Abstract
Herpes simplex virus 1 (HSV-1) immediate-early protein ICP0 is required for efficient lytic infection and productive reactivation from latency and induces derepression of quiescent viral genomes. Despite being unrelated at the sequence level, ICP0 and human cytomegalovirus proteins IE1 and pp71 share some functional similarities in their abilities to counteract antiviral restriction mediated by components of cellular nuclear structures known as ND10. To investigate the extent to which IE1 and pp71 might substitute for ICP0, cell lines were developed that express either IE1 or pp71, or both together, in an inducible manner. We found that pp71 dissociated the hDaxx-ATRX complex and inhibited accumulation of these proteins at sites juxtaposed to HSV-1 genomes but had no effect on the promyelocytic leukemia protein (PML) or Sp100. IE1 caused loss of the small ubiquitin-like modifier (SUMO)-conjugated forms of PML and Sp100 and inhibited the recruitment of these proteins to HSV-1 genome foci but had little effect on hDaxx or ATRX in these assays. Both IE1 and pp71 stimulated ICP0-null mutant plaque formation, but neither to the extent achieved by ICP0. The combination of IE1 and pp71, however, inhibited recruitment of all ND10 proteins to viral genome foci, stimulated ICP0-null mutant HSV-1 plaque formation to near wild-type levels, and efficiently induced derepression of quiescent HSV-1 genomes. These results suggest that ND10-related intrinsic resistance results from the additive effects of several ND10 components and that the effects of IE1 and pp71 on subsets of these components combine to mirror the overall activities of ICP0.
INTRODUCTION
The concept of intrinsic immunity, or intrinsic antiviral defense, has emerged in recent years as a means of cellular restriction of viral infections. First defined in retroviral systems (1), intrinsic resistance is mediated by constitutively expressed cellular proteins that in many cases are also involved in regulating cellular pathways. In contrast to acquired and innate immunity, intrinsic defense functions at the single-cell level and includes a wide diversity of mechanisms by which different viruses can be targeted at various stages of their replication cycles. Other defining characteristics of intrinsic resistance are (i) that it is often countered by viral regulatory proteins that target the relevant cellular repressors, (ii) that even in the absence of the viral countermeasures the restriction can be overcome by a high input of viral genomes, and (iii) that the efficiency of restriction varies between different cell types. Therefore, the defective phenotype of a mutant virus that does not express a relevant regulatory protein is often multiplicity and cell type dependent.
In the case of herpesviruses, and in particular herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV), there is accumulating evidence that one aspect of intrinsic resistance is mediated by components of cellular nuclear substructures known as promyelocytic leukemia (PML) nuclear bodies (also known as ND10). The consequence of intrinsic resistance activity to HSV-1 and HCMV is the repression of viral gene expression, such that a cell infected at low multiplicity has a low probability of entering the lytic cycle. Work from several laboratories has established that HSV-1 immediate-early (IE) protein ICP0 relieves intrinsic resistance, and in the case of HCMV, the tegument protein pp71 and the IE protein IE1 (ie72) have analogous roles. Indeed, all three of these proteins target ND10 in some manner. For example, ICP0 is a RING finger E3 ubiquitin ligase (2) that induces the degradation of PML itself, particularly those forms of the protein that are modified by members of the small ubiquitin-like modifier (SUMO) family of proteins (2–5). ICP0 also abrogates SUMO modification of Sp100 (5, 6), another major ND10 component, and causes the dispersal of the remaining forms of Sp100 and also of hDaxx and ATRX, two other important components of ND10 (7). HCMV tegument protein pp71 interacts with hDaxx (8), induces its degradation (9), and disperses ATRX from ND10 prior to any discernible effect on hDaxx (10). IE1 disrupts ND10 (11–13) and causes the loss of SUMO conjugates of PML and Sp100 without degrading the unmodified proteins (5, 14–17).
The biological significance of the effects of HSV-1 and HCMV regulatory proteins on ND10 components has been substantiated by the discoveries that mutant viruses that lack ICP0, pp71, or IE1 replicate more efficiently in cells in which selected ND10 components have been depleted using RNA interference (9, 10, 15, 17–25). Therefore, there are considerable parallels between the effects of ICP0, pp71, and IE1 on ND10 components and the underlying consequences of these phenomena. However, there are no obvious sequence similarities between these three proteins, and their biochemical mechanisms of action are likely to differ. This study was stimulated by the hypothesis that the actions of ICP0, pp71, and IE1 on ND10, albeit by varied mechanisms, are functionally analogous. If so, it follows that IE1 and pp71 should augment the replication efficiency of ICP0-null mutant HSV-1. There is evidence that HCMV coinfection and more specifically pp71 can at least partially complement ICP0-null mutant HSV-1 (26, 27). Potential complementation of ICP0-null mutant HSV-1 by IE1, however, has not previously been addressed. Here, we describe an inducible cell line expression system to compare a selection of functions of ICP0, pp71, and IE1. Because the question to be addressed concerns complementation of ICP0-null mutant HSV-1 rather than HCMV, we chose to adapt a well-characterized system based on HepaRG cells (7). We found that IE1 stimulates ICP0-null mutant HSV-1 infection by nearly two orders of magnitude, while pp71 does so to a lesser extent. Simultaneous expression of IE1 and pp71 almost completely complemented the plaque-forming efficiency of ICP0-null mutant HSV-1 and also stimulated derepression of quiescent HSV-1 gene expression. We propose that ICP0 combats the repressive effects of all ND10 components tested here, while IE1 and pp71 target subsets of these proteins such that, when expressed in combination, they recapitulate the core ICP0 activities in stimulating lytic infection and reactivation from quiescence.
MATERIALS AND METHODS
Viruses and cells.
HSV-1 strain 17+ was the wild-type (wt) strain used, from which the ICP0-null mutant dl1403 was derived (28). Viruses in1863 and dl1403/CMVlacZ are wt and ICP0-null mutant derivatives of the above-described virus that contain the lacZ gene under the control of the HCMV promoter/enhancer inserted into the tk gene (kindly provided by Chris Preston). HSV-1 mutant virus in1374 contains the tsK temperature-sensitive lesion in ICP4, a deletion of the ICP0 gene, and a mutation within VP16 that inactivates its ability to stimulate IE gene expression (29). All viruses were grown in BHK cells and titrated in U2OS cells, in which ICP0 is not required for efficient HSV-1 replication (30). Virus in1374 was propagated at the permissive temperature of 31°C in the presence of 2.5 mM HMBA (29). Human diploid fibroblasts (HFs), U2OS, and HEK-293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS). BHK cells were grown in Glasgow modified Eagle's medium supplemented with 10% newborn calf serum and 10% tryptose phosphate broth. HepaRG hepatocyte cells (31) were grown in William's medium E supplemented with 10% fetal bovine serum Gold (PAA Laboratories Ltd.), 2 mM glutamine, 5 μg/ml insulin, and 500 nM hydrocortisone. All cell growth media were supplemented with 100 units/ml penicillin and 0.1 mg/ml streptomycin. Lentivirus-transduced cells were maintained with continuous antibiotic selection, as appropriate.
Plasmids.
Lentivirus vector plasmids expressing the tetracycline repressor linked to a nuclear localization signal and enhanced green fluorescent protein (EGFP; pLKOneo.EGFPnlsTetR) and ICP0 from a tetracycline-inducible promoter (pLKO.DCMV.TetO.cICP0) have been described previously (7). The IE1 cDNA was isolated by PCR from a plasmid gifted by John Sinclair using primers that generated a SalI site immediately 5′ of the initiation codon and an EcoRI site just 3′ of the stop codon and then inserted in place of the ICP0 cDNA in the above-described plasmid to generate pLDT.IE1. Variants of these plasmid-expressing mutant forms of IE1 were constructed by PCR splicing using mutagenic oligonucleotides. The DNA sequence of the complete IE1 coding region was confirmed in each case. Plasmid pLDT.myc.pp71 (kindly provided by Steven McFarlane and Chris Preston) was isolated using a similar strategy, except that in this case the 5′ flanking region includes a NotI site and encodes a Myc tag. Plasmid pLBDT.myc.pp71 is a derivative of the above-described plasmid that expresses blasticidin rather than puromycin resistance. Plasmid pLDT.myc.pp71.DID2/3 includes the DID2/3 deletion mutation of pp71 (residues 324 to 331) that affects its interaction with hDaxx (8). This was also constructed by PCR splicing using mutagenic oligonucleotides, followed by DNA sequence analysis of the complete open reading frame.
