Abstract
Bacillus cereus G9241, the causative agent of anthrax-like disease, harbors virulence plasmids encoding anthrax toxins as well as hyaluronic acid (HA) and B. cereus exopolysaccharide (BPS) capsules. B. cereus G9241 also harbors S-layer genes, including homologs of Bacillus anthracis surface array protein (Sap), extractable antigen 1 (EA1), and the S-layer-associated proteins (BSLs). In B. anthracis, S-layer proteins and BSLs attach via their S-layer homology domains (SLH) to the secondary cell wall polysaccharide (SCWP) in a manner requiring csaB, a predicted ketalpyruvate transferase. Here we used a genetic approach to analyze B. cereus G9241 S-layer assembly and function. Variants lacking the csaB gene synthesized SCWP but failed to retain Sap, EA1, and BSLs in the bacterial envelope. The B. cereus G9241 csaB mutant assembled capsular polysaccharides but displayed an increase in chain length relative to the wild-type strain. This phenotype is likely due to its inability to deposit BslO murein hydrolase at divisional septa. During growth under capsule-inducing conditions, B. cereus G9241 assembled BSLs (BslA and BslO) and the Sap S-layer protein, but not EA1, in the envelope. Finally, csaB-mediated assembly of S-layer proteins and BSLs in B. cereus G9241 contributes to the pathogenesis of anthrax-like disease in mice.
INTRODUCTION
Gram-positive bacteria synthesize a thick cell wall peptidoglycan envelope, which serves as an assembly scaffold for the surface display of polypeptides, capsular polymers, and wall teichoic acids (1). These surface molecules mediate the interactions between microbes and their environments, notably the tissues of infected hosts. Bacillus anthracis, the causative agent of anthrax (2), decorates its peptidoglycan with poly-d-γ-glutamate (PDGA) capsule (3), whose synthesis is encoded by the capBCADE operon, located on the pXO2 virulence plasmid (4, 5). B. anthracis lacks wall teichoic acids; however, this organism synthesizes a secondary cell wall polysaccharide (SCWP) linked via murein linkage units, GlcNAc-ManNAc, to the C-6 hydroxyl of N-acetylmuramic acid (MurNAc) in the repeating MurNAc-GlcNAc disaccharide structure of peptidoglycan (6). The SCWP consists of a repeating trisaccharide [→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→6)-α-GlcNAc-(1→]n, where α-GlcNAc is substituted with α-Gal and β-Gal at O-3 and O-4, respectively, and the β-GlcNAc is substituted with α-Gal at O-3 (7).
Attached to B. anthracis SCWP is an S-layer, a two-dimensional paracrystalline lattice comprised of two S-layer proteins, Sap and EA1 (8). The S-layer also harbors 22 B. anthracis S-layer-associated proteins (BSLs) that contribute to the uptake of nutrients (BslK), the adhesion to host tissues (BslA), and the separation of cells within chains of vegetative bacilli (BslO) (9–12). S-layer proteins and BSLs contain S-layer homology domains (SLHs), which fold into a three-pronged spindle structure for association with the SCWP (13). SLH domain association and subsequent S-layer assembly absolutely require ketalpyruvate modification of the SCWP by the csaB gene product of B. anthracis (6, 14). The csaB mutants are viable but lack an S-layer and form long chains of incompletely separated vegetative cells (6, 14). Assembly of the S-layer and that of the PDGA capsule of B. anthracis are thought to occur as independent yet compatible events: PDGA capsule strands traverse the paracrystalline S-layer formed from Sap and EA1 proteins (15).
Bacillus anthracis belongs to the Bacillus cereus sensu lato group, whose other members are Bacillus cereus and Bacillus thuringiensis (16). B. thuringiensis is a pathogen of insects (17, 18). The species designation B. cereus includes (i) environmental isolates not associated with disease, (ii) strains causing noninvasive gastrointestinal disease in humans, (iii) strains associated with periodontal disease, (iv) pathogens that are opportunistic in immunocompromised patients receiving chemotherapy, and (v) virulent isolates that cause invasive human disease (19–23). B. cereus G9241 is a member of the latter group and has been isolated from anthrax-like respiratory disease (24). The strain is endowed with two virulence plasmids: pBCXO1, which harbors the anthrax toxin genes that are also found on the pXO1 virulence plasmid of B. anthracis (24), and pBC218, which harbors genes for the B. cereus exopolysaccharide (BPS) (25). B. cereus G9241 forms a large capsule from two polysaccharides, BPS and hyaluronic acid (HA) (25). The hasACB genes are responsible for the synthesis of HA and are also located on pBCXO1 (25). Each capsular structure (HA and BPS) contributes to the pathogenesis of B. cereus G9241 anthrax-like disease in mice (25). Variants lacking both hasACB and bpsX-H are highly sensitive to phagocytic killing by macrophages and are unable to cause anthrax disease in animal challenge experiments (25).
Similar to B. anthracis, B. cereus G9241 elaborates the SCWP (26, 27), and its chromosome harbors genes for S-layer assembly (csaB-sap-eag) and for BSLs (24) (Fig. 1). The presence of csaB-sap-eag and bsl genes in B. cereus G9241 is in agreement with the general hypothesis that the S-layer may be an important feature of B. cereus strains that are pathogenic to humans and animals (including B. anthracis) but are otherwise absent from environmental B. cereus isolates lacking the capacity to cause disease (28). Here we asked whether S-layer formation and HA/BPS capsule synthesis occur in B. cereus G9241 as independent, compatible events and whether assembly of the S-layer contributes to the pathogenesis of anthrax-like disease.
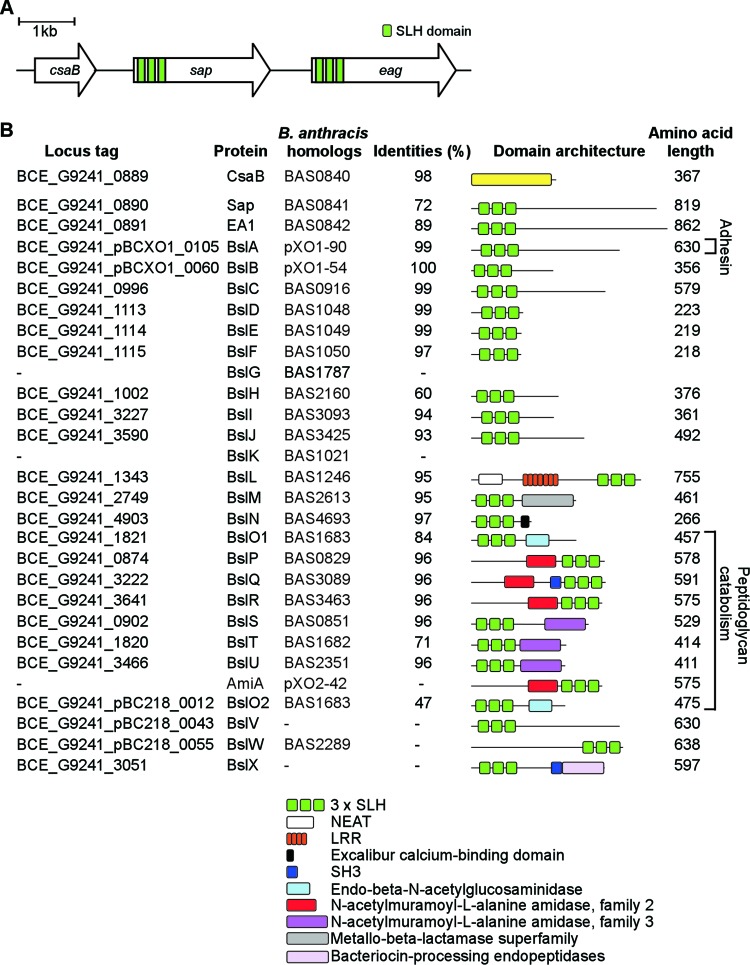
Fig 1.
