Abstract
Nitrous oxide (N2O) is a major radiative forcing and stratospheric ozone-depleting gas emitted from terrestrial and aquatic ecosystems. It can be transformed to nitrogen gas (N2) by bacteria and archaea harboring the N2O reductase (N2OR), which is the only known N2O sink in the biosphere. Despite its crucial role in mitigating N2O emissions, knowledge of the N2OR in the environment remains limited. Here, we report a comprehensive phylogenetic analysis of the nosZ gene coding the N2OR in genomes retrieved from public databases. The resulting phylogeny revealed two distinct clades of nosZ, with one unaccounted for in studies investigating N2O-reducing communities. Examination of N2OR structural elements not considered in the phylogeny revealed that the two clades differ in their signal peptides, indicating differences in the translocation pathway of the N2OR across the membrane. Sequencing of environmental clones of the previously undetected nosZ lineage in various environments showed that it is widespread and diverse. Using quantitative PCR, we demonstrate that this clade was most often at least as abundant as the other, thereby more than doubling the known extent of the overall N2O-reducing community in the environment. Furthermore, we observed that the relative abundance of nosZ from either clade varied among habitat types and environmental conditions. Our results indicate a physiological dichotomy in the diversity of N2O-reducing microorganisms, which might be of importance for understanding the relationship between the diversity of N2O-reducing microorganisms and N2O reduction in different ecosystems.
Keywords: denitrification, functional gene diversity, nitrous oxide reductase, protein translocation pathway
Introduction
The relationship between biogeochemical process rates and the diversity of organisms that mediate them is of fundamental interest in microbial ecology. In the nitrogen cycle, the denitrification pathway is performed by a wide range of foremost bacterial taxa that reduce nitrate in the biosphere to nitrogen gas (N2) through a series of soluble (nitrite, NO2−) and gaseous intermediates (nitric oxide (NO) and nitrous oxide (N2O)). In terrestrial ecosystems, this process is a main contributor to emissions of N2O (Mosier, 1998), which is a major greenhouse gas and currently the single most dominant ozone-depleting substance (Ravishankara et al., 2009). However, as N2O is either an intermediate or end product in denitrification, this process may potentially act as both a source and a sink for N2O (Chapuis-Lardy et al., 2007).
The denitrification pathway can be described as modular in nature, as organisms may perform only a subset of the steps in the pathway (Zumft, 1997). This is substantiated by genomic evidence showing that denitrifying organisms can have different combinations of genes involved in the denitrification pathway (Zumft and Kroneck, 2007; Jones et al., 2008). For example, genome sequencing of Agrobacterium tumefaciens first revealed that denitrifying bacteria can lack the nosZ gene, which encodes the N2O reductase (N2OR) that catalyzes the reduction of N2O to N2 (Wood et al., 2001). In fact, of genomes that possess the nir genes encoding the nitrite reductases necessary to catalyze the dissimilatory reduction of NO2− to NO, approximately one-third is currently known to lack nosZ (Jones et al., 2008). A greater discrepancy is observed in studies comparing the abundance of the nir and nosZ genes in various environments, where the number of nir genes can exceed that of nosZ by up to an order of magnitude (Babić et al., 2008; Hallin et al., 2009; Philippot et al., 2009a, 2011; Garcia-Lledo et al., 2011). Manipulation of the soil microbial community demonstrated that the proportion of denitrifiers lacking nosZ may be a critical factor in determining the N2O emission potential in different soils and, interestingly, one of the manipulated soils was potentially a sink of N2O (Philippot et al., 2011). Others have also reported net negative fluxes of N2O from soils in the field, indicating N2O consumption by the microbial community (Chapuis-Lardy et al., 2007). Whether shifts in denitrifier community composition, particularly among the proportion of denitrifiers capable of reducing N2O (Cavigelli and Robertson, 2000; Cheneby et al., 2004), will adversely affect greenhouse gas emissions (Avrahami and Bohannan, 2009; Philippot et al., 2009a, 2009b) is unclear. Understanding the link between ecosystem scale denitrification rates and the ecology of organisms that mediate this pathway has therefore been identified as a critical research goal for mitigation of climate change (Richardson et al., 2009).
Environmental studies investigating the diversity of denitrifiers commonly use the nosZ gene as a marker for N2O-reducing communities. However, such studies only retrieve nosZ sequences that are similar to those from alpha-, beta- or gamma-proteobacteria. Recent reports (Green et al., 2010; Jones et al., 2011) show that nosZ is present in a much larger range of archaeal and bacterial phyla, some of which include taxa that are commonly found in high abundance in different ecosystems (Roesch et al., 2007; Kirchman et al., 2010; Wessén et al., 2010; Newton et al., 2011) or have unique structural variants of the N2OR (Simon et al., 2004). Of greater significance for N2O emissions is the observation that Desulfitobacterium hafniense and Anaeromyxobacter spp., which are found in soil and sediments, do not possess either of the two nir genes, yet they have the full genetic potential to produce functional N2OR (Sanford et al., 2002; Simon et al., 2004; Zumft and Kroneck, 2007). Thus, the abundance of these organisms may also influence potential N2O emissions from different environments and could be a key for understanding why some act as N2O sinks.
The objective of this study was to examine the extent of N2OR sequence diversity among prokaryotic genomes, and determine the presence and abundance of previously undetected N2O-reducing genotypes in different environments. As two major lineages of nosZ were observed, we examined N2OR structural elements that may explain the differentiation of genotypes into the two clades, including regions outside the core N2OR domains that were not used in the phylogenetic analysis. By specifically targeting the previously undetected clade in environmental samples using molecular probes designed in this study, we could get a first insight of the unknown diversity of N2OR genes in the environment and the relative abundance of the two clades.
