Abstract
Transposable elements possess specific patterns of integration. The biological impact of these integration profiles is not well understood. Tf1, a long-terminal repeat retrotransposon in Schizosaccharomyces pombe, integrates into promoters with a preference for the promoters of stress response genes. To determine the biological significance of Tf1 integration, we took advantage of saturated maps of insertion activity and studied how integration at hot spots affected the expression of the adjacent genes. Our study revealed that Tf1 integration did not reduce gene expression. Importantly, the insertions activated the expression of 6 of 32 genes tested. We found that Tf1 increased gene expression by inserting enhancer activity. Interestingly, the enhancer activity of Tf1 could be limited by Abp1, a host surveillance factor that sequesters transposon sequences into structures containing histone deacetylases. We found the Tf1 promoter was activated by heat treatment and, remarkably, only genes that themselves were induced by heat could be activated by Tf1 integration, suggesting a synergy of Tf1 enhancer sequence with the stress response elements of target promoters. We propose that the integration preference of Tf1 for the promoters of stress response genes and the ability of Tf1 to enhance the expression of these genes co-evolved to promote the survival of cells under stress.
INTRODUCTION
Transposons are mobile genetic elements that are widespread among eukaryotes and comprise a substantial portion of eukaryotic genomes. Yet transposon integration has the potential to interrupt coding sequences throughout the genome of the host cell. They are often said to be ‘junk’ DNA or parasites of the host genome, because they make no obvious contribution to the cell (1–3). To defend against these deleterious effects, the host cells have evolved multiple mechanisms to silence transposons including DNA methylation and small RNA inhibition mechanisms (4–7). However, accumulating evidence demonstrates that the transcription of transposons can be activated when cells are under stress (8–10). Environmental stress has been shown to induce integration events (8). The possibility that transposon insertions triggered by conditions of stress may benefit the host by improving survival is an intriguing hypothesis put forward by McClintock (11). However, this model remains unsubstantiated.
In Saccharomyces cerevisiae, the long-terminal repeat (LTR) retrotransposons integrate into gene-free positions of the chromosomes. Ty1 has a strong preference for integrating into regions spanning 700 bp upstream of pol III transcribed genes, whereas Ty3 integrates one or two nucleotides upstream of the transcription start sites of tRNA genes (12–15). Although the impact of Ty1 and Ty3 integration on the transcription of pol III genes has not been explored genome wide, these elements do increase expression of the limited number of pol III transcribed genes tested (16,17).
In contrast to the integration of Ty1 and Ty3 upstream of pol III transcribed genes, the LTR retrotransposon Tf1 of Schizosaccharomyces pombe integrates into the promoters of pol II transcribed genes (18–21). Saturated profiles of insertion sites revealed that Tf1 integrates with a preference for pol II promoters that are induced by environmental stresses (21). To determine the biological impact of integration, we examined the effect of Tf1 integration on the expression of adjacent genes. We studied 32 genes often targeted by Tf1 and found that integration did not reduce their expression. In six cases, Tf1 insertion actually increased the expression of adjacent genes by enhancing the levels of the native transcripts. In other cases, host factors that participate in genome surveillance such as Upf1 and Abp1 were found to restrict the expression of genes that would otherwise have been enhanced by Tf1 insertion. We found that Tf1 transcription was induced by heat treatment, and interestingly, only genes that themselves were induced by heat could be activated by Tf1 integration. We propose that it is the synergy of Tf1 enhancer sequence with the stress response elements of target promoters that results in gene activation. We speculate that Tf1 integration has the potential to improve the survival of individual cells exposed to environmental stress.
MATERIALS AND METHODS
Media and growth of S. pombe
Selective media consisted of Edinburgh minimal 2 media (EMM) (22) with 2 g of a dropout mix, which contained all amino acids, 250 µg/ml adenine and uracil but lacked nutrients that were absent for selection. A liter of rich media, Yeast Extract Plus Supplements (YES), contained 2 g of dropout powder, 30 g glucose and 5 g Difco yeast extract. Vitamin B1 (10 µM) was added to repress the nmt1 promoter when indicated. EMM-5-fluoroorotic acid (5-FOA) plates contained 1 mg/ml 5-FOA (#F5050, United States Biologicals, Swampscott, MA). YES-5-FOA-G418 plates contained 500 µg/ml (corrected for purity) of Geneticin (#11811-031, Life Technologies, Rockville, MD) and 1 mg/ml 5-FOA. All S. pombe strains were grown at 32°C. Cells were grown under stress conditions similar to those described previously (9). For heat stress, cells were transferred from 32°C to a pre-warmed flask in a 39°C water bath, and the cultures were incubated for 10 min to allow the media to reach 39°C. The cultures were then incubated for the designated time intervals. To generate oxidative stress, hydrogen peroxide (H2O2, Sigma) was added to a final concentration of 0.5 mM. To create osmotic stress, a final concentration of 1 M sorbitol (Sigma) was added to the cultures. For heavy metal stress, cadmium sulfate (CdSO4, Sigma) was added to a final concentration of 0.5 mM.
Yeast strains and plasmid construction
The yeast strains and plasmids used in this study are listed in Supplementary Tables S1 and S2, respectively. The oligonucleotide sequences are listed in Supplementary Table S3. The upf1 gene was deleted from YHL1101 using the drug-resistance marker, nat1, encoding nourseothricin (NAT) acetyltransferase (23). A DNA fragment containing nat1 flanked by 80 bp of upf1 homologous sequences was created by polymerase chain reaction (PCR) using pCR2.1-nat (pHL2621) as the template and HL2252 and HL2253 as the primers. To induce homologous recombination between upf1- and the nat1-containing fragment, 5 µg of this nat1 fragment was introduced to YHL1101 with the lithium acetate transformation method (22). NAT-resistant colonies were isolated on YES plates containing 100 µg/ml NAT (Werner BioAgents, Germany). DNA blots and PCR were performed to confirm the single gene deletion. The abp1 deletion strain was a gift from Shiv Grewal. Strains containing Tf1 insertions and lacking abp1 were obtained by standard genetic crosses.
To create a plasmid with wild-type upf1+, a 3.8 kb fragment was amplified by high-fidelity DNA polymerase pfx (Invitrogen) with HL2417 and HL2418 and ligated into pHL1288 digested with SpeI-SalI. The open reading frame (ORF) of upf1 in the constructed plasmid was confirmed by DNA sequencing.
Transposition assay
The transposition assay of Tf1 was described previously (24). The strains were transformed with the Tf1 plasmid pHL2541 by selection on EMM lacking uracil. Strains were arranged on EMM-Ura+B1 plates in 2 cm2 patches. After 2 days of incubation, these plates were replica printed to EMM-Ura-B1 to induce transcription of Tf1. After 4 days of incubation, the strains were replica printed to EMM+Ura+B1+FOA to select against the Tf1 donor plasmid. Following 2 days of incubation, transposition events were selected by replica printed to YES+FOA+G418.
DNA preparations and blots
Genomic DNA was extracted from cells using glass beads and phenol (25). Ten micrograms of genomic DNA isolated from cells (OD600 = 1.0) were digested by HindIII, and the products were separated on a 1.0% agarose gel. The gel was transferred to Genescreen Plus (Perkin Elmer), and the filters were hybridized with a 1.0-kb neo probe derived from a BamHI digest of pHL765.
Determination of Tf1 insertion sites by inverse PCR
Five micrograms of genomic DNA were digested by HindIII. To circularize the fragments, the DNA was diluted to 1 ng/µl, and 17 µl was ligated with 1 µl containing 20 U of T4 DNA ligase (Invitrogen) and 2 µl buffer at 18°C overnight. Several parallel ligations were performed and extracted with phenol–chloroform. One hundred nanograms of ligated DNA was amplified by Titanium Taq (Clontech) with primers HL2347 and HL1104. After gel electrophoresis, the desired products, whose sizes were predicted by DNA blots, were gel purified (Qiagen), ligated into pCR2.1-TOPO (Invitrogen) and sequenced. After determining the sequence of the genomic DNA downstream of the 3′ LTR, primers were designed to PCR amplify the junction between the genomic DNA and the 5′ LTR. In all cases, 5 bp target site duplications were found, indicating that the Tf1 transposons were introduced by bona fide integration.
RNA preparations
RNA was extracted from 40 ml of cells that were harvested at OD600 = 1.0 and resuspended in 400 µl of buffer AE (50 mM NaOAc, pH 5.3 and 10 mM EDTA) and 40 µl of 10% sodium dodecyl sulfate. The resuspended cells were combined with 400 µl of acid phenol (Ambion) and 400 µl of acid-washed glass beads (Sigma, G-8772). The cells were lysed by 10 min of bead beating (Mini Beadbeater, Biospec Products) in cycles of 30 sec on and 30 sec off. The cell lysates were cooled at −80°C for 10 min and then centrifuged for 5 min at 14 000rpm to separate the aqueous phase from the beads, phenol and cell debris. Phenol–chloroform extractions were repeated until the interfaces were no longer white. After adding 40 µl 3 M NaOAc and 1 ml 100% ethanol, RNA was precipitated and stored at −80°C until use. For cells exposed to stress, cultures with OD600 = 1.0 were harvested quickly before and at 15 and 60 min after exposed to stress.