S-layer and S-layer-associated proteins of Bacillus cereus G9241. (A) Illustration of the S-layer genes of B. cereus G9241, including csaB (proposed ketalpyruvate transferase gene), sap (encoding Sap [for surface array protein]), and eag (encoding extractable antigen 1). Three green bars in sap and eag coding sequences signify the three S-layer homology domains of the corresponding gene products. (B) Drawing to summarize the B. cereus genes for CsaB, S-layer (Sap and EA1) and S-layer-associated (BSLs) proteins predicted from genome sequence, their locus tag, B. anthracis homologs and their percent amino acid identities. Three B. anthracis BSLs (BslG, BslK and AmiA) are absent from B. cereus G9241. Further, B. cereus G9241 harbors two homologs of BslO (BslO and BslO2) and three unique BSLs (BslV, BslW, and BslX). The legend identifies relevant domain structures for B. cereus G9241 S-layer and S-layer-associated proteins.
MATERIALS AND METHODS
Bacterial growth and capsule production.
B. cereus G9241 and its variants were cultured in brain heart infusion (BHI) broth. Escherichia coli was grown in Luria-Bertani broth. When necessary, growth media were supplemented with spectinomycin (200 μg ml−1) or kanamycin (50 μg ml−1) to maintain plasmid selection. B. cereus strains were sporulated during growth in modified G medium (29). Spore suspensions were germinated by inoculation into BHI and grown at 30°C. For capsule production, spores of B. cereus G9241 or its variants were inoculated into 50% (vol/vol) heat-inactivated fetal bovine serum (FBS) containing BHI. Bacterial growth, genetic manipulation, and animal experiments involving B. cereus G9241 and its variants were carried out with approved protocols in biological safety level 3 containment laboratories under supervision of the institutional biosafety committee of the University of Chicago.
Construction of plasmids and Bacillus cereus G9241 strains.
B. cereus G9241 csaB mutants were constructed by allelic replacement of coding sequences with an omega element conferring spectinomycin resistance (Ω-Sp), using the temperature-sensitive plasmid pLM4 (30). Two fragments of approximately 1-kb DNA sequences flanking the csaB coding region (BCE_G9241_0889 and NZ_AAEK01000002.1) were amplified by PCR using two primers pairs, csaB-UPF (TTTCCCGGGGATACTGCAGGTTTCGATTCCA)/csaB-UPR (TTTGGTACCCTTAATCTCCTCCAACATTTCGC) and csaB-DNF (TTTGGATCCGAGGACATCCTCTTTTTTATTTTTTG)/csaB-UPR (TTTGAATTCGCCCATGAATCTTGAGCATC). PCR fragments were ligated into the pLM4 XmaI and EcoRI restriction sites. In this construct, a KpnI restriction site was inserted between the two fragments, where the blunted Ω-Sp cassette from pJRS312 plasmid (31) was inserted to generate pYT19. pYT19 was isolated from E. coli K1077 (dcm/dam) (32) prior to electroporation into B. cereus. Cultures of B. cereus G9241 harboring pYT19 were diluted 1:100, plated on BHI agar (50 μg ml−1 kanamycin) and incubated at 43°C overnight (restrictive temperature). Single colonies were inoculated into LB broth without antibiotics and cultures passaged every 12 h at 30°C to ensure loss of plasmid. DNA from kanamycin-sensitive colonies was isolated and analyzed by PCR for the presence or absence of mutant alleles. Nucleic acid sequences of wild-type and mutant alleles were verified by DNA sequencing. Plasmid pcsaB was derived from pLM4 and used for complementation. The csaB gene as well as its upstream flanking region including the native promoter was amplified using the primers csaBF (TTTCCCGGGATGAGTCCATAAATGAGTCCATAAATG) and csaBR (TTTGGTACCTTGATATATTTCCTCCTTAGGAATGTAA). Mutations were transduced into B. cereus ΔhasACB, ΔbpsX-H, and ΔhasACB ΔbpsAB mutants (25) to generate double or triple mutants (33). Briefly, bacteriophage CP-51 was used to infect the B. cereus G9241 csaB::Ω-Sp strain and generate a lysate (34). Filtered CP-51 phages from such lysates were used to transduce the csaB::Ω-Sp allele into B. cereus capsular mutants. Spectinomycin-resistant transductants were screened by PCR and mutants were confirmed by DNA sequencing.
Immunoblotting.
B. cereus overnight cultures were diluted 1:100 into fresh BHI and grown to an A600 of 2.5. Where indicated, spores were inoculated into 50% FBS to induce B. cereus capsule production. One-milliliter aliquots were removed from the culture and centrifuged at 13,000 ×g for 10 min to separate supernatant (culture medium) and sediment (vegetative forms). Proteins in the supernatant were precipitated by addition of trichloroacetic acid (TCA; final concentration, 10%). Vegetative bacilli were washed in PBS and proteins precipitated with 10% TCA. All protein precipitates were sedimented by centrifugation at 13,000 × g for 10 min, washed with ice-cold acetone, dried, and suspended in sample buffer. Samples were heated at 95°C for 10 min and subjected to electrophoresis on SDS-PAGE. Proteins were electrotransferred to polyvinylidene difluoride (PVDF) membrane and probed with specific polyclonal rabbit IgG antibodies (anti-Sap, anti-EA1, anti-BslA, anti-BslO, or anti-L6) as well as anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase, followed by chemiluminescence detection.
Scanning electron microscopy.
Bacilli were washed in H2O, fixed for 30 min in 2% glutaraldehyde–PBS at room temperature, and then postfixed for 30 min on freshly prepared poly-l-lysine coated glass coverslips. Samples were washed twice with PBS and serially dehydrated by consecutive incubations in 25% and 50% ethanol–PBS (vol/vol), 75% and 90% ethanol–H2O, and 100% ethanol (twice), followed by 50% ethanol–hexamethyldisilazane (HDMS) and finally 100% HDMS. After overnight evaporation of HDMS at room temperature, samples were incubated for 3 h at 37°C, mounted onto specimen mounts (Ted Pella, Inc., Redding, CA), and coated with 80% Pt–20% Pd to 8 nm using a Cressington 208HR sputter coater at 20 mA prior to examination in a Fei Nova NanoSEM 200 scanning electron microscope (FEI Co., Hillsboro, OR). The microscope was operated with an acceleration voltage of 5 kV, and samples were viewed at a distance of 5 mm.
Purification of Bacillus anthracis SCWP.
B. anthracis Sterne (34F2) was grown overnight at 37°C on BHI agar, and bacteria were scraped off the plates, suspended and washed in water. Bacilli were boiled for 30 min in 4% SDS, washed and suspended in water, and then broken in a bead beater instrument with 0.1 mm glass beads. Murein sacculi were sedimented by centrifugation and suspended in 100 mM Tris-HCl (pH 7.5). Samples were incubated for 4 h at 37°C with 10 μg/ml DNase and 10 μg/ml RNase supplemented with 20 mM MgSO4 and then incubated for 16 h at 37°C with 10 μM trypsin supplemented with 10 mM CaCl2. Enzymes were inactivated by boiling for 30 min in 1% SDS. The detergent was removed by repeated centrifugation and water-washing steps. Murein sacculi were washed with successive steps in water, 100 mM Tris-HCl (pH 8.0), water, 100 mM EDTA (pH 8.0), water, and acetone and finally twice with water. Murein sacculi were suspended in a small volume (5 to 7 ml) of water, and 25 ml of 48% hydrofluoric acid was added for overnight incubation on ice. The SCWP-containing supernatant was added to ice-cold ethanol at a ratio of 1:5. The SCWP precipitate was sedimented by centrifugation and washed extensively with ice-cold ethanol.
SCWP-specific antiserum.
