Abstract
The recombinant canarypox vector, ALVAC-HIV, together with human immunodeficiency virus (HIV) gp120 envelope glycoprotein, has protected 31.2% of Thai individuals from HIV acquisition in the RV144 HIV vaccine trial. This outcome was unexpected, given the limited ability of the vaccine components to induce CD8+ T-cell responses or broadly neutralizing antibodies. We vaccinated macaques with an immunization regimen intended to mimic the RV144 trial and exposed them intrarectally to a dose of the simian immunodeficiency virus SIVmac251 that transmits few virus variants, similar to HIV transmission to humans. Vaccination induced anti-envelope antibodies in all vaccinees and CD4+ and CD8+ T-cell responses. Three of the 11 macaques vaccinated with ALVAC-SIV/gp120 were protected from SIVmac251 acquisition, but the result was not significant. The remaining vaccinees were infected and progressed to disease. The magnitudes of vaccine-induced SIVmac251-specific T-cell responses and binding antibodies were not significantly different between protected and infected animals. However, sera from protected animals had higher avidity antibodies to gp120, recognized the variable envelope regions V1/V2, and reduced SIVmac251 infectivity in cells that express high levels of α4β7 integrins, suggesting a functional role of antibodies to V2. The current results emphasize the utility of determining the titer of repeated mucosal challenge in the preclinical evaluation of HIV vaccines.
INTRODUCTION
To date, there have been only four large-scale human immunodeficiency virus (HIV) vaccine efficacy trials (1–4). Of these four trials, only the RV144 trial, the largest HIV vaccine trial so far concluded in humans, showed a limited but significant protection (31.2%) from HIV acquisition (P = 0.04) in the 16,395 participants (4, 5). This result has engendered cautious optimism about the feasibility of a vaccine for HIV. The RV144 trial was conducted in cohorts of Thai men and women primarily at risk for HIV infection via heterosexual exposure. The vaccine regimen included four inoculations of the recombinant avian poxvirus live vector ALVAC-HIV (vCP1521), expressing the Gag-Pro of HIV clade B; a membrane-anchored clade E gp120; and two simultaneous inoculations of the gp120 proteins AIDSVAX B/E, a bivalent recombinant gp120 of clades B and E. The result of the trial was unexpected (6–8), in part because in two prior trials in Thailand and the United States, the AIDSVAX B/E or B/B vaccines alone failed to protect from HIV acquisition (2, 3). The fourth HIV vaccine efficacy study, the STEP trial, tested three inoculations of an adenovirus 5-based vaccine (MRKAd5) containing HIV gag, pol, and nef inserts in multicenter cohorts from North, Central, and South America and Australia (1). Despite the ability of the Ad5-based vaccine platform to elicit stronger T-cell responses than the combination of vCP1521 and AIDVAX, no protection against HIV acquisition was observed.
The mode of HIV transmission and HIV incidence differed among these trials. Heterosexual exposure (female to male) was the predominant mode of transmission in the RV144 cohort, and the HIV incidence was less than one infection per 100 people yearly. In contrast, HIV transmission occurred mostly by sexual exposure among men who have sex with men (MSM) in the STEP trial and the AIDSVAX B/B trial and by needle sharing in the AIDSVAX B/E trial (2, 3). The HIV incidence was between three to four infections per 100 people yearly, in both the STEP and the two AIDSVAX trials (1–3). Thus, whether the nature of the vaccine-elicited immune responses (9) and/or differences in the mode or risk of exposure to HIV account for the differential outcome in these trials remains unclear. The reported efficacy of vaccine modalities, similar to those used in the RV144 trial, varied in different preclinical studies using animals of different ages and viral challenges varying in identity, coreceptor usage, dose, and route (10–18). Recent evidence suggests that the dose of challenge exposure to the CCR5-tropic simian immunodeficiency virus SIVmac251 affects vaccine efficacy: at higher doses of challenge exposure, multiple virus variants were transmitted and vaccine protection was diminished (19) (M. Vaccari, B. F. Keele, S. E. Bosinger, M. N. Doster, J. Zhong-Min Ma Pollara, A. Hryniewicz, G. Ferrari, G. Yongjun, D. N. Forthal, D. Venzon, C. Fenizia, T. Morgan, D. C. Montefiori, J. Lifson, C. Miller, G. Silvestri, M. Rosati, B. K. Felber, G. Pavlakis, J. Tartaglia, G. Franchini, submitted for publication). It is estimated that in humans, the risk of HIV transmission by different exposures ranges between 1:10 and 1:1,000 per encounter (20–23). For most heterosexual transmissions, when infection occurs, a single viral variant, or only few variants, initiate systemic infection (24). Here, we explored the ability of using titers in intrarectal challenge of Indian rhesus macaques with SIVmac251 to model vaccine efficacy observed in humans by using vaccines similar to those used in the AIDSVAX and the RV144 trials and by using a dose of SIVmac251 intended to transmit few viral variants (24). We found that, using these conditions, this macaque model recapitulated the reported efficacy of this vaccine in humans. As in humans, antibody (Ab) to the variable region V2 of the envelope may be important for protection.
MATERIALS AND METHODS
Animals and study design.
We used 34 colony-bred Indian rhesus macaques (Macaca mulatta) obtained from Covance Research Products (Alice, TX). The animals were housed and maintained in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). All rhesus macaques were tested for simian retrovirus, simian T-cell leukemia virus type 1, and herpesvirus B before the study. The major histocompatibility typing for selected alleles was performed as previously described (25) and is depicted in Table 1. Macaques were immunized with ALVAC-SIV and gp120 in alum (11 animals) or with gp120 in alum alone (12 animals). The remaining 11 animals received alum only as a control. Immunization with 108 PFU of ALVAC-SIV Gag, Pol, and Env (16) was performed at 0, 4, 12, and 24 weeks by the intramuscular route in the thigh and the gp120 immunization (200 mg of SIV-gp120 protein [26] formulated in alum) intramuscularly in the contralateral thigh, at weeks 12 and 24. The control group received alum intramuscularly at weeks 12 and 24. All macaques were exposed to a repeated dose of a 1:500 dilution of SIVmac251 (120 50% tissue culture infective doses [TCID50] on cells) starting at week 28, as previously described (27).
Table 1.
Mamu class I genotypes of macaques
| Controls |
ALVAC-SIV/gp120 |
gp120 |
|||
|---|---|---|---|---|---|
| Animal no. | Class I allele(s) | Animal no. | Class I allele(s) | Animal no. | Class I allele(s) |
| P065a | A01/A08/B01/B17 | ||||
| P064a | A01/A02 | P062a | A01/A02 | P122 | A08/B01 |
| P146 | A08 | P063 | A02 | P123 | A08/B01 |
| P149 | Negb | P067 | A02/A08/B01 | P124 | A02/A08 |
| P158 | A02/B03 | P112a | A01/A08 | P126 | A08 |
| P161 | A02 | P147 | Neg | P127 | A02/B01 |
| P258 | Neg | P148 | A08 | P132 | A08/B01 |
| P262 | B01 | P150 | B03/B08 | P133 | A08/B01 |
| M381a | A01 | P172 | B01 | P135 | A08 |
| M385a | A01/A08 | P250 | A08 | P137 | Neg |
| M895 | B17 B017/B01/BO3/A02 | P254 | A02/B04 | P139a | A01/A08 |
| M903 | A02/A11/B03 | M624a | A01 | M927a | A01/B01 |
MamuA01+.
Neg, animals negative for all major histocompatibility complex alleles tested: MamuA01, A02, A08, A11, B01, B03, B04, B08, B17.
Viral load quantification–single-genome amplification (SGA) analysis and tripartite motif-containing protein 5 alpha (TRIM5α) genotyping.
Plasma SIV RNA was quantified by nucleic acid sequence-based amplification (NASBA), as previously described (28). SIV DNA was quantified in the blood and tissue of macaques by quantitative PCR, as previously described (29).
Transmitted/founder viruses and their progeny were identified by single-genome amplification (SGA) of SIV RNA from plasma or rectal pinches. SIV RNA was extracted, and limiting-dilution PCR of newly synthesized cDNA was performed. Transmitted/founder virus lineages were identified by low-diversity sequence and by single sequences with unique mutations. Phylogenetic trees were generated using ClustalW.
The TRIM5α genotype was analyzed for all of the macaques included in the study and was previously described (30). Briefly, genomic DNA was isolated from peripheral blood mononuclear cells (PBMCs), and the 526-nucleotide PCR product of the B30.2/SPRY domain of TRIM5α was direct sequenced. The sequences of the primers utilized both for PCR and for the sequencing reaction were CAGTGCTGACTCCTTTGCTTG for the forward primer and GCTTCCCTGATGTGATAC for the reverse primer. The sequences were then aligned with the genomic DNA sequence of the rhesus macaque (accession number DQ842021.1) to characterize the polymorphisms at nucleic acid positions 997, 1,015 to 1020, and 1,022 of TRIM5α.
ELISpot assays.
