Abstract
The environmental arylamine mutagens are implicated in the etiology of various sporadic human cancers. Arylamine-modified dG lesions were studied in two fully paired 11-mer duplexes with a -G*CN- sequence context, in which G* is a C8-substituted dG adduct derived from fluorinated analogs of 4-aminobiphenyl (FABP), 2-aminofluorene (FAF) or 2-acetylaminofluorene (FAAF), and N is either dA or dT. The FABP and FAF lesions exist in a simple mixture of ‘stacked’ (S) and ‘B-type’ (B) conformers, whereas the N-acetylated FAAF also samples a ‘wedge’ (W) conformer. FAAF is repaired three to four times more efficiently than FABP and FAF. A simple A- to -T polarity swap in the G*CA/G*CT transition produced a dramatic increase in syn-conformation and resulted in 2- to 3-fold lower nucleotide excision repair (NER) efficiencies in Escherichia coli. These results indicate that lesion-induced DNA bending/thermodynamic destabilization is an important DNA damage recognition factor, more so than the local S/B-conformational heterogeneity that was observed previously for FAF and FAAF in certain sequence contexts. This work represents a novel 3′-next flanking sequence effect as a unique NER factor for bulky arylamine lesions in E. coli.
INTRODUCTION
Structural and conformational damage in specific areas of the genome can trigger tumorigenesis. For example, disruption of a gene that encodes the tumor suppressor p53 protein has been found in the majority of sporadic human cancer (1). Although human cells are equipped with repair pathways to safeguard the genome from various DNA damage, some lesions may go unrepaired, thereby serving as a faulty template to produce a complex array of mutations and genomic instability, ultimately leading to cancer initiation (2).
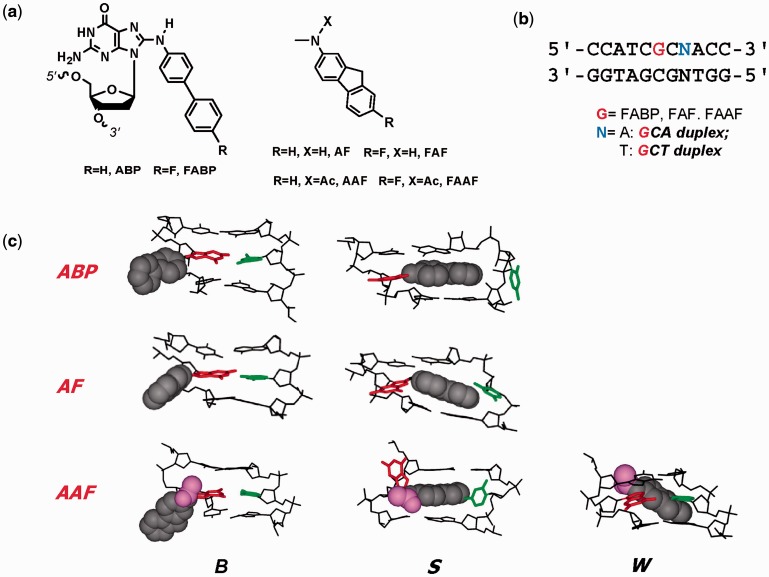
Arylamines and heterocyclic amines are well-known environmental mutagens/carcinogens, which have been implicated in the etiology of breast, liver and bladder cancers in humans (3). Metabolic activation of these amines in vivo produces C8-substituted dG as the major DNA adducts (4). For example, the human bladder carcinogen 4-aminobiphenyl produces ABP (Figure 1a). Similarly, AF and AAF are the major DNA adducts derived from 2-aminofluorene, 2-nitrofluorene and 2-acetylaminofluorene (Figure 1a). The ABP and AF adducts in fully paired duplex DNA have been shown to adopt an equilibrium of two prototype conformers: ‘B-type’, in which the carcinogen resides in the major groove of a relatively unperturbed double helical DNA, and ‘stacked (S)’, in which the carcinogen is base displaced and the glycosidic linkage to the modified guanine is syn (Figure 1c) (5,6). The aromatic moieties of ABP are not coplanar as in AF, which results in a much lower S-state population than AF. AF-induced S/B-heterogeneity is dependent on the flanking sequence, which modulates mutational and repair outcomes (6,7). AAF is chemically identical to AF except for a single acetyl group on the central nitrogen (Figure 1a), leading to sampling of an additional W-conformation, in which the fluorene moiety is in the minor groove along with a syn glycosidic linkage (Figure 1c) (7,8). The B and S conformers exhibited by AAF are similar to those obtained for ABP and AF.
Figure 1.
(a) Structures of ABP [N-(2′-deoxyguanosin-8-yl)-4-aminobiphenyl], AF [N-(2′-deoxyguanosin-8-yl)-2-aminofluorene] and AAF [N-(2′-deoxyguanosin-8-yl)-2-acetylaminofluorene] and their fluoro models, FABP [N-(2′-deoxyguanosin-8-yl)-4-fluoro-4-aminobiphenyl], FAF [N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-aminofluorene] and FAAF [N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-acetylaminofluorene]; (b) 11-mer GCA and GCT duplexes used in this study; (c) Major groove views of the B, S and W-conformers of ABP, AF and AAF. Modified-dG (red), dC (green) opposite the lesion site (orphan C), fluorene (grey CPK), acetyl (AAF only, magenta).
Nucleotide excision repair (NER) is the major cellular pathway for removing bulky DNA lesions in cells. Accumulated evidence suggests that efficiency of NER is governed by various structural, cellular and biological factors (9–11). Sequence context, in particular, plays an important role in NER of bulky DNA lesions (7,12). The most notable sequence effects were observed in the NarI sequence (5′-…CG1G2CG3CC…-3′), which is well known for inducing higher frequency of −2 deletion mutations when adducted by AAF at G3 position despite the similar chemical reactivities of three guanines (13,14). Fuchs and coworkers have shown that AAF in duplex is an excellent substrate for Escherichia coli UvrABC and human exonuclease repair systems (15–18). They reported that in E. coli, the relative repair efficiencies of AAF at G1, G2 and G3 were in a ratio of 100:18:66, respectively, whereas the human exonuclease exhibited 38:100:68 ratio (17,18). Mu et al. (19) have recently carried out a human NER study of these lesions in HeLa cell extracts and found similar sequence-dependent NER efficiencies. Their molecular dynamics (MD) simulation data indicated that the greater NER efficiencies are correlated with base sequence–dependent local untwisting and minor groove opening together with weaker stacking interactions (19). Recently, we conducted E. coli UvrABC NER studies on the NarI sequence duplexes (5′–G1G2CG3CC–3′), in which guanines are modified by either AF or AAF (7). Results showed that the bulky AAF adducts repair in a conformation-specific manner, with the highly S/W-conformeric G3 and G1 duplexes incised considerably more efficiently than the highly B-conformeric G2 duplex (G3 ∼ G1 > G2). Conversely, the repair rate of N-deacetylated AF was 2- to 3-fold lower than AAF, and the order of incision efficiencies was opposite of that observed for the AAF case. We have coined the term ‘N-acetyl factor’ to describe the complexity of NER recognition of AF versus AAF (7).
Here, we describe an unusual 3′-next flanking base effect on the conformational properties and E. coli NER efficiencies of three prototype arylamine adducts in the G*CN sequence context (Figure 1b: G*=ABP, AF, or AAF; N=A or T). Results from spectroscopy (19F NMR and induced circular dichroism [ICD]), thermodynamic quantification (differential scanning calorimetry [DSC]/ultraviolet (UV)-melting experiment) and gel electrophoresis, as well as MD/potential of mean force (PMF) calculations, show that sequence-dependent lesion-induced DNA bending coupled with thermodynamic destabilization is responsible for the altered repair recognition of bulky arylamine-DNA adducts in E. coli. This work represents a novel 3′- next flanking sequence effect as a unique NER factor for bulky arylamine lesions in E. coli.
MATERIALS AND METHODS
Adduct synthesis
Modified duplexes were prepared following the published procedures (7,8,20–23). The modified oligos were characterized by electrospray time-of-flight mass spectrometry analysis as reported previously (24). An identical set of unmodified duplexes was also prepared as controls.
Differential scanning calorimetry
DSC measurements were performed using a VP-DSC Micro-calorimeter from Microcal Inc. (Northampton, MA) according to the procedures published previously (22). All sample solutions were 0.12 mM concentration. Tm was the temperature at half the peak area. ΔG and ΔS values were determined by the procedures of Chakrabarti et al. (25). The uncertainties in the values of Tm, ΔH, ΔG and ΔS represent the random errors inherent in the DSC measurements.