Purified SCWP was dialyzed against water overnight and lyophilized. One mg of dried polysaccharide was dissolved in 90 μl of 0.15 M HEPES buffer (pH 7.4). Subsequently, 90 μl of 45 mg/ml of 1-cyano-4-dimethylaminopyridinium tetrafluoroborate (CDAP) in acetonitrile and 120 μl of aqueous 0.3 M triethylamine as well as 4 mg bovine serum albumin (BSA) dissolved in 350 μl 0.01 M PBS (pH 7.4) were added (26). The solution was stirred at 4°C for 18 h, and carbohydrate conjugation to BSA was quenched by adding 120 μl of 0.5 M ethanolamine in 0.75 M HEPES buffer (pH 7.4). Nonconjugated sugars were removed by filtration. The conjugate was emulsified by mixing 500 μg SCWP-BSA conjugate in 500 μl complete Freund's adjuvant (Difco). The samples were injected subcutaneously into female New Zealand White rabbits. Antibody production was stimulated at 21-day intervals with two booster injections of SCWP-BSA antigen emulsified in incomplete Freund's adjuvant.
Light and fluorescence microscopy.
Digital micrographs of bacterial samples fixed with PBS buffered 4% formalin were captured with a CCD camera on an Olympus IX81 microscope with 100×, 40×, 20× or 4× objectives. Length data were measured directly from micrographs using ImageJ software. To visualize capsules, india ink was added to samples prior to imaging. To probe for Sap, BslO, BslA, as well as SCWP, immunofluorescence microscopy experiments were performed. Briefly, fixed bacilli were sedimented by centrifugation at 10,000 × g, washed in PBS and blocked with 3% bovine serum albumin (wt/vol in PBS) or 3% horse serum (vol/vol in PBS). Specific rabbit antisera were diluted 1:1,000 into PBS and incubated with the cells for 1 h followed by three washes in PBS and labeling with Alexa-Fluor 594 conjugated goat anti-rabbit IgG (Fisher) prior to microscopy.
Animal challenge.
Protocols for animal experiments were reviewed, approved and supervised by the Institutional Animal Care and Use Committee at the University of Chicago (IACUC). All infections were carried out in animal biological safety level 3 containment laboratories at the Howard Taylor Ricketts Laboratory. Female C57BL/6 mice (Jackson Laboratory) were challenged by intraperitoneal injection of B. cereus spores as described previously (25). Moribund animals were euthanized and subjected to necropsy, and their spleens, livers, kidneys, and lungs were removed. Organs were immediately fixed by submersion in 10% neutral buffered formalin and embedded in paraffin. Samples were submitted to the University of Chicago Animal Pathology Core for serial 4-μm thin sectioning and stained with hematoxylin-eosin. Tissue samples were viewed by light microscopy. Organ samples isolated during necropsy were also homogenized in phosphate-buffered saline, serially diluted, and plated on tryptic soy agar (TSA) to enumerate bacterial loads (in CFU). Alternatively, samples were fixed with neutral buffered formalin and stained with india ink to visualize the capsules of bacilli.
Animals were managed by the University of Chicago Animal Resource Center. Animals judged to be moribund were euthanized using cervical dislocation followed by midline incision of the thorax. Acquired data were processed using GraphPad PRISM 5.0 software to generate graphs for statistical analyses. Data on bacterial load were analyzed with the unpaired two-tailed Student's t test. Comparisons of animal survival between cohorts challenged with different strains were examined with the log-rank test.
RESULTS
S-layer genes of Bacillus cereus G9241.
The BLAST search algorithm was used to identify the S-layer genes of B. cereus G9241, using nucleotide sequences of B. anthracis Sterne genes as queries (35, 36). The csaB-sap-eag genes of B. cereus G9241 are 98%, 72%, and 89% identical to the corresponding genes of B. anthracis (Fig. 1B) (9, 24). Queries of the B. cereus G9241 genome with the SLH Pfam domain (PF00395) identified 25 genes whose predicted products contain N-terminal signal peptides as well as SLH domains, i.e., the two S-layer proteins and 23 BSLs (Fig. 1). The B. cereus G9241 genome lacks the bslG, bslK, and amiA genes of B. anthracis yet harbors three unique bsl genes not found in the genomes of other B. anthracis isolates (bclV, bslW, and bslX) (Fig. 1).
Isolation and characterization of B. cereus G9241 csaB mutants.
The omega-spectinomycin cassette (Ω-Sp) and DNA sequences flanking csaB (BCE_G9241_0889, GI:47558208) were used to generate a csaB mutant of B. cereus G9241 harboring the csaB::Ω-Sp allele. When propagated on agar, the csaB mutant formed small colonies that lacked the ground-glass-like appearance of wild-type colonies (12) (Fig. 2A). Scanning electron microscopy experiments revealed B. cereus G9241 vegetative bacilli as short chains of incompletely separated cells, typically 2 to 8 vegetative bacilli in length (Fig. 2B). In contrast, the chain length of the csaB mutant was exaggerated; on some electron microscopy images chains appeared to be >50 vegetative cells in length. Transformation of the csaB mutant with pLM4 (empty plasmid vector) did not affect the chain length phenotype of vegetative forms. On the other hand, expression of wild-type csaB from a plasmid (pcsaB) restored the chain length to wild-type levels (Fig. 2B). Growth and chain length of vegetative bacilli were quantified following germination of wild-type and mutant spores in fresh media. The csaB mutant with or without the complementing plasmid (pcsaB) grew at the same rate as wild-type bacilli (Fig. 2C). Culture aliquots were removed at timed intervals and observed by light microscopy, and average chain lengths of bacilli were quantified. Two hours after inoculation of spores into fresh nutrient broth, the average chain length of the csaB mutant (12.69 ± 0.3391 μm; n = 109) was greater than that of wild-type bacilli (9.889 ± 0.2099 μm; n = 109) (Fig. 2D; wild-type versus csaB mutant, P < 0.0001). A similar result was observed at the 3-h interval (wild-type, 36.00 ± 1.693 μm, versus csaB mutant, 41.41 ± 1.880; P = 0.0336). At later intervals (4, 5, 6, and 7 h), the chain lengths of the wild-type bacilli gradually decreased, whereas those of the csaB mutant did not (Fig. 2D). The chain length phenotype of the csaB mutant is due to the csaB::Ω-Sp mutation, as plasmid-encoded wild-type csaB (pcsaB) reduced the chain length of the mutant to wild-type levels (Fig. 2D).
Fig 2.
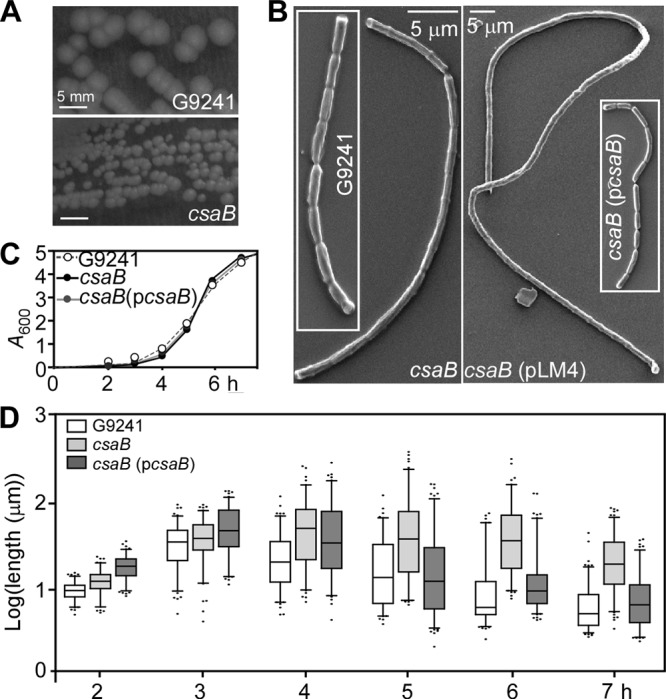
Increased chain length of vegetative forms derived from B. cereus G9241 csaB mutants. (A) B. cereus G9241 wild-type and csaB mutant colonies on BHI agar. The csaB mutant colonies are smaller, irregular in size and without a ground-glass-like appearance. (B) B. cereus G9241 wild-type and csaB mutant bacilli without or with control (pLM4) and complementing plasmids (pcsaB) were grown on BHI agar and analyzed by scanning electron microscopy (SEM). (C) The growth of B. cereus G9241 wild-type and csaB mutant without or with pcsaB in BHI was monitored by measuring absorbance at 600 nm (A600) over time. (D) Spores of B. cereus G9241 wild-type and its csaB mutant without plasmid or with pcsaB were inoculated into BHI, and the lengths of vegetative forms (n > 100) were determined by differential interference contrast microscopy (DIC) at variable time points. Data are displayed as a box-and-whisker plot, where each box bounds the 5th and 95th percentiles. Black bars in the boxes indicate the sample median, and black dots represent the first and last 5 percentiles. Data represent one of three independent experimental replicates.
