Abstract
Alternative lengthening of telomeres (ALT) is one of the two known telomere length maintenance mechanisms that are essential for the unlimited proliferation potential of cancer cells. Existing methods for detecting ALT in tumors require substantial amounts of tumor material and are labor intensive, making it difficult to study prevalence and prognostic significance of ALT in large tumor cohorts. Here, we present a novel strategy utilizing telomere quantitative PCR to diagnose ALT. The protocol is more rapid than conventional methods and scrutinizes two distinct characteristics of ALT cells concurrently: long telomeres and the presence of C-circles (partially double-stranded circles of telomeric C-strand DNA). Requiring only 30 ng of genomic DNA, this protocol will facilitate large-scale studies of ALT in tumors and can be readily adopted by clinical laboratories.
INTRODUCTION
Telomeres, specialized structures of repetitive DNA sequence (5′-TTAGGG-3′) located at the ends of chromosomes, undergo progressive shortening in replicating cells, which prevents the unlimited proliferation of normal somatic cells. Cancer cells evade this barrier and become immortalized by activating one of the two known telomere length (TL) maintenance mechanisms. These are the telomerase enzyme and alternative lengthening of telomeres (ALT), which synthesize new telomeric DNA from an RNA template via reverse transcription and from a DNA template via homologous recombination-mediated DNA replication, respectively (1). Telomerase activity (TA) is most commonly measured by the PCR-based telomere repeat amplification protocol, and a large number of cancers have been screened for TA by this method (2). In the absence of an enzyme activity assay for ALT, ALT has been detected in tumors by observing telomere-related phenotypic characteristics that have previously been documented in immortalized TA-negative cell lines (3). However, detecting ALT in tumors is significantly more challenging than in cell lines as most assays used to identify these phenotypic features in cell lines cannot be applied to tumors. The ability to diagnose ALT efficiently in clinical settings will facilitate the appropriate selection of telomere maintenance mechanism-targeted drugs in the clinic.
There are currently two main methods for detecting ALT in tumors. The first of these is terminal restriction fragment (TRF) Southern-blot analysis of TL profile. In ALT cells, telomeres range from very short to extremely long in individual cells, with mean TL (>20 kb) being about twice of that of TA-positive or normal somatic cells (5–10 kb) (4). ALT cells are therefore described as having long and heterogeneous telomeres as compared to the shorter and more homogeneous telomeres of ALT-negative cells. The second method is detection by a combined promyelocytic leukemia (PML) immunofluorescence/telomere fluorescence in situ hybridization (FISH) analysis of tumor sections for ALT-associated PML bodies (APBs) (5). APBs are PML bodies that contain telomeric DNA and telomere binding proteins (6). Both of these conventional assays are labor-intensive, making screening of large numbers of tumors challenging. Recently, a variation of the APB assay in which telomere FISH was used to detect ultra-bright telomeric signals was used to analyze more than 6000 tumor specimens, mostly in tissue microarray format (7).
An assay, which reliably detects ALT in cell lines by quantitating C-circles (CC) (8), is currently being assessed for its suitability for detecting ALT in tumors. CC are extra-chromosomal circles of telomeric DNA that are partially single-stranded, where the C-rich strand is complete and the G-rich strand is gapped. The presence of abundant CC is ALT-specific, and CC levels were shown to have a quantitative relationship to ALT activity (8). The CC assay is an isothermic polymerase reaction, where CC act as self-priming DNA templates in rolling circle amplification, producing long linear telomeric single-stranded DNA (ssDNA) products. These products are then quantitated by dot-blot analysis using a 32P-labeled telomeric probe (8).
In this study, a strategy utilizing TL measurement by quantitative PCR (qPCR) was developed and evaluated for detecting ALT in cancer cell lines and tumors. While telomere qPCR has been widely applied as a high throughput technique in population studies to measure TL in leukocyte DNA and is capable of detecting small differences (9–11), it has not been used previously to detect ALT. Here, we demonstrated that telomere qPCR could be employed to quantify both TL and CC concurrently with just 30 ng of DNA and in significantly less time than is required for TRF and APB analysis. Furthermore, we showed that examining two ALT characteristics (TL and CC) concurrently in this assay significantly improved ALT detection in tumors, compared with evaluating just one feature, as 50% of the ALT tumors were found to be positive for only one of the two characteristics.
MATERIALS AND METHODS
Samples
Frozen tumor samples were acquired with institution ethics committee approval. The 23 cell lines include CHLA-90, CHP-100, CHP-134, G-292, GM847, IIICF/c, LAN-2, LAN-5, MCF-7, MDA-MB-231, MeT-4A, MG-63, NB69, NM39, NM179, SK-LU-1, SK-N-AS, SK-N-FI, SH-SY5Y, SK-N-BE2, SK-N-DZ, U-2 OS, and W-V. SK-N-FI, U-2 OS, and G-292 used for calibration measurements were purchased from ECACC at passage 37, +4 and 24, respectively. The cell lines are described in the Supplementary Table S1.
DNA extraction and quantitation
Genomic DNA from both cell lines and tumors was extracted by lysis at 37°C with 2% sodium dodecyl sulfate (SDS) buffer containing 50 mM Tris, 20 mM ethylenediaminetetraacetic acid (EDTA) and 200 µg/ml Pronase protease (Sigma), followed by precipitation with 5 M sodium chloride and ethanol. DNA was quantitated with a Qubit Fluorometer (Invitrogen).
