Biosciences (Merck Chemicals, Nottingham, UK). Collection of the purified DNA glycosylases and AP endonucleases was from the laboratory stock (33,34). The activities of various DNA repair proteins were tested using their principal substrates and were checked just before use (data not shown).
Oligonucleotides
All oligonucleotides containing modified bases and their complementary strands were purchased from Eurogentec (Seraing, Belgium) including the following: 40 mer d(AATTGCTATCTAGCTCCGCXCGCTGGTACCCATCTCATGA) where X is either 8oxoA or εC and 40 mer d(AATTGCTATCTAGCTCCGCYGGCTGGTACCCATCTCATGA) where Y is either U or T. Sequences of the oligonucleotide duplexes used in the present work are shown in Supplementary Table S1.
The 40 mer single-stranded oligonucleotide was 5′-end labelled with [γ-32P]-ATP (PerkinElmer, France) using T4 polynucleotide kinase (New England Biolabs). Radioactively labelled oligonucleotides were desalted with a Sephadex G25 column, equilibrated in water and then annealed with corresponding complementary strands for 3 min at 65°C in a buffer containing 20 mM HEPES–KOH (pH 7.6), 50 mM KCl as previously described (34).
Escherichia coli strains and mammalian cell cultures
Escherichia coli JM105 [F’ traD36 lacIqΔ(lacZ)M15 proA+B+/thi rpsL (Strr) endA sbcB15 sbcC201 hsdR4 (
 ) Δ(lac-proAB)] [wild-type (‘WT’)] and its isogenic derivatives MS50 (mug-1::KnR) were from laboratory stock (35). To prepare cell-free extracts, E. coli cultures were grown overnight and harvested by centrifugation, washed in cold extraction buffer [0.5 M KCl, 50 mM Tris–HCl (pH 7.5), 0.1 mM ethylenediaminetetraacetic acid (EDTA) and protease inhibitor cocktail] and stored at −20°C. The frozen cells were thawed in an ice water bath and disrupted by sonication. Cell debris were removed by centrifugation at 20 000g for 15 min at 4°C, and supernatants were aliquoted and stored at −80°C until use.
) Δ(lac-proAB)] [wild-type (‘WT’)] and its isogenic derivatives MS50 (mug-1::KnR) were from laboratory stock (35). To prepare cell-free extracts, E. coli cultures were grown overnight and harvested by centrifugation, washed in cold extraction buffer [0.5 M KCl, 50 mM Tris–HCl (pH 7.5), 0.1 mM ethylenediaminetetraacetic acid (EDTA) and protease inhibitor cocktail] and stored at −20°C. The frozen cells were thawed in an ice water bath and disrupted by sonication. Cell debris were removed by centrifugation at 20 000g for 15 min at 4°C, and supernatants were aliquoted and stored at −80°C until use.
Mouse embryonic fibroblast (MEF)–WT and MEF–Tdg−/− cell lines were obtained as previously described (36). MEF–WT and MEF–Tdg−/− cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in the presence of 5% CO2. To prepare whole cell-free extracts, MEF cells were washed with ice-cold phosphate buffered saline and incubated 30 min in lysis buffer containing 80 mM HEPES–KOH (pH 7.6), 1 M KCl, 2 mM DTT, 0.1 mM EDTA, 0.3% non-ionic detergent Nonidet P-40 (NP-40) and protease inhibitor cocktail (Complete EDTA-free, Roche). Extracts were clarified by centrifugation at 20 000g for 15 min at 4°C.
DNA repair assays
The standard reaction mixture (20 μl) used for screening of DNA repair activities contained 10 nM [32P]-labelled U•G, G•T, εC•G and 8oxoA•T oligonucleotide duplexes and limited amounts of enzymes for 10 min at 37°C, unless otherwise stated. Assays for the AlkA, Nth, MUG, UDG and Nfo proteins were performed in buffer containing 20 mM HEPES–KOH (pH 7.6), 50 mM KCl, 0.1 mM EDTA, 0.1 mg•ml−1 BSA and 1 mM DTT. Assays for the Fpg, Nei, NEIL1 and NEIL2 proteins were performed in buffer containing 25 mM HEPES–KOH (pH 7.6), 100 mM KCl, 1 mM EDTA, 5 mM β-mercaptoethanol and 6% glycerol. Assays for the APE1, alkyl-N-purine-DNA glycosylase (ANPG), human single-strand-selective monofunctional uracil-DNA glycosylase 1 (SMUG1), human uracil-DNA glycosylase (hUNG) and N-terminally tagged A/G-specific Adenine-DNA Glycosylase, MutY Homolog, (hMYH) proteins were performed in buffer containing 20 mM HEPES–KOH (pH 7.6), 50 mM KCl, 5 mM MgCl2, 0.1 mg•ml−1 BSA and 1 mM DTT. For the MutY protein, we used buffer containing 20 mM HEPES–KOH (pH 7.0), 10 mM EDTA, 5 µM ZnCl2, 75 µg•ml−1 BSA, 0.5 mM DTT and 1.45% glycerol. Finally, assays for the hTDG, hTDGcat and human methyl-binding domain protein 4 (MBD4) proteins were performed in buffer containing 20 mM Tris–HCl (pH 8.0), 1 mM EDTA, 1 mM DTT, 100 mg•ml−1 BSA and 100 mM NaCl. Reactions were stopped by adding 10 µl of a solution containing 0.5% sodium dodecyl sulphate (SDS) and 20 mM EDTA. For the mono-functional DNA glycosylases, the abasic sites left after excision of damaged bases were cleaved either by co-incubation with the purified AP endonuclease or by light piperidine treatment [10% (v/v) piperidine at 37°C for 40 min].
The standard reaction mixture (20 μl) used for detailed kinetic characterization of DNA glycosylase activities contained 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 1 mM DTT and 100 µg•ml−1 BSA, 50 nM [32P]-labelled duplex oligonucleotide and either 5 µM MUG or 500 nM hTDG and hTDGcat proteins. The mixture was incubated at 37°C for a varied period, and AP sites were cleaved by light piperidine treatment, unless otherwise stated.
The standard reaction mixture (100 µl) for repair assays in whole-cell extracts contained 2.5 nM [32P]-labelled εC•G or 8oxoA•N oligonucleotide duplexes in 50 mM KCl, 20 mM HEPES–KOH (pH 7.6), 0.1 mg•ml−1 BSA, 1 mM DTT, 1 mM EDTA and either 30 µg of MEF protein extract or 20 µg of E. coli protein extract or 50 nM purified hTDG, unless otherwise stated. The reaction mixtures were incubated for 60 min at 37°C and stopped by adding 0.5% SDS, 0.1 mg•ml−1 proteinase K followed by incubation for 10 min at 50°C.
After treatments, the reaction samples were desalted using Sephadex G25 column (Amersham Biosciences) and equilibrated in 7.5 M urea. Purified reaction product were analysed by electrophoresis through denaturing 20% (w/v) polyacrylamide gels (7.5 M urea, 0.5× Tris/Borate/EDTA (TBE), 42°C). The wet gel was wrapped in plastic wrap and exposed to Fuji FLA-3000 Phosphor Screen and quantified by Image Gauge V3.12 Software. The release of U, T, εC and 8oxoA base adducts was measured by the cleavage of the oligonucleotide containing a single lesion at a defined position.