Lentivirus transductions and induction of protein expression.
Lentivirus transduction, selection of transduced cells, and maintenance of cell lines were as described previously (20). Selection during routine culture used puromycin at 500 ng/ml, G418 at 0.5 mg/ml, and blasticidin (Invitrogen) at 1 μg/ml. The antibiotics were omitted from cells seeded for and during experimentation. For induction of protein expression, cells were treated with medium containing doxycycline (BD Biosciences) at 100 ng/ml for various times as indicated in the text and was maintained in the medium throughout the duration of an experiment.
Virus plaque and reactivation assays.
Cells were seeded for plaque assays into 24-well dishes at 1 × 105 cells per well and then infected the following day with appropriate sequential 3-fold dilutions of in1863 or dl1403/CMVlacZ. After virus adsorption, the cells were overlaid with medium containing 1% human serum, and then the cells were stained for β-galactosidase-positive plaques 24 h later (32). For reactivation (derepression) assays, cells in 24-well dishes were infected with in1374 at a multiplicity of infection (MOI) of 5 PFU per cell and at nonpermissive temperature (NPT; 38.5°C) and then incubated at NPT for 24 h. Derepression of the lacZ marker gene in the in1374 genome was assayed after by treatment with doxycycline (100 ng/ml) for 24 h to induce expression of ICP0, IE1, or pp71 from the integrated lentiviral vector. Cells were stained for β-galactosidase activity the following day, using the blue plaque detection method noted above.
Infections and Western blot analysis.
Cells were seeded into 24-well dishes at 1 × 105 cells per well. After the relevant experimental manipulations, the cells were washed twice with PBS before harvesting in SDS-PAGE loading buffer. Proteins were resolved on 7.5% SDS gels and then transferred to nitrocellulose membranes by Western blotting. The following antibodies were used: anti-IE1/2 mouse monoclonal antibody (MAb) E13 (Serotec), anti-myc tag MAb 9E10 (Source Bioscience), anti-actin MAb AC-40 (Sigma-Aldrich), anti-tubulin MAb T4026 (Sigma-Aldrich), anti-PML rabbit polyclonal (rAb) A301-167A (Bethyl Laboratories) or MAb 5E10 (33), anti-Sp100 rAb SpGH (a gift from Hans Will, Hamburg, Germany), anti-hDaxx MAb Daxx-01 (Santa Cruz Biotechnology), anti-hDaxx rAb D7810 (Sigma), anti-ATRX rAb H-300 (Santa Cruz Biotechnology), and anti-ATRX MAb 39F (a gift from Richard Gibbons, University of Oxford).
Immunofluorescence and confocal microscopy.
Cells on 13-mm glass coverslips were fixed and prepared for immunofluorescence as described previously (34). PML was detected with rAb AB1370 (Chemicon) or MAb 5E10 (33), Sp100 with rAb SpGH, hDaxx with rAb 07-471 (Upstate), and ATRX with rAb H-300 or MAb 39F. The secondary antibodies used were Alexa 555-conjugated goat anti-mouse IgG and Alexa 633-conjugated goat anti-rabbit IgG (Invitrogen). The samples were examined using a Zeiss LSM 510 confocal microscope, with 488-nm, 543-nm, and 633-nm laser lines, scanning each channel separately under image capture conditions that eliminated channel overlap. The images were exported as TIF files, minimally adjusted using Photoshop, and then assembled into figures using Illustrator.
Immunoprecipitation.
Cells were seeded into 90-mm plates at 4.5 × 106 cells per plate and then treated as relevant with doxycycline for 24 h to induce protein expression. The cells were washed twice with phosphate-buffered saline on ice, and then 1.5 ml of lysis buffer (Pierce product number 87787; supplemented with complete protease inhibitor cocktail [Roche Diagnostics]) was added directly onto the cells on the plate. After incubation on ice for 30 min, the plate was scraped with a syringe plunger and the harvested lysates were centrifuged at 13,000 rpm for 30 min at 4°C. The supernatant was precleared by incubation with protein G agarose beads (Millipore; 16-201) for 30 min at 4°C, mixing end over end, and then the beads were removed by centrifugation at 13,000 rpm for 10 min. Aliquots of the clarified supernatant (0.5 ml) were incubated at 4°C overnight, with end-over-end mixing, with 1 μg antibodies as follows: nonspecific preimmune rabbit IgG, anti-ATRX H-300, or anti-hDaxx 07-471 antibodies. Mixtures were then incubated with 30 μl fresh protein G agarose beads for 60 min at 4°C with continuous mixing. The beads were washed four times with a buffer containing 10 mM Tris [pH 7.6], 1.5 mM MgCl2, 300 mM NaCl, 5% glycerol, 0.2 mM EDTA, 0.1% NP-40, and protease inhibitors, and then the beads were pelleted and resuspended in 30 μl gel loading mix. Proteins were separated using 6.75% SDS-polyacrylamide Tris-glycine gels. After electroblotting overnight (50 mA, at 4°C), ATRX was detected using MAb 39F, hDaxx by MAb Daxx-01, and Myc-tagged pp71 with MAb 9E10.
RESULTS
Expression of HCMV proteins IE1 and pp71 in an inducible cell line system.
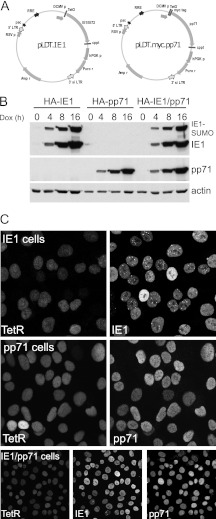
We constructed lentiviral vectors for inducible expression of pp71 and IE1 as previously described for ICP0 (7). HepaRG cells were transduced with a lentivirus vector expressing G418 resistance and the tetracycline repressor linked to EGFP and a nuclear localization signal, and then these cells were transduced with a second lentivirus expressing puromycin resistance and either IE1 or Myc-tagged pp71 from a tetracycline-inducible promoter (Fig. 1A). This approach for inducible IE1 expression proved successful previously (35). A derivative of the pp71 lentiviral vector was constructed expressing blasticidin resistance, allowing cells that express IE1 to be transduced a third time to enable simultaneous inducible expression of both pp71 and IE1. Neither protein was expressed at levels detectable by Western blotting in the absence of induction, whereas after induction both were expressed very efficiently within a few hours (Fig. 1B). Two IE1 bands were detected, with the more slowly migrating band being the SUMO-modified form identified in previous studies (36–38) (see also below). Immunofluorescence staining indicated that, after induction, a very high proportion of cells expressed either IE1 or pp71, or in the case of the double-expressing cells, both simultaneously (Fig. 1C). Prior to induction, a small percentage of IE1 cells (about 5%) expressed low levels of IE1, while pp71 expression was undetectable by this method (data not shown).
Fig 1.
Expression of HCMV IE1 and pp71 proteins in an inducible cell line system. (A) Maps of the lentivirus expression vectors for inducible expression of IE1 and pp71, showing the core features. A derivative of the pp71 vector expressing blasticidin instead of puromycin was also constructed. (B) Analysis of IE1 and pp71 expression by HepaRG cells transduced to express the tetracycline repressor linked to a nuclear localization signal and EGFP, then with the IE1 (HA-IE1 cells) and Myc-tagged pp71 (HA-pp71 cells) expression vectors, or both (HA-IE1/pp71 cells, using the puromycin IE1 vector and the blasticidin pp71 vector). A time course of expression after the indicated times of induction is shown. (C) Analysis of IE1 and pp71 expression by immunofluorescence 24 h after induction in the IE1, pp71, and double-expressing cells. TetR indicates the nuclear-located EGFPnlsTetR fusion protein. Each pair or set of three images shows the same field of cells.