Materials and methods
Acquisition, alignment and analysis of nosZ sequences from genomes
Full-length nosZ nucleotide sequences from genomes were obtained from the Functional Gene Repository (http://fungene.cme.msu.edu/index.spr) and aligned by amino acid using the MAFFT alignment algorithm with the ‘ginsi' refinement settings (Katoh and Toh, 2008). The resulting alignment was then imported into the ARB software (Ludwig et al., 2004) and manually corrected guided by the secondary structure of the nosZ product from Paracoccus denitrificans (Protein Database entry 1FWX). The N-terminal region of the alignment containing the signal peptide as well as C-terminal regions extending beyond the known crystal structure of P. denitrificans was removed before analysis. An amino-acid phylogeny was generated that included insertion/deletions within the alignment by binary coding of gap sites in a second partition in the final analysis file. The LG+Γ substitution model (Le and Gascuel, 2008) with empirically determined amino-acid frequencies (+F) was used based on results from the Prottest program (Abascal et al., 2005), whereas the binary partition was analyzed using a binary substitution model with a Γ rate heterogeneity parameter. The maximum likelihood tree was calculated using RAxML v. 7.2.8 (Stamatakis, 2006) with 20 independent tree searches, and node support was determined using 500 bootstrap replicates. Detection of signal peptides was performed using the PRED-TAT algorithm (Bagos et al., 2010), which differentiates between signal peptides for the secretory (Sec) and twin-arginine translocation (Tat) pathways.
Primer design and validation using isolates and environmental samples
The phylogeny generated from the full-length amino-acid alignment was used as a reference tree for clade-specific primer design using the corresponding nucleotide alignment. Several sets of degenerate primers specific to nosZ sequences within clade II were designed using a combination of ARB and the CODEHOP algorithm (Rose et al., 1998) and tested on isolates known to possess the nosZ gene from clade II based on genomes in the NCBI database (Supplementary Table S1), as well as isolates that possess the clade I nosZ. Apart from those that were received as DNA extracts, all isolates were obtained from either the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany) or the American Type Culture Collection (LGC Standards AB, Boras, Sweden), and cultivated according to collection recommendations. Genomic DNA from two non-denitrifying bacteria that do not possess the nosZ gene, Arthrobacter chlorophenolicus A6 and Lactobacillus gasseri, were used as negative controls. Genomic DNA was extracted using either the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), or by lysing cells in 0.5 μg μl−1 Proteinase K and 0.5% SDS overnight at 37 °C, followed by isopropanol precipitation (Supplementary Table S1). Candidate primer pairs were used to amplify a 690–720 bp fragment of nosZ within clade II, and PCR reaction conditions were optimized using all isolates. All reactions were performed using DreamTaq Green PCR Master Mix (Fermentas, St Leon-Rot, Germany), with an initial concentration of 4.5 mℳ MgCl2, 1.0 μg μl−1 bovine serum albumin, 0.8 μℳ for each primer and 25–50 ng of genomic DNA. Thermal cycling conditions were an initial 5 min denaturing step at 95 °C, followed by 35 cycles of 95 °C for 30 s, a gradient annealing of 48–63 °C for 60 s, 72 °C extension for 60 s and a final extension at 72 °C for 10 min. Amplicon size was determined by electrophoresis using 2% agarose in 1 × Tris-acetate-EDTA buffer.
On the basis of the results of gradient PCR trials with the isolates, the primers nosZ-II-F (5′-CTI GGI CCI YTK CAY AC-3′) and nosZ-II-R (5′-GCI GAR CAR AAI TCB GTR C-3′), corresponding to positions 1162–1178 and 1889–1907 of the nosZ gene of P. denitrificans PD1222, were the best candidates. To test their specificity and extent of nosZ sequence diversity captured, clone libraries were generated from environmental samples obtained from either a single unique site or from multiple sites within the same environmental type (Supplementary Table S2). Environmental DNA was extracted using the FastDNA SPIN for Soil Kit (MP Biomedicals, Santa Ana, CA, USA). The same PCR reagent concentrations and thermal cycling program was used as described above for clade II nosZ genes from isolates, except that the MgCl2 concentration was reduced to 3.0 mℳ and the annealing temperature was set at 54 °C. The PCR products were purified by electrophoresis in a 2% agarose gel in Tris-acetate-EDTA buffer at 2.5 V cm−2 for 5 h, and the band was excised from the gel whereupon DNA was extracted using the MinElute Gel Extraction Kit (Qiagen) according to the manufacturer's instructions. Cloning was performed using the TOPO-TA cloning kit for sequencing with TOP10 chemically competent cells (Invitrogen—Life Technologies, Stockholm, Sweden). Overnight colonies from each library were lysed in nuclease-free water at 98 °C for 5 min, and the insert was amplified using M13 forward and reverse sequencing primers. PCR reactions contained DreamTaq PCR Buffer (Fermentas-Thermo Scientific, Waltham, MA, USA), 0.2 mℳ deoxyribonucleotide triphosphate, 0.5 mℳ each of M13 forward and reverse sequencing primers and 1.25 U DreamTaq DNA Polymerase. Thermocycler conditions were an initial denaturation of 95 °C for 5 min followed by 30 cycles of 95 °C for 30 s, 47 °C for 60 s and 72 °C for 60 s, with a final extension step of 72 °C form 10 min. Final PCR products were then sequenced by capillary sequencing (Macrogen Europe, Amsterdam, The Netherlands), and electrophoresis trace inspection was performed using Geneious v 5.5 (BioMatters, Ltd, Auckland, NZ, USA).