Expression and purification of hTDG
The expression vectors pET28c–hTDG (for full-length protein) and pET28c–TDGcat (for catalytic domain protein) were previously described (37). Rosetta 2 (DE3) cells transformed with a pET28c–hTDG and pET28c–hTDGcat were grown at 37°C in Luria Broth medium, supplemented with 50 µg•ml−1 kanamycin, on an orbital shaker to OD600 nm = 0.6–0.8. Then temperature was reduced to 30°C, and the proteins expression was induced by 0.2 mM isopropyl β-d-galactopyranoside (Sigma-Aldrich), and the cells were further grown either for 2 h for induction of the hTDG or 15 h for induction of the hTDGcat proteins. Bacteria were harvested by centrifugation, and cell pellets were lysed using a French press at 18 000 psi in buffer containing 20 mM HEPES–KOH pH 7.6, 50 mM KCl supplemented with Complete™ Protease Inhibitor Cocktail (Roche Diagnostics, Switzerland). Lysates were cleared by centrifugation at 40 000g for 30 min at 4°C, the resulting supernatant was adjusted to 500 mM NaCl and 20 mM imidazole and loaded onto HiTrap Chelating HP column (Amersham Biosciences, GE Health). All purification procedures were carried out at 4°C. The column was washed with buffer A (20 mM HEPES, 500 mM NaCl and 20 mM imidazole), and the bound proteins were eluted with a linear gradient of 20–500 mM imidazole in buffer A. Eluted fractions were analysed by SDS–polyacrylamide gel electrophoresis (PAGE), and fractions containing the pure His-tagged hTDG and hTDGcat proteins were stored at −80°C in 50% glycerol. The concentration of purified proteins was determined by the method of Bradford.
Single turnover kinetics
Here, we used single turnover kinetics under large excess of enzyme over substrate (Kd<[E]≫[S]) to obtain rate constants (kobs) that are not affected by enzyme–substrate association or by product inhibition, such that kobs reflects the maximal base excision rate (kobs ≈ kmax) (38–40). The data were fitted by non-linear regression to one phase exponential association Equation 1 using GraphPad Prism 5 software.
| (1) |
where A is the amplitude, kmax is the rate constant and t is the reaction time (in seconds).
The enzymatic assays were performed in large volume reaction mixture containing 50 nM duplex oligonucleotide and 5 µM MUG or 500 nM hTDG for varying periods at 37°C. At each time point, 20 µl of sample was withdrawn, and reaction was stopped by light piperidine treatment. Reaction products were analysed as described earlier in the text.
In vitro reconstitution of 8oxoA•T repair by hTDG
5 nM 8oxoA•T unlabelled duplex was incubated in the presence of 20 nM hTDG, 5 nM APE1, 2 nM flap-structure endonuclease 1 (FEN1), 0.1 U of DNA polymerase-β (POL-β) and 5 nM T4 DNA Ligase, 50 µM dNTPs, 20 µCi of [α-32P]dATP in buffer containing 50 mM HEPES–KOH (pH 7.6), 30 mM NaCl, 0.1 mg•ml−1 BSA, 2 mM DTT, 2 mM ATP and 3 mM MgCl2 for 5 and 30 min at 37°C, unless otherwise stated. Reaction products were analysed as described earlier in the text.
Proliferation assay of MEF cells exposed to γ-irradiation
When the exponentially growing WT and Tdg−/− MEFs cell density reached ≈80% confluence, cells were trypsinized, harvested and resuspended in phosphate buffered saline at a concentration of 106 cells/ml. The cell suspensions were exposed to a single dose ranging from 0, 1, 2, 3, 5, 10, 15 and 20 Gy using a 137Cs source (IBL 637, Institut Gustave Roussy, Villejuif, France) with a dose rate of 2.9 Gy/min. Cells were plated in triplicate for each dose (100 000 cells per well in 24-well plate) with fresh Dulbecco’s modified Eagle’s medium medium; the non-irradiated control cells were plated in the same way. Cells were incubated at 37°C in humidified atmosphere containing 5% of CO2 for indicated period; at regular intervals, cells were trypsinized, stained with trypan blue (Sigma-Aldrich) and counted under a microscope. At each time point, number of viable WT and Tdg−/− MEFs cells after irradiation were reported to number of the corresponding viable non-irradiated control cells for each radiation dose. The results were expressed as the percentage of viable cells in irradiated samples relative to that in corresponding control non-irradiated cells. Kinetics of WT and Tdg−/− MEFs proliferation were presented as fold increase in cell numbers depending on time. The data were calculated from three independent experiments and presented as the mean ± standard error. Statistical analyses were performed using GraphPad Prism 5 software.
RESULTS
Mono-functional mismatch-specific uracil/thymine DNA glycosylases MUG and hTDG excise 8oxoA residue paired with thymine
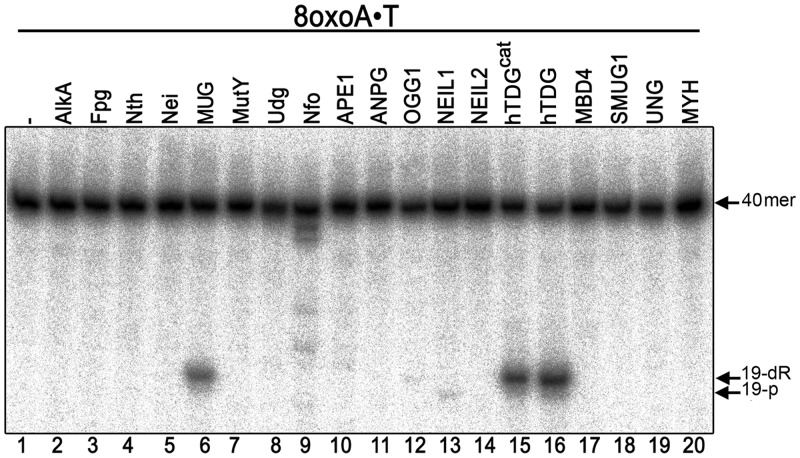
We examined whether 8oxoA•T pair is a substrate for previously characterized BER enzymes from E. coli and human. For this, a 5′-[32P]-labelled 40 mer oligonucleotide duplex containing a single 8oxoA•T pair was challenged with a variety of highly purified AP endonucleases and DNA glycosylases. As not all DNA glycosylases possess an AP lyase activity, the samples incubated with mono-functional DNA glycosylases were treated with light piperidine to cleave the potential abasic sites generated by the excision of 8oxoA. As shown in Figure 1, only incubation with E. coli MUG, human full-length TDG (hTDG) and catalytic domain TDG (hTDGcat) led to the cleavage of the labelled oligonucleotide at the position of 8oxoA and generation of 19 mer product fragment (lanes 6, 15 and 16). As expected, human OGG1 and NEIL1 cleave the 8oxoA•T duplex with extremely low efficiency (lanes 12 and 13). It should be noted that 19 mer cleavage fragments generated by MUG/hTDG coupled to light piperidine treatment and OGG1 contain a 3′-phosphoraldehyde terminus and migrate at the same position (lanes 6, 12 and 15,16), whereas that of NEIL1 generated fragment contains 3′-phosphate terminus and migrates faster than that of OGG1 (lane 13 versus 12). Despite being used in excess amount, the pure AlkA, Fpg, Nth, Nei, MutY, UDG, Nfo, APE1, ANPG, NEIL2, MBD4, SMUG1, UNG2 and MYH proteins did not act on 8oxoA•T (lanes 2–5, 7–11, 14 and 17–20).
