Abstract
Eukaryotic ribosome biogenesis requires more than 150 auxiliary proteins, which transiently interact with pre-ribosomal particles. Previous studies suggest that several of these biogenesis factors function together as modules. Using a heterologous expression system, we show that the large ribosomal subunit (LSU) biogenesis factor Noc1p of Saccharomyces cerevisiae can simultaneously interact with the LSU biogenesis factor Noc2p and Rrp5p, a factor required for biogenesis of the large and the small ribosomal subunit. Proteome analysis of RNA polymerase-I-associated chromatin and chromatin immunopurification experiments indicated that all members of this protein module and a specific set of LSU biogenesis factors are co-transcriptionally recruited to nascent ribosomal RNA (rRNA) precursors in yeast cells. Further ex vivo analyses showed that all module members predominantly interact with early pre-LSU particles after the initial pre-rRNA processing events have occurred. In yeast strains depleted of Noc1p, Noc2p or Rrp5p, levels of the major LSU pre-rRNAs decreased and the respective other module members were associated with accumulating aberrant rRNA fragments. Therefore, we conclude that the module exhibits several binding interfaces with pre-ribosomes. Taken together, our results suggest a co- and post-transcriptional role of the yeast Rrp5p–Noc1p–Noc2p module in the structural organization of early LSU precursors protecting them from non-productive RNase activity.
INTRODUCTION
Ribosome biogenesis is a complex and energy consuming process in eukaryotic cells. It requires the synthesis of the structural and functional components, i.e. the ribosomal proteins and four ribosomal RNAs (rRNAs) (18S, 5S, 5.8S and 25S rRNAs) to form the small 40S (SSU) and the large 60S (LSU) ribosomal subunits. In addition, more than 70 small nucleolar (sno) RNAs and more than 150 non-ribosomal proteins termed ribosome biogenesis factors interact transiently with pre-ribosomal particles to assure the generation of functional ribosomes [for reviews see (1,2)].
Ribosome biogenesis starts with the transcription of the genes encoding 18S, 5.8S and 25S rRNA (rDNA) by RNA polymerase I (Pol-I) in the nucleolus, yielding a common polycistronic precursor transcript, termed 35S pre-rRNA in the yeast Saccharomyces cerevisiae. Within the subsequent biogenesis events, the mature rRNAs are generated by endo- and exonucleolytic processing via well-described pre-rRNA intermediates (Supplementary Figure S1).
The nascent 35S rRNA is co-transcriptionally decorated with pre-40S biogenesis factors, that drive early pre-rRNA processing steps 5′ and 3′ of the 18S rRNA sequence (A0, A1, A2 processing in S. cerevisiae; Supplementary Figure S1). They form a huge U3 snoRNA-containing complex termed SSU processome that can be observed in electron micrographs of rDNA chromatin spreads on 5′-ends of nascent rDNA transcripts as terminal ball structures, referred to as SSU knobs (3–8). In S. cerevisiae, early pre-rRNA processing at sites A0, A1 and A2 can occur co-transcriptionally (9,10), resulting in the loss of the SSU knobs and formation of new terminal structures on the rDNA transcripts, which probably represent the earliest LSU precursor particles (9). A few LSU biogenesis factors like Nop15p and Nop53p were suggested to interact co-transcriptionally with the nascent pre-rRNA (8,11), but otherwise, the nature of the pre-60S terminal knobs remains obscure. In S. cerevisiae, ∼30% of Pol-I transcripts are completed before pre-rRNA processing has been initiated by cleavage at positions A0, A1 and A2 (10). In this case, a common precursor of large and small subunits is formed, which among others contain unprocessed 35S pre-rRNA, U3 snoRNA and SSU processome components (5,6,12).
Several SSU processome subcomplexes could be isolated from yeast cellular extracts: the UTP-A (which resembles t-UTP for transcription UTP), UTP-B (or PWP2), UTP-C, Mpp10p and the Noc4p/Nop14p modules, which were shown to be recruited to the pre-rRNA in a hierarchical manner (7,13–17). Rrp5p, a further component of early 90S particles is an atypical trans-acting factor as it is not only required for the SSU maturation, but also involved in LSU maturation (18). Notably, the requirement of Rrp5p for the LSU and SSU pathway could be separated into its N- and C-terminal parts, respectively (19–21). The N-terminal fragment contains nine copies of a S1-type RNA binding motif, whereas the C-terminal part consists of three additional S1-type RNA binding motifs and seven tetratricopetide repeats (TPR) that often mediate protein–protein interactions (22). Both the N- and the C-terminal parts of Rrp5p were shown to be involved in its in vitro interaction with internal transcribed spacer 1 (ITS1) pre-RNA sequences separating 18S rRNA and 5.8S rRNA (23,24). Co-expression of both fragments complements the essential functions of Rrp5p in vivo (19,20).
More recently, Rrp5p was co-purified with the two LSU-biogenesis factors Noc1p and Noc2p from extracts of yeast cells in which rRNA synthesis has been shut down (25). This indicated that these three proteins together might form a protein module acting in LSU synthesis. Both Noc1p and Noc2p are required for early pre-60S maturation steps and were suggested to affect the intranuclear transport of pre-60S particles from the nucleolus to the nucleoplasm (26–28). Notably, all of the three putative module components have homologues in higher eukaryotes, all of which localize to the nucleolus (29) and a function of Rrp5p and Noc2p in ribosome biogenesis is conserved from yeast to human (26,27,30–32). Besides, the human homologues apparently adopted additional functions in the course of evolution. Human Noc1p/CEBPζ/CBF was described to stimulate transcription from the hsp70 promoter (33–35). Rrp5p/NKBP was shown to interact with NF-κB (36,37) and Noc2p/NIR was described to act as an inhibitor of histone acetyl transferases and to modulate p53 and TAp63 activity (38,39), adding a possible supplementary link between ribosome biogenesis and the p53 stress response pathway in higher eukaryotes [reviewed in (40,41)].
The goal of this study was to prove the existence of the potential Rrp5p–Noc1p–Noc2p module of S. cerevisiae and to pursue a more detailed characterization of its role in ribosome biogenesis. Insights into its molecular architecture were obtained through reconstitution of the complex from recombinantly expressed proteins. Ex vivo analyses of associated proteins, pre-rRNA and rDNA chromatin suggested that Rrp5p, Noc1p and Noc2p can already co-transcriptionally be recruited to nascent pre-rRNA together with other LSU biogenesis factors and that the module tightly interacts with early LSU precursor particles. In light of the observed mutual independent binding of Rrp5p, Noc1p and Noc2p to early and aberrant pre-rRNA processing products in yeast mutants, we discuss the potential physiological function of the Rrp5p–Noc1p–Noc2p module in structural organization and stabilization of early pre-ribosomal particles.
MATERIALS AND METHODS
Yeast cell culture and strain construction
Yeast strains employed in this study are listed in Supplementary Figure S3. For cultivation and transformation of yeast, standard protocols were followed (42). Unless otherwise stated, yeast strains were grown at 30°C in rich medium (10 g/l yeast extract, 20 g/l peptone, 100 mg/l adenine hemi-sulphate, 20 g/l sugar) containing galactose (YPAG) or glucose (YPAD) as carbon source.
Yeast strains expressing endogenously encoded Protein A or TAP-tag fusion proteins were constructed by transformation of PCR-based tagging cassettes and homologous recombination as described (43,44). The plasmids and oligonucleotides used are listed in Supplementary Figures S4 and S5, respectively.
Double shuffle strains (Y3668, Y4207, Y3715) were generated as described (45).
Strains expressing NOC1, NOC2 or RRP5 under control of the galactose inducible/glucose repressible GAL1/10 promoter were obtained by plasmid shuffling (46) using the respective shuffle strains and selection on galactose containing minimal medium [yeast nitrogen base (YNB), Sunrise Science] supplemented with the required amino acids and 1 g/l 5-FOA (Toronto research). These strains were normally grown in YPAG medium. To deplete the respective proteins, strains were shifted to YPAD medium and grown for 10, 18 or 24 h to an OD600 of 0.8–1.1.
SF21 insect cell culture, generation of recombinant baculo viruses and heterologous protein expression
Maintenance cultures of SF21 insect cells were cultivated in suspension in Sf900 II SFM medium (life technologies) at 27°C on an orbital shaker (Heidolph Unimax 2010, 100 rpm) and diluted daily to a density of 0.5 × 106 cells per ml.
Recombinant baculo viruses encoding combinations of yeast NOC1, NOC2 and RRP5 were constructed using the MultiBac system essentially as described (47,48). Briefly, the coding regions of the respective yeast genes were amplified by PCR and inserted into the plasmids pUCDM, pFL, pSPL or derivatives thereof by standard cloning techniques (49). The plasmids and oligonucleotides used are listed in Supplementary Figures S4 and S5, respectively. Fusion plasmids containing combinations of genes were obtained by in vitro Cre-Lox recombination of the respective plasmids using Cre-recombinase (NEB). The fusion plasmids were transformed into Escherichia coli DH10-MultiBac-eYFP cells to integrate the plasmids into the viral genome. Recombinant Bacmid DNA was isolated and transfected into adherently growing SF21 insect cells using FuGene transfection reagent (Fugene HD, Promega E2312) to generate recombinant baculo viruses (V0 stock). The resulting viruses were amplified in 50 ml SF21 cultures over a period of 3–5 days (V1 stock) and subsequently used for expression of the recombinant proteins. Therefore, 200-ml SF21 cell culture (1 × 106 cells/ml) in 1 l Erlenmeyer flasks were infected with 5 ml of V1 virus and incubated for 48 h at 27°C. Cells were harvested in aliquots of 50 × 106 cells. After centrifugation (130 g, 10’ min, room temperature), cell pellets were flash-frozen in liquid nitrogen and stored at −20°C.
Affinity purification of recombinantly expressed FLAG-tag fusion proteins
Cell pellets according to 50 × 106 infected SF21 cells were thawed on ice and re-suspended in 40-ml ice-cold A100+ buffer [20 mM Tris–HCl pH 8, 100 mM KCl, 5 mM Mg(OAc)2, 2 mM Benzamidine, 1 mM PMSF, 0.5% Triton X-100, 0.1% Tween-20]. Cells were lysed by sonication using Branson Sonifier 250 (output 5, duty cycle 40%, 30 s pulse, 30 s cooling, six repeats) and cell debris was removed by centrifugation (4°C, 20 min, 3300g). The cleared cell lysate was incubated with 50 µl anti-FLAG M2 affinity gel (Sigma-Aldrich), equilibrated with buffer A100+, for 2 h at 4°C on a turning wheel. After centrifugation (4°C, 1 min, 130g), the supernatant was removed and the beads were washed with buffer A100+ in batch mode (3 × 10 ml, 3 × 1 ml). To elute the FLAG-tag fusion protein, the beads were incubated with 100 µl buffer A100+ containing 300 µg/ml FLAG peptide (Sigma-Aldrich) for 2 h at 4°C on a turning wheel. Finally, the beads were removed from the eluate by centrifugation (4°C, 1 min, 16 000g) through a MobiCol microspin column (MoBiTec).
Affinity-purified protein complexes were analysed using the Smart System (Pharmacia Biotech) and a Superose6 PC 3.2/30 gel filtration column (GE Healthcare) equilibrated with buffer A200 [20 mM Tris–HCl pH 8, 200 mM KCl, 5 mM Mg(OAc)2].
Electron microscopy
Protein complex (∼5 µg/ml) was adsorbed to glow-discharged carbon film for 10 s, followed by staining with a 1% (w/v) uranyl acetate solution for 10 s two times. Images of the complex were recorded with a Philips CM12 (FEI Electron Optics) transmission electron microscope (120 keV, magnification 28 000), using a slow-scan-CCD-Kamera (1024 × 1024 pixels; Model 0124, TVIPS Tietz, Gauting, Germany).
Western blotting analysis
Expression and purification of proteins in/from yeast cells or SF21 insect cells was monitored by Western blotting. FLAG-tag and HA-tag fusion proteins were detected with anti-FLAG (Sigma-Aldrich, F7425) and anti-HA antibodies (Roche, 3F10), respectively. Recombinantly expressed Noc1p was detected with a polyclonal anti-serum from rabbit (27). HRP-conjugated secondary antibodies were from Dianova (111-035-144, 112-035-068). TAP-tag fusion proteins were detected with PAP detection reagent (Sigma, P1291) (Figure 4) or with anti-ProtA antibody (P3775, Sigma-Aldrich) and a fluorophor-coupled secondary antibody (LICOR, 926-32211) (Figure 8). Protein signals were visualized using BM chemiluminescence blotting reagent (Roche) and a LAS-3000 Image Reader (Fujifilm) or by fluorescence using Odyssey Infrared Imaging System (LI-COR). Data were quantified using Multi-Gauge V3.0 (Fujifilm).
Figure 4.