TRF analysis
TL measurement by TRF Southern-blot analysis was performed as previously described (12). Briefly, genomic DNA was digested with HinfI and RsaI restriction enzymes (4 U/µg DNA) [New England Biolabs (NEB)] and 25 ng/µg RNase (Roche) and resolved on a 1% agarose gel using pulsed-field gel electrophoresis. The dried gel was denatured, neutralized and hybridized to an end-labeled 32P-(CCCTAA)4 telomeric probe. The phosphor screen was scanned on a Typhoon Trio Variable Mode Imager (GE Healthcare) and signal intensity quantitated using the LAS 4000 Multi Gauge software.
CC assay
Rolling circle amplification of CC was performed as described (8). A 10 μl CC reaction contains 0.2 µg/µl bovine serum albumin, 0.1% Tween, 4 µM dithiothreitol (DTT), 1 mM each dATP, dCTP, dGTP, dTTP, φ29 DNA polymerase (φ29, 3.75 U/16 ng DNA) (NEB), 1× φ29 buffer and 16 ng genomic DNA, and was incubated at 30°C for 8 h then at 65°C for 20 min. For each sample, the assay was done with and without φ29.
To quantitate CC assay products by slot-blot analysis, 32 ng of DNA was used for each CC assay. The 20 µl CC assay product was then diluted with 100 µl of 2× saline-sodium citrate (SSC) and slot-blotted in duplicate onto a Biodyne B nylon membrane (Pall). After UV-cross-linking, the membrane was hybridized overnight at 37°C with end-labeled 32P-(CCCTAA)3 telomeric probe in PerfectHyb Plus buffer (Sigma) in native (non-denatured) condition. The membrane was then washed four times at 37°C in 0.5× SSC/0.1% SDS buffer. The phosphor screen was scanned on a Typhoon Trio Variable Mode Imager (GE Healthcare) scanner and signal intensity was quantitated using the ImageQuant software.
Quantitative PCR
The CC assay was performed with and without φ29. For each sample, 10 µl of the CC assay product containing 16 ng of input genomic DNA was diluted with 30 µl of Tris–EDTA (10 mM Tris, 0.1 mM EDTA, pH 7.6), of which 5 µl containing 2 ng of input genomic DNA was used for each PCR. Each sample required 12 PCRs: triplicate telomere PCR of CC assay/φ29+, triplicate telomere PCR of CC assay/φ29−, triplicate single copy gene (SCG) PCR of CC assay/φ29+ and triplicate SCG PCR of CC assay/φ29−. To generate standard curves, serially diluted genomic DNA of the ALT+ CHLA-90 cell line was subjected to the CC assay with φ29 for telomere PCR and without φ29 for SCG PCR. Six concentrations of duplicate standards, ranging from 0.0013 to 1 ng/µl for telomere PCR and from 0.0063 to 2 ng/µl for SCG PCR, were included.
Each 25 µl qPCR consisted of 1× QuantiTECT SYBR Green master mix (Qiagen), 10 mM DTT, 0.5 µl dimethyl sulfoxide, 5 µl DNA template and primer sets. The final primer concentrations were: (i) telomere: forward 300 nM and reverse 400 nM, (ii) VAV2: forward 700 nM and reverse 400 nM and (iii) 36B4: forward 300 nM and reverse 500 nM. The primer sequences (5′ to 3′) were: (i) telomere: forward GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT and reverse TCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA (9), (ii) 36B4: forward CAGCAAGTGGGAAGGTGTAATCC and reverse CCATTCTATCATCAACGGGTACAA (9) and (iii) VAV2: forward TGGGCATGACTGAAGATGAC and reverse ATCTGCCCTCACCTTCTCAA. The 36B4 (RPLP0) gene, located on chromosome 12q24, encodes acidic ribosomal phosphoprotein P0 and VAV2, located on chromosome 9q34, encodes guanine nucleotide exchange factor. 36B4, which has been applied as a SCG for both cancer (13) and non-cancer cells (9,11), was used for melanoma tumors and cell lines, as well as glioblastoma multiforme (GBM) tumors and soft tissue sarcomas (STS). Comparative Genomic Hybridization studies indicate that 36B4 is an appropriate SCG for melanoma (14) and GBM (15). VAV2 was used for neuroblastoma (NB) tumors and all cell lines except for melanoma. VAV2 has previously been validated as a SCG for NB (16), whereas 36B4 is in a region of the genome where a low level of gene amplification has been reported in NB (17).
All PCRs were performed with the Rotor-Gene Q (Qiagen) real-time cycler. PCR conditions were: (i) telomere: 95°C for 15 min, 33 cycles of 95°C for 15 s and 54°C for 2 min, (ii) VAV2: 95°C for 15 min, 40 cycles of 95°C 15 s, 57°C 30 s and 72°C 1 min and (iii) 36B4: 95°C 15 min, 35 cycles of 95°C 15 s and 58°C 1 min. Rotor-Gene Q series software was used to obtain data and generate standard curves.
Data analysis
The unweighted mean TRF length was calculated as ∑(ODi × Li)/∑(ODi), where ODi is the signal intensity above background within interval i and Li is the molecular weight (kb) at the mid-point of interval i. A sample was considered TRF+ (ALT+ by TRF) when mean TRF length was ≥16 kb and semi-interquartile range (SIR) was ≥4 kb (5). SIR of the TRF lengths was calculated as (3rd quartile–1st quartile)/2. For CC assay product quantitation by slot-blot analysis, CC assay level was calculated by subtracting the 32P-probe signal of the CC assay without φ29 from that of the CC assay with φ29. Results were expressed as average of duplicates and as arbitrary units (AU).
qPCR data were analyzed using the two standard curves method as previously described (9). Telomere product was normalized (norm-TEL) with the SCG product. The relative telomeric DNA content (TC) of a sample was the norm-TEL obtained from the CC assay without φ29 (norm-TEL/φ29−) and was relative to the TC of the ALT-positive U-2 OS cell line with an arbitrary value of 40. The relative CC assay level of a sample was calculated as [(norm-TEL/φ29+) − (norm-TEL/φ29−)] and was relative to the CC assay level of U-2 OS cell line with an arbitrary value of 170. A sample was only considered positive for CC when the norm-TEL value of each individual φ29(+) triplicate was higher than the norm-TEL of all three of the φ29(−) replicates and the calculated CC assay level was ≥cut-off value. All PCR results were expressed as mean of triplicate reactions ±SEM.