Figure 1.
Activity of various E. coli and human BER enzymes on 8oxoA•T oligonucleotide duplex. 10 nM 5′-[32P]-labelled 40 mer 8oxoA•T oligonucleotide was incubated with 20 nM of enzyme (except 160 nM of TDGcat and 0.6 unit of MYH were used) for 10 min at 37°C (except for MUG, TDGs and MBD4 incubation was for 1 h at 30°C). When using mono-functional DNA glycosylases, the reaction products were subjected to light piperidine treatment to cleave DNA fragments at AP sites resulting from base excision. For details see ‘Materials and Methods’ section.
Kinetic parameters of the excision of 8oxoA, U, T and εC residues when present in duplex DNA by hTDG and MUG
Previously, it was demonstrated that MUG and hTDG are product inhibited; they remain bound to the produced AP site opposite guanine with a higher affinity than for the substrate U•G and T•G DNA duplexes (21,39). As MUG and hTDG fail to turnover during reaction, which implies that each molecule of enzyme can only remove one uracil or thymine residue, comparing the steady-state kinetic parameters, such as KM and kcat of these DNA glycosylases for different DNA substrates is not useful. Therefore, we further characterized substrate specificity of hTDG, hTDGcat and MUG by measuring the cleavage rates of 8oxoA•T, U•G, T•G and εC•G duplexes under single-turnover conditions, using a molar excess of enzyme over DNA substrate, which provide the maximal rate of base excision (kmax) for a given substrate (38–40) (Supplementary Figure S1 and Supplementary Table S1). It should be noted that the kinetic parameter kmax determined under single-turnover conditions measures the maximal rate of AP site product formation after DNA binding and before product release; therefore, it does not reflect product release as in case of kcat (40). The time course of the cleavage product generation shows that U•G is the most preferred substrate for hTDG followed by εC•G and then by 8oxoA•T and T•G, which are cleaved with similar efficiency (Supplementary Figure S1A and B). Importantly, MUG reaches a plateau of only 15% of 8oxoA•T cleaved after 2 h of incubation, indicating that the bacterial DNA glycosylase has low affinity for the DNA substrate (Supplementary Figure S1C). Under the experimental condition used, the truncated version of human enzyme hTDGcat exhibits lower activity on all DNA substrate tested (Supplementary Figure S1B), which is in agreement with a previous report (37). The kmax constants were determined by fitting the single-turnover data to one phase exponential association using non-linear regression (38–40). As shown in Table 1, the kmax value of hTDG-catalysed excision of 8oxoA (0.35 min−1) is similar to that for εC and thymine excision (0.36 and 0.16 min−1, respectively), but 17-fold lower than the kmax for uracil excision (6.1 min−1). Compared with the full-length protein, the kmax values for U, T and εC excision by hTDGcat were 32, 2.9 and 1.3-fold lower, respectively (Table 1). Because of low activity of hTDGcat on 8oxoA, we were not able to determine kmax value for this substrate–enzyme combination. The rate constant of MUG for 8oxoA excision (kmax = 0.085 min−1) is >170-fold lower as compared with that of εC (15.0 min−1), indicating that bacterial enzyme repairs oxidized adenines with low efficiency compared with εC residues. It should be noted that the reaction rates of hTDG and MUG on U and εC residues, respectively, quickly reach a plateau within the first minute of incubation. Therefore, the kmax values measured under the experimental conditions we used may represent an underestimate for these DNA substrates.
Table 1.
Pre-steady-state kinetic parameters of the hTDG, hTDGcat and MUG proteins acting on various DNA substrates
|
kmax (min−1)a |
|||
|---|---|---|---|
| Substrate | hTDG | hTDGcat | MUGb |
| 8oxoA•T | 0.35 ± 0.05 | ND | 0.085 ± 0.003 |
| U•G | 6.1 ± 3.7 | 0.19 ± 0.04 | 0.101 ± 0.003 |
| T•G | 0.16 ± 0.01 | 0.056 ± 0.003 | ND |
| εC•G | 0.36 ± 0.05 | 0.27 ± 0.09 | 15.0 ± 5.8 |
akmax constants were calculated by one phase exponential association equation using GraphPad Prism 5.
bkmax constants for MUG were determined using only data collected during the first 15 min of reaction because of the accelerated enzyme inactivation at 37°C (39). ND, not determined.
Role of opposite base and sequence context in the excision of 8oxoA
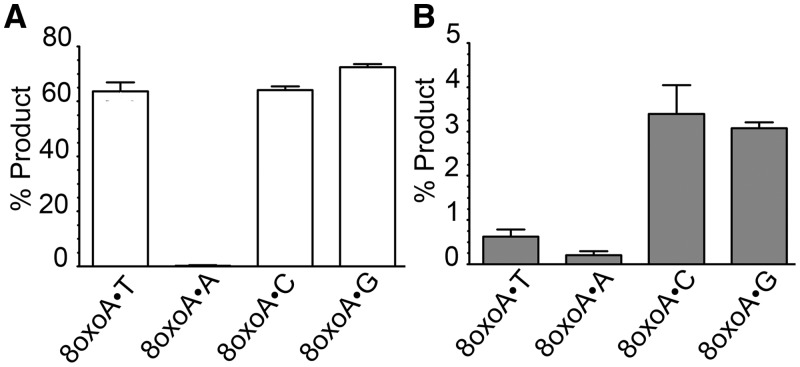
To further characterize 8oxoA repair, we examined the base pair specificity of MUG and hTDG DNA glycosylase activities using 8oxoA•T, 8oxoA•A, 8oxoA•C and 8oxoA•G oligonucleotide duplexes. Similar excision efficiencies were observed for full-length hTDG when 8oxoA was paired with T, C or G, whereas no activity was observed on 8oxoA•A (Figure 2A). For E. coli MUG, excision of 8oxoA was more efficient when paired to either C or G than to T or A (Figure 2B). In summary, the relative order for opposite base-dependent incision of 8oxoA•N oligonucleotide duplexes was G ≥ T ≈ C ≫ A for hTDG and C ≥ G ≫ T > A for MUG (Supplementary Table S1).
Figure 2.
Cleavage of oligonucleotide duplex containing 8oxoA paired with one of the four regular DNA bases by the hTDG and MUG proteins. 5 nM 5′-[32P]-labelled 40 mer 8oxoA•N duplex oligonucleotide (where N is T, A, C or G) was incubated in the presence of the 50 nM purified protein for 15 min at 37°C. The reaction products were subjected to light piperidine treatment to cleave DNA fragments at AP sites. (A) Graphic representation of the mean values of hTDG activity; (B) graphic representation of the mean values of MUG activity. Each bar represents the mean ± standard deviation (SD) of three independent experiments. For details see ‘Materials and Methods’ section.
