Abstract
Despite the eradication of smallpox, orthopoxviruses (OPV) remain public health concerns. Efforts to develop new therapeutics and vaccines for smallpox continue through their evaluation in animal models despite limited understanding of the specific correlates of protective immunity. Recent monkeypox virus challenge studies have established the black-tailed prairie dog (Cynomys ludovicianus) as a model of human systemic OPV infections. In this study, we assess the induction of humoral immunity in humans and prairie dogs receiving Dryvax, Acam2000, or Imvamune vaccine and characterize the proteomic profile of immune recognition using enzyme-linked immunosorbent assays (ELISA), neutralization assays, and protein microarrays. We confirm anticipated similarities of antigenic protein targets of smallpox vaccine-induced responses in humans and prairie dogs and identify several differences. Subsequent monkeypox virus intranasal infection of vaccinated prairie dogs resulted in a significant boost in humoral immunity characterized by a shift in reactivity of increased intensity to a broader range of OPV proteins. This work provides evidence of similarities between the vaccine responses in prairie dogs and humans that enhance the value of the prairie dog model system as an OPV vaccination model and offers novel findings that form a framework for examining the humoral immune response induced by systemic orthopoxvirus infection.
INTRODUCTION
Over 30 years after the eradication of smallpox (1), orthopoxviruses (OPVs) remain relevant public health threats. Variola virus remains a priority of biodefense preparedness research (2), and other OPVs, such as vaccinia virus (VACV) (3), cowpox virus (4), and monkeypox virus (MPXV) (5), remain global emerging infectious disease threats. Important characteristics of VACV smallpox vaccines are their ability to induce cross-protective antibodies against other OPVs and to provide a vaccine backbone for recombinant vaccines against diseases such as rabies (6). While 1st-generation vaccines were used to eradicate smallpox, they can cause multiple adverse effects, including eczema vaccinatum, progressive vaccinia, and myopericarditis (7–9). Continued concerns over vaccine safety and fears of smallpox release have fueled research on safer, better-characterized vaccines. These concerns have advanced development of 2nd- and 3rd-generation vaccines, such as Acam2000 (Acambis) (10) and Imvamune (11). However, questions remain regarding the breadth and quality of response (12), protection of immunocompromised individuals (13), and proper utilization strategies (14). In addition, development of first-generation vaccines used to eradicate smallpox occurred in the absence of sophisticated techniques to define specific and detailed immune responses. Contemporary techniques have provided the means to characterize in more detail vaccine (1st, 2nd, and 3rd generation)-induced immunity and factors associated with protection (15).
The animal models used for smallpox vaccine testing were a macaque model of variola virus infection (16), an MPXV-infected macaque model (17, 18), multiple mouse models (19–21), the rabbitpox-infected rabbit model (22), and others (2). Each of these models has salient differences from human infections with systemic OPVs that may include the route of infection, disease pathogenesis, or a compressed disease incubation period. The 2003 U.S. monkeypox outbreak was caused when imported exotic animals carrying MPXV were sold throughout the Midwest, subsequently infecting cohoused black-tailed prairie dogs and ultimately humans (23). Following this outbreak, an experimental infection model of the black-tailed prairie dog (Cynomys ludovicianus) was characterized using monkeypox virus. Aspects of clinical disease similar to systemic human OPV infection were found using a monkeypox virus intranasal (i.n.)-infection model, including a protracted asymptomatic incubation phase similar to that of human smallpox (24). Further refinement of the model allowed testing of vaccine efficacy using 1st-, 2nd-, and 3rd-generation smallpox vaccines (25).
Animal models for OPV have also been used to clarify the immune response and requirements for protection from pathogenic OPV infections. Murine models have indicated roles for cellular- and humoral-based responses, although the requirement for cellular responses varies by model system. Vaccination with Dryvax (Wyeth) via tail scratch was shown to provide protection in mice deficient in B cells when later challenged intranasally with the VACV Western Reserve strain (VACV WR) (26), indicating overlapping protection between the immune responses. Similarly, mice vaccinated with either modified Ankara vaccinia virus (MVA) or a recombinant VACV via skin scarification (s.s.) were protected from respiratory challenge as long as either antibody or T cells were present (27). However, other models have shown that protection against secondary infection was dependent on antibody, but not CD4 or CD8 T cell function. For example, when mice were primed with avirulent ectromelia virus (ECTV) intraperitoneally and later challenged with virulent ECTV, only antibody was necessary (28, 29). Experiments done with Dryvax s.s.-vaccinated rhesus macaques have also demonstrated a necessity for antibodies by showing that protection from lethal MPXV challenge required the presence of B cells (30) and that passive treatment with vaccinia immune globulin (VIG) was sufficient to prevent death. Thus, interest in understanding in greater detail the requirements for protection and the protective components of the humoral response remains.
Most of the protective antigens for OPV infection that have been studied are surface proteins from the two primary virion forms, the intracellular mature virions (MV) and extracellular enveloped virions (EV). These immune response targets include MV proteins L1, A17, A27, D8 H3, A13, and A28 and EV proteins A33 and B5 (31–38). The protective nature of antibodies reactive to these MV proteins, especially when combined with those against EV, indicate the possibility that a select subset of proteins may be sufficient to provide protective immunity. However, protein targets of humoral immunity upon vaccination are known to differ between individuals (39), and many viral proteins have not been implicated in protective responses despite being effective at eliciting host immune induction (31, 40–42). Furthermore, recent analysis of CD8 T cell responses in mice have indicated that larger than expected major histocompatibility complex (MHC)-dependent differences were seen between VACV-infected outbred and inbred populations (43), which raises the question of whether these observations are also true for B cell responses in an outbred model. The mechanism of protection may include all of these factors (specific antigen recognition, cellular responses, and diversity of neutralizing antibody response); however, a better understanding of the mechanisms of protection in newly developed smallpox vaccines will be crucial for understanding novel-vaccine efficacy.
We present research to further explore components of vaccine- and OPV challenge-induced immunity in the prairie dog model of OPV infection, which emulates human systemic OPV infections. However, it is important to show that vaccination of prairie dogs induces a similar immune response and not just similar levels of response relative to humans. Thus, we utilize antigenic profiling to assess similarities and differences. Our results add value to the use of this model of systemic OPV vaccination and infection and may be utilized to better understand the humoral correlates of protection in human vaccination against smallpox. Protein microarrays have been previously utilized to show that immune response profiles in MVA- and Dryvax-vaccinated macaques, humans, and rabbits are comparable (44). We performed proteome microarray comparative analysis of humoral immunity in Dryvax- and Acam2000-vaccinated humans versus Dryvax-, Acam2000-, and Imvamune-vaccinated prairie dogs. We examined this novel model system for biomarkers that may be associated with protective immunity. We then extended the analysis to characterizing immune response following MPXV challenge of prairie dogs by examining changes that occur in vaccinated and unvaccinated animals. Our results show that there are salient differences in the magnitude and breadth of immune response after vaccination and those after challenge. These differences may be important as we transition to safer, and likely further attenuated, vaccine strains.
MATERIALS AND METHODS
Protein arrays.
Protein arrays were fabricated as described previously (40, 45). Briefly, individual open reading frames (ORFs) of VACV WR were amplified and cloned into a T7 expression vector by homologous recombination. Of 210 cloned genes, 62 were selected based on previous known reactivity to serologic samples (see Table S1 in the supplemental material) for further testing. Proteins were produced using Escherichia coli-based cell-free coupled transcription/translation reactions (RTS 100 kits; Roche) according to the manufacturer's instructions and printed without further purification on FAST nitrocellulose-coated glass slides (Whatman) using an OmniGrid 100 microarrayer (Gene Machines). Protein expression was monitored by using hemagglutinin (HA) and His tags present on the protein termini, which were detected in 77% and 99% of expressed proteins, respectively; however, quantification of the amount of protein spotted was not possible. No-DNA control spots containing the reaction mixture and no template DNA were included throughout the array to correct for background binding to E. coli proteins found in the transcription-translation mixture.
Prairie dogs utilized and vaccinations.
Prairie dogs were captured, screened, and housed as described previously (46). The animals ranged between 9 and 12 months of age during vaccination and challenge. Sera from a series of studies described previously (25) were utilized and are summarized in Table 1. The animals in study 1 (n = 9 animals) were divided into 2 groups of 3 animals each and inoculated with Dryvax or Acam2000 with 2 × 105 PFU of vaccine via multiple puncture (m.p.). All vaccine doses were human doses and were administered on day −30 prior to challenge. Study 2 (n = 32 animals; n = 29 with sera available for microarray testing) animals were grouped as follows: Dryvax (n = 8 animals), Acam2000 (n = 9 animals), Imvamune (n = 10 animals), and phosphate-buffered saline (PBS) (n = 1 animal). Dryvax and Acam2000 animals were vaccinated with 2 × 105 PFU of vaccine, while Imvamune animals were vaccinated twice with 1 × 108 PFU subcutaneously (s.c.) on day −60 and day −30 prior to challenge (day 0). Day −60 serum samples (naïve) from animals given Imvamune were unavailable, so day −30 was used as the baseline for determining postvaccination results. Antibody responses after a single dose of Imvamune were minimally reactive, except for protein D8, which produced fluorescence slightly higher than that seen in other naïve animals but significantly below the post-two-dose time point (see Fig. S1 in the supplemental material). Blood samples were taken every 3 or 4 days for 30 days to monitor disease progression. Sera were not available for each animal at every time point.
Table 1.
