Abstract
First-generation adenovirus vectors (FG AdVs) are widely used in basic studies and gene therapy. However, virus-associated (VA) RNAs that act as small-interference RNAs are indeed transcribed from the vector genome. These VA RNAs can trigger the innate immune response. Moreover, VA RNAs are processed to functional viral miRNAs and disturb the expressions of numerous cellular genes. Therefore, VA-deleted AdVs lacking VA RNA genes would be advantageous for basic studies, both in vitro and in vivo. Here, we describe an efficient method of producing VA-deleted AdVs. First, a VA RNA-substituted “pre-vector” lacking the original VA RNA genes but alternatively possessing an intact VA RNA region flanked by a pair of FRTs was constructed. VA-deleted AdVs were efficiently obtained by infecting 293hde12 cells, which highly express FLP, with the pre-vector. The resulting transduction titers of VA-deleted AdVs were sufficient for practical use. Therefore, VA-deleted AdVs may be substitute for current FG AdV.
E1- and E3-deleted adenovirus vectors (AdVs) developed in the middle of 1990's1,2 are commonly known as first-generation (FG) AdVs, and have been extensively used not only for the basic studies of various gene functions in vitro and in vivo but also for preclinical and clinical gene therapy. Because FG AdVs lack E1A gene, which is an essential transactivator in all other viral promoters driven by RNA polymerase II. Therefore, FG AdVs were usually considered that they do not express any viral gene products. However, FG AdVs, in fact, express VA RNAs that are transcribed by RNA polymerase III when using these vectors both in vitro and in vivo, because its activity is probably independent of RNA polymerase II.
VA RNAs, VAI and VAII, located at about 30 map units on adenovirus 5 (Ad5), are non-coding RNAs consisting of 157–160 nucleotides (nt). These VA RNAs are extremely abundant during the late phase of infection and inhibit cellular RNA-interference pathways by saturating Exportin 5 and Dicer3. Also, VAI inhibits protein kinase R (PKR) activity and, consequently, eliminates the block of the cellular translation machinery to allow the efficient production of viral proteins. Moreover, VA RNAs were processed and generate miRNAs4,5, known as mivaRNAI and mivaRNAII that disturb the expression of many cellular genes, with the probable result of blocking cellular antiviral machinery. Therefore, there is no doubt that the AdVs lacking VA RNA genes are superior to current FG AdVs.
VAI and VAII are also transcribed not only during the early phase of Ad5 but also in E1A-deleted FG AdV. An E1-containing Ad5 mutant virus is reported which lack the expressions of both VAI and VAII and can proliferate in human cells, but its titers are approximately 60-fold lower than that of wild-type Ad56. Therefore, VA RNAs are not essential, but play an important role in efficient viral growth by overcoming cellular antiviral machinery. Although the E1-containing mutant virus lacking expression of VA RNAs can slightly grow, efficient systems for producing E1-, E3- and VA-deleted AdVs (simply denoted VA-deleted AdVs) are extremely difficult to develop. One system for generating VA-deleted AdV expressing GFP using a 293 cell line that inducibly expresses the VAI gene has been reported7. However, the transduction titer produced using this system is approximately 1,000-fold lower than that of current FG AdVs. Furthermore, 18 days (including a secondary passage) are required before VA-deleted AdV are first observed, and the aid of expression markers, such as GFP or luciferase fluorescence, may be necessary to isolate the VA-deleted AdV. Therefore, this production system using a VA-expressing cell line is impractical for general use.
We hypothesized that, although VA RNAs are not essential for viral growth, VA-deleted AdV cannot grow during the initial step of vector generation, where only a few copies of the viral genome are present per cell, possibly because viral genes other than VA RNAs that block the cellular antiviral machinery may not be sufficiently expressed. The amount of VA RNAs expressed from the chromosomes of the established cell lines may be insufficient, probably because abundant VA RNAs are necessary in the late phase of viral replication. Therefore, we adopted a strategy in which a VA-containing FG AdV (denoted pre-vector) is extensively amplified using 293 cells and the pre-vector is then converted to VA-deleted AdV through the removal of the VA genes using recombinase-expressing 293 cells to obtain a large amount of VA-deleted AdVs sufficient for the simultaneous introduction of numerous viral copies to the cells. However, this strategy appeared unrealizable because it requires the 293 cell line highly expressing the recombinase sufficient to remove VA RNA genes completely from rapidly replicating pre-vector genomes. We have established the 293 cell line, 293hde128, that contains the codon-humanized FLPe (hFLPe) that highly expresses thermo-stable FLPe recombinase9. We here showed that the VA genes present in the pre-vector genome were virtually completely removed using this cell line and high-titer VA-deleted AdVs were efficiently obtained.