UV-Melting (Cary100 Bio, Beckman) and Circular Dichroism (CD) (J-810, Jasco) experiments were performed using the previously reported procedures (7,20,23,26).
Dynamic 19F NMR
Duplex samples (about 20–30 ODS) were dissolved in 300 μl of pH 7.0 buffer (100 mM NaCl, 10 mM Na3PO4 and 100 μM EDTA in 10% D2O/90% H2O) and filtered into through a Shigemi tube using a 0.2-μm membrane filter. All 1H and 19F NMR experiments were conducted using a dedicated 5-mm 19F/1H dual probe on a Bruker DPX400 Avance spectrometer operating at 400.0 and 376.5 MHz, respectively. Imino proton spectra were obtained using phase-sensitive jump-return sequences at 5°C. 19F NMR spectra were acquired in the 1H-decoupled mode and referenced to CFCl3 by assigning external C6F6 in C6D6 at −164.90 ppm. Temperature dependence spectra were processed as reported previously (20,27).
EMSA assay
The N-(2′-deoxyguanosin-8-yl)-4-fluoro-4-aminobiphenyl (FABP), N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-aminofluorene (FAF) and N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-acetylaminofluorene (FAAF)-modified 19-mer GCT and GCA sequences were each (100 nM) annealed with an equimolar complementary sequence, in which the 5′-end was γ-32P-labeled using T4 polynucleotide kinase and [γ-32P] ATP (Perkin-Elmer radiochemical, Boston, MA) in a buffer containing NaCl (25 mM) and Tris–HCl (25 mM). The mixture was heated at 95°C for 5 min and then cooled to room temperature overnight. The duplexes were subjected to 15% non-denaturing polyacrylamide (acrylamide:bisacrylamide: 29:1, w/w) gel electrophoresis at 1800 V, and the temperature was maintained at 4–8°C by regularly replacing the running buffer with the ice-cold Tris/Borate/EDTA (TBE) buffer. Gel was exposed to Kodak phosphor imaging screen overnight and scanned on Typhoon 9410.
Nucleotide excision assay
DNA substrates of 58 bp containing a single FABP, FAF or FAAF, each adducted at either G*CT or G*CA sequences, were constructed as described previously (28,29). UvrA, UvrB and UvrC proteins were overexpressed in E. coli and then purified as described previously (30). The 5′-terminally labeled DNA substrates were incubated and incised by UvrABC as described previously (28,29). Briefly, the DNA substrates (2 nM) were incubated in the UvrABC reaction buffer (50 mM Tris–HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT) at 37°C in the presence of UvrABC (10 nM UvrA, 250 nM UvrB and 100 nM UvrC). The Uvr proteins were diluted and premixed in Uvr storage buffer before addition to the reaction. Aliquots were collected at 0, 5, 10, 15 and 20 min into the reaction. The reaction was terminated by heating at 95°C for 5 min. The products were denatured by addition of formamide loading buffer and heating to 95°C for 5 min, followed by quick chilling on ice. The incision products were then analyzed by electrophoresis on a 12% polyacrylamide sequencing gel under denaturing conditions with Tris/Borate/EDTA (TBE) buffer.
To quantify the incision products, radioactivity was measured using a Fuji FLA-5000 Image Scanner with MultiGauge V3.0 software. The DNA incised (in femtomoles) by UvrABC was calculated based on the total molar amount of DNA used in each reaction and the ratio of the radioactivity of incision products to total radioactivity of DNA. At least three independent experiments were performed for determination of the rates of incision.
MD and PMF calculations
PMF calculations were performed on the GCA and GCT 11-mers initiated from the canonical B form of DNA for ‘anti’ simulations where the glycosidic bond is in the anti form. ‘Syn’ simulations were initiated from models based on an NMR structure (PDB: 1C0Y) in which the glycosidic bond is in the syn form. MD simulations were performed with the programs CHARMM and NAMD, using the CHARMM27 additive nucleic acid force field. Modified G* lesions were created based on the CHARMM General Force Field followed by additional optimization of the dihedral parameters linking the G base to the adduct. Determination of the PMFs followed the protocol of Banavali and MacKerell (31) with details of the simulations included in the supporting information.
Results
Model systems
As model systems, 11-mer DNA duplexes [d(5′-CCATCG*CNACC-3′).d(5′-GGTNGCGATGG-3′)] were prepared, in which G* is FABP, FAF or FAAF and N is either dA or dT (designated as G*CA and G*CT duplexes, respectively) (Figure 1b). The two sequences are chemically isomeric, differing only on the polarity of the 3′-next flanking A:T versus T:A. The utility of fluorine-tagged lesions as effective structure probes has been documented (32). Both the G*CA and G*CT sequences have been used previously for the studies of bulky adducts (22,33).
19F NMR spectroscopy
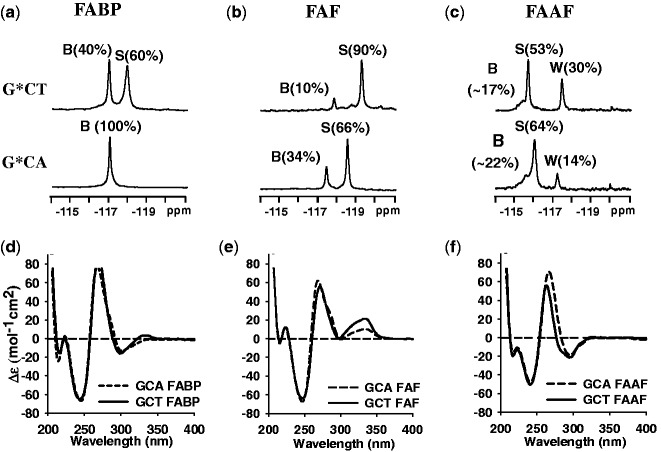
Figure 2a–c compares the 19F NMR spectra (−114 to −121 ppm) of modified DNA duplexes at 20°C for the G*CT and G*CA sequence contexts (see Supplementary Figure. S1 for full temperature ranges). 19F signal assignments were made based on the H/D solvent effect, exchange spectroscopy, adduct-induced CD (ICD290–350 nm) and chemical shifts as previously described (6,26,32,34).
Figure 2.
19F NMR (a–c) and CD (d–f) spectra of FABP-, FAF- and FAAF-modified 11-mer duplexes, respectively, in the G*CA and G*CT sequence contexts at 20°C. The B, S and W notation used in the 19F NMR (a–c) signal assignments represent major groove ‘B’, base displaced stacked ‘S’ and minor groove ‘W’ conformers, respectively (see Figure 1c legend). The CD spectra (d–f) of modified duplexes show the typical B-DNA characteristic (positive and negative ellipticity at 275 and 245 nm, respectively) and different ICD290–350 nm patterns represent the lesion conformation in the duplex (see CD in Results).
FABP-duplexes
A clear conformational difference exists between the two isomeric FABP-modified G*CA and G*CT duplexes (Figure 2a). The single signal at −116.9 ppm for FABP-G*CA has been previously assigned to the B-conformer (22). In contrast, FABP-G*CT exhibited two signals at −116.9 (B) and −118.0 (S) ppm in a 40:60% ratio and adopted a two-site exchange (EXSY spectra at 5 and 17°C, inset, Supplementary Figure S1a). A large chemical shift gap (∼1 ppm) between the two signals suggests differences of their electronic environments. In addition, the −116.9 ppm signal revealed a large H/D effect (+0.24 ppm) compared to the −118.0 ppm signal (+0.14 ppm) (data not shown) on increasing the D2O content from 10 to 100%. The results indicate the exposed fluorine atom in the B-conformer, as observed in the MD/PMF simulations (Supplementary Figure S2).
FAF-duplexes
Although not as dramatic, a similar sequence effect was observed for FAF. The FAF-G*CA duplex showed signals at −117.4 and −118.6 ppm in a 34:66% ratio (Figure 2b), which have been assigned to B- and S-conformers, respectively (20,22,23,26,35). The S conformer population was increased to 90% in the FAF-G*CT duplex. Consistent with this assignment, the downfield −117.4 ppm signal revealed a larger H/D effect (+0.19 ppm) compared with the −118.8 ppm signal (+0.03 ppm) (data not shown), again consistent with the MD/PMF simulations (Supplementary Figure S2).