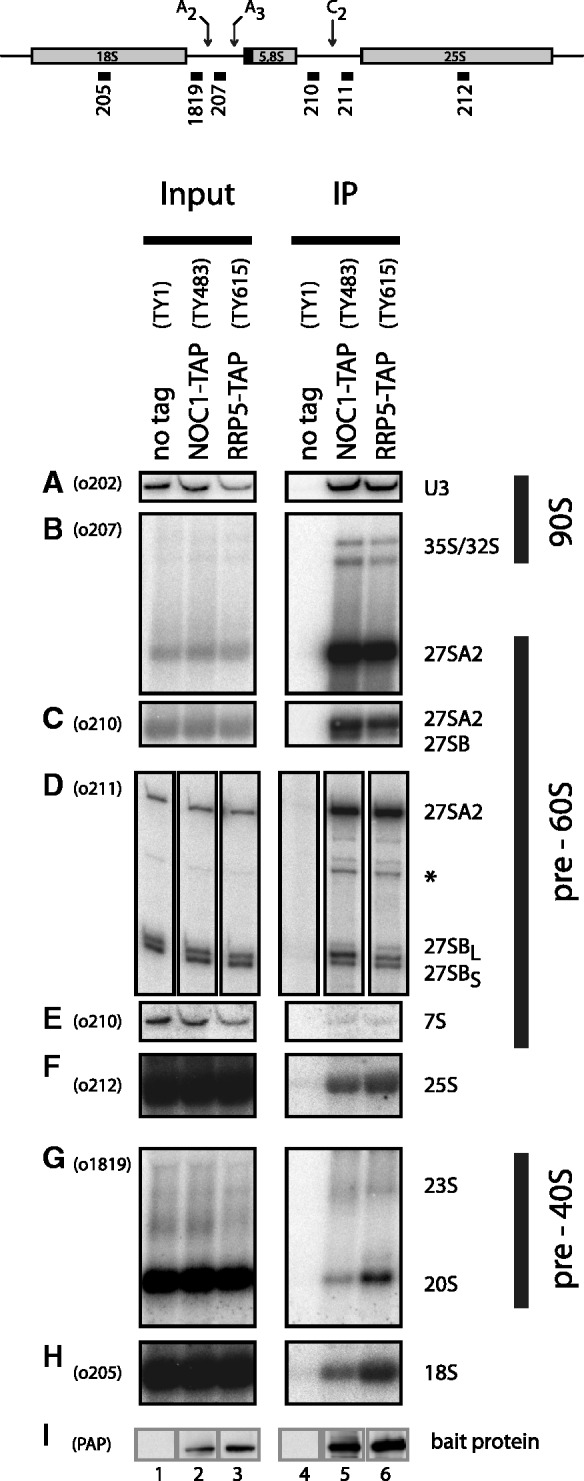
Comparison of pre-ribosomal particles associated with Noc1p or Rrp5p. Yeast strains expressing chromosomally encoded Noc1p-TAP (TY483) or Rrp5p-TAP (TY615) and an untagged control strain (TY1) were grown to exponential phase (OD600 = 0.8) in rich medium. TAP-tagged proteins were affinity purified from cell extracts using IgG-coupled magnetic beads. After washing, the beads were split for the analysis of co-purified RNAs and proteins. RNA isolated from aliquots of cell extracts (Input) and precipitates (IP) was separated on acryl amide (A, E) or agarose (B, C, F, G, H) gels and analysed by Northern blotting using the indicated probes (upper panel, o202–o1819). Alternatively, the isolated RNA was used as template in primer extension reactions (D). Purification of the bait proteins was controlled by Western blotting (I) against the Protein A moiety of the TAP tag using PAP detection reagent. Equal signal intensities in Input and IP correspond to 2 and 11% precipitation efficiency in Northern (0.067% In, 3.33% IP) and Western blot (0.11% In, 1% IP) analysis, respectively. (signal potentially arising from 27SA3 pre-rRNA is indicated by asterisk).
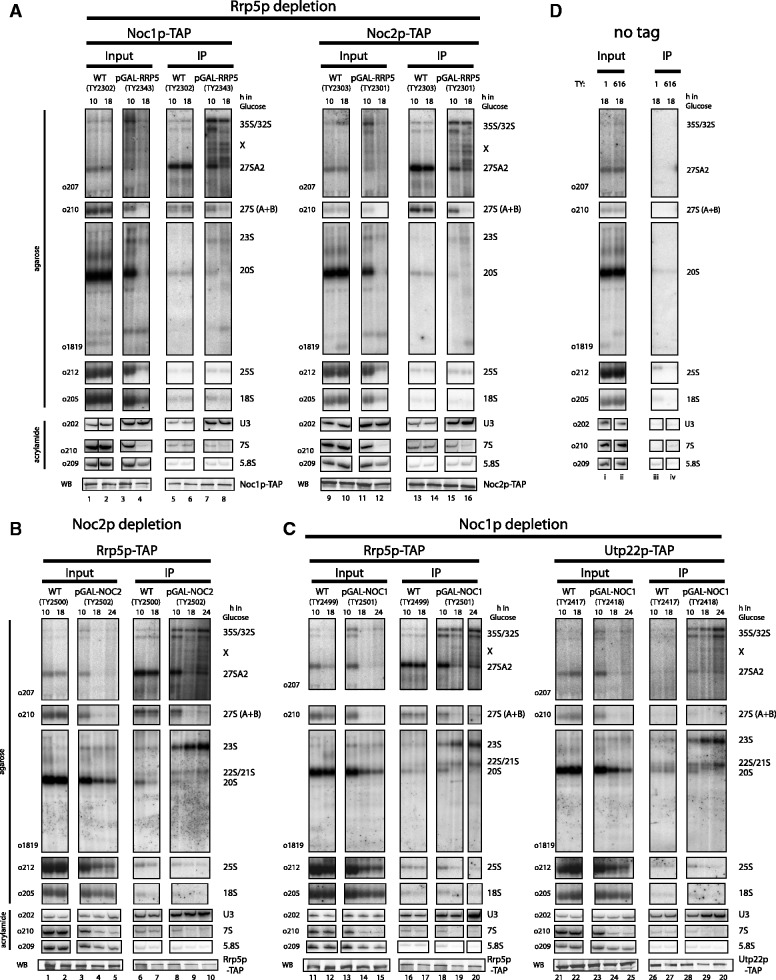
Figure 8.
Analysis of the binding hierarchy of biogenesis factors to pre-ribosomal particles. Yeast strains expressing chromosomally encoded Noc1p-TAP (TY2302/TY2343) or Noc2p-TAP (TY2303/TY2301) in which the RRP5 gene is either under control of the endogenous or the inducible GAL 1/10 promoter were cultivated for 10 and 18 h in glucose containing rich medium (final OD600 = 0.5–1). Analogous experiments were carried out with strains expressing chromosomally encoded Rrp5p-TAP (TY2499/TY2501) or Utp22p-TAP (TY2417/TY2418) with NOC1 under the control of its endogenous or the GAL 1/10 promoter, and for strains that express chromosomally encoded Rrp5p-TAP (TY2500/TY2502) with NOC2 under the control of its endogenous or the GAL1/10 promoter. These strains were cultivated for 10,18 and 24 h in glucose containing rich medium (final OD600 = 0.5–1). The respective background strains expressing no tagged protein (TY1, TY616), which served as controls, were cultivated for 18 h in glucose containing rich medium. TAP-tagged proteins were affinity purified from cell extracts using IgG sepharose. After washing, the beads were split for the analysis of co-purified RNAs and proteins. RNAs isolated from aliquots of cell extracts (Input) and immuno-purified fractions (IP) were separated on acrylamide or agarose gels and analysed by Northern blotting by subsequent hybridization using the indicated probes (o202–o1819; binding sites are depicted in Supplementary Figure S1A). In general, all input and IP samples of the purification of one bait protein in the different strains were analysed on the same gel [see also entire blots shown in the Supplementary Material (Supplementary Figure S7)]. Purification of the bait proteins was controlled by Western blotting (WB) against the Protein A moiety of the TAP tag using anti-Protein A antibody. (A) Depletion of Rrp5p; (B) depletion of Noc2p; (C) depletion of Noc1p; (D) control strains. Equal signal intensities in Input and IP correspond to 2 and 1.5% purification efficiency in Northern blots of agarose and acrylamide gels, respectively. In Western blots, equal signal intensities in Input and IP correspond to 20% (Rrp5-TAP), 33% (Noc1-TAP), 17% (Noc2-TAP) and 50% (UTP22-TAP) purification efficiencies. Aberrant pre-rRNA fragments resulting from depletion of biogenesis factors are indicated by multiplication symbol.
Analysis of Noc1p-associated proteins
Purification of Noc1p-TAP from yeast cell extracts and analysis of co-purified proteins was performed as described (50).
Affinity purification of pre-ribosomal particles
Affinity purification of pre-ribosomal particles was performed essentially as described (51) with the following modifications. The cell pellet corresponding to 1 l yeast culture with OD600 = 0.8–1.2 was re-suspended in 1.5 ml of cold A100 buffer [20 mM Tris–HCl pH 8, 100 mM KCl, 5 mM Mg(OAc)2, 2 mM Benzamidine, 1 mM PMSF], supplemented with 0.04 U/ml RNasin, per gram of cell pellet. 800 μl aliquots of this cell suspension were divided into 2 ml reaction tubes containing 1.4 ml glass beads (Ø 0.75–1 mm). Cells were disrupted on an IKA Vibrax VXR basic shaker with 2200 rpm at 4°C for 15 min, followed by 5 min on ice. This procedure was repeated twice. The cell lysate was cleared from cell debris by two centrifugation steps (4°C, 5 min, 16 000g; 4°C, 10 min, 16 000g). The protein concentration of the cleared lysate was determined using the Bradford assay. Triton X-100 (0.5% final concentration) and Tween-20 (0.1% final concentration) was added to the cell lysate.
For subsequent comparative mass spectrometric analysis of Noc1p-TAP- and Rrp5p-TAP-associated pre-ribosomal particles (Figure 4), IgG-coupled magnetic beads were used for affinity purification. Equal protein amounts of cell lysates (typically, 1.1 ml with 30–40 mg of total protein) were incubated for 1 h at 4°C with 100 µl of IgG (rabbit serum, I5006-100MG, Sigma) coupled magnetic beads slurry (1 mm BcMag, FC-102, Bioclone) equilibrated in A100+ buffer (A100 buffer supplemented with 0.5% Triton X-100 and 0.1% Tween). The beads were washed four times with 700 µl cold A100+ buffer. After the fourth washing step, an aliquot of 20% of the beads was separated for the analysis of co-purified RNA. The remaining beads were washed twice with 700 µl AC buffer (100 mM NH4OAc pH 7.4, 0.1 mM MgCl2) to remove the remaining salt from the sample. Bound proteins were eluted twice with 500 µl of freshly prepared 500 mM NH4OH solution for 20 min at RT. Both eluate fractions were pooled, an aliquot of 10% was separated for Sodium dodecyl sulphate (SDS)–polyacrylamide gel electrophoresis (PAGE) analysis and the remaining eluate was lyophilized over night.
For analysis of pre-ribosomal particles associated with Noc1p-TAP, Noc2p-TAP, Rrp5p-TAP or Utp22p-TAP in presence or absence of other biogenesis factors (Figure 8), IgG sepharose was used for affinity purification. Equal protein amounts of cell lysates (typically 800 µl with 10 mg of total protein) were incubated for 1 h at 4°C with 60 µl of IgG sepharose slurry (GE Healthcare, 52-2083-00 AH) equilibrated in A100+ buffer. The beads were washed twice with 1 ml, five times with 2 ml and twice with 10 ml cold A100+ buffer in a 10-ml column. After washing, the beads were split for analysis of purified protein (25%) and co-purified RNA (75%).
Gel-based mass spectrometric analysis of proteins
Mass spectrometric (MS) analysis of Coomassie-stained protein bands was done as described (52). Peptide mass fingerprinting and tandem MS (MS/MS) analyses were performed with a 4800 Proteomics Analyzer MALDI-TOF/TOF instrument (ABI), operated in positive-ion reflector mode. The data were evaluated by searching the NCBI non-redundant (nr) protein sequence database using the Mascot module implemented in the GPS Explorer, version 3.5, software (ABI).
Comparative MALDI TOF/TOF analyses (cMS)
Comparative mass spectrometric analysis using iTRAQ reagents were performed as described previously (51).
Hierarchical clustering analysis of datasets derived from several experiments was performed with cluster 3.0 software (53) using the ‘log2 transform data’ and the ‘median center arrays’ settings for data adjustment and the Euclidean distance and centroid linkage settings for gene and array clustering. Java Treeview was used for cluster visualization (see http://www.eisenlab.org/eisen/?page_id=42).