T-cell responses: the enzyme-linked immunosorbent spot assays (ELISpot) were performed using monkey gamma interferon (IFN-γ) ELISpot kits from Mabtech in triplicate on freshly isolated PBMCs. A total of 3 × 105 cells/well was used with or without stimulation with the SIVmac251 Gag or Env overlapping peptides as described previously (29). Concanavalin A (ConA; Sigma-Aldrich) was used as a positive control (1 μg/ml final concentrations). The cells were then incubated at 37°C in a 5% CO2 atmosphere for 24 h and washed. The plates were assayed for IFN-γ-producing cells as per the manufacturer's protocol (MABTECH ELISpot plus for the monkey IFN-γ kit). Spot-forming cells (SFC) were counted on an ELISpot reader. The results are expressed as the number of spots per 106 cells after subtracting the number of spots in unstimulated wells.
B-cell responses.
SIV Env-specific or total IgG or IgA antibody-secreting cells were analyzed by ELISpot as described previously (31). Briefly, MultiScreen 96-well plates (Millipore MAIPS4510) were incubated with 70% ethanol, rinsed four times with 1× phosphate-buffered saline (PBS), and then coated with 500 ng/well of SIVmac251 gp120 (Advanced BioScience Laboratories, Inc.) in PBS or 10 μg/ml of goat anti monkey IgG or IgA (KPL). Coated plates were incubated at 4°C overnight. Plates were washed twice with 1× PBS and blocked with RPMI with 10% fetal bovine serum (FBS) and 3% bovine serum albumin (BSA) for 2 h at 37°C. Peripheral blood mononuclear cells were stimulated with 1 μg/ml CpG (ODN-2006; Operon), 0.5 μg/ml CD40L, and 50 ng/ml interleukin 21 (IL-21) (Peprotech) in complete medium for 3 days at 37°C in 24-well plates. Stimulated PBMCs were then harvested and washed, and 3 × 105 cells were plated in 200 μl of RPMI containing 10% FBS and incubated overnight at 37°C. Plates were then washed five times in PBS with 0.05% Tween 20 (Sigma) (PBS-T) and incubated with 100 μl of 1 μg/ml of biotinylated goat anti-monkey IgG or IgA (Rockland) in PBS-T containing 1% FBS. Plates were then washed and incubated for 1 h with 5 μg/ml horseradish peroxidase (HRP)-avidin D conjugate (Vector Laboratories, CA) in PBS-T and 1% FBS. Plates were washed a total six times with PBS-T, the final three washes were done with PBS, and plates were developed using 3-amino-9-ethyl-carbazole (AEC; Sigma). Spot quantitation was performed by using an ELISpot reader.
Lymphocyte proliferation assays.
Proliferation was performed using as a readout [3H]thymidine incorporation as described elsewhere (29). Briefly, fresh PBMCs were cultured at 105 cells/well in triplicate for 3 days in the absence or the presence of native high-performance liquid chromatography (HPLC)-purified SIVmac251 p27Gag and gp120 Env proteins (Advanced BioScience Laboratories) or ConA as a positive control. The cells were then pulsed overnight with 1 μCi of [3H]thymidine before harvest. The stimulation index was calculated as the fold thymidine incorporation into cellular DNA over medium control.
As an alternative approach to measure lymphocyte proliferation, carboxyfluorescein succinimidyl ester (CFSE) staining was performed on cryopreserved peripheral blood lymphocytes. These cells were incubated with 5 mM CFSE (Invitrogen) in PBS for 5 min at room temperature (RT), and cells were washed twice with RPMI containing 10% FBS and 1% antibiotics (R10). Cells were enumerated, and 1 × 106 cells were plated per ml of R10 in 24-well plates containing 5 μg/ml of SIV Gag, Env, or ConA used as a positive control or left unstimulated. Plates were incubated for 5 days at 37°C, and cells were harvested and stained for 30 min at 4°C with CD3 (clone SP34-2), CD8 (clone RPA-T8), CD4 (clone L200), or CD95 (clone DX2), all from BD Biosciences, in addition to the live/dead aqua dye to discriminate dead cells (Invitrogen). After staining, samples were washed with PBS, fixed with 1% paraformaldehyde (PFA), and acquired on an LSRII flow cytometer using FACS DIVA 6.0 software (BD Biosciences). The results are expressed as the frequency of Gag- or Env-specific memory (CD95+) CD28+ CD4+ or CD8+ T cells after background subtraction (% CFSE low unstimulated cells). Data analysis was performed using FlowJo software (TreeStar), and data were graphed using GraphPad Prism.
Binding antibody assay and peptide mapping.
To detect anti-SIVmac251-binding antibodies, serial dilutions of plasma were incubated with the lysate of SIVmac251 spiked with native purified gp120 Env protein of SIVmac251 bound to microtiter enzyme-linked immunosorbent assay (ELISA) plates, as described previously (32). Endpoint titers were defined as the reciprocal of the highest serum dilution that gave an optical absorbency at 450 nm at least two standard deviations greater than average values obtained with negative-control serum. For peptide mapping, the sera were diluted to 1:20 and added to plates coated with peptides encompassing the entire SIVK6W gp120 amino acid sequence (33).
Avidity assay.
A format of previously described capture ELISA for HIV-1 gp120 (34, 35), in which HIV-1 gp120 is captured through its C terminus by goat antibody D7324, was adapted for detecting antibodies against SIV gp120. Recombinant SIV gp120 protein made from codon-optimized SIVmac239 gp120 fused to the C-terminal tag of HIV-1 gp120 is used as the antigen for the capture ELISA to detect SIV Abs against a conformational epitope.
Ab avidity is determined by parallel ELISA as previously described (27). Brief, heat-inactivated plasma samples were serially diluted and applied to a 96-well-plate-captured SIVmac239 gp120 as described above in parallel duplicates. After 1 h of incubation, the plate was washed and half of the samples were treated with Tris-buffered saline (TBS), while the paired samples were treated with 1.5 M sodium thiocyanate (NaSCN; Sigma-Aldrich) for 10 min at room temperature. The plate was washed and detected with goat anti-monkey IgG-detecting Ab (Fitzgerald). The plate was then developed for optical density (OD) readout of Ab binding as described in reference 34. The avidity index (%) was calculated by taking the ratio of the NaSCN-treated plasma dilution giving an OD of 0.5 to the TBS-treated plasma dilution giving an OD of 0.5 and multiplying by 100. The plasma of uninfected normal macaque served as the negative control. A high-avidity macaque monoclonal antibody (MAb) of 3.11H (36) was included on every plate as the standard.
Surface plasmon resonance.
Surface plasmon resonance (SPR) measurements were conducted with a Biacore T100 using CM5 chips prepared as described below. The immobilization wizard packaged within the T100 control software was used to immobilize 1 uM streptavidin in 20 mM sodium acetate, pH 4.2 (10-min contact time), to each flow cell (7,000 response units [RU]). Biotinylated cyclic V2 peptides (Fig. 5E) were prepared at 1 μM in 20 mM Tris, pH 7.4, and injected manually until 4,000 RU of peptide was captured by the streptavidin-coated surface. Following two 30-s injections of 50 mM HCl, 1 mM biotin in 20 mM Tris, pH 7.4, was then injected twice (60-s contact time) over the reference and analyzed flow cells.
Fig 5.
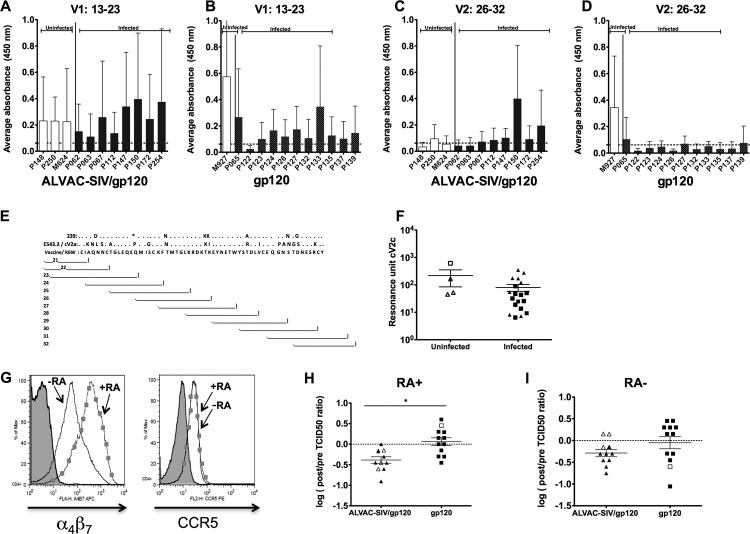
Evaluation of antibody responses targeting the V1/V2 loops of gp120. Serum antibody binding to peptides spanning the V1 region, amino acids 13 to 23 of the SIVmac251K6W gp120, in animals vaccinated with ALVAC-SIV/gp120 (A) or gp120 (B). Peptide binding is represented as average absorbance, and uninfected animals are on the left in white bars. Binding to peptides spanning the V2 region, amino acids 26 to 32 of gp120, in the serum of ALVAC-SIV/gp120-vaccinated (C) or gp120-vaccinated (D) animals. (E) The amino acid sequence (in single-letter amino acid code) of the linear V2 region of SIVmac239 and SIVSME543.3, is presented in comparison to the SIVmacK6W sequence used in the ALVAC-SIV construct (33). Dots represent identical amino acid sequence. The asterisk signifies one amino acid deletion. Each line represents a peptide whose number is depicted on the left. (F) Surface plasmon resonance with sera (week 27) from uninfected and infected macaques assayed against the cV2c-truncated peptide. (G) Surface expression of α4β7 and CCR5 on CD4+ T cells untreated (line with squares) or treated with retinoic acid (RA; solid line). Reduction of SIVmac251 infectivity mediated by the sera of animals immunized with ALVAC-SIV/gp120 and gp120 in RA-treated (H) or untreated (I) animals. (H and I) The log-transformed change in TCID50 pre- and postvaccination is shown with uninfected animals in open symbols and infected animals in closed.