Means were compared using unpaired two-tailed Student’s t-test and medians compared by the Mann–Whitney test. Linear regression was analyzed using Prism 4 software (GraphPad). Sensitivity was the number of samples that tested positive divided by the number of samples that were classified as positive by the ‘gold standard’. Specificity was the number of samples that were tested negative divided by the number of samples that were classified as negative by the gold standard. TRF analysis was the gold standard when the performance of telomere qPCR was compared with TRF and 32P detection method the gold standard when telomere qPCR was compared with 32P for quantitation of CC assay level. Cut-off values were chosen to provide the highest concordance rate (both tests in agreement) and the best separation between the test-positive and test-negative group.
RESULTS
Distinguishing ALT+ and ALT− samples by TC
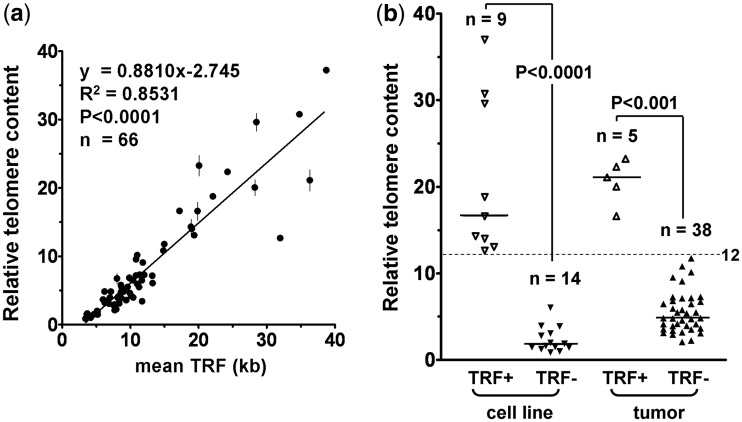
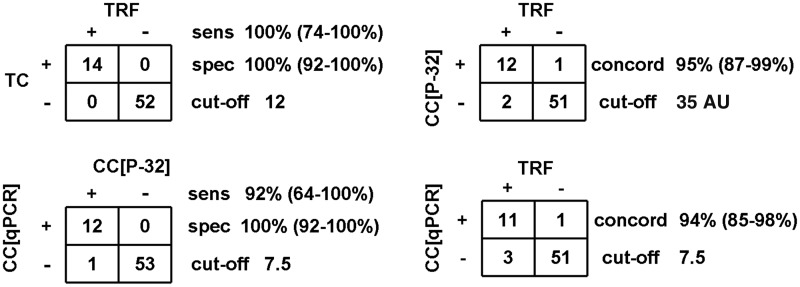
In this study, 66 samples, consisting of 23 cell lines and 43 tumors (18 NB, 15 GBM and 10 melanomas), were analyzed initially. The telomere maintenance mechanism status has previously been published for 16 of the cell lines (Supplementary Table S1). TRF Southern-blot analysis was used to measure TL. As previously described, a sample was classified as ALT+ by TRF when the mean TRF length is ≥16 and SIR ≥4 kb (5). TC was measured by telomere qPCR. We found a very good correlation (R2 = 0.8531; P < 0.0001) between TC and mean TRF (Figure 1a). Importantly, samples that were ALT+ by TRF (n = 14) had significantly higher relative TC than TRF− (ALT− by TRF) samples (n = 52; median 19.4 versus 4.1; P < 0.0001). Using a cut-off of 12.0, TC separated the samples distinctly into TRF+ and TRF− groups (Figure 1b), with a sensitivity of 100% (95% CI: 74–100%) and specificity of 100% (95% CI: 92–100%) (Figure 2).
Figure 1.
Analysis of telomeric DNA content (TC) detects the long telomeres of ALT+ samples. Relative TC was determined by telomere qPCR and mean TRF length by TRF analysis in 23 cell lines and 43 tumor samples. (a) Linear regression demonstrates correlation between relative TC (mean ± SEM, n = 3) and mean TRF length. The TC of a sample is relative to the TC of the ALT+ U-2 OS cell line (arbitrary value of 40). (b) Relative TC levels are plotted according to whether or not TRF analysis determined that the samples are ALT-like (TRF+ or TRF−). The median TC (horizontal bar) is significantly higher in TRF+ than in TRF− cell lines (16.6 versus 1.7) and tumors (21.1 versus 4.8). Dotted lines indicate the relative TC cut-off (12.0) for distinguishing ALT+ from ALT− samples.
Figure 2.
Sensitivity, specificity and concordance of telomeric DNA content (TC) and CC assay level for ALT detection. Sensitivity (sens) and specificity (spec) of qPCR for TC and CC detection were determined using TRF and CC[P-32] assay as the gold standard, respectively. A sample was regarded as TRF+ for ALT when mean TRF ≥16 kb and SIR ≥4 kb and CC+ when CC[P-32] ≥35AU. The indicated cut-off values for TC and CC[qPCR] separated samples into test-positive and test-negative groups. The concordance (concord) rate refers to the proportion of samples where both tests were in agreement.