The csaB gene is required for S-layer assembly in the envelope of B. cereus G9241.
We sought to characterize the assembly of S-layer (Sap and EA1) and S-layer-associated (BslO) proteins in the envelope of wild-type and csaB mutant B. cereus G9241. BslO is a peptidoglycan hydrolase, which localizes to the septal region of B. anthracis vegetative chains (12, 37). B. cereus G9241 cultures were centrifuged, and the extracellular medium was separated with the supernatant (S) from the bacterial sediment (pellet [P]). Proteins in both fractions were analyzed by immunoblotting. B. cereus G9241 retained Sap, EA1, and BslO in its envelope (Fig. 3A). In contrast, the csaB mutant released all of these proteins into the extracellular medium (Fig. 3A). As a control, ribosomal protein L6 was found in the bacterial sediment of all strains tested (Fig. 3A). The ability of the csaB mutant to retain Sap, EA1, and BslO in the bacterial envelope was restored when the mutant was transformed with plasmid pcsaB but not with the vector control (pLM4) (Fig. 3A).
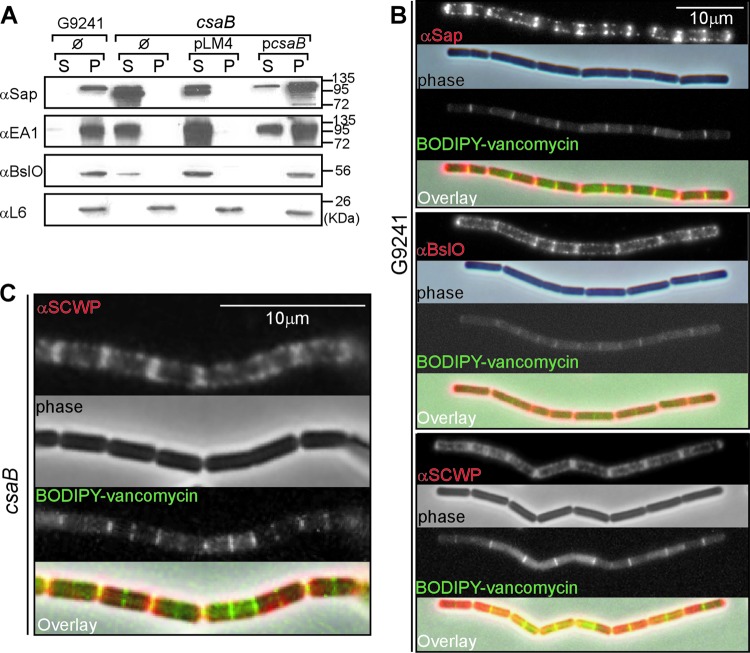
Fig 3.
The B. cereus G9241 csaB mutant cannot assemble S-layer proteins and S-layer-associated proteins into the bacterial envelope. (A) B. cereus G9241 wild-type and csaB mutant were grown to exponential phase in BHI broth. Bacilli harbored no plasmid (Ø), the pLM4 vector, or the complementing plasmid pcsaB. Cultures were centrifuged to sediment vegetative bacilli (pellet [P]) and separate them from the extracellular medium (supernatant [S]). Proteins in both fractions were precipitated with TCA and analyzed by immunoblotting with rabbit antisera raised against purified recombinant Sap (αSap), EA1 (αEA1), BslO (αBslO), and ribosomal protein L6 (αL6). The migratory positions of molecular mass markers on SDS-PAGE are indicated. (B) Immunofluorescence and phase-contrast microscopy images of wild-type B. cereus G9241 grown to exponential phase in BHI. Bacilli were stained with antisera specific for Sap, BslO, or SCWP and with secondary antibody conjugated to Alexa-Fluor 594 (red). To reveal the cell wall septa between adjacent bacilli, chains were stained with BODIPY-vancomycin (green), which binds the peptidoglycan precursor lipid II. (C) Immunofluorescence and phase-contrast microscopy images of csaB mutant B. cereus G9241 grown to exponential phase in BHI. Bacilli were stained with antisera specific for the SCWP (red) or with BODIPY-vancomycin (green). Data are representative of three independent experimental determinations.
Immunofluorescence microscopy of vegetative chains was used to detect Sap, BslO, and the secondary cell wall polysaccharide (SCWP) in the bacterial envelope. To visualize the septal region, images of bacilli were captured by phase-contrast and fluorescence microscopy after staining with BODIPY-vancomycin, which reveals newly synthesized peptidoglycan in the septa of dividing bacilli (30, 38). Sap and BslO were detected mostly at the septal regions of B. cereus G9241 vegetative chains but were also distributed throughout the envelope (Fig. 3B). A similar distribution was observed for the SCWP (Fig. 3B). As expected, immunofluorescence microscopy failed to detect Sap in the envelope of csaB mutant bacilli (see Fig. S1 in the supplemental material). In contrast, distribution and abundance of the SCWP in the bacterial envelope were not affected by the csaB mutation (Fig. 3C).
Capsule formation in wild-type and csaB mutant B. cereus G9241.
The production of HA and BPS capsule is induced when B. cereus G9241 is grown in medium containing blood products, for example BHI supplemented with 50% fetal bovine serum (BHI-FBS) (25). Capsule production in B. cereus G9241 was detected by india ink exclusion staining and light microscopy (Fig. 4A). Disruption of the csaB gene did not alter capsule formation (Fig. 4A, top right). Similarly, transformation of bacilli with pLM4 or pcsaB did not affect the production of capsular polysaccharides (Fig. 4A, bottom). To examine whether HA and BPS were expressed in wild-type and csaB mutant strains, we introduced the csaB::Ω-Sp allele into bacilli carrying deletions in the genes required for HA (ΔhasACB) or BPS (ΔbpsX-H) capsule formation. Spores of the mutant strains were inoculated into BHI-FBS, and replicating vegetative forms were analyzed by microscopy. As previously reported, the ΔbpsX-H and ΔhasACB strains formed capsular polysaccharides that were somewhat thinner than that of the wild-type, whereas the ΔhasACB ΔbpsAB mutant was unable to produce any capsule (Fig. 4B) (25). Introduction of the csaB::Ω-Sp allele into the ΔbpsX-H, ΔhasACB, or ΔhasACB ΔbpsAB mutant strains did not affect capsule production. These data suggest that the S-layer proteins Sap and EA1 as well as the S-layer-associated proteins are not required for HA and BPS capsule production in B. cereus G9241.
Fig 4.