To examine whether excision of 8oxoA residues depends on the neighbouring sequence context, we used DNA duplexes with varying bases 5′ and 3′ to the oxidized adenine (Supplementary Table S2). At variance to the previously described context specificity for thymine excision (41), the rate of 8oxoA excision by hTDG varied little among seven sequence contexts tested (Supplementary Figure S2), the maximum difference being only 2-fold between the most (5′-CpA*pC-3′/5′-GpTpG-3′) and least (5′-GpA*pC-3′/5′-GpTpC-3′) preferred one (Supplementary Figure S2).
In vitro reconstitution of the BER pathway for 8oxoA paired with thymine
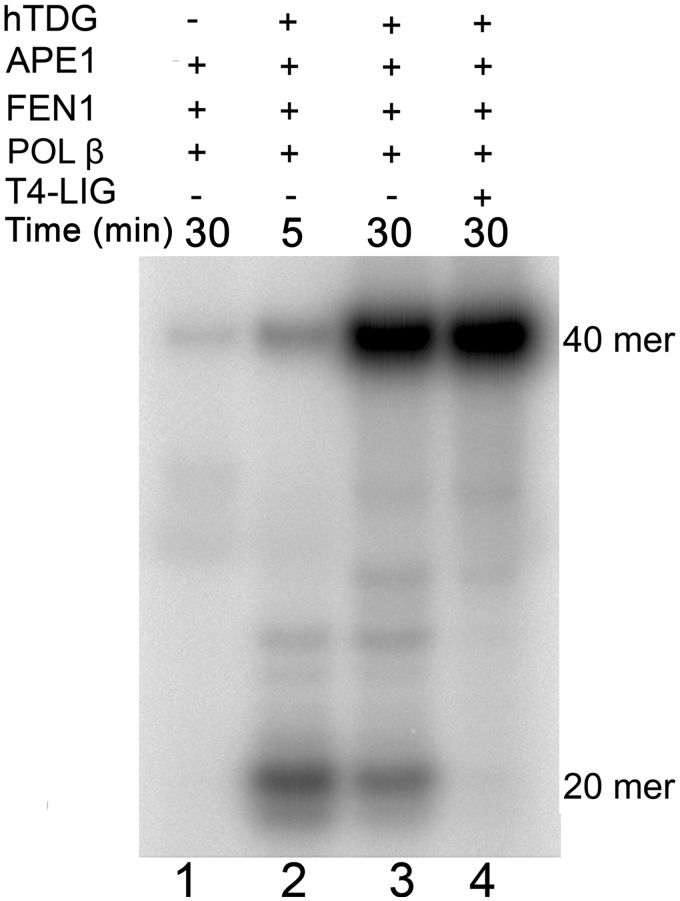
As shown earlier in the text, the hTDG-catalysed excision of 8oxoA generates an AP site, which may be further processed by either the short-patch or the long-patch BER pathway. To examine the contribution of both downstream BER processes, we have reconstituted the BER pathway for the 8oxoA•T pair in vitro using purified proteins (Figure 3 and Supplementary Figure S3). Incubation of a 40 mer 8oxoA•T oligonucleotide duplex in the presence of hTDG, APE1, FEN1, DNA POL-β, all dNTPs except for dATP and [α-32P]-dATP for 5 min generated labelled major 20 mer and minor 40 mer DNA products (lane 2). Continued incubation of the reaction mixture resulted in accumulation of the labelled full-length 40 mer DNA product (lane 3). These results indicate that APE1 cleaves AP sites generated by hTDG and allows POL-β to remove the 5′-deoxyribose phosphate residue and either insert one nucleotide (lane 2) and/or initiate strand-displacement (lane 3). Addition of T4 DNA ligase completes the restoration of the full-length 40 mer duplex and results in disappearance of 20 mer cleavage product (lane 4). In addition, we have studied in vitro reconstitution of 8oxoA repair when any one of these five proteins was omitted from the reaction (Supplementary Figure S3). As expected, no significant incorporation of [α-32P]-dATP was observed in the absence of either hTDG or APE1 or POL-β. Although in the absence of either DNA ligase or FEN1, POL-β-catalysed strand-displacement synthesis can efficiently generate the full-length 40 mer DNA repair product (Supplementary Figure S3). These data demonstrate that 8oxoA residues can be processed by both the short-patch and long-patch BER pathways.
Figure 3.
In vitro reconstitution of the BER pathway using 8oxoA•T duplex DNA substrate. 5 nM 40 mer 8oxoA•T duplex was incubated in the presence of 20 nM hTDG, 5 nM APE1, 2 nM FEN1, 0.1 U POL-β and 5 nM T4 DNA ligase in buffer containing 20 µCi of [α-32P]dATP, 50 µM dNTPs, 50 mM HEPES–KOH (pH 7.6), 30 mM NaCl, 0.1 mg/ml BSA, 2 mM DTT, 2 mM ATP and 3 mM MgCl2 for 5 and 30 min at 37°C. Lane 1, 30 min in the absence of hTDG and T4 DNA ligase; lane 2, 5 min in the absence of T4 DNA ligase; lane 3, same as 2, but 30 min; lane 4, 30 min in the presence of all proteins. For details see ‘Materials and Methods’ section.