RNA analysis
RNA was extracted by hot acidic phenol–chloroform treatment essentially as described (54,25), except that before ethanol precipitation the RNA fraction was split to obtain samples in MOPS- and TBE-based solubilization buffer (49). The respective RNA samples were separated on formaldehyde/MOPS agarose gels or Urea/TBE/polyacrylamide gels as described (49). The transfer from agarose gels onto the positively charged membrane (Positive TM, MP-Biomedicals) was performed in 10x SSC buffer by applying a vacuum of 5 bar for 90 min using a vacuum blotter (Biorad). The transfer from acrylamide gels onto the positively charged membrane (Positive TM, MP-Biomedicals) was performed in 0.5x TBE by applying a voltage of 50 V for 90 min using an electro blotter (Biorad).
Hybridization was performed in 50% formamide/5x SSC/0.5% SDS/5x Denhardt’s solution at 30°C with 5′ 32P-labelled probes. The oligonucleotides used are listed in Supplementary Figure S5 and their binding sites are indicated in Supplementary Figure S1A. Prior to hybridization with a new probe, the membranes were washed two times for 15 min in hot 1% SDS solution at 65°C. Labelled (pre-) rRNA signals were detected using a Phosphor Imager FLA3000 (Fujifilm) and data were quantified using MultiGauge V3.0 (Fujifilm). Digital adjustment of brightness and contrast was applied to the whole blot images using Adobe Photoshop (Adobe).
Primer extension analyses were performed as described (55) using RNAs dissolved in TBE-based buffers as templates.
Chromatin immunoprecipitation and PCR-based analysis of co-purified DNA
Cultures of yeast strains expressing TAP-tagged ribosome biogenesis factors (50 ml, YPAD) were treated with formaldehyde as described (56) and chromatin immunoprecipitation (ChIP) was performed as described (57) with the following modifications. Formaldehyde crosslinked cells were re-suspended in 350 µl of lysis buffer and cells were disrupted on an IKA Vibrax VXR basic shaker for 15 min with 2200 rpm at 4°C followed by 10 min incubation on ice. This procedure was repeated twice. After sonication and centrifugation, the soluble chromatin fraction was split into three aliquots. A total of 40 µl of each aliquot served as input control and 200 µl of each aliquot was incubated for 120 min at 4°C with 125 µl IgG-Sepharose slurry (GE Healthcare, 52-2083-00 AH) to enrich the TAP-tagged proteins.
Relative DNA amounts present in input samples and purified fractions (IP) were determined by quantitative PCR using SYBR green I dye (Roche) for DNA detection with a Rotor-Gene 3000 system (Corbett Life Science/Qiagen) and the comparative analysis software module. Primer pairs used for amplification are listed in Supplementary Figure S5. Input DNA was diluted 1:500 and IP DNA was diluted 1:20 prior to analysis. All samples were run in triplicate to ensure accuracy of the data. For each amplicon in each purification, the precipitation efficiencies [percent IP (rDNA)] were calculated and normalized to the PDC1 precipitation efficiencies [percent IP (rDNA)/percent IP (PDC1)]. The mean values and error bars are derived from three independent ChIP experiments.
ChIP and analysis of co-purified proteins
pChIP was adapted from a previous study (58). Yeast cells were grown at 30°C in 500 ml YPD medium to mid-exponential phase (OD600 = 0.5–0.8) and treated with formaldehyde (0.5% final concentration, 10 min, 30°C). After quenching of excess formaldehyde with Tris–glycine solution (10 mM final concentration, 5 min, 30°C), cells were harvested (10 min, 4°C, 2200g) and washed twice with 40 ml of ice cold PBS (4°C, 5 min, 2000g) and once with cold lysis buffer [50mM HEPES–KOH (pH 7.5), 200 mM NaCl, 0.5 mM EDTA, 1% Triton, 0.1% Na-Deoxycholate, 0.1% SDS, 2 mM Benzamidine, 1 mM PMSF]. The cell pellet was re-suspended in 1.5 ml lysis buffer per gram of cells. 500 μl aliquots were mixed with 500 μl glass beads (diameter 0.75 to 1.0 mm; Roth) and cells were disrupted on an IKA Vibrax VXR basic shaker (4°C, 2200 rpm) for four cycles of 10 min with a 2-min cooling step in between. Extracts were centrifuged (5 min, 4°C, 850g), the supernatant was collected and adjusted to 1 ml final volume with lysis buffer. Sonication was performed using a Branson Sonifier 250 for five cycles of 10 pulses (Microtip limit 3, 90% Duty cycle) with a 30-s cooling step in between. Sonicated extract was centrifuged (10 min, 4°C, 18000g) and the supernatant was collected and used in the affinity purification.
Equal volumes of cell lysates were incubated over night at 4°C on a turning wheel with 200 µl of IgG (rabbit serum, I5006-100MG, Sigma) coupled magnetic beads slurry (1 mm BcMag, FC-102, Bioclone) equilibrated in lysis buffer. The beads were washed four times with lysis buffer and two times with AC buffer (100 mM NH4OAc pH 7.4, 0.1 mM MgCl2) for 5 min each at 4°C on a turning wheel. Bound proteins were eluted twice with 500 µl of freshly prepared 500 mM NH4OH solution for 20 min at RT. Eluates were combined and lyophilized and subjected to comparative mass spectrometric analysis (cMS).
RESULTS
Evidence for a ribosome biogenesis factor module consisting of Noc1p, Noc2p and Rrp5p
Previous studies indicated that yeast Noc1p and Noc2p directly interact with each other and may form together with Rrp5p a protein module involved in ribosome maturation (25,27).
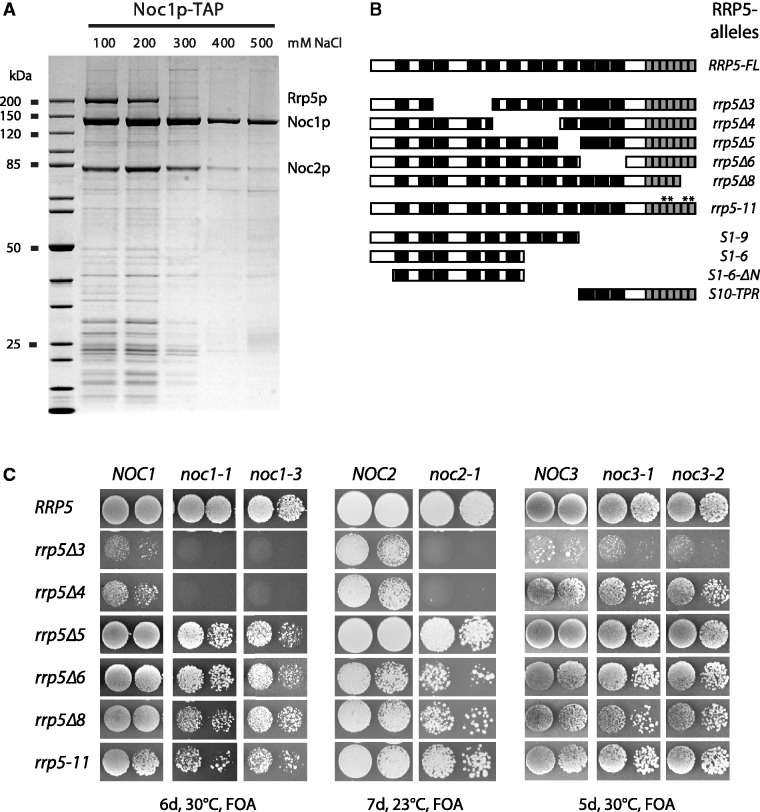
To investigate whether in vivo major populations of Noc1p are more stably associated with Noc2p and Rrp5p than with other biogenesis factors and to further characterize the stability of this potential Rrp5p–Noc1p–Noc2p protein module, we affinity purified chromosomally encoded Noc1p-TAP from yeast extracts using buffers containing varying salt concentrations.
SDS–PAGE analyses of the purified fractions followed by Coomassie staining and mass spectrometric identification of proteins showed that significant amounts of Noc2p and Rrp5p were co-purified with tagged Noc1p when applying moderate salt concentrations (Figure 1A, 100–200mM NaCl). In contrast, other biogenesis factors were co-purified with significant lower yields. Salt concentrations of 300 mM NaCl strongly reduced the amounts of co-purifying Rrp5p, whereas co-purification of Noc2p was substantially diminished when salt concentrations of 400–500 mM NaCl in the affinity purification buffers were applied.
Figure 1.
The ribosome biogenesis factors Noc1p, Noc2p and Rrp5p interact in vivo. (A) Noc1p-TAP efficiently co-purifies Noc2p and Rrp5p from yeast extracts. Noc1-TAP was isolated from extracts of strain Y3572 applying the tandem affinity purification method under increasing concentration of NaCl. The composition of the Noc1p containing particles was analysed on a 4–12% gradient SDS–PAGE gel, stained with Coomassie Blue. Prominent co-purified proteins are indicated. (B) Overview of the rrp5 alleles analysed in this study and schematic presentation of the corresponding protein variants. Black bars illustrate the S1 RNA binding motifs, grey bars the tetratricopeptide repeats, respectively [adapted from (20); see ‘Introduction’ section]. Point mutations in the temperature sensitive rrp5-11 allele are indicated by asterisk. (C) Genetic interaction studies show synthetic lethal effects between rrp5 alleles with deletions of N-terminal S1-repeats and noc1-ts or noc2-ts mutants. The respective double shuffle strains (Y3668, Y4207 and Y3715) were transformed with plasmids carrying the indicated wild-type and mutant alleles of ProtA-RRP5, ProtA-NOC1, NOC2 or ProtA-NOC3 (Supplementary Figure S4). Transformants were spotted in a 10-fold dilution series on SDC + FOA plates and incubated at the indicated temperature and for the indicated time to analyse genetic interactions.
These findings were in agreement with the existence of a ribosome biogenesis factor module in yeast cells consisting of Noc1p and Noc2p, which can establish salt labile interactions with Rrp5p.
Next, we tested whether RRP5 mutants [Figure 1B; (20)] are genetically linked with mutants of NOC1 or NOC2 (27). We created double shuffle strains, which were transformed with various plasmids coding for the indicated alleles. The resulting growth phenotypes were analysed by counterselection on FOA-containing plates against the plasmids carrying the corresponding wild-type alleles (Figure 1C). As a specificity control, we used mutants of the LSU biogenesis factor Noc3p. Noc3p was shown to co-purify with Noc2p, but is stably associated with later pre-60S particles than Noc1p (27,28,59). We observed that two mutant RRP5 alleles (rrp5Δ3, rrp5Δ4), are synthetic lethal with temperature sensitive alleles of NOC1 and NOC2 (noc1-1, noc1-3; noc2-1). This is consistent with earlier studies, which had discovered that rrp5Δ3, Δ4 exhibit 60S/LSU maturation defects, whereas rrp5Δ6, Δ8 and rrp5-11 mutants are impaired in 40S/SSU biogenesis (20,60) (Supplementary Figure S2). Taken together, these findings further indicated that Rrp5p together with Noc1p and Noc2p build up a protein module that is essential for LSU biogenesis in the yeast S. cerevisiae.
Recombinantly expressed Noc1p, Noc2p and Rrp5p form a large protein complex
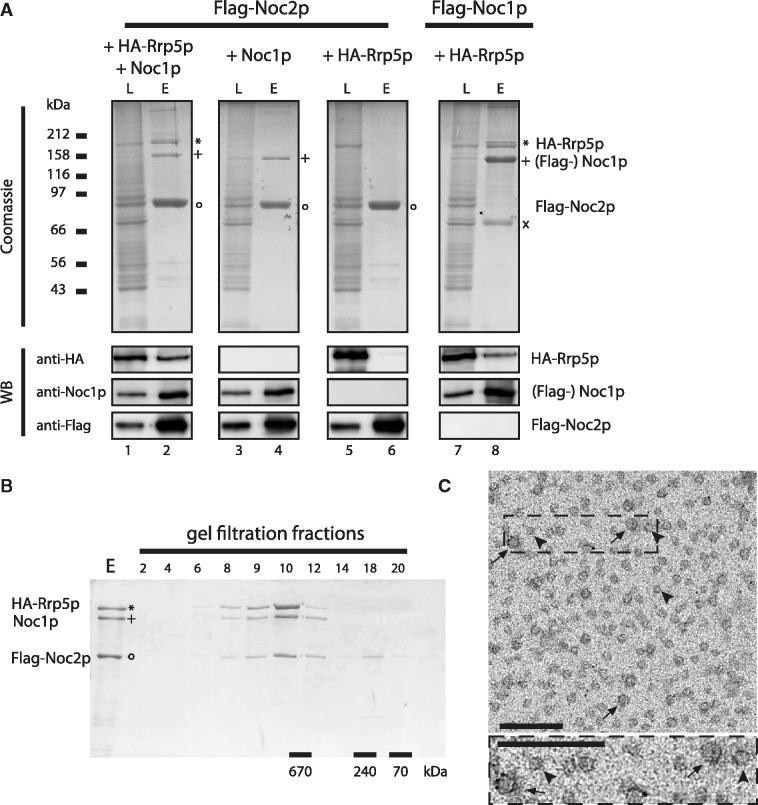
In order to investigate, if Rrp5p can directly interact with the Noc1p–Noc2p complex, we co-expressed yeast Noc1p, Noc2p and Rrp5p in SF21 insect cells using recombinant baculo viruses (47,48). Different components were tagged with the FLAG epitope and affinity purified. The final eluates were analysed by SDS–PAGE stained with Coomassie blue, Western blot analysis and subsequent mass spectrometric (MS) analysis (Figure 2A).