Detection of CC assay products by radioactive telomeric probe
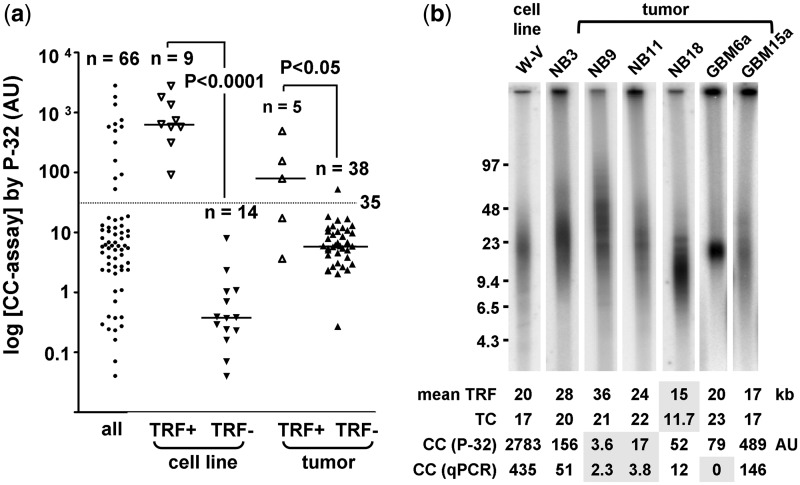
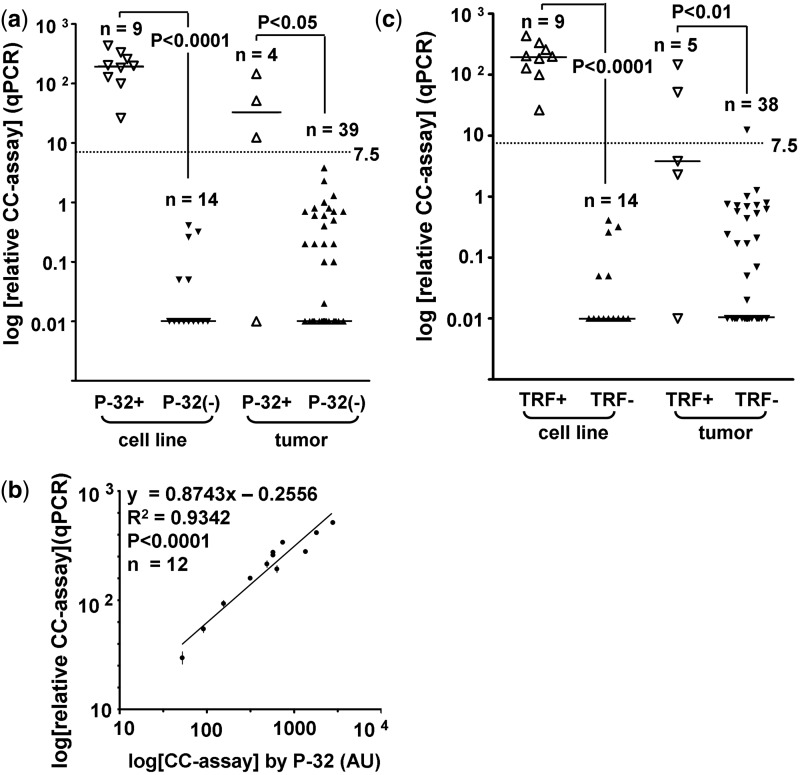
The CC assay is the first ALT assay that appears to have a quantitative correlation with ALT activity levels (8). The CC assay utilizes φ29 DNA polymerase to produce long G-rich telomeric ssDNA through rolling circle amplification of telomeric CC that are abundant in ALT cells. CC assay products are routinely detected by a 32P-labeled telomeric probe in a dot-blot analysis. We found that TRF+ samples (n = 14) had significantly higher CC assay levels than the TRF− samples (n = 52) (median 532 versus 5.1 AU; P < 0.0001). The CC assay levels of all samples were distributed into two clusters that corresponded well to the TRF status (Figure 3a). Ninety-two percent (12 of 13) of the samples in the upper cluster were TRF+ and 96% (51 of 53) in the lower cluster were TRF−. The cut-off for ALT detection was set at 35 AU, which represents the mid-point of the interval between the smallest value of the upper cluster and the largest value of the lower cluster (Figure 3a). With a cut-off of 35.0 AU, the CC assay by 32P detection (CC[32P]) had a 95% concordance with the TRF assay (95% CI: 87–99%; Figures 2 and 3a). The three discordant samples were NB tumors. NB9 and NB11 that were TRF+/CC− had CC assay levels in the range of TRF− samples, despite having high relative TC (average 21.7) and a high mean TRF (average 30.3 kb) (Figure 3b). This suggests that not all TRF+ samples are CC+. The one TRF−/CC+ sample (NB18) had borderline mean TRF (14.9 kb)/TC (11.7) and CC[32P] assay level of 52.4 AU (Figure 3b).
Figure 3.
Correlation between TRF and CC assays. (a) Genomic DNA of 66 samples was subjected to CC assay followed by detection with 32P labeled telomeric probe in slot-blot analysis. CC assay level was plotted on a logarithmic scale according to TRF status. The median CC assay level (horizontal bar) was significantly higher in TRF+ than in TRF− cell lines (636 versus 0.4 AU) and tumors (79 versus 6.0 AU). The cut-off (35.0 AU) is indicated by the dotted line. (b) TRF analysis of samples with different telomere length and CC profile. Numbers that are shaded are values below the cut-off. The cut-off is 16 kb for mean TRF, 12 for relative telomeric DNA content (TC), 35AU for CC[32P] and 7.5 for CC[qPCR].