Repair activities on DNA containing εC•G and 8oxoA•N pairs in E. coli and mouse cell-free extracts
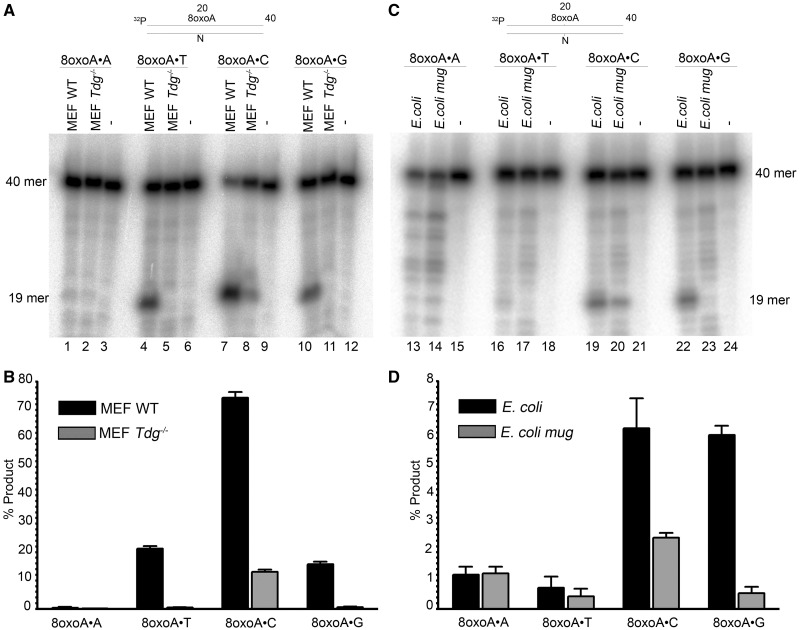
Data obtained with the purified DNA repair proteins show that 8oxoA paired with thymine, cytosine and guanine is mainly repaired by MUG and hTDG. To ascertain the role of these DNA glycosylases in vivo, we examined εC– and 8oxoA–DNA glycosylase activities in cell-free extracts from E. coli and MEFs cells lacking MUG and TDG, respectively. It should be recalled that we and others have previously demonstrated that E. coli mug strain completely lacks εC–DNA glycosylase activity and that hTDG is the most efficient εC–DNA glycosylase among other mammalian repair enzymes (16,35). To measure DNA glycosylase activities in the extracts, we used 5′-[32P]-labelled εC•G and 8oxoA•N oligonucleotide duplexes as DNA substrates. As expected, we observed robust εC–DNA glycosylase activities in cell extracts from WT MEFs and E. coli (Supplementary Figure S4), whereas no εC-activity was observed in cell extracts from TDG knockout MEFs (Tdg−/−) and the E. coli mug strain (Supplementary Figure S4), indicating that MUG and TDG are major εC–DNA glycosylases in E. coli and MEF, respectively. As shown in Figure 4, cell extracts from Tdg−/− MEFs completely lack 8oxoA•T- and 8oxoA•G-specific DNA glycosylase activity (Figure 4A, lanes 5 and 11 and B), whereas extracts from WT MEFs exhibit robust cleavage activity (lanes 4 and 7). Interestingly, cleavage of the 8oxoA•C duplex is dramatically reduced in the Tdg−/− MEF extract (Figure 4A, lane 8) when compared with the WT MEF extract (lane 7), suggesting that TDG also significantly contributes to the repair of 8oxoA when paired with C (Figure 4B). The 8oxoA•A duplex is poorly processed in both MEF extracts, and the marginal cleavage activity is not dependent on TDG (Figure 4A, lanes 1–2, and B). Taken together, these results suggest that TDG represents the main 8oxoA–DNA glycosylase activity in mammalian cells. Unlike mouse cell extracts, E. coli WT and mug cell-free extracts did not exhibit significant 8oxoA•T-specific cleavage activities under the experimental condition used here (Figure 4C, lanes 16–17). However, strong and moderate dependences on presence of MUG were detected for cleavage of 8oxoA•G and 8oxoA•C, respectively (Figure 4C lanes 22–23, 19–20 and D). Only weak processing activities were found for 8oxoA•A, which were independent from the MUG status (Figure 4C lanes 13–14 and D). These results are in agreement with data obtained with the purified protein and suggest that MUG can efficiently excise 8oxoA when paired with C and G.
Figure 4.
DNA repair activities towards 8oxoA containing duplex oligonucleotides in extracts from E. coli and mouse cells. 2.5 nM 5′-[32P]-labelled 40 mer 8oxoA•N oligonucleotide duplexes was incubated with either 30 µg of MEFs extract or 20 µg of E. coli cell extract. The repair assay (volume 100 µl) was performed in BER + EDTA buffer containing 50 mM KCl, 20 mM HEPES–KOH (pH 7.6), 0.1 mg/ml BSA, 1 mM DTT and 1 mM EDTA, for 1 h at 37°C. The reactions were stopped by adding SDS and proteinase K. (A) Denaturing PAGE analysis of the cleavage products after incubation of 8oxoA•N duplexes with mouse cell extracts. (B) Graphic representation of the mean values of cleavage activities in mouse cell extracts. (C) Denaturing PAGE analysis of the cleavage products after incubation of 8oxoA•N duplexes with E. coli cell extracts. (D) Graphic representation of the mean values of cleavage activities in E. coli cell extracts. For details see ‘Materials and Methods’ section.
Role of the TDG protein in the cellular response to ionizing radiation
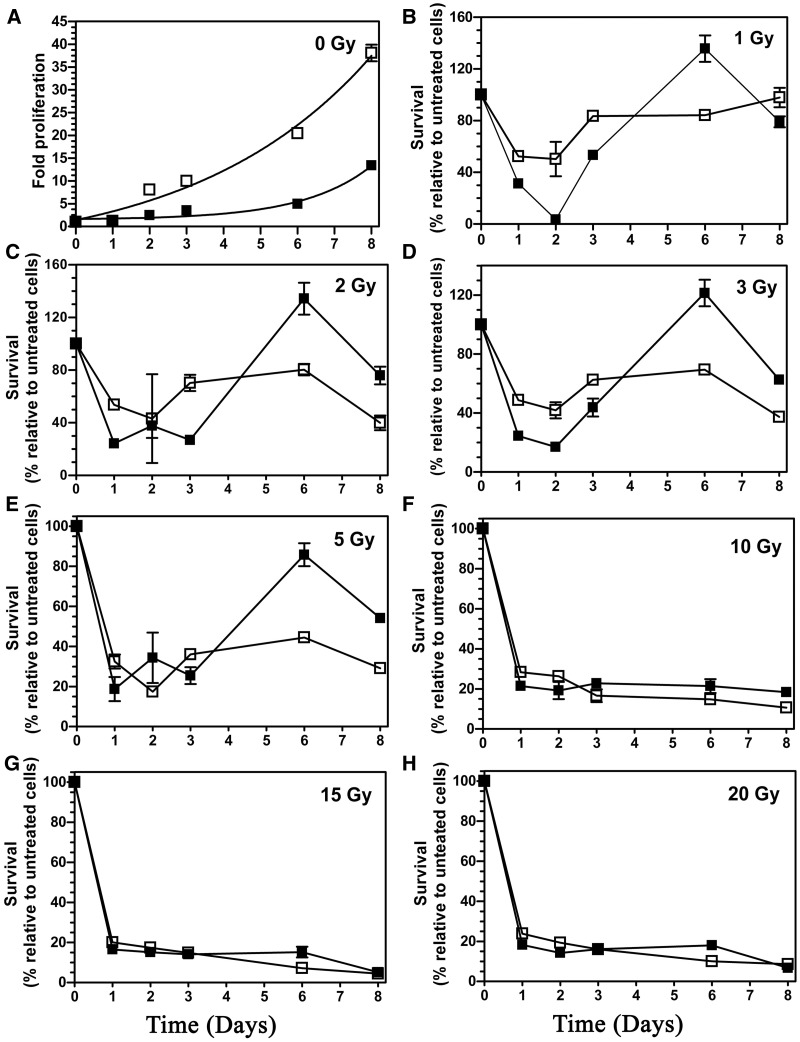
A previous study has demonstrated that the TDG status did not affect cell survival after ionizing radiation (IR) and H2O2 exposures (25). We, therefore, used the cell proliferation assay as a functional end point to investigate whether absence of TDG would affect the proliferation rate after IR exposure. The cells were irradiated with single doses ranging from 1 to 20 Gy of γ-irradiation and then analysed on days 1, 2, 3, 6 and 8 post-irradiation. It should be noted that under unchallenged conditions (no irradiation) the growth rate was much higher for Tdg−/− MEFs than for WT MEFs (Figure 5A). Compared with the respective untreated cells, proliferation rates of both WT and Tdg−/− MEFs were drastically reduced (<20% of control cells) by high doses (10–20 Gy) of IR even until the 8th day after exposure (Figure 5F–H). Importantly, no effect of the TDG status on the proliferation rate was detected at high doses of IR. At lower doses of 1–5 Gy, Tdg−/− MEFs showed reduced growth rates over an 8-day period after IR exposure (Figure 5B–E), whereas this reduction was only transient for treated WT MEFs and recovered after 6 and 8 days post-irradiation (Figure 5C–E). Note that we observed an abnormal increase in the number of WT MEFs at the 6th day after irradiation for doses of 1–3 Gy (Figure 5B–D). However, this increase in the growth rate of irradiated WT MEFs was not observed at the 8th day. Taken together, these data suggest that the TDG status may affect growth rate of MEFs for a long period after exposure to low doses of IR.