Effects of IE1 and pp71 on ND10 components.
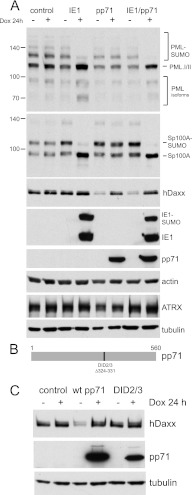
We next analyzed the effects of IE1 and pp71 on a number of ND10 components by Western blotting. Although these questions have been addressed previously in several publications, it was necessary to authenticate the inducible cell system used here. Our results confirm and extend this previous work. As expected (5, 14–17), IE1 caused the loss of the SUMO-conjugated forms of both PML and Sp100 (Fig. 2A), whereas pp71 had no effect on these proteins. In both IE1 and IE1/pp71 cells, the loss of the SUMO-conjugated forms of PML isoforms I and II, and of those of Sp100-A, led to concomitant increases in the corresponding unmodified forms of these proteins (Fig. 2A, upper two panels). There was also an increase in the intensity of the faster-mobility isoforms of PML in the presence of IE1, which was sometimes clearer than in the example presented (see also Fig. 7C). This suggests that these less-abundant PML isoforms are also subjected to SUMO modification, as would be expected, but low abundance of these modified forms renders them difficult to detect.
Fig 2.
Western blot analysis of the effects of IE1 and pp71 on major ND10 components PML, Sp100, hDaxx, and ATRX. (A) Control and IE1, pp71, and IE1/pp71 double cells were analyzed for PML, Sp100, hDaxx, and actin before and after induction of expression of the viral proteins for 24 h with 100 ng/ml doxycycline, as indicated. The lowest two panels of this section show ATRX and tubulin levels in samples from an independent but otherwise identical experiment. (B) A representation of pp71 showing the position of the DID2/3 deletion. (C) Expression of hDaxx is reduced in a third independent batch of pp71 cells, but the DID2/3 mutant has no effect.
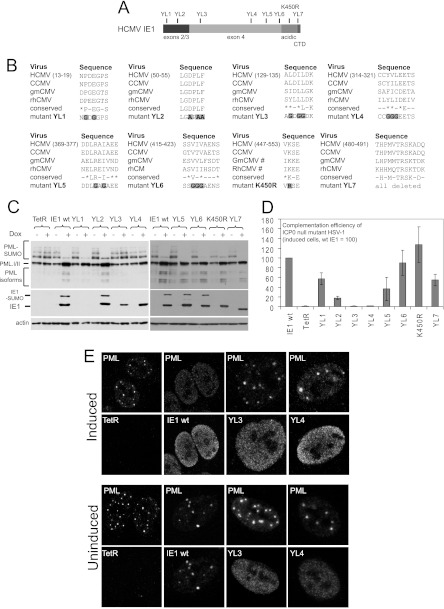
Fig 7.
Analysis of the properties of selected IE1 mutants in the inducible cell line system. (A) A map of the IE1 coding sequence, showing the locations of the exon2 and exon3 sequences common with IE2, the positions of the mutations analyzed, the acidic C-terminal region, and the chromatin tethering domain (CTD). (B) The sequences of the regions chosen for mutagenesis, showing the aligned sequences of human, chimpanzee, green monkey, and rhesus macaque CMVs (HCMV, CCMV, gmCMV, and rhCMV). The conserved residues are indicated, with an asterisk indicating similar residues rather than precise conservation in all sequences. The lowermost row shows the sequences of the mutants, with the mutated residues highlighted. (C) Western blot analysis of the expression of wt and mutant IE1 proteins, prior to and after induction, compared to the control (TetR), and their effects on PML. The SUMO-modified forms of IE1 and PML are indicated. Actin provides the loading control. Mutant YL1 is not detected by western blotting because the mutation affects the epitope for the E13 antibody 48). (D) Complementation of plaque forming ability of ICP0-null mutant HSV-1 in induced IE1 cells, with that in the mutant IE1 cells being expressed as a percentage of wt IE1. (E) Effects of wt and selected mutant forms of IE1 on the distribution of PML in induced and uninduced cells (upper and lower pairs of rows, respectively). Wt IE1 colocalizes with PML when low-level expression can be detected in a small subset of uninduced cells but disperses PML and becomes disperse itself in induced cells. Mutants YL3 and YL4 are diffuse when detectable in uninduced cells and do not disperse PML after induction. All other mutants of this set behaved as wt IE1 in this assay.
Surprisingly, hDaxx levels were significantly decreased in pp71 and IE1/pp71 cells prior to induction compared to those in control cells or those after pp71 induction (Fig. 2A). These are not chance results, as the single pp71 and double IE1/pp71 cell lines were generated independently using different pp71 lentiviral vectors. It is especially surprising since pp71 expression was undetectable by Western blotting or immunofluorescence prior to induction (Fig. 1). The most likely explanation is that inducible cell line systems are subjected to a certain level of leakiness in the uninduced state. Therefore, trace amounts of pp71 are likely to be expressed prior to induction, and these are sufficient to destabilize hDaxx. This hypothesis was investigated by construction of a vector expressing the DID2/3 mutant form of pp71, which does not interact with or induce the degradation of hDaxx (8). A further batch of cells transduced with the wt pp71 vector was isolated, alongside cells expressing the DID2/3 mutant. This third, independent batch of wt pp71 cells again exhibited reduced hDaxx levels in the uninduced state, while normal levels were reestablished after induction. In contrast, hDaxx levels were normal in both uninduced and induced DID2/3 cells (Fig. 2B and C). These results provide very strong support for the model that pp71 is expressed prior to induction at levels that are sufficient to destabilize hDaxx yet are undetectable by Western blotting. It is pertinent that hDaxx levels are reduced in very low-multiplicity HCMV infections in which pp71 is also present below the level of detection (9). Further support for the expression of biologically active levels of pp71 prior to induction is provided by the functional analyses presented below.
That hDaxx levels recover to normal levels after induction is initially surprising. The situation is reminiscent, however, of the observation that hDaxx levels are reduced by tegument-associated pp71 during the early stages of HCMV infection (when only small amounts of pp71 will be present), but they recover when pp71 is expressed in greater quantities as infection progresses (9). This recovery is substantial at 24 h after infection and complete after 72 h (9). The high levels of pp71 expressed in the induced cell system may accelerate the recovery process. The importance of the relative abundance of pp71 to the fate of hDaxx provides a plausible explanation for inconsistencies in detail in the literature concerning the ability of pp71 to degrade hDaxx (9, 39). The current result indicates that, in this expression system in this cell type, high levels of pp71 do not cause loss of hDaxx. However, as presented below, pp71 has a very important consequence for the hDaxx-ATRX complex.
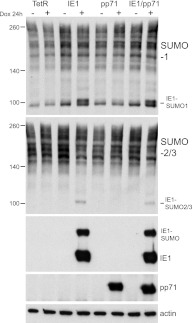
Neither IE1 nor pp71 had any effect on the expression level of ATRX (Fig. 2) or, in contrast to ICP0 (3, 4), on the abundance of SUMO-conjugated proteins in general (Fig. 3). IE1 is conjugated to both SUMO-1 and SUMO-2/3 in the induced cells, as novel SUMO-conjugated bands of the appropriate gel mobility were detected in the IE1- and IE1/pp71-induced cell samples (Fig. 3).
Fig 3.
Neither IE1 nor pp71 affect the abundance of SUMO-conjugated proteins in general. Extracts were prepared as described for Fig. 2, and then Western blots were analyzed with antibodies specific for SUMO-1 or SUMO-2/3. Novel bands that correspond to SUMO-modified forms of IE1 are clearly visible in the relevant induced extracts.