Correlation between detection of CC assay products by 32P and telomere qPCR
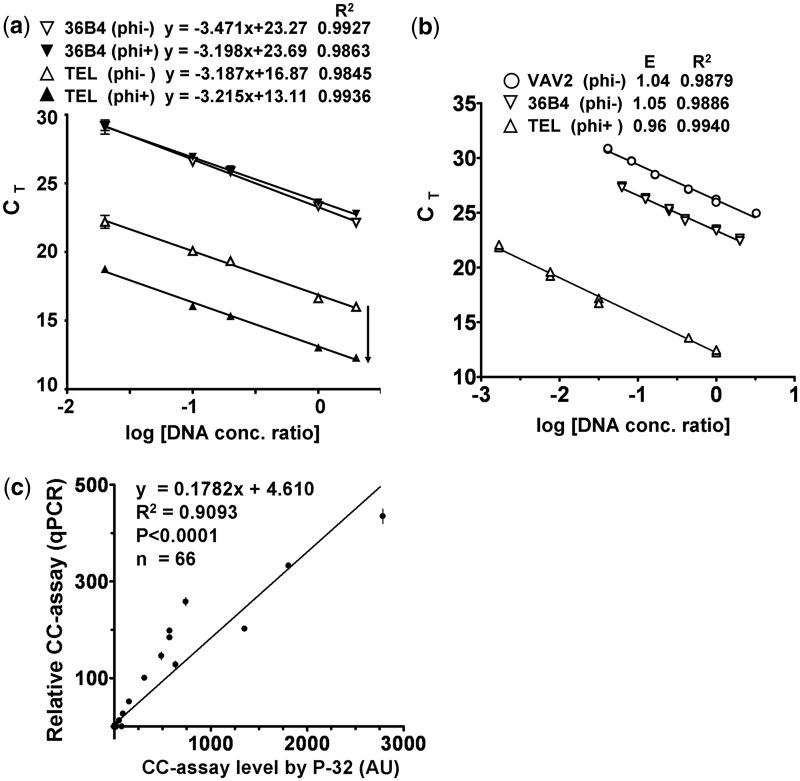
We next examined whether CC assay products could be measured by telomere qPCR (CC[qPCR]). Genomic DNA was subjected to CC assay with and without φ29. The TC present in the CC assay with no added φ29 reflects the TC of the input genomic DNA since there is no rolling circle amplification. In comparison, the TC derived from the CC assay with φ29 reflects both TC from the input genomic DNA and the G-rich linear telomeric ssDNA synthesized from rolling circle amplification of CC. Therefore, the difference in TC between the paired φ29(+) and φ29(−) samples represents CC assay products. As shown by the standard curves in Figure 4a, the presence of φ29 results in the same reduction in CT value over a 100-fold range of DNA concentrations for the telomere qPCR, but not for the 36B4 SCG qPCR. The robust standard curves demonstrating PCR amplification efficiencies close to 100% with excellent correlation (Figure 4a and b) support the use of telomere qPCR for measuring TC and CC assay products. Furthermore, there was very good correlation between CC[32P] and CC[qPCR] measurements (R2 = 0.9093; P < 0.0001; Figure 4c).
Figure 4.
Correlation between CC assay levels detected by 32P-labeled probe or by telomere qPCR. (a) Five concentrations (0.01, 0.05, 0.1, 0.5, 1 ng/µl) of DNA from the ALT+ cell line CHLA-90 were subjected to the CC assay with and without φ29 DNA polymerase (φ29), followed by telomere and 36B4 qPCR. The addition of φ29 to the CC assay resulted in a parallel downward shift of the telomere (TEL) standard curves, but not for the 36B4 single copy gene. Each data point of the standard curves is the average ± range (n = 2). (b) qPCR standard curves for telomere and the two single copy genes (36B4 and VAV2), as well as the corresponding PCR efficiency (E) and R2 are shown. (c) A total of 66 samples were subjected to the CC assay, followed by detection with 32P or telomere qPCR. The relative CC assay level is the difference in telomeric DNA content between the φ29+ and φ29− paired samples and is relative to that of the ALT+ U-2 OS cell line (arbitrary value of 170). Linear regression demonstrates correlation between the two detection methods. The relative CC assay level by qPCR is plotted as mean ± SEM, n = 3.
Telomere qPCR detects CCs in ALT cells
For CC assay level measurements, we compared the performance of CC[qPCR] with CC[32P] in identifying TRF+ samples. Samples that were CC[32P]+ (CC assay level ≥35 AU) had significantly higher CC[qPCR] assay levels than CC[32P]− samples (median 146 versus 0; P < 0.0001) (Figure 5a). The only discrepancy between the two methods occurred with a GBM tumor (GBM 6a) that was CC[qPCR]−, but had a CC[32P] assay level of 79 AU and mean TRF of 20.1 kb/SIR 8.0 kb (Figure 3b). The good correlation (R2 = 0.9342) between the two methods was maintained when the analysis was applied to the 12 samples that were CC[qPCR]+ (CC assay level ≥7.5) (Figure 5b). In addition, samples that were TRF+ had significantly higher CC[qPCR] assay levels than TRF− samples (median 137 versus 0; P < 0.0001) (Figure 5c). Applying a cut-off of 7.5, CC[qPCR] has a concordance of 94% (95% CI: 85–98%) with the TRF assay for ALT, which is similar to CC[32P] detection [concordance of 95% (95% CI: 87–99%); Figure 2].