Prairie dog samples utilized for data analysis
| Dose | Vaccination |
Challenge |
||||
|---|---|---|---|---|---|---|
| Virus | Dosage | Route | Virus | Dosage | Route | |
| Low | DVX (n = 3) | 2E5 PFU | m.p. | MPXV (n = 3) | 2E5 PFU | i.n. |
| Acam (n = 3) | 2E5 PFU | m.p. | MPXV (n = 3) | 2E5 PFU | i.n. | |
| PBS (n = 3) | NAa | m.p. | MPXV (n = 3) | 2E5 PFU | i.n. | |
| High | DVX (n = 8) | 2E5 PFU | m.p. | MPXV (n = 4) | 2E6 PFU | i.n. |
| Acam (n = 9) | 2E5 PFU | m.p. | MPXV (n = 7) | 2E6 PFU | i.n. | |
| 2X Imvamune (n = 10) | 1E8 TCID/1E8 TCID | s.c. | MPXV (n = 8) | 2E6 PFU | i.n. | |
| PBS (n = 1) | NA | m.p. | MPXV (n = 1) | 2E6 PFU | i.n. | |
‖NA, not applicable.
Prairie dog monkeypox virus challenge.
In a low-dose challenge, animals from vaccination study 1 (Dryvax, n = 3; Acam2000, n = 3; and PBS, n = 3) were challenged i.n. with 105 PFU of Congo Basin MPXV-ROC-2003-385. In the second experiment (high-dose challenge), the dose was increased to 106 PFU of MPXV. Vaccinated animals from each group were challenged with sera available for testing from Dryvax (n = 4), Acam2000 (n = 7), and Imvamune (n = 8) animals. One PBS group animal (n = 1) was unvaccinated and used as a virus challenge control. As described by Keckler et al. (25), the resulting rash burdens and mortalities were not statistically different between the low-dose and high-dose Dryvax- and Acam2000-vaccinated animals, nor were the microarray responses here statistically different (data not shown). Thus, we combined high- and low-dose data for appropriate groups in our analyses.
Human vaccinee sera and VIGIV.
Human sera were collected from primary vaccinees via venipuncture as part of a smallpox vaccination study involving laboratory workers that is approved and monitored by the CDC Institutional Review Board (IRB) to ensure the use of approved protocols, properly trained personnel, and appropriate personal protective equipment (PPE). The vaccinees, aged 23 to 34 and 25 to 30, were vaccinated with Dryvax or Acam2000, respectively. Sera were collected prior to vaccination and at approximately 7-day intervals thereafter from day 7 to day 49 postvaccination. Vaccinia immune globulin intravenous (VIGIV) was received from the Strategic National Stockpile (CDC, Atlanta, GA) and was produced by Cangene (Cangene Corporoation, Winnipeg, Canada). It is an anion-exchange column-purified globulin fraction from VACV (Dryvax)-vaccinated and boosted plasma donors (47). Lot 1730203, used here, had total protein (IgG) of 55 mg/ml.
ELISA.
A modified version of an enzyme-linked immunosorbent assay (ELISA) was used for analysis of prairie dog anti-OPV (25). Briefly, microtiter plates (Immulon II; Dynatech) were coated with crude VACV or BSC-40 cell lysate by overnight incubation and subsequently inactivated, blocked, and washed prior to incubation with dilutions of prairie dog sera and then ImmunoPure A/G-horseradish peroxidase (HRP) conjugate (Pierce). The BSC-40 cell lysate half of each plate was used to generate a cutoff value (COV) for each plate by averaging all the values of the BSC-40 lysate half and adding 2 standard deviations (SD). Specimens were considered positive if the test sample's value was above the COV. The endpoint titer for each animal was determined based on the highest dilution that was positive. Study 1 animals were used for time course analysis of ELISA and microarray signals. ELISA of human samples was performed as previously described (48). Briefly, microtiter plates were coated with crude VACV overnight, inactivated, blocked, and washed prior to incubation with human sera and then goat anti-human IgG-HRP (KPL, Gaithersburg, MD). After development, a COV was determined using the mean optical density (OD) from known naïve standards plus 3 SD. Sera were considered positive if the test sample's value was above the COV. The endpoint titer was determined based on the highest dilution that was positive. For comparison with microarray fluorescence, reciprocal values (log transformed) were used.
HCS GFP neutralization assay.
Neutralizing antibody (NAb) titers postvaccination were measured using the ArrayScan high-content screening (HCS) reader to detect green fluorescent protein (GFP) introduced by infection with a GFP-expressing VACV WR strain (49). This assay detects the percentage of GFP-producing cells, which is then normalized to control wells to obtain a relative percent responder (RPR) value. Reported 50% RPR values correlate with the serum inhibitory concentration required to neutralize 50% of viral infection (50% infective dose [ID50]) in a traditional plaque reduction neutralization titer (PRNT) assay.
Array probing and signal calculation.
Sera were probed on the arrays at 1:100 dilution in protein array blocking buffer (Whatman) plus 20% E. coli lysate to block antibodies reactive to E. coli proteins present in the transcription-translation mixture. The remaining steps were carried out in blocking buffer with 10% E. coli lysate. VIG was probed on the arrays at 1:500 dilution. For human sera and VIG, a secondary donkey anti-human IgG-biotin conjugate (Jackson Immuno) at 1:200 was used, followed by a streptavidin-Surelight P-3 (Columbia Biosciences) conjugate. Prairie dog serum antibodies were visualized using protein G-biotin (Pierce) secondary antibody at 1:200, followed by streptavidin-Surelight P-3 conjugate at 1:200. The microarrays were scanned using a Gene Pix 4100A scanner (Molecular Devices, CA) with laser settings at 100% and a photomultiplier (PMT) gain of 350. Image analysis was performed with Genepix Pro 5.0 software (Molecular Devices). The spot intensity was calculated as the median spot value minus local spot background. A secondary correction for background binding to E. coli proteins in the reaction mixture was done by subtracting an average of the no-DNA spots from the background-corrected spot value.
Array data analysis.
Prevaccination/preinoculation sera or post-single vaccination sera for Imvamune animals (see “Prairie dogs utilized and vaccinations” above) were used to determine naïve seroreactivity. Due to differences between the assays for human and prairie dog samples (e.g., use of anti-human IgG for human samples or protein G with prairie dog samples), the cutoff value for a positive hit was determined empirically for each conjugate. This cutoff value was defined by the threshold where 1% false positives occur when calculated from non-orthopox virus proteins included on the array. These proteins included varicella zoster virus (VZV) and herpes simplex virus (HSV). A fluorescence cutoff value of 150 arbitrary fluorescence units (AU) for prairie dogs or 1,000 AU for human samples after subtraction of prevaccination values, combined with a greater-than-2-fold rise in reactivity, were used to determine positive hits. The 2-fold rise was included to account for array-to-array variability. For VIG and human sera for which no prevaccination samples were available, a collection of naïve donor sera was used to calculate an average value of previous reactivity to orthopox virus proteins.
Heat map normalization of human and prairie dog data.
Heat map intensity values were normalized between human and prairie dog samples by running human samples with protein G-biotin secondary conjugate and comparing the resultant signals with data obtained using donkey anti-human IgG-biotin (both assays utilized streptavidin-Surelight P3 label). Differences in fluorescence intensities when comparing the individual serum reactivities with individual protein spots between the two assays provided correlative intensities. Equivalent intensity ranges were then established beginning at 1/2 of the cutoff value (500 in the anti-human assay or 75 in the protein G assay) and continuing with approximately 2-fold increases up to the maximum fluorescence intensity measurable by the array scanner (65,000). Correlative values between assays were assigned the same shading and used to show similar responses within the heat maps.
Statistical analysis.
SPSS was used for statistical analyses. The Wilcoxon rank sum test was utilized to compare responses to proteins with known neutralizing antigenic sites and neutralizing capacities of vaccinee sera. The Wilcoxon rank sum test is a nonparametric test to compare related samples and determine if their population mean ranks are different; it was utilized given the variability found in some array responses. Data were compiled by identifying microarray fluorescence values above the appropriate cutoff value (150 or 1,000) but did not include the 2-fold rise in reactivity otherwise utilized in order to allow comparison of naïve samples.
RESULTS
Vaccine-induced humoral immunity.