FAAF-duplexes
At least three major signals are present (Figure 2c) for FAAF in the G*CA and G*CT sequences. These signals are shifted considerably (∼2 ppm) to the downfield compared with FAF, a phenomenon associated with the N-acetyl factor. We reported previously S/B/W-conformer assignment of FAAF-modified 12- and 16-mer duplexes in several sequence contexts (i.e. TG*A, CG*C, CG*G and GG*C) (7,8). The results revealed that 19F signals of B-, S- and W-conformers appear consistently going from downfield to upfield in order of −115.0 to −115.5, −115.5 to −117.0 and −116.5 to −118.0 ppm, respectively. The major 19F signals in Figure 2c fit that pattern. In particular, the signal patterns of the FAAF-modified G*CA/G*CT duplexes (5′-CCATCG*CNAC-3′) match well with those observed for 16-mer (5′-CTCTCG1G2CG3*CCATCAC-3′) (7) and 12-mer (5′-CTTCTCG*CCCTC-3′) duplexes (8), both of which have the CG*C sequence context (underlined). In line with this observation, their proton spectra displayed a mixture of broad imino signals arising not only from those involved in Watson–Crick hydrogen bonds (12–14 ppm) but also from the lesion site and its vicinity (11–12 ppm) (Supplementary Figure S3). Although the two sequences are similar in the total syn conformation (S + W) (i.e. 83 vs. 78%, respectively, for G*CT and G*CA), the W-conformer population appears to be significantly greater for G*CT (30%) compared with G*CA (14%) (Supplementary Figure S4).
Induced circular dichroism
Figure 2d–f show the CD of the modified G*CA and G*CT duplexes. We reported that B- and S-conformers are characterized by positive and negative ICD290–350 nm, respectively (26). Accordingly, the B-conformeric FABP-G*CA displayed a strongly negative ICD290–350 nm, whereas an S-shape curve was observed for the S/B-mixture G*CT duplex (Figure 2d). These results are in good agreement with the 19F NMR results (Figure 2a). Unlike FABP, FAF on both sequences exhibited a strong positive ICD290–350 nm with the effect much greater for G*CT (Figure 2e), consistent with the greater S-conformer population determined by 19F NMR (Figure 2b). The ICD of FAAF (Figure 2f), which is confined in the narrow 290–320 nm range, has not been defined as clearly as FAF (8).
In addition, the modified duplexes displayed a significant blue shift relative to their respective control duplexes (Supplementary Figure S5a and b and Table 1). All except for FAF-G*CT exhibited significant blue shifts up to 8 nm. The bulky N-acetylated FAAF exhibited greater shifts (ΔG*-G = 4–8 nm) than FAF and FABP (ΔG*-G = 0–4 nm). GCA sequences, which are prone to the B-conformer, displayed greater blue shift (ΔG*CA-G*CT = 2–4 nm, Table 1) compared with GCT. It is well known that protein-induced DNA bending exhibits significant CD shift at 275 nm of regular B-type DNA (36–38). For instance, the HMG box protein SOX-5 bends DNA by ∼74° on binding, which resulted in a significant blue CD shift (37). These reports suggest that the blue shifts observed in this study result from the distortion of the DNA backbone, particularly bending.
Table 1.
Lesion-induced CD blue shifts
| Lesion | GCA (nm) | GCT (nm) | Blue Shift Δa(G*–G) (nm) | Blue shift Δb(G*CA–G*CT) (nm) | |
|---|---|---|---|---|---|
| Control | 271 | 271 | GCA | GCT | |
| FABP | 267 | 269 | 4 | 2 | 2 |
| FAF | 268 | 271 | 3 | 0 | 3 |
| FAAF | 263 | 267 | 8 | 4 | 4 |
aDifference in the wavelength of positive band between the modified and control duplexes.
bDifference in the wavelength of positive band between the G*CA and G*CT duplexes.
Gel mobility assay
Two 19-mer sequences (5′-CTTACCATCGCNACCATTC-3′, N = T or A) were used to investigate the impact of the A/T polarity swap at the N position on the gel mobility of the modified duplexes. Initially, the abovementioned 11-mer sequences were used but they denatured in the 15% native polyacrylamide gel at 1800 V (data not shown). Figure 3 compares the electrophoretic mobility of the 19-mer GCA and GCT sequences with and without modifications. Differential mobility between the single strand and double strand (ds) oligonucleotides confirmed the integrity of the duplexes (Figure 3). The modified duplexes exhibited retardation in the mobility in a lesion-dependent manner. In both sequences, major retardation effect was observed for FAAF followed by FABP, whereas no retardation was observed for FAF, results consistent with the CD blue shift data above (Table 1). It should be noted that the magnitude of retardation in mobility observed in this study is significantly lower than what was previously observed in benzo[a]pyrene modified duplexes (39–41). We cannot rule out a possibility of C8-subsitituted dG (this study) versus N2-substituted dG (benzo[a]pyrene) binding. However, the rationale behind this small difference in mobility could be well due to the significantly longer oligonucleotides (19-mer) used in this study as opposed to the aforementioned benzo[a]pyrene cases (11 and 15-mer). The utilization of longer sequences will reduce the number of adducts per helical turn, which might result into diminishing of the lesion-induced bending effect. Tsao et al. reported similar effects of oligonucleotide length on the electrophoretic mobility of benzo[a]pyrene-modified duplexes (41).
Figure 3.
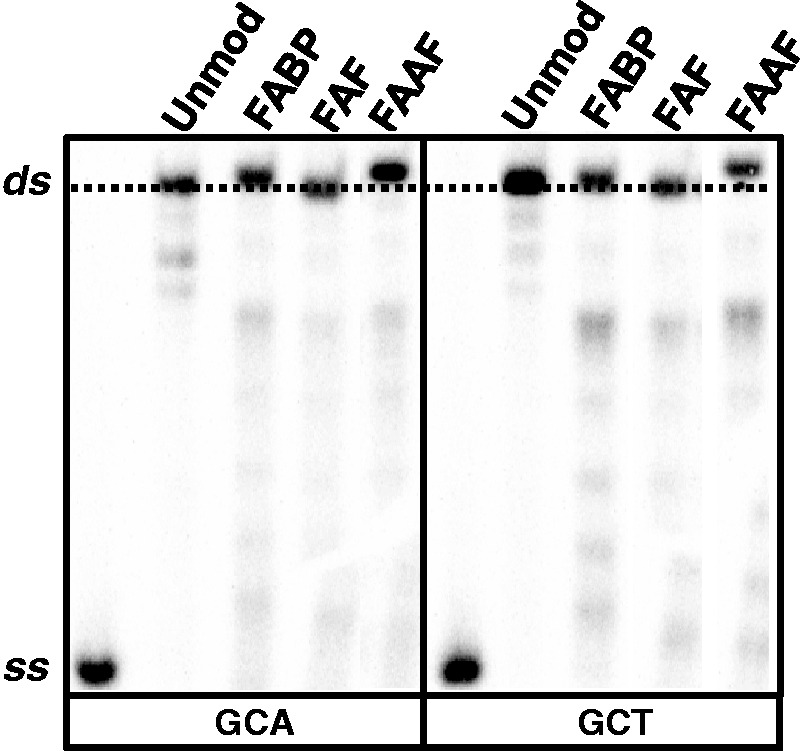
Autoradiograph of 15% (w/v) native polyacrylamide gel (acrylamide/bis-acrylamide 29:1, w/w) showing the relative mobility of ss and GCA/GCT 19-mer ds; both unmodified and single site specifically modified by FABP, FAF and FAAF.
Thermodynamics
Thermodynamics results from UV-optical melting (Supplementary Table S1) and DSC, which is not dependent on melting patterns and stoichiometry (22) (Table 2), are comparable. Supplementary Figure S6 shows the DSC thermograms of modified duplexes in the G*CA and G*CT sequences relative to the unmodified controls. These curves were transformed into the corresponding thermodynamic charts (Figure 4a and b), and the results are tabulated in Table 2. According to the NMR results (Figure 2), FABP and FAF display a S/B-equilibrium, whereas FAAF produces a complex S/B/W-equilibrium; thus, they will be compared separately.
Table 2.