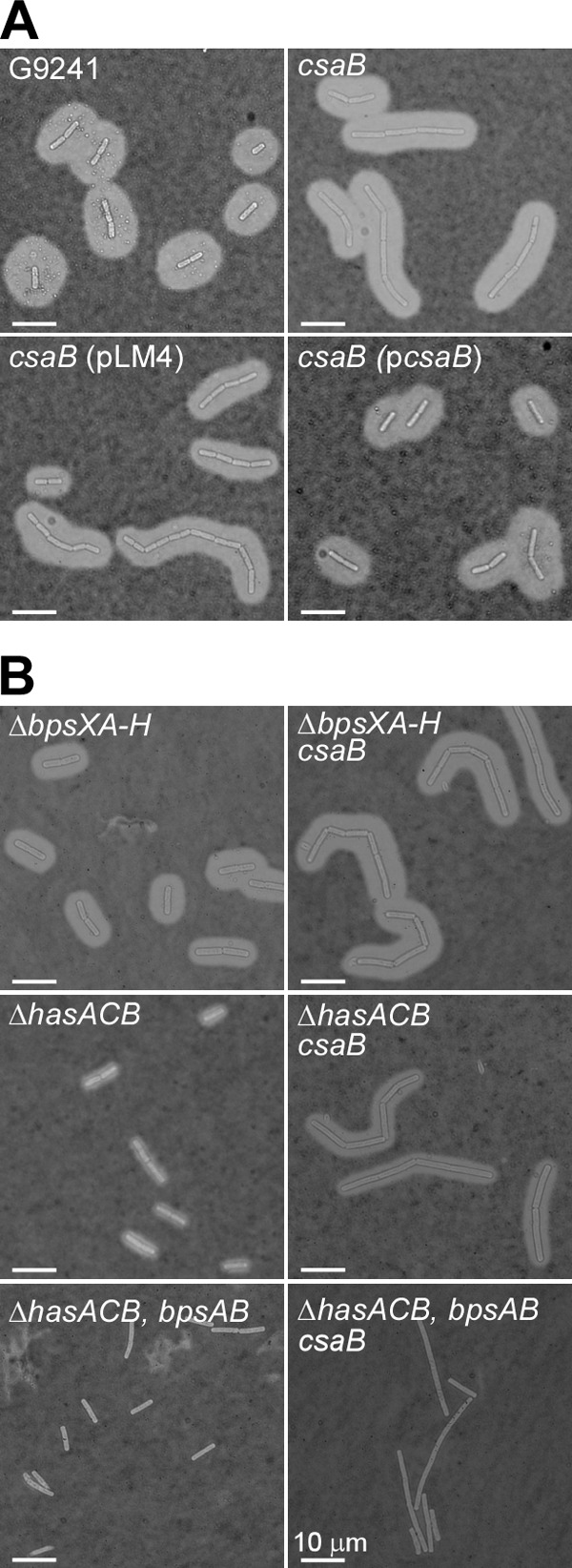
Capsule assembly in wild-type and csaB mutant B. cereus G9241. (A) B. cereus G9241 wild-type and csaB mutant were grown at 30°C for 5 h in BHI broth with 50% fetal bovine serum (BHI-FBS). Mutant bacilli harbored no plasmid, the pLM4 vector, or the complementing plasmid pcsaB. Capsule formation was detected by microscopy of india ink-stained samples. Data are representative of three independent experimental determinations. (B) B. cereus G9241 mutants lacking capsular polysaccharide operons responsible for BPS (ΔbpsX-H), hyaluronic acid (HA; ΔhasACB), or BPS and HA production (ΔhasACB ΔbpsAB) were transduced with the csaB::Ω-Sp allele. Capsule formation was detected by microscopy of india ink-stained samples. Data are representative of three independent experimental determinations.
S-layer production during B. cereus G9241 growth.
We sought to determine whether B. cereus G9241 cultures induced for capsule production also express S-layer and S-layer-associated proteins. During exponential growth (A600 = 1) either with (BHI-FBS) or without (BHI) fetal bovine serum, bacillus gene expression was analyzed by immunoblotting of the corresponding protein product (Fig. 5A). During exponential growth in BHI-FBS, B. cereus G9241 synthesized the S-layer protein Sap but not the EA1 polypeptide (Fig. 5A). Further, BslO and BslA, the pBCXO1 (pXO1)-encoded homolog of the S-layer-associated protein and adhesin of B. anthracis (9), were readily detected (Fig. 5A). When sampled during exponential growth in BHI without FBS, B. cereus G9241 produced neither capsular polysaccharides (data not shown) nor BslA (Fig. 5A). Similar to B. anthracis variants lacking PDGA capsule (39), exponential-phase B. cereus G9241 cultures with FBS induction expressed the S-layer protein Sap but not EA1 (Fig. 5A). During late exponential/stationary phase in BHI-FBS or BHI cultures (A600 = 4), B. cereus G9241 synthesized both Sap and EA1 but not BslA (Fig. 5B). The murein hydrolase BslO was detected in exponential- and stationary-phase cultures (Fig. 5). As a control, the peptidylprolyl isomerase PrsA was found in bacilli that had been grown in the presence or absence of serum (Fig. 5). During all growth stages under both capsule-inducing and noninducing conditions, the B. cereus G9241 csaB variant failed to retain S-layer and S-layer-associated proteins in the bacterial envelope (Fig. 5). This defect was complemented by pcsaB (Fig. 5). Deletion of capsule genes did not affect the expression or retention of S-layer and S-layer-associated proteins, as both wild-type and ΔhasACB ΔbpsAB mutant bacilli harbored similar amounts of S-layer and S-layer-associated proteins (Fig. 5).
Fig 5.
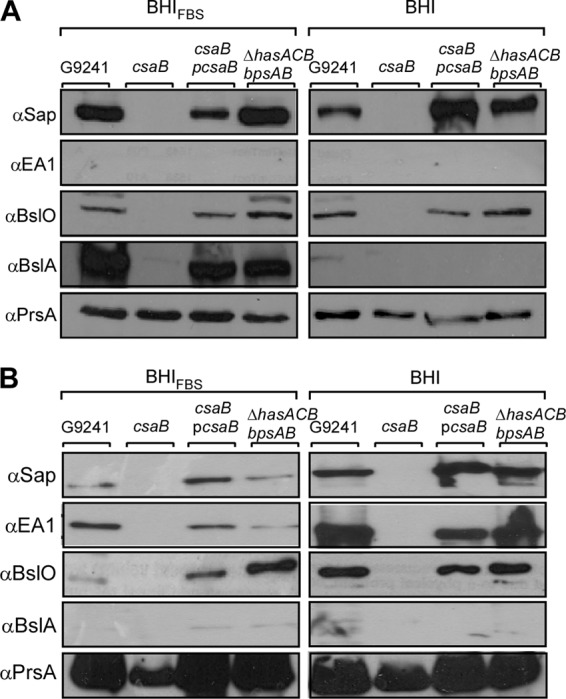
Expression of B. cereus G9241 S-layer proteins and S-layer-associated proteins under capsule-inducing conditions. B. cereus G9241 was grown under capsule-inducing (BHI-FBS) or noninducing (BHI) conditions to (A) exponential (A600 = 1) or (B) late exponential/stationary phase (A600 = 4). Cultures were centrifuged to sediment vegetative bacilli, and the extracellular medium was removed. Proteins were precipitated with TCA and analyzed by immunoblotting with rabbit antisera raised against purified recombinant Sap (αSap), EA1 (αEA1), BslA (αBslA), BslO (αBslO), and membrane protein PrsA (αPrsA). The abundance of S-layer proteins and S-layer-associated proteins in the envelopes of B. cereus G9241 wild-type, csaB, csaB(pcsaB), and ΔhasACB ΔbpsAB strains was compared. Data are representative of three independent experimental determinations.