Plasma samples were heat inactivated (56°C, 45 min) and centrifuged (5 min, 4°C, 14,500 rpm), and the supernatant was filtered prior to use. The plasma was diluted 1:50 in Tris-buffered saline, pH 7.4, and passed over the chip surface at 30 μl/min for 3 min followed by a 5-min dissociation period. At the end of the 5-min period, a 20-μg/ml solution of affinity-purified gamma chain-specific goat anti-monkey IgG antibody (Rockland Inc., Gilbertsville, PA) was passed over the flow cells for 2 min at a flow rate of 10 μl/min. After a 70-s dissociation period, the peptide and reference surfaces were regenerated using a 30-s pulse of 50 mM HCl, a 30-s pulse of 100 mM EDTA in 20 mM Tris, pH 7.4, and another 30-s pulse of 50 mM HCl followed by a 1-min injection of Tris-buffered saline, pH 7.4. Nonspecific binding was subtracted, and data analysis was performed using the BIA evaluation 4.1 software. The reported response units for the IgG-specific values are the difference between the average value of a 5-s window taken 60 s after the end of the anti-IgG injection and the average value of a 5-s window taken 10 s before the beginning of the anti-IgG injection. The data (RU) are presented as dot plots for individual plasma samples.
Standard neutralization assays.
Neutralization was measured as a reduction in luciferase reporter gene expression after a single round of infection in either TZM-bl or 5.25.EGFP.Luc.M7 (M7-Luc) cells as described previously (10, 37). TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu. M7-Luc cells were kindly provided by Nathaniel R. Landau. For the TZM-bl assay, 200 TCID50 of virus were incubated with serial 3-fold dilutions of test sample in duplicate, in a total volume of 150 μl for 1 h out of 18 h at 37°C, as indicated, in 96-well flat-bottom culture plates. Freshly trypsinized cells (10,000 cells in 100 μl of growth medium containing 75 μg/ml DEAE-dextran) were added to each well. One set of control wells received cells and virus (virus control), and another set received cells only (background control). After the 48-h incubation, 100 μl of cells was transferred to 96-well black solid plates (Costar) for measurements of luminescence using the Britelite luminescence reporter gene assay system (PerkinElmer Life Sciences). Neutralization titers are the dilution at which relative luminescence units (RLU) were reduced by 50% compared to that in virus control wells after subtraction of background RLUs.
M7-Luc cells were maintained in RPMI 1640 containing 12% heat-inactivated fetal bovine serum, 50 μg gentamicin/ml, 0.5 μg puromycin/ml, 300 μg G418/ml, and 200 μg hygromycin/ml to preserve CCR5 and reporter gene plasmids. This is a CEMx174 cell clone that was produced by retroviral vector transduction to express CCR5 (CD4 and CXCR4 are expressed naturally) and transfection to contain Tat-responsive Luc and green fluorescence protein (GFP) reporter genes. For neutralization assays, 500 TCID50 of virus were incubated with serial dilutions of serum samples in triplicate, in a total volume of 150 μl for 1 h at 37°C in 96-well flat-bottom culture plates. M7-Luc cells were suspended at a density of 5 × 105/ml in growth medium containing DEAE-dextran (10 μg/ml) but lacking puromycin, G418, and hygromycin. Cells (100 μl) were added to each well. One set of control wells received cells and virus (virus control), and another set received cells only (background control). Assay plates were incubated until approximately 10% of cells, in virus control wells, were positive for GFP expression by fluorescence microscopy (approximately 3 days). At this time, a 100-μl suspension of cells was transferred to a 96-well black solid plate (Costar) for measurements of luciferase activity as described above. Neutralization titers are the serum dilution at which the number of RLUs was reduced by 50% compared to virus control wells after subtraction of background RLUs.
Assay stocks of molecularly cloned Env-pseudotyped viruses (SIVmac251 CS.41, SIVmac239 0.23, SIVmac251 WY:30) were prepared by transfection in 293T cells and were titrated in TZM-bl cells as described previously (38, 39). Assay stocks of uncloned TCLA-SIVmac251 and SIVmac251 CS/2002 letvin were produced in H9 and human PBMCs, respectively, and were titrated in M7-Luc and TZM-bl cells, respectively.
ADCC assay.
Antibody-dependent cell-mediated cytotoxicity (ADCC) activity mediated by plasma samples was detected by the GranToxiLux (GTL) procedure as previously described (40). Briefly, CEM.NKRCCR5 target cells (41) were coated with recombinant SIVmac251 gp120 (Advanced Bioscience Laboratories Inc., Kensington, MD) and labeled with a fluorescent target-cell marker (TFL4; OncoImmunin, Inc., Gaithersburg, MD) and a viability marker (NFL1; OncoImmunin, Inc.). Labeled target cells were washed and plated at 1 × 104 viable cells/well. Cryopreserved human PBMCs from an HIV-seronegative donor served as effectors and were added to the assay wells at an effector/target ratio of 30:1. Fluorogenic granzyme B (GzB) substrate (OncoImmunin, Inc.) was added to each well to a final concentration of 0.25×. After incubation for 5 min at RT, serially diluted serum/plasma samples (4-fold; starting at 1:100) were added to the assay wells. The plates were incubated for 15 min at RT, centrifuged for 1 min at 300 × g, and incubated for 1 h at 37°C and 5% CO2. The plates were then washed, and cells were resuspended in 200 μl of PBS containing 1% FBS. A minimum of 2,500 events representing viable target cells was acquired for each well using an LSRII flow cytometer (BD Biosciences, San Jose, CA). Data analysis was performed using FlowJo 8.8.4 software (Tree Star Inc., Ashland, OR). The percentage of GzB activity was defined as the percentage of cells positive for proteolytically active GzB out of the total viable target cell population. The final results are expressed after subtracting the background, represented by the percentage of GzB activity observed in wells containing effector and target T cells in the absence of serum/plasma. The results were considered positive if the percentage of GzB activity after background subtraction was >8%.
Virus reduction assay in the presence or absence of retinoic acid.
PBMC was isolated from EDTA blood pooled from four naïve Indian rhesus macaques by Ficoll-hypaque centrifugation and depleted of CD8+ T cells by magnetic beads (Dynal). The enriched CD4+ T cells were cultured in vitro with tosyl-activated magnetic beads (Dynal) coated with equal amounts of anti-CD3 (clone FN18: NIH nonhuman primate reagent resource) and anti-CD28 (BD Biosciences) and 50 U rIL2 for 5 days in a CO2 incubator in the absence or presence of 10 nM all-trans retinoic acid (RA). Activated cells were subsequently washed, examined for expression of both α4β7 and CCR5 by flow cytometry (FACS Calibur), and used as targets in infection assays.
For infection (TCID50) assay, plasma or serum samples were heat inactivated at 56°C for 30 min, filtered using a 0.22-μm membrane, and diluted 1:10 in 100 μl of medium in a 96-well plate. SIVmac251 grown in rhesus PBMC with a titer of 8.2 × 104 TCID50/ml was serially diluted 4-fold in 50 μl medium and added to the diluted serum/plasma. After incubation for 1 h at 37°C, 2 × 105 CD3/CD28-activated CD4+ T-cell enriched PBMCs were added to each well in 50 μl medium, making the final serum dilution 1:20. Wells were thoroughly washed after 120 h to remove residual virus and then replenished with fresh medium. Cells were cultured for an additional 5 days with occasional replenishing of fresh medium. To score infection, 100 μl of culture medium was removed from each well for quantitation of SIV p27 level using the SIV antigen micro-ELISA kit (ABL). TCID50 values were calculated using Spearman-Karber calculations.
Mucosal antibodies.