Results
Efficient production of VA-deleted AdVs
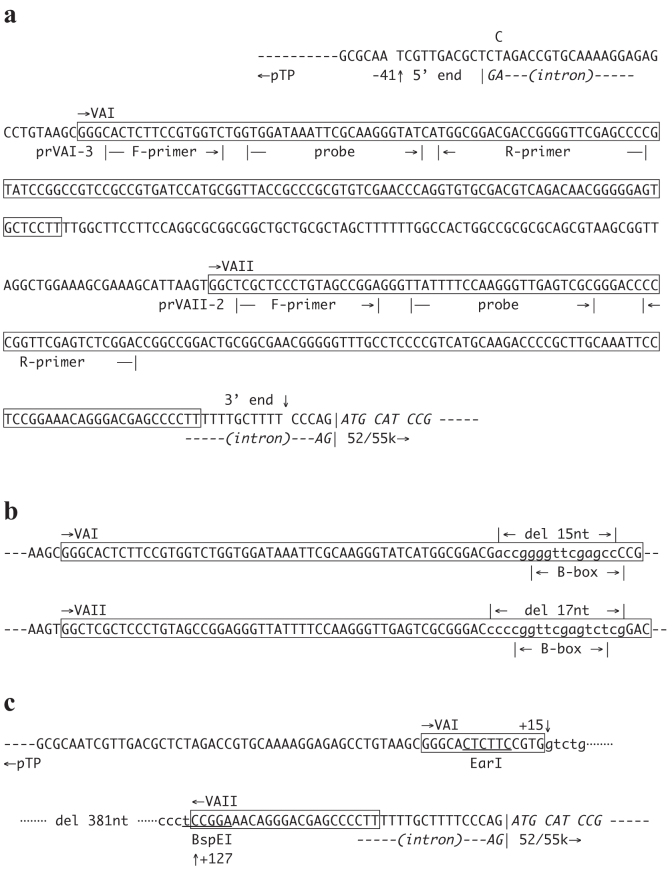
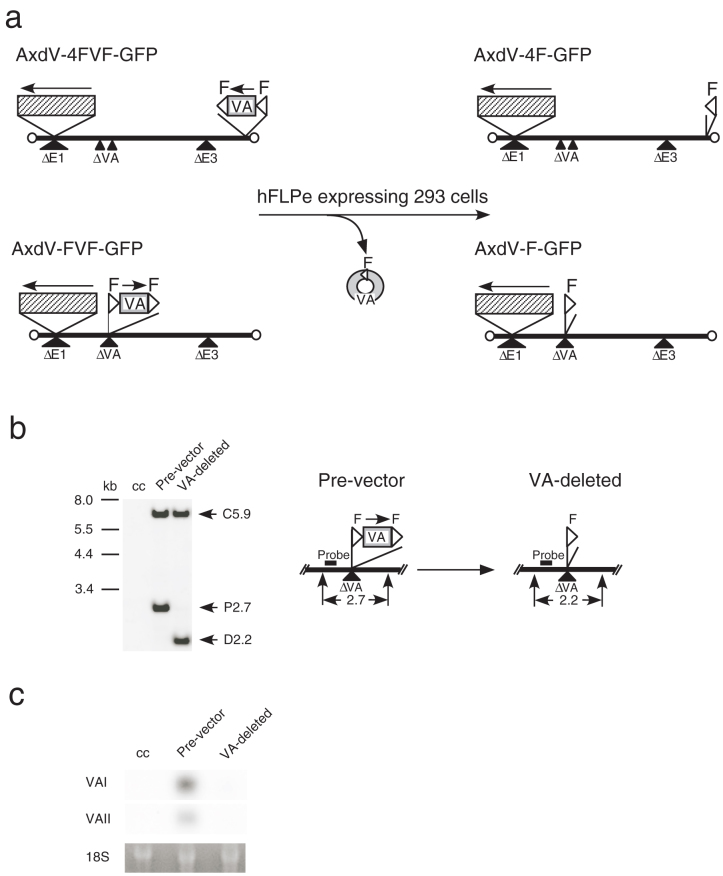
We first constructed a VA-expressing DNA fragment, FVF, that contains intact VAI and VAII genes (Fig. 1a) flanked with a couple of FRTs (F refers to FRT in this study). The splicing acceptor site present between the initiation codon of the terminal protein precursor (pTP) and the 5′ end of VAI was disrupted by replacing T with C to prevent possible aberrant splicing. Then, we constructed two structurally different EF-AdV pre-vectors expressing GFP under the control of the EF1α promoter, AxdV-4FVF-GFP and AxdV-FVF-GFP, in which the original VAI and VAII genes were disrupted and, instead, the functional FVF fragment was inserted at different positions. The former pre-vector lacks both B-box sequences of the VAI and VAII that are essential for the activity of the internal polymerase-III promoter because of the deletions of 15 nt and 17 nt, respectively, (Fig. 1b), and instead bears the FVF fragment at the E4 insertion site near the right end of the viral genome10 (Fig. 1a, upper left). The latter pre-vector lacks most of the VAI and VAII regions because of a deletion of 381 nt (Fig. 1c) and contains the FVF fragment within this region (Fig. 2a, lower left). Both pre-vectors grew well in the 293 cells, and their transduction titers were only slightly lower than that of the control FG AdV (Table 1, 37 × 107 and 60 × 107 versus 83 × 107 copies/mL).
Figure 1. Nucleotide sequences for VAI-VAII region.