Quantitative reverse transcription PCR
Quantitative reverse transcription PCR (qRT-PCR) was conducted and analyzed as described previously (26). Briefly, 44 µl of total RNA (lower than 200 ng/µl) was treated with 1 µl of TURBO DNase (Ambion) in 5 µl buffer to remove any residual genomic DNA. Following incubation for 30 min at 37°C, the reaction was mixed with DNase inactivation reagent (Ambion). Then, 500 ng of total RNA was reverse transcribed into cDNA using random primers with the high-capacity cDNA reverse transcription kit (Applied Biosystems). As a control for contamination by genomic DNA, the reaction lacking reverse transcriptase was also subjected to qRT-PCR. After cDNA was synthesized and diluted to about 10 ng/µl, 6 µl of this cDNA was added into 1 well of a 96-well plate and mixed with 10 µl of 2X Power SYBR green PCR master mix (Applied Biosystems) and 4 µl of gene specific primer sets (each primer is 500 nM). The sample was run in the Applied Biosystems StepOnePlus Real-Time PCR System according to their instructions. All primer sets were validated for their PCR efficiency by serial dilution of standard cDNA. Efficiency values of 1.8–2.2 were obtained. Each sample from three independent yeast colonies was quantified and averaged as triplicates. Expression of genes in all conditions was normalized to act1. Relative changes in expression of the genes were determined by the comparative Ct (2−Ct) method (26). For the strand-specific qRT-PCR, total RNA was reverse transcribed into cDNA using strand-specific primer with MultiScribe reverse transcriptase (Applied Biosystems). The conditions of other reactions were the same as the normal qRT-PCR. The sense or antisense RNA of tested genes was normalized to sense RNA of act1 to determine the presence and relative levels. The reaction without reverse transcriptase was conducted for the control of genomic DNA, and the reaction without any primer was a control of self-priming of RNA during RT. A strand-specific control was also performed using the specific primer for sense or antisense RNA of act1.
RNA blots
RNA blots were performed as described previously (20). Ten micrograms of total RNA was separated on a 1.0% agarose–formaldehyde denaturing gel. After the RNA was transferred to a nylon membrane, it was hybridized with random primed DNA probes (Roche), oligonucelotide probes or RNA probes synthesized by in vitro transcription (Ambion). As a loading control, the blots were either stained with ethidium bromide for the ribosomal RNA (rRNA) or hybridized to a probe of act1. The act1 probe was produced from a 1 kb Eco RI-Bam HI fragment of pHL859-1. The levels of RNA from three independent yeast colonies were quantified with phosphoimaging and averaged. Expression of genes was normalized to rRNA in the stress conditions and normalized to act1 in other conditions.
Rapid amplification of cDNA ends
The 5′ and 3′ rapid amplification of cDNA ends were assayed by the RNA ligase–mediated RACE kit (Ambion) according to the manufacturer’s instruction. After nested PCR and separation in a 2% agarose gel, the desired and specific DNA products were gel purified and ligated with pCR2.1-TOPO vectors (Invitrogen) and sequenced from both ends. Only high-quality and specific sequences were analyzed.
RESULTS
Integration of Tf1 into Pol II transcribed promoters did not reduce promoter function
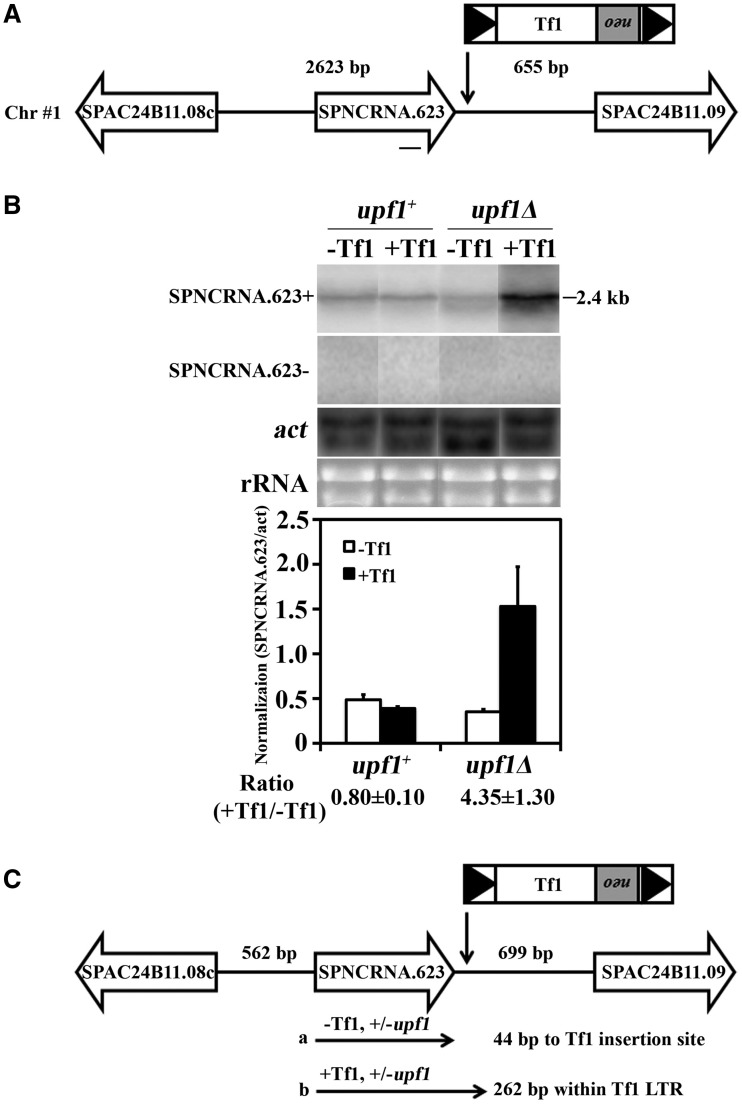
The integration of Tf1 into pol II transcribed promoters has the potential to disrupt the expression of the genes on either side of the insertions (20). To investigate the effect of Tf1 integration on the expression of flanking genes, we generated a collection of strains that had single copy insertions of Tf1. To create these strains, Tf1 containing a neo gene (Tf1-neo) was expressed from a plasmid, and cells that obtained inserts were isolated on a medium containing G418 (24,27). The insertions were generated in a strain that had no pre-existing copies of Tf1. For reasons explained in a later section, this strain contained a deletion of upf1 (upf1Δ) and a plasmid copy of upf1+ to complement the deletion.
From the strains that became resistant to G418, 14 were selected at random and analyzed for the presence of Tf1-neo insertions. DNA blots showed that each strain contained a single copy of Tf1-neo (Supplementary Figure S1). Inverse PCR was used to sequence the position of the insertions, and these data revealed that each integration mapped to a unique site in the genome. All inserts of Tf1 were flanked by 5 bp duplications, a signature structure demonstrating that each insertion was catalyzed by the Tf1 integrase (28). Table 1 lists each insertion, its orientation and the genes on either side of the integration.
Table 1.