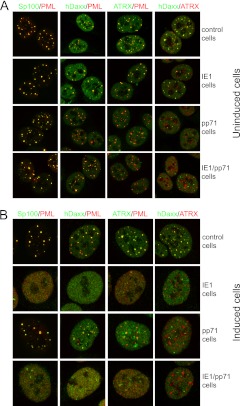
The distributions of major ND10 components in the majority of IE1 cells prior to induction were indistinguishable from the controls (Fig. 4A, upper two rows). A small proportion of uninduced cells expressed IE1 that accumulated in foci larger than normal ND10, and these contained PML but not hDaxx or ATRX (data not shown). Although pp71 was not itself detectable in uninduced pp71 or IE1/pp71 cells, there were some significant differences compared to the controls. The PML foci were less regular in size and Sp100 content (although these differences were only subtle), the overall hDaxx fluorescence signal was weaker (consistent with Fig. 2), and there were fewer hDaxx foci. ATRX was more dispersed and rarely colocalized with PML, and any ATRX foci were separate from those of hDaxx (Fig. 4A, lower two rows). It appears that the implied minor amount of pp71 that is expressed in these cells prior to induction is sufficient to change to the relative localizations of hDaxx and ATRX. The separated channels of the images in Fig. 4A are presented in Fig. S1 in the supplemental material.
Fig 4.
Immunofluorescence analysis of control, IE1, pp71, and IE1/pp71 cells. (A) Before induction of viral protein expression. (B) After induction with 100 ng/ml doxycycline for 24 h. In both cases, samples were stained as indicated for PML (MAb 5E10) and rabbit antibodies for Sp100, hDaxx, and ATRX, except for the hDaxx-ATRX images, which were stained with rabbit anti-hDaxx and anti-ATRX MAb 39F. Secondary antibodies were anti-mouse Alexa 555 and anti-rabbit Alexa 633 antibodies (shown in the green channel here). Analogous samples were stained for the viral proteins and the ND10 components, indicating the high proportion of cells positive for viral protein expression after induction (see Fig. 1) and the mostly nuclear diffuse distribution of both IE1 and pp71 after induction (images not shown).
After a 24-h induction with doxycycline, IE1 was mostly diffuse within the nucleus, with a few punctate foci (Fig. 1C, top row), and all ND10 components tested were dispersed (Fig. 4B, second row). The overall hDaxx fluorescence signal was restored in induced pp71 cells, with foci of increased intensity, some of which were PML associated. These hDaxx foci were, however, clearly separate from those of ATRX. The DID2/3 mutant of pp71 had no effect on either hDaxx or ATRX in either induced or uninduced cells (data not shown). Coexpression of IE1 and pp71 had the expected additive effects (Fig. 4B). The separated channels of the images in Fig. 4B are presented in Fig. S2 in the supplemental material. Previously, analyses found that pp71 initially colocalizes with hDaxx in ND10, prior to hDaxx and ATRX being dispersed (8, 10, 40). This was not generally observed in the pp71 cells used here at the 24-h induction time point, probably because the long-term trace expression of pp71 in the uninduced cultures disrupted the interactions that normally enable efficient ND10 localization of pp71 in naïve cells. The high level of induced expression and diffuse localization of the bulk of pp71 might also mask any weak association with hDaxx foci. However, at very early times after pp71 induction, faint foci of pp71 that colocalized with hDaxx could be detected (data not shown).
The hDaxx-ATRX complex is disrupted by pp71.
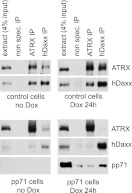
ATRX and hDaxx form a complex that is connected to chromatin modification and histone chaperone pathways (41–44). This complex is required for the repressive effects of hDaxx during HSV-1 infection (22). One of the earliest events that occurs during HCMV infection is the pp71-mediated dispersal of ATRX from ND10, and this occurs prior to any effect of pp71 on the localization of hDaxx (10). The hypothesis arises, therefore, that pp71 might disrupt the hDaxx-ATRX complex. The inducible cell line system provides an amenable means of testing this hypothesis, because pp71 can be expressed in the absence of other viral proteins in virtually all cells in the population. Extracts were prepared from uninduced and induced control, and pp71 cells were then subjected to immunoprecipitation with control, anti-ATRX, and anti-hDaxx antibodies. In the control cell extracts, hDaxx was readily detected in the ATRX immune precipitate and vice versa, whereas neither protein was present in the control precipitate (Fig. 5). In the induced pp71 cell experiment, both ATRX and hDaxx were efficiently precipitated by their cognate antibodies, but neither protein was coprecipitated with the other (Fig. 5). These results are consistent with the immunofluorescence analysis, and they indicate that a major function of pp71 is to disrupt the hDaxx-ATRX complex. We also detected pp71 in the hDaxx immunoprecipitate, confirming the previously reported interaction between the two proteins (8), whereas pp71 was not present above background levels in the ATRX precipitate (Fig. 5).
Fig 5.
Immunoprecipitation of hDaxx and ATRX from control and pp71 cell samples. Extracts from uninduced and induced (100 ng/ml doxycycline for 24 h) control and pp71 cells were used for immunoprecipitation with control nonspecific or anti-ATRX and anti-hDaxx rabbit antibodies, as indicated. Samples of the clarified extract and the immune precipitates were analyzed by Western blotting for ATRX, hDaxx, and Myc-tagged pp71 using MAbs 39F, Daxx-01, and 9E10, respectively.
The results were less clear cut using the uninduced pp71 cell extract. As expected from Fig. 2 and 4, hDaxx was much less abundant in the extract, and although the amounts of hDaxx in the ATRX precipitation and ATRX in the hDaxx precipitation were reduced compared to those in the controls, it is not possible to conclude whether this is because the complex is disrupted by trace amounts of pp71 (which would be consistent with Fig. 4) or simply because the amount of hDaxx is greatly reduced anyway. The unequivocal conclusions of these experiments are that trace amounts of pp71 are sufficient to reduce the steady-state levels of hDaxx in the cell, while larger amounts clearly dissociate the hDaxx-ATRX complex.
IE1 and pp71 stimulate plaque formation efficiency of ICP0-null mutant HSV-1.
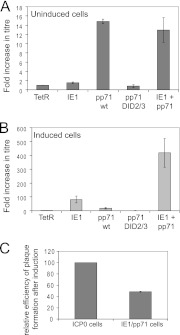
The inducible cell system allows reliable quantification of the extent to which particular ICP0 mutant proteins complement ICP0-null HSV-1 plaque formation efficiency (PFE) (7). Therefore, analogous assays were carried out in IE1, pp71, and IE1/pp71 cells before and after induction. The PFE of ICP0-null mutant HSV-1 was similar in control and uninduced IE1 cells (Fig. 6A). Surprisingly, but consistent with the observed presumed effects of trace amounts of pp71 on ATRX and hDaxx, the PFE of the mutant increased around 15-fold in uninduced pp71 cells and to a similar degree in uninduced IE1/pp71 cells. This did not occur in either uninduced or induced DID2/3 mutant pp71 cells, confirming the deleterious effect of this mutation (Fig. 6A). After induction, the PFE of ICP0-null mutant HSV-1 was increased by almost 100-fold in IE1 cells and by about 20-fold in pp71 cells (Fig. 6B). The degree of stimulation of ICP0-null mutant PFE in the induced pp71 cells is similar to that observed after preexpression of pp71 using a viral vector (26). Strikingly, complementation was of the order of 400-fold in induced IE1/pp71 cells (Fig. 6B), which was within about 2-fold of that achieved in ICP0-inducible cells analyzed in parallel (Fig. 6C). Therefore, IE1 and pp71, when expressed together, have cooperative effects that together can substitute for the great majority of ICP0's functions in a simple plaque assay. The titers of wt HSV-1 were not significantly different in any of the cell lines either before or after induction (data not shown).
Fig 6.