Mucosal IgG and IgA binding antibodies were measured by binding antibody multiplex assay as previously described (42, 43) and SPR as previously described (44). Antibodies were eluted from rectal Weck-cel in cold elution buffer (1× protease inhibitor cocktail [Calbiochem], 0.25% BSA) by spinning 2 times at 16,000 × g for 15 to 20 min at 4°C. Total and SIV-specific immunoglobulin levels were measured by ELISA according to the manufacturer's instructions (monkey IgG ELISA, monkey IgA ELISA; Alpha Diagnostics, San Antonio, TX) and expressed as μg/ml to calculate specific activity for the binding responses. The SIV antigens utilized in both the multiplex binding antibody assay and SPR assay are SIVmac239 p55 Gag (Protein Sciences), SIV p27 Gag (ImmunoDiagnostics, Woburn, MA), rgp41 (Immunodiagnostics), SIVmac251 rgp130 (ImmunoDiagnostics, Woburn, MA), and SIV gp140 (provided by Bing Chen, Harvard). For analysis of SIV-specific IgG by multiplex binding antibody assay, SIV proteins were coupled to microspheres (Bio-Rad) and incubated with serial dilutions of samples, and specific antibody binding was detected by biotinylated anti-monkey IgG (Rockland) by mean fluorescent intensity (with background and blank bead subtracted). Positive and negative monkey serum controls were used in each assay, and the midpoint titer (EC50) of each sample was calculated using a four-parameter logistic fit (4-PL). The avidity of antibody binding was measured on a BIAcore 4000 instrument (BIAcore/GE Healthcare) using the multiplex array format (1 by 16) in which samples were flowed over duplicate spots of 8 different antigen surfaces on a series S CM5 chip (BIAcore/GE Healthcare). Proteins (pFB SIV gp140 and recombinant gp130 SIV Mac 251) were immobilized to about 5 to 10,000 RU, and peptides were immobilized to about 1 to 3,000 RU. The following peptides were also used in this study: SIVmac251 V2 linear, SIVmac251 V2 S-S, cyclic DLV SIVmac251, SIVmac251 LDV gp41, cyclic LDV. There were 7 animals with both rectal and vaginal samples and 27 rectal samples included in this study. Antigen surface activity was monitored using DBM5 IgG, HIVIG, and serum samples as positive controls and Synagis MAb as the negative control. DBM5 IgG was titrated (0 to 100 μg/ml) to generate a standard curve for calculating Weck sample IgG concentration. All Weck-cel extraction samples were run undiluted and injected over each of the flow cells with replicate spots (2×) at 10 μl/min for an association time of 120 s and a dissociation time of 450 s. Following each binding cycle, surfaces were regenerated with a short injection (20 s) of glycine, pH 2.0. Each surface activity was monitored by including DBM5 IgG (100 μg/ml) injection at regular interval of every 20 cycles. Bulk effect from a buffer injection was subtracted from each IgG sample binding data. Data analyses were performed with BIAevaluation 4000 and BIAevaluation 4.1 software (BIAcore/GE Healthcare). Kinetic binding responses were measured by averaging postinjection response units (RU) over a 10-s window and dissociation rate constant, Kd(s−1), was measured during the postinjection/buffer wash phase (after the first 30 s to allow stabilization of signal) following curve fitting to a Langmuir dissociation equation. A relative avidity binding score was calculated for each IgG sample as follows: RU.s = (RU)/Kd(s − 1), where RU.s is the avidity score and (RU)/Kd(s − 1) is the binding response. Higher levels of binding responses and a lower Kd are indicators of higher-affinity interactions. For normalized responses, binding responses of a DBM5 IgG titration at known concentrations on an anti-monkey IgG chip surface were plotted to generate a calibration curve. Equivalent IgG concentrations for each Weck sample were calculated using the slope of the standard curve. Weck samples with binding responses above the highest DBM5 concentration of 100 μg/ml were reported as 150 μg/ml. Kinetic binding responses were normalized according to calculated concentrations.
Statistics.
Differences between groups were assessed using the exact Wilcoxon rank sum test, with P values corrected by the Hochberg method for multiple antigens tested and multiple pairwise group comparisons.
Nucleotide sequence accession numbers.
All 1,116 envelope sequences were deposited in GenBank under accession numbers JX497793 to JX498908.
RESULTS
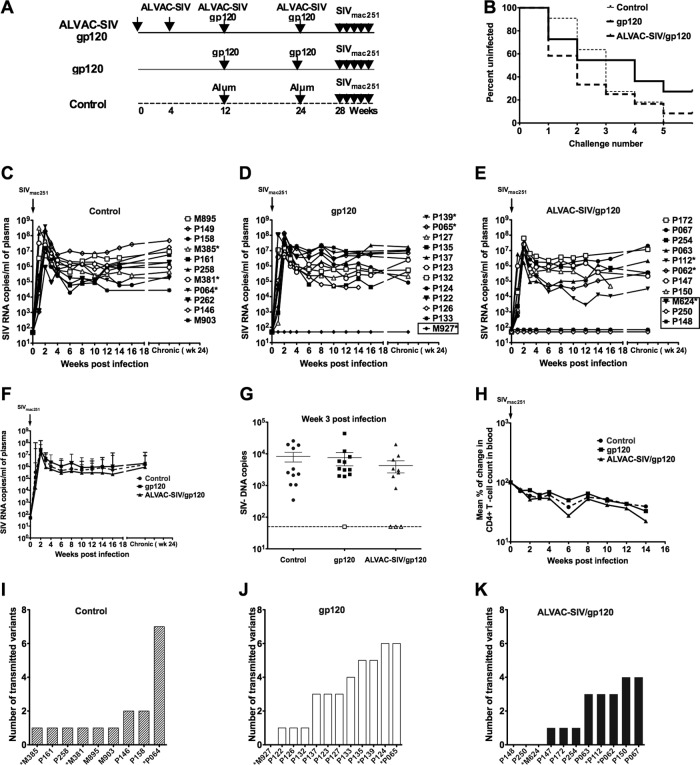
Relative vaccine efficacy following mucosal exposure to a dose of SIVmac251 that transmits few virus variants.
We immunized 11 adult Indian rhesus macaques, in a prime boost regimen, with four intramuscular immunizations of the ALVAC-SIV recombinant virus expressing the SIVK6W gag, pol, and env genes (15) and with two coimmunizations with the SIVgp120 protein alum adjuvanted. A second group of 12 macaques were immunized with gp120 protein adjuvanted in alum and a third group of 11 animals with the alum adjuvant alone (Fig. 1A). Animal groups were randomized for the presence of protective class I alleles (Table 1). At the end of the immunization regimen, all animals were exposed weekly by the intrarectal route at a dose of 120 TCID50 of SIVmac251, shown previously to result in transmission of only a few virus variants (27). Infected animals were not rechallenged once viremia was detected, and new challenges with SIVmac251 were terminated when all 11 animals in the control group became viremic (five exposures). The choice to terminate the challenge was supported by the notion that further challenges would not have added significance to vaccine efficacy. We observed no significant differences in the time of acquisition of SIVmac251 in the three groups of animals (Fig. 1B). However, while all control animals became infected (Fig. 1C), three (P148, P250, and M624) of the 11 macaques immunized with the ALVAC-SIV/gp120 (27.3%) and one (M927) of the 12 macaques immunized with gp120 (8.3%) remained uninfected (Fig. 1D and 1E). There were no significant differences in viral loads between vaccinated animals and controls; however, the ALVAC-SIV-vaccinated group demonstrated transiently lower viremia in the acute phase than the control animals (Fig. 1F). Similarly, we observed no significant difference in the levels of SIV DNA in the rectum of vaccinated animals and controls 3 weeks postinfection (Fig. 1G). SIV DNA was undetectable in the rectal mucosa collected at 3 weeks from the last exposure of uninfected animals P148, P250, M624, and M927 (Fig. 1G). We observed no significant differences in CD4+ T-cell loss between infected vaccinated and control groups (Fig. 1H). Although the viral SIVmac251 stock at the 120 TCID50 dose used for challenge was previously shown to transmit few virus variants (27), to assess the number of viral variants that established systemic infections, we performed single-genome amplification (SGA) and direct sequencing of the env gene to enumerate the number of transmitted/founder variants. Unique transmitted lineages were identified by phylogenetic analysis within the context of the inoculum sequences, as described elsewhere (19). Control and animals vaccinated with the combination of ALVAC-SIV and gp120 had a median number of one variant, whereas the animals immunized with gp120 alone had a median number of three variants (Fig. 1I to K). The difference in the number of transmitted variants did not differ significantly among the groups. Since the number of transmitted variants was greater in the gp120 group and animals infected after the first or second challenge appeared to have more transmitted viral variants, we evaluated a potential correlation between the number of transmitted variants and the number of challenges. We found an inverse correlation between the number of transmitted variants and the time of acquisition when we analyzed all the animals in the study (R = −0.70, P < 0.0001). When this analysis was performed separately among the two vaccine groups and the controls, the inverse correlation remained significant for both the vaccinated groups (ALVAC-SIV/gp120, R = −0.84, P = 0.0025; gp120, R = −0.65, P = 0.028) but not the control group (R = −0.31, P = 0.42).
Fig 1.
Mucosal challenge exposure of macaques to low repeated doses of SIVmac251 dose. (A) Schematic representation of the study design. Four weeks after last immunization (week 28), macaques were exposed to a total of five repeated doses of SIVmac251 (120 TCID50 each) given weekly by the intrarectal route. (B) The rate of infection in the three groups is depicted; ALVAC-SIV/gp120, solid black line; gp120, dark dotted line; and control, light dotted line. SIV RNA levels in plasma of control (C), gp120-immunized (D), and ALVAC-SIV/gp120-immunized (E) animals. The animals that remained uninfected in the ALVAC-SIV/gp120 (M624, P250, and P148) group and the one in the gp120 group (M927) are boxed. (F) Geometric mean SIV RNA levels ± standard error in plasma of ALVAC-SIV/gp120 (triangle), gp120 (square), and control (circle) animals. (G) Viral DNA copies in rectal biopsy specimens collected at day 19 after infection or after the fifth exposure to a low dose of SIVmac251 in the animals that remained uninfected (open symbols). (H) Mean percentage of change in blood CD4+ T cell number following SIVmac251 infection. The uninfected animals are excluded from the analysis. Number of virus variants transmitted in control (I), gp120-immunized (J), and ALVAC-SIV/gp120-immunized (K) macaques.