Effect of Tf1 integration on expression of adjacent genes in the strains with and without upf1
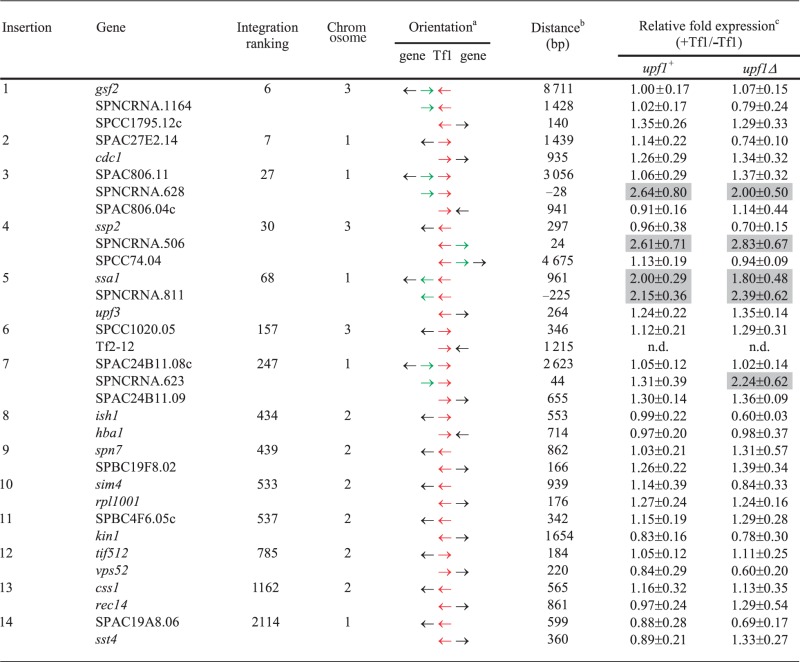 |
aRed arrow represents Tf1; black arrow represents protein-coding gene; green arrow represents non-coding RNA gene. The strand of the non-coding RNA genes depicted (SPNCRNA.628, SPNCRNA.506 and SPNCRNA.811) was based on strand-specific qRT-PCR. For SPNCRNA.623, the transcribed strand was determined with a strand-specific RNA blot.
bDistances between Tf1 inserts and coding genes were measured from the end of Tf1 to the ORF. The distances to non-coding RNA genes were measured to the ends of the non-coding transcripts. The locations of the ORFs, SPNCRNA.1164 and SPNCRNA.811 were based on the PomBase annotations (31). The distances between the non-coding RNA genes SPNCRNA.628, SPNCRNA.506, SPNCRNA.623 and Tf1 were based on the results of 5′ and 3′ RACE (Supplementary Figure S3B, S3C and S3H). The negative numbers of distances indicates that the Tf1 insertion was located inside of the gene.
cValues are results of qRT-PCR and are the average of three independent experiments (mean ± SD).
Of 73 125 insertions of Tf1 that were sequenced previously, 93% occurred in intergenic regions containing pol II transcribed promoters (21). Approximately 1000 promoter regions are preferred for integration, and based on their specific level of integration, each intergenic region was assigned a ranking. All 14 insertions in this study occurred in intergenic sequences containing pol II promoters, and 12 of the 14 were positioned in intergenic regions that were ranked among the top 1000 targets of integration in the S. pombe genome (Table 1). As a result, the impact of these insertions on the expression of the adjacent genes has particular relevance.
To test systematically the effect of Tf1 insertion on the expression of adjacent genes, we used qRT-PCR and measured the relative expression of genes adjacent to Tf1-neo and compared this with the expression of the genes in the parent strain that lacked the insertions. In this study, we measured the mRNA from the coding and non-coding genes on either side of the insertions except for that of Tf2-12 (insertion #6), which as a repeat element could not be independently analyzed. Five of the intergenic sequences with insertions are reported to produce non-coding RNAs (ncRNAs) (29,30). Although it is not known whether these ncRNAs possess function, we measured these transcripts as well. As a result, we monitored a total of 32 genes (Table 1). Although the insertion of 6 kb of transposon DNA into promoter regions might be expected to damage promoter function, none of the 32 genes exhibited reduced expression resulting from the insertion of Tf1 (Table 1, relative fold expression, upf1+). The lack of genes with reduced expression is unlikely due to negative selection during isolation of the insertion strains because these intergenic sequences were also common insertion sites in diploid strains (21).
Integration of Tf1 can enhance the expression of adjacent genes
Insertion of Tf1-neo did increase the expression of four genes adjacent to integration sites. In the case of ssa1, a gene encoding heat-shock protein 70 (Hsp70), an insertion 961 bp upstream of the ORF enhanced its expression by 2.0-fold. In addition, the expression of three ncRNA genes SPNCRNA.628, SPNCRNA.506, and SPNCRNA.811 were enhanced 2.6, 2.6 and 2.1-fold, respectively (Table 1). The results of strand-specific qRT-PCR revealed that SPNCRNA.628 and SPNCRNA.506 are transcribed on the top strand, and contrary to annotations of the reference genome, SPNCRNA.811 was transcribed on the bottom strand of the reference genome (Supplementary Table S4) (31). Together, these results demonstrate that Tf1 insertion had the capacity to stimulate the expression of adjacent genes.
Host surveillance mechanisms can restrict the impact of Tf1 integration on the expression of adjacent genes
The finding that Tf1 insertion enhanced the expression of 4 adjacent genes led us to question why the other 28 were not activated. One possibility we considered was that the expression of ORFs adjacent to Tf1 could potentially be enhanced by mRNA originating from Tf1 but that much of the mRNA was degraded by the nonsense-mediated decay (NMD) pathway, the system that degrades aberrant mRNA (32,33). Indeed, mRNA originating from a closely related transposon Tf2 does read into adjacent sequence and in some cases activates the expression of neighboring genes (10). This type of read-through transcript could have premature stop codons that would trigger NMD. To make it efficient to test whether NMD might restrict the expression of ORFs adjacent to Tf1, the original insertions were created in a strain that had a deletion of upf1, the gene encoding a key factor required for NMD. To complement the deletion, the strain also contained a plasmid-encoded copy of upf1. qRT-PCR revealed that the plasmid copy of upf1 produced levels of mRNA similar to that produced by a single genomic copy of upf1 (data not shown). The expression studies described above were all done with strains containing the deletion of upf1 and the plasmid encoded upf1.
We tested NMD for a potential role in restricting the expression of the genes adjacent to Tf1 by measuring the impact of Tf1 insertion on mRNA levels in strains that lacked both chromosomal and plasmid copies of upf1. Surprisingly, qRT-PCR revealed that the expression of 31 of the adjacent genes did not change when upf1 was absent (Table 1). Although NMD may not play a frequent role in reducing the RNA of genes next to Tf1, we found that one Tf1 insertion did enhance RNA levels of SPNCRNA.623 2.2-fold with an increase that was specific for cells lacking upf1 (Table 1, insertion #7).
Another host factor that may limit the ability of Tf1 to enhance expression of adjacent genes is the CENP-B homolog Abp1. The previous work showed that Abp1 plays a role in clustering Tf1 and Tf2 elements into nuclear bodies and inhibiting the expression of the transposons by recruiting histone deacetylases (HDACs) (34). In at least one case, Abp1 inhibits the expression of a gene next to an existing LTR (34). To investigate directly whether Abp1 limits the ability of de novo insertions to increase the expression of adjacent genes, we crossed six strains containing Tf1 insertions with a strain lacking abp1. We then used qRT-PCR to measure whether Tf1 insertion in strains lacking abp1 enhanced the expression of the adjacent genes. These strains all contained the single chromosomal allele of upf1. Interestingly, we found that in cells lacking abp1, Tf1 insertion did increase the expression of spn7, css1 and SPBC4F6.05 c, whereas Tf1 integration had no effect on expression of these genes in cells containing abp1 (Table 2). The activation of ssa1 and SPNCRNA.811 observed in cells with abp1 was not significantly changed in the cells that lacked abp1. These results indicated that Abp1 did play a role in inhibiting the ability of Tf1 to enhance the expression of some adjacent genes.
Table 2.
The effect of Tf1 integration on expression of adjacent genes in strains with and without abp1
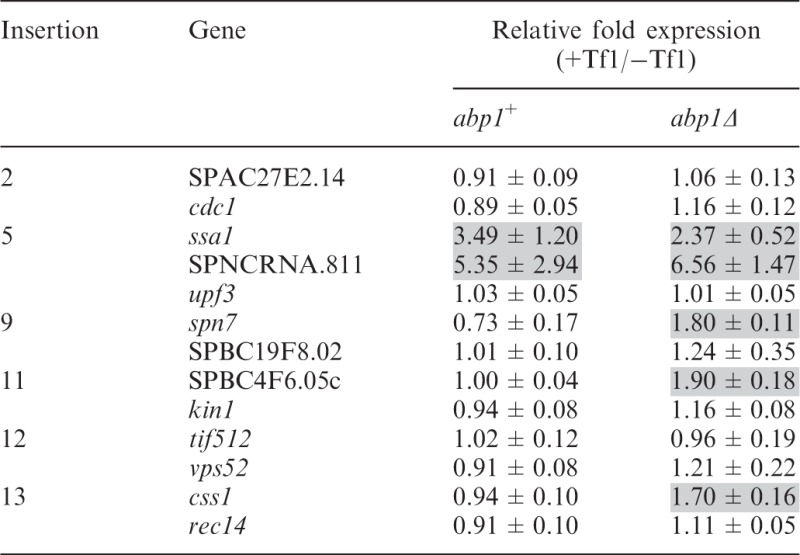 |
The RNA levels were measured by qRT-PCR from three independent strains and averaged.