All reads were compared with the non-redundant amino-acid database in Genbank using BLASTX. After removal of poor-quality reads (low-quality base-calling and frameshift errors) from a total of 535 reads, sequences were analyzed for chimeras using the Belleropheron algorithm (Huber et al., 2004). All potential chimeric sequences (50 in total) were removed from each library before phylogenetic analysis, and the final set of 403 sequences were appended to the full-length nosZ alignment by translating to amino acid and aligning with HMMER 3.0 (Eddy, 1998), using the alignment of full-length nosZ amino-acid sequences as a reference alignment. Regions of the alignment that were outside the amplified fragment were excluded from the analysis, and phylogenetic analysis based on amino-acid sequences was performed using RAxML with the same model and run specifications as the full-length sequence tree stated above, with the exception that gaps were not binary coded. Nucleotide sequences have been submitted to Genbank under accession numbers JQ513977-JQ514066 and JQ647516-JQ647829.
Comparison of clade I and clade II nosZ abundance in different environments
A quantitative PCR assay was developed to compare the abundance of nosZ sequences from clades I and II among replicate samples from a range of different environments (Supplementary Table S3), and values were normalized across samples by calculating the ratio of nosZ and 16S rRNA gene abundance in each sample. Extraction of DNA from environmental samples was performed either by using the FastDNA SPIN Kit for Soil, the DNeasy Blood and Tissue Kit modified for extraction of DNA from Gram-positive bacteria per manufacturer instructions, or by using the ISO standardized methods for extraction of genomic DNA from soil (Philippot et al., 2010; see Supplementary Table S3). The abundance of 16S rRNA and clade I and II nosZ genes was determined using a StepOnePlus quantitative PCR platform (Applied Biosystems, Villebon Sur Yvette, France). Quantification of 16S rRNA genes was performed in 15 μl reactions that included Absolute QPCR SYBR Green Rox 2 × mastermix (ABGene—Thermo Scientific, Surry, UK), 250 ng of T4 gp32 (Qbiogene, Illkirch Cedex, France), and a 0.4 μℳ concentration of 16S rRNA-specific primers described in Lopez-Gutierrez et al. (2004). Thermal cycling conditions consisted of an initial denaturing step of 95 °C 15 min, followed by 40 cycles of 95 °C 15 s, 60 °C 30 s, 72 °C 30 s and a final step of 80 °C 30 s at which SYBR green fluorescence was measured. Reaction volume, components and concentrations for quantification of clade II were the same as described for the 16S rRNA genes, with the exception that a primer concentration of 1 μℳ was used with an annealing temperature of 54 °C. Quantification of nosZ clade I was performed as described in Henry et al. (2006) using primers 1840F and 2090R, which do not amplify clade II nosZ sequences. Standard curves for each assay were generated from linearized plasmids containing either cloned 16S rRNA genes from Pseudomonas aeruginosa PAO1, or cloned nosZ genes from either Bradyrhizobium japonicum USDA 110 or Gemmatimonas aurentica 27-T for nosZ clades I and II, respectively. Two independent quantitative PCR assays were performed for each gene and three no-template controls were run for each quantitative PCR assay, which gave null or negligible values. The presence of PCR inhibitors in DNA extracted from soil was estimated by mixing a known amount of standard DNA with environmental DNA extracts before quantitative PCR. No inhibition was detected, and efficiencies for 16S rRNA, clade I nosZ and clade II nosZ amplifications were estimated at 96%, 98% and 75%, with an r2 of >0.99 for each gene.
Results
nosZ phylogeny reflects differences in signal peptide motif
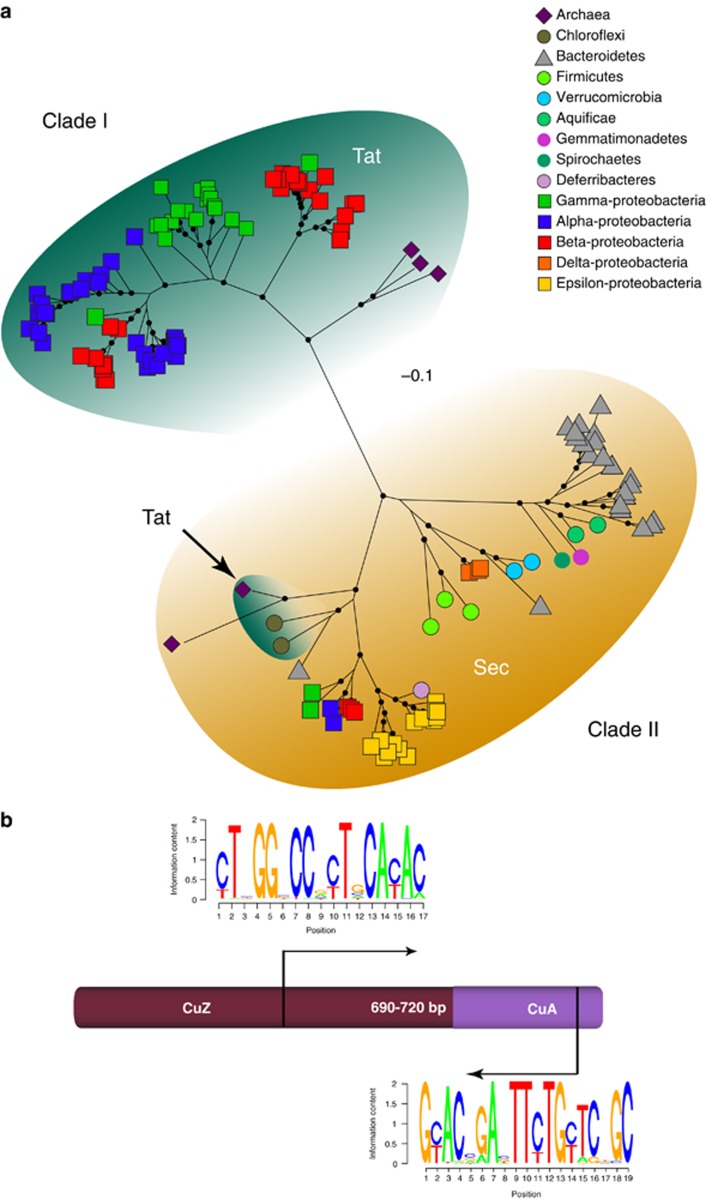
Of the ∼2000 completed prokaryotic genome projects available in the NCBI microbes database at the time of sequence retrieval (October 2011), we found 216 genomes with the nosZ gene, as well as a few large contigs from isolates and fosmid libraries with complete nosZ operons within the FUNGENE database. This was reduced to a data set of 142 full-length sequences after removal of highly similar nosZ sequences (>99% amino-acid similarity) from within the same genera. The N-terminal region encoding the signal peptide, which identifies the nascent translated protein as a substrate for different protein translocation mechanisms in the cell, were removed from the alignment along with C-terminal regions that may contain lineage-specific extensions or protein fusions. Subsequent phylogenetic analysis of the core catalytic region of the N2OR containing the CuZ and CuA active sites (Figure 1b) revealed two distinct clades with high bootstrap support (>70%), hereafter referred to as clades I and II (Figure 1a; see Supplementary Figure S1 for species names).