Figure 5.
Proliferation assay of irradiated MEFs cells. The MEF WT (filled square) and MEF Tdg−/− (open square) cells were treated with a single dose ranging from 0 Gy to 20 Gy γ irradiation, and the viable cells were scored at the indicated day. (A) Proliferation rates of non-irradiated WT and Tdg−/− MEFs. (B) Proliferation rates of WT and Tdg−/− MEFs after exposure to 1 Gy. (C) 2 Gy. (D) 3 Gy. (E) 5 Gy. (F) 10 Gy. (G) 15 Gy. (H) 20 Gy. The values are presented as the mean ± SD of three independent determinations of at least three sets of experiments. For details see ‘Materials and Methods’ section.
DISCUSSION
Our results show for the first time that E. coli and human mismatch-specific uracil/thymine DNA glycosylases can excise 8oxoA residues in duplex DNA when paired with T, C and G (Figures 1 and 2). Analysis of kinetic parameters for the full-length hTDG protein showed that it excises 8oxoA, εC and T residues from 8oxoA•T, εC•G and G•T base pairs, respectively, with similar efficiencies (Table 1), suggesting that 8oxoA is a physiologically relevant substrate for the human DNA glycosylase. The catalytic domain of hTDG (111–308), hTDGcat, has extremely low activity on 8oxoA•T, but it excises εC with an efficiency similar to the full-length protein, suggesting that the N-terminal and C-terminal parts of hTDG are essential for the recognition of damaged purines but not of exocyclic DNA adducts (Table 1). In agreement with our previous observation, hTDGcat exhibits lower activity on U•G and T•G duplexes as compared with the full-length protein (Supplementary Figure S1 and Table 1) (37). Importantly, both hTDGcat and bacterial MUG, which shares sequence homology with the core domain of human enzyme, exhibit similar DNA substrate preference.
It should be stressed that 8oxoA residues can arise in DNA either by direct oxidation of chromosomal DNA (1) or by incorporation into the nascent strand during replication as 8-oxo-2′-deoxyadenosine-5′-monophosphate (8oxodAMP) (12). Recently, it has been shown that 8oxodATP can be misincorporated opposite guanine and adenine by various DNA polymerases, resulting in the generation of 8oxoA•G and 8oxoA•A base pairs (42). Study of the base pair specificity of hTDG in the present work revealed that the DNA glycosylase can also excise 8oxoA from 8oxoA•C with similar efficiency and from 8oxoA•G with ∼5-fold higher rate as compared with 8oxoA•T, but it has no activity on 8oxoA•A pair (Figure 2 and Supplementary Table S1). Interestingly, the hTDG-catalysed excision of 8oxoA is barely dependent on the neighbouring base pairs context (Supplementary Figure S3). Taken together, these data demonstrate that the substrate specificity of MUG and hTDG depends more on the base pairing partner rather than on the sequence context of the lesion. This is in contrast to the observations made on family-1 UDGs for which the sequences flanking the target uracil have a greater effect on DNA glycosylase activity than the partner base of the uracil (43,44). Overall, the effects of sequence context and opposite base on excision of 8oxoA by hTDG are similar to that of εC and are dramatically different from that of T and U, suggesting that the human DNA glycosylase can recognize 8oxoA and εC in a global genome context. Finally, the removal of 8oxoA paired with T can be fully reconstituted in vitro using the purified BER proteins (Figures 3 and Supplementary Figure S3).
Crystal structures of MUG and hTDGcat in complex with DNA substrates have been resolved (45–47). The structure of MUG with a non-hydrolysable deoxyuridine analogue reveals an essentially non-specific pyrimidine-binding pocket that could accommodate bulky exocyclic DNA adducts, thus explaining wide substrate specificity of the bacterial DNA glycosylase (45). The structures of hTDGcat in complex with DNA containing an AP site, 5caC or its fluorinated analogue have also been solved (46,47). The structure of hTDGcat bound to DNA containing an abasic site reveals that hTDGcat crystallized in a 2:1 complex with 22 mer duplex oligonucleotide DNA. One molecule of protein was bound at the AP site, whereas the other was bound to an undamaged DNA site (46). Apparently, formation of 2:1 complex has no effect on DNA glycosylase activity of hTDG and hTDGcat. The structures of hTDGcat with 5caC containing DNA showed that the DNA glycosylase specifically recognizes 5caC in its well organized carboxyl binding active site pocket (47). Based on these DNA-liganded structures, it is still not clear how hTDG can accommodate oxidized purine residues in the active site pocket. Previously, we and others have demonstrated that TDG enzymes can excise hypoxanthine paired with guanine, suggesting that compared with other mismatch-specific uracil-DNA glycosylases, TDG has a spacious active site pocket, able to accommodate bulky purine lesions (20,48). Yet, the precise structural basis of 8oxoA recognition by MUG/TDG enzymes needs further investigations.
Data obtained in cell-free extracts from E. coli WT and mug mutant cells indicate that MUG is the main DNA glycosylase involved in the repair of 8oxoA•G and 8oxoA•C in E. coli (Figure 4). On the other hand, MUG-dependent activity on 8oxoA•T is barely detectable in these extracts, which is in agreement with results obtained with the purified protein. In addition, mutagenesis studies in vitro and in vivo have shown that 8oxoA is weakly mutagenic in E. coli (4,6,7). Consistent with these observations, E. coli mug mutant does not exhibit a mutator phenotype under normal growth conditions (49). Interestingly, when measuring spontaneous mutation frequencies based on Miller’s lacZ reversion assay system, we observed in average a 3-fold increase in A•T→C•G transversions in E. coli mug mutant when compared with WT strain; however, this increase was not statistically significant under the experimental condition used (35). Subsequent studies have shown that MUG expression strongly depends on growth phase, being specifically expressed in stationary phase cells (50). Furthermore, its inactivation elevated the frequency of rifampicin-resistant mutants by 6-fold when cells were maintained in stationary phase for >2 days (50). Taken together, these observations suggest a potential role of MUG-catalysed removal of 8oxoA residues from DNA in preventing mutations in non-dividing E. coli cells.
The εC residues in DNA can be excised by three mammalian TDG, MBD4 and SMUG1 DNA glycosylases, although the two latter enzymes exhibit much weaker activity as compared with TDG (16,51,52). Repair of εC and 8oxoA residues in whole cell extracts from MEFs showed that cleavage of εC•G, 8oxoA•G and 8oxoA•T duplexes have an absolute requirement for the intact Tdg gene, indicating that in mouse cells TDG is the main DNA glycosylase excising εC and 8oxoA paired to G and T, respectively (Figure 4 and Supplementary Figure S4). In agreement with a previous study, we detected the cleavage of 8oxoA•C duplex in the cell extracts from MEFs Tdg−/−, suggesting that the mouse OGG1 excises 8oxoA paired with C (13) (Figure 4A and B). We also observed barely detectable cleavage of 8oxoA•A duplex in both WT and Tdg−/− cell extracts, which could be because of unidentified DNA glycosylase activities (Figure 4A). Overall, studies of the 8oxoA–DNA glycosylase activities with purified enzymes and mammalian cell-free extracts support the major role of TDG in the repair of 8oxoA residues paired with T and G in vivo.