Conditions of heat and oxidative stress were required for Tf1 to increase the expression of some adjacent genes
Tf1 integration favors promoters of stress response genes, and several of the genes targeted by Tf1 in this study are reported to be induced by heat or oxidative stress. As a result, we tested whether conditions of stress would reveal or enhance the ability of Tf1 sequences to increase the expression of adjacent genes. All 14 strains with a Tf1 insertion were incubated at 39°C for 15 and 60 min, and qRT-PCR of their RNA revealed the expression of four genes, SPNCRNA.506, SPNCRNA.811, spn7 and css1, was enhanced by adjacent Tf1 inserts (Table 3). Although expression of SPNCRNA.506 and SPNCRNA.811 was enhanced by Tf1 in cells grown in regular conditions (Tables 1 and 3), their enhancements were increased by the heat treatments, 2.2- and 2.7-fold, respectively. In contrast, the increase of spn7 and css1 expression by Tf1 sequence required heat stress (Tables 1 and 3). An antisense ncRNA gene (SPNCRNA.1588) has been predicted to overlap spn7 (29). However, we used strand-specific qRT-PCR and found that it was the sense RNA of spn7 that was enhanced by adjacent Tf1 sequence after heat treatment (Table 3). Despite being enhanced by Tf1 in cells grown at regular temperature, the expression of genes ssa1 (Supplementary Figure S2) and SPNCRNA.628 was not increased by Tf1 in cells exposed to heat treatment (Table 3).
Table 3.
Effect of Tf1 integration on expression of adjacent genes under heat stress
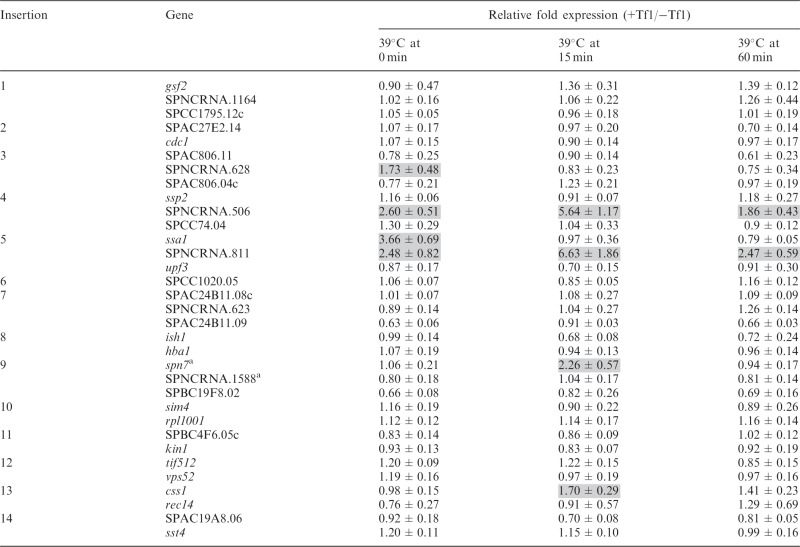 |
aResult of strand specific qRT-PCR.
Many genes of S. pombe induced by heat treatment are also induced by oxidative stress (9). Therefore, we tested whether the genes activated by adjacent Tf1 sequence by heat treatment were also induced by oxidative stress. The expression of genes enhanced by Tf1 insertion when exposed to heat was also enhanced when cells were treated with H2O2 (Table 4). For SPNCRNA.506 and SPNCRNA.811, the levels were significantly greater when cells were treated with H2O2 compared with heat treatment.
Table 4.
Effect of Tf1 integration on expression of adjacent genes under oxidative stress
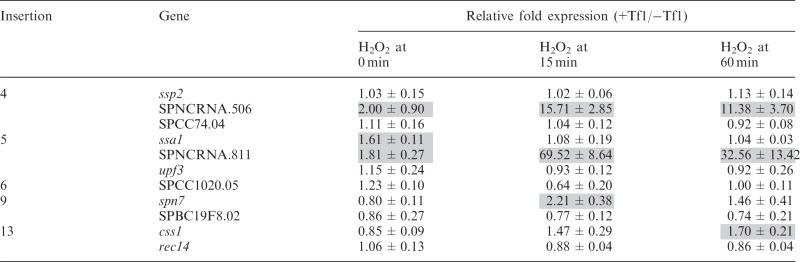 |
Although environmental stress allowed Tf1 sequences to increase the expression of 4 of the genes studied, the expression of the other 28 genes adjacent to Tf1 insertions was unchanged with these conditions (Tables 3 and 4). We tested whether the expression of genes unaffected by Tf1 tested during stress might be limited by NMD. Nevertheless, the expression of SPNCRNA.623, vps52 and rec14 in upf1Δ cells remained unchanged by Tf1 insertion in cells treated with heat (data not shown). Collectively, these data indicate that the ability of Tf1 to increase the expression of adjacent genes can be mediated by environmental stress.
The transcription of Tf1 is induced by environmental stress
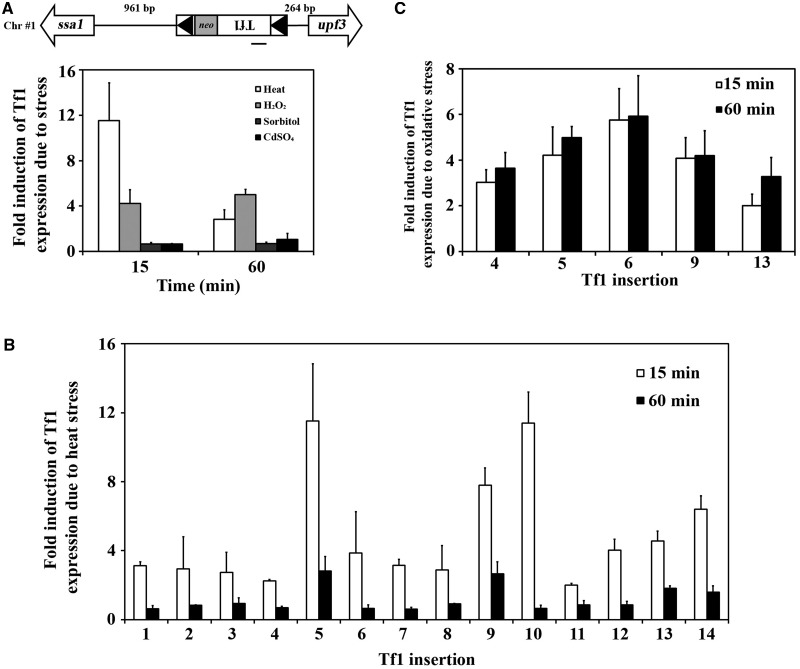
To understand how Tf1 sequences might enhance the expression of adjacent genes, we tested whether the promoter of Tf1 itself responded to environmental stress. Using qRT-PCR, we measured the expression of Tf1 in the strain that contained a single-copy Tf1 adjacent to ssa1 (insertion #5). This copy of Tf1 was induced substantially when cells were heat treated and to a lesser extent when subjected to oxidative stress (Figure 1A, heat vs. H2O2). In comparison, osmotic stress and exposure to a heavy metal did not induce this Tf1 element. To determine whether the stress-mediated induction of Tf1 mRNA was dependent on the location of the transposon, we surveyed Tf1 mRNA levels produced from all 14 insertion positions. Although the expression of Tf1 at all 14 positions was induced by heat treatment, Tf1 mRNA from insertions #5 and #10 increased more than 10-fold (Figure 1B). In the five different insertion positions tested, Tf1 mRNA was also induced by oxidative stress, indicating that both heat and oxidative stress triggered increased transcription of Tf1 regardless of the position of the transposon (Figure 1C).
Figure 1.
Tf1 transcription is induced by environmental stress. (A) The Tf1-neo of insertion #5 occurred in Chr #1 between divergent genes ssa1 and upf3. The LTRs are indicated by black triangles and the orientation of Tf1 transcription is from right to left. The distance between the insertion site and the ATGs of the adjacent ORFs is shown. The relative change of Tf1 mRNA in response to environmental stresses for 15 and 60 min was measured by qRT-PCR. The amplicon is indicated by the bar under Tf1. The fold change is represented in the histogram. In all RNA measurements here and in all other experiments except in Figure 4, Table 2 and Supplementary Figure S5, the fold change from three independent plasmid transformants is averaged. One standard deviation is represented with error bars. (B) The response of Tf1 expression to heat (15 and 60 min) in each of the 14 insertion strains was measured by qRT-PCR and is shown in histograms. (C) qRT-PCR was used to measure the response of Tf1 expression to oxidative stress in a representative group of the insertion strains. The inductions of Tf1 mRNAs after 15 and 60 min of treatment are shown.