Complementation of ICP0-null mutant HSV-1 in plaque formation assays. The titer of a stock of ICP0-null mutant HSV-1 dl1403/CMVlacZ was determined in control, IE1, pp71, pp71.DID2/3, and IE1/pp71 cells before (A) and after (B) induction (100 ng/ml of doxycycline for 24 h prior to infection). Doxycycline was maintained in the medium throughout in the induced samples, and plaques were counted the day after infection by staining for expression of the β-galactosidase marker gene in dl1403/CMVlacZ. The results are expressed as fold-increase in absolute titer (PFU per ml) in each cell type over that in control cells. (C) Relative plaque-forming efficiency of ICP0-null mutant HSV-1 in induced ICP0 and IE1/pp71 cells, expressed as a percentage of that in induced ICP0 cells.
Analysis of mutant forms of IE1 in the inducible cell line system.
The data presented above indicate that the inducible cell line system provides a convenient means of monitoring IE1 function, and therefore we extended the study to analyze the properties of a number of mutant IE1 proteins. The mutations selected target the SUMO modification site (K450) (37, 38), the chromatin tethering domain (CTD) in the C-terminal end of the protein (45), and a number motifs throughout the protein that show a degree of conservation between closely related IE1 proteins (HCMV and chimpanzee CMV) and more divergent orthologs (green monkey and rhesus macaque CMVs) (Fig. 7A and B). The mutant IE1 proteins were expressed in inducible cells and then analyzed for expression and effects on SUMO-modified PML (Fig. 7C), complementation of ICP0-null mutant HSV-1 (Fig. 7D), and intracellular distribution and effects on ND10 (Fig. 7E).
As expected, mutant K450R was not SUMO modified, but in addition mutations YL3, YL4, and YL7 (of which the latter deletes the CTD) also greatly compromised the level of SUMO modification. Although the K450R mutation reduces viral replication when in the context of the viral genome (46), it was not found previously to affect IE1 activity in a variety of assays, including complementation of IE1 mutant HCMV when expressed in transduced cells (37, 38). These latter two studies are consistent with our observations, and they support the idea that there may be parallels between complementation of IE1-deficient HCMV and ICP0-null mutant HSV-1 when IE1 is expressed in this type of system. Similarly, the CTD deletion mutant (YL7) retained considerable complementation activity and the ability to induce loss of the SUMO-modified forms of PML (Fig. 7C and D), while a similar deletion mutation did not decrease HCMV replication in cultured cells (47). Mutations YL1 and YL2 affect highly conserved motifs in the region of IE1 that is in common with IE2. Detection of the YL1 mutant protein (P14G, E16G) was very inefficient, probably because the epitope of the E13 monoclonal antibody is within residues 1 to 24 (48), but it retained close to wt activity in all assays. YL2 induced loss of SUMO-conjugated PML but was about 4-fold less active than wt IE1 in complementation assays (Fig. 7C and D). This indicates that the YL2 mutation affects a function that is distinct from the effects of IE1 on ND10.
The most interesting mutants that we identified are YL3 and YL4. These mutants were defective in SUMO modification, induction of the loss of SUMO-conjugated PML, and colocalization with PML when expressed in low quantity in uninduced cells (Fig. 7C and E). Furthermore, both YL3 and YL4 were completely unable to complement ICP0-null mutant HSV-1 (Fig. 7D). The properties of YL3 and YL4 are in several respects similar to a previously described mutant, L174P (38), implying that large sections of the central portion of IE1 are required for its effects on PML. These data provide further evidence that these effects of IE1 on ND10 correlate well with its ability to stimulate virus infection.
IE1 and pp71, acting in concert, stimulate derepression of quiescent HSV-1 gene expression.
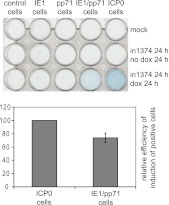
The inducible cell system also allows assay of the ability of ICP0 to reactivate, or more properly derepress, expression of a marker gene in quiescent HSV-1 infections (7). This assay involves infection of cells with mutant virus in1374, which carries the β-galactosidase coding region linked to the HCMV IE promoter/enhancer in a genome that carries a temperature-sensitive lesion in ICP4, a deletion of ICP0, and a mutation in VP16 that inactivates its ability to stimulate HSV-1 IE gene expression. This virus can be used to infect cells at high multiplicity at the nonpermissive temperature to establish monolayers in which a high proportion of the cells carry a quiescent viral genome. Subsequent expression of ICP0 stimulates efficient derepression of marker gene expression (7, 49, 50). Quiescent infections were established by in1374 infection of control, IE1, pp71, and IE1/pp71 cells, which exhibited negligible numbers of β-galactosidase-positive cells prior to induction (Fig. 8). After quiescence had been established for 24 h, expression of IE1, pp71, or both together was induced, and then the cells were stained a further 24 h later. Surprisingly, given the strong complementation of IE1 on ICP0-null mutant PFE (Fig. 6) and the role of pp71 in stimulating the HCMV IE promoter/enhancer (51–53), neither protein by itself induced derepression of marker gene activity (Fig. 8). In contrast, when both were induced together, derepression occurred efficiently, within 2-fold of the activity of ICP0 itself in this assay (Fig. 8). We conclude that, despite the individual activities of IE1 and pp71 in complementing ICP0-null mutant PFE (Fig. 6), the combined activities of the two HCMV proteins are required to replicate the role of ICP0 in derepression of gene expression from quiescent HSV-1 genomes in this system.
Fig 8.
Derepression of marker gene expression in cells quiescently infected with HSV-1. Cell lines as indicated were either mock infected (upper row) or infected with in1374 at an MOI of 3 for 24 h at the nonpermissive temperature of 38.5°C. The cells in the lowest row were then treated with doxycycline (100 ng/ml) to induce expression of the viral proteins, and all cells were incubated for a further 24 h at 38.5°C prior to staining for β-galactosidase expression. The histogram shows the mean and standard deviation of the proportion of IE1/pp71 cells that were β-galactosidase positive, normalized as a percentage of that in the ICP0 cells after induction. Positive cells were counted in three random views using a 20× objective (about 600 cells total per field of view, of which around 40% of the ICP0-expressing cells were positive).
Consistent with the results presented here, a previous study found that pp71 expressed from a viral vector did not stimulate marker gene expression from quiescent HSV-1 genomes during the first 3 days of pp71 expression (29). However, extended pp71 expression for longer times enabled reactivation of expression from the quiescent genomes (29). We did not investigate these longer time points after pp71 induction in the current study.
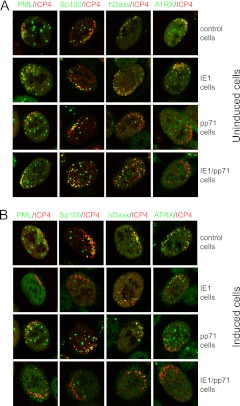
IE1 and pp71 inhibit the recruitment of subsets of ND10 components to sites associated with incoming HSV-1 genomes.
One of the most striking aspects of the behavior of ND10 components such as PML, Sp100, hDaxx, and ATRX is their rapid relocation from preexisting ND10 to novel sites that are closely associated with HSV-1 parental genomes and early replication compartments (34). This process is very short-lived in wt virus infections because it is countered by the effects of ICP0, but in ICP0-null mutant infections it is easily visualized, particularly in cells at the periphery of developing plaques, because of the manner in which the virus spreads to newly infected cells (34, 54, 55). The effects of mutations in ICP0 on its core biological activities closely correlate with efficiency of inhibition of ND10 component recruitment to the HSV-1-induced ND10-like foci (7). Furthermore, the repressive effects of hDaxx and PML isoform I are dependent on their SUMO interaction motifs that are required for recruitment into these novel foci (56). Therefore, it appears that the recruitment process is linked with ND10-mediated restriction of HSV-1 gene expression in the absence of ICP0. Because ND10 component recruitment to the viral genomes does not require viral gene expression or transcription (34), it is a reasonable hypothesis that similar events occur during other large DNA virus infections. However, this is not easily tested in HCMV infections because of the inefficiency of plaque formation of mutants that lack IE1, which might be predicted to be defective in inhibition of ND10 component recruitment. On the other hand, it has long been known that HCMV genomes are associated with ND10 during the early stages of infection (57, 58). The inducible cell system described here provides an alternative approach to testing whether IE1 and pp71 influence the efficiency of recruitment of ND10 components to large DNA virus genomes, because it is straightforward to examine this process in ICP0-null mutant HSV-1-infected cells. Therefore, recruitment assays were performed using the cell lines characterized above.