Thermal and thermodynamic parameters derived from DSC curves
| CCATCG*CAACC | CCATCG*CTACC | |||||||
|---|---|---|---|---|---|---|---|---|
| GGTAGCGTTGG |
GGTAGCGATGG |
|||||||
| −ΔH (kcal/mol) | −ΔS (eu) | −ΔG37 (kcal/mol) | Tm (°C) | −ΔH (kcal/mol) | −ΔS (eu) | −ΔG37 (kcal/mol) | Tm (°C) | |
| Controla | 79.1 | 214.9 | 12.4 | 64.5 | 75.0 | 203.9 | 11.8 | 63.0 |
| FABPa | 76.4 | 214.1 | 10.0 | 54.3 | 64.2 | 178.3 | 8.9 | 51.8 |
| FAFa | 65.7 | 180.7 | 9.7 | 55.4 | 58.6 | 160.1 | 8.9 | 53.4 |
| FAAFa | 39.7 | 106.1 | 6.8 | 42.7 | 33.9 | 88.8 | 6.4 | 39.7 |
| ΔΔH (kcal/mol)b | ΔΔS (eu)c | ΔΔG37 (kcal/mol)d | ΔTm (°C)e | ΔΔH (kcal/mol)b | ΔΔS (eu)c | ΔΔG37 (kcal/mol)d | ΔTm (°C)e | |
| FABPa | 2.7 | 0.8 | 2.4 | −10.2 | 10.8 | 25.6 | 2.9 | −11.2 |
| FAFa | 13.4 | 34.2 | 2.7 | −9.1 | 16.4 | 43.8 | 2.9 | −9.6 |
| FAAFa | 39.4 | 108.8 | 5.6 | −21.8 | 41.1 | 115.1 | 5.4 | −23.3 |
The average standard deviations for ΔG, ΔH, and Tm are ±0.2, ±2.0, and ±0.4, respectively.
aThe results were calculated from integration of the DSC curve directly. ΔG and ΔH represent the heat absorbed during duplex melting at 0.12 mM.
bΔΔH = ΔH (modified duplex) − ΔH (control duplex).
cΔΔS = ΔS (modified duplex) − ΔS (control duplex).
dΔΔG = ΔG (modified duplex) − ΔG (control duplex).
eΔTm = Tm (modified duplex) − Tm (control duplex).
Figure 4.
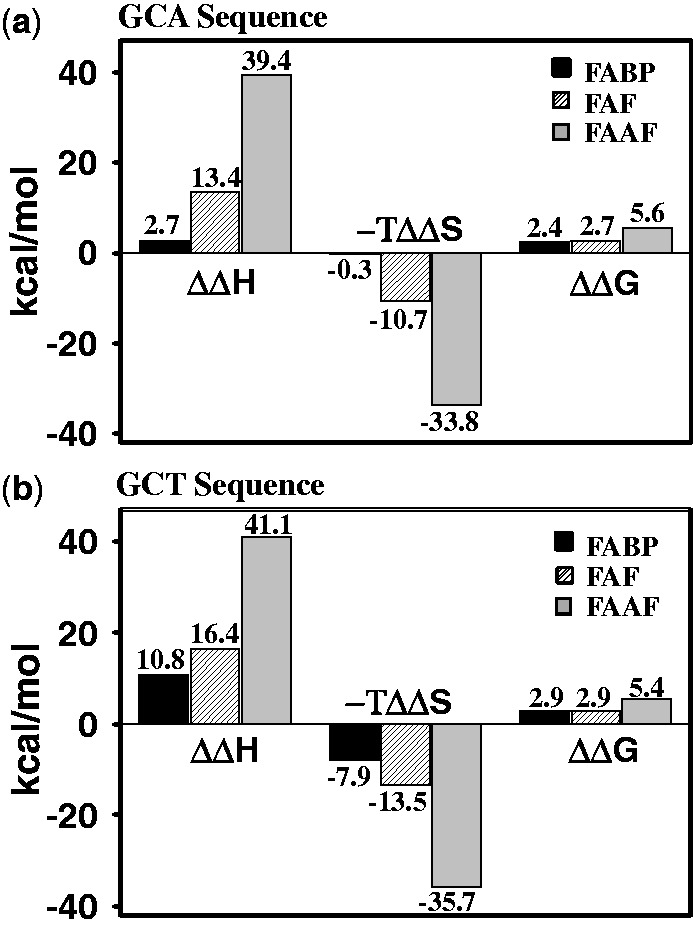
Comparative thermodynamic parameters histogram of FABP (black), FAF (hatched) and FAAF (gray) in (a) G*CA duplexes and (b) G*CT duplexes. The ΔΔ values represent: ΔΔH = ΔH (modified duplex) − ΔH (control duplex), ΔΔS = ΔS (modified duplex) − ΔS (control duplex) and ΔΔG = ΔG (modified duplex)− ΔG (control duplex).
FABP/FAF-DNA duplexes
Both FABP and FAF resulted in destabilization (Figure 4 and Table 2). FABP reduced Tm for the G*CA and G*CT duplexes by −10.2 and −11.2°C and ΔΔG37°C by 2.4 and 2.9 kcal/mol, respectively. The G*CA/G*CT transition produced major effect on ΔΔH (2.7 vs. 10.8 kcal/mol) and ΔΔS (0.8 vs. 25.6 eu), consistent with a significant increase in the S-conformer population from 0 to ∼60% (Figure 2a). The B-conformer FABP-G*CA is expected to lead to small entropy compensation, and consequently, the enthalpy reduction dominated the free-energy destabilization (Figure 4a). As expected, the structural disturbance caused by the S/B mixture FABP-G*CT duplex leads to a considerable reduction of melting enthalpy; however, most of it is compensated by entropy (Figure 4b).
FAF modification resulted in a similar destabilization effect: ΔTm by −9.1 and −9.6°C and ΔΔG37°C by 2.7 and 2.9 kcal/mol, respectively, for G*CA and G*CT. However, compared with FABP, FAF in both sequences yielded significantly larger reduction in enthalpy (ΔΔH = 13.4 and 16.4 kcal/mol) and entropy (ΔΔS = 34.2 and 43.8 eu) compensation (Figure 4 and Table 2). FAF exhibits more S-conformer than FABP in both sequences because of stronger stacking effect. As a result, sequence dependence on the thermodynamics was not as dramatic as in FABP. It is clear from Figure 4 that FAF (over FABP) and G*CT (over G*CA) combinations produce consistently greater enthalpy/entropy compensation, that is, FAF/G*CT >FAF/G*CA > FABP/G*CT > FABP/G*CA from the highest to the lowest. As expected the N-deacetylated FABP-G*CT and FAF-G*CA exhibited two site exchange (Supplementary Table S2).
FAAF-duplexes
FAAF modification resulted in the most significant reduction of ΔTm by −21.8 and −23.3°C and ΔΔH by 39.4 and 41.1 kcal/mol, respectively, for G*CA and G*CT sequence contexts (Figure 4a and b and Table 2). This is due to the bulky acetyl group in FAAF (Supplementary Figure S2). Like FABP and FAF, however, entropy compensation contributed a stabilization, that is, ΔΔG = 5.6 and 5.4 kcal/mol, respectively, for the G*CA and G*CT.
Molecular dynamics/potential mean force calculations
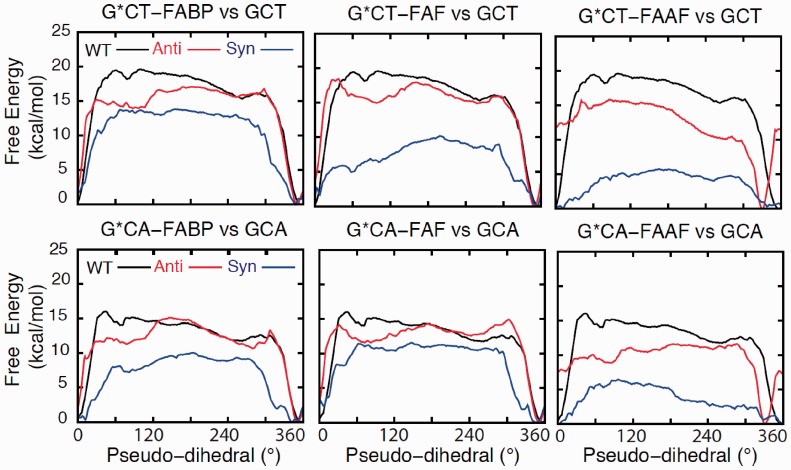
To further understand the impact of lesion modification on the G*CA and G*CT duplexes, MD simulations were performed in combination with potential mean force (PMF) calculations. PMF calculations yield the free energy as a function of the extent of flipping of the modified G* base (Supplementary Figure S7). Individual PMFs were determined with G* in the either anti or syn orientations, whereas only the anti orentation was studied for the unmodified duplexes.