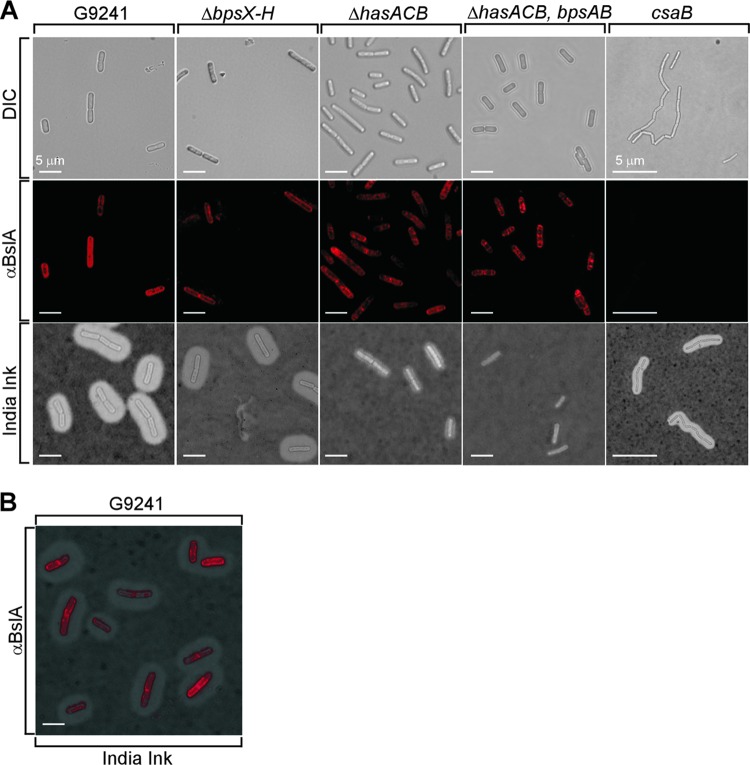
Surface display of BslA by encapsulated B. cereus G9241.
As BslA, but not Sap or EA1, is expressed in B. cereus G9241 during capsule production, we examined the distribution and surface display of BslA in encapsulated bacilli (Fig. 6). When grown in BHI-FBS and stained with india ink, B. cereus G9241 vegetative forms were found to be fully encapsulated. The BslA adhesin was detected by immunofluorescence microscopy and was found to be distributed throughout the bacterial envelope (Fig. 6). Deletion of the csaB gene abolished BslA display on the surface of B. cereus (Fig. 6A). In contrast, deletion of HA (ΔhasACB), BPS (ΔbpsX-H), or HA and BPS (ΔhasACB ΔbpsAB) capsule genes did not affect the display of BslA on the surface of B. cereus (Fig. 6A).
Fig 6.
Assembly of capsule and S-layer-associated proteins in the envelope of B. cereus G9241. (A) B. cereus G9241 wild-type, ΔbpsX-H, ΔhasACB, ΔhasACB ΔbpsAB, and csaB strains were grown at 30°C for 5 h in BHI-FBS. Vegetative bacilli were viewed by differential interference contrast microscopy (DIC). Immunofluorescence microscopy with BslA-specific antiserum revealed the assembly of BslA in the envelopes of wild-type but not csaB bacilli. Capsule formation was detected via microscopy of india ink-stained samples. (B) Overlay of DIC, BslA immunofluorescence, and india ink microscopy images of wild-type B. cereus G9241. Data are representative of three independent experimental determinations.
The S-layer of B. cereus G9241 contributes to the pathogenesis of anthrax-like disease.
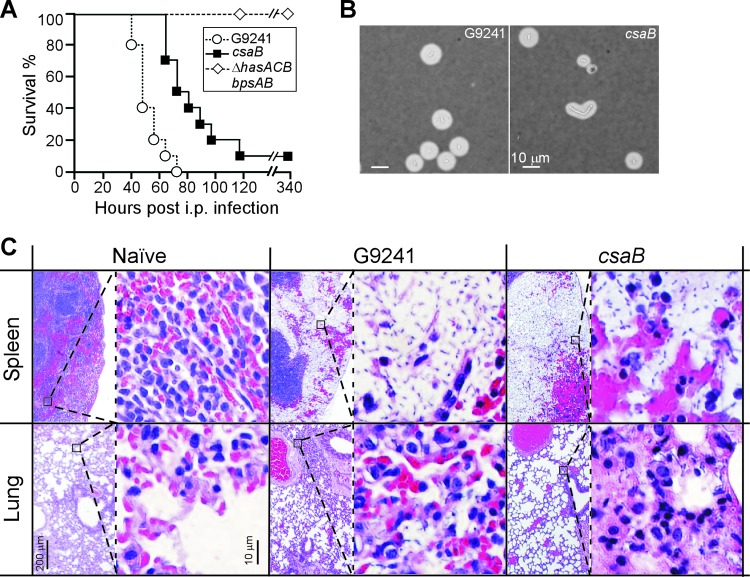
Cohorts of C57BL/6 mice (n = 10) were injected into the peritoneal cavity with 1 × 105 spores derived from either wild-type G9241 (30 mean lethal dose equivalents) as well as its ΔhasACB ΔbpsAB or csaB mutant vegetative forms (25). All animals challenged with wild-type spores succumbed to infection within 40 to 80 h (median time of survival, 48.5 h) (Fig. 7A). Animals challenged with the csaB mutant displayed a delayed time to death (median time of survival, 76.5 h), even though most animals succumbed to infection within 65 to 120 h. When these data were analyzed with the log-rank test, a significant difference in survival between wild-type and csaB mutant challenge groups was seen (P = 0.0002).
Fig 7.
Contribution of the csaB mediated S-layer assembly pathway to the pathogenesis of B. cereus G9241 anthrax-like disease in mice. (A) Survival of cohorts (n = 10) of C57BL/6 mice following intraperitoneal inoculation of (1 × 105) spores derived from B. cereus G9241 wild-type as well as ΔhasACB ΔbpsAB and csaB mutant strains. When analyzed with the log-rank test, the survival of animals infected with the csaB mutant is significantly increased compared to animals infected with the wild-type strain (P = 0.0002). The ΔhasACB ΔbpsAB mutant was avirulent in this model (wild-type versus ΔhasACB ΔbpsAB, P < 0.0001). Data are representative of two independent experimental determinations. (B) Spleen tissue homogenates from animals infected with wild-type B. cereus G9241 or the csaB mutant were stained with india ink and viewed by microscopy. Images are representative of vegetative forms identified in spleen tissues from five different animals analyzed for each group. (C) Naïve (uninfected) or moribund animals (infected with either wild-type B. cereus G9241 or its csaB mutant) were euthanized and necropsied. Lung and spleen tissues were fixed, thin sectioned, stained with hematoxylin-eosin, and analyzed for histopathology features via light microscopy at magnifications of ×40 (left) and ×400 (right). Images are representative of histopathology found in each of five animals per cohort. See the text for details.
All animals that were infected with spores of the nonencapsulated variant (ΔhasACB ΔbpsAB mutant) survived the challenge, similar to experiments detailed in an earlier report (25). Moribund animals were euthanized and necropsied, and spleen as well as lung tissue was stained with hematoxylin-eosin and examined for the histopathology features of anthrax-like disease. The spleens of animals that had been challenged with wild-type B. cereus G9241 spores displayed the characteristic displacement of red pulp tissue with massive numbers of vegetative bacilli (Fig. 7B). Large numbers of leukocytes, irregularly scattered throughout the bacterial population, were also associated with these lesions (Fig. 7B). India ink staining of spleen tissue revealed the encapsulated vegetative forms of B. cereus G9241 (Fig. 7C). As reported earlier (25), lung tissue from animals infected with B. cereus G9241 exhibited areas of focal inflammation and bacterial replication suggestive of a secondary pneumonia. Microcolonies of bacilli were observed in capillaries and occasionally in alveolar spaces but without significant immune cell infiltrates of the parenchyma (Fig. 7B). Mice that had been infected with the csaB mutant spores also displayed the displacement of the red pulp in the spleen with vegetative bacilli (Fig. 7B). As revealed by india ink staining, csaB mutant vegetative forms were fully encapsulated (Fig. 7C). In contrast to animals infected with wild-type spores, mice challenged with the csaB mutant developed remarkably few features of B. cereus G9241 lung disease (Fig. 7B).