Uninduced control cells exhibited recruitment of PML, Sp100, hDaxx, and ATRX to sites associated with HSV-1 genomes and early replication compartments (detected by staining for ICP4) (Fig. 9A, top row). The recruitment is visualized by the asymmetric distributions of ICP4 and the ND10 component, giving partially overlapping (red/green/yellow) foci that are distinct from the apparently random distribution of ND10 in normal cells. Similar results were obtained in uninduced IE1 cells (Fig. 9A). While recruitment of PML and Sp100 occurred in all uninduced pp71 and IE1/pp71 cells, recruitment of ATRX was absent in at least 75% of cells in both cell lines (Fig. 9A). This mirrors the effects of the trace levels of expression of pp71 in the cells that is implied by the results of Fig. 2 and 4 to 6. Recruitment of hDaxx occurred in the majority (at least 85%) of uninduced pp71 and IE1/pp71 cells (Fig. 9A, lower two rows), although it was less intense because of the reduced levels of hDaxx expression in these cells prior to induction (Fig. 2). The DID2/3 mutant of pp71 had no effect on recruitment of hDaxx or ATRX in either induced or uninduced cells (data not shown). The separated channels of the images in Fig. 8A are presented in Fig. S3 in the supplemental material.
Fig 9.
Immunofluorescence analysis of the recruitment of ND10 components to sites associated with HSV-1 genomes in control, IE1, pp71, and IE1/pp71 cells. Cells were infected with dl1403/CMVlacZ and then stained for ICP4 and the indicated ND10 proteins at 24 h after infection. (A) The infections were performed in uninduced cells at an MOI of 0.5, 0.2, 0.05, and 0.05 for control, IE1, pp71, and IE1/pp71 cells, respectively. The MOI was varied between cell lines to ensure a convenient number of plaques for analysis. (B) The infections were performed after induction with 100 ng/ml doxycycline for 24 h, using an MOI of 0.5, 0.01, 0.02, and 0.005 for control, IE1, pp71, and IE1/pp71 cells, respectively. Samples were stained as indicated for ICP4 (MAb 58S) and rabbit antibodies for PML, Sp100, hDaxx, and ATRX. Secondary antibodies were anti-mouse Alexa 555 and anti-rabbit Alexa 633 (shown in the green channel here). Cells at the periphery of developing plaques with asymmetric distributions of HSV-1 genomes and early replication compartments were identified visually and then scanned to determine the distributions of the ND10 components. The quantitative data given in the text were determined by examination of 20 cells in each case.
When cells were infected after 24 h of treatment with doxycycline, recruitment of this group of proteins to ICP0-null mutant HSV-1 genomes occurred in all control cells (Fig. 9B, top row), while that of PML and Sp100 was absent in IE1 cells (Fig. 9B, second row). On the other hand, and despite the disperse location of hDaxx and ATRX in uninfected cells expressing IE1 (Fig. 4B), both of these proteins were recruited to the asymmetric ICP4 foci in over 90% of IE1 cells (Fig. 9B). After induction of pp71, recruitment of both PML and Sp100 occurred normally in all cells, but recruitment of ATRX was absent in 95% of cells. Recruitment of hDaxx still occurred in over 90% of induced pp71 cells and was more prominent than in the uninduced cells (Fig. 9B, third row). Given previous work on pp71 and hDaxx, this particular result might seem surprising, but it is consistent with the increased expression of hDaxx in the presence of high levels of pp71 (Fig. 2). Therefore, although pp71 interacts with and affects the stability of hDaxx (at least when expressed at low levels) (8, 9), in the system described here the consequence is disruption of the hDaxx-ATRX complex (Fig. 5) and inhibition of recruitment of ATRX to sites near HSV-1 genomes. It is significant that both ATRX recruitment and the repressive effect of hDaxx on ICP0-null mutant plaque formation efficiency require an intact hDaxx-ATRX complex (22).
The results in induced IE1/pp71 cells reflect additive effects of the expression of both IE1 and pp71: no recruitment of PML, Sp100, or ATRX and weak but positive recruitment of hDaxx (Fig. 9B, bottom row), correlating with the combined effects of the proteins in both plaque and derepression assays (Fig. 6 and 8). The separated channels of the images in Fig. 9B are presented in Fig. S4 in the supplemental material.
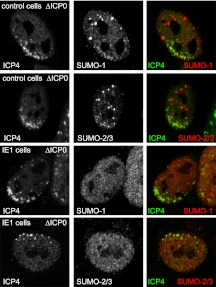
In addition to the specific ND10 components analyzed above, SUMO-conjugated proteins are also recruited to the HSV-1 genome foci in the absence of ICP0 (56). Because ICP0 inhibits the recruitment of such proteins (56), we investigated the effects of IE1 and pp71 in this assay. The latter had no effect (data not shown). While recruitment of SUMO-1 to locations associated with ICP4 foci was detectable in all control cells examined (Fig. 10, top row), this did not occur in induced IE1 cells (Fig. 9, third rows). Recruitment of SUMO-2/3 was also prominent in all control cells but was not detected in two-thirds of cells expressing IE1 (Fig. 9, rows 2 and 4, and data not shown). Although the recruited SUMO conjugates will include PML and Sp100 under normal circumstances, recruitment of SUMO-2/3 is independent of PML and SUMO-conjugated Sp100 (56). Therefore, it appears that, despite the lack of any effect of IE1 on the abundance of SUMO-2/3 conjugates (Fig. 3), IE1 affects the behavior of a subset of these protein species in response to herpesvirus infection. The ability of IE1 to influence recruitment to viral genomes of not only PML and Sp100 but also components of the SUMO modification pathway may be an important factor in its ability to stimulate ICP0-null mutant plaque formation so efficiently.
Fig 10.
Inhibition by HCMV IE1 of the recruitment of SUMO-modified species to sites associated with HSV-1 genomes. The upper two panels show the presence of SUMO-1- and SUMO-2-marked proteins in close vicinity of ICP0-null mutant HSV-1 genomes (detected by staining for ICP4) in control cells. The third row shows the absence of local concentrations of SUMO-1 species associated with the ICP4 signal in cells induced to express IE1. The bottom row shows an IE1-expressing cell negative for prominent SUMO-2/3 recruitment. Inductions and infections were performed as noted in the legend to Fig. 9. Samples were stained for ICP4 (MAb 58S) and rabbit antibodies for SUMO-1 or SUMO-2/3 as indicated. Secondary antibodies were anti-mouse Alexa 555 (shown in green) and anti-rabbit Alexa 633 (shown in the red). The quantitative data given in the text were determined by examination of 20 cells in each case.
DISCUSSION
The inducible cell line system provides a powerful approach to the study of viral regulatory proteins. We previously used this system to compare several members of the alphaherpesvirus ICP0 family of proteins (59), and here we applied the method to HCMV proteins IE1 and pp71. Although these HCMV proteins have been analyzed extensively in many previous publications, the motivation for this study was to determine whether, despite the lack of relatedness at the primary sequence level, the HCMV proteins could substitute for ICP0 during HSV-1 infection. The core finding was that IE1 and pp71 have additive effects that together substitute almost completely for the effects of ICP0 in stimulating lytic HSV-1 infection and the derepression of gene expression from quiescent HSV-1 genomes. An analogous inducible cell system was used to demonstrate that IE1 expressed in this manner complemented IE1 mutant HCMV to wt levels of replication (35).