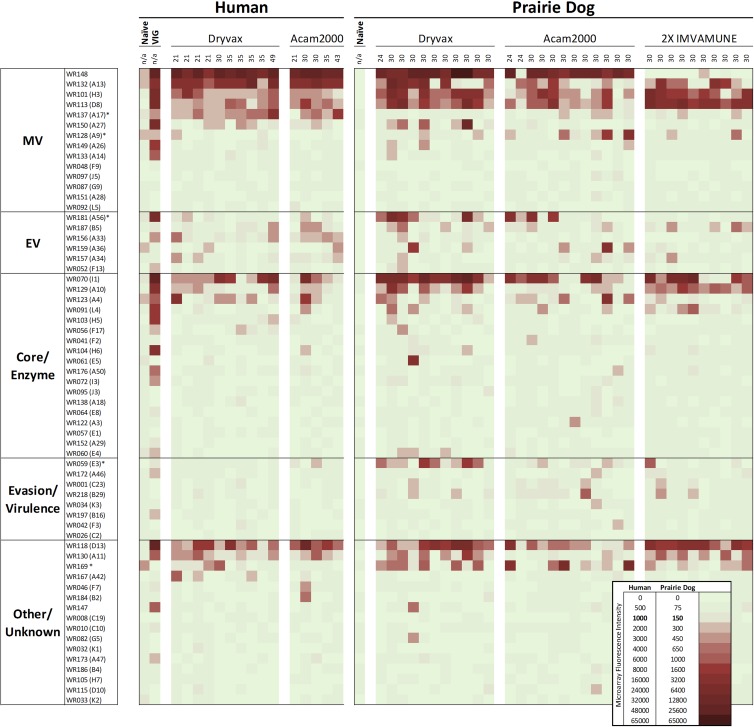
We first set out to provide a global view of reactivity and to identify major trends between days 21 and 49 after vaccination of a group of human subjects and prairie dogs. A core group of antigens elicit antibodies following vaccination with Dryvax and Acam2000 in both species, as well as in Imvamune-vaccinated prairie dogs (Fig. 1). As standards for immune reactivity on the array, naïve sera were used to show common background reactivity, and VIG was used to define frequently recognized proteins. Interrogation with VIG revealed immune recognition of 26 of 62 antigens (Fig. 1). The highest reactivity measured by fluorescence intensity on this array was observed to MV proteins WR148, A13, H3, D8, and A27; EV protein A56; core proteins I1 and A10; and H6 and D13 (other functions). As a group, MV and core proteins were most frequently recognized by VIG (Fig. 1). In comparing vaccine-induced immunity in humans and prairie dogs regardless of the vaccine strain, commonly reactive antigens, defined those as having greater than 50% reactivity within a vaccination group, were observed in both host species and included WR148, A13, H3, D8, I1, and D13. Prominent differences observed include absence of a response to MV protein A17 in prairie dogs, but also more frequent recognition of MV protein A9, EV protein A56, evasion protein E3, and WR169 (unknown function) (Fig. 1, asterisks, and Table 2). Similar to the VIG profile, MV proteins account for the majority of reactive proteins in vaccinees and for 44% of the total signal. Within individual serum samples, one or more EV proteins were frequently recognized in each vaccine group, but rarely was the same protein commonly recognized. One exception was reactivity to A56 in prairie dog Dryvax and Acam2000 vaccinees, but not those receiving Imvamune. Reactivity to EV protein F13 was also predominantly absent in all groups. Responses to core proteins and enzymes accounted for 22% of the chip signal. Host immune evasion and virulence factor recognition were absent in human vaccinees and were seen only minimally in prairie dogs, with a predominance of reactivity to E3 in the Dryvax group. Overall, an average reactivity to 8.5 of 62 antigens was found in both humans and prairie dogs. Additionally, while there were many commonly reactive antigens, no antigen was reactive in 100% of human and prairie dog vaccinees.
Fig 1.
Normalized proteome view associated with vaccination using Dryvax, Acam2000, or two-dose Imvamune in humans and prairie dogs. The proteins are sorted by function and location (if known), with signal intensities represented by pale green for no response and red for increasingly intense reactions. Average raw values for naïve human and prairie dog serum profiles were included to provide visual controls for background proteome-wide antibody reactivity, whereas VIG and the remaining sample profiles are shown after subtraction of naive reactivity. Each column is an average of measurements for individual vaccinees, with the time point postvaccination indicated above each column. The normalization procedure for comparing human and prairie dog responses is described in Materials and Methods.
Table 2.
Frequently recognized antigens in vaccinated subjectsa
| Protein category | Protein | Positive responseb |
||||
|---|---|---|---|---|---|---|
| Humans |
Prairie dogs |
|||||
| DVX (n = 10) | Acam (n = 5) | DVX (n = 11) | Acam (n = 12) | Imvamune (n = 10) | ||
| MV | WR148 | +++ | +++ | +++ | +++ | + |
| WR132 (A13) | +++ | +++ | +++ | +++ | +++ | |
| WR101 (H3) | +++ | +++ | +++ | +++ | +++ | |
| WR113 (D8) | +++ | +++ | +++ | +++ | +++ | |
| WR137 (A17) | +++ | +++ | − | ++ | + | |
| WR150 (A27) | ++ | + | ++ | − | − | |
| WR128 (A9) | − | − | − | ++ | − | |
| EV | WR181 (A56) | − | − | +++ | ++ | − |
| WR187 (B5) | − | ++ | ++ | + | ++ | |
| WR156 (A33) | + | +++ | − | − | − | |
| WR159 (A36) | − | + | + | ++ | − | |
| WR157 (A34) | + | + | − | − | − | |
| Core | WR070 (I1) | +++ | +++ | +++ | +++ | +++ |
| WR129 (A10) | ++ | ++ | +++ | + | +++ | |
| WR123 (A4) | ++ | + | ++ | ++ | − | |
| WR091 (L4) | − | + | ++ | + | ++ | |
| Evasion | WR059 (E3) | − | + | +++ | + | − |
| Other/unknown | WR118 (D13) | +++ | +++ | +++ | +++ | +++ |
| WR130 (A11) | +++ | +++ | +++ | − | +++ | |
| WR169 | + | − | ++ | ++ | ++ | |
Proteins with greater than 5 hits over all samples are shown.
+, >10% of subjects with a positive response; ++, >25% of subjects with a positive response; +++, >50% of subjects with a positive response; −, no subjects with a positive response.
Human response to vaccination.
We then established a baseline for comparison by examining human antibody profiles after Dryvax and Acam2000 vaccination. Responses were highly similar between vaccine strains (Fig. 1 and Table 2) and included recognition of MV proteins WR148, A13, H3, D8, and A17 and, to a lesser extent, A27L. EV proteins A56, B5, A33, A36, and A34 were recognized by at least one Dryvax- or Acam2000-vaccinated individual, and A33 was recognized by more than 50% of Acam2000 vaccinees. Human responses to core proteins I1L, A10, and A4 were observed in Dryvax and Acam2000 vaccinees, while responses to proteins categorized as other/unknown function included D13 and A11. Ranked mean fluorescence intensities of proteins with positive hits revealed that WR148 was the most immunogenic protein in human vaccinees, followed by A13 and then I1 and D13 for both Dryvax and Acam2000 vaccinees (Fig. 1).
Prairie dog response to vaccination.
We then set out to examine the prairie dog response in detail and to assess similarities and differences between species. When given Dryvax and Acam2000, prairie dogs showed responses highly similar to human vaccinee responses (Fig. 1 and Table 2). Of the animals vaccinated with Dryvax and Acam2000, 70% reacted to MV proteins WR148, H3, and D8, and 45% reacted to each of those plus A13. In contrast to humans, A13 responses were less frequently seen, with only a 56% response rate versus 100% in humans. A17 responses were also predominantly absent in prairie dogs, and EV protein A56 was commonly recognized in Dryvax- and Acam2000-vaccinated animals. Core proteins I1L, A10L, and A4L were also recognized in both live-vaccine groups at levels slightly higher than those seen in humans. E3L (evasion) protein was found to be reactive in Dryvax-vaccinated prairie dogs, though much less so in Acam2000-vaccinated prairie dogs (as in humans). Similar to human responses, prairie dogs vaccinated with Dryvax or Acam2000 react to scaffold protein D13L and protein of unknown function WR169, but reactivity to morphogenesis protein All was absent in Acam2000. For prairie dogs vaccinated with Imvamune, highly reactive proteins were significantly more limited and included MV proteins A13L, H3L, and D8. Except for one low-level binding event (less than 2 times the cutoff), no response was observed to WR148, which is known to be deleted in Imvamune. Imvamune also induced lower responses overall to EV proteins than were seen with Dryvax and Acam2000, and a comparable reaction to core proteins I1L and A10L versus live vaccines was observed. Overall, a predominant absence of response to A17L in this assay was consistent in all vaccinated prairie dogs. Of interest, evasion protein E3L was reactive almost exclusively in Dryvax-immunized animals. Reactive proteins in Imvamune vaccinees were comparable to those in Acam2000 vaccinees, and both had fewer recognized antigens than Dryvax vaccinees. Ranked mean fluorescence intensities of proteins with positive hits revealed that WR148 (except in Imvamune vaccinees) and I1L were the most immunogenic proteins for prairie dogs (Fig. 1).
Immune responses to proteins targeted by neutralizing antibodies after vaccination.
To more specifically analyze antibody profiles related to neutralizing capacity, the reactivities of sera to known neutralizing targets A13, A17, D8, H3, A27, and B5 and protective targets A33, A4, and A10 were evaluated (Fig. 2a). Since neutralizing capacity has been shown to be redundant (39), reactivities to these eight proteins were combined to determine if a minimum number of recognition events was associated with a neutralizing titer. Neutralizing target L1 was excluded from this array, as it failed to be reactive when produced from this E. coli system (45), and A28 (36), although included on the array, was excluded from the analysis as response was absent in all but one vaccinee. In all groups, postvaccination data showed increased reaction to this subset of proteins. Higher reactivity in prevaccination responses for humans was due to background binding to A4. Additionally, since prairie dog Imvamune prevaccination sera were unavailable for testing, higher initial reactivity was due to increased recognition of D8 following the first dosing of Imvamune, which is presented here as prevaccination. The lower overall response postvaccination in prairie dogs was in part due to the absence of reactivity to A17, an observation shown in Fig. 1. While humans responded strongly to A17 after live vaccination, prairie dogs typically failed to mount a response to the protein following live or attenuated vaccination. No statistical difference in the total number of responses was observed between human vaccinees and prairie dogs given Dryvax or Imvamune (P = 0.56 to 1; Wilcoxon rank sum). However, the total number of known neutralizing targets recognized in prairie dogs given Acam2000 was statistically lower than in Dryvax- or Imvamune-vaccinated animals (P = 0.04 and 0.03, respectively; Wilcoxon rank sum test). Sera were also evaluated using the HCS-GFP neutralization assay (49) to observe the NAb titer (Fig. 2b). Neutralization increased postvaccination, with NAb titers (50% RPR titers) of 4.8 × 102 and 4.2 × 102 observed in Dryvax- and Acam2000-vaccinated humans. Postvaccination NAb titers for each prairie dog group were 3.4 ×102, 1.9 × 102, and 7.3 × 102, respectively. Insufficient prevaccination sera were available for prairie dog vaccinees for HCS-GFP testing; however, we recently showed that NAb titers of unvaccinated prairie dogs were typically below 1.0 × 102 (25) and thus were comparable to human prevaccination values. No correlation was seen when comparing individual vaccinee responses to known neutralizing antibody targets and NAb titers (data not shown), and thus, the reduced number of neutralizing targets in Acam2000-vaccinated prairie dogs was not biologically significant.