Figure 2.
Reconstitution of the Rrp5p–Noc1p–Noc2p module from proteins co-expressed in insect cells. The indicated combinations of Flag-tagged bait proteins and potential interaction partners were co-expressed in SF21 insect cells infected with recombinant baculo viruses containing the plasmids TK1232, TK1504, TK1658 or TK1677 (Supplementary Figure S4). (A) The bait proteins were purified with anti-Flag affinity matrix from lysates of 50 × 106 infected cells and eluted with buffer containing Flag peptide. Aliquots of the lysates (L) and eluates (E) were analysed by coomassie stained SDS–PAGE (upper panel; 0.05% L, 23% E) and MS (HA-Rrp5p is indicated by “*”; Flag-Noc2p by “°”; (Flag-) Noc1p by “+”; N-terminal Flag-Noc1p fragment by “×”) or Western blotting using anti-HA, anti-Noc1p and anti-Flag antibodies (lower panel; 0.006% L, 1.2% E). (B) Affinity purified Flag-Noc2p–Noc1p–Rrp5p complex was fractionated on a Superose 6 gel filtration column. Aliquots of the eluate (E; 20%) and the fractions (2–20; 40%) were analysed by Coomassie stained SDS–PAGE. Elution of marker proteins in independent gel filtration runs is indicated at the bottom. (C) Electron micrograph of an uranyl acetate stained aliquot of the Flag-Noc2p–Noc1p–Rrp5p complex after Superose 6 gel filtration column from an independent purification (corresponding to fraction 10 in Figure 2B). Arrows and arrowheads indicate particles of 12 nm and 8 nm diameter, respectively. The lower panel shows an enlarged view of the boxed area. Scale bars are 70 nm.
First, we expressed all three proteins together and aimed to purify the complex via Flag-Noc2p as bait protein (Figure 2A, lanes 1 and 2). Here, anti-Flag affinity purification significantly enriched the bait protein and co-purified efficiently Noc1p and Rrp5p (Figure 2A, lane 2), strongly suggesting direct interactions between the three proteins. Next, we aimed to determine the topology of the Rrp5p–Noc1p–Noc2p module. Therefore, we co-expressed pairs of the three proteins and tested for individual interactions (Figure 2A, lanes 3–8). When the Flag-Noc2p bait protein was co-expressed with Noc1p or HA-Rrp5p, respectively, we observed only co-purification of Noc1p, but not of Rrp5p (Figure 2A, lanes 4 and 6). Therefore, we conclude that Rrp5p does not, or only weakly, bind to Noc2p and predict a stable interaction between Rrp5p and Noc1p. Indeed, co-expression of Flag-Noc1p and HA-Rrp5p, followed by purification of Flag-Noc1p yielded a significant co-purification of Rrp5p (Figure 2A, lane 8). Control experiments using IgG-agarose beads showed no unspecific binding of the proteins to the resin material (data not shown). These experiments not only confirmed that Noc1p interacts with Noc2p in a robust and specific way (27), but also suggested that Noc1p can directly interact with Rrp5p. In this way, Noc1p bridges between Noc2p and Rrp5p to form a hetero-trimeric protein complex, which might act as a functional entity in ribosome biogenesis.
Next, we determined the apparent size of the affinity-purified Flag-Noc2p–Noc1p–Rrp5p complex on a gel filtration column (Figure 2B). This analysis showed that the bait protein Flag-Noc2p was present in over-stoichiometric amounts in the purified fractions. Excess Noc2p was separated from the other complex components during gel filtration (Figure 2B, fractions 18 + 20). A subpopulation of Flag-Noc2p [theoretical MW (Noc2p) = 81 kDa] co-eluted together with Noc1p (theoretical MW = 116 kDa) and HA-Rrp5p [theoretical MW (Rrp5p) = 193 kDa] with an apparent molecular weight of >670 kDa, which could indicate a higher order stoichiometry of some of the proteins within the complex. Electron micrographs of negatively stained Rrp5p–Noc1p–Noc2p complexes showed particles of ∼8 nm and ∼12 nm in diameter (Figure 2C), the latter of which is well compatible with the apparent molecular weight of 670 kDa, estimated by size exclusion chromatography. However, it remains unclear whether the differently sized particles represent different orientations of the complex on the grid or particles differing in protein composition.
In summary, these experiments provide strong evidence that Rrp5p, Noc1p and Noc2p form a large hetero-oligomeric protein complex, but further experiments are necessary to determine in detail the structure of the complex and the stoichiometry of its components.
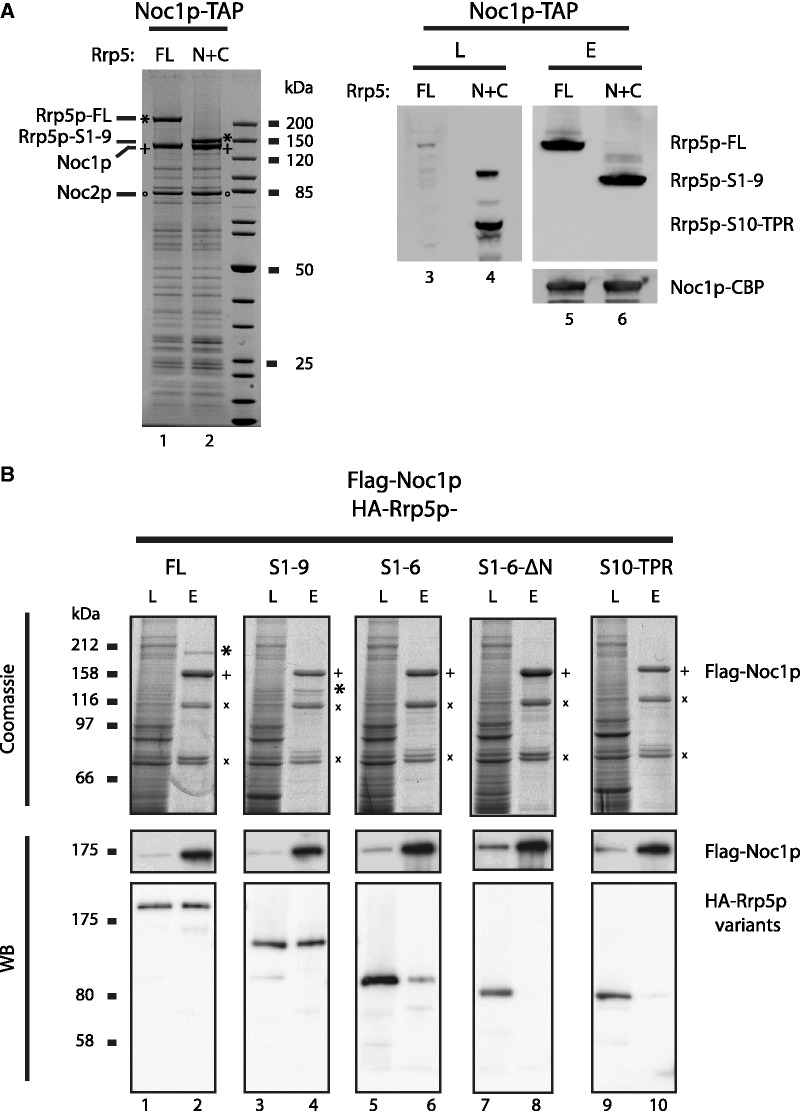
The N-terminus of Rrp5p mediates stable interaction with Noc1p
Because N-terminal RRP5 mutants that predominantly affect LSU biogenesis (Supplementary Figure S2) (19,20,60,61), are genetically linked to NOC1 mutants (Figure 1C), we speculated that Noc1p could interact with the N-terminus of Rrp5p. To test this idea, we constructed yeast NOC1-TAP strains in which the deletion of the essential RRP5 gene was complemented by either ectopically expressed full-length Rrp5p fused to green fluorescent protein (Rrp5p-FL-GFP) or by co-expression of an N- and a C-terminal fragment of Rrp5p fused to GFP (Rrp5p-S1-9-GFP and Rrp5p-S10-TPR-GFP; Figure 1B). Finally, the chromosomally encoded Noc1p-TAP fusion protein was affinity purified from cellular extracts of these strains. The soluble fraction of these cell lysate contained significantly enriched levels of Rrp5p-S1-9-GFP and Rrp5p-S10-TPR-GFP when compared with Rrp5p-FL-GFP (Figure 3A, right panel). This upregulation may be induced in order to compensate for bisection of Rrp5p. However, despite similar levels of Rrp5p-S1-9-GFP and Rrp5p-S10-TPR-GFP within the supernatant, only Rrp5p-FL-GFP and Rrp5p-S1-9-GFP could be efficiently co-purified with Noc1p-TAP, as shown by Coomassie stained SDS–PAGE and Western blot analyses (Figure 3A). Accordingly, these results strongly supported our assumption that Noc1p interacts with the N-terminal part of Rrp5p.
Figure 3.
The N-terminal part of Rrp5p is required for stable association with Noc1p. (A) Co-precipitation of full-length and truncated Rrp5p variants with Noc1p from yeast cells. Genomic NOC1-TAP, rrp5Δ yeast strains harbouring plasmids encoding Rrp5p-FL-GFP (FL) or Rrp5p-S1-9-GFP and Rrp5p-S10-TPR-GFP (N + C; Figure 1B and Supplementary Figure S4) were generated by plasmid shuffling from strain Y3716 and used for tandem affinity purification of Noc1p. The calmodulin eluates were analysed by coomassie stained SDS–PAGE (left panel), whereas the supernatant of the cell lysate and calmodulin eluates were analysed by western blotting (right panel) using anti-GFP (upper panel) and anti-CBP antibodies (lower panel) to detect Rrp5-GFP variants or Noc1-CBP, respectively. (B) Heterologous reconstitution of complexes containing Flag-Noc1p and HA-Rrp5p variants. The indicated combinations of Flag-Noc1p and truncated HA-Rrp5p variants (Figure 1B) were co-expressed in SF21 insect cells infected with recombinant baculo viruses containing the plasmids TK1658, TK1727, TK1728, TK1729 or TK1732 (Supplementary Figure S4). Flag-Noc1p was purified with anti-Flag affinity matrix from lysates of 50 × 106 infected cells and eluted with buffer containing Flag peptide. Aliquots of the lysates (L) and eluates (E) were analysed by SDS–PAGE and coomassie staining (upper panel; 0.04% L, 20% E) or western blotting using anti-HA and anti-Flag antibodies (lower panels; 0.006% L, 1% E). Coomassie-stained proteins were identified by mass spectrometry (HA-Rrp5p variants are indicated by “*”; Flag-Noc1p by “+”; N-terminal Flag-Noc1p fragments by “×”).
To further confirm this conclusion, we analysed the interaction between yeast Flag-Noc1p and different Rrp5p variants in the heterologous SF21 insect cell co-expression system. We co-expressed Flag-Noc1p together with HA-Rrp5p, HA-Rrp5p-S1-S9, HA-Rrp5p-S1-S6, HA-Rrp5p-S1-S6-ΔN and HA-Rrp5p-S10-TRP, respectively (Figure 1B), and performed anti-Flag affinity purification. Subsequent SDS–PAGE, Western blot analysis and MS analysis (Figure 3B) revealed that full-length Rrp5p and Rrp5p-S1-9 (1–1087 aa) co-purified with equal efficiency with the bait protein Flag-Noc1p (Figure 3B, lanes 2 and 4). Furthermore, we observed that the co-purification efficiency of Rrp5 S1-6 (1–777 aa) with Flag-Noc1p was significantly reduced, whereas HA-Rrp5p-S1-S6-ΔN (106–777 aa) and Rrp5p-S10-TPR (1082–1729 aa) could not be detected in the eluate fractions (Figure 3C, lanes 6, 8 and 10). Altogether, we conclude that the N-terminal part of Rrp5p, which contains the S1 repeat motifs S1–9, is sufficient and required to establish a specific physical interaction with Noc1p.
Noc1p and Rrp5p are stably associated with similar 90S and pre-60S particles
After having established the topology of the Rrp5p–Noc1p–Noc2p module, we aimed to determine in more detail into which kind of ribosomal precursor particles the module is incorporated in vivo. Previous proteomic approaches indicated that Noc1p and Rrp5p are constituents of early LSU precursors purified via Ssf1p or Nsa3p (28,59,62), but absent from intermediate pre-60S particles purified via Nsa1p (59). Moreover, Rrp5p was shown to be part of U3 snoRNA and 35S pre-rRNA containing pre-ribosomal particles termed 90S pre-ribosome or SSU processome (see ‘Introduction’ section). It remained unclear whether the function of Rrp5p in LSU and SSU biogenesis (18,20) is achieved by interaction of Rrp5p with different populations of LSU and SSU precursor particles, respectively. Since detailed knowledge on the composition of Rrp5p- or Noc1p-bound pre-ribosomes was still lacking, we decided to perform a thorough comparative characterization of pre-ribosomes associated with Noc1p or Rrp5.