It was inevitable that the initial characterization of the system involved some replication of extensive previous work on IE1 and pp71. In the course of this groundwork, however, we noted some significant new findings. For example, the presence of trace amounts of pp71 prior to induction can be inferred from a reduction in hDaxx expression (Fig. 2), changes to hDaxx and ATRX distribution (Fig. 4), and complementation activity (Fig. 6). The amount of pp71 entering a cell during a natural low MOI infection will be very limited, and therefore it makes sense that only tiny amounts should be required for biological activity. This principle may extend to other herpesvirus regulatory proteins that are components of the tegument. It remains a puzzle why hDaxx levels recover when pp71 accumulates in larger amounts, both during HCMV infection (9) and in this system (Fig. 2). It has been established that hDaxx is degraded via a proteasome-dependent mechanism during HCMV infection (9). To explore the basis of the reduced hDaxx levels in uninduced pp71 cells, we tested the effect of MG132. Although we found that this did not cause a marked increase in hDaxx levels, this does not imply that pp71-mediated degradation of hDaxx does not occur via proteasome-dependent degradation in this system, only that the rate of resynthesis of hDaxx is insufficient to reestablish normal expression levels over the 6-h time course of the experiment. The major hDaxx band also becomes less intense during HSV-1 infection, although in this case it occurs through inefficient detection of extensively phosphorylated forms (22). Treatment with phosphatase did not affect the intensity of the hDaxx band in uninduced pp71 cell extracts (data not shown), and therefore increased phosphorylation does not appear to be involved in this instance. Whatever the reason, the current results help explain variations in the effects of pp71 on hDaxx in previous studies, because the outcome will be influenced by the method, efficiency, and time of pp71 expression.
A notable effect of pp71 is the complete disruption of the hDaxx-ATRX complex (Fig. 5). This complex is involved in chromatin remodeling (44, 60), has histone chaperone activity (41, 43), and interacts with histone deacetylases (42). It is clear from extensive studies with HCMV that the role of pp71 is to counteract the assembly of a repressive viral chromatin structure (9, 19, 25). Similar events are likely to occur in the case of HSV-1 infection. From the degree of complementation of ICP0-null mutant virus infection imparted by pp71, it appears that these activities increase the PFE of the mutant virus by 15- to 20-fold, which is only 2- to 3-fold greater than the increase in ICP0-null mutant infectivity in cells depleted of hDaxx or ATRX (22). A key finding of this paper is that pp71 overcomes the repressive effects of the hDaxx-ATRX complex not only by reducing expression of hDaxx but also by dissociating the complex and by eliminating the recruitment of ATRX to sites associated with viral genomes.
ICP0 also inhibits recruitment of the hDaxx-ATRX complex to HSV-1 genome-associated sites but does so by a completely different mechanism. It does not affect expression of hDaxx or disrupt the hDaxx-ATRX complex either during infection (22) or in the inducible cell line system (data not shown). Instead, it inhibits the SIM-dependent recruitment of hDaxx, and therefore the hDaxx-ATRX complex, to the viral genomes (22, 56). The fact that two very different viral regulatory proteins target the same repressive complex but by different mechanisms provides strong evidence not only that the hDaxx-ATRX complex is a significant factor in regulating herpesvirus infections but also that a key issue is their ability to respond to the invading viral genome by accumulating at associated sites.
A similar principle of a related outcome but via different mechanisms concerns the effects of ICP0 and IE1 on PML and Sp100. ICP0 is a ubiquitin ligase that targets PML and its SUMO-modified forms for proteasome-dependent degradation through both SUMO-dependent mechanisms and a direct interaction with PML.I, the most abundant PML isoform (2–4, 61). IE1 also targets SUMO-modified PML, but although the mechanism is unknown, it is not through the proteasome pathway (5, 14, 16). IE1 itself does not appear to have SUMO protease activity (14), and its ability to disrupt ND10 and disperse PML is independent of SUMO modification of either IE1 or PML, although it likely requires the ability of IE1 to interact with PML (5, 14, 37, 38, 46). Despite their obvious differences in mechanisms of action, both ICP0 and IE1 inhibit the recruitment of PML and Sp100 to sites associated with HSV-1 genomes. PML lacking its major SUMO modification sites or its SIM is not recruited to the HSV-1-induced foci, while recruitment of Sp100 requires its SIM but not SUMO modification and is independent of PML (20, 21, 56). It appears therefore that both ICP0 and IE1 affect the behavior of PML and Sp100 independently, and whatever the mechanisms used, the result is the same in terms of inhibition of recruitment of these proteins to sites that are closely associated with HSV-1 genomes.
Although an interaction between IE1 and hDaxx has been detected (62), and despite the dispersal of hDaxx in uninfected cells expressing IE1 (Fig. 4B), IE1 did not eliminate the recruitment of either hDaxx or ATRX to the HSV-1-induced foci (Fig. 9B). This is consistent with the recruitment of hDaxx being PML independent (21). It is reasonable to suggest that IE1 is not as effective as ICP0 in stimulating HSV-1 plaque formation, because it does not counteract the response of the hDaxx-ATRX complex to nuclear entry of the viral genome.
Considering the available data, a plausible hypothesis is that the repressive effects of ND10 are modular, with various components each contributing to the full repressive effect. Thus, IE1 affects PML and Sp100, and pp71 affects the hDaxx-ATRX complex, and together they are almost as powerful in stimulating HSV-1 infection as ICP0, which affects all four proteins. In a similar vein, RNA interference (RNAi)-mediated depletion of any single ND10 component results in only a modest improvement of ICP0-null mutant HSV-1 replication (21, 22), but depletion of both PML and Sp100 simultaneously has a greater effect (20). Analogous observations have been made with HCMV (18, 24, 63). Recent results from our laboratory indicate that simultaneous depletion of three major ND10 components gives the greatest improvement in ICP0-null mutant plaque formation yet achieved by the RNAi approach (64). This group of ND10 proteins appears to be targeted by an ever-increasing list of important herpesvirus regulatory proteins (for recent examples, see references 65 and 66), and the repressive effects of hDaxx are also counteracted by adenovirus (67). Given that a common feature between ICP0, IE1, and pp71 is to impede the response of these ND10 components to nuclear entry of the viral genome, an increasingly compelling picture is emerging that herpesviruses have developed various mechanisms that target this cellular response in order to overcome the effects of ND10-mediated intrinsic antiviral resistance.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Medical Research Council.
We thank Chris Preston and Steven McFarlane (MRC-GU CVR), John Sinclair (University of Cambridge), Roel van Driel (University of Amsterdam), Hans Will (Heinrich-Pette Institute, Hamburg), and Richard Gibbons (University of Oxford) for gifts of reagents. We are grateful for an alignment of IE1 sequences provided by Andrew Davison and for constructive comments on the manuscript from Chris Boutell and Mandy Glass.
Footnotes
Published ahead of print 7 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01103-12.