Fig 2.
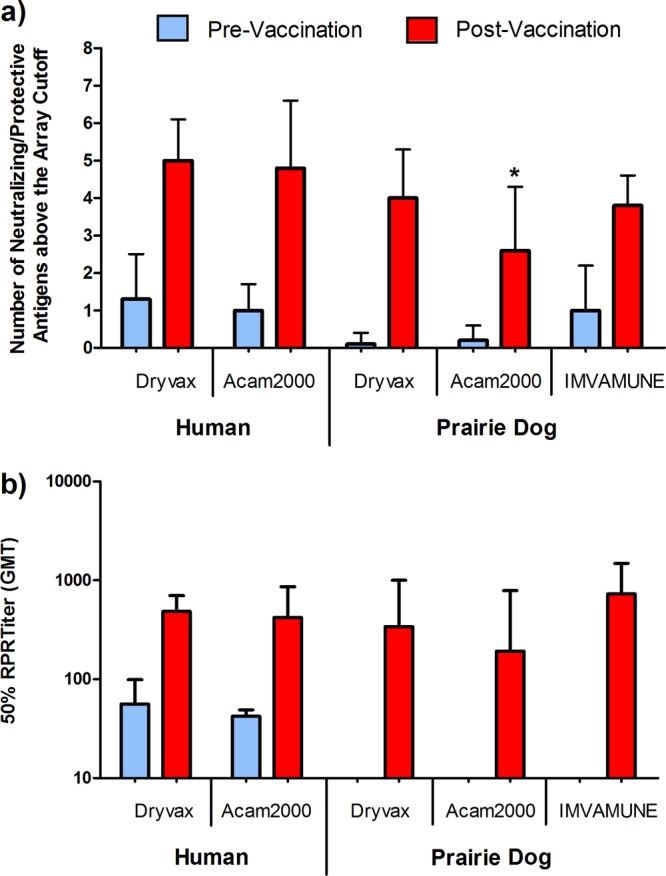
Antibody response to known neutralizing or protective antigenic targets and induction of neutralizing antibodies after Dryvax, Acam2000, or two-dose Imvamune vaccination. (a) Fluorescence intensities above the microarray cutoff for human and prairie dog sera in response to eight known neutralizing targets, H3, D8, A17, A27, and B5 and protective targets A33, A4, and A10, were scored positive or negative, counted for each vaccinee, and averaged (Wilcoxon rank sum; *, P < 0.05). (b) Neutralizing antibody titers for vaccinees were evaluated by an HCS-GFP neutralization assay and are displayed as the 50% RPR titer (50% neutralization). Human sera were not heat inactivated prior to analysis. Insufficient quantities of prevaccination sera were available for prairie dog NAb testing with the HCS-GFP assay. Time points for both assays are as shown in Fig. 1 and range from 21 to 49 (median = 30) days postvaccination. The error bars indicate standard deviation (SD).
Temporal evaluation of antibody responses after vaccination and comparison with ELISA responses.
We next looked at the dynamics of the antibody responses over a 32-day period by protein microarray and ELISA endpoint titer (EPT). Since ELISA is the study's gold standard for confirming antibody reactivity to MV protein, our array results should produce similar responses and show increasing response to those antigens over time. Six prairie dogs and six humans were evaluated at approximately 7-day intervals post-Dryvax or -Acam2000 vaccination. Representative individual responses for one Dryvax-vaccinated human and one Acam2000-vaccinated prairie dog (Fig. 3) and aggregate results for all time-course analyzed vaccinees (Fig. 4) showed correlation between the two assays. In observing individual responses (Fig. 3), ELISA EPTs typically rose by day 14 in both species. Human EPTs then showed modest increases at later time points; however, prairie dogs typically showed no further increases. In both species, the fluorescence responses to individual proteins on our array were predominantly below the cutoff initially and steadily increased at later time points. As previously noted in Fig. 1, WR148, A13, H3, and D8 were among the most antigenic proteins. Response to A17 was seen in humans but remained below the cutoff in prairie dogs. To compare the microarray and ELISAs, results for all time points were combined and displayed graphically (Fig. 4). Microarray fluorescence with human samples was better correlated with the ELISA EPT than that with prairie dogs, although both species showed increases in fluorescence associated with higher titers. Human samples reached a maximal ELISA titer of 103, whereas prairie dog sera, on average, could be diluted to 104.
Fig 3.
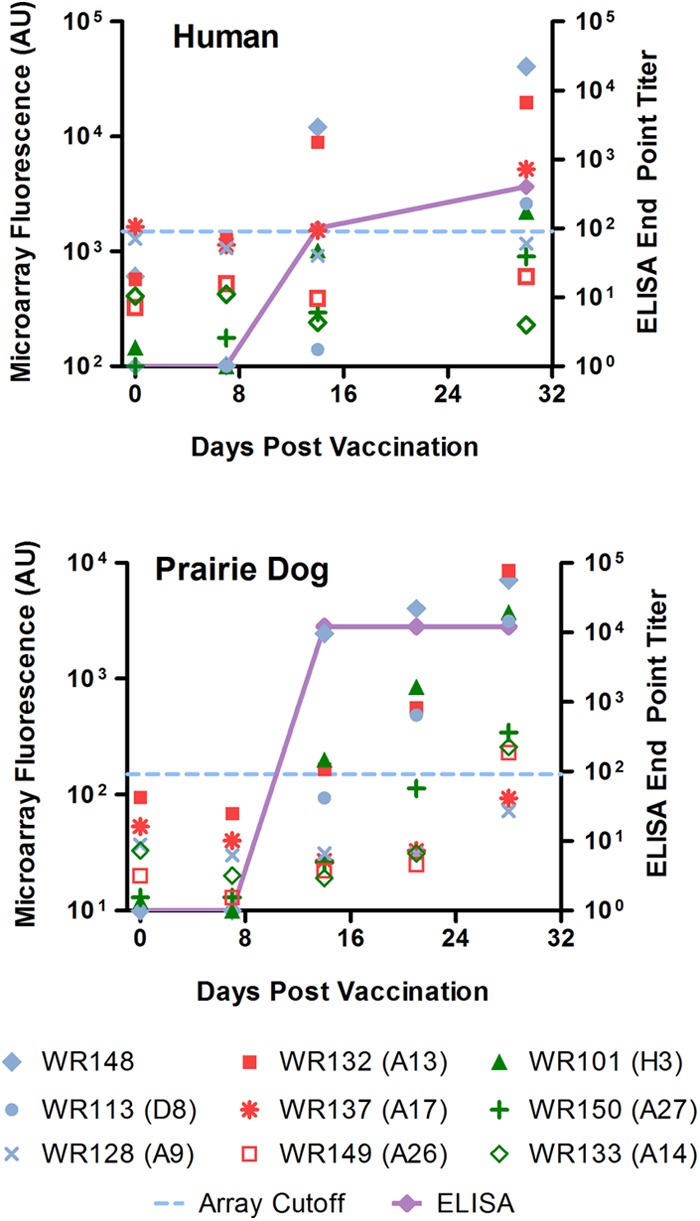
Kinetic view of responses to MV proteins at days 0, 7, 14, 21, and 28 or 30 and correlation with ELISA results; shown are data for an Acam200-vaccinated human and a Dryvax-vaccinated prairie dog. Raw microarray signal intensities for select MV surface proteins are plotted, along with the array cutoff (dashed blue lines) and endpoint titer ELISA values (solid purple lines). The data are typical for each species regardless of the vaccination strain.
Fig 4.
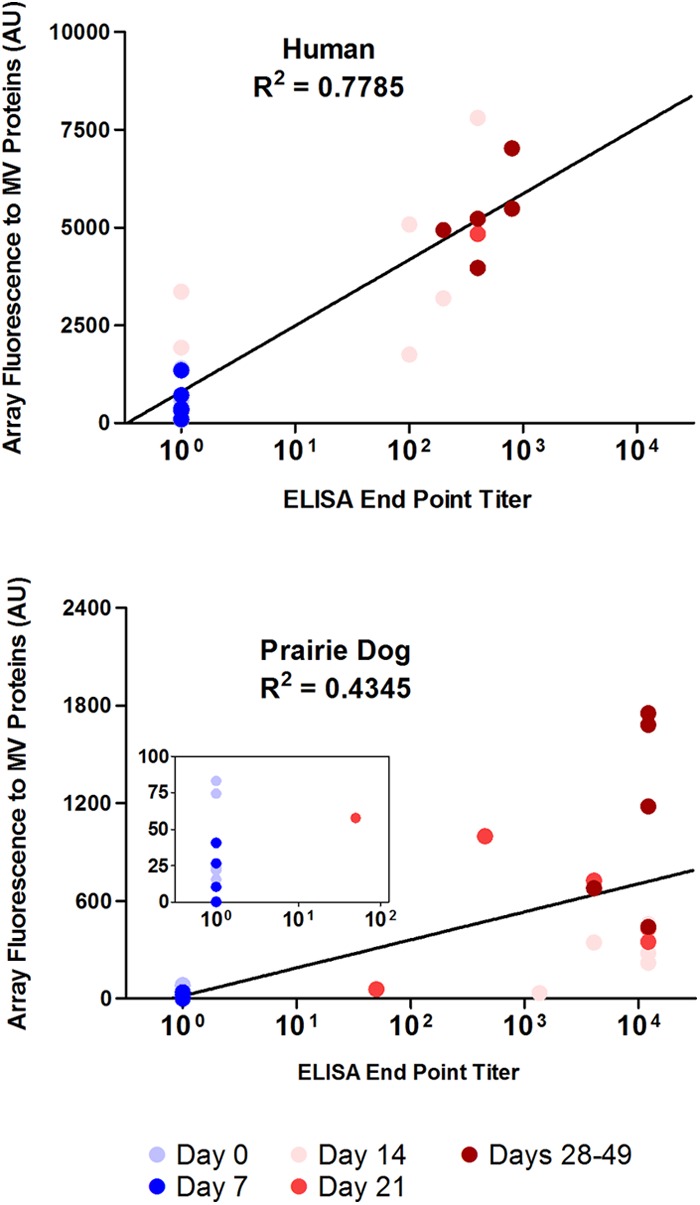
Microarray fluorescence in response to MV proteins correlates with the ELISA endpoint antibody titer. The intensities for all MV proteins present on the array were analyzed at days 0, 7, 14, and 21 to 49 after Dryvax or Acam2000 vaccination of humans (n = 6) and prairie dogs (n = 6) and compared with the corresponding ELISA titers. The data points are shown in color and represent the date of sampling, i.e., days 0, 7, 14, 21, and 29 to 49. The inset in the prairie dog graph shows a close-up view of the data with a 100 ELISA endpoint titer. Data for all time points were not available for all individuals.