Figure 1.
(a) Unrooted maximum likelihood phylogeny of full-length nosZ amino-acid sequences obtained from genomes. The distribution of signal peptide motif detected for each clade is indicated. Symbols on tree tips specify major taxonomic groups, and scale bar indicates corrected substitutions per site (LG+Γ+F for amino acids; Binary+Γ for indel sites). Nodes with >70% bootstrap support (n=500) are denoted by dots. (b) Schematic representation of nosZ gene indicating the center multinuclear copper catalytic site (CuZ) and the C-terminal cupredoxin active site (CuA). Arrows indicate primer binding sites and amplicon size, and the consensus sequence of clade II for each site is shown as SeqLogo diagrams.
Clade I consisted entirely of sequences from Alpha-, Beta-, and Gamma-proteobacteria, generally following the species phylogeny as previously observed (Jones et al., 2008; Palmer et al., 2009). However, some exceptions are notable especially among sequences from Beta-proteobacteria, which grouped with those from Alpha-proteobacteria. Despite having no influence on the nosZ phylogeny, inspection of signal peptides showed that all sequences within clade I possessed a Tat signal peptide motif. Sequences from the halophilic archaeal genera Haloarcula, Halorubrum and Halogeometricum formed a distinct cluster that was more closely related to sequences in clade I. We therefore placed them in the clade I group, as nosZ genes from these species also encode a Tat signal peptide. No unique C-terminal domains were observed among clade I sequences.
Clade II consisted of sequences found among a diverse range of bacterial and archaeal phyla (Figure 1a). The majority of nosZ sequences in this clade encoded a Sec signal recognition motif with the exception of those from the hyperthermophilic archaea Ferroglobus placidus, as well as Thermomicrobium roseum and Sphaerobacter thermophilum, both of which are characterized as thermophilic bacteria within the phylum Chloroflexi (Hugenholtz and Stackebrandt, 2004). Interestingly, the Bacteroidetes Rhodothermus marinus and the archaea Pyrobaculum calidifontis had a Sec signal peptide motif, despite their relatively close position to other thermophiles in clade II with a Tat motif. The nosZ sequences from the Epsilon-proteobacteria formed a distinct, well-supported group within clade II, with all sequences possessing a C-terminal heme c-binding domain, which is characteristic of N2OR from this group (Simon et al., 2004). A similar, yet shorter extension was also observed in the few nosZ sequences from Alpha-, Beta- and Gamma-proteobacteria that grouped within clade II. Sequences originating from Gemmatimonadetes, Spirochaetes and Aquifaceae grouped with nosZ sequences from Bacteroidetes. The amino-acid alignment revealed an insert in the CuZ region encoding an alpha helix for all members of this group, as well as a short (11–36 aa) C-terminal extension with a relatively conserved amino-acid motif corresponding to a beta-strand structure, based on secondary structure prediction using the PSI-PRED server (McGuffin et al., 2000).
Capturing the unexplored diversity of clade II nosZ
The clear separation of the two clades led us to develop PCR primers that specifically target the clade II nosZ sequences, which have been previously undetected using currently available primer sets. For the forward primer, a single region was identified that was highly conserved among all sequences in the nosZ phylogeny (Figure 1b). This region corresponds to one of the coordinating sites for the CuZ active center of the N2OR, containing a histidine that binds the 4Cu:2S catalytic site (Pomowski et al., 2011). The reverse primer targets a region of the nosZ gene that encodes amino acids binding the CuA active site within the cytochrome c oxidase II (COXII) domain. This region is more variable among the complete set of nosZ sequences than the site targeted by the forward primer; however, it is highly conserved within clade II (Supplementary Figure S1). The optimal primer configuration amplified the targeted 690–720 bp nosZ fragment in 9 out of the 11 clade II representative species with varying band intensity (Supplementary Table S1). P. calidifontis and Anaeromyxobacter dehalogens did not produce visible amplification products, which may be due to mismatches in the 3′ end of the reverse primer for both P. calidifontis and A. dehalogens. Only one single clade I representative, Psychromonas ingrahamii, showed a band of the expected size.