Tf1 increases the expression of adjacent genes by providing enhancer activity
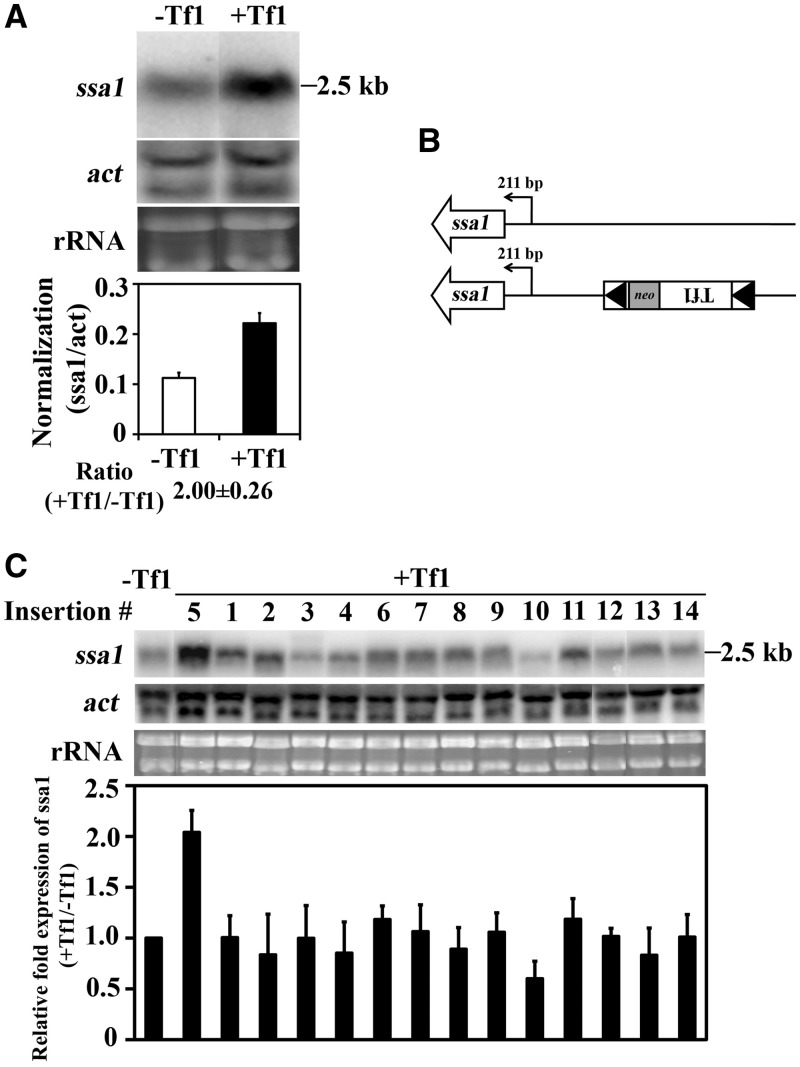
The enhanced expression of genes by adjacent copies of Tf1 may have resulted from transcription that initiated from within the transposon. Alternatively, Tf1 may contain enhancer elements that could have increased the activity of promoters that pre-existed the insertion event. To distinguish between these possibilities, we ran RNA blots and tested whether mRNAs with levels enhanced by Tf1 insertions were the same size as those produced in the absence of Tf1. An RNA blot of insertion strain #5 and the parent lacking the insertion confirmed that the insertion of Tf1 increased the expression of ssa1 by 2.0-fold (Figure 2A). More importantly, the ssa1 mRNA produced by insertion strain #5 co-migrated with the ssa1 mRNA from the strain that lacked the Tf1 insertion. The results of 5′ RACE experiments and the sequencing of the RACE products revealed that the ssa1 mRNA increased by the Tf1 insertion initiated from the same nucleotide as the mRNA produced by the strain lacking the insertion, 211 bp upstream of the ORF (Figure 2B and Supplementary Figure S3A). This result indicated that the insertion of Tf1 increased the transcription of ssa1 from its natural start site.
Figure 2.
Tf1 insertion increased the expression of ssa1. (A) The levels of ssa1 mRNA from cells lacking Tf1 and from cells with Tf1 upstream of ssa1 was measured on RNA blots. The amount of ssa1 mRNA relative to actin mRNA is shown in the histogram. (B) The transcription start site of ssa1 mRNA from cells with and without the Tf1 insertion at ssa1 was mapped by 5′ RACE and sequencing of the products. (C) The levels of ssa1 mRNA from cells without Tf1 and from all the 14 strains with Tf1 insertions were measured on an RNA blot. The histogram shows for each insertion strain the change in the expression of ssa1 mRNA caused by the insertion of Tf1.
Ssa1 is a member of the Hsp70 superfamily of stress-induced protein chaperones. This raised the possibility that ssa1 expression was increased not because it was adjacent to Tf1 sequence but because insertion of the transposon resulted in expression of Tf1 protein that in turn triggered induction of stress response proteins such as Ssa1. If true, increases of ssa1 expression would occur independent of the position of Tf1 integration. To test this possibility, we measured the ssa1 mRNA produced by all 14 of the insertion strains. RNA blots revealed that integration adjacent to ssa1 (insertion #5) was the only position of Tf1 that increased ssa1 expression (Figure 2C). This result demonstrated that the enhanced expression of ssa1 was the direct result of the proximity of Tf1 sequence to ssa1.
To identify the mechanism that enhanced the expression of ncRNA, we analyzed RNA from the insertion strain #3 on blots. This RNA blot confirmed the qRT-PCR result that insertion of Tf1 did increase the expression of SPNCRNA.628 (Supplementary Figure S4A and S4B). The blot showed that the increase was 3.7-fold. The enhanced SPNCRNA.628 RNA co-migrated with the SPNCRNA.628 RNA expressed by the parent strain that lacked the insertion. This indicates that Tf1 sequence can also increase the expression of ncRNA genes by providing enhancer activity. Our RACE analysis of the 5′ start of transcription revealed that the SPNCRNA.628 RNA enhanced by Tf1 initiated from its natural start site (Supplementary Figures S3B and S4C). Interestingly, 3′ RACE assays revealed that Tf1 inserted within SPNCRNA.628 28 bp from the end of the gene (Supplementary Figures S3B and S4C). The result of this insertion was the SPNCRNA.628 RNA terminated at base 262 of the Tf1 LTR.
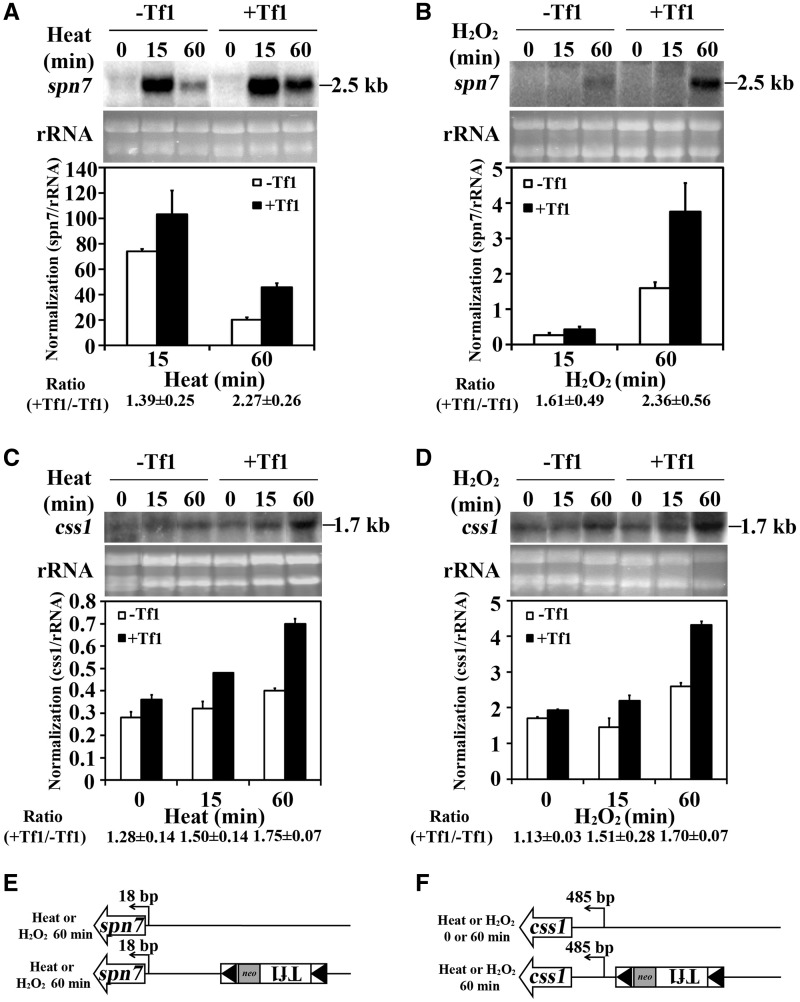
To determine how Tf1 enhances the expression of adjacent genes in response to environmental stress, we again characterized mRNAs. RNA blots of cells heat treated for 60 min showed that the spn7 mRNA increased by Tf1 co-migrated with spn7 mRNA produced by the strain that lacked the insertion (Figure 3A). The same cells exposed to oxidative stress generated increased levels of spn7 mRNA that co-migrated with the spn7 mRNA from cells lacking the insertion (Figure 3B). In cells exposed to heat or oxidative stress, the css1 mRNA enhanced by insertion of Tf1 also co-migrated with css1 from cells lacking the insertion (Figure 3C and D). In each case, regardless whether cells were treated with heat or oxidative stress, 5′ RACE followed by sequencing showed that the spn7 and css1 mRNAs with levels increased by Tf1 started from their natural initiation sites (Figure 3E and F and Supplementary Figures S3D and S3E).