Figure 5 shows the free energies from the PMFs. In the unmodified and anti-G* PMFs, there is a deep minimum in 330–360° corresponding to the Watson-Crick (WC)-base paired state (31), which in case of lesions corresponds to the B-state. Deep minima are also present in 330–360° in the syn-G* PMFs, which corresponds to the S-state. The conformer assignments were made based on the published experimental NOE data (Supplementary Figures S8–S11 and Supplementary Tables S3–S6) (42–45). The relative energies of the flipped states are highest in the unmodified duplexes in all cases indicating that the lesions lower the relative energies of the flipped state and/or destabilize the low-energy B- or S-states. Notably, the free-energy surfaces show the relative energies of the flipped states to be lower in the syn PMFs, consistent with the conformational thermodynamic data discussed above. As such, the lowest energies of the flipped state occur with FAAF (Table 2) indicating, that the syn FAAF may sample a wider range of conformations as compared with FABP and FAF, consistent with the 19F NMR data (Figure 2). Further validation of the PMFs is the energies of the minima being about 15 kcal/mol, which is in good agreement with the experimental ΔG≠ of 14.1 kcal/mol required for a B/S conversion. Representative B/S/W structures from the NOE-based PMFs are shown in Supplementary Figures S12–S14. For all three lesions, the presence of WC base pairing in the B-state and the stacking of the adduct into the duplex in the S-state are evident. In contrast, the structure of the W-state, which is only populated by FAAF, is highly distorted.
Figure 5.
Free-energy profiles as a function of the pseudo-dihedral angle phi (Supplementary Figure S7) from PMF calculations over the sampling range of 0.5–3 ns for G*CT-FABP, G*CA-FABP, G*CT-FAF, G*CA-FAF, G*CT-FAAF and G*CA-FAAF modified sequences (red and blue) and the unmodified GCT and GCA (black).
In the anti-G* PMFs, the average solvent accessible surface areas (SASAs) of both the lesion and fluorine atom are high in the vicinity of the B state, consistent with the location adduct in the major groove (Supplementary Figure S2). For the syn PMFs, the SASA values are low in the regions corresponding to the S-state, consistent with adducts being stacked inside the helices. However, the SASA values are higher with FAAF compared with FABP and FAF, suggesting an altered environment for FAAF. In addition, the syn PMFs of FAAF exhibits SASA minima in the 60–120° region, which encompasses the W-state. These suggest a fundamental difference in conformational properties of FAF/FABP versus FAAF, consistent with the significant difference in the 19F chemical shifts for the FAAF species and thermodynamic data.
Shown in Supplementary Figure S15 are bending probability distributions for the B-, S- and W-states for the three lesions in both the G*CA and the G*CT contexts. In the B- and S-states, the extent of bending is significantly larger with FAAF (cyan) versus FABP (red) and FAF (blue). These results are consistent with the experimental data obtained from the greater blue shift in CD (Supplementary Figure S5 and Table 1), although the changes in G*CA occur only in the S-state (Supplementary Figure S15). In addition, the simulations indicate the extent of bending for FABP to be similar to that of FAF. The significant increase in bending in FAAF is consistent with the greater destabilization of the duplexes caused by the FAAF lesion (Table 2). Concerning the bending, calculation of local helicoidal parameters revealed significant differences in twist and tilt for base pair 8, where the A/T switch occurs (Supplementary Table S7). For example, twist values are systematically larger and tilt values are less negative in GCT versus GCA sequences. These differences suggest that the local structural alteration associated with the A/T switch is being propagated to the overall helix.
Escherichia coli UvrABC incision
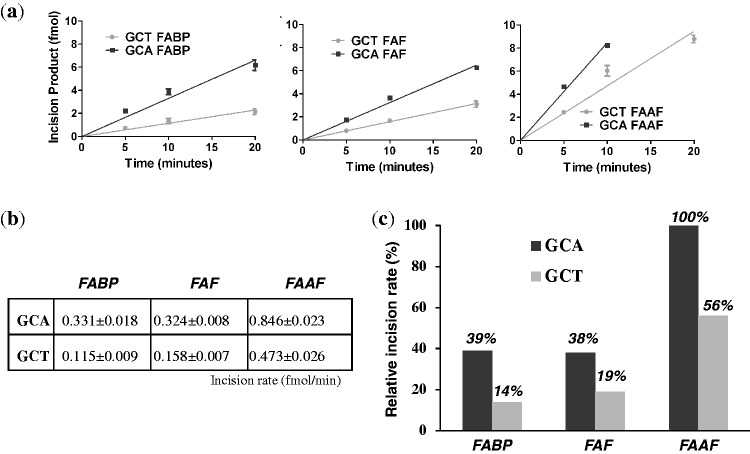
DNA substrates containing lesions in the defined sequences were subjected to incisions by the E. coli UvrABC system in a kinetic assay. These substrates were radioactively labeled at the 5′-end of the adducted strand and the major incision products separated on a urea-PAGE gel running under denaturing conditions (Supplementary Figure S16). The incision occurred at the eighth phosphate bond 5′ to G*, which is consistent with the currently accepted mechanisms of UvrABC-based NER (28,29). The substrates were incised at differing efficiencies depending on not only the type of DNA adduct but also the sequence context (Figure 6). Specifically, the G*CA sequences were incised at higher rates by ∼2-fold than G*CT, while the order of incision rate of adducts is FAAF > FAF ≈ FABP for both sequences, with FAAF being incised with 2- to 3-fold greater efficiency than the other lesions.
Figure 6.
Absolute incision rates (a and b) of FABP, FAF and FAAF-modified 55-mer substrates in the G*CA and G*CT sequence contexts. (c) Relative percent incision rate histograms with respect to G*CA-FAAF modified duplex.
It should be noted that the 5′-incision products appeared as doublet bands (Supplementary Figure S16). Similar incision products of this type of lesion have been observed previously (35,46,47). This is likely either due to the type of arylamine lesions or due to the structural heterogeneity exhibited by this type of lesions as demonstrated in this study and previous studies, suggesting that UvrABC may make the 5′-incision at the site different by one nucleotide for the different conformers of arylamine lesion.
DISCUSSION
Conformational and thermodynamic effects on the G*CA/G*CT transition
The NMR/ICD results indicate that lesion stacking is affected considerably by a polarity swap at the 3′-next flanking base (GCA → GCT). The effect was most significant for FABP, which produced a dramatic increase in S-conformer (0–60%) (Figure 2a). This is an extraordinary DNA sequence effect. A similar trend was observed for FAF, although the S-conformer was only 24% greater in G*CT than in G*CA (Figure 2b). Unlike FABP and FAF, the impact of the A/T swap on FAAF was minimal; specifically, the syn-glycosidic S- and W-conformers remained relatively unchanged (∼78 to ∼83%) (Figure 2c). Interestingly, the increase of W-conformer (14–30%) appeared to be compensated by a concomitant decrease of S-conformer (64–53%). These data indicate that the N-acetyl group in FAAF can push the low-energy syn-S/W-equilibrium toward W (see ‘N-acetyl factor’). Overall, these results indicate that the A/T swap has the largest impact on the most stable system, whereas the least stable FAAF lesion is the least impacted.
As expected, all modified duplexes were consistently destabilized compared with the controls (Figure 4 and Table 2): FAAF > FAF ≈ FABP. The G*CA/G*CT transition led to further destabilization, which was associated with increases in lesion stacking (greater S/W) for all three lesions. Obviously, a higher population of the syn-S-/W-conformer states is expected to disrupt the double helical structure, which would significantly reduce the enthalpy, accompanied by a compensatory increase in entropy (22).
Lesion-induced DNA bending as a major NER recognition factor
For each lesion, a greater proportion of B-conformer was observed in G*CA (FABP: 100%, FAF: 34% and FAAF: 22%) than G*CT (FABP: 40%, FAF: 10% and FAAF: 17%). Moreover, good correlation between the magnitudes of change in conformer populations and incision efficiencies was found among the lesions. In FABP, the A/T polarity swap caused a 60% change in S-conformer proportion and a 3-fold reduction in repair efficiency. The changes were significantly lower in FAF and FAAF (24 and 5–15%, respectively), as were the repair efficiencies (2.0- and 1.8-fold, respectively). At a glance, the results seem to suggest B-conformers have greater reparability than S-conformers. This feature is in clear contrast to the trend that has been observed previously for AF and AAF in certain sequence contexts, that is, the S-conformer is more reparable than the B-conformer (7,35). This type of conformation-specific repair is not only restricted to arylamines but also applied to other bulky lesions. For instance, Geacintov et al. have reported that the base-displaced cis-N2-dG adducts of benzo[a]pyrene are incised more efficiently than the minor groove–orientated trans-N2-dG adducts (33).