First, we analysed the rRNA composition of pre-ribosomes associated with Rrp5p-TAP and Noc1p-TAP particles. Noc1p-TAP and Rrp5p-TAP were affinity purified from yeast cell extracts in a one-step procedure in mild buffer conditions to preserve pre-ribosomal particles (Figure 4, panel IP) and co-purified (pre-) ribosomal RNA was analysed by Northern blotting and primer extension reactions (Figure 4). The bait proteins Noc1p-TAP and Rrp5p-TAP were purified with equal efficiency as judged by Western blotting (Figure 4I, compare signal intensities in lanes 2/5 and 3/6).
Consistent with a direct interaction of Rrp5p and Noc1p during ribosome biogenesis, both purifications contained a highly similar pattern of co-purified rRNA. In both purifications, the most efficiently enriched pre-rRNA was the 27SA2 rRNA intermediate (Figure 4B), which is the first specific LSU precursor RNA directly formed by pre-rRNA processing at site A2 (Supplementary Figure S1B). In contrast, the alternative downstream intermediates 27SBL and 27SBS pre-rRNAs were specifically, but much less efficiently co-purified with both Noc1p and Rrp5p, as shown by Northern blot (Figure 4C) and primer extension analyses (Figure 4D). This is in agreement with previous studies, which indicated that Noc1p and Rrp5p are part of early pre-60S particles (23,59), but depleted in intermediate pre-60S particles purified via Nsa1p (59). Interestingly, Noc1p-TAP preferentially co-purified the less abundant 27SBL pre-rRNA from cellular extracts (Figure 4D; compare 27SBL/27SBS ratios in lanes 2, 3, 5 and 6, respectively), indicating that different release mechanisms might play a role in the alternative B1L and B1S processing pathways.
Furthermore, 35S and 32S pre-rRNA, as well as U3 snoRNA were efficiently enriched with both Rrp5p and Noc1p, consistent with previous studies identifying Rrp5p as a component of 90S/SSU processome particles (5,6,63). In contrast, later pre-LSU or pre-SSU RNA species co-purified only with low efficiency with Noc1p-TAP or Rrp5p-TAP (Figure 4E and G, compare input lanes 2 and 3 with IP lanes 5 and 6). Quantitation of these co-purification efficiencies showed that they were in the range or below the ones of 18S rRNA and 25S rRNA (Figure 4F and H, compare input lanes 2 and 3 with IP lanes 5 and 6), which were analysed as measures for the internal background of the experimental setup.
Altogether, these analyses indicated that Noc1p and Rrp5p are associated with highly overlapping populations of early pre-ribosomes. In S. cerevisiae, a significant pool of the common precursor transcript (35S pre-RNA) is cleaved co-transcriptionally into 20S and 27SA2 pre-rRNA (9,10). Thus, co-precipitation of 27SA2 and 35S pre-rRNA implies that Noc1p and Rrp5p are recruited to the earliest LSU precursor particles.
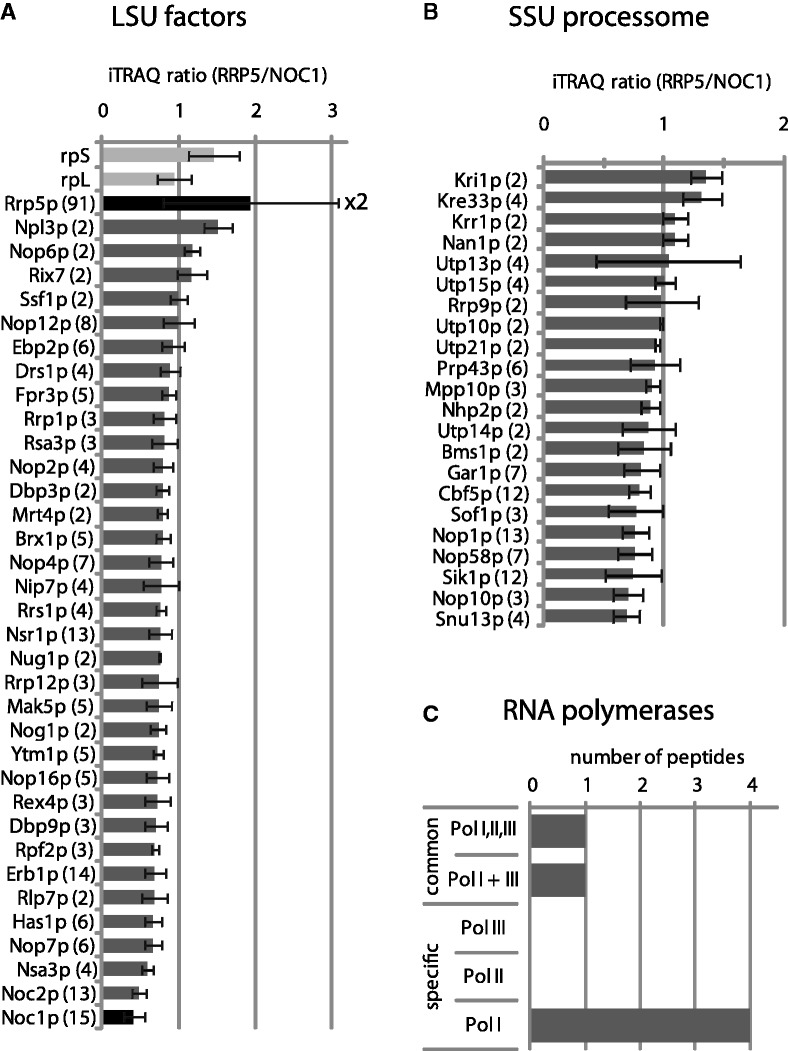
To further characterize the composition of these early LSU precursors, the proteins contained in pre-ribosomes purified via Noc1p-TAP and Rrp5p-TAP were identified and relative amounts of components detected in the two purifications were compared by MS using iTRAQ reagents (25,64). In these analyses, a set of 33 large subunit biogenesis factors was identified by two or more peptides in the combined affinity-purified fractions (Figure 5A). Most of these factors were, similar to Noc1p and Rrp5p, previously shown to be required for early steps in LSU maturation, as the processing of the 5′-end of 5.8S rRNA and production and/or stabilization of 27SB pre-rRNA [e.g. Rix7p (65), Ssf1p (62), Ebp2p, Rrp1p, Ytm1p, Erb1p, Rlp7p, Nop7p, Nsa3p (66–71), Brx1p (72), Nop4p (73), Rrs1p (74), Dbp9p (75)]. In agreement with the low levels of 7S pre-rRNA and 20S pre-rRNA detected in the fractions co-purified with Rrp5p and Noc1p (Figure 4E and G), no factors indicative for later 60S or 40S precursors, like Rea1p, Arx1p, Nmd3p, Rix1p, Ipi1p, Ipi3p, Drg1p, Lsg1p or Efl1p, Rio2p or Ltv1p were identified. In contrast, and in agreement with the 35S pre-rRNA and U3 co-purification observed above (Figure 4A and B), 22 components of the SSU processome, including CD box and H/ACA box snoRNP components, were identified by two or more peptides in the affinity-purified fractions (Figure 5B). In addition to that, in total, six peptides of subunits of Pol-I, which synthesizes the primary pre-rRNA transcript, were detected (Figure 5C, see below). When the relative amounts of biogenesis factors specifically co-purifying with Noc1p-TAP and Rrp5p-TAP were compared, for nearly none of them a clear indication for preferential co-purification with either Noc1p-TAP or Rrp5p-TAP could be observed [Figure 5A and B, iTRAQ ratios (Rrp5p/Noc1p) between 0.5 and 1.5]. We noticed, however, that Rrp5p was clearly over-represented in Rrp5p-TAP fractions (Figure 5A). This might be due to partial release of the labile Rrp5p–Noc1p interaction (Figure 1A) or due to the existence of a cellular pool of Rrp5p, which is not involved in Noc1p-related interactions with pre-ribosomes.
Figure 5.
Comparative proteome analysis of Noc1p and Rrp5p containing pre-ribosomes. Proteins from aliquots of Noc1p-TAP and Rrp5p-TAP purifications (Figure 4) were digested using trypsin and the resulting peptides were labelled with iTRAQ(R) reagents 116 (Noc1p purification) and 117 (Rrp5p purification), subsequently pooled, separated by reversed phase nano HPLC and spotted on a MALDI–MS/MS target. The top eight peptides of the MS run in each spot were selected for fragmentation in MS/MS mode to determine the sequence of the peptide and the ratio of the iTRAQ reporter group signal intensities (117/116). For proteins identified with more than one peptide [ion score confidence interval (CI) >95%] the average iTRAQ ratio (117/116 = RRP5/NOC1) was calculated. LSU biogenesis factors (A) and SSU processome components (B) identified in both purifications are listed according to their average iTRAQ ratio (error bars are standard deviations of the average ratio; numbers of identified peptides are indicated in brackets). The average iTRAQ ratios of ribosomal proteins of the large (rpL) and the small (rpS) subunit identified with more than one peptide are shown in (A). (C) Peptide count analysis of RNA polymerase subunits identified in both purifications. Only peptides of Pol-I-specific subunits (A12.2, A49, A135, A190) and of common subunits (AC40, ABC23) were identified with a score >95% CI (one peptide each).
In summary, these results showed that the particles associated with Noc1p and Rrp5p are very similar in their RNA composition (U3 snoRNA; 35S, 32S, 27SA2, 27SBS pre-rRNAs), as well as in their content of SSU processome components and early LSU ribosome biogenesis factors. Accordingly, we conclude that Noc1p and Rrp5p interact as a protein complex with the common 90S precursor of the large and the small ribosomal subunits and with subsequent early pre-60S particles.
Evidence for co-transcriptional recruitment of Rrp5p, Noc1p and Noc2p to pre-ribosomes
In the MS analysis of proteins co-purifying with Noc1p-TAP and Rrp5p-TAP, similar abundant peptides of six subunits of RNA polymerases were identified (iTRAQ ratios ∼1, data not shown). These subunits were either specific for Pol-I (A12.2, A49, A135 and A190) or common to Pol-I, II and III (AC40, ABC23) (Figure 5C). Since Pol-I synthesizes the primary pre-rRNA transcripts, these data gave first evidence for a co-transcriptional recruitment of Rrp5p and Noc1p to nascent pre-ribosomal particles.
To further test this hypothesis, we analysed whether Rrp5p, Noc1p or Noc2p are specifically associated with rDNA chromatin transcribed by Pol-I. To better distinguish specific protein components of Pol-I chromatin from unspecific co-purified proteins, we compared Pol-I with Pol-II-associated chromatin. Since both polymerases are chromatin-associated transcription machineries, which have common subunits and transcription factors, but which should also differ in other co-transcriptionally associated factors, we considered this as a reasonable specificity control.
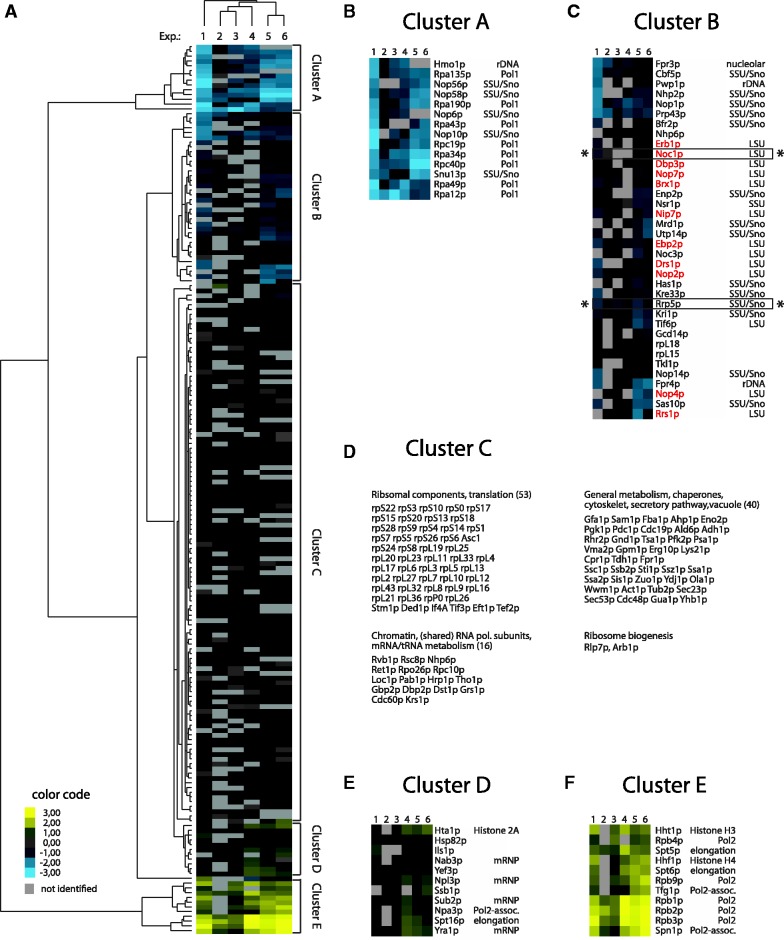
Pol-I and Pol-II were affinity purified from chromatin fractions of formaldehyde-treated yeast cells via Protein A tagged subunits Rpa135p and Rpb2p, respectively and co-purified proteins were analysed by comparative mass spectrometry using iTRAQ reagents. The results from six independent experiments were subjected to statistical analysis to determine groups of proteins specifically associated with Pol-I- or Pol-II-transcribed chromatin. In this way, five main groups showing different interaction patterns could be identified (Figure 6). Two groups (Figure 6, clusters A and B) consisted of proteins enriched in the Pol-I purification, whereas two other groups (Figure 6, clusters D and E) consisted of proteins preferentially enriched in the Pol-II purification. A fifth group (Figure 6, cluster C) contained proteins, which are similarly abundant in both purifications, like common components of RNA Pol-I and Pol-II chromatin (e.g. shared polymerase subunits Rpo26p, Rpc10p), general chromatin components (Rvb1p, Rsc8p) and a large number of obviously unspecific contaminants (e.g. r-proteins, chaperones, etc.).