Vaccine-elicited T-cell responses.
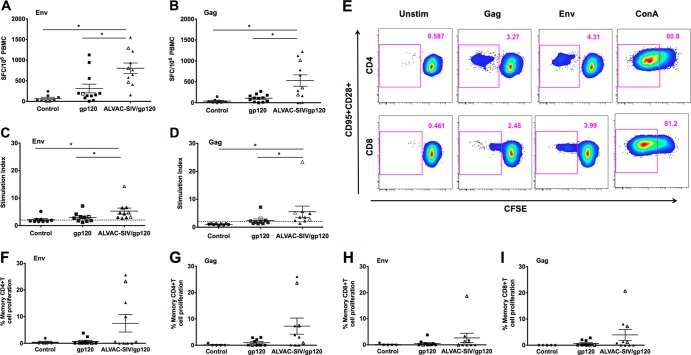
The immunogenicities of the two vaccine regimens were assessed by comparing T cell responses prechallenge (week 27, 3 weeks after the last immunization) to unvaccinated controls (Fig. 2). IFN-γ, measured by ELISpot using PBMCs stimulated with SIV Gag and Env overlapping peptides, demonstrated nonsignificant differences between control and gp120-immunized macaques (P = 0.071 for each) but were significantly different for both antigens between ALVAC-SIV/gp120-immunized animals and control (P = 0.0069 and P = 0.0001, respectively) (Fig. 2A and B). Macaques immunized with the ALVAC-SIV/gp120 developed significantly higher IFN-γ ELISpot responses to Gag and Env than macaques immunized with gp120 protein alone (P = 0.011 and P = 0.016, respectively) (Fig. 2A and B). Lymphocyte-proliferative responses were measured by thymidine incorporation (Fig. 2C and D) or CFSE staining (Fig. 2E to I) following in vitro stimulation with SIV Gag- and Env-purified proteins. Proliferation measured by thymidine incorporation showed a significant increase of stimulation indexes in ALVAC-SIV/gp120 compared to gp120 or control groups after Env (P = 0.043 and P < 0.0001) or Gag stimulations (P = 0.035 and P = 0.0009) (Fig. 2C and D). However, when the proliferative ability of CD4+ and CD8+ T cells were measured with CFSE on cryopreserved PBMCs, the differences between the two vaccinated groups only approached statistical significance (Fig. 2E to I).
Fig 2.
Vaccine-induced T-cell responses. All the data were obtained from PBMCs harvested at 3 weeks after the last immunization (week 27). All the data from animals protected from SIVmac251 acquisition are depicted with open symbols. IFN-γ-secreting cells measured by ELISpot in response to SIV Env (A) or Gag (B). Results are shown as the number of spots per 106 cells, and each symbol represents one animal. Lymphoproliferative responses (LPR) measured by thymidine incorporation to SIV Env (C) or Gag (D). The horizontal bars in the figures represent the average and standard error for the vaccinated and control animals. Gating strategy for CD4+ and CD8+ CFSE+ memory (CD95+) T cells (E). (F to I) Percentage of CD95+ CFSE dim or negative CD8+ or CD4+ T cells that proliferate following Env or Gag stimulation.
Vaccine-elicited B-cell responses.
The envelope-specific B-cell response was measured at week 27, 3 weeks after the last immunization, by B-cell ELISpot, binding titers in blood, specific activity in mucosal secretions, avidity of binding antibodies to gp120, and the ability of antibodies to mediate ADCC. There was a trend for a higher IgG to SIV gp120 in animals immunized with ALVAC-SIV/gp120 than in control animals (P = 0.072), and no significant difference between the gp120-immunized and control macaques (Fig. 3A). The mean binding antibody titers to the gp120 envelope protein detected by ELISA in the sera were higher in the ALVAC-SIV/gp120 than in the gp120 group (P = 0.018, Fig. 3B), but the levels of gp140 IgG-specific activity, measured in rectal secretions, were not significantly different (Fig. 3C). The avidity of antibodies to gp120 did not differ significantly in the two immunized groups but was high in all the animals that were protected from infection (depicted as open symbols in Fig. 3D). The avidity of mucosal IgG was tested by surface plasmon resonance (44) against gp120 proteins or the V2 peptides, depicted in Fig. S1A in the supplemental material, and did not differ significantly in the two vaccinated groups (see Fig. S1A and B). The neutralizing titers in the M7-Luc assay were significantly higher in the ALVAC-SIV/gp120 group than in controls and in gp120-vaccinated animals (P = 0.0005 and P < 0.0001) (Fig. 3E), whereas the mean titers of neutralizing antibodies in the TZM-bl cell assay did not differ between the two immunized groups (Fig. 3F). ADCC was measured by calculating the maximum granzyme activity, which was not significantly different in the two vaccinated groups (Fig. 3G), and ADCC titers, which were higher in the ALVAC-SIV/gp120 group (P = 0.028, Fig. 3H).
Fig 3.
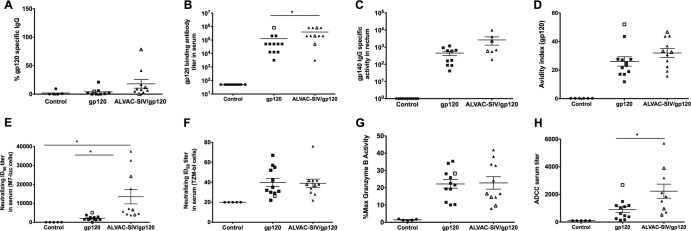
Vaccine-induced B-cell responses. All data presented in the figure were obtained from sera collected at week 27, 3 weeks after the last immunization. In all the figures, the animals protected from SIV acquisition are depicted with open symbols, and the horizontal bars in the figures represent the average values and standard errors. (A) Percentages of gp120-specific IgG of the total cell-producing IgG at week 27. (B) Endpoint titers of SIV Env-specific IgG in serum of animals vaccinated with ALVAC-SIV/gp120, SIV gp120, or controls. (C) Gp140 IgG-specific activity in rectal secretions from vaccinated animals and controls. (D) Avidity of antibodies to the SIVmac239 Env protein in sera of the animals vaccinated with either ALVAC-SIV/gp120 or SIV gp120 or controls. SIV-specific neutralization activity measured in animal sera using SIV pseudotyped lentiviruses and either the M7-luc cell line (E) or TZM-bl cells (F) as targets. ADCC capacity of antibodies in serum of all animals represented as the percent maximum granzyme activity (G) or serum ADCC titer (H).
Vaccine-elicited T- and B-cell responses in protection from SIVmac251 acquisition or plasma virus level.
We investigated the immune responses that contributed to protection, delayed SIVmac251 acquisition, or decreased plasma virus levels by analyzing the vaccine-induced immune responses at week 27, 3 weeks after the last immunization and 1 week prior to challenge exposure to SIVmac251. T-cell responses to the Gag and envelope antigens measured by IFN-γ ELISpot or proliferation assays did not correlate with time of acquisition or blood virus levels. Neither did the number of IgG-producing B cells to envelope measured by ELISpot nor envelope binding antibodies in blood or mucosal secretions correlate with SIVmac251 acquisition. The only trend of note was in the binding antibody titers to gp120, which were apparently inversely correlated with the virus levels at week 3 (R = −0.65, P = 0.036, not corrected for multiple comparisons). No correlation was found with prechallenge neutralizing antibody titers to SIVmac251 in both the M7-luc and the TZM-bl assays or ADCC, using maximum granzyme levels, ADCC titers, avidity and time of acquisition, or blood virus levels. However, the antibody in the sera of the four immunized, uninfected macaques had significantly higher antibody avidity to gp120 than those from macaques that became infected (P = 0.012) (Fig. 4A). The antibody avidity to gp120 remained significantly higher over time (3 and 6 weeks postchallenge) in the uninfected animals (P = 0.0018 and P = 0.0060, respectively) than SIV-infected animals (Fig. 4B and C). Interestingly, the significant difference in avidity of antibodies to gp120 among uninfected or infected macaques was not observed when the sera of these animals were tested against a gp120 deleted in the V1 and V2 regions (Fig. 4D), suggesting that antibody responses to the V1 and V2 regions of gp120 might be of importance.
Fig 4.