Figure 5.
Detection of CC by φ29 and telomere qPCR compared to 32P-labeled probe. (a and c) Mean relative CC[qPCR] assay level was plotted according to 32P and TRF status. A sample was regarded as CC[32P]+ when the level was ≥35AU. For the purpose of plotting negative data points on a logarithmic scale, negative or zero values were assigned a value of 0.01. The median CC assay level (horizontal bar) was significantly higher in CC[32P]+ than in CC[32P]− cell lines (198 versus 0) and tumors (32 versus 0). The mean CC[qPCR] assay level is significantly higher in TRF+ than in TRF− cell lines (198 versus 0) and tumors (3.8 versus 0). The cut-off (7.5) is indicated by the dotted line. (b) Linear regression demonstrates good correlation between CC assay levels by qPCR and by 32P in the 12 CC[qPCR]+ samples (level ≥7.5). Data were plotted as mean ± SEM, n = 3.
Validation and calibration of the assay
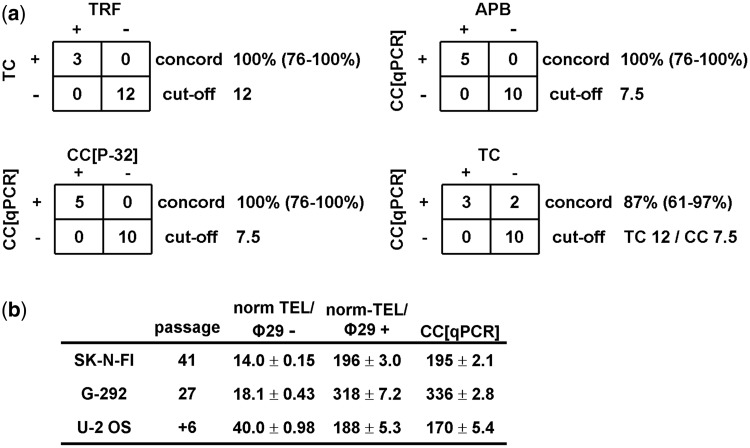
To validate the assay, we applied the cut-offs for relative TC and CC[qPCR] to an independent cohort of 15 STS. The ALT status of these STS samples has previously been determined in frozen sections using the APB assay (5). There was 100% (95% CI: 76–100%) concordance between TRF and TC, as well as between CC[32P] and CC[qPCR] (Figure 6a). Of these 15 samples, 5 were CC+ with 100% (95% CI: 76–100%) concordance between APB and CC[qPCR] status (Figure 6a). Of note, the sample that was found to have low frequency of APBs also had the lowest CC[qPCR] assay level of 9.4 in this study. Of the five STS that were CC+, three were TRF/TC+ and the remaining two were TRF/TC− (TRF 14.0 kb/TC 10.3/CC 261 and TRF 7.4 kb/TC 4.9/CC 9.4). This suggests not all CC+ samples have very long telomeres.
Figure 6.
Validation and calibration of the ALT detection assay. (a) Concordance of telomeric DNA content (TC) and CC assay level for ALT detection in the validation cohort of 15 soft tissue sarcomas (STS). The cut-offs for TRF, TC and CC assay levels are the same as in Figure 2. The concordance (concord) rate refers to the proportion of samples where both tests were in agreement. 95% confidence intervals are shown in parentheses. (b) Calibration measurements including norm-TEL/φ29+, norm-TEL/φ29− and the calculated CC[qPCR] level (as described in ‘Materials and Methods’ section) for U-2 OS, SK-N-FI and G-292 cell lines are shown. VAV2 was used as the single copy gene to normalize telomere PCR product. PCRs were done in triplicate and three independent experiments were performed. Results for norm-TEL/φ29+ and norm-TEL/φ29− are expressed as the mean of the three independent experiments ±SEM. For CC[qPCR], the CC assay level relative to U-2 OS was first calculated for each of the independent experiments and the mean of the three experiments ±SEM was then obtained.
Furthermore, we have provided calibration parameters derived from three commercially available ALT cell lines (SK-N-FI, U-2 OS and G-292) at specific passages to facilitate this qPCR-based assay being used by other laboratories (Figure 6b).
DISCUSSION
ALT is one of the two known telomere maintenance mechanisms, which play an essential role in the immortalization of cancer cells. While the prevalence and prognostic implications of TA have been reported for a large number of human cancers (2), equivalent data are not yet available for ALT. This is primarily because conventional methods for detecting ALT in tumors are labor-intensive. In addition, the quantity of DNA required for TRF analysis may exceed what is available, especially when the tumor of interest is not treated initially by resection and only a small amount of biopsy material is obtained for clinical diagnosis. A less laborious detection method that requires significantly less tumor material is needed for studying ALT in human cancers.
In this study, we developed a new strategy based on the well-established telomere qPCR and the more recently described CC assay to detect two characteristics of ALT in a cohort of 81 samples. It is a two-step procedure whereby genomic DNA is first subjected (or not) to rolling circle amplification by φ29, followed by telomere qPCR analysis of the paired amplified/non-amplified samples. By this means, both the TC and the CC[qPCR] assay level can be obtained from 30 ng genomic DNA. We demonstrated good correlation between TC and mean TRF length, as has been reported in population studies (9). We use the term TC, instead of TL, in this context because in ALT cells the abundant extra-chromosomal telomeric repeats also serve as templates for the qPCR. TC distinguished TRF+ and TRF− samples with 100% sensitivity and specificity and therefore can substitute for conventional TRF Southern blots. In addition to consuming large amounts of DNA (1–2 µg), TRF analysis can yield a falsely high mean TRF if care is not taken to obtain adequate restriction enzyme digestion of the genomic DNA.