DISCUSSION
B. anthracis, the causative agent of anthrax disease (2), elaborates PDGA capsule (3), thereby endowing vegetative forms with the ability to escape phagocytic clearance in tissues of infected hosts (5). B. anthracis also elaborates an S-layer via the assembly of two secreted products, Sap and EA1, which are arrayed into a two-dimensional paracrystalline lattice enveloping vegetative cells. In addition to Sap and EA1, the S-layer accommodates S-layer-associated proteins that have a wide spectrum of functions, including cell separation (BslO) (12), bacterial adhesion to host tissues (BslA) (9) and nutrient uptake (BslK) (11). Electron microscopy studies revealed the envelope structure of B. anthracis, in which linear PDGA strands traverse the bacterial peptidoglycan and S-layer (15). Retention of secreted S-layer proteins in the bacterial envelope requires the synthesis of the SCWP (14), which is immobilized via murein linkage units to the cell wall (6). Modification of the terminal β-ManNAc residue of the SCWP with ketalpyruvate in a reaction that is presumably catalyzed by the csaB gene product is a prerequisite for S-layer assembly (6, 40). The three-pronged spindle structure derived from the three SLH domains in S-layer and S-layer-associated proteins binds to pyruvylated SCWP, thereby enabling the assembly of the S-layer in the envelope of B. anthracis (13). Analysis of the contribution of the S-layer to anthrax disease, for example through the analysis of B. anthracis csaB mutants in animal challenge experiments, has not yet been carried out. Nevertheless, as the pXO1-encoded BslA adhesin of B. anthracis contributes to pathogenesis in guinea pigs (10), it seems plausible that bacterial S-layer assembly may also be an important feature of anthrax.
B. cereus G9241 is the causative agent of respiratory anthrax-like disease in humans, which is most frequently observed in welders (23). The genome sequence of B. cereus G9241 revealed its close homology to other B. cereus strains isolated from human cases with fatal pneumonia as well as the presence of three plasmids, designated pBCXO1, pBC218, and pBClin29 (24). When tested in mice with a peritoneal challenge model, B. cereus G9241 causes anthrax-like disease in a manner requiring genes for protective antigen (pagA, located on pBCXO1), BPS (bpsX-H on pBC218), and HA polysaccharides (hasACB on pBCXO1). Further, B. cereus G9241 causes respiratory anthrax-like disease in rabbits (41). Of note, B. cereus G9241 does not harbor the capBCDAE genes, which are present on B. anthracis pXO2 (24, 42). B. anthracis lacks the bpsX-H genes of B. cereus G9241 and harbors a frameshift mutation in the hasA gene on pXO1, which abolishes the synthesis of HA capsule (25). Thus, the structure of capsules elaborated by B. anthracis and B. cereus G9241 is fundamentally different, raising the possibility that assembly and function of S-layer and S-layer-associated proteins may also differ between these two closely related species.
We show here that B. cereus G9241 elaborates two S-layer proteins, Sap and EA1, which are conserved relative to the S-layer proteins of B. anthracis but not identical to them (Fig. 1B). The S-layer and S-layer-associated proteins (BslA and BslO) of B. cereus G9241 are retained in the bacterial envelope in a manner requiring csaB, a gene whose sequence is virtually identical to that of B. anthracis. Together with the observation that B. cereus G9241 elaborates SCWP with a structure similar to that of B. anthracis (27), we presume that the overall mechanism of envelope assembly from S-layer and S-layer-associated proteins is likely conserved between the two species. Furthermore, B. cereus G9241 capsule production is induced via CO2 (bicarbonate) and serum signals, as occurs for B. anthracis (43). Under such conditions, the B. cereus G9241 S-layer protein Sap is synthesized, while EA1 cannot be detected during exponential growth. Further, the S-layer-associated proteins BslA and BslO are both produced and assembled into the bacterial envelope. Thus, similar to B. anthracis, capsule (BPS as well as HA) and S-layer proteins are compatible structures in B. cereus G9241 that likely require a coordinated assembly process (15).
The finding that B. cereus G9241 csaB mutants cannot retain S-layer proteins and display a concomitant decrease in virulence suggests that S-layer assembly is important for the pathogenesis of anthrax-like disease. In addition to the observed delay in time to death, we noted altered histopathological features in lung but not in spleen tissues infected by the csaB mutant. These observations are in agreement with the conjecture that S-layers and S-layer-associated proteins have many different functions during infection. One of these functions is the control of the chain length of vegetative forms (12, 37, 44), which represents a mechanism for bacterial escape from opsonophagocytic killing. Simply put, if bacillus chain length exceeds the size of macrophages or granulocytes, bacteria cannot be engulfed (45). During infection of experimental animals, B. anthracis elaborates vegetative forms with and without PDGA capsule (5). It is conceivable that a similar phenomenon exists for B. cereus G9241, which could help explain the need for S-layer and S-layer-associated protein assembly controlling chain length during infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of our laboratory for discussion, Justin Kern for experimental guidance and Hwan Keun Kim for help in generating anti-SCWP serum.
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (NIAID), Infectious Disease Branch AI069227 to O.S. We acknowledge membership in and support from the Region V ‘Great Lakes’ Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (GLRCE, NIAID award 1-U54-AI-057153).
We declare no conflicts of interests.
Footnotes
Published ahead of print 30 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02005-12.
REFERENCES
- 1. Navarre WW, Schneewind O. 1999. Surface proteins of Gram-positive bacteria and the mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63: 174–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koch R. 1876. Die Ätiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschichte des Bacillus anthracis. Beitr. Biol. Pflanzen 2: 277–310 [Google Scholar]
- 3. Bruckner V, Kovacs J, Denes G. 1953. Structure of poly-d-glutamic acid isolated from capsulated strains of B. anthracis. Nature 172: 508. [DOI] [PubMed] [Google Scholar]
- 4. Candela T, Fouet A. 2006. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 60: 1091–1098 [DOI] [PubMed] [Google Scholar]
- 5. Richter GS, Anderson VJ, Garufi G, Lu L, Joachimiak A, He C, Schneewind O, Missiakas D. 2009. Capsule anchoring in Bacillus anthracis occurs by a transpeptidation mechanism that is inhibited by capsidin. Mol. Microbiol. 71: 404–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kern J, Ryan C, Faull K, Schneewind O. 2010. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J. Mol. Biol. 401: 757–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choudhury B, Leoff C, Saile E, Wilkins P, Quinn CP, Kannenberg EL, Carlson RW. 2006. The structure of the major cell wall polysaccharide of Bacillus anthracis is species specific. J. Biol. Chem. 281: 27932–27941 [DOI] [PubMed] [Google Scholar]
- 8. Mesnage S, Tosi-Couture E, Mock M, Gounon P, Fouet A. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23: 1147–1155 [DOI] [PubMed] [Google Scholar]
- 9. Kern JW, Schneewind O. 2008. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol. Microbiol. 68: 504–515 [DOI] [PubMed] [Google Scholar]