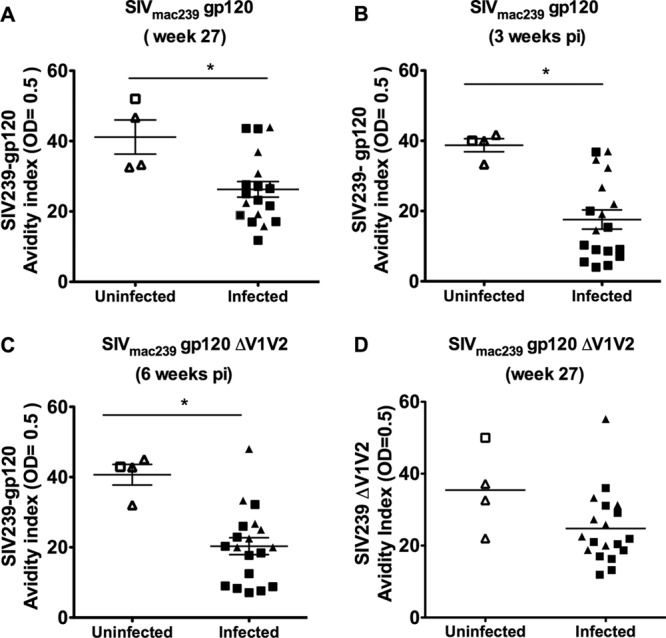
Avidity of antibodies to gp120 and protection from SIVmac251 acquisition. Avidity index of antibody to the entire SIVmac239 gp120 in vaccinated animals that were protected or SIV infected measured at week 27, 3 weeks after the last vaccination (A) or at three (B) or six (C) weeks after SIV infection. (D) Avidity of antibody to the gp120 protein deleted in the V1 and V2 regions, in protected and infected animals.
Protection from HIV acquisition, in the RV144 human trial (4), correlated significantly with the presence of antibody response to V1/V2 (45). Since the data presented in Fig. 4A to D suggested that immune responses to V1/V2 are also induced by this vaccine regimen in macaques, we mapped the antibody response induced by our vaccines. The sera of all macaques collected at week 27 were tested for their ability to recognize 89 linear overlapping peptides (15-mers), derived from the SIVK6W gp120 and the gp41 (peptide 90 to 141) envelope amino acid sequence (33) included in the ALVAC SIVgpe vaccine. Most of the macaques in both vaccinated groups recognized linear V1 peptides 13 to 23 (Fig. 5A and B); some recognized the V2 peptides 23 to 32 (Fig. 5C and D) and V3 peptides 50 to 55 (data not shown). Antibodies directed to the V2 region encompassed between peptides 26 and 32 were significantly higher in infected ALVACgp120-vaccinated animals than in infected gp120 vaccinees (P = 0.031). However, we did not observe a specific pattern of linear peptide recognition to V2 using this approach (Fig. 5C and D; see also Fig. S1C in the supplemental material). Next, we evaluated serum recognition using the cV2a cyclic peptide that encompasses the entire amino acid sequence of the SIVSME660, as well as the cV2c peptide, that has a truncation of the first 17 amino acids of V2 (Fig. 5E). Both peptides were tested in a surface plasmon resonance assay. Surface plasmon resonance demonstrated that the sera, collected at week 27 from the uninfected animals, had higher activity than those that become infected, but this difference approached statistical significance only when the cyclic cv2c-truncated peptide was used in the assay and was based mainly on the result of one single animal (Fig. 5F). The cV2c peptide is 75% identical to the V2 region of the SIVK6W envelope gene inserted in the ALVAC backbone used in this study. Importantly, the reactivity to the cV2c peptide was also found in the assay in the mucosal samples of two of the four protected macaques (data not shown).
The sera of ALVAC/SIV/gp120-immunized macaque reduce SIVmac251 infectivity in vitro.
Since our data suggested that the antibody response and in particular the responses to the V1/V2 loop of the gp120 was higher in the ALVAC-SIV/gp120 group, we developed a functional assay to investigate the ability of sera of the immunized macaque to inhibit SIVmac251 infection in vitro.
The V2 has been shown to interact with the integrin α4β7 that is expressed on the surface of activated CD4+ T cells (37). Even though blocking α4β7 does not inhibit infection, overexpression of α4β7 has been demonstrated to increase viral infectivity in standard infectivity assays, possibly by favoring clustering of CD4 and CCR5 on the cell surface (46). We tested whether the preimmune and immune sera of vaccinated macaques decreased SIVmac251 infection in cells, whereby α4β7 expression was enhanced by retinoic acid (RA) treatment. Activated (CD8+-T-cell-depleted) PBMCs, cultured in the presence of RA, had an upregulation of α4β7 expression but similar levels of CCR5 expression (Fig. 5G). Sera from animals before and after vaccination were mixed with serial dilutions of SIVmac251, added to activated cells, and cultured for 10 days. TCID50 titers were then measured as the amount of virus necessary to infect 50% of the cells in the presence of sera. Interestingly, sera from ALVAC/SIVgp120 animals reduced SIVmac251 infectivity better in RA-treated cells than the sera of gp120-vaccinated animals (P = 0.0027) (Fig. 5H). This trend was also seen in cells not treated with RA, but the difference did not reach statistical significance (P = 0.13) (Fig. 5I). The sera of the gp120-immunized macaques did not affect SIVmac251 infectivity in RA-treated or untreated cells (Fig. 5H and I).
DISCUSSION
A validated animal model could hasten progress in the development of an effective vaccine for HIV. Here, we demonstrate, in a small pilot study, that immunizations of macaques with vaccines similar to those used in the HIV vaccine RV144 trial in Thailand, which resulted in protection of one-third of the vaccinees from HIV acquisition (4), also protected few vaccinated macaques from acquisition of the highly pathogenic SIVmac251 using a dose of challenge virus that transmits few virus variants (24). However, because the macaque study was not powered sufficiently for scoring protection, further studies will be needed to properly investigate the correlates of protection. Restrictive TRIM5α alleles for SIVmac251 replication did not account for the lack of SIVmac251 acquisition in macaques P148, P250, M624, and M927, as demonstrated in previously published work with this virus stock (30). Interestingly, of the four vaccinated animals that resisted five challenge exposures, two carried the MamuA01 protective allele (animals M624 and M927) (Table 1). However, because the control group also had animals that carried the same protective allele (Table 1) and nevertheless acquired SIVmac251 following mucosal challenge, it is likely that vaccination, in combination with the genotypic status, contributed to the protection from infection, as also observed earlier with these vaccine modalities (15). In the same conditions, vaccination with gp120 alone did not protect from infection, except a single animal that carried the protective MamuA01 allele and was also able to mount high titers of gp120 antibody with high avidity (Fig. 4A). Similar to humans vaccinated with an equivalent vaccine regimen (4), ALVAC-SIV/gp120 immunization did not protect macaques from CD4+ T-cell loss or high virus plasma levels over time. The ALVAC-SIV/gp120 vaccines in macaques elicited limited CD4+ T-cell responses and negligible CD8+ T-cell responses, as in humans (4). Similarly, all the vaccinated macaques developed binding antibodies to the envelope protein (4) and neutralizing antibodies to lab-adapted SIVmac251. Because the study was not powered to assess correlates of protection, we considered the results that follow as exploratory. Vaccine-induced protection from SIVmac251 acquisition was significantly associated with antibody with high avidity for gp120 but not with neutralizing activity in vitro, ADCC, or T-cell responses measured by ELISpot, intracellular cytokine staining, and T-cell proliferation. Interestingly, the significant difference in antibody avidity to gp120 was lost when the gp120 protein, used in the assay, had a deletion in V1 and V2. These data suggest that either the V1/V2 could be among target epitopes of antibodies with high avidity or that the V1 and/or V2 may be necessary to maintain the appropriate conformation of the gp120 protein for the optimal binding of high-avidity antibodies, directed to regions other than V1/V2. The results obtained with the cyclic cV2c peptide (Fig. 5F), although only approaching statistical significance, suggest that V2 may be a target of high-avidity antibodies. In addition, our findings with the virus reduction assay, following retinoic acid stimulation of primary CD4+ cells, suggest that the ALVAC-SIV priming may be important for the elicitation of antibodies able to reduce viral infectivity under conditions whereby α4β7 is increased by retinoic acid since the sera from animals immunized with gp120 alone did not have this activity (Fig. 5H and I).
Studies by others also have found that antibody avidity to the envelope was associated with reduced viral load following SHIV and SIV challenge exposure (47), and conversely in another system, low antibody avidity has been associated with poor protective efficacy with an RSV vaccine (48).
The similarity of the results obtained in macaques here with those reported in humans using equivalent vaccines, as well as in those by others using different vaccine modalities (49, 50), is encouraging but needs to be confirmed by properly powered studies since the present study confirms the usefulness of this macaque model despite differences in the immunogens used. The ALVAC-HIV vaccine used in humans expressed the gag-pro genes, whereas the ALVAC-SIV vaccine used in the present study expressed the gag-pol genes. The ALVAC-HIV in humans expressed a fused HIV clade E gp120 to the gp41 transmembrane domain of the HIVLAI strain, whereas the ALVAC-SIV expressed the entire envelope protein. The envelope protein boost in humans consisted of two proteins: HIV clade E and clade B fused to the gD HSV signal peptide and were produced in CHO cells (4). In macaques, we used only the native monomeric SIVmac251 gp120 produced in T cells. In addition, the challenge used in the macaque study here was homologous (approximately 1% diversity in the envelope gene of the challenge and the vaccine strain), whereas the vaccinated volunteers in the RV144 trial were likely exposed to more genetically diverse clade E and AE strains (4). Hopefully, SIVmac251-based vaccines, constructed as the human vaccines, may be proven, in properly powered animal studies, to confer significant protection from SIVmac251 as observed within 3 years from vaccination in the Thai volunteers (4). Animal models have accelerated the development of effective vaccines for human diseases and significantly facilitated the identification of correlates of protection (51–55), but the models should reflect conditions of the human infection and disease as accurately as possible (56). Notably, in the present study, the low repeated doses of SIVmac251 used to challenge the macaques resulted in the transmission of few virus variants, as typically seen in heterosexual transmission of HIV-1 to humans. However, we noticed a significant correlation between time of acquisition and number of variants transmitted in vaccinated but not control animals, suggesting that in animals that are not protected from infection, vaccination may be associated with early transmission of a higher number of virus variants. Further studies will be necessary to confirm these results. Our data demonstrate that the low repeated dose challenge of rhesus macaques with SIVmac251 may be a relevant model for potentially defining correlates of protection, for demonstrating the effectiveness of other candidate vaccines, and ultimately for the improvement and optimization of vaccines against HIV.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Guroff for sharing the unpublished data reported in reference 27 and N. Miller for providing the SIVmac251 virus stock. We are grateful to the investigators at ABL: J. Treece, D. Weiss, and P. Markham for animal husbandry and care; H. K. Chang and E. Lee for quantitative analysis of viral RNA and DNA; L. Ajayi and V. Kalyanaraman for binding-antibody measurement, and Teresa Habina for editorial assistance.