REFERENCES
- 1. Bieniasz PD. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109–1115 [DOI] [PubMed] [Google Scholar]
- 2. Boutell C, Sadis S, Everett RD. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boutell C, Cuchet-Lourenco D, Vanni E, Orr A, Glass M, McFarlane S, Everett RD. 2011. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 7:e1002245 doi:10.1371/journal.ppat.1002245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muller S, Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parkinson J, Everett RD. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006–10017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Everett RD, Parsy ML, Orr A. 2009. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J. Virol. 83:4963–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hofmann H, Sindre H, Stamminger T. 2002. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 76:5769–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saffert RT, Kalejta RF. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lukashchuk V, McFarlane S, Everett RD, Preston CM. 2008. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J. Virol. 82:12543–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ahn JH, Hayward GS. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155–158 [DOI] [PubMed] [Google Scholar]
- 13. Wilkinson GW, Kelly C, Sinclair JH, Rickards C. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79:1233–1245 [DOI] [PubMed] [Google Scholar]
- 14. Kang H, Kim ET, Lee HR, Park JJ, Go YY, Choi CY, Ahn JH. 2006. Inhibition of SUMO-independent PML oligomerization by the human cytomegalovirus IE1 protein. J. Gen. Virol. 87:2181–2190 [DOI] [PubMed] [Google Scholar]
- 15. Kim YE, Lee JH, Kim ET, Shin HJ, Gu SY, Seol HS, Ling P, Lee CH, Ahn JH. 2011. Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J. Virol. 85:11928–11937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tavalai N, Adler M, Scherer M, Riedl Y, Stamminger T. 2011. Evidence for a dual antiviral role of the major nuclear domain 10 component Sp100 during the immediate-early and late phases of the human cytomegalovirus replication cycle. J. Virol. 85:9447–9458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adler M, Tavalai N, Muller R, Stamminger T. 2011. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J. Gen. Virol. 92:1532–1538 [DOI] [PubMed] [Google Scholar]
- 19. Cantrell SR, Bresnahan WA. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80:6188–6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lukashchuk V, Orr A, Everett RD. 2010. Regulation of ICP0 null mutant HSV-1 infection by ND10 components ATRX and hDaxx. J. Virol. 84:4026–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Preston CM, Nicholl MJ. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 87:1113–1121 [DOI] [PubMed] [Google Scholar]
- 24. Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woodhall DL, Groves IJ, Reeves MB, Wilkinson G, Sinclair JH. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652–37660 [DOI] [PubMed] [Google Scholar]
- 26. Marshall KR, Rowley KV, Rinaldi A, Nicholson IP, Ishov AM, Maul GG, Preston CM. 2002. Activity and intracellular localization of the human cytomegalovirus protein pp71. J. Gen. Virol. 83:1601–1612 [DOI] [PubMed] [Google Scholar]
- 27. Stow EC, Stow ND. 1989. Complementation of a herpes simplex virus type 1 Vmw110 deletion mutant by human cytomegalovirus. J. Gen. Virol. 70:695–704 [DOI] [PubMed] [Google Scholar]
- 28. Stow ND, Stow EC. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571–2585 [DOI] [PubMed] [Google Scholar]
- 29. Preston CM, Nicholl MJ. 2005. Human cytomegalovirus tegument protein pp71 directs long-term gene expression from quiescent herpes simplex virus genomes. J. Virol. 79:525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yao F, Schaffer PA. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. U. S. A. 99:15655–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jamieson DRS, Robinson LH, Daksis JI, Nicholl MJ, Preston CM. 1995. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus Vmw65 mutants. J. Gen. Virol. 76:1417–1431 [DOI] [PubMed] [Google Scholar]
- 33. Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773–784 [DOI] [PubMed] [Google Scholar]
- 34. Everett RD, Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Knoblach T, Grandel B, Seiler J, Nevels M, Paulus C. 2011. Human cytomegalovirus IE1 protein elicits a type II interferon-like host cell response that depends on activated STAT1 but not interferon-gamma. PLoS Pathog. 7:e1002016 doi:10.1371/journal.ppat.1002016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ahn JH, Xu Y, Jang WJ, Matunis MJ, Hayward GS. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 75:3859–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spengler ML, Kurapatwinski K, Black AR, Azizkhan-Clifford J. 2002. SUMO-1 modification of human cytomegalovirus IE1/IE72. J. Virol. 76:2990–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu Y, Ahn JH, Cheng M, CMapRhys Chiou CJ, Zong J, Matunis MJ, Hayward GS. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J. Virol. 75:10683–10695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tavalai N, Kraiger M, Kaiser N, Stamminger T. 2008. Insertion of an EYFP-pp71 (UL82) coding sequence into the human cytomegalovirus genome results in a recombinant virus with enhanced viral growth. J. Virol. 82:10543–10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ishov AM, Vladimirova OV, Maul GG. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76:7705–7712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24:1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 115:3319–3330 [DOI] [PubMed] [Google Scholar]
- 43. Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U. S. A. 107:14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. 2003. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. U. S. A. 100:10635–10640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reinhardt J, Smith GB, Himmelheber CT, Azizkhan-Clifford J, Mocarski ES. 2005. The carboxyl-terminal region of human cytomegalovirus IE1491aa contains an acidic domain that plays a regulatory role and a chromatin-tethering domain that is dispensable during viral replication. J. Virol. 79:225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nevels M, Brune W, Shenk T. 2004. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J. Virol. 78:7803–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shin HJ, Kim YE, Kim ET, Ahn JH. 2012. The chromatin-tethering domain of human cytomegalovirus immediate-early (IE) 1 mediates associations of IE1, PML and STAT2 with mitotic chromosomes, but is not essential for viral replication. J. Gen. Virol. 93:716–721 [DOI] [PubMed] [Google Scholar]
- 48. Mazeron MC, Jahn G, Plachter B. 1992. Monoclonal antibody E-13 (M-810) to human cytomegalovirus recognizes an epitope encoded by exon 2 of the major immediate early gene. J. Gen. Virol. 73(Pt 10):2699–2703 [DOI] [PubMed] [Google Scholar]
- 49. Preston CM, Mabbs R, Nicholl MJ. 1997. Construction and characterization of herpes simplex virus type 1 mutants with conditional defects in immediate early gene expression. Virology 229:228–239 [DOI] [PubMed] [Google Scholar]
- 50. Preston CM, Nicholl MJ. 1997. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J. Virol. 71:7807–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bresnahan WA, Shenk TE. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. U. S. A. 97:14506–14511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Homer EG, Rinaldi A, Nicholl MJ, Preston CM. 1999. Activation of herpesvirus gene expression by the human cytomegalovirus protein pp71. J. Virol. 73:8512–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu B, Stinski MF. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66:4434–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Everett RD. 2011. The use of fluorescence microscopy to study the association between herpesviruses and intrinsic resistance factors. Viruses 3:2412–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Everett RD, Murray J, Orr A, Preston CM. 2007. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J. Virol. 81:10991–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cuchet-Lourenco D, Boutell C, Lukashchuk V, Grant K, Sykes A, Murray J, Orr A, Everett RD. 2011. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 7:e1002123 doi:10.1371/journal.ppat.1002123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maul GG. 2008. Initiation of cytomegalovirus infection at ND10. Curr. Top. Microbiol. Immunol. 325:117–132 [DOI] [PubMed] [Google Scholar]
- 58. Maul GG. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660–667 [DOI] [PubMed] [Google Scholar]
- 59. Everett RD, Boutell C, McNair C, Grant L, Orr A. 2010. Comparison of the biological and biochemical activities of several members of the alphaherpesvirus ICP0 family of proteins. J. Virol. 84:3476–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X. 2004. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 279:20369–20377 [DOI] [PubMed] [Google Scholar]
- 61. Cuchet-Lourenco D, Vanni E, Glass M, Orr A, Everett RD. 2012. Herpes simplex virus 1 ubiquitin ligase ICP0 interacts with PML isoform I and induces its SUMO-independent degradation. J. Virol. 86:11209–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reeves M, Woodhall D, Compton T, Sinclair J. 2010. Human cytomegalovirus IE72 protein interacts with the transcriptional repressor hDaxx to regulate LUNA gene expression during lytic infection. J. Virol. 84:7185–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tavalai N, Papior P, Rechter S, Stamminger T. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82:126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Glass M, Everett RD. Components of PML nuclear bodies (ND10) act cooperatively to repress herpesvirus infection. J. Virol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Full F, Reuter N, Zielke K, Stamminger T, Ensser A. 2012. Herpesvirus Saimiri antagonizes nuclear domain 10-instituted intrinsic immunity via an ORF3-mediated selective degradation of cellular protein Sp100. J. Virol. 86:3541–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tsai K, Thikmyanova N, Wojcechowskyj JA, Delecluse HJ, Lieberman PM. 2011. EBV tegument protein BNRF1 disrupts DAXX-ATRX to activate viral early gene transcription. PLoS Pathog. 7:e1002376 doi:10.1371/journal.ppat.1002376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schreiner S, Martinez R, Groitl P, Rayne F, Vaillant R, Wimmer P, Bossis G, Sternsdorf T, Marcinowski L, Ruzsics Z, Dobner T, Wodrich H. 2012. Transcriptional activation of the adenoviral genome is mediated by capsid protein VI. PLoS Pathog. 8:e1002549 doi:10.1371/journal.ppat.1002549 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.