To obtain sufficient sensitivity, CC assay product (long linear telomeric ssDNA) has previously been detected with 32P-labeled probes (8) and with this technique assaying duplicate samples with and without φ29 amplification requires 120 ng of DNA, compared with 30 ng for the CC[qPCR] detection method. The correlation between the two quantitation methods was very good. There was one discrepant result in the entire cohort: a GBM tumor that was considered to be CC[32P]+ and CC[qPCR]−. This tumor had a 9-fold lower CC[32P] assay level than the mean CC[32P] level of TRF+ samples (79 versus 693 AU), while its relative TC of 23.2 was similar to the mean TC (21.6) of TRF+ samples. This discrepancy between the two detection methods may be related to the difficulty in telomere PCR detecting very low levels of CC due to interference from the high levels of double-stranded TC. The low amount of CC assay telomeric product representing only a very small proportion of the high TC may lead to insignificant differences between the paired φ29(+) and φ29(−) samples. It is also relevant to note that this tumor had a somewhat atypical TRF profile with a mean TRF of 20 kb, but with much less heterogeneity (lowest SIR of 8 kb) than the other 13 ALT-positive samples in this study which had a mean SIR of 17.7 kb (range: 11.1–23.4 kb) (Figure 3b). Overall, qPCR is a reliable quantitation method for CC assay levels and has advantages over the original CC assay protocol (8) as this assay requires less DNA, takes less time and does not involve radio-isotope.
To validate the cut-offs for TC and CC[qPCR], we applied them to an independent cohort of 15 STS with known APB status (5). There was 100% concordance between TRF and TC, between CC[32P] and CC[qPCR] and between APB and CC[qPCR]. This verifies these cut-offs were appropriately set. We found discrepancies between TRF/TC and CC assays for ALT in both the original and validation cohort. Such discrepancies have also been reported in cell line studies. Telomerase overexpression in ALT− cells can cause long heterogeneous telomeres (18) while short telomeres have been observed in an ALT+ cell line (19). In both cases, the CC assay was the more accurate determinant of ALT status (8). This study is the first report of tumors with long and heterogeneous TL but without elevated levels of CC. We found this in two of the four TRF+ NB tumors and both of these TRF+/CC− NB were APB negative. Conversely, we found a STS in the validation cohort that was CC+ without long telomeres (TRF 7.4 kb/TC 4.9/CC 9.4). This STS was previously reported to have low APB frequency, which correlates well with the low CC[qPCR] assay level. There appears to be a good correlation between APB and CC for ALT detection; however, the APB assay is significantly more labor intensive. The CC assay has been studied extensively in cell lines, but validation of the accuracy of the CC assay in a variety of tumor types is needed.
By combining two tests evaluating different ALT characteristics in this protocol, the problem of equivocal results can also be minimized. For example, in this study, two tumors (NB and STS) with equivocal mean TRF/TC have unequivocally positive CC[qPCR] levels of 52 and 261, respectively. Assessing embryonal tumors such as NB for ALT by TRF or TC can be particularly challenging as the separation between the two groups is often not as distinct as in adult tumors, possibly because less telomere attrition occurs during the genesis of embryonal tumors. In NB, we found that ALT− samples had an average TC of 7.3 and TRF of 11.2 kb, compared with GBM and melanoma that had much lower average TC (4.4) and TRF (8.5 kb). Furthermore, of the 58 tumor samples analyzed, six have TC in the mid-range (TC 8–12), of which two were CC-positive and four were CC-negative. The CC assay is therefore required to segregate tumors with mid-TC levels, which can represent 10% of tumor samples.
Evaluating two characteristics simultaneously also increases both the sensitivity and specificity for ALT detection. In this study, we identified 11 tumors that had TC and/or CC[qPCR] levels above the cut-off for ALT (six from initial cohort and five from validation cohort). However, only five samples had both TC and CC[qPCR] levels above the cut-off. The other six samples had only one parameter above the cut-off (three TC+/CC−; three TC−/CC+). Although the significance of only one ALT characteristic being positive is not yet clear, the data suggest that examining only one ALT characteristic can potentially miss 25–30% of ALT+ tumors.
The accuracy of qPCR using genomic DNA as the DNA template relies on appropriate choice of SCG. SCG normalizes for input DNA amount and PCR efficiency. 36B4, albumin and β globin are the most widely used SCG in non-cancerous cells (10,20). However, selecting SCG in cancer cells can be challenging as gene copy number variations are common in cancer cells and there are significant differences in affected chromosome regions among different cancers. Fortunately, comparative genomic hybridization array data are now widely available for most tumor types and can direct the selection of SCG. Care must also be taken when evaluating the quantity of SCG product in each sample to ensure the results are consistent with the behavior of a SCG.
Furthermore, qPCR requires normalization to a reference sample. In the setting of this assay, the cut-offs can only be applied when normalization is accurate. We therefore provided relative qPCR measurements obtained from three commercially available ALT cell lines at specified passage. This allows the user of this qPCR-based assay to perform multi-point calibration for both TC and CC measurements prior to testing unknown samples and to ensure that the cut-off values, normalized to U-2 OS, can be accurately applied.