Figure 3.
Tf1 insertion increased the expression of spn7 and css1 when cells were exposed to stress. (A and B) The increase in spn7 mRNA caused by Tf1 insertion upstream of spn7 was measured on RNA blots. Cells were heat treated for 0, 15 and 60 min or (B) treated with peroxide for 0, 15 and 60 min. (C and D) The increase in css1 mRNA caused by Tf1 insertion upstream of css1 was measured on RNA blots. The cells were either treated with (C) heat or (D) hydrogen peroxide. (E and F) The transcription start site of (E) spn7 mRNA or (F) css1 mRNA in cells with or without upstream insertions of Tf1 was mapped by 5′ RACE followed by sequencing of the products. The cells were treated with heat and peroxide.
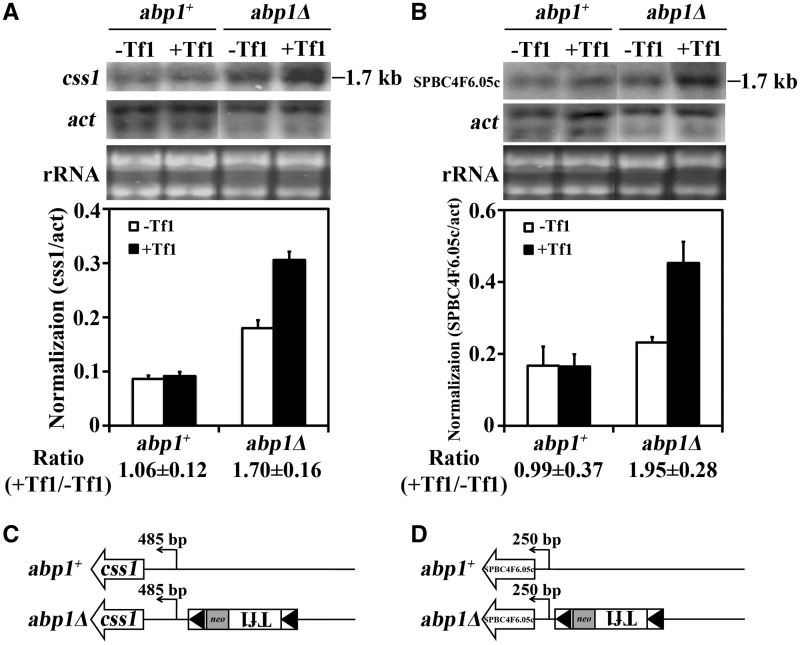
In experiments described earlier, Abp1 was found to restrict the Tf1-mediated increase in the expression of spn7, SPBC4F6.05c and css1 (Table 2). To determine what type of enhancement mechanisms were restricted by Abp1, we again characterized the mRNAs. In the absence of abp1, the css1 and SPBC4F6.05c mRNAs increased by adjacent copies of Tf1 co-migrated with the RNAs from the strain lacking Tf1 (Figure 4A and B). Again, the results of 5′ RACE demonstrated that the enhanced mRNAs start from the same nucleotides as the mRNAs from the strain that lacked Tf1 (Figure 4C and D and Supplementary Figure S3F and S3G). These data indicate that the enhanced expression of genes by adjacent copies of Tf1 results from increased activity of the genes’ promoters. This was true regardless of whether the increases were mediated by environmental stress or the absence of the genome surveillance protein Abp1.
Figure 4.
Abp1 restricted the enhancer activity of Tf1 and limited the expression of adjacent genes. (A) The effect of the Tf1 insertion on levels of css1 mRNA in cells with and without abp1 was measured on RNA blots. The strains analyzed lacked Tf1 or contained a Tf1 insertion upstream of css1. RNA from cells lacking abp1 was analyzed revealing that Abp1 restricted the activation of css1. The levels of css1 mRNA relative to actin mRNA were averaged from three independent strains in the histogram. (B) The effect of Tf1 integration on the expression of SPBC4F6.05c was analyzed as described in (A). The transcription start sites for (C) css1 and (D) SPBC4F6.05c were determined by 5′ RACE followed by sequencing of the products.
The ability of Tf1 to increase the activity of adjacent promoters suggests that the transposon possesses enhancer elements. Previously published data indicated that Abp1 recruits HDACs to LTRs of Tf1 and Tf2, and to full-length Tf2s, retrotransposons that are closely related to Tf1 and reside in laboratory strains of S. pombe (34). The deacetylases, Clr3 and Clr6, inhibit the transcription of the Tf2s in the laboratory strain and a de novo inserted Tf1 at SPAC7D4.08. If Tf1 increases the expression of adjacent genes by providing enhancer activity, the inhibitory activity of Abp1 on spn7, SPBC4F6.05c and css1 would likely act on the promoters of the Tf1 adjacent to these genes. We did find that expression of the Tf1 copies adjacent to spn7, SPBC4F6.05c and css1 was significantly restricted by Abp1 (Supplementary Figure S5, insertions #9, #11 and #13). Three other positions of Tf1 insertion were also tested, and in each case, Abp1 inhibited Tf1 expression (Supplementary Figure S5).
Upf1 degrades SPNCRNA.623 RNA enhanced by Tf1 insertion
One possible mechanism that could cause Tf1 insertions to increase the expression of adjacent genes would be the transcription of RNA that initiates in Tf1 continues into the coding sequences of neighboring genes. Although this mechanism was not observed, we tested whether these read-through RNAs might be generated by Tf1 but degraded by the NMD pathway. Although NMD did not play a general role in restricting the enhancement of genes by Tf1, the expression of 1 of the 32 genes studied, SPNCRNA.623, was enhanced by an adjacent copy of Tf1 when upf1 was deleted (Table 1, insertion #7). RNA blots demonstrated that SPNCRNA.623 was increased 4.3-fold by the adjacent copy of Tf1, and this increase was only observed in the absence of upf1 (Figure 5A and B). Although this result suggested that Upf1 had degraded an RNA initiated in Tf1 that continued into SPNCRNA.623, strand-specific probes of the RNA blot demonstrated that SPNCRNA.623 is a plus strand transcript and therefore could not have been encoded by an RNA initiated from the downstream Tf1 (Figure 5A and B).
Figure 5.
Upf1 restricted the increased expression of SPNCRNA.623 by the enhancer activity of Tf1. (A) The orientation and position of the Tf1 insertion between SPNCRNA.623 and SPAC24B11.09 is shown. The sequence of the probe is represented by a line underneath SPNCRNA.623. The distances from the Tf1 insertion to the adjacent ORFs is shown above the line. (B) The levels of SPNCRNA.623 RNA were measured from strains that lacked Tf1 or contained Tf1 inserted downstream of SPNCRNA.623. The RNA blots were hybridized with strand-specific probes to determine the orientation of SPNCRNA.623 transcription. In addition, RNA from cells lacking upf1 was analyzed to determine whether NMD limited the enhanced expression of SPNCRNA.623 by Tf1. The levels of SPNCRNA.623 RNA are shown in the histogram. (C) The 5′ and 3′ termini of SPNCRNA.623 RNA were determined by RACE followed by sequencing. The distance from the 5′ and 3′ termini of SPNCRNA.623 to the adjacent ORFs is shown above the line. The termini are represented by the arrows labeled a and b.
To understand how Tf1 increased transcription of SPNCRNA.623 and why the NMD pathway degraded this RNA, the start and termination sites of this transcript were mapped with 5′ and 3′ RACE. The sequences of the 5′ RACE products revealed that the RNA of SPNCRNA.623 started at the same site regardless of whether Tf1 was present or whether upf1 was deleted (Figure 5C and Supplementary Figure S3H). However, 3′ RACE assays indicated that the adjacent copy of Tf1 caused the SPNCRNA.623 RNA to extend past its normal termination site and end 262 bp into the 5′ LTR (Figure 5C and Supplementary Figure S3H). This termination site in the LTR corresponded to the same position of the 3′ end of the SPNCRNA.628 RNA that continued into the Tf1 sequence (Supplementary Figure S4C), indicating that the termination is mediated by a signal in the LTR. The extended version of the SPNCRNA.623 RNA had premature termination codons that would be recognized by the NMD pathway. The termination codons would explain why Upf1 degraded the extended version of the SPNCRNA.623 transcript. Despite the 3′ extension of the transcript, the RNA with levels enhanced by Tf1 initiated from its natural start site, indicating that the insertion increased the amount of the RNA by providing enhancer activity.