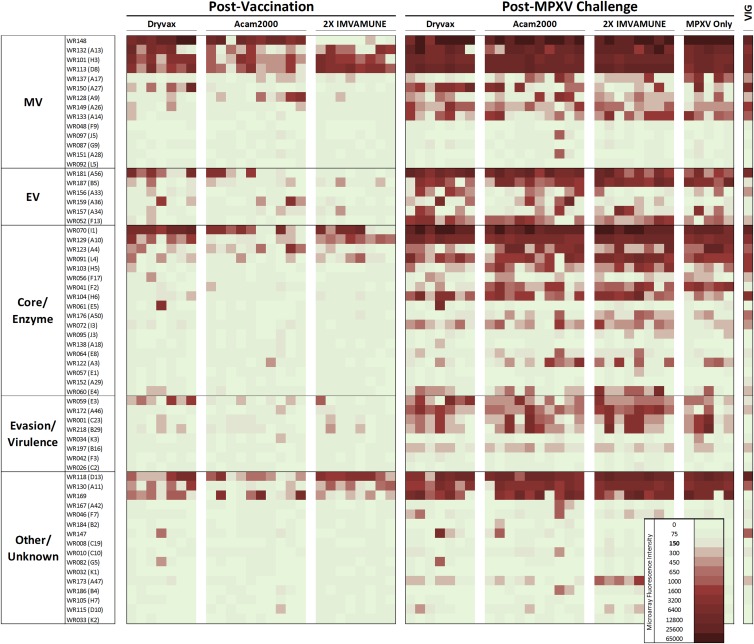
Immune response profiles in monkeypox virus-challenged vaccinated prairie dogs.
To determine how antibody profiles were modified upon MPXV challenge, Dryvax-, Acam2000-, and Imvamune-vaccinated prairie dogs were challenged with monkeypox virus 30 days postvaccination (60 days post-first-dose Imvamune vaccination). At 24 to 30 days post-MPXV challenge, overall reactivity to the proteins was boosted compared to prechallenge levels; antibodies to commonly recognized antigens postvaccination became universally recognized and produced higher-intensity array signals (Fig. 5). Among these were MV proteins WR148 (except in Imvamune-vaccinated animals), A13, H3, and D8; core proteins I1 and A10; and virion scaffold protein (“other”) D13 (Table 3). MPXV challenge also induced strong primary antibody response to proteins that were minimally or not recognized after vaccination (Table 3, underlined proteins), i.e., MV proteins A17, A27, A26, and A14; EV proteins A56, B5, and F13; core proteins A4, H5, and I3; enzymatic proteins L4, F2, H6, and E4; immunomodulatory proteins E3, A46, C23, and B29; virion assembly protein A11 (other); and WR169, whose function is unknown. Of the antigens commonly recognized postchallenge, four were rarely seen postvaccination in prairie dogs and only minimally reactive to human VIG. Proteins F2 and A3 (core/enzyme) were most unique, as they were each reactive only once postvaccination but were recognized by 73% and 42% of subjects once challenged (Fig. 5). The immunomodulatory protein C23 and its gene duplicate B29 (evasion/virulence) were also unrecognized by VIG and initially recognized in only 8% and 13% of vaccinees, respectively, but were reactive in 50% and 55% of samples postchallenge. Three additional proteins also rarely reactive in vaccinated prairie dogs are DNA binding protein I3 (core/enzyme), phospholipase protein F13 (EV), and interleukin 1 (IL-1) inhibitor A46 (evasion/virulence). Postchallenge responses for these proteins ranged from 72 to 100%. Although I3, F13, and A46 were reactive with VIG, the fluorescence intensities were each less than 2 times the positive cutoff value.
Fig 5.
Heat map of reactivities to microarray proteins of prairie dog sera postvaccination and post-MPXV challenge. The animals were vaccinated with Dryvax, Acam2000, or Imvamune; allowed to seroconvert; and then challenged with MPXV. The animals providing MPXV-only samples were not vaccinated. The samples were collected between 27 and 30 days postchallenge and had prevaccination reactivity subtracted. VIG is shown for visual comparison with the human response to vaccination.
Table 3.
Changes in antibody targets in vaccinated prairie dogs after challenge with MPXVa
| Protein category | Protein |
|
|---|---|---|
| Prechallenge | Postchallengeb | |
| MV | WR148c, A13, H3, D8 | WR148, A13, H3, D8, A17, A27, A26, A14 |
| EV | None | A56, B5R, F13 |
| Core/enzyme | I1L, A10L | I1, A10, A4, H5, I3, L4, F2, H6, E4 |
| Evasion/virulence | None | E3, A46, C23, B29 |
| Other/unknown | D13L | D13, A11, WR169 |
Reactive in greater than 50% of subjects.
Underlined proteins were not significantly recognized postvaccination.
Absent in Imvamune- but dominant in Dryvax- and Acam2000-vaccinated prairie dogs.
DISCUSSION
Validation of animal models for use as surrogates for vaccination studies is of paramount interest when human experimentation is not feasible. Monkeypox virus infection of prairie dogs produces disease that is highly similar to that in humans after systemic OPV infection, and protection from disease occurs when animals are vaccinated prior to challenge with monkeypox virus (24, 25). Additionally, since vaccination in humans produces an antibody response correlated with protection, a highly similar response in prairie dogs would further support their use as a surrogate animal model of OPV vaccination. This in turn would allow testing of additional smallpox vaccines in this model for efficacy against systemic OPV infections. Here, we used a protein microarray spotted with select VACV WR proteins and compared the antibody response profiles from VACV-vaccinated humans and prairie dogs. The profiles were highly heterogeneous, with one or more individuals in each group reacting to 15 of the 61 proteins on the chip, yet also contained strong and consistent reactions to several immunodominant and known neutralizing proteins. We further characterized the immune response after MPXV challenge of naïve and vaccinated prairie dogs. Upon challenge, the response profiles became much more homogeneous, with an average of 27.4 antigens recognized by each prairie dog and 21 of those antigens recognized by more than 75% of all challenged animals.
One concern when using nonclonal and unpurified proteins was the potential for background reactivity due to aberrant protein production or the complexity of the crude mixture spotted. A high degree of correlation between our results and those of others using quantified or purified protein arrays would indicate that this is not an issue. In one of the more complete efforts to date, 193 of 273 vaccinia virus Copenhagen strain proteins were expressed and purified from a baculovirus protein system; it was found that <10% of the proteome was reactive to VIG or vaccinated individuals (42). Using a rabbit reticulocyte system, Duke-Cohan et al. (41) selected 25 surface-exposed or known antigenic proteins of the 218 ORFs of the WR proteome to produce proteins in a micro-ELISA format. Reactivity was found to 17 proteins, although only 10 were strongly reactive. The correlations of vaccinated individuals and VIG profiles were similar in both sets of experiments. In our experiments, 13 proteins were recognized by more than one vaccinee, and 26 proteins were recognized by VIG. Overall, among all sets of results, there was good concordance when looking at strongly reactive proteins, such as D13, H3, D8, I1, A33, A27, and B5. When comparing less-reactive proteins, although there was variability as to which individual proteins were recognized, our experimental method did not produce significantly different hits than either the Schmid or Duke-Cohan data sets. A comparative table of reactivity observed by each system can be found in Table S2 in the supplemental material.
Our results show significant reactivity to membrane proteins in vaccinated humans and prairie dogs, with WR148, A13, H3, D8, and A17 commonly seen. The last four MV proteins have all been shown to be effective targets for virus neutralization and thus critical targets of the immune system to inactivate and limit virus spread (50). However, A17 was recognized less frequently in prairie dogs, with only 6 of 34 animals responding to it, and four of those were in the Acam2000 group. A17 is an MV protein that is associated with viral morphogenesis and is essential for replication (51). The N-terminal region has been identified as the target of neutralizing antibodies, as well as the region that interacts with A14 and A27 (52). As such, reduced reactivity may be indicative of subtle changes that limit the accessibility of the external N-terminal region, or the region may simply be less immunogenic in the prairie dog. A fifth MV neutralizing target, A27, was seen in nearly 50% of Dryvax-vaccinated individuals of both species but failed to react with any Acam2000 or Imvamune vaccinee. Protein L1 was excluded from the chip, even though it has historically been shown to produce neutralizing antibodies (53, 54). At the time of writing, L1 failed to produce a response in human sera and was only minimally reactive in concentrations higher than normal (data not shown). L1 is notable for significant disulfide bonding; thus, the limitation is likely structural, and some gains have since been realized (44). With baculovirus expression, detection ranged from 0 to 30% recognition (42, 50), and in a rabbit reticulocyte lysate expression system, recognition was <25%, although a strong correlation with neutralization in 4 samples with anti-L1 antibodies was noted (41).