However, the E. coli repair results in this study seem to match well with events of adduct-induced DNA bending/distortion, as evidenced by blue shifts in CD (Table 1) and retardation of mobility in electrophoretic mobility shift assay (Figure 3). The slowed mobility indicates flexibility at the lesion site as observed by Tsao et al. for (+)-trans-anti-[BP]-N2-dG lesion in the TG*T sequence context with concomitant thermal destabilization (41,48). Similarly, the bulky N-acetyl FAAF exhibited significantly slower electrophoretic mobility compared with FAF and FABP within the same sequence context. In case of sequence, the G*CA duplex exhibited consistently greater bending than its G*CT counterpart, with the effect being significantly greater for FAAF than FAF and FABP. A similar CD pattern has been reported for AAF-modified NarI duplexes related to the formation of a B-Z junction (49). Clearly, the A/T swap alters the conformational equilibrium anti (B-) to syn (S- or W-). It should be noted that the G*CA (–TCG*CAA–) sequence contains a stretch of alternating pyrimidine:purine bases, which are predisposed to DNA bending (50–52). In contrast, such a stretch is interrupted in the highly S-conformeric G*CT (–TCG*CTA–) sequence. It is possible that the B-conformer may facilitate DNA bending, due to the exposure of the carcinogen moiety to the major groove’s hydrophilic environment. In both sequences, a major effect was observed with FAAF, followed by FABP and FAF (Figure 3). However, MD/PMF simulations indicate that the major changes in G*CA occur in the S-state. Also, unlike the CD data, there were no significant differences in electrophoretic mobility between the two sequences (Figure 3). The reason for the inconsistency in the mobility, CD and MD data demonstrating the sequence effect is not apparent, but the greater bending and flexibility of FAAF over FABP or FAF is in good agreement with the observed repair efficiencies (FAAF >> FAF ≈ FABP; Figure 6).
The repair results in Figure 6 along with previously reported work on polycyclic aromatic hydrocarbons (41,53) and arylamines (7) indicate that lesion-induced destabilization of DNA is a major determining factor for repair. However, these lesions were consistently repaired two to three times more efficiently in G*CA than in G*CT, which was not consistent with relative thermodynamic stabilities observed for each. The inconsistence is likely due to the second step of damage recognition (54) that becomes much more significant for FAAF versus FABP and FAF within a given sequence. Unlike the initial step of damage recognition by UvrA2, which depends on DNA conformation and sequence, the second step of recognition is well known to be characterized by the direct interaction of UvrB with adduct itself on DNA strand opening (47,54–56). In other words, the structure and chemistry of the lesions matter more with UvrB than UvrA2. Recently, Liu et al. reported the NER incision efficiencies of the bulky benzo[a]pyrene and equine estrogen substrates using human HeLa cell extracts and bacterial UvrABC proteins (53). They demonstrated that despite having differences in the prokaryotic and eukaryotic NER proteins, XPC-RAD23B and UvrB, respectively, they exhibit common feature of β-hairpin intrusion for damage recognition. In addition, it was found that local thermodynamic destabilization near the lesion site assists the insertion of β-hairpin, thus recognition.
Clearly, this study shows that the thermodynamic destabilization of the DNA duplex along with lesion flexibility promotes strand opening and thus the second step of damage recognition. The presence of the N-acetyl group (see below) may make FAAF more efficiently recognized than FAF and FABP at the second step due to its flexible nature and greater destabilization of the DNA double helix. As for the G*CA/G*CT transition, the initial recognition step conducted by UvrA2 should be a major determinant factor as the same efficiency of recognition at the second step is expected for the same type of lesion. Thus, bending appears to be an important factor for the DNA damage recognition. Indeed, a recent crystal study by Jaciuk et al. (10) found that in the active site of UvrA, the fluorescein-modified duplexes were bent by ∼15° and the structure was related to the kinked structure of psoralen and PAH adducts according to NMR (57). They concluded that the UvrA2 protein does not have direct chemical contacts with a lesion per se, but indirectly senses the overall helical distortion (unwinding and bending) (10). Because energy is required for the bending, formation of the pre-bent DNA induced by bulky lesions would likely enhance the UvrA2 binding and thus damage recognition.
Cai et al. have reported a similar repair trend for the 5′-CACACCG*CACAC-3′ sequence versus 5′-CCATCCG*CTACC-3′, in which G* is the major mutagenic lesion derived from the environmental carcinogen benzo[a]pyrene, 10S (+)-trans-anti-B[a]P-N2-dG (39). A greater repair (1.6-fold) of the CG*CA duplex over the CG*CT counterpart was attributed to its higher bending of the distant 5′-end sequences, as evidenced by gel experiments and MD simulations; these findings are consistent with the bending argument made in this study. Although the sequence contexts (underlined above) near the lesion site, including the 3′-next flanking base, are identical to those used in this study, they did not consider the structural and repair consequences of the GC*A/G*CT swap.
N-acetyl factor
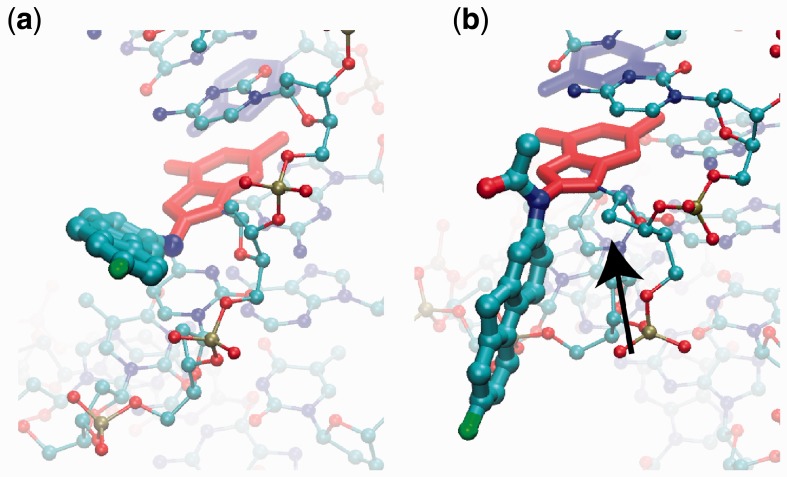
Although a relatively small modification, the N-acetyl group has an important structural consequence. As shown in Figure 7, the lack of the acetyl moiety in G*CT-FAF allows the G* moiety (red licorice representation) to point away from the sugar and stay in the plane of the GC base pair, where the N-H bond is directed toward the sugar. However, in G*CT-FAAF, the acetyl group will have a steric clash with the sugar moiety of G* (identified with a black arrow), thereby leading the fluorene moiety (cyan) to be perpendicular to the G* ring system. This persistent ‘perpendicular’ lesion orientation is predicted to lead to more disruption of the DNA duplex. A similar observation regarding the differences in the orientation of AF and AAF was reported by Mu et al. (19) who have conducted a NER study of these lesions in human HeLa cells. MD simulations in that work indicate that the greater repair susceptibility of AAF stems from steric hindrance effects of the acetyl group, which significantly diminish the adduct base stabilizing van der Waals stacking interactions relative to AF. The persistent ‘perpendicular’ FAAF mentioned earlier could raise barriers between conformations of FAAF modified DNA, resulting in the overall lower free energy of the syn-G* PMFs for FAAF, compared with FABP and FAF. In other words, the N-acetyl group in FAAF could act as a ‘conformational locker’ (7) that orients the adduct in a position that will lead to greater destabilization of the DNA duplex (Figure 4 and Table 2), as well as the increased bending observed in CD (Table 1) and mobility assays (Figure 3). As a result, FAAF lesions are repaired at significantly greater rate compared with the FABP and FAF lesions (7).
Figure 7.
Image of the (a) G*CT-FAF adduct in the B state (350°) and (b) G*CT-FAAF adduct in the B state (330°). The flipping G6 base is red licorice, the orphan C6 is blue licorice, adducts are atom-colored thick CPK and the reminder of the DNA is atom-colored thin CPK. (b) The arrow indicates the sugar moiety with which the acetyl or aryl moiety on FAAF is suggested to form a steric clash.
CONCLUSION
The A to T polarity swap in the arylamine-modified G*CA/G*CT transition produced a dramatic increase in destabilized stacked conformation but resulted in unexpected 2- to 3-fold lower NER efficiencies. These results are consistent with lesion-induced DNA bending/distortion. As for lesions, FAAF was repaired three to four times more efficiently than FABP and FAF lesions, which is consistent with the extent of bending and helix destabilization, as well as the steric constraint in the duplex (‘N-acetyl factor’) (7). A number of different damage recognition parameters have been implicated in the molecular mechanisms of NER (9,55,58). However, it is known that thermal/thermodynamic destabilization and DNA distortion/bending are important factors for damage recognition by repair proteins (9,39,53). The results of this study show that lesion-induced DNA bending/thermodynamic destabilization is a more important NER factor than the usual S/B conformational heterogeneity, as has been observed previously for AF and AAF in certain sequence contexts (7,35). This work represents a novel 3′-next flanking sequence effect as a unique NER factor for bulky arylamine lesions in E. coli. Taken together, the results of this study demonstrate the complexity in DNA recognition factors for repair of bulky arylamine lesions in E. coli.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–7, Supplementary Figures 1–16, Supplementary Method and Supplementary References [59–79].