Figure 6.
A specific set of LSU and SSU biogenesis factors is part of Pol-I-transcribed chromatin. Yeast strains expressing chromosomally encoded Protein A fusion proteins of the Pol-I subunit Rpa135p (TY2423) or the Pol-II subunit Rpb2p (TY2424) were grown in rich medium to exponential phase (OD600 ∼0.5–0.8) and cross linked using formaldehyde. Chromatin fractions were prepared and the bait proteins were purified using IgG coupled magnetic beads. The co-purified proteins were subjected to comparative MS analysis using iTRAQ reagents. For proteins identified with at least one peptide with an ion score of >95% CI, the (average) iTRAQ ratio (Pol-II versus Pol-I purification) was calculated to determine the relative abundance of the respective protein in the Pol-I purification compared with the Pol-II purification. The results of six independent experiments were subjected to statistical analysis using clustering algorithms to determine groups of proteins specifically associated with Pol-I or Pol-II-transcribed chromatin. Only proteins identified in at least four out of six experiments were included. (A) Overview of the cluster analysis of six independent comparative Pol-II/Pol-I purifications with the five main clusters indicated on the right. The colour code for the log2 transformed iTRAQ ratios of Pol-II versus Pol-I purifications are indicated (for details, see ‘Materials and Methods’ section). grey: protein not identified in this experiment; blue: enriched in Pol-I purifications; yellow: enriched in Pol-II purifications; black: equally abundant in Pol-I and Pol-II purifications. (B, C, E, F) Detailed view of the clusters A, B, D and E, respectively, including a classification of the identified proteins. rDNA: shown to co-immunoprecipitate rDNA; Pol1: Pol-I subunit; SSU/Sno: part of SSU-processome/90S pre-ribosome or snoRNPs involved in ribosome biogensis; LSU: Large ribosomal subunit biogenesis factor; mRNP: part of co-transcriptionally formed mRNP; elongation: Pol-II elongation factor; Pol2-assoc.: shown to associate with Pol-II. LSU biogenesis factors are shown in red if they were already identified in the Noc1p/Rrp5p proteome analyses shown in Figure 4. Identified components of the Rrp5p–Noc1p–Noc2p module are indicated by asterisk. (D) Proteins in cluster C are classified and listed according to their physiological function. The numbers of identified proteins are indicated in brackets.
Clusters D and E predominantly contained Pol-II-specific subunits, as well as Pol-II elongation factors (e.g. Spt5p, Spt6p, Figure 6E and F) and proteins involved in co-transcriptional mRNA metabolism (e.g. Yra1p, Sub2p, Figure 6E and F), together with a few putative contaminants like chaperones (Hsp82p, Ssb1p). Many of these proteins are expected to be part of Pol-II-transcribed chromatin, underlining the specificity of the purifications.
Strikingly, cluster A contained Pol-I-specific subunits, a known rDNA chromatin component [Hmo1p; (56)], as well as some components of the SSU processome and small nucleolar RNPs (snoRNPs) supposed to be co-transcriptionally recruited to nascent pre-rRNA (Figure 6B) (5,76). Cluster B contained a larger group of proteins specifically enriched in Pol-I-transcribed rDNA chromatin. Among them were two proteins previously shown to bind to rDNA chromatin [Pwp1p (77), Fpr4p (78)], several components of the SSU processome and snoRNPs, as well as several LSU biogenesis factors (Figure 6C), but only few putative contaminants (e.g. Tkl1p, rpL15, rpL18). Interestingly, the LSU factors included Rrp5p and Noc1p, and 11 out of 13 of them were already identified in the experiments shown in Figure 5A as constituents of Noc1p- and Rrp5p-associated early LSU precursor particles. Possibly due to limitations in sensitivity, Noc2p and several factors thought to be co-transcriptionally recruited to rDNA chromatin, e.g. SSU processome components, as well as several RNA polymerase subunits, could not be detected using this approach. Nevertheless, these results clearly indicated that Noc1p, Rrp5p and a specific set of LSU biogenesis factors are part of Pol-I-associated chromatin, and therefore most likely co-transcriptionally recruited to nascent pre-(60S) rRNA.
Taken together, these findings support our hypothesis that Noc1p and Rrp5p are components of the earliest pre-60S particles.
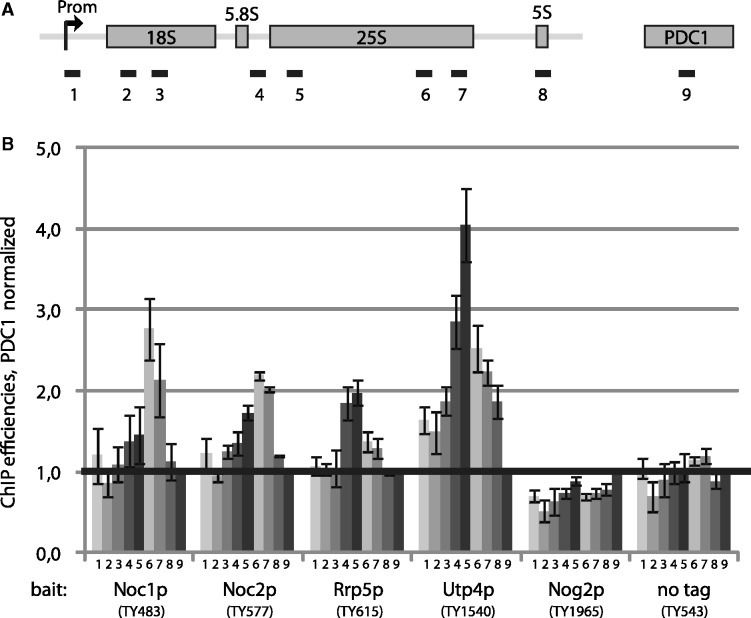
To analyse more directly the association of Rrp5p, Noc1p or Noc2p with Pol-I-transcribed rDNA chromatin, we performed ChIP experiments using yeast strains expressing TAP tag fusion proteins of Rrp5p, Noc1p or Noc2p. Additionally, ChIP was carried out with extracts from yeast strains expressing TAP tag fusion proteins of Utp4p, a SSU processome component reported to be co-transcriptionally recruited to nascent pre-ribosomes (5,8), and of Nog2p, a LSU biogenesis factor predominantly associated with LSU precursors of later maturation state (79). The DNA co-purified with tagged factors from extracts of formaldehyde-treated cells was measured by quantitative PCR. In addition to several amplicons distributed over the Pol-I-transcribed region of rDNA, amplicons spanning regions of the Pol-III-transcribed 5S rRNA gene and the Pol-II-transcribed PDC1 gene were included in the analyses as internal background controls (Figure 7A). In purifications from cell extracts of strains expressing no tagged protein or Nog2p-TAP, Pol-I-transcribed regions of rDNA were not significantly enriched compared with DNA from the PDC1 locus or the 5S rRNA gene locus (Figure 7B, compare amplicons 1–7 with 8 and 9). Consistent with analyses performed by Wery et al. (8), in the Utp4p-TAP purification DNA spanning the internal transcribed region 2 (ITS2) and the 5′-end of the 25S rRNA coding region of rDNA were specifically enriched over the internal background (Figure 7B, compare amplicons 4–7 with 8 and 9). Note that 5S rDNA is significantly more enriched than PDC1 DNA, possibly due to increased non-specific crosslinking of Utp4p to nucleolar chromatin.
Figure 7.
Analysis of co-transcriptional recruitment of Noc1p, Noc2p and Rrp5p to the 35S rRNA gene. (A) Primer pair positions on the rRNA gene to perform quantitative PCR (qPCR) analysis of ChIP experiments. The relative positions of the rDNA amplicons analysed by qPCR (1–8) are indicated. For normalization, an amplicon in the PDC1 gene (9) was used. (B) ChIP experiments were performed with yeast strains in which Noc1p (TY483), Noc2p (TY577), Rrp5p (TY615), Utp4p (TY1540) or Nog2p (TY1965) are expressed as tandem affinity purification (TAP) tag fusion proteins, and with a control strain expressing no tagged protein (TY543). Cells were grown in rich medium at 30°C to exponential phase (OD600 = 0.5–0.7) and cross-linked using formaldehyde (final concentration 1%) for 15’ at 30°C. ChIP analysis was performed as described in ‘Materials and Methods’ section. The amounts of specific DNA fragments present in the input and retained on the beads were determined by qPCR with primer pairs amplifying the regions 1–8 of the rDNA depicted in the schematic representation and of the PDC1 gene (primer pair 9). In each experiment the precipitation efficiencies [percent IP (rDNA)] for the respective amplified DNA regions were calculated and normalized to the PDC1 precipitation efficiencies [percent IP (rDNA)/percent IP (PDC1)]. The graph shows the average of three biological replicates, including standard deviations. A black line depicts the internal background as a result of the normalization to the precipitated PDC1 DNA.
A similar result was obtained when analyzing the DNA co-purified with Rrp5p-TAP. However, enrichment of ITS2 and the 5′-end of the 25S rRNA coding regions with regard to the internal background of co-purifying PDC1 DNA in Rrp5p-TAP purifications was less than the enrichment observed in Utp4p-TAP purifications. On the other hand, enrichment of ITS2 and 25S rRNA coding regions over 5S rDNA was very comparable in both cases. Analyses of DNA co-purified with Noc1p-TAP and Noc2p-TAP indicated specific association of both proteins with rDNA chromatin regions coding for the 3′ region of 25S rRNA (Figure 7B, compare amplicons 4–7 with 8 and 9.).
Recruitment of UTP-A and UTP-B components to rDNA chromatin might be mediated by nascent (pre-r)RNA (8). To address this point for the components of the Rrp5p–Noc1p–Noc2p module, we performed ChIP experiments as described above, however with or without treatment of the chromatin fractions with RNase prior to chromatin IP (Supplementary Figure S6). In a similar approach we analysed if RNase treatment affects the protein composition of chromatin co-purifying with Pol-I. The results of both analyses were in agreement with the assumption that the association of sub-groups of early acting LSU biogenesis factors with rDNA chromatin is mediated by pre-rRNA. However, we note that interpretation of these analyses was complicated by the partial interference of RNase treatment with the purification of Pol-I-associated chromatin and of the TAP-tagged Noc1p and Noc2p proteins (see comments in figure legend of Supplementary Figure S6 for more details).
Analysis of the binding hierarchy to pre-ribosomes
Next, we wanted to elucidate the binding hierarchy of the Rrp5p–Noc1p–Noc2p module members to pre-ribosomes. First, we tested whether Rrp5p, which directly interacts with specific sites in the pre-rRNA (23,24), affects recruitment of Noc1p and Noc2p to pre-ribosomes. Therefore, chromosomally encoded TAP-tag fusion proteins of Noc1p and Noc2p were affinity purified from extracts of cells expressing Rrp5p under control of the galactose inducible/glucose repressible GAL1/10- or the endogenous RRP5-promoter after cultivation in glucose containing medium.