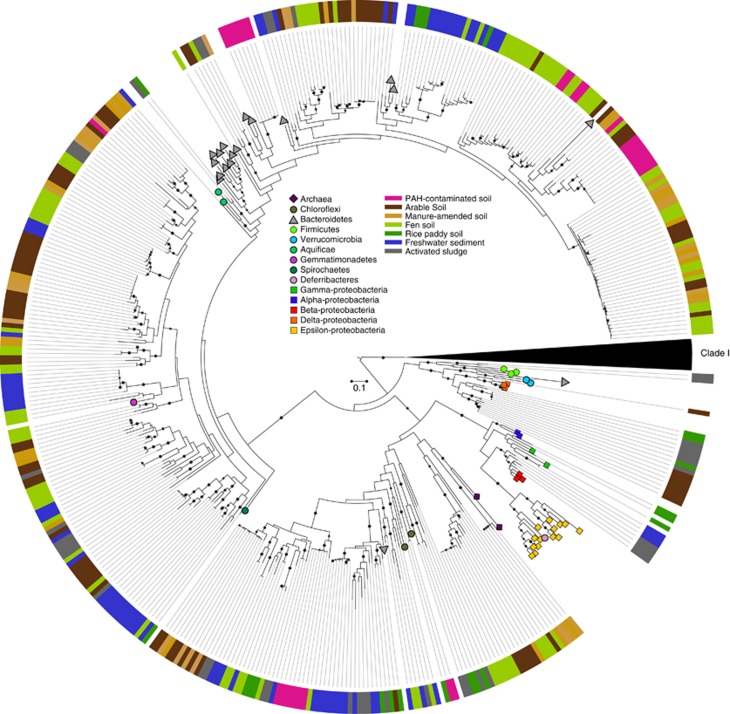
Of the 21 environmental samples used for generating clone libraries, 20 produced a single amplification product of the expected size (Supplementary Table S2). Comparison of all sequences from the clone libraries to the non-redundant protein database in Genbank using BLASTX indicated that only three sequences returned significant (E<10−6) hits to proteins other than nosZ, whereas all others were similar to nosZ sequences within clade II. Phylogenetic analysis demonstrated a wide range of environmental sequence diversity within clade II (Figure 2). Approximately 37% of all sequences were most similar (60–88% amino-acid similarity) to nosZ sequences from Bacteroidetes and were detected in all samples. Nearly 33% of the sequences were most closely related to Gemmatimonas aurentiaca nosZ, the majority of which were from soil and fen samples. Interestingly, 18% of all clones grouped with nosZ sequences from thermophilic Bacteroidetes and Chloroflexi, and were predominantly from sediment, sludge, fen and rice paddy samples. We also detected 22 nosZ sequences from fen, arable and manure-amended soil samples that clustered with nosZ from thermophilic archaea, although amino-acid similarities were lower (47–55%) than that observed between other environmental clones and reference sequences. Ten sequences, all from rice paddy, sediment or sludge samples grouped closely to nosZ sequences from Candidatus accumulibacter, Dechloromonas aromatica, Dechlorosoma sullum and Thiocapsa marina, and 17 sequences from sludge, rice paddy and arable soil environments were most similar to Anaeromyxobacter spp. Finally, two sequences from sludge samples were most similar to the clade of Gram-positive bacteria, which included D. hafniense and Geobacillus thermodenitrificans.
Figure 2.
Maximum likelihood phylogeny of clade II nosZ amino-acid sequences obtained from environmental samples listed in Supplementary Table S2 and reference sequences from genomes. Full-length clade I nosZ reference sequences (n=82) have been collapsed to a single black wedge. Symbols on tree tips specify major taxonomic affiliations of reference sequences, and the outer color strip shows the source of environmental clones. Scale bar indicates corrected amino-acid substitutions per site (LG+Γ+F), and nodes with >70% bootstrap support (n=500) are indicated by gray dots.
nosZ sequences from clade II are at least as abundant as those from clade I
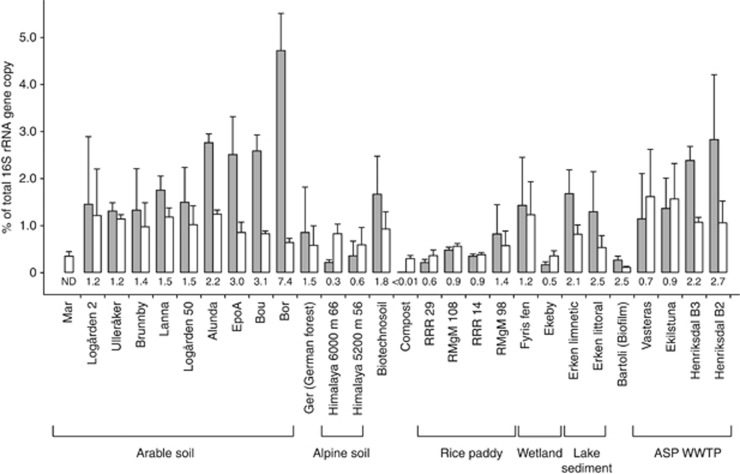
The relative abundance of clade I and II nosZ genes in various environments was determined by calculating the ratio of nosZ gene copies to the total bacterial 16S rRNA gene copies (Figure 3). nosZ sequences from both clades were quantifiable in all samples except one arable soil, in which clade II nosZ sequences were below the detection limit. The ratio of clade II nosZ to 16S rRNA gene copies ranged from below detection limit to 4.9%, whereas that of clade I nosZ to 16S rRNA copies was 0.1 to 1.7%. Across all samples, the mean relative abundance of clade I nosZ (0.8%) was significantly lower than clade II nosZ (1.4% bootstrap t-test P value <0.001, n=10 000); however, the clade II to clade I ratio was variable among samples within environmental categories. In arable soils, the ratio was typically >1, where in a vineyard soil clade II nosZ gene copies exceeded that of clade I by a factor of seven. By contrast, compost, alpine soils and rice paddy soils were generally lower, often <1. There was no general pattern among activated sludge processes as the ratio of clade II to clade I nosZ varied between 0.7 and 2.7.