There were fewer and more varied responses to EV proteins after vaccination regimens in both species with serorecognition of A33 and sometimes B5, as indicated by low-level but significant fluorescence intensity in humans and serorecognition of A56 and B5 in prairie dogs. Interestingly, upon MPXV challenge, prairie dogs showed nearly universal responses to A56, B5, and F13 and lower responses to A34 and A36. This finding is interesting, since EV virions are thought to aid in virus dissemination in natural infection (55) and typical vaccine strains produce only a single localized lesion. However, it is unclear whether this difference in reactivity to EV proteins is due to limited spread from the vaccination site or simply due to less immunologic insult than the systemic spread commonly found with monkeypox virus infection. It is also possible that since EV proteins would likely be posttranslationally modified, this low reactivity is an artifact of the E. coli system. However, a low frequency of reactivity to EV proteins after immunization was also noted in similar assays using different expression systems (41, 42). Reactivity to EV proteins using single-antigen ELISA showed increases in response after vaccination (56), but the absence of individual pre- and postvaccination sera makes comparison difficult. It is also notable that while very few sera were reactive with multiple EV proteins, 8 of 15 human vaccinees and 20 of 34 prairie dogs were reactive to at least one EV protein, and over 40% of all vaccinees were reactive to two or more EV proteins. This variability at the whole-protein level correlates with recent studies of antibody response after smallpox vaccination, where redundant neutralizing antibodies targeted a multitude of viral epitopes as opposed to a single immunodominant epitope, and shows that the specificities of the response may differ substantially by individual (39).
Antibody responses to core proteins after vaccination were highly similar between virus strains and species. As core proteins, the accessibility of each would be limited until lysis of infected cells and then would be expected to be somewhat correlated with their relative abundances and the availability of immunogenic epitopes. Reports of relative abundances in MV virions (57, 58) showed that A4 and A10 were predominant proteins, with I1 representing as little as 1/100 of their total mass. This relationship of antigen mass to antibody recognition was reversed here, as I1 was immunodominant.
The recognition of E3 by Dryvax-vaccinated prairie dogs, but not humans, was also interesting. The protein was recognized only weakly in one human vaccinee and was previously seen with equivocal recognition by humans and no recognition by primate vaccinees (40, 59). In comparison, strong recognition of E3 was seen in mice and rabbits given the pathogenic VACV WR strain (40, 45), and primates challenged with MPXV show reactivity to E3, and in particular the truncated MPXV homologue of E3 (59). Similar to the absence of reactivity to A17, reactivity to E3 after vaccination may result from species-specific sensitivity to available epitopes, and only upon challenge (VACV WR or MPXV) is the response sufficient to ensure that it is measurable. It is also possible that stronger recognition is at least partially a function of the pathogenicity of the challenge virus or the degree of systemic spread of virus during disease.
There were three commonly recognized proteins after vaccination that did not fit into one of the above-mentioned groups. D13 is a scaffold protein that is lost prior to MV maturation (60). However, its necessity as a structural protein in crescent formation and its ubiquity confirm it as a likely immunologic target. A11 is a nonstructural protein also associated with crescent formation (61). Its near absence in Acam2000-vaccinated prairie dogs was somewhat surprising but, upon examination of signal intensities, may simply be a function of low levels of response making it indistinguishable from background. While A11 was recognized in all other vaccination groups, it typically had an average intensity of only 2 to 3 times the cutoff value and for all groups was in the bottom third of reactive proteins by ranked average intensity.
Consistent with previous reports of vaccine immunogenicity (62), we saw no difference between Dryvax and Acam2000 array responses in humans, with an average of 10.0 and 9.3 array hits per sample, respectively. However, in prairie dogs, there was a trend of lower response after Acam2000 than after Dryvax, including many instances where Dryvax vaccinees responded to a given protein 100% of the time whereas Acam2000 vaccinees were less frequently recognized but still greater than the 50% designation shown in Table 2. A similar overall reduction was observed after Imvamune vaccination, as well, only part of which can be accounted for by the loss of reactivity to WR148. Relative to Dryvax, reduced antibody response has also been seen in Imvamune vaccination of humans (63) and included absent or reduced reactivity to WR148 and A17 but also significantly lower responses to A56, A27, and A33. We observed no reactivity to each of these antigenic targets in Imvamune-vaccinated prairie dogs, as well. The lower overall response between viruses resulted in a drop in the number of recognized antigens from an average of 11.7 hits per sample after Dryvax vaccination to 7.8 and 7.9 hits per sample after Acam2000 or Imvamune vaccination. This difference accounts for an ∼15% loss in positive hits over the entire proteome between the vaccines. ELISA geometric mean titers (GMTs) also showed this reduced response, with both Acam2000 and Imvamune having between 2- and 10-fold-reduced peak GMTs relative to Dryvax.
After MPXV challenge, the response to a much greater number of antigens (n = 27.4) than after vaccination (n = 9.1) may reflect the more systemic presentation of the virus. In Dryvax revaccination cases, only a modest increase in the number of recognized targets has been seen (40). We also recently showed that disease severity, as judged by weight loss, lesion count, and nasal involvement, was significantly limited upon challenge after vaccination, particularly after Dryvax or Acam2000 vaccination (25). Although the limited number of immunodominant antigens after vaccination may have been expected to continue dominating the profile, along with the addition of a few novel targets that may have been recognized during vaccination but failed to generate a detectable antibody response, the large increase here suggests that differences in virus pathogenicities or life cycles affect immune recognition. It was observed that pathogenic WR virus infection of naïve rabbits induced a large and robust response to a broad range of proteins compared to vaccination alone (44), and a recent report showed that the antibody response to monkeypox virus infection of naïve macaques also targeted a larger number of antigens than human vaccination (59). Thus, dramatic increases in the frequency and intensity of responses to multiple targets upon challenge, but not repeat vaccination, provides evidence that increased pathogenicity drives the stronger immune response. Added support for pathogenicity affecting immune response is shown in the shift from intermediate- and late-promoter genes (64–66), which are typical of structural virion core and membrane proteins and are present on the surfaces of infected cells (67), to early-promoter genes. In the present study, all seven antigens reactive in greater than 50% of vaccinated prairie dogs were intermediate- or late-promoter driven. Of 27 commonly reactive antigens after challenge, 13 were classified as intermediate or late promoter, but 6 early/immediate-early and 8 early/late promoters were seen. One final piece of evidence supporting virus pathogenesis driving the response to novel antigens includes mounting of a secondary IgM response noted in vaccinated individuals during the 2003 monkeypox outbreak in the United States (48). The IgM assay utilized whole VACV virions (Dryvax) to assess post-monkeypox IgM induction, and as such, a response would indicate new immune priming by previously unrecognized proteins and epitopes after vaccination. An increase in pathogenicity between highly similar yet different viruses, such as VACV and then MPXV, would allow for the large number of additional antigenic targets to highly homologous proteins recognized here and the observed secondary IgM response. One additional correlative observation was the relationship between antigens recognized by proteome analysis and disease severity upon challenge of the vaccinated animals. Prairie dogs given Imvamune responded to fewer antigens by our analysis and showed some signs of morbidity upon challenge (25). No morbidity was observed when challenging Dryvax-vaccinated animals. Although there were fewer targets overall, there was no consistently absent antibody target that correlated with protective or known neutralizing targets, nor were there statistically significant differences in the average numbers of neutralizing targets recognized. However, we also saw fewer responses during Acam2000 vaccination yet no increase in morbidity after challenge, suggesting that multiple factors are involved in immunogenicity and protection. It would also be of interest to gain greater insight into the full spectrum of antibody responses before and after challenge using an MPXV proteomic approach. Keasey et al. observed greater increases in antibody responses to several MPXV proteins than to VACV proteins by aerosol MPXV-challenged macaques (59), indicating that although broad cross-reactivity exists, a stronger response may be directed toward MPXV-specific proteins. Differences in neutralization titers have also been observed, depending on the strain of OPV utilized in the PRNT assays (68, 69). While a direct comparison between titration methods is not possible using the vaccinia virus-based HCS-GFP assay, future experiments that shed light on the virus specificity of this broader response and the virus-specific neutralization capacity after vaccination and challenge would be of interest.
In conclusion, we have shown that the antibody response in prairie dog vaccination closely resembles that in human vaccination when examined at the whole-proteome level. While some differences were observed, including variability in the recognition of proteins A17, A9, A56, E3, and WR169 after vaccination, a clear set of proteins recognized by all vaccinees in both species remained. Of the proteins known to be targeted by neutralizing antibodies, only the absence of A17 recognition in prairie dogs was observed. However, there was no significant difference in neutralizing capacity, as demonstrated by similar 50% RPR titers in our HSC-GFP assay, suggesting this loss of reactivity alone is not critical in protection, nor is it a salient difference in the animal model system. We observed fewer responses after vaccination with the attenuated Imvamune strain, which correlated with increases in morbidity when the prairie dogs were challenged with MPXV. We have also shown that challenge with pathogenic MPXV induced a larger-magnitude response to a greater number of targets than vaccination, regardless of prechallenge status (i.e., vaccinated or naïve). This dramatic increase in the frequency of antigen recognition after challenge may be relevant as safer, and likely more attenuated, vaccines are developed. The increase in the breadth of response after challenge also supports observations of lifelong immunity following smallpox but allows for wide variations in waning immunity years after vaccination (70, 71).