P.P., M.V., S.G., and G.F. designed and coordinated the study; P.P and M.D. performed T and B cell ELISpot, ICS, and proliferation assays; B.F.K. and J.D.L. analyzed virus variants; Y.G. analyzed antibody avidity; G. Ferrari and D.M. analyzed ADCC and virus neutralization, respectively; C.F. analyzed the TRIM5α gene; R.P., M.G.F., and L.H. performed the virus reduction assay; M.R. and E.B. performed the serum logical assay on cyclic V2 peptide; J.T. generated the ALVAC-SIV; N.M. and J.K. contributed to the study design; D.V. and D.S. performed statistical analysis; and G. Franchini, together with all coauthors, wrote the paper.
This research was supported by the Intramural Research Program of the NIH, NCI, and under NCI contract HHSN2662004000088C.
Footnotes
Published ahead of print 21 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02544-12.
REFERENCES
- 1. Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del RC, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191:654–665 [DOI] [PubMed] [Google Scholar]
- 3. Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, GFvan Hu D, Tappero JW, Choopanya K. 2006. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 194:1661–1671 [DOI] [PubMed] [Google Scholar]
- 4. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, SMde Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 5. Gilbert PB, Berger JO, Stablein D, Becker S, Essex M, Hammer SM, Kim JH, Degruttola VG. 2011. Statistical interpretation of the RV144 HIV vaccine efficacy trial in Thailand: a case study for statistical issues in efficacy trials. J. Infect. Dis. 203:969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belshe R, Franchini G, Girard MP, Gotch F, Kaleebu P, Marthas ML, McChesney MB, McCullough R, Mhalu F, Salmon-Ceron D, Sekaly RP, van Rompay K, Verrier B, Wahren B, Weissenbacher M. 2004. Support for the RV144 HIV vaccine trial. Science 305:177–180 [DOI] [PubMed] [Google Scholar]
- 7. Burton DR, Desrosiers RC, Doms RW, Feinberg MB, Gallo RC, Hahn B, Hoxie JA, Hunter E, Korber B, Landay A, Lederman MM, Lieberman J, McCune JM, Moore JP, Nathanson N, Picker L, Richman D, Rinaldo C, Stevenson M, Watkins DI, Wolinksky SM, Zack JA. 2004. Public health. A sound rationale needed for phase III HIV-1 vaccine trials. Science 303:316. [DOI] [PubMed] [Google Scholar]
- 8. McNeil JG, Johnston MI, Birx DL, Tramont EC. 2004. Policy rebuttal. HIV vaccine trial justified. Science 303:961. [DOI] [PubMed] [Google Scholar]
- 9. McMichael AJ. 2006. HIV vaccines. Annu. Rev. Immunol. 24:227–255 [DOI] [PubMed] [Google Scholar]
- 10. Andersson S, Makitalo B, Thorstensson R, Franchini G, Tartaglia J, Limbach K, Paoletti E, Putkonen P, Biberfeld G. 1996. Immunogenicity and protective efficacy of a human immunodeficiency virus type 2 recombinant canarypox (ALVAC) vaccine candidate in cynomolgus monkeys. J. Infect. Dis. 174:977–985 [DOI] [PubMed] [Google Scholar]
- 11. Benson J, Chougnet C, Robert-Guroff M, Montefiori D, Markham PD, Shearer GM, Gallo RC, Cranage MP, Paoletti E, Limbach K, Venzon D, Tartaglia J, Franchini G. 1998. Recombinant vaccine-induced protection against the highly pathogenic SIVmac251: dependence on route of challenge exposure. J. Virol. 72:4170–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franchini G, Gurunathan S, Baglyos L, Plotkin S, Tartaglia J. 2004. Poxvirus-based vaccine candidates for HIV: two decades of experience with special emphasis on canarypox vectors. Expert. Rev. Vaccines 3(Suppl 1):S75–S88 [DOI] [PubMed] [Google Scholar]
- 13. Franchini G, Robert-Guroff M, Tartaglia J, Aggarwal A, Abimiku AG, Benson J, Markham PD, Limbach K, Hurteau G, Fullen J, Aldrich K, Miller N, Sadoff J, Paoletti E, Gallo RC. 1995. Highly attenuated HIV type 2 recombinant poxviruses, but not HIV-2 recombinant Salmonella vaccines, induce long-lasting protection in rhesus macaques. AIDS Res. Hum. Retroviruses 11:909–920 [DOI] [PubMed] [Google Scholar]
- 14. Myagkikh M, Alipanah S, Markham PD, Tartaglia J, Paoletti E, Gallo RC, Franchini G, Robert-Guroff M. 1996. Multiple immunizations with attenuated poxvirus HIV type 2 recombinants and subunit boosts required for protection of rhesus macaques. AIDS Res. Hum. Retroviruses 12:985–992 [DOI] [PubMed] [Google Scholar]
- 15. Pal R, Venzon D, Letvin NL, Santra S, Montefiori DC, Miller NR, Tryniszewska E, Lewis MG, Vancott TC, Hirsch V, Woodward R, Gibson A, Grace M, Dobratz E, Markham PD, Hel Z, Nacsa J, Klein M, Tartaglia J, Franchini G. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pal R, Venzon D, Santra S, Kalyanaraman VS, Montefiori DC, Hocker L, Hudacik L, Rose N, Nacsa J, Edghill-Smith Y, Moniuszko M, Hel Z, Belyakov IM, Berzofsky JA, Parks RW, Markham PD, Letvin NL, Tartaglia J, Franchini G. 2006. Systemic immunization with an ALVAC-HIV-1/protein boost vaccine strategy protects rhesus macaques from CD4+ T-cell loss and reduces both systemic and mucosal simian-human immunodeficiency virus SHIVKU2 RNA levels. J. Virol. 80:3732–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patterson LJ, Peng B, Abimiku AG, Aldrich K, Murty L, Markham PD, Kalyanaraman VS, Alvord WG, Tartaglia J, Franchini G, Robert-Guroff M. 2000. Cross-protection in NYVAC-HIV-1-immunized/HIV-2-challenged but not in NYVAC-HIV-2-immunized/SHIV-challenged rhesus macaques. AIDS 14:2445–2455 [DOI] [PubMed] [Google Scholar]
- 18. Van Rompay KK, Abel K, Lawson JR, Singh RP, Schmidt KA, Evans T, Earl P, Harvey D, Franchini G, Tartaglia J, Montefiori D, Hattangadi S, Moss B, Marthas ML. 2005. Attenuated poxvirus-based simian immunodeficiency virus (SIV) vaccines given in infancy partially protect infant and juvenile macaques against repeated oral challenge with virulent SIV. J. Acquir. Immune Defic. Syndr. 38:124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, Mascola JR, Nabel GJ, Haynes BF, Bhattacharya T, Perelson AS, Korber BT, Hahn BH, Shaw GM. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gray RH, Wawer MJ, Brookmeyer R, Sewankambo NK, Serwadda D, Wabwire-Mangen F, Lutalo T, Li X, VanCott T, Quinn TC. 2001. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357:1149–1153 [DOI] [PubMed] [Google Scholar]
- 21. Hladik F, Hope TJ. 2009. HIV infection of the genital mucosa in women. Curr. HIV/AIDS Rep. 6:20–28 [DOI] [PubMed] [Google Scholar]
- 22. Mofenson LM. 2003. Advances in the prevention of vertical transmission of human immunodeficiency virus. Semin. Pediatr. Infect. Dis. 14:295–308 [DOI] [PubMed] [Google Scholar]
- 23. Winkelstein W, Jr, Lyman DM, Padian N, Grant R, Samuel M, Wiley JA, Anderson RE, Lang W, Riggs J, Levy JA. 1987. Sexual practices and risk of infection by the human immunodeficiency virus. The San Francisco Men's Health Study. JAMA 257:321–325 [PubMed] [Google Scholar]
- 24. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knapp LA, Lehmann E, Piekarczyk MS, Urvater JA, Watkins DI. 1997. A high frequency of Mamu-A*01 in the rhesus macaque detected by PCR-SSP and direct sequencing. Tissue Antigens 50:657–661 [DOI] [PubMed] [Google Scholar]
- 26. Kalyanaraman VS, Rodriguez V, Veronese F, Rahman R, Lusso P, DeVico AL, Copeland T, Oroszlan S, Gallo RC, Sarngadharan MG. 1990. Characterization of the secreted, native gp120 and gp160 of the human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 6:371–380 [DOI] [PubMed] [Google Scholar]
- 27. Xiao P, Patterson LJ, Kuate S, Brocca-Cofano E, Thomas MA, Venzon D, Zhao J, DiPasquale J, Fenizia C, Lee EM, Kalisz I, Kalyanaraman VS, Pal R, Montefiori D, Keele BF, Robert-Guroff M. 2012. Replicating adenovirus-SIV recombinant priming and envelope protein boosting elicits localized, mucosal IgA immunity in rhesus macaques correlated with delayed acquisition following a repeated low dose rectal SIVmac251 challenge. J. Virol. 86:4644–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Romano JW, Williams KG, Shurtliff RN, Ginocchio C, Kaplan M. 1997. NASBA technology: isothermal RNA amplification in qualitative and quantitative diagnostics. Immunol. Invest. 26:15–28 [DOI] [PubMed] [Google Scholar]