The biological role of TDG in cellular response to oxidative stress conditions is supported by its ability to remove various oxidized bases generated by reactive oxygen species, including 5-hydroxymethyluracil (20), thymine glycol (17), 5-hydroxycytosine (18) and 8oxoA (present study) residues when present in DNA. However, in the present work, we showed that the cellular response of WT and Tdg−/− MEFs to high doses (10–20 Gy) of γ-irradiation was not dependent on the TDG status (Figure 5). Yet, compared with Tdg−/− MEFs, WT MEFs exhibited a somewhat better recovery after lower doses of γ-irradiation (1–5 Gy) after a prolonged recovery time, although this difference was not seen during the first 3 days after irradiation and also at higher doses of IR (Figure 5B–E).
Given the major role of TDG in the repair of mismatched T, εC and 8oxoA residues, one might expect that TDG deficiency would be associated with higher rate of C→T and A→C mutations and increased cancer risk. However, a recent study on the embryonic lethal phenotype of TDG knockout mice demonstrated that mutation frequencies in a Big Blue transgenic MEFs Tdg−/− were similar to that of WT MEFs, suggesting that the biological role of TDG in the repair of oxidized and/or deaminated DNA bases is minor (25). To explain the inconsistencies between biochemical data and genetic evidences, we may propose that MBD4, a second mammalian T•G-specific DNA glycosylase, can efficiently back up TDG for the repair of G•U and G•T mismatches, which originate in CpG contexts through deamination of C and 5mC, respectively. However, it should be stressed that there is no efficient back up enzyme to repair εC•G and 8oxoA•T pairs in the absence of TDG, and this might lead to accumulation of these lesions in cellular DNA of Tdg−/− MEFs. Interestingly, recently, it has been demonstrated that loss of TDG in tumour cells of mismatch-repair deficient patient resulted in high number of somatic base substitution mutations, providing the first in vivo evidence for the role of TDG in mutation prevention (53). In conclusion, we suggest that the physiological role of TDG in the defence against the mutagenic and toxic effects of εC and 8oxoA residues in DNA might be more important at tissue, organ and whole body level rather than in cultured cell lines. Further studies are required to elucidate the role of various DNA repair activities of TDG in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–4.
FUNDING
Agence Nationale pour la Recherche [ANR Blanc 2010 Projet ANR- 09-GENO-000 to M.S.] (http://www.agence-nationale-recherche.fr); Centre National de la Recherche Scientifique (http://www.cnrs.fr) [PICS N5479-Russie, CNRS-INCA-MSHE Franco-Pologne #3037987 to M.S.]; Electricité de France (http://www.edf.fr) Contrat Radioprotection [RB 2011 to M.S.]; Fondation de France (http://www.fondationdefrance.org) [#2012 00029161 to A.A.I.]; Fondation ARC pour la recherche sur le cancer (http://www.arc-cancer.net) (to I.T.) and Fondation pour la Recherche Medicale (http://www.frm.org) (to S.C.). Funding for open access charge: Agence Nationale pour la Recherche [ANR Blanc 2010 Projet ANR-09-GENO-000 to M.S.] (http://www.agence-nationale-recherche.fr).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Jacques Laval and Narayanan Venkatesan for critical reading of the manuscript and thoughtful discussions.
Footnotes
This paper was updated to remove a spelling error in the title.
REFERENCES
- 1.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Dizdaroglu M. Oxidatively induced DNA damage: mechanisms, repair and disease. Cancer Lett. 2012;327:26–47. doi: 10.1016/j.canlet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Bonicel A, Mariaggi N, Hughes E, Teoule R. In vitro gamma irradiation of DNA: identification of radioinduced chemical modifications of the adenine moiety. Radiat. Res. 1980;83:19–26. [PubMed] [Google Scholar]
- 4.Guschlbauer W, Duplaa AM, Guy A, Teoule R, Fazakerley GV. Structure and in vitro replication of DNA templates containing 7,8-dihydro-8-oxoadenine. Nucleic Acids Res. 1991;19:1753–1758. doi: 10.1093/nar/19.8.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonard GA, Guy A, Brown T, Teoule R, Hunter WN. Conformation of guanine-8-oxoadenine base pairs in the crystal structure of d(CGCGAATT(O8A)GCG) Biochemistry. 1992;31:8415–8420. doi: 10.1021/bi00151a004. [DOI] [PubMed] [Google Scholar]
- 6.Shibutani S, Bodepudi V, Johnson F, Grollman AP. Translesional synthesis on DNA templates containing 8-oxo-7,8-dihydrodeoxyadenosine. Biochemistry. 1993;32:4615–4621. doi: 10.1021/bi00068a019. [DOI] [PubMed] [Google Scholar]
- 7.Wood ML, Esteve A, Morningstar ML, Kuziemko GM, Essigmann JM. Genetic effects of oxidative DNA damage: comparative mutagenesis of 7,8-dihydro-8-oxoguanine and 7,8-dihydro-8-oxoadenine in Escherichia coli. Nucleic Acids Res. 1992;20:6023–6032. doi: 10.1093/nar/20.22.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamiya H, Miura H, Murata-Kamiya N, Ishikawa H, Sakaguchi T, Inoue H, Sasaki T, Masutani C, Hanaoka F, Nishimura S, et al. 8-Hydroxyadenine (7,8-dihydro-8-oxoadenine) induces misincorporation in in vitro DNA synthesis and mutations in NIH 3T3 cells. Nucleic Acids Res. 1995;23:2893–2899. doi: 10.1093/nar/23.15.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan X, Grollman AP, Shibutani S. Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2′-deoxyadenosine and 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA lesions in mammalian cells. Carcinogenesis. 1999;20:2287–2292. doi: 10.1093/carcin/20.12.2287. [DOI] [PubMed] [Google Scholar]
- 10.Girard PM, D’Ham C, Cadet J, Boiteux S. Opposite base-dependent excision of 7,8-dihydro-8-oxoadenine by the Ogg1 protein of Saccharomyces cerevisiae. Carcinogenesis. 1998;19:1299–1305. doi: 10.1093/carcin/19.7.1299. [DOI] [PubMed] [Google Scholar]
- 11.Grin IR, Dianov GL, Zharkov DO. The role of mammalian NEIL1 protein in the repair of 8-oxo-7,8-dihydroadenine in DNA. FEBS Lett. 2010;584:1553–1557. doi: 10.1016/j.febslet.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujikawa K, Kamiya H, Yakushiji H, Fujii Y, Nakabeppu Y, Kasai H. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem. 1999;274:18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- 13.Jensen A, Calvayrac G, Karahalil B, Bohr VA, Stevnsner T. Mammalian 8-oxoguanine DNA glycosylase 1 incises 8-oxoadenine opposite cytosine in nuclei and mitochondria, while a different glycosylase incises 8-oxoadenine opposite guanine in nuclei. J. Biol. Chem. 2003;278:19541–19548. doi: 10.1074/jbc.M301504200. [DOI] [PubMed] [Google Scholar]