Supplementary Material
ACKNOWLEDGMENTS
We thank Gregory Langham, Pauline Cates, and Christopher Meshida of the Lawrenceville Animal Resources Branch (ARB) and Allison Williams, Brock Martin, and Eddie Jackson of the Roybal ARB for animal husbandry. We acknowledge Jason Abel for human serum collection, Erin McDowell for ELISA processing, Matthew Johnson for HCS-GFP neutralization assay development and sample processing, and Christine Hughes for statistical analysis. We also thank Douglas M. Molina for microarray printing.
This work was supported by program funds from the Centers for Disease Control and Prevention.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
Footnotes
Published ahead of print 7 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02089-12.
REFERENCES
- 1. Breman JG, Arita I. 1980. The confirmation and maintenance of smallpox eradication. N. Engl. J. Med. 303:1263–1273 [DOI] [PubMed] [Google Scholar]
- 2. Chapman JL, Nichols DK, Martinez MJ, Raymond JW. 2010. Animal models of orthopoxvirus infection. Vet. Pathol. 47:852–870 [DOI] [PubMed] [Google Scholar]
- 3. Trindade GS, Emerson GL, Carroll DS, Kroon EG, Damon IK. 2007. Brazilian vaccinia viruses and their origins. Emerg. Infect. Dis. 13:965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vorou RM, Papavassiliou VG, Pierroutsakos IN. 2008. Cowpox virus infection: an emerging health threat. Curr. Opin. Infect. Dis. 21:153–156 [DOI] [PubMed] [Google Scholar]
- 5. Rimoin AW, Mulembakani PM, Johnston SC, Lloyd Smith JO, Kisalu NK, Kinkela TL, Blumberg S, Thomassen HA, Pike BL, Fair JN, Wolfe ND, Shongo RL, Graham BS, Formenty P, Okitolonda E, Hensley LE, Meyer H, Wright LL, Muyembe JJ. 2010. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U. S. A. 107:16262–16267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jacobs BL, Langland JO, Kibler KV, Denzler KL, White SD, Holechek SA, Wong S, Huynh T, Baskin CR. 2009. Vaccinia virus vaccines: past, present and future. Antivir. Res. 84:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention 2003. Supplemental recommendations on adverse events following smallpox vaccine in the pre-event vaccination program: recommendations of the Advisory Committee on Immunization Practices. JAMA 289:2064. [DOI] [PubMed] [Google Scholar]
- 8. Cassimatis DC, Atwood JE, Engler RM, Linz PE, Grabenstein JD, Vernalis MN. 2004. Smallpox vaccination and myopericarditis: a clinical review. J. Am. Coll. Cardiol. 43:1503–1510 [DOI] [PubMed] [Google Scholar]
- 9. Lane JM, Ruben FL, Neff JM, Millar JD. 1969. Complications of smallpox vaccination, 1968. N. Engl. J. Med. 281:1201–1208 [DOI] [PubMed] [Google Scholar]
- 10. Monath TP, Caldwell JR, Mundt W, Fusco J, Johnson CS, Buller M, Liu J, Gardner B, Downing G, Blum PS, Kemp T, Nichols R, Weltzin R. 2004. ACAM2000 clonal Vero cell culture vaccinia virus (New York City Board of Health strain)—a second-generation smallpox vaccine for biological defense. Int. J. Infect. Dis. 8(Suppl. 2):S31–S44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vollmar J, Arndtz N, Eckl KM, Thomsen T, Petzold B, Mateo L, Schlereth B, Handley A, King L, Hulsemann V, Tzatzaris M, Merkl K, Wulff N, Chaplin P. 2006. Safety and immunogenicity of IMVAMUNE, a promising candidate as a third generation smallpox vaccine. Vaccine 24:2065–2070 [DOI] [PubMed] [Google Scholar]
- 12. Midgley CM, Putz MM, Weber JN, Smith GL. 2008. Vaccinia virus strain NYVAC induces substantially lower and qualitatively different human antibody responses compared with strains Lister and Dryvax. J. Gen. Virol. 89:2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edghill-Smith Y, Bray M, Whitehouse CA, Miller D, Mucker E, Manischewitz J, King LR, Robert-Guroff M, Hryniewicz A, Venzon D, Meseda C, Weir J, Nalca A, Livingston V, Wells J, Lewis MG, Huggins J, Zwiers SH, Golding H, Franchini G. 2005. Smallpox vaccine does not protect macaques with AIDS from a lethal monkeypox virus challenge. J. Infect. Dis. 191:372–381 [DOI] [PubMed] [Google Scholar]
- 14. Grandpre LE, Duke-Cohan JS, Ewald BA, Devoy C, Barouch DH, Letvin NL, Reinherz EL, Baden LR, Dolin R, Seaman MS. 2009. Immunogenicity of recombinant modified vaccinia ankara following a single or multi-dose vaccine regimen in rhesus monkeys. Vaccine 27:1549–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Riedmann EM. May 2010. FDA Fast Track status for IMVAMUNE. Hum. Vaccin. 6:368–372 http://dx.doi.org/10.4161/hv.6.5.12214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jahrling PB, Hensley LE, Martinez MJ, Leduc JW, Rubins KH, Relman DA, Huggins JW. 2004. Exploring the potential of variola virus infection of cynomolgus macaques as a model for human smallpox. Proc. Natl. Acad. Sci. U. S. A. 101:15196–15200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Earl PL, Americo JL, Wyatt LS, Eller LA, Whitbeck JC, Cohen GH, Eisenberg RJ, Hartmann CJ, Jackson DL, Kulesh DA, Martinez MJ, Miller DM, Mucker EM, Shamblin JD, Zwiers SH, Huggins JW, Jahrling PB, Moss B. 2004. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature 428:182–185 [DOI] [PubMed] [Google Scholar]
- 18. Zaucha GM, Jahrling PB, Geisbert TW, Swearengen JR, Hensley L. 2001. The pathology of experimental aerosolized monkeypox virus infection in cynomolgus monkeys (Macaca fascicularis). Lab. Invest. 81:1581–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karupiah G, Buller RM, Van Rooijen N, Duarte CJ, Chen J. 1996. Different roles for CD4+ and CD8+ T lymphocytes and macrophage subsets in the control of a generalized virus infection. J. Virol. 70:8301–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martinez MJ, Bray MP, Huggins JW. 2000. A mouse model of aerosol-transmitted orthopoxviral disease: morphology of experimental aerosol-transmitted orthopoxviral disease in a cowpox virus-BALB/c mouse system. Arch. Pathol. Lab. Med. 124:362–377 [DOI] [PubMed] [Google Scholar]
- 21. Schriewer J, Buller RM, Owens G. 2004. Mouse models for studying orthopoxvirus respiratory infections. Methods Mol. Biol. 269:289–308 [DOI] [PubMed] [Google Scholar]
- 22. Adams MM, Rice AD, Moyer RW. 2007. Rabbitpox virus and vaccinia virus infection of rabbits as a model for human smallpox. J. Virol. 81:11084–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reed KD, Melski JW, Graham MB, Regnery RL, Sotir MJ, Wegner MV, Kazmierczak JJ, Stratman EJ, Li Y, Fairley JA, Swain GR, Olson VA, Sargent EK, Kehl SC, Frace MA, Kline R, Foldy SL, Davis JP, Damon IK. 2004. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350:342–350 [DOI] [PubMed] [Google Scholar]
- 24. Hutson CL, Olson VA, Carroll DS, Abel JA, Hughes CM, Braden ZH, Weiss S, Self J, Osorio JE, Hudson PN, Dillon M, Karem KL, Damon IK, Regnery RL. 2009. A prairie dog animal model of systemic orthopoxvirus disease using West African and Congo Basin strains of monkeypox virus. J. Gen. Virol. 90:323–333 [DOI] [PubMed] [Google Scholar]
- 25. Keckler MS, Carroll DS, Gallardo-Romero NF, Lash RR, Salzer JS, Weiss SL, Patel N, Clemmons CJ, Smith SK, Hutson CL, Karem KL, Damon IK. 2011. Establishment of the black-tailed prairie dog (Cynomys ludovicianus) as a novel animal model for comparing smallpox vaccines administered preexposure in both high- and low-dose monkeypox virus challenges. J. Virol. 85:7683–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Belyakov IM, Earl P, Dzutsev A, Kuznetsov VA, Lemon M, Wyatt LS, Snyder JT, Ahlers JD, Franchini G, Moss B, Berzofsky JA. 2003. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. Proc. Natl. Acad. Sci. U. S. A. 100:9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu L, Zhong Q, Tian T, Dubin K, Athale SK, Kupper TS. 2010. Epidermal injury and infection during poxvirus immunization is crucial for the generation of highly protective T cell-mediated immunity. Nat. Med. 16:224–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Panchanathan V, Chaudhri G, Karupiah G. 2008. Correlates of protective immunity in poxvirus infection: where does antibody stand? Immunol. Cell Biol. 86:80–86 [DOI] [PubMed] [Google Scholar]
- 29. Panchanathan V, Chaudhri G, Karupiah G. 2006. Protective immunity against secondary poxvirus infection is dependent on antibody but not on CD4 or CD8 T-cell function. J. Virol. 80:6333–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edghill-Smith Y, Golding H, Manischewitz J, King LR, Scott D, Bray M, Nalca A, Hooper JW, Whitehouse CA, Schmitz JE, Reimann KA, Franchini G. 2005. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat. Med. 11:740–747 [DOI] [PubMed] [Google Scholar]
- 31. Davies DH, McCausland MM, Valdez C, Huynh D, Hernandez JE, Mu Y, Hirst S, Villarreal L, Felgner PL, Crotty S. 2005. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 79:11724–11733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fogg CN, Americo JL, Earl PL, Resch W, Aldaz-Carroll L, Eisenberg RJ, Cohen GH, Moss B. 2008. Disparity between levels of in vitro neutralization of vaccinia virus by antibody to the A27 protein and protection of mice against intranasal challenge. J. Virol. 82:8022–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heraud JM, Edghill-Smith Y, Ayala V, Kalisz I, Parrino J, Kalyanaraman VS, Manischewitz J, King LR, Hryniewicz A, Trindade CJ, Hassett M, Tsai WP, Venzon D, Nalca A, Vaccari M, Silvera P, Bray M, Graham BS, Golding H, Hooper JW, Franchini G. 2006. Subunit recombinant vaccine protects against monkeypox. J. Immunol. 177:2552–2564 [DOI] [PubMed] [Google Scholar]