FUNDING
National Institutes of Health (NIH) [CA098296 to B.C., GM051501 to A.D.M. and CA86927 to Y.Z.]; the National Science Foundation/RI-EPSCoR [0554548] and the RI-INBRE core facility supported by National Institutes of Health [P20 RR016457]. Funding for open access charge: NIH/NCI.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Greenbaltt MS, Bennett WP, Hollstein MH, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 2. Friedberg, E.C., Walker, G.C., Siede, W., Wood, R.D., Schultz, R.A., Ellenberger, T. (eds) (2006) DNA Repair and Mutagenesis (2nd edn). ASM Press, Washington.
- 3.Luch A. Nature and nurture—lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 4.Beland FA, Kadlubar FF. Handbook of Experimental Pharmacology. Spring-Verlag, Heidelberg: 1990. pp. 267–325. [Google Scholar]
- 5.Cho B. Structure-function characteristics of aromatic amine-DNA adducts. In: Geacintov NE, Broyde S, editors. The Chemical Biology of DNA Damage. Germany: Wiley-VCH, Weinheim; 2010. pp. 217–233. [Google Scholar]
- 6.Meneni SR, Shell SM, Gao L, Jurecka P, Lee W, Sponer J, Zou Y, Chiarelli MP, Cho BP. Spectroscopic and theoretical insights into sequence effects of aminofluorene-induced conformational heterogeneity and nucleotide excision repair. Biochemistry. 2007;46:11263–11278. doi: 10.1021/bi700858s. [DOI] [PubMed] [Google Scholar]
- 7.Jain V, Hilton B, Patnaik S, Zou Y, Chiarelli MP, Cho BP. Conformational and thermodynamic properties modulate the nucleotide excision repair of 2-aminofluorene and 2-acetylaminofluorene dG adducts in the NarI sequence. Nucleic Acids Res. 2012;40:3939–3951. doi: 10.1093/nar/gkr1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patnaik S, Cho BP. Structures of 2-acetylaminofluorene modified DNA revisited: insight into conformational heterogeneity. Chem. Res. Toxicol. 2010;23:1650–1652. doi: 10.1021/tx100341u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel D. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 10.Jaciuk M, Nowak E, Skowronek K, Tanska A, Nowotny M. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat. Struct. Mol. Biol. 2011;18:191–197. doi: 10.1038/nsmb.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 12.Cai Y, Patel DJ, Broyde S, Geacintov NE. Base sequence context effects on nucleotide excision repair. J. Nucleic Acids. 2010:2010. doi: 10.4061/2010/174252. Article ID 174252, doi:10.4061/2010/174252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broschard TH, Koffel-Schwartz N, Fuchs RP. Sequence-dependent modulation of frameshift mutagenesis at NarI-derived mutation hot spots. J. Mol. Biol. 1999;288:191–199. doi: 10.1006/jmbi.1999.2667. [DOI] [PubMed] [Google Scholar]
- 14.Burnouf D, Koehl P, Fuchs RP. Single adduct mutagenesis: strong effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Proc. Natl Acad. Sci. USA. 1989;86:4147–4151. doi: 10.1073/pnas.86.11.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delagoutte E, Bertrand-Burggraf E, Lambert IB, Fuchs RP. Binding and incision activities of UvrABC excinuclease on slipped DNA intermediates that generate frameshift mutations. J. Mol. Biol. 1996;257:970–976. doi: 10.1006/jmbi.1996.0216. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs RP, Seeberg E. pBR322 plasmid DNA modified with 2-acetylaminofluorene derivatives: transforming activity and in vitro strand cleavage by the Escherichia coli uvrABC endonuclease. EMBO J. 1984;3:757–760. doi: 10.1002/j.1460-2075.1984.tb01880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mu D, Bertrand-Burggraf E, Huang JC, Fuchs RP, Sancar A, Fuchs BP. Human and E. coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 1994;22:4869–4871. doi: 10.1093/nar/22.23.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeberg E, Fuchs RP. Acetylaminofluorene bound to different guanines of the sequence -GGCGCC- is excised with different efficiencies by the UvrABC excision nuclease in a pattern not correlated to the potency of mutation induction. Proc. Natl Acad. Sci. USA. 1990;87:191–194. doi: 10.1073/pnas.87.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mu H, Kropachev K, Wang L, Zhang L, Kolbanovskiy A, Kolbanovskiy M, Geacintov NE, Broyde S. Nucleotide excision repair of 2-acetylaminofluorene- and 2-aminofluorene-(C8)-guanine adducts: molecular dynamics simulations elucidate how lesion structure and base sequence context impact repair efficiencies. Nucleic Acids Res. 2012;40:9675–9690. doi: 10.1093/nar/gks788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain N, Li Y, Zhang L, Meneni SR, Cho BP. Probing the sequence effects on NarI-induced -2 frameshift mutagenesis by dynamic 19F NMR, UV, and CD spectroscopy. Biochemistry. 2007;46:13310–13321. doi: 10.1021/bi701386f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain N, Meneni S, Jain V, Cho BP. Influence of flanking sequence context on the conformational flexibility of aminofluorene-modified dG adduct in dA mismatch DNA duplexes. Nucleic Acids Res. 2009;37:1628–1637. doi: 10.1093/nar/gkn1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang F, Cho BP. Enthalpy-entropy contribution to carcinogen-induced DNA conformational heterogeneity. Biochemistry. 2010;49:259–266. doi: 10.1021/bi901629p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meneni SR, D'Mello R, Norigian G, Baker G, Gao L, Chiarelli MP, Cho BP. Sequence effects of aminofluorene-modified DNA duplexes: thermodynamic and circular dichroism properties. Nucleic Acids Res. 2006;34:755–763. doi: 10.1093/nar/gkj480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao L, Zhang L, Cho BP, Chiarelli MP. Sequence verification of oligonucleotides containing multiple arylamine modifications by enzymatic digestion and liquid chromatography mass spectrometry (LC/MS) J. Am. Soc. Mass Spectrom. 2008;19:1147–1155. doi: 10.1016/j.jasms.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakrabarti MC, Schwarz FP. Thermal stability of PNA/DNA and DNA/DNA duplexes by differential scanning calorimetry. Nucleic Acids Res. 1999;27:4801–4806. doi: 10.1093/nar/27.24.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang F, Meneni S, Cho BP. Induced circular dichroism characteristics as conformational probes for carcinogenic aminofluorene-DNA adducts. Chem. Res. Toxicol. 2006;19:1040–1043. doi: 10.1021/tx0601253. [DOI] [PubMed] [Google Scholar]
- 27.Cho BP, Zhou L. Probing the conformational heterogeneity of the acetylaminofluorene-modified 2′-deoxyguanosine and DNA by 19F NMR spectroscopy. Biochemistry. 1999;38:7572–7583. doi: 10.1021/bi990182d. [DOI] [PubMed] [Google Scholar]
- 28.Luo C, Krishnasamy R, Basu AK, Zou Y. Recognition and incision of site-specifically modified C8 guanine adducts formed by 2-aminofluorene, N-acetyl-2-aminofluorene and 1-nitropyrene by UvrABC nuclease. Nucleic Acids Res. 2000;28:3719–3724. doi: 10.1093/nar/28.19.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou Y, Liu TM, Geacintov NE, Van Houten B. Interaction of the UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG-(-)-anti-BPDE. Biochemistry. 1995;34:13582–13593. doi: 10.1021/bi00041a038. [DOI] [PubMed] [Google Scholar]
- 30.Zou Y, Van Houten B. Strand opening by the UvrA(2)B complex allows dynamic recognition of DNA damage. EMBO J. 1999;18:4889–4901. doi: 10.1093/emboj/18.17.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banavali NK, MacKerell AD., Jr Free energy and structural pathways of base flipping in a DNA GCGC containing sequence. J. Mol. Biol. 2002;319:141–160. doi: 10.1016/S0022-2836(02)00194-8. [DOI] [PubMed] [Google Scholar]
- 32.Meneni S, Liang F, Cho BP. Examination of the long-range effects of aminofluorene-induced conformational heterogeneity and its relevance to the mechanism of translesional DNA synthesis. J. Mol. Biol. 2007;366:1387–1400. doi: 10.1016/j.jmb.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 34.Zhou L, Rajabzadeh M, Traficante DD, Cho BP. Conformational heterogeneity of arylamine-modified DNA: 19F NMR evidence. J. Am. Chem. Soc. 1997;119:5384–5389. [Google Scholar]
- 35.Meneni S, Shell SM, Zou Y, Cho BP. Conformation-specific recognition of carcinogen-DNA adduct in escherichia coli nucleotide excision repair. Chem. Res. Toxicol. 2007;20:6–10. doi: 10.1021/tx600273h. [DOI] [PubMed] [Google Scholar]