- 14.Neddermann P, Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 15.Hang B, Medina M, Fraenkel-Conrat H, Singer B. A 55-kDa protein isolated from human cells shows DNA glycosylase activity toward 3,N4-ethenocytosine and the G/T mismatch. Proc. Natl Acad. Sci. USA. 1998;95:13561–13566. doi: 10.1073/pnas.95.23.13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saparbaev M, Laval J. 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc. Natl Acad. Sci. USA. 1998;95:8508–8513. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon JH, Iwai S, O’Connor TR, Pfeifer GP. Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg:G mispair. Nucleic Acids Res. 2003;31:5399–5404. doi: 10.1093/nar/gkg730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neddermann P, Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc. Natl Acad. Sci. USA. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters TR, Swann PF. Kinetics of the action of thymine DNA glycosylase. J. Biol. Chem. 1998;273:20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- 22.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 23.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 25.Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 26.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 30.Hang B, Downing G, Guliaev AB, Singer B. Novel activity of Escherichia Coli mismatch uracil-DNA glycosylase (Mug) excising 8-(hydroxymethyl)-3,N4-ethenocytosine, a potential product resulting from glycidaldehyde reaction. Biochemistry. 2002;41:2158–2165. doi: 10.1021/bi011542b. [DOI] [PubMed] [Google Scholar]
- 31.Saparbaev M, Langouet S, Privezentzev CV, Guengerich FP, Cai H, Elder RH, Laval J. 1,N(2)-ethenoguanine, a mutagenic DNA adduct, is a primary substrate of Escherichia Coli mismatch-specific uracil-DNA glycosylase and human alkylpurine-DNA-N-glycosylase. J. Biol. Chem. 2002;277:26987–26993. doi: 10.1074/jbc.M111100200. [DOI] [PubMed] [Google Scholar]
- 32.O’Neill RJ, Vorob’eva OV, Shahbakhti H, Zmuda E, Bhagwat AS, Baldwin GS. Mismatch uracil glycosylase from Escherichia Coli: a general mismatch or a specific dna glycosylase? J. Biol. Chem. 2003;278:20526–20532. doi: 10.1074/jbc.M210860200. [DOI] [PubMed] [Google Scholar]
- 33.Couve-Privat S, Mace G, Rosselli F, Saparbaev MK. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007;35:5672–5682. doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gelin A, Redrejo-Rodriguez M, Laval J, Fedorova OS, Saparbaev M, Ishchenko AA. Genetic and biochemical characterization of human AP endonuclease 1 mutants deficient in nucleotide incision repair activity. PLoS One. 2010;5:e12241. doi: 10.1371/journal.pone.0012241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jurado J, Maciejewska A, Krwawicz J, Laval J, Saparbaev MK. Role of mismatch-specific uracil-DNA glycosylase in repair of 3,N(4)-ethenocytosine in vivo. DNA Repair (Amst.) 2004;3:1579–1590. doi: 10.1016/j.dnarep.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Kunz C, Focke F, Saito Y, Schuermann D, Lettieri T, Selfridge J, Schar P. Base excision by thymine DNA glycosylase mediates DNA-directed cytotoxicity of 5-fluorouracil. PLoS Biol. 2009;7:e91. doi: 10.1371/journal.pbio.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 38.Morgan MT, Bennett MT, Drohat AC. Excision of 5-Halogenated uracils by human thymine DNA glycosylase: robust activity for dna contexts other than CpG. J. Biol. Chem. 2007;282:27578–27586. doi: 10.1074/jbc.M704253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Neill RJ, Vorob’eva OV, Shahbakhti H, Zmuda E, Bhagwat AS, Baldwin GS. Mismatch uracil glycosylase from Escherichia Coli: a general mismatch or a specific dna glycosylase? J. Biol. Chem. 2003;278:20526–20532. doi: 10.1074/jbc.M210860200. [DOI] [PubMed] [Google Scholar]
- 40.Fitzgerald ME, Drohat AC. Coordinating the initial steps of base excision repair. Apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex. J. Biol. Chem. 2008;283:32680–32690. doi: 10.1074/jbc.M805504200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sibghat U, Gallinari P, Xu YZ, Goodman MF, Bloom LB, Jiricny J, Day RS., 3rd Base analog and neighboring base effects on substrate specificity of recombinant human G:T mismatch-specific thymine DNA-glycosylase. Biochemistry. 1996;35:12926–12932. doi: 10.1021/bi961022u. [DOI] [PubMed] [Google Scholar]
- 42.Grin IR, Vasilyeva SV, Dovgerd AP, Silnikov VN, Zharkov DO. Human and bacterial DNA polymerases discriminate against 7,8-dihydro-8-oxo-2′-deoxyadenosine-5′-triphosphate. Biopolym. Cell. 2012;28:306–309. [Google Scholar]
- 43.Eftedal I, Guddal PH, Slupphaug G, Volden G, Krokan HE. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993;21:2095–2101. doi: 10.1093/nar/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellamy SR, Baldwin GS. A kinetic analysis of substrate recognition by uracil-DNA glycosylase from herpes simplex virus type 1. Nucleic Acids Res. 2001;29:3857–3863. doi: 10.1093/nar/29.18.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett TE, Scharer OD, Savva R, Brown T, Jiricny J, Verdine GL, Pearl LH. Crystal structure of a thwarted mismatch glycosylase DNA repair complex. EMBO J. 1999;18:6599–6609. doi: 10.1093/emboj/18.23.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc. Natl Acad. Sci. USA. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong L, Mi R, Glass RA, Barry JN, Cao W. Repair of deaminated base damage by Schizosaccharomyces pombe thymine DNA glycosylase. DNA Repair (Amst.) 2008;7:1962–1972. doi: 10.1016/j.dnarep.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 49.Lutsenko E, Bhagwat AS. The role of the Escherichia Coli mug protein in the removal of uracil and 3,N(4)-ethenocytosine from DNA. J. Biol. Chem. 1999;274:31034–31038. doi: 10.1074/jbc.274.43.31034. [DOI] [PubMed] [Google Scholar]
- 50.Mokkapati SK, Fernandez de Henestrosa AR, Bhagwat AS. Escherichia Coli DNA glycosylase Mug: a growth-regulated enzyme required for mutation avoidance in stationary-phase cells. Mol. Microbiol. 2001;41:1101–1111. doi: 10.1046/j.1365-2958.2001.02559.x. [DOI] [PubMed] [Google Scholar]
- 51.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Genuardi M, Karbowski M, Yeung AT, Matsumoto Y, Bellacosa A. Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): fundamental role of the catalytic domain. J. Cell. Physiol. 2000;185:473–480. doi: 10.1002/1097-4652(200012)185:3<473::AID-JCP19>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 52.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 53.Vasovcak P, Krepelova A, Menigatti M, Puchmajerova A, Skapa P, Augustinakova A, Amann G, Wernstedt A, Jiricny J, Marra G, et al. Unique mutational profile associated with a loss of TDG expression in the rectal cancer of a patient with a constitutional PMS2 deficiency. DNA Repair (Amst.) 2012;11:616–623. doi: 10.1016/j.dnarep.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.