- 10. Kern JW, Schneewind O. 2010. BslA, the S-layer adhesin of Bacillus anthracis, is a virulence factor for anthrax pathogenesis. Mol. Microbiol. 75: 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tarlovsky Y, Fabian M, Solomaha E, Honsa E, Olson JS, Maresso AW. 2010. A Bacillus anthracis S-layer homology protein that binds heme and mediates heme delivery to IsdC. J. Bacteriol. 192: 3503–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson VJ, Kern JW, McCool JW, Schneewind O, Missiakas DM. 2011. The SLH domain protein BslO is a determinant of Bacillus anthracis chain length. Mol. Microbiol. 81: 192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kern JW, Wilton R, Zhang R, Binkowski A, Joachimiak A, Schneewind O. 2011. Structure of the SLH domains from Bacillus anthracis surface array protein. J. Biol. Chem. 286: 26042–26049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19: 4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mesnage S, Tosi-Couture E, Gounon P, Mock M, Fouet A. 1998. The capsule and S-layer: two independent and yet compatible macromolecular structures in Bacillus anthracis. J. Bacteriol. 180: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen GB, Hensen BM, Eilenberg J, Mahillon J. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5: 631–640 [DOI] [PubMed] [Google Scholar]
- 17. Han CS, Xie G, Challacombe JF, Altherr MR, Bhotika SS, Brown N, Bruce D, Campbell CS, Campbell ML, Chen J, Chertkov O, Cleland C, Dimitrijevic M, Doggett NA, Fawcett JJ, Glavina T, Goodwin LA, Green LD, Hill KK, Hitchcock P, Jackson PJ, Keim P, Kewalramani AR, Longmire J, Lucas S, Malfatti S, McMurry K, Meincke LJ, Misra M, Moseman BL, Mundt M, Munk AC, Okinaka RT, Parson-Quintana B, Reilly LP, Richardson P, Robinson DL, Rubin E, Saunders E, Tapia R, Tesmer JG, Thayer N, Thompson LS, Tice H, Ticknor LO, Wills PL, Brettin TS, Gilna P. 2006. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 188: 3382–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kolstø AB, Lereclus D, Mock M. 2002. Genome structure and evolution of the Bacillus cereus group. Curr. Top. Microbiol. Immunol. 264: 95–108 [PubMed] [Google Scholar]
- 19. Rasko DA, Rosovitz MJ, Økstad OA, Fouts DE, Jiang L, Cer RZ, Kolstø AB, Gill SR, Ravel J. 2007. Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J. Bacteriol. 189: 52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D'Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423: 87–91 [DOI] [PubMed] [Google Scholar]
- 21. Klee SR, Brzuszkiewicz EB, Nattermann H, Brüggemann H, Dupke S, Wollherr A, Franz T, Pauli G, Appel B, Liebl W, Couacy-Hymann E, Boesch C, Meyer FD, Leendertz FH, Ellerbrok H, Gottschalk G, Grunow R, Liesegang H. 2010. The genome of a Bacillus isolate causing anthrax in chimpanzees combines chromosomal properties of B. cereus with B. anthracis virulence plasmids. PLoS One 5: e10986 doi:10.1371/journal.pone.0010986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drobniewski FA. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6: 324–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoffmaster AR, Hill KK, Gee JE, Marston CK, De BK, Popovic T, Sue D, Wilkins PP, Avashia SB, Drumgoole R, Helma CH, Ticknor LO, Okinaka RT, Jackson PJ. 2006. Characterization of Bacillus cereus isolates associated with fatal pneumonias: strains are closely related to Bacillus anthracis and harbor B. anthracis virulence genes. J. Clin. Microbiol. 44: 3352–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, Maiden MC, Priest FG, Barker M, Jiang L, Cer RZ, Rilstone J, Peterson SN, Weyant RS, Galloway DR, Read TD, Popovic T, Fraser CM. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. U. S. A. 101: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oh SY, Budzik JM, Garufi G, Schneewind O. 2011. Two capsular polysaccharides enable Bacillus cereus G9241 to cause anthrax-like disease. Mol. Microbiol. 79: 455–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leoff C, Saile E, Rauvolfova J, Quinn C, Hoffmaster AR, Zhong W, Mehta AS, Boons GJ, Carlson RW, Kannenberg EL. 2009. Secondary cell wall polysaccharides of Bacillus anthracis are antigens that contain specific epitopes which cross-react with three pathogenic Bacillus cereus strains that caused severe disease, and other epitopes common to all the Bacillus cereus strains tested. Glycobiology 19: 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Forsberg LS, Choudhury B, Leoff C, Marston CK, Hoffmaster AR, Saile E, Quinn CP, Kannenberg EL, Carlson RW. 2011. Secondary cell wall polysaccharides from Bacillus cereus strains G9241, 03BB87 and 03BB102 causing fatal pneumonia share similar glycosyl structures with the polysaccharides from Bacillus anthracis. Glycobiology 21: 934–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mignot T, Denis B, Couture-Tosi E, Kosto AB, Mock M, Fouet A. 2001. Distribution of S-layers on the surface of Bacillus cereus strains: phylogenetic origin and ecological pressure. Environ. Microbiol. 3: 493–501 [DOI] [PubMed] [Google Scholar]
- 29. Kim HU, Goepfert JM. 1974. A sporulation medium for Bacillus anthracis. J. Appl. Bacteriol. 37: 265–267 [DOI] [PubMed] [Google Scholar]
- 30. Marraffini LA, Schneewind O. 2006. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol. Microbiol. 62: 1402–1417 [DOI] [PubMed] [Google Scholar]
- 31. Saile E, Koehler TM. 2002. Control of anthrax gene expression by the transition state regulator abrB. J. Bacteriol. 184: 370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fulford W, Model P. 1984. Specificity of translational regulation by two DNA-binding proteins. J. Mol. Biol. 173: 211–226 [DOI] [PubMed] [Google Scholar]
- 33. Skaar EP, Gaspar AH, Schneewind O. 2006. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J. Bacteriol. 188: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sastalla I, Rosovitz MJ, Leppla SH. 2010. Accidental selection and intentional restoration of sporulation-deficient Bacillus anthracis mutants. Appl. Environ. Microbiol. 76: 6318–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- 37. Kern VJ, Kern JW, Theriot JA, Schneewind O, Missiakas DM. 2012. Surface (S)-layer proteins Sap and EA1 govern the binding of the S-layer associated protein BslO at the cell septa of Bacillus anthracis. J. Bacteriol. 194: 3833–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marraffini LA, Schneewind O. 2007. Sortase C-mediated anchoring of BasI to the cell wall envelope of Bacillus anthracis. J. Bacteriol. 189: 6425–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Etienne-Toumelin I, Sirard JC, Duflot E, Mock M, Fouet A. 1995. Characterization of the Bacillus anthracis S-layer: cloning and sequencing of the structural gene. J. Bacteriol. 177: 614–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Forsberg LS, Abshire TG, Friedlander A, Quinn CP, Kannenberg EL, Carlson RW. 2012. Localization and structural analysis of a conserved pyruvylated epitope in Bacillus anthracis secondary cell wall polysaccharides and characterization of the galactose deficient wall polysaccharide from avirulent B. anthracis CDC 684. Glycobiology 22: 1103–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson MK, Vergis JM, Alem F, Palmer JR, Keane-Myers AM, Brahmbhatt TN, Ventura CL, O'Brien AD. 2011. Bacillus cereus G9241 makes anthrax toxin and capsule like highly virulent B. anthracis Ames but behaves like attenuated toxigenic non-encapsulated B. anthracis Sterne in rabbits and mice. Infect. Immun. 79: 3012–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okinaka R, Cloud K, Hampton O, Hoffmaster A, Hill K, Keim P, Koehler T, Lamke G, Kumano S, Manter D, Martinez Y, Ricke D, Svensson R, Jackson P. 1999. Sequence, assembly and analysis of pX01 and pX02. J. Appl. Microbiol. 87: 261–262 [DOI] [PubMed] [Google Scholar]
- 43. Mock M, Fouet A. 2001. Anthrax. Annu. Rev. Microbiol. 55: 647–671 [DOI] [PubMed] [Google Scholar]
- 44. Nguyen-Mau S-M, Oh SY, Kern V, Missiakas D, Schneewind O. 2012. Secretion genes as determinants of Bacillus anthracis chain length. J. Bacteriol. 194: 3841–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruthel G, Ribot WJ, Bavari S, Hoover T. 2004. Time-lapse confocal imaging of development of Bacillus anthracis in macrophages. J. Infect. Dis. 189: 1313–1316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.