- 29. Vaccari M, Mattapallil J, Song K, Tsai WP, Hryniewicz A, Venzon D, Zanetti M, Reimann KA, Roederer M, Franchini G. 2008. Reduced protection from simian immunodeficiency virus SIVmac251 infection afforded by memory CD8+ T cells induced by vaccination during CD4+ T-cell deficiency. J. Virol. 82:9629–9638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fenizia C, Keele BF, Nichols D, Cornara S, Binello N, Vaccari M, Pegu P, Robert-Guroff M, Ma ZM, Miller CJ, Venzon D, Hirsch V, Franchini G. 2011. TRIM5alpha does not affect simian immunodeficiency virus SIV(mac251) replication in vaccinated or unvaccinated Indian rhesus macaques following intrarectal challenge exposure. J. Virol. 85:12399–12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brocca-Cofano E, McKinnon K, Demberg T, Venzon D, Hidajat R, Xiao P, Daltabuit-Test M, Patterson LJ, Robert-Guroff M. 2011. Vaccine-elicited SIV and HIV envelope-specific IgA and IgG memory B cells in rhesus macaque peripheral blood correlate with functional antibody responses and reduced viremia. Vaccine 29:3310–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hel Z, Tsai WP, Tryniszewska E, Nacsa J, Markham PD, Lewis MG, Pavlakis GN, Felber BK, Tartaglia J, Franchini G. 2006. Improved vaccine protection from simian AIDS by the addition of nonstructural simian immunodeficiency virus genes. J. Immunol. 176:85–96 [DOI] [PubMed] [Google Scholar]
- 33. Franchini G, Gurgo C, Guo HG, Gallo RC, Collalti E, Fargnoli KA, Hall LF, Wong-Staal F, Reitz MS., Jr 1987. Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature 328:539–543 [DOI] [PubMed] [Google Scholar]
- 34. Guan Y, Sajadi MM, Kamin-Lewis R, Fouts TR, Dimitrov A, Zhang Z, Redfield RR, DeVico AL, Gallo RC, Lewis GK. 2009. Discordant memory B cell and circulating anti-Env antibody responses in HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 106:3952–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore JP, Wallace LA, Follett EA, McKeating JA. 1989. An enzyme-linked immunosorbent assay for antibodies to the envelope glycoproteins of divergent strains of HIV-1. AIDS 3:155–163 [DOI] [PubMed] [Google Scholar]
- 36. Cole KS, Alvarez M, Elliott DH, Lam H, Martin E, Chau T, Micken K, Rowles JL, Clements JE, Murphey-Corb M, Montelaro RC, Robinson JE. 2001. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology 290:59–73 [DOI] [PubMed] [Google Scholar]
- 37. Arthos J, Cicala C, Martinelli E, Macleod K, Van RD, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. 2008. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat. Immunol. 9:301–309 [DOI] [PubMed] [Google Scholar]
- 38. Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12:Unit 12.11 [DOI] [PubMed] [Google Scholar]
- 40. Pollara J, Hart L, Brewer F, Pickeral J, Packard BZ, Hoxie JA, Komoriya A, Ochsenbauer C, Kappes JC, Roederer M, Huang Y, Weinhold KJ, Tomaras GD, Haynes BF, Montefiori DC, Ferrari G. 2011. High-throughput quantitative analysis of HIV-1 and SIV-specific ADCC-mediating antibody responses. Cytometry A 79:603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trkola A, Matthews J, Gordon C, Ketas T, Moore JP. 1999. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J. Virol. 73:8966–8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bolton DL, Song K, Wilson RL, Kozlowski PA, Tomaras GD, Keele BF, Lovingood RV, Rao S, Roederer M. 2012. Comparison of systemic and mucosal vaccination: impact on intravenous and rectal SIV challenge. Mucosal Immunol. 5:41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, Cohen M, Eron J, Hicks CB, Liao HX, Self SG, Landucci G, Forthal DN, Weinhold KJ, Keele BF, Hahn BH, Greenberg ML, Morris L, Karim SS, Blattner WA, Montefiori DC, Shaw GM, Perelson AS, Haynes BF. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Flynn BJ, Kastenmuller K, Wille-Reece U, Tomaras GD, Alam M, Lindsay RW, Salazar AM, Perdiguero B, Gomez CE, Wagner R, Esteban M, Park CG, Trumpfheller C, Keler T, Pantaleo G, Steinman RM, Seder R. 2011. Immunization with HIV Gag targeted to dendritic cells followed by recombinant New York vaccinia virus induces robust T-cell immunity in nonhuman primates. Proc. Natl. Acad. Sci. U. S. A. 108:7131–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cicala C, Martinelli E, McNally JP, Goode DJ, Gopaul R, Hiatt J, Jelicic K, Kottilil S, Macleod K, O'Shea A, Patel N, Van RD, Wei D, Pascuccio M, Yi L, McKinnon L, Izulla P, Kimani J, Kaul R, Fauci AS, Arthos J. 2009. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc. Natl. Acad. Sci. U. S. A. 106:20877–20882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao J, Lai L, Amara RR, Montefiori DC, Villinger F, Chennareddi L, Wyatt LS, Moss B, Robinson HL. 2009. Preclinical studies of human immunodeficiency virus/AIDS vaccines: inverse correlation between avidity of anti-Env antibodies and peak postchallenge viremia. J. Virol. 83:4102–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Delgado MF, Coviello S, Monsalvo AC, Melendi GA, Hernandez JZ, Batalle JP, Diaz L, Trento A, Chang HY, Mitzner W, Ravetch J, Melero JA, Irusta PM, Polack FP. 2009. Lack of antibody affinity maturation due to poor Toll-like receptor stimulation leads to enhanced respiratory syncytial virus disease. Nat. Med. 15:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qureshi H, MA ZM, Huang Y, Hodge G, Thomas MA, DiPasquale J, DeSilva V, Fritts L, Bett AJ, Casimiro DR, Shiver JW, Robert-Guroff M, Robertson MN, McChesney MB, Gilbert PB, Miller CJ. 2012. Low-dose penile SIVmac251 exposure of rhesus macaques infected with adenovirus type 5 (Ad5) and then immunized with a replication-defective Ad5-based SIV gag/pol/nef vaccine recapitulates the results of the phase IIb step trial of a similar HIV-1 vaccine. J. Virol. 86:2239–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reynolds MR, Weiler AM, Piaskowski SM, Piatak M, Jr, Robertson HT, Allison DB, Bett AJ, Casimiro DR, Shiver JW, Wilson NA, Lifson JD, Koff WC, Watkins DI. 2012. A trivalent recombinant Ad5 gag/pol/nef vaccine fails to protect rhesus macaques from infection or control virus replication after a limiting-dose heterologous SIV challenge. Vaccine 30:4465–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Edghill-Smith Y, Golding H, Manischewitz J, King LR, Scott D, Bray M, Nalca A, Hooper JW, Whitehouse CA, Schmitz JE, Reimann KA, Franchini G. 2005. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat. Med. 11:740–747 [DOI] [PubMed] [Google Scholar]
- 52. Ogata N, Cote PJ, Zanetti AR, Miller RH, Shapiro M, Gerin J, Purcell RH. 1999. Licensed recombinant hepatitis B vaccines protect chimpanzees against infection with the prototype surface gene mutant of hepatitis B virus. Hepatology 30:779–786 [DOI] [PubMed] [Google Scholar]
- 53. Sabin AB. 1985. Oral poliovirus vaccine: history of its development and use and current challenge to eliminate poliomyelitis from the world. J. Infect. Dis. 151:420–436 [DOI] [PubMed] [Google Scholar]
- 54. Salk D. 1980. Eradication of poliomyelitis in the United States. III. Poliovaccines—practical considerations. Rev. Infect. Dis. 2:258–273 [DOI] [PubMed] [Google Scholar]
- 55. Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, Govindarajan S, Purcell RH, Chisari FV. 2002. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. U. S. A. 99:15661–15668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Feinberg MB, Moore JP. 2002. AIDS vaccine models: challenging challenge viruses. Nat. Med. 8:207–210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.