Figure 3.
Relative abundance of clade I (open bars) and clade II (gray bars) nosZ gene copies in samples from different environmental types, calculated as proportion of total bacterial 16S rRNA gene copies (mean±s.d.; see Supplementary Table S2 for number of replicates per site). The ratio of clade II to clade I mean relative abundance is shown for each sample below the bars. ASP/WWTP denotes samples from activated sludge processes in municipal wastewater treatment plants with nitrogen removal processes.
Discussion
The need to mitigate N2O emissions, particularly in managed ecosystems with high nitrogen inputs, has been highlighted in various reports on climate change. The N2OR is likely to have a central role in any management strategy designed to attenuate N2O release from different environments (Richardson et al., 2009). However, our knowledge of the extant diversity and abundance of the only enzyme known to date that utilizes N2O as a primary substrate remains limited. In this study we demonstrate that the overall phylogenetic diversity of nosZ sequences can be divided into two physiologically meaningful groups that, with few exceptions, differ in how the N2OR is translocated across the cell membrane. The extent of the detected phylogenetic diversity of nosZ clade II and the fact that it was present in nearly all tested environments in equal or higher abundance as nosZ clade I suggests that at least half of the diversity of nosZ has been unaccounted for in previous studies. This hitherto unexplored lineage of N2OR is significant for understanding the relationship between diversity and functioning in N2O-reducing microbial communities.
Given the relative coherence of the nosZ and 16S rRNA phylogenies (Jones et al., 2008; Palmer et al., 2009), the phylogenetic placement of environmental sequences in combination with the quantitative PCR results indicate that organisms within the Bacteroidetes, Gemmatimonadetes and Delta-proteobacteria make up a significant proportion of N2O-reducing communities in different environments. This is consistent with several studies showing these phyla to be among the more abundant bacterial groups in soil and freshwater lakes and sediments (Lauber et al., 2009; Newton et al., 2011). For example, Bacteroidetes is often the second or third most abundant group in arable soils (Roesch et al., 2007; Youssef and Elshahed, 2009; Wessén et al., 2010). This may explain the higher relative abundance of clade II type nosZ sequences in several of the arable soil samples (Figure 3), as the majority of sequences from soil clone libraries grouped with nosZ from Bacteroidetes. The same pattern was observed for the lake sediment samples, where the two-fold difference in abundance between clade I and clade II corresponds to previous reports that have identified Bacteroidetes as being highly abundant members of bacterial communities and freshwater lakes, and can dominate in sites with high DOC or humic matter loading (Fierer et al., 2007; Newton et al., 2011). No representatives from the Epsilon-proteobacterial nosZ clade were detected, which is in agreement with 16S rRNA-based surveys of proteobacterial diversity in soils and limnetic sediments that commonly do not detect Epsilon-proteobacteria sequences (Spain et al., 2009), or find them in very low abundance (Roesch et al., 2007). This is not surprising as nearly all cultured representatives of Epsilon-proteobacteria have been isolated from animal intestinal tracts or deep-sea hydrothermal and cold seep vents.
Given that the sequences detected in this study were from a somewhat limited range of samples, we extrapolated the sequencing results in a quantitative manner by determining the abundance of both clades I and II nosZ on samples from a more diverse set of environments. The quantification of nosZ and other denitrification genes in elucidating the relative importance of abiotic environmental factors versus denitrifier diveristy in determining ecosystem denitrification rates has been advocated in recent studies (Hallin et al., 2009; Petersen et al., 2012). A key interest to studies relating denitrification rates to community structure is the ratio between the abundance of nitrite reductase genes and nosZ, as changes in this ratio may directly influence potential N2O emissions (Garcia-Lledo et al., 2011; Philippot et al., 2011). Environmental studies have commonly shown a 2–10 times lower abundance of denitrifiers harboring the nosZ gene compared with those having the genes encoding the preceding denitrifying reductases in the pathway (Henry et al., 2006; Hallin et al., 2009; Bru et al., 2010). We observed that the mean relative abundance of clade I and clade II nosZ across the quantitative PCR sample set was quite similar; however, the ratio of the two clades was variable among samples within different environment categories. Quantification for the first time of the abundance of clade II nosZ indicates that N2O-reducing communities have been underestimated in previous studies, which only targeted clade I nosZ. However, as the abundance of clade II is in the same range as clade I, the sum of the abundance of nosZ genes from both clades remains substantially lower than the abundance of the nitrite reductase genes reported in different field studies (Henry et al., 2006; Kandeler et al., 2006; Yergeau and Kowalchuk, 2008; Hallin et al., 2009; Bru et al., 2010). Although we cannot exclude that we still do not account for the entire nosZ diversity, these results indicate that even taking into account the newly characterized nosZ clade II, a significant proportion of denitrifiers in the environment is genetically unable to reduce N2O, which is supported by denitrifier genome analysis (Jones et al., 2008).