- 36.Blazy B, Culard F, Maurizot JC. Interaction between the cyclic AMP receptor protein and DNA. Conformational studies. J. Mol. Biol. 1987;195:175–183. doi: 10.1016/0022-2836(87)90334-2. [DOI] [PubMed] [Google Scholar]
- 37.Connor F, Cary PD, Read CM, Preston NS, Driscoll PC, Denny P, Crane-Robinson C, Ashworth A. DNA binding and bending properties of the post-meiotically expressed Sry-related protein Sox-5. Nucleic Acids Res. 1994;22:3339–3346. doi: 10.1093/nar/22.16.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meierhans D, Sieber M, Allemann RK. High affinity binding of MEF-2C correlates with DNA bending. Nucleic Acids Res. 1997;25:4537–4544. doi: 10.1093/nar/25.22.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai Y, Kropachev K, Xu R, Tang Y, Kolbanovskii M, Kolbanovskii A, Amin S, Patel DJ, Broyde S, Geacintov NE. Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion. J. Mol. Biol. 2010;399:397–409. doi: 10.1016/j.jmb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suh M, Ariese F, Small GJ, Jankowiak R, Liu TM, Geacintov NE. Conformational studies of the (+)-trans, (-)-trans, (+)-cis, and (-)-cis adducts of anti-benzo[a]pyrene diolepoxide to N2-dG in duplex oligonucleotides using polyacrylamide gel electrophoresis and low-temperature fluorescence spectroscopy. Biophys. Chem. 1995;56:281–296. doi: 10.1016/0301-4622(95)00055-3. [DOI] [PubMed] [Google Scholar]
- 41.Tsao H, Mao B, Zhuang P, Xu R, Amin S, Geacintov NE. Sequence dependence and characteristics of bends induced by site-specific polynuclear aromatic carcinogen-deoxyguanosine lesions in oligonucleotides. Biochemistry. 1998;37:4993–5000. doi: 10.1021/bi980291c. [DOI] [PubMed] [Google Scholar]
- 42.Abuaf P, Hingerty BE, Broyde S, Grunberger D. Solution conformation of the N-(deoxyguanosin-8-yl)aminofluorene adduct opposite deoxyinosine and deoxyguanosine in DNA by NMR and computational characterization. Chem. Res. Toxicol. 1995;8:369–378. doi: 10.1021/tx00045a008. [DOI] [PubMed] [Google Scholar]
- 43.Mao B, Hingerty BE, Broyde S, Patel DJ. Solution structure of the aminofluorene [AF]-intercalated conformer of the syn-[AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry. 1998;37:81–94. doi: 10.1021/bi972257o. [DOI] [PubMed] [Google Scholar]
- 44.O'Handley SF, Sanford DG, Xu R, Lester CC, Hingerty BE, Broyde S, Krugh TR. Structural characterization of an N-acetyl-2-aminofluorene (AAF) modified DNA oligomer by NMR, energy minimization, and molecular dynamics. Biochemistry. 1993;32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 45.Eckel LM, Krugh TR. Structural characterization of two interchangeable conformations of a 2-aminofluorene-modified DNA oligomer by NMR and energy minimization. Biochemistry. 1994;33:13611–13624. doi: 10.1021/bi00250a012. [DOI] [PubMed] [Google Scholar]
- 46.Yang Z, Colis LC, Basu AK, Zou Y. Recognition and incision of gamma-radiation-induced cross-linked guanine-thymine tandem lesion G[8,5-Me]T by UvrABC nuclease. Chem. Res. Toxicol. 2005;18:1339–1346. doi: 10.1021/tx050147+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, Zou Y, Lader J, Malkani B, Amin S, et al. Sequence context- and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxide-guanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry. 2007;46:7006–7015. doi: 10.1021/bi700294k. [DOI] [PubMed] [Google Scholar]
- 49.Koehl P, Valladier P, Lefevre JF, Fuchs RP. Strong structural effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Nucleic Acids Res. 1989;17:9531–9541. doi: 10.1093/nar/17.23.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lankas F, Sponer J, Langowski J, Cheatham TE., III DNA basepair step deformability inferred from molecular dynamics simulations. Biophys. J. 2003;85:2872–2883. doi: 10.1016/S0006-3495(03)74710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McConnell KJ, Beveridge DL. Molecular dynamics simulations of B '-DNA: sequence effects on A-tract-induced bending and flexibility. J. Mol. Biol. 2001;314:23–40. doi: 10.1006/jmbi.2001.4926. [DOI] [PubMed] [Google Scholar]
- 52.Lyubchenko YL, Shlyakhtenko LS, Appella E, Harrington RE. CA runs increase DNA flexibility in the complex of lambda Cro protein with the OR3 site. Biochemistry. 1993;32:4121–4127. doi: 10.1021/bi00066a038. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, et al. Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst) 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou Y, Luo C, Geacintov NE. Hierarchy of DNA damage recognition in Escherichia coli nucleotide excision repair. Biochemistry. 2001;40:2923–2931. doi: 10.1021/bi001504c. [DOI] [PubMed] [Google Scholar]
- 55.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 56.Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. ‘Close-fitting sleeves’: DNA damage recognition by the UvrABC nuclease system. Mutat. Res. 2005;577:92–117. doi: 10.1016/j.mrfmmm.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 57.Van Houten B, Snowden A. Mechanism of action of the Escherichia coli UvrABC nuclease: clues to the damage recognition problem. Bioessays. 1993;15:51–59. doi: 10.1002/bies.950150108. [DOI] [PubMed] [Google Scholar]
- 58.Malta E, Verhagen CP, Moolenaar GF, Filippov DV, van der Marel GA, Goosen N. Functions of base flipping in E. coli nucleotide excision repair. DNA Repair (Amst) 2008;7:1647–1658. doi: 10.1016/j.dnarep.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Brooks BR, Brooks CL, III, MacKerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J. Comp. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacKerell AD, Jr, Banavali NK. All-atom empirical force field for nucleic acids: 2) Application to solution MD simulations of DNA. J. Comp. Chem. 2000;21:105–120. [Google Scholar]
- 61.Foloppe N, MacKerell AD., Jr All-atom empirical force field for nucleic acids: 1) Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comp. Chem. 2000;21:86–104. [Google Scholar]
- 62.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comp. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levitt M, Lifson S. Refinement of protein conformations using a macromolecular energy minimization procedure. J. Mol. Biol. 1969;46:269–279. doi: 10.1016/0022-2836(69)90421-5. [DOI] [PubMed] [Google Scholar]
- 65.Fletcher R, Reeves CM. Function minimization by conjugate gradients. Comput. J. 1964;7:149–154. [Google Scholar]
- 66.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 67.Allen MP, Tildesley DJ. Computer Simulation of Liquids. New York: Oxford University Press; 1989. [Google Scholar]
- 68.Darden TA, York D, Pedersen LG. Particle mesh Ewald: an Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 69.Steinbach PJ, Brooks BR. New spherical-cutoff methods of long-range forces in macromolecular simulations. J. Comp. Chem. 1994;15:667–683. [Google Scholar]
- 70.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comp. Phys. 1977;23:327–341. [Google Scholar]
- 71.Nose S, Klein ML. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 72.Hoover WG. Canonical dynamics—equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 73.Feller SE, Zhang Y, Pastor RW, Brooks RW. Constant pressure molecular dynamics simulation: the Langevin Piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 74.Zheng L, Chen M, Yang W. Random walk in orthogonal space to achieve efficient free-energy simulation of complex systems. Proc. Natl Acad. Sci. USA. 2008;105:20227–20232. doi: 10.1073/pnas.0810631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song K, Campbell AJ, Bergonzo C, de los Santos C, Grollman AP, Simmerling C. An improved reaction coordinate for nucleic acid base flipping studies. J.Chem. Theory Comput. 2009;5:3105–3113. doi: 10.1021/ct9001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The Weighted Histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comp. Chem. 1992;13:1011. [Google Scholar]
- 77.Grossfield A. WHAM: an implementation of the Weighted Histogram analysis method. 2011 http://dasher.wustl.edu/alan/wham ed (1 April 2011, date last accessed) [Google Scholar]
- 78.Ponder JW, Richards FM. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J. Mol. Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 79.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.