Genes with expression increased by Tf1 insertions are stress response genes
Of the 32 genes analyzed in this study, 6 had expression enhanced by Tf1 sequence, in cells grown at 32°C or in response to heat treatment (Tables 1 and 3). Results of RNA blots and RACE assays indicated that the transcription of these genes was increased by enhancer activity carried in Tf1. One key question was why did the enhancer activity in Tf1 not increase the transcription of the other 26 genes. One possibility was that the chromatin context of each insertion imposed varying degrees of access to transcription factors. If the enhancer function of some Tf1 copies were impaired, these insertions would not be expected to increase expression of adjacent genes. To test this possibility, we asked whether there was a correlation between the genes with expression increased by Tf1 sequences and the transcription levels of the adjacent copies of Tf1. qRT-PCR revealed that the copies of Tf1 of insertion #7 and #10 were expressed 10 and 7 times higher than the other inserts, respectively (Supplementary Figure S6A). None of the six genes with expression enhanced by Tf1 were adjacent to inserts #7 or #10. This poor correlation suggests that it was not a variation in the enhancer activities of the Tf1 inserts that determined which adjacent genes had increased expression. In addition, the transcription levels of the cellular genes in the strain that lacked Tf1 insertions were no higher for the genes that had expression increased by Tf1 (Supplementary Figure S6B), indicating that it was not the chromatin context of the insertion sites that determined which genes had expression increased by Tf1 insertions. We also recognized that it was not solely the Tf1 elements with the greatest response to stress (Figure 1B, insertions #5 and #10) that determined which genes in stress conditions had increased expression when adjacent to Tf1 (Table 3). Thus, the Tf1 promoters were not the only factors responsible for increased expression of adjacent genes.
The increased expression of adjacent genes by Tf1 enhancer elements may require interactions between the transcription factors bound to the Tf1 enhancer and those bound to the adjacent promoter. This interaction would be more likely to occur if the transcription factors bound at the two sites were the same protein or closely related. The promoters of all 14 Tf1 copies were induced by heat (Figure 1B). To determine which of the 32 genes flanking Tf1 insertions might be regulated by the same stress response factors as Tf1, we asked which genes were induced by heat or oxidative stress independently of Tf1. In the strain lacking Tf1 insertion, 8 of the 32 genes were induced 2-fold or more by heat treatment (Supplementary Table S5). Most of the genes tested were also induced by oxidative stress. Importantly, all six genes that had expression increased by adjacent Tf1 sequences were among the eight genes activated by heat. The correlation between the genes with expression enhanced by Tf1 and those that are stress induced is strong as all the genes with expression increased by Tf1 were also stress induced. (The Fisher's exact test two-tailed P-value is less than 0.0001.) The strength of this correlation argues that inserts of Tf1 can enhance the transcription of adjacent genes only if the genes have stress response promoters.
To examine the promoters that had expression enhanced by Tf1 for stress response elements, we analyzed their sequences for common motifs. The motif-based sequence analysis tool MEME (35) identified a motif present in all six promoters that had activity increased by Tf1 and in the Tf1 promoter itself (Supplementary Figure S7A–S7C). Importantly, the search tool FIMO found that this motif was not present in 25 of the 26 genes not affected by Tf1 insertion (36). Although there is little information about the sequences bound by the transcription factors of S. pombe, we used the motif comparison tool TOMTOM (37) to compare the motif present in Tf1 and the promoters activated by Tf1 to specific sequences bound by proteins in S. cerevisiae. We found the motif was significantly similar to the sequence bound by Sko1 of S. cerevisiae, a ATF/CREB stress response factor (P = 0.038) (Supplementary Figure S7D). What is more, the sequence bound by Sko1 in S. cerevisiae (TGACGT, Supplementary Figure S7D) is very similar to the sequence (TGACGT) (38) bound by the ATF/CREB stress response factor Atf1 of S. pombe. Together these analyses indicate that the promoters enhanced by Tf1 insertions and the Tf1 promoter itself share a common motif that may be recognized by an ATF/CREB stress response factor.
DISCUSSION
Integration of transposons has the potential to destroy coding sequences throughout the genome of the host cell. However, because the fate of transposons is inexorably tied to that of the cell, a variety of mechanisms have evolved that direct integration to regions of the genome that lack coding sequences. In S. cerevisiae, Ty5 integrates into regions of heterochromatin and Ty1 and Ty3 insert into the gene-free sequences upstream of pol III transcribed genes (12,14,39,40). In plants, many transposons accumulate in heterochromatin, and in the case of the chromovirus transposons and Tal1, this distribution likely resulted from integration mechanisms that recognize histone modifications (41,42). In light of the mechanisms that direct integration to gene free sequences, it is surprising that Tf1 integrates in the promoters of pol II transcribed genes (20,43). Integration in promoters was expected to disrupt recognition elements and reduce transcription. Also, Tf1 insertions were expected to reduce expression of adjacent genes because the binding of Abp1 to Tf LTRs recruits HDACs that not only reduces transcription of the LTRs but also has been found to inhibit expression of an adjacent gene (34). Indeed, we confirmed that Abp1 inhibited Tf1 transcription, and in three cases, Abp1 reduced the transcription of genes next to de novo inserted copies of Tf1. Nevertheless, our study revealed that insertion of Tf1 next to 32 different genes did not reduce the expression of any of those genes. Saturated profiles of integration levels at each promoter of S. pombe revealed that the 32 promoters studied here are among the most common sites of Tf1 integration (21). As a result, our findings indicate that the bulk of Tf1 integration events do not reduce the expression of adjacent genes.
Tf1 integration increased the expression of 6 of 32 genes studied, and if not, for the restrictive action of the cellular genes upf1 and abp1, Tf1 would have enhanced the expression of 2 additional genes. The LTRs of Tf2 are transcribed (30,34,44–46), and in rare cases, the RNAs can continue into adjacent sequence and read-through neighboring genes (10). However, in the examples studied here, the increases in mRNA of genes caused by Tf1 insertion were not the result of read-through transcripts. Instead, the results of RNA blots and 5′ RACE assays revealed that Tf1 carried enhancer activity that increased the promoter activity of genes adjacent to Tf1.
One significant question is why did Tf1 insertion enhance the expression of some genes and not others. We found that genes induced by heat were the only genes with expression that was increased by Tf1 insertion. This together with our finding that the transcription of Tf1 itself was induced by heat suggests that the Tf1 enhancer and the stress response genes were recognized by the same or similar activators of transcription. Indeed, the identification of a motif common to the promoters of Tf1 and the genes that had expression increased by Tf1 insertions support this model. Similar factors bound to adjacent sequences often multimerize and cause synergistic increases in transcription. With this model, the insertion of Tf1 next to a stress response gene would be effectively increasing the number of binding sites for the same transcription activators and thus stimulate transcription.
Four hundred twenty-seven ncRNA genes have been detected in S. pombe (30). The significance of these genes is an important question that is being actively studied. It is interesting that of the 14 Tf1 insertions chosen at random for this study, 5 were adjacent to ncRNA genes. And these five were associated with the most common positions of integration (Table 1). We also found that the expression of three of the five non-coding genes was increased by Tf1 insertion. These observations together with the result that the expression of these three ncRNA genes was induced by heat indicates that the non-coding genes are among the stress response genes that are targeted by Tf1. These data suggest that some ncRNA genes may have a function related to stress response and the ability of Tf1 to increase the expression of these genes may have a biological impact.
Although retrotransposons and retroviruses have previously been shown to enhance expression of genes positioned at insertion sites, there are no estimates for what fraction of insertions cause changes in expression of genes at the target sites. In the cases where retrovirus or retrotransposons increase expression of adjacent genes, the events are rare insertions isolated with strong conditions of selection and obtained only after many generations of cell division. For example, after long latency periods of infection, mouse mammary tumor virus induces adenocarcinomas of the mammary gland by activating the expression of oncogenes at insertion sites (47–50). In S. cerevisiae, insertions of Ty1 occurs upstream of tRNA genes (51–54). But in rare cases, Ty1 insertions that activate or inactivate the expression of pol II transcribed genes can be isolated with strong positive selection where growth requires increased or decreased expression of a specific gene product (55–58). The availability of a genome-wide profile of integration sites and the finding that integration of Tf1 commonly leads to increased expression of adjacent genes demonstrate that a large proportion of Tf1 integration results in the enhanced expression of stress response genes. This detailed survey of integration events provides a unique understanding of how integration impacts the biology of the host cell.