Northern blot analyses of total RNA isolated from cell extracts indicated that 18 h after shut down of Rrp5p expression, the previously described pre-rRNA processing phenotype was well established (18). The 35S pre-rRNA was still detectable in similar levels as in the non-depleted control strain, but levels of 32S, 27SA2, 27SB, 20S and 7S pre-rRNAs were strongly reduced. Accordingly, Rrp5p-depletion caused a general destabilization of the canonical rRNA precursors (Figure 8A, compare lanes 2/4, 10/12), whereas persistence of 35S pre-rRNA could be either due to ongoing transcription, impaired pre-rRNA processing or to both. Additionally, several aberrant pre-rRNA fragments [most likely resembling the described 31S′, 30S′, 24S, 17S′, 12S′ RNAs (18)] were detected and levels of mature 25S, 18S and 5.8S were reduced (Figure 8A, compare lanes 2/4, 10/12). As described above (Figure 4), Noc1p-TAP co-purified U3 snoRNA, 35S, 32S, 27SA2 and less efficiently 27SB pre-rRNAs from lysates of cells expressing Rrp5p (Figure 8A, lanes 5 and 6). Noc1p-TAP still co-purified U3 snoRNA with similar and 35S pre-rRNA with even increased efficiency after in vivo depletion of Rrp5p (Figure 8A, compare lanes 2/6 and 4/8). Besides, significant amounts of aberrant pre-rRNA fragments detected by oligo o207 (Supplementary Figure S1A), were specifically enriched in affinity-purified Noc1p-TAP fractions (Figure 8A, lane 8, see fragments running between 35S and 27SA2 pre-rRNA marked by a cross).
Analysis of the Noc2p-TAP purifications showed virtually the same results. Noc2p specifically co-purified U3 snoRNA, 35S, 32S, 27SA2 and 27SB pre-rRNAs and some amounts of 7S pre-rRNA from extracts of cells expressing Rrp5p (Figure 8A, lanes 13 and 14). After in vivo depletion of Rrp5p, U3 snoRNA and 35S pre-rRNA were co-purified even more efficiently with Noc2p-TAP, and also the above mentioned aberrant pre-rRNA fragments were specifically enriched in the affinity-purified Noc2p-TAP fractions (Figure 8A, compare lanes 10/14 and 12/16, fragments marked by a cross).
These experiments indicated that Noc1p and Noc2p can bind to nascent, destabilized pre-ribosomes when Rrp5p expression is shut down.
Next, we investigated whether Noc1p or Noc2p depletion affects the recruitment of Rrp5p to pre-ribosomes. Besides, we addressed the influence of Noc1p on the recruitment of the UTP-C component Utp22p to pre-ribosomes, which was found to depend on the function of Rrp5p (16). Therefore, analogous experiments as above (Figure 8A) were performed with yeast strains in which NOC1 or NOC2 genes are under the control of the GAL1/10 promoter and which express TAP-tag fusion proteins of Rrp5p or Utp22p (Figure 8B and C). In the corresponding strains protein levels of Noc1p and Noc2p were reduced to ∼25%, <5% and <2% of endogenous levels after 10, 18 and 24 h shift to glucose containing medium (data not shown).
The pre-rRNA processing phenotypes in these mutant strains were virtually identical and largely resembled the ones observed in yeast strains carrying temperature sensitive alleles of NOC1 and NOC2 (27). Levels of 35S/32S and 23S pre-rRNA were slightly elevated compared with the wild-type control. This delay in early pre-rRNA processing events at A0, A1 and A2 was observed before in numerous mutants affecting the LSU maturation pathway [(80), and discussion therein; see also (81,82)]. The 27SA2, 27SB and 7S pre-rRNAs were drastically reduced, whereas decrease in 20S pre-rRNA was less severe. Consequently, levels of mature LSU components 25S and 5.8S rRNA were significantly diminished, whereas levels of the mature SSU component 18S rRNA showed only slight reduction (Figure 8B and C, compare lanes 2/4/5, 12/14/15, 22/24/25). Altogether, the data indicated a pronounced destabilization of most LSU precursors after depletion of Noc1p and Noc2p.
Rrp5p-TAP co-purified the same RNA species as observed in the experiments shown in Figure 4 from extracts of cells normally expressing Noc1p and Noc2p (Figure 8B and C, lanes 6, 7, 16 and 17). After in vivo depletion of Noc1p or Noc2p, co-purification efficiency of U3 snoRNA, 35S, 32S and 23S pre-rRNAs with Rrp5p-TAP increased (Figure 8B and C, lanes 9, 10, 19 and 20), indicating Noc1p/Noc2p independent recruitment of Rrp5p to pre-ribosomes and a possible prolonged dwelling time of Rrp5p in the corresponding RNPs. Remarkably, in this situation a significantly increased population of Rrp5p was found to be associated with particles containing 23S pre-rRNA, possibly by direct interactions with a pre-rRNA region between processing sites A2 and A3 (23,24). Besides, a quite heterogeneous set of non-canonical pre-rRNA fragments migrating between 32S and 27SA2 pre-rRNA were specifically enriched with Rrp5p (Figure 8B and C, lanes 9, 10, 19, 20, 29 and 30), underlining that in the absence of Noc1p or Noc2p, early ribosomal precursors are not properly processed or are destabilized and finally get degraded.
As expected, the UTP-C component Utp22p-TAP co-purified most efficiently U3 snoRNA, 35S, 32S, as well as 23S, 22S and 21S pre-rRNAs from cells expressing Noc1p (Figure 8C, lanes 26 and 27). After in vivo depletion of Noc1p, co-purification efficiency of U3 snoRNA, 35S, 32S and 23S pre-rRNAs with Utp22p-TAP increased (Figure 8C, lanes 29 and 30), as observed for Rrp5p-TAP (see above). These results indicated a Noc1p independent recruitment of Utp22p to pre-ribosomes and a possible prolonged dwelling time of Utp22p in the corresponding RNPs.
Altogether, these data suggested that LSU precursors that are depleted of either Noc1p, Noc2p or Rrp5p are specifically sensitive to pre-rRNA degradation pathways. Interestingly, after depletion of one complex component the respective other module members were still associated with the residual detectable particles.
DISCUSSION
Here, we present a detailed analysis of the ribosome biogenesis factor module constituted of yeast Noc1p, Noc2p and Rrp5p. We show evidence that Noc1p can simultaneously establish specific contacts with Rrp5p and Noc2p and thus enables the formation of a bridged hetero-oligomeric Rrp5p–Noc1p–Noc2p protein complex. In agreement with Rrp5p and Noc1p acting in vivo as part of one protein module on pre-ribosomes, we found that the RNA and protein compositions of ex vivo purified Rrp5p and Noc1p containing RNPs are very similar. Both proteins are predominantly associated with the first detectable specific pre-60S particle, which result in yeast from pre-rRNA cleavage in the ITS1 pre-rRNA region at site A2 and the subsequent separation of the LSU and SSU precursors [Figure 4; see also (23)]. According to studies of de Boer et al. (23) and our analyses, dissociation of Noc1p and Rrp5p from pre-ribosomes is tightly linked to the removal of residual ITS1 sequences from LSU pre-rRNAs downstream of A2.
Our data indicate that the Rrp5p–Noc1p–Noc2p module is already co-transcriptionally recruited to nascent pre-rRNA. Congruently, Pol-I subunits were detected together with 35S pre-rRNA and U3 snoRNA and numerous other co-transcriptionally recruited SSU processome components (5–8,14,16) in affinity-purified Noc1p and Rrp5p fractions. Assembly of Noc1p and Noc2p into the earliest ribosomal precursors is consistent with the previously observed co-sedimentation of Noc1p with 35S pre-rRNA in sucrose gradients (27) and the co-precipitation of U3 snoRNA with human Noc2p/NIR (32). Strikingly, chIP analyses showed that Rrp5p, Noc1p and Noc2p associate specifically with the 25S rRNA coding region of the 35S rDNA chromatin. In agreement with this, many of the SSU and LSU biogenesis factors found in early pre-ribosomal particles together with Rrp5p and Noc1p were detected in Pol-I-associated rDNA chromatin. Altogether, there is good evidence that recruitment of the Rrp5p–Noc1p–Noc2p module into pre-ribosomes can occur already co-transcriptionally in yeast, similar to the recruitment of SSU processome components. But in clear contrast to SSU processome components, the Rrp5p–Noc1p–Noc2p module predominantly associates with LSU precursors after separation of LSU and SSU pre-rRNAs through cleavage in the ITS1.
The previously observed apparent lack of LSU factors in 90S/SSU processome particles (6) can be in part explained by the existence of different SSU processome particles in the cell (83). Accordingly, one population contains the common 35S pre-rRNA, SSU biogenesis factors and LSU biogenesis factors as the Rrp5p–Noc1p–Noc2p module. After cleavage in the ITS1 region of the rRNA precursor, which occurs in yeast with fast kinetics (10), the assembled factors stay, at least in part, still associated with the resulting pre-40S and pre-60S particles. As a consequence, pre-ribosomal particles purified via tagged SSU processome components can comprise a mixture of common 90S and predominantly, specific pre-40S pre-ribosomes. Analogously, Noc1p and Rrp5p co-purify predominantly LSU precursors together with LSU biogenesis factors and smaller amounts of 35S rRNA and SSU biogenesis factors containing common precursor particles.
Co-transcriptional assembly of the SSU processome and the concomitant gradual compaction of the pre-rRNA is thought to result in the formation of ‘terminal ball’ structures or ‘SSU knobs’ seen in chromatin spreads of rRNA genes (3–5). Appearance of new terminal structures, called ‘LSU knobs’, was observed on nascent rDNA transcripts of S. cerevisiae, from which SSU knobs are thought to be cleaved off in some cases through a co-transcriptional cut in the rRNA–ITS1 region (9,10). We propose that the Rrp5p–Noc1p–Noc2p module assembles either into these early LSU knobs or, in case cleavage in the ITS1 pre-rRNA region does not occur during ongoing transcription, into 90S pre-ribosomes containing 35S pre-rRNA and SSU processome components. The size of the reconstituted Rrp5p–Noc1p–Noc2p module (∼12 nm; Figure 2C) is smaller than the size of early LSU knobs (15–20 nm) (9). This could mean that other LSU biogenesis factors, which co-purify with Noc1p and Rrp5p (Figure 5) and which are part of the Pol-I-associated chromatin like Erb1p, Dbp3p, Nop7p, Brx1p (Figure 6C), or which show (weak) association with 35S pre-rRNA-like Ssf1p (62), Nop7p (84), Rlp7p, Nsa3p and Ytm1p (71) are also constituents of early LSU knobs, as it was recently suggested for Nop53p (11).
Although the protein compositions of affinity purified Rrp5p and Noc1p particles are comparable, Rrp5p shows features and functions different from those of Noc1p and Noc2p: (i) the C-terminal part of Rrp5p, which does not bind to Noc1p (Figure 3), is required for pre-40S biogenesis (18–20); (ii) Rrp5p, not Noc1p, is required for the recruitment of the UTP-C complex to 35S pre-rRNA (Figure 8C); (16); (iii) the rDNA ChIP profiles of Rrp5p are shifted more towards 18S rRNA coding regions when compared with the ones of Noc1p and Noc2p, similar as for the SSU component Utp4p (Figure 7); (8). All of them are consistent with an additional temporally and/or physically separated role of Rrp5p in ribosome biogenesis. It is possible that Rrp5p acquires these pre-40S-specific features in the context of the Rrp5p–Noc1p–Noc2p module, potentially involving its C-terminus. Alternatively, a Noc1p–Noc2p-free fraction of Rrp5p could exist that interacts with pre-40S particles either weakly or very transiently. In either case, this implies that Rrp5p could have another interaction interface with SSU rRNA precursors upstream of the A2 processing site, as suggested by Young et al. (24).
The experiments analyzing the binding hierarchy of Rrp5p–Noc1p–Noc2p module members to 60S subunit precursors (Figure 8) point toward important functional characteristics of factors acting early in the LSU maturation pathway. Individual depletion of each module component leads to a drastic reduction of canonical rRNA precursors (Figure 7, input lanes) and to the appearance of a variety of aberrant pre-rRNAs, which are still bound by the other module components, respectively (Figure 7, IP lanes). We see two major implications of these results: first, each of the module members is individually required to protect pre-ribosomal particles from aberrant RNA processing and, most likely, degradation events. Second, the interaction interface with pre-ribosomes is not provided by one single module member. Taken together, it is likely that the Rrp5p–Noc1p–Noc2p module establishes a rather extended interaction interface within nascent pre-ribosomal particles during rDNA transcription. This in turn might protect pre-LSUs from inappropriate and non-productive nucleolytic activities and might be involved in early folding events of the rRNA. Depletion of other LSU biogenesis factors found in the early LSU precursors purified via Rrp5p and Noc1p causes a similar pronounced destabilization of pre-rRNAs [e.g. Rix7p (65), Ssf1p (62), Nop4p (73), Rrs3p (74), Dbp9p (75)]. Therefore, it is possible that either each of these factors has similar functional properties or represents an essential unit of one common functional entity in early LSU biogenesis. A common function of these factors is also supported by the observation that in corresponding yeast mutant strains residual non-degraded pre-60S particles fail to leave the nucleolar compartment (27,70,85). Future in vivo and in vitro studies will help to reveal how the Rrp5p–Noc1p–Noc2p module stimulates folding and compaction of pre-rRNA into LSU knob-like structures and how this affects downstream assembly events and the action of nucleases involved in pre-ribosome processing and turnover.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7, Supplementary Methods and Supplementary References [86–89].