Although previous studies examining the translocation of N2OR in different organisms have shown this enzyme to be exported to the periplasm by either the Tat or Sec translocation pathways (Simon et al., 2004; Zumft and Kroneck, 2007; Van Spanning et al., 2011), the phylogenetic analysis presented in this study revealed a distinct clustering of N2OR sequences into two clades that strongly corresponds with the inferred N2OR translocation pathway among N2O reducers. The coherence between the nosZ and 16S rRNA phylogenies makes it difficult to determine whether the preference toward one translocation pathway or the other is coincident with the overall evolution of the organisms harboring nosZ, or whether the mechanism of N2OR translocation in different organisms had a role in the evolution of nosZ. The difference in energetic cost of protein translocation between the two systems is substantial; protein export via the Sec pathway requires ∼1 molecule of ATP per 20 amino acids translocated across the membrane, whereas the energy use of Tat translocation is equivalent to 10 000 molecules of ATP (Lee et al., 2006). It is puzzling that the N2OR would be exported by two disparate mechanisms unless a selective pressure exists that causes one pathway to be preferred over the other among different organisms, or in different environments. The need for holoenzyme assembly to occur in the cytoplasm has been proposed as one possible selective force for using the Tat system, and is commonly observed for metalloproteins involved in electron transport (Pohlschröder et al., 2005). However, physiological evidence indicates that insertion of the copper and sulfur ligands into the folded N2OR precursor occurs in the periplasm for the N2OR, regardless of which translocation system is utilized (Zumft and Bothe, 2007). Other possible selective forces include ecological constraints on protein folding outside the cytoplasm, which seems reasonable for organisms that thrive in harsh conditions such as the Haloarcula and the other halophilic archaea in clade I, as well as thermophiles such as Ferroglobus and some Chloroflexi in clade II that have a Tat signal peptide. However, we find exceptions to this pattern, as nosZ in P. calidifontidis and Salinibacter ruber do not encode a Tat signal peptide, yet both thrive in extreme environments. Physiological tests comparing the fitness of denitrifiers with either Sec- or Tat-translocated nosZ under varying denitrifying conditions may help to elucidate the selective forces that have lead to the preference of one system over another. Differences in the efficiency of N2O reduction could potentially explain why the two N2OR lineages vary between habitat types or environmental conditions. Further studies should also address whether the predominance of organisms with clade I or clade II type nosZ in the environment could have consequences for net N2O emissions, particularly if a given ecosystem acts as a source or sink for N2O.
Acknowledgments
We gratefully acknowledge K McMahon, E Pelve and R Bernander for providing genomic DNA from Candidatus accumulibacter, Haloarcula marismortui and Pyrobaculum calidifontidis, J Andert for the Magnetospirillum magneticum culture and A Welsh for technical assistance. This work was supported by the European Commission within EcoFINDERS project (FP7-264465), the Swedish Research Council Formas (contract 2007-1197) and the Oscar and Lili Lamm foundation (2010-11-13).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Avrahami S, Bohannan BJM. N2O emission rates in a California meadow soil are influenced by fertilizer level, soil moisture and the community structure of ammonia-oxidizing bacteria. Global Change Biol. 2009;15:643–655. [Google Scholar]
- Babić KH, Schauss K, Hai B, Sikora S, Redzepović S, Radl V, et al. Influence of different Sinorhizobium meliloti inocula on abundance of genes involved in nitrogen transformations in the rhizosphere of alfalfa (Medicago sativa L.) Environ Microbiol. 2008;10:2922–2930. doi: 10.1111/j.1462-2920.2008.01762.x. [DOI] [PubMed] [Google Scholar]
- Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics. 2010;26:2811–2817. doi: 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- Bru D, Ramette A, Saby NPA, Dequiedt S, Ranjard L, Jolivet C, et al. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 2010;5:532–542. doi: 10.1038/ismej.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavigelli M, Robertson G. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology. 2000;81:1402–1414. [Google Scholar]
- Chapuis-Lardy L, Wrage N, Metay A, Chotte J, Bernoux M. Soils, a sink for N2O? A review. Global Change Biol. 2007;13:1–17. [Google Scholar]
- Cheneby D, Perrez S, Devroe C, Hallet S, Couton Y, Bizouard F, et al. Denitrifying bacteria in bulk and maize-rhizospheric soil: diversity and N 2 O-reducing abilities. Can J Microbiol. 2004;50:469–474. doi: 10.1139/w04-037. [DOI] [PubMed] [Google Scholar]
- Eddy S. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Garcia-Lledo A, Vilar-Sanz A, Trias R, Hallin S, Baneras L. Genetic potential for N2O emissions from the sediment of a free water surface constructed wetland. Water Res. 2011;45:5621–5632. doi: 10.1016/j.watres.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Green SJ, Prakash O, Gihring TM, Akob DM, Jasrotia P, Jardine PM, et al. Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixed-waste contamination. Appl Environ Microb. 2010;76:3244–3254. doi: 10.1128/AEM.03069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin S, Jones CM, Schloter M, Philippot L. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 2009;3:597–605. doi: 10.1038/ismej.2008.128. [DOI] [PubMed] [Google Scholar]
- Henry S, Bru D, Stres B, Hallet S, Philippot L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microb. 2006;72:5181–5189. doi: 10.1128/AEM.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Stackebrandt E. Reclassification of Sphaerobacter thermophilus from the subclass Sphaerobacteridae in the phylum Actinobacteria to the class Thermomicrobia (emended description) in the phylum Chloroflexi (emended description) Int J Syst Evol Micr. 2004;54:2049–2051. doi: 10.1099/ijs.0.03028-0. [DOI] [PubMed] [Google Scholar]