In addition to research applications, this ALT detection protocol can be readily adopted by clinical laboratories. Many clinical laboratories routinely perform qPCR now, but are not equipped for Southern-blot analysis and for techniques involving radiation. The ability to diagnose ALT efficiently in both research and clinical settings will be crucial for the development of ALT inhibitors as anticancer therapies and for choosing the appropriate telomere maintenance mechanism-targeted drugs in the clinic. As more tumor types are screened for ALT, the prognostic implications of ALT will also be better known. Moreover, CC have been detected in the serum of patients with ALT+ tumors (8), indicating that CC circulate as tumor DNA in the blood stream and are therefore a potential biomarker for ALT activity.
In conclusion, this novel protocol is the first to detect ALT by telomere qPCR and is feasible in clinical pathology laboratories. This radio-isotope-free assay has advantages over the original CC assay (8), which requires more DNA for the detection of one characteristic (CC), uses radioactive probe and is more labor-intensive. The concurrent evaluation of two distinct ALT characteristics increases the sensitivity and specificity of ALT detection and the format of the assay together with its requirement for only a small amount of DNA will facilitate medium-throughput screening of large tumor cohorts for ALT.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1 and Supplementary References [21–25].
FUNDING
Cancer Institute of New South Wales (Australia) [07/CRF/1-05 to L.M.S.L., 09/CDF/2-25 to J.D.H., 07/ECF/1-01 to A.Y.M.A.]; National Health and Medical Research Council (Australia) [APP1012500 to L.M.S.L.]; Cure Cancer Australia Foundation (to L.M.S.L. and J.D.H.); Cancer Society of New Zealand (to J.A.R.) and Cancer Council of New South Wales (Australia) [PG11-08 to R.R.R.]. Funding for open access charge: Children's Medical Research Institute.
Conflict of interest statement. J.D.H. and R.R.R. have applied for a patent regarding the CC assay.
Supplementary Material
ACKNOWLEDGEMENTS
We thank The Children’s Hospital at Westmead Tumour Bank, Professor Graham Mann and Professor Raphael Pollock for tumor samples.
REFERENCES
- 1.Reddel RR. The role of senescence and immortalization in carcinogenesis. Carcinogenesis. 2000;21:477–484. doi: 10.1093/carcin/21.3.477. [DOI] [PubMed] [Google Scholar]
- 2.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 3.Henson JD, Reddel RR. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–3811. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 5.Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton SB, Jellinek DA, Arbuckle SM, Yoo J, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin. Cancer Res. 2005;11:217–225. [PubMed] [Google Scholar]
- 6.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 7.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011;179:1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 9.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farzaneh-Far R, Lin J, Epel ES, Harris WS, Blackburn EH, Whooley MA. Association of marine omega-3 fatty acid levels with telomeric aging in patients with coronary heart disease. JAMA. 2010;303:250–257. doi: 10.1001/jama.2009.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das B, Saini D, Seshadri M. Telomere length in human adults and high level natural background radiation. PLoS One. 2009;4:e8440. doi: 10.1371/journal.pone.0008440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol. 2001;21:3862–3875. doi: 10.1128/MCB.21.12.3862-3875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulay JL, Reuter J, Ritschard R, Terracciano L, Herrmann R, Rochlitz C. Gene dosage by quantitative real-time PCR. Biotechniques. 1999;27:228–230, 232. doi: 10.2144/99272bm03. [DOI] [PubMed] [Google Scholar]
- 14.Bastian BC, Olshen AB, LeBoit PE, Pinkel D. Classifying melanocytic tumors based on DNA copy number changes. Am. J. Pathol. 2003;163:1765–1770. doi: 10.1016/S0002-9440(10)63536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misra A, Pellarin M, Nigro J, Smirnov I, Moore D, Lamborn KR, Pinkel D, Albertson DG, Feuerstein BG. Array comparative genomic hybridization identifies genetic subgroups in grade 4 human astrocytoma. Clin. Cancer Res. 2005;11:2907–2918. doi: 10.1158/1078-0432.CCR-04-0708. [DOI] [PubMed] [Google Scholar]
- 16.Mosse YP, Diskin SJ, Wasserman N, Rinaldi K, Attiyeh EF, Cole K, Jagannathan J, Bhambhani K, Winter C, Maris JM. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer. 2007;46:936–949. doi: 10.1002/gcc.20477. [DOI] [PubMed] [Google Scholar]
- 17.Wolf M, Korja M, Karhu R, Edgren H, Kilpinen S, Ojala K, Mousses S, Kallioniemi A, Haapasalo H. Array-based gene expression, CGH and tissue data defines a 12q24 gain in neuroblastic tumors with prognostic implication. BMC Cancer. 2010;10:181. doi: 10.1186/1471-2407-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickett HA, Cesare AJ, Johnstone RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerone MA, Autexier C, Londono-Vallejo JA, Bacchetti S. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene. 2005;24:7893–7901. doi: 10.1038/sj.onc.1208934. [DOI] [PubMed] [Google Scholar]
- 20.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 22.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fasching CL, Bower K, Reddel RR. Telomerase-independent telomere length maintenance in the absence of alternative lengthening of telomeres-associated promyelocytic leukemia bodies. Cancer Res. 2005;65:2722–2729. doi: 10.1158/0008-5472.CAN-04-2881. [DOI] [PubMed] [Google Scholar]
- 24.Binz N, Shalaby T, Rivera P, Shin-ya K, Grotzer MA. Telomerase inhibition, telomere shortening, cell growth suppression and induction of apoptosis by telomestatin in childhood neuroblastoma cells. Eur. J. Cancer. 2005;41:2873–2881. doi: 10.1016/j.ejca.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 25.Goueli BS, Janknecht R. Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene. 2003;22:8042–8047. doi: 10.1038/sj.onc.1206847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.