FUNDING
The Deutsche Forschungsgemeinschaft (DFG) [SFB 960 and SFB 638]. Funding for open access charge: DFG [SFB960].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Drs Olivier Gadal (Université Toulouse) and Jan Vos (Vrije Universiteit Amsterdam) for providing us with plasmids.
REFERENCES
- 1.Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 4.Mougey EB, O’Reilly M, Osheim Y, Miller OL, Jr, Beyer A, Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- 5.Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 7.Gallagher JEG, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wery M, Ruidant S, Schillewaert S, Leporé N, Lafontaine DLJ. The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA. 2009;15:406–419. doi: 10.1261/rna.1402709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 10.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granato DC, Machado-Santelli GM, Oliveira CC. Nop53p interacts with 5.8S rRNA co-transcriptionally, and regulates processing of pre-rRNA by the exosome. FEBS J. 2008;275:4164–4178. doi: 10.1111/j.1742-4658.2008.06565.x. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein KA, Gallagher JEG, Mitchell BM, Granneman S, Baserga SJ. The small-subunit processome is a ribosome assembly intermediate. Eukaryotic Cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watkins NJ, Ségault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Lührmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 14.Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- 15.Krogan NJ, Peng W-T, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 16.Pérez-Fernández J, Román A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kühn H, Hierlmeier T, Merl J, Jakob S, Aguissa-Touré A-H, Milkereit P, Tschochner H. The Noc-domain containing C-terminus of Noc4p mediates both formation of the Noc4p-Nop14p submodule and its incorporation into the SSU processome. PLoS ONE. 2009;4:e8370. doi: 10.1371/journal.pone.0008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 19.Torchet C, Jacq C, Hermann-Le Denmat S. Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA. 1998;4:1636–1652. doi: 10.1017/s1355838298981511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eppens NA, Rensen S, Granneman S, Raué HA, Venema J. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA. 1999;5:779–793. doi: 10.1017/s1355838299990313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eppens NA, Faber AW, Rondaij M, Jahangir RS, van Hemert S, Vos JC, Venema J, Raué HA. Deletions in the S1 domain of Rrp5p cause processing at a novel site in ITS1 of yeast pre-rRNA that depends on Rex4p. Nucleic Acids Res. 2002;30:4222–4231. doi: 10.1093/nar/gkf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 23.de Boer P, Vos HR, Faber AW, Vos JC, Raué HA. Rrp5p, a trans-acting factor in yeast ribosome biogenesis, is an RNA-binding protein with a pronounced preference for U-rich sequences. RNA. 2006;12:263–271. doi: 10.1261/rna.2257606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young CL, Karbstein K. The roles of S1 RNA-binding domains in Rrp5’s interactions with pre-rRNA. RNA. 2011;17:512–521. doi: 10.1261/rna.2458811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merl J, Jakob S, Ridinger K, Hierlmeier T, Deutzmann R, Milkereit P, Tschochner H. Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes. Nucleic Acids Res. 2010;38:3068–3080. doi: 10.1093/nar/gkp1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edskes HK, Ohtake Y, Wickner RB. Mak21p of Saccharomyces cerevisiae, a homolog of human CAATT-binding protein, is essential for 60 S ribosomal subunit biogenesis. J. Biol. Chem. 1998;273:28912–28920. doi: 10.1074/jbc.273.44.28912. [DOI] [PubMed] [Google Scholar]
- 27.Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell. 2001;105:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 28.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen JS, Lyon CE, Fox AH, Leung AKL, Lam YW, Steen H, Mann M, Lamond AI. Directed Proteomic Analysis of the Human Nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 30.Sweet T, Yen W, Khalili K, Amini S. Evidence for involvement of NFBP in processing of ribosomal RNA. J. Cell. Physiol. 2008;214:381–388. doi: 10.1002/jcp.21204. [DOI] [PubMed] [Google Scholar]
- 31.Li N, Yuan L, Liu N, Shi D, Li X, Tang Z, Liu J, Sundaresan V, Yang W-C. SLOW WALKER2, a NOC1/MAK21 homologue, is essential for coordinated cell cycle progression during female gametophyte development in Arabidopsis. Plant Physiol. 2009;151:1486–1497. doi: 10.1104/pp.109.142414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Zhang Y, Wang Y, Kong R, Hu L, Schuele R, Du X, Ke Y. Transcriptional repressor NIR functions in the ribosome RNA processing of both 40S and 60S subunits. PLoS ONE. 2012;7:e31692. doi: 10.1371/journal.pone.0031692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lum LS, Sultzman LA, Kaufman RJ, Linzer DI, Wu BJ. A cloned human CCAAT-box-binding factor stimulates transcription from the human hsp70 promoter. Mol. Cell. Biol. 1990;10:6709–6717. doi: 10.1128/mcb.10.12.6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lum LS, Hsu S, Vaewhongs M, Wu B. The hsp70 gene CCAAT-binding factor mediates transcriptional activation by the adenovirus E1a protein. Mol. Cell. Biol. 1992;12:2599–2605. doi: 10.1128/mcb.12.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imbriano C, Bolognese F, Gurtner A, Piaggio G, Mantovani R. HSP-CBF Is an NF-Y-dependent Coactivator of the Heat Shock Promoters CCAAT Boxes. J. Biol. Chem. 2001;276:26332–26339. doi: 10.1074/jbc.M101553200. [DOI] [PubMed] [Google Scholar]
- 36.Sweet T, Khalili K, Sawaya BE, Amini S. Identification of a novel protein from glial cells based on its ability to interact with NF-kappaB subunits. J. Cell. Biochem. 2003;90:884–891. doi: 10.1002/jcb.10701. [DOI] [PubMed] [Google Scholar]
- 37.Sweet T, Sawaya BE, Khalili K, Amini S. Interplay between NFBP and NF-kappaB modulates tat activation of the LTR. J. Cell. Physiol. 2005;204:375–380. doi: 10.1002/jcp.20419. [DOI] [PubMed] [Google Scholar]
- 38.Hublitz P, Kunowska N, Mayer UP, Müller JM, Heyne K, Yin N, Fritzsche C, Poli C, Miguet L, Schupp IW, et al. NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes Dev. 2005;19:2912–2924. doi: 10.1101/gad.351205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heyne K, Willnecker V, Schneider J, Conrad M, Raulf N, Schüle R, Roemer K. NIR, an inhibitor of histone acetyltransferases, regulates transcription factor TAp63 and is controlled by the cell cycle. Nucleic Acids Res. 2010;38:3159–3171. doi: 10.1093/nar/gkq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 41.Chakraborty A, Uechi T, Kenmochi N. Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip. Rev. RNA. 2011;2:507–522. doi: 10.1002/wrna.73. [DOI] [PubMed] [Google Scholar]
- 42.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. 2005 edn. New York: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 43.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 44.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 45.Strässer K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Meth. Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 47.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 48.Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I. Protein complex expression by using multigene baculoviral vectors. Nat. Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Vol. 3. New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 50.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 51.Jakob S, Ohmayer U, Neueder A, Hierlmeier T, Perez-Fernandez J, Hochmuth E, Deutzmann R, Griesenbeck J, Tschochner H, Milkereit P. Interrelationships between yeast ribosomal protein assembly events and transient ribosome biogenesis factors interactions in early pre-ribosomes. PLoS One. 2012;7:e32552. doi: 10.1371/journal.pone.0032552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiter A, Steinbauer R, Philippi A, Gerber J, Tschochner H, Milkereit P, Griesenbeck J. Reduction in ribosomal protein synthesis is sufficient to explain major effects on ribosome production after short-term TOR inactivation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2011;31:803–817. doi: 10.1128/MCB.01227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milkereit P, Strauss D, Bassler J, Gadal O, Kühn H, Schütz S, Gas N, Lechner J, Hurt E, Tschochner H. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- 55.Venema J, Planta RJ, Raué HA. In vivo mutational analysis of ribosomal RNA in Saccharomyces cerevisiae. Methods Mol. Biol. 1998;77:257–270. doi: 10.1385/0-89603-397-X:257. [DOI] [PubMed] [Google Scholar]
- 56.Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008;22:1190–1204. doi: 10.1101/gad.466908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goetze H, Wittner M, Hamperl S, Hondele M, Merz K, Stoeckl U, Griesenbeck J. Alternative chromatin structures of the 35S rRNA genes in Saccharomyces cerevisiae provide a molecular basis for the selective recruitment of RNA polymerases I and II. Mol. Cell. Biol. 2010;30:2028–2045. doi: 10.1128/MCB.01512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tardiff DF, Abruzzi KC, Rosbash M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc. Natl Acad. Sci. USA. 2007;104:19948–19953. doi: 10.1073/pnas.0710179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kressler D, Roser D, Pertschy B, Hurt E. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J. Cell Biol. 2008;181:935–944. doi: 10.1083/jcb.200801181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vos HR, Faber AW, de Gier MD, Vos JC, Raué HA. Deletion of the three distal S1 motifs of Saccharomyces cerevisiae Rrp5p abolishes pre-rRNA processing at site A(2) without reducing the production of functional 40S subunits. Eukaryot. Cell. 2004;3:1504–1512. doi: 10.1128/EC.3.6.1504-1512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torchet C, Hermann-Le Denmat S. Bypassing the rRNA processing endonucleolytic cleavage at site A2 in Saccharomyces cerevisiae. RNA. 2000;6:1498–1508. doi: 10.1017/s1355838200000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fatica A, Cronshaw AD, Dlakić M, Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 63.Vos HR, Bax R, Faber AW, Vos JC, Raué HA. U3 snoRNP and Rrp5p associate independently with Saccharomyces cerevisiae 35S pre-rRNA, but Rrp5p is essential for association of Rok1p. Nucleic Acids Res. 2004;32:5827–5833. doi: 10.1093/nar/gkh904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Gadal O, Strauss D, Braspenning J, Hoepfner D, Petfalski E, Philippsen P, Tollervey D, Hurt E. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 2001;20:3695–3704. doi: 10.1093/emboj/20.14.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huber MD, Dworet JH, Shire K, Frappier L, McAlear MA. The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J. Biol. Chem. 2000;275:28764–28773. doi: 10.1074/jbc.M000594200. [DOI] [PubMed] [Google Scholar]
- 67.Pestov DG, Stockelman MG, Strezoska Z, Lau LF. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 2001;29:3621–3630. doi: 10.1093/nar/29.17.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oeffinger M, Leung A, Lamond A, Tollervey D, Lueng A. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA. 2002;8:626–636. doi: 10.1017/s1355838202020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horsey EW, Jakovljevic J, Miles TD, Harnpicharnchai P, Woolford JL., Jr Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 2004;10:813–827. doi: 10.1261/rna.5255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL., Jr Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL., Jr Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 2011;30:4020–4032. doi: 10.1038/emboj.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaser A, Bogengruber E, Hallegger M, Doppler E, Lepperdinger G, Jantsch M, Breitenbach M, Kreil G. Brix from xenopus laevis and brx1p from yeast define a new family of proteins involved in the biogenesis of large ribosomal subunits. Biol. Chem. 2001;382:1637–1647. doi: 10.1515/BC.2001.199. [DOI] [PubMed] [Google Scholar]
- 73.Bergès T, Petfalski E, Tollervey D, Hurt EC. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsuno A, Miyoshi K, Tsujii R, Miyakawa T, Mizuta K. RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:2066–2074. doi: 10.1128/mcb.20.6.2066-2074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Daugeron MC, Kressler D, Linder P. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA. 2001;7:1317–1334. doi: 10.1017/s1355838201010640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.García-Gómez JJ, Babiano R, Lebaron S, Froment C, Monsarrat B, Henry Y, de la Cruz J. Nop6, a component of 90S pre-ribosomal particles, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. RNA Biol. 2011;8:112–124. doi: 10.4161/rna.8.1.14143. [DOI] [PubMed] [Google Scholar]
- 77.Suka N, Nakashima E, Shinmyozu K, Hidaka M, Jingami H. The WD40-repeat protein Pwp1p associates in vivo with 25S ribosomal chromatin in a histone H4 tail-dependent manner. Nucleic Acids Res. 2006;34:3555–3567. doi: 10.1093/nar/gkl487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuzuhara T, Horikoshi M. A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat. Struct. Mol. Biol. 2004;11:275–283. doi: 10.1038/nsmb733. [DOI] [PubMed] [Google Scholar]
- 79.Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Beekvelt CA, de Graaff-Vincent M, Faber AW, van’t Riet J, Venema J, Raué HA. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:5001–5008. doi: 10.1093/nar/29.24.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pöll G, Braun T, Jakovljevic J, Neueder A, Jakob S, Woolford JL, Jr, Tschochner H, Milkereit P. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One. 2009;4:e8249. doi: 10.1371/journal.pone.0008249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 84.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 85.Gadal O, Strauss D, Petfalski E, Gleizes P-E, Gas N, Tollervey D, Hurt E. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell Biol. 2002;157:941–951. doi: 10.1083/jcb.200111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 87.Cadwell C, Yoon HJ, Zebarjadian Y, Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.