- 34. Hooper JW, Ferro AM, Golden JW, Silvera P, Dudek J, Alterson K, Custer M, Rivers B, Morris J, Owens G, Smith JF, Kamrud KI. 2009. Molecular smallpox vaccine delivered by alphavirus replicons elicits protective immunity in mice and non-human primates. Vaccine 28:494–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaufman DR, Goudsmit J, Holterman L, Ewald BA, Denholtz M, Devoy C, Giri A, Grandpre LE, Heraud JM, Franchini G, Seaman MS, Havenga MJ, Barouch DH. 2008. Differential antigen requirements for protection against systemic and intranasal vaccinia virus challenges in mice. J. Virol. 82:6829–6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shinoda K, Wyatt LS, Moss B. 2010. The neutralizing antibody response to the vaccinia virus A28 protein is specifically enhanced by its association with the H2 protein. Virology 405:41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wallengren K, Risco C, Krijnse-Locker J, Esteban M, Rodriguez D. 2001. The A17L gene product of vaccinia virus is exposed on the surface of IMV. Virology 290:143–152 [DOI] [PubMed] [Google Scholar]
- 38. Xu C, Meng X, Yan B, Crotty S, Deng J, Xiang Y. 2011. An epitope conserved in orthopoxvirus A13 envelope protein is the target of neutralizing and protective antibodies. Virology 418:67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benhnia MR, McCausland MM, Su HP, Singh K, Hoffmann J, Davies DH, Felgner PL, Head S, Sette A, Garboczi DN, Crotty S. 2008. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J. Virol. 82:3751–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, Pablo J, Unal B, Nakajima-Sasaki R, Liang X, Crotty S, Karem KL, Damon IK, Ahmed R, Villarreal L, Felgner PL. 2007. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics 7:1678–1686 [DOI] [PubMed] [Google Scholar]
- 41. Duke-Cohan JS, Wollenick K, Witten EA, Seaman MS, Baden LR, Dolin R, Reinherz EL. 2009. The heterogeneity of human antibody responses to vaccinia virus revealed through use of focused protein arrays. Vaccine 27:1154–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmid K, Keasey SL, Pittman P, Emerson GL, Meegan J, Tikhonov AP, Chen GX, Schweitzer B, Ulrich RG. 2008. Analysis of the human immune response to vaccinia by use of a novel protein microarray suggests that antibodies recognize less than 10% of the total viral proteome. Proteomics Clin. Appl. 2:1528–1538 [DOI] [PubMed] [Google Scholar]
- 43. Flesch IE, Woo WP, Wang Y, Panchanathan V, Wong YC, La Gruta NL, Cukalac T, Tscharke DC. 2010. Altered CD8(+) T cell immunodominance after vaccinia virus infection and the naive repertoire in inbred and F(1) mice. J. Immunol. 184:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davies DH, Wyatt LS, Newman FK, Earl PL, Chun S, Hernandez JE, Molina DM, Hirst S, Moss B, Frey SE, Felgner PL. 2008. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus Ankara is comparable to that of Dryvax. J. Virol. 82:652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. U. S. A. 102:547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Keckler MS, Gallardo-Romero NF, Langham GL, Damon IK, Karem KL, Carroll DS. 2010. Physiologic reference ranges for captive black-tailed prairie dogs (Cynomys ludovicianus). J. Am. Assoc. Lab. Anim. Sci. 49:274–281 [PMC free article] [PubMed] [Google Scholar]
- 47. Cangene 2005. Highlights of prescribing information: CNJ-016, vaccinia immune globulin intravenous (human). Cangene, Winnipeg, Canada [Google Scholar]
- 48. Karem KL, Reynolds M, Braden Z, Lou G, Bernard N, Patton J, Damon IK. 2005. characterization of acute-phase humoral immunity to monkeypox: use of immunoglobulin M enzyme-linked immunosorbent assay for detection of monkeypox infection during the 2003 North American outbreak. Clin. Diagn. Lab. Immunol. 12:867–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johnson MC, Damon IK, Karem KL. 2008. A rapid, high-throughput vaccinia virus neutralization assay for testing smallpox vaccine efficacy based on detection of green fluorescent protein. J. Virol. Methods 150:14–20 [DOI] [PubMed] [Google Scholar]
- 50. Lawrence SJ, Lottenbach KR, Newman FK, Buller RML, Bellone CJ, Chen JJ, Cohen GH, Eisenberg RJ, Belshe RB, Stanley SL, Frey SE. 2007. Antibody responses to vaccinia membrane proteins after smallpox vaccination. J. Infect. Dis. 196:220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wolffe EJ, Moore DM, Peters PJ, Moss B. 1996. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 70:2797–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kochan G, Escors D, Gonzalez JM, Casasnovas JM, Esteban M. 2008. Membrane cell fusion activity of the vaccinia virus A17-A27 protein complex. Cell Microbiol. 10:149–164 [DOI] [PubMed] [Google Scholar]
- 53. Sakhatskyy P, Wang S, Zhang C, Chou TH, Kishko M, Lu S. 2008. Immunogenicity and protection efficacy of subunit-based smallpox vaccines using variola major antigens. Virology 371:98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wolffe EJ, Vijaya S, Moss B. 1995. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology 211:53–63 [DOI] [PubMed] [Google Scholar]
- 55. Payne LG, Kristensson K. 1985. Extracellular release of enveloped vaccinia virus from mouse nasal epithelial cells in vivo. J. Gen. Virol. 66:643–646 [DOI] [PubMed] [Google Scholar]
- 56. Putz MM, Midgley CM, Law M, Smith GL. 2006. Quantification of antibody responses against multiple antigens of the two infectious forms of Vaccinia virus provides a benchmark for smallpox vaccination. Nat. Med. 12:1310–1315 [DOI] [PubMed] [Google Scholar]
- 57. Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80:2127–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Resch W, Hixson KK, Moore RJ, Lipton MS, Moss B. 2007. Protein composition of the vaccinia virus mature virion. Virology 358:233–247 [DOI] [PubMed] [Google Scholar]
- 59. Keasey S, Pugh C, Tikhonov A, Chen G, Schweitzer B, Nalca A, Ulrich RG. 2010. Proteomic basis of the antibody response to monkeypox virus infection examined in cynomolgus macaques and a comparison to human smallpox vaccination. PLoS One 5:e15547 doi:10.1371/journal.pone.0015547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sodeik B, Griffiths G, Ericsson M, Moss B, Doms RW. 1994. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J. Virol. 68:1103–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Condit RC, Moussatche N, Traktman P. 2006. In a nutshell: structure and assembly of the vaccinia virion. Adv. Virus Res. 66:31–124 [DOI] [PubMed] [Google Scholar]
- 62. Handley L, Buller RM, Frey SE, Bellone C, Parker S. 2009. The new ACAM2000 vaccine and other therapies to control orthopoxvirus outbreaks and bioterror attacks. Expert Rev. Vaccines 8:841–850 [DOI] [PubMed] [Google Scholar]
- 63. Hermanson G, Chun S, Felgner J, Tan X, Pablo J, Nakajima-Sasaki R, Molina DM, Felgner PL, Liang X, Davies DH. 2012. Measurement of antibody responses to modified vaccinia virus Ankara (MVA) and Dryvax((R)) using proteome microarrays and development of recombinant protein ELISAs. Vaccine 30:614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. 2008. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc. Natl. Acad. Sci. U. S. A. 105:2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Broyles SS. 2003. Vaccinia virus transcription. J. Gen. Virol. 84:2293–2303 [DOI] [PubMed] [Google Scholar]
- 66. Yang Z, Reynolds SE, Martens CA, Bruno DP, Porcella SF, Moss B. 2011. Expression profiling of the intermediate and late stages of poxvirus replication. J. Virol. 85:9899–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM. 1995. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J. Biol. Chem. 270:15974–15978 [DOI] [PubMed] [Google Scholar]
- 68. Hooper JW, Custer DM, Thompson E. 2003. Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology 306:181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hughes CM, Newman FK, Davidson WB, Olson VA, Smith SK, Holman RC, Yan L, Frey SE, Belshe RB, Karem KL, Damon IK. 2012. Analysis of variola and vaccinia virus neutralization assays for smallpox vaccines. Clin. Vaccine Immunol. 19:1116–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hammarlund E, Lewis MW, Hanifin JM, Mori M, Koudelka CW, Slifka MK. 2010. Antiviral immunity following smallpox virus infection: a case-control study. J. Virol. 84:12754–12760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Karem KL, Reynolds M, Hughes C, Braden Z, Nigam P, Crotty S, Glidewell J, Ahmed R, Amara R, Damon IK. 2007. Monkeypox-induced immunity and failure of childhood smallpox vaccination to provide complete protection. Clin. Vaccine Immunol. 14:1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.