- Jones CM, Stres B, Rosenquist M, Hallin S. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol Biol Evol. 2008;25:1955–1966. doi: 10.1093/molbev/msn146. [DOI] [PubMed] [Google Scholar]
- Jones CM, Welsh A, Throbäck IN, Dorsch P, Bakken LR, Hallin S. Phenotypic and genotypic heterogeneity among closely related soil-borne N2—and N2O-producing Bacillus isolates harboring the nosZ gene. Fems Microbiol Ecol. 2011;76:541–552. doi: 10.1111/j.1574-6941.2011.01071.x. [DOI] [PubMed] [Google Scholar]
- Kandeler E, Deiglmayr K, Tscherko D, Bru D, Philippot L. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl Environ Microb. 2006;72:5957–5962. doi: 10.1128/AEM.00439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- Lauber C, Hamady M, Knight R. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microb. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lee PA, Tullman-Ercek D, Georgiou G. The bacterial twin-arginine translocation pathway. Annu Rev Microbiol. 2006;60:373–395. doi: 10.1146/annurev.micro.60.080805.142212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gutierrez J, Henry S, Hallet S, Martin-Laurent F, Catroux G, Philippot L. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J Microbiol Meth. 2004;57:399–407. doi: 10.1016/j.mimet.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L, Bryson K, Jones D. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mosier AR. Soil processes and global change. Biol Fert Soils. 1998;27:221–229. [Google Scholar]
- Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer K, Drake HL, Horn MA. Genome-derived criteria for assigning environmental narG and nosZ sequences to operational taxonomic units of nitrate reducers. Appl Environ Microb. 2009;75:5170–5174. doi: 10.1128/AEM.00254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen DG, Blazewicz SJ, Firestone M, Herman DJ, Turetsky M, Waldrop M. Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ Microbiol. 2012;14:993–1008. doi: 10.1111/j.1462-2920.2011.02679.x. [DOI] [PubMed] [Google Scholar]
- Philippot L, Abbate C, Bispo A, Chesnot T, Hallin S, Lemanceau P, et al. Soil microbial diversity: an ISO standard for soil DNA extraction. J Soil Sediment. 2010;10:1344–1345. [Google Scholar]
- Philippot L, Andert J, Jones CM, Bru D, Hallin S. Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Global Change Biol. 2011;17:1497–1504. [Google Scholar]
- Philippot L, Cuhel J, Saby NPA, Cheneby D, Chronakova A, Bru D, et al. Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ Microbiol. 2009a;11:1518–1526. doi: 10.1111/j.1462-2920.2009.01879.x. [DOI] [PubMed] [Google Scholar]
- Philippot L, Hallin S, Borjesson G, Baggs EM. Biochemical cycling in the rhizosphere having an impact on global change. Plant Soil. 2009b;321:61–81. [Google Scholar]
- Pohlschröder M, Hartmann E, Hand NJ, Dilks K, Haddad A. Diversity and evolution of protein translocation. Annu Rev Microbiol. 2005;59:91–111. doi: 10.1146/annurev.micro.59.030804.121353. [DOI] [PubMed] [Google Scholar]
- Pomowski A, Zumft WG, Kroneck PMH, Einsle O. N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase. Nature. 2011;477:234–237. doi: 10.1038/nature10332. [DOI] [PubMed] [Google Scholar]
- Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- Richardson D, Felgate H, Watmough N, Thomson A, Baggs E. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle—could enzymic regulation hold the key. Trends Biotechnol. 2009;27:388–397. doi: 10.1016/j.tibtech.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose T, Schultz E, Henikoff J, Pietrokovski S, McCallum C, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford R, Cole J, Tiedje J. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl Environ Microb. 2002;68:893–900. doi: 10.1128/AEM.68.2.893-900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Einsle O, Kroneck PMH, Zumft WG. The unprecedented nos gene cluster of Wolinella succinogenes encodes a novel respiratory electron transfer pathway to cytochrome c nitrous oxide reductase. FEBS Lett. 2004;569:7–12. doi: 10.1016/j.febslet.2004.05.060. [DOI] [PubMed] [Google Scholar]
- Spain AM, Krumholz LR, Elshahed MS. Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 2009;3:992–1000. doi: 10.1038/ismej.2009.43. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Van Spanning RJM.2011Structure, function, regulation and evolution of the nitrite and nitrous oxide reductases: denitrification enzymes with a beta-propeller FoldIn: Moir JWB, (ed)Nitrogen Cycling in Bacteria: Molecular Analysis Calister Academic Press: Norwich UK; 135–161. [Google Scholar]
- Wessén E, Hallin S, Philippot L. Differential responses of bacterial and archaeal groups at high taxonomical ranks to soil management. Soil Biol Biochem. 2010;42:1759–1765. [Google Scholar]
- Wood D, Setubal J, Kaul R, Monks D, Kitajima J, Okura V, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- Yergeau E, Kowalchuk GA. Responses of Antarctic soil microbial communities and associated functions to temperature and freeze-thaw cycle frequency. Environ Microbiol. 2008;10:2223–2235. doi: 10.1111/j.1462-2920.2008.01644.x. [DOI] [PubMed] [Google Scholar]
- Youssef NH, Elshahed MS. Diversity rankings among bacterial lineages in soil. ISME J. 2009;3:305–313. doi: 10.1038/ismej.2008.106. [DOI] [PubMed] [Google Scholar]
- Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W, Bothe H.2007Nitrous oxide reductasesIn: Bothe H, Ferguson SJ, Newton WE, (eds)Biology of the Nitrogen Cycle Elsevier: Amsterdam; 67–81. [Google Scholar]
- Zumft W, Kroneck P. Respiratory transformation of nitrous oxide (N2O) to dinitrogen by Bacteria and Archaea. Adv Microb Physiol. 2007;52:107–227. doi: 10.1016/S0065-2911(06)52003-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.