Her discovery and pioneering studies of transposable elements in maize led McClintock to propose that transposons provide the host with important genetic diversity. Her observation that transposon mobility was induced by environmental stress led her to formulate the model that transposons reorganize the host genome as a means of responding to stress (11). The ability of Tf1 to increase the expression of stress response genes is consistent with McClintock’s intriguing hypothesis because Tf1 insertion may protect a fraction of cells subjected to environmental stress. Our finding that heat and oxidative stress increased expression of Tf1 provides additional support for this model because increased transcription of Tf1 can lead to de novo integration (24,28). These observations together with the finding that Tf1 insertions are directed to stress response genes suggest that Tf1 may have evolved its transcription and integration mechanisms to improve the survival of cells exposed to environmental stress (1,21). Although this ability of Tf1 to increase expression of stress response genes is an intriguing process, additional studies are necessary to test whether these transposon-mediated increases in expression actually benefit cells when exposed to stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–7.
FUNDING
Intramural Research Program of the National Institutes of Health from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Funding for open access charge: NIH intramural program and NICHD.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Shiv Grewal for the gift of the strain that lacks abp1.
REFERENCES
- 1.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore SP, Liti G, Stefanisko KM, Nyswaner KM, Chang C, Louis EJ, Garfinkel DJ. Analysis of a Ty1-less variant of Saccharomyces paradoxus: the gain and loss of Ty1 elements. Yeast. 2004;21:649–660. doi: 10.1002/yea.1129. [DOI] [PubMed] [Google Scholar]
- 3.Scheifele LZ, Cost GJ, Zupancic ML, Caputo EM, Boeke JD. Retrotransposon overdose and genome integrity. Proc. Natl Acad. Sci. USA. 2009;106:13927–13932. doi: 10.1073/pnas.0906552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 5.Ooi SK, O'Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J. Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461:423–426. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]
- 7.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 8.Todeschini AL, Morillon A, Springer M, Lesage P. Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:7459–7472. doi: 10.1128/MCB.25.17.7459-7472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen DR, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bahler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sehgal A, Lee CY, Espenshade PJ. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 2007;3:e131. doi: 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 12.Sandmeyer S. Integration by design. Proc. Natl Acad. Sci. USA. 2003;100:5586–5588. doi: 10.1073/pnas.1031802100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bushman FD. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. doi: 10.1016/s0092-8674(03)00760-8. [DOI] [PubMed] [Google Scholar]
- 14.Lesage P, Todeschini AL. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- 15.Ebina H, Levin HL. Stress management: how cells take control of their transposons. Mol. Cell. 2007;27:180–181. doi: 10.1016/j.molcel.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Bolton EC, Boeke JD. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res. 2003;13:254–263. doi: 10.1101/gr.612203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinsey PT, Sandmeyer SB. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991;19:1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singleton TL, Levin HL. A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of insertion. Eukaryot. Cell. 2002;1:44–55. doi: 10.1128/EC.01.1.44-55.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowen NJ, Jordan I, Epstein J, Wood V, Levin HL. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements derived from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leem YE, Ripmaster TL, Kelly FD, Ebina H, Heincelman ME, Zhang K, Grewal SIS, Hoffman CS, Levin HL. Retrotransposon Tf1 is targeted to pol II promoters by transcription activators. Mol. Cell. 2008;30:98–107. doi: 10.1016/j.molcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, Levin HL. High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res. 2010;20:239–248. doi: 10.1101/gr.099648.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- 24.Levin HL. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol. Cell. Biol. 1995;15:3310–3317. doi: 10.1128/mcb.15.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atwood A, Choi J, Levin HL. The application of a homologous recombination assay revealed amino acid residues in an LTR-retrotransposon that were critical for integration. J. Virol. 1998;72:1324–1333. doi: 10.1128/jvi.72.2.1324-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 27.Levin HL. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol. Cell. Biol. 1996;16:5645–5654. doi: 10.1128/mcb.16.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin HL, Boeke JD. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J. 1992;11:1145–1153. doi: 10.1002/j.1460-2075.1992.tb05155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 31.Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2011;40:D695–D699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 33.Amrani N, Dong S, He F, Ganesan R, Ghosh S, Kervestin S, Li C, Mangus DA, Spatrick P, Jacobson A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2006;34:39–42. doi: 10.1042/BST20060039. [DOI] [PubMed] [Google Scholar]
- 34.Cam HP, Noma K, Ebina H, Levin HL, Grewal SIS. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:U431–U432. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 35.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eshaghi M, Lee JH, Zhu L, Poon SY, Li J, Cho KH, Chu Z, Karuturi RK, Liu J. Genomic binding profiling of the fission yeast stress-activated MAPK Sty1 and the bZIP transcriptional activator Atf1 in response to H2O2. PLoS One. 2010;5:e11620. doi: 10.1371/journal.pone.0011620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y, Dai J, Fuerst PG, Voytas DF. From the cover: controlling integration specificity of a yeast retrotransposon. Proc. Natl Acad. Sci. USA. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai J, Xie W, Brady TL, Gao J, Voytas DF. Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Mol. Cell. 2007;27:289–299. doi: 10.1016/j.molcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008;18:359–369. doi: 10.1101/gr.7146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsukahara S, Kawabe A, Kobayashi A, Ito T, Aizu T, Shin IT, Toyoda A, Fujiyama A, Tarutani Y, Kakutani T. Centromere-targeted de novo integrations of an LTR retrotransposon of Arabidopsis lyrata. Genes Dev. 2012;26:705–713. doi: 10.1101/gad.183871.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Majumdar A, Chatterjee AG, Ripmaster TL, Levin HL. The determinants that specify the integration pattern of retrotransposon Tf1 in the fbp1 promoter of Schizosaccharomyces pombe. J. Virol. 2011;85:519–529. doi: 10.1128/JVI.01719-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat. Genet. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson HE, Wardle J, Korkut SV, Murton HE, Lopez-Maury L, Bahler J, Whitehall SK. The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol. Cell. Biol. 2009;29:5158–5167. doi: 10.1128/MCB.00698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woolcock KJ, Gaidatzis D, Punga T, Buhler M. Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2010; 18:94–99. doi: 10.1038/nsmb.1935. [DOI] [PubMed] [Google Scholar]
- 47.Nusse R, van Ooyen A, Rijsewijk F, van Lohuizen M, Schuuring E, van't Veer L. Retroviral insertional mutagenesis in murine mammary cancer. Proc. Roy Soc. Lond. B Biol. Sci. 1985;226:3–13. doi: 10.1098/rspb.1985.0075. [DOI] [PubMed] [Google Scholar]
- 48.Ross SR. Mouse mammary tumor virus molecular biology and oncogenesis. Viruses. 2010;2:2000–2012. doi: 10.3390/v2092000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shackleford GM, MacArthur CA, Kwan HC, Varmus HE. Mouse mammary tumor virus infection accelerates mammary carcinogenesis in Wnt-1 transgenic mice by insertional activation of int-2/Fgf-3 and hst/Fgf-4. Proc. Natl Acad. Sci. USA. 1993;90:740–744. doi: 10.1073/pnas.90.2.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, Jonkers J, Hilkens J. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat. Genet. 2007;39:759–769. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 51.Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 52.Ji H, Moore DP, Blomberg MA, Braiterman LT, Voytas DF, Natsoulis G, Boeke JD. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 53.Baller JA, Gao J, Stamenova R, Curcio MJ, Voytas DF. A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res. 2012;22:704–713. doi: 10.1101/gr.129585.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mularoni L, Zhou Y, Bowen T, Gangadharan S, Wheelan SJ, Boeke JD. Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res. 2012;22:693–703. doi: 10.1101/gr.129460.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scherer S, Mann C, Davis RW. Reversion of a promoter deletion in yeast. Nature. 1982;298:815–819. doi: 10.1038/298815a0. [DOI] [PubMed] [Google Scholar]
- 56.Natsoulis G, Thomas W, Roghmann MC, Winston F, Boeke JD. Ty1 transposition in Saccharomyces cerevisiae is nonrandom. Genetics. 1989;123:269–279. doi: 10.1093/genetics/123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Errede B, Cardillo TS, Wever G, Sherman F, Stiles JI, Friedman LR. Studies on transposable elements in yeast. I. ROAM mutations causing increased expression of yeast genes: their activation by signals directed toward conjugation functions and their formation by insertion of Ty1 repetitive elements. II. deletions, duplications, and transpositions of the COR segment that encompasses the structural gene of yeast iso-1-cytochrome c. Cold Spring Harb. Symp. Quant. Biol. 1981;45(Pt 2):593–607. [PubMed] [Google Scholar]
- 58.Roeder GS, Farabaugh PJ, Chaleff DT, Fink GR. The origins of gene instability in yeast. Science. 1980;209:1375–1380. doi: 10.1126/science.6251544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.