(a) VAI and VAII genes in the FVF fragment. VAI and VAII are boxed. prVAI-3 and prVAII-2 are the primers used for qPCR for the detection and quantification of VAI and VAII, respectively. Note that the actual R-primers are complementary to the sequences shown in this figure. pTP, terminal protein precursor; 52/55 k, 52 k and 55 k proteins. (b) B-box deletions of internal promoter in AxdV-4FVF-GFP and AxdV-4F-GFP. The deleted nucleotides are shown in small letters. (c) VAI-VAII deletion in AxdV-FVF-GFP and AxdV-F-GFP. The deleted region is shown in small letters. The deletion of 381 nt starts at 15 nt and 127 nt downstream from the 5′ end of VAI and VAII, respectively.
Figure 2. Structures of pre-vectors and VA-deleted AdVs.
(a) Strategy for the generation of VA-deleted vectors. F, FRT. Hatched box, GFP expression unit including EF1α promoter. An arrow shows the orientation of the transcription. (b) Removal of FVF fragment from the pre-vector AxdV-FVF-GFP genome. Total DNAs of infected HuH-7 cells were digested with KpnI, and the DNA fragment containing FVF and F was detected using a Southern blot technique using the probe shown in the figure (enzyme, positions). C5.9 refers to the 5.9-kb fragment derived from outside of this region (1.7-kb BglII fragment, nt 30823-82495), showing that the amounts of the vector genomes were almost the same. cc, uninfected control cells; P, pre-vector; D, VA-deleted AdV. (c) Detection of expressed VA RNAs. Total RNAs of the same infected HuH-7 cells were analyzed using a northern blot technique. 18 S, 18 S ribosomal RNA.
Table 1. Transduction titers of VA-deleted AdVs.
| AdV | Type | Transduction titer copies/mL (×107) | Ratio (%) |
|---|---|---|---|
| AxdV-4FVF-GFP | pre-vector | 37 | 100 |
| AxdV-4F-GFP | VA-deleted | 5 | 14 |
| AxdV-FVF-GFP | pre-vector | 60 | 100 |
| AxdV-F-GFP | VA-deleted | 7 | 12 |
| AxdV-4FVF-NCre | pre-vector | 47 | 100 |
| AxdV-4F-NCre | VA-deleted | 4 | 8 |
| AxdV-FVF-SRChe | pre-vector | 20 | 100 |
| AxdV-F-SRChe | VA-deleted | 2 | 12 |
The transduction titer of the control FG AdV, AxEFGFP, was 83 × 107 copies/mL. The transduction titer was called the relative vector titer (rVT)12, which is about five-times lower than the conventional TCID50 titer of the pre-virus measured using 293 cells (see Methods).
Then, 293hde12 cells that constitutively and highly express codon-humanized FLPe recombinase8,11 were infected with pre-vectors to obtain the first stock of VA-deleted AdV. The vector lacking VA RNA genes grew efficiently in the 293hde12 cells, similar to the pre-vector genome in 293 cells, because the VA RNAs were supplied from the excised circular DNAs consisting of VA RNA genes and one copy of FRT (Fig. 2a, middle) and from the pre-vector genome prior to the FLP-mediated excision. Then, five times more volume of the first stock than usual was used to infect the 293hde12 cells to create the second stock. The vector replicated in the 293hde12 cells, probably because a large amount of VA-deleted AdV were infected: the titer of the vector in the first stock was very high, and a much larger volume of stock was used for infection. The resulting vectors were named AxdV-4F-GFP and AxdV-F-GFP, respectively (Fig. 2a, upper and lower right). When the vectors in the second stock were used to infect 293hde12 cells, a minimal degree of replications was observed. Using a high multiplicity of infection (MOI), however, the slight replication was observed though the replication progressed slowly. Vectors from the second stock were used for further characterization.
The transduction titers of VA-deleted AdVs were sufficient for practical use
Because VA-deleted AdV is difficult to grow in 293 cells, the conventional titration method for measuring viral growth cannot be used. Therefore, the transduction titers12 of the VA-deleted AdV and the pre-vector were measured; these titers show the copy number of viral genomes successfully transduced into infected target cells as evaluated using real-time PCR. The titers of VA-deleted AdV, pre-vector, and commonly used FG AdV can be compared using this method. The transduction titers of both AxdV-4F-GFP and AxdV-F-GFP were only one order lower than those of their pre-vectors (14% and 12%, respectively, Table 1), indicating that the titers of the VA-deleted vectors were sufficient for practical use as an alternative to FG AdVs. Moreover, using the same strategy, we produced other VA-deleted AdVs, such as AxdV-4F-NCre expressing Cre recombinase under the control of EF1α promoter and AxdV-F-SRChe expressing the Cherry marker under the control of the SRα promoter, and the resulting transduction titers were similar to those for AxdV-4F-GFP and AxdV-F-GFP (Table 1). Of note, the titer of Cre-expressing VA-deleted AdV was successfully obtained without trouble and was also quantitatively sufficient for general use, since Cre-expressing FG AdVs often exhibit a low titer and vector expansion is sometimes difficult.
Viral stocks of VA-deleted AdV likely contain a small amount of pre-virus that escaped the removal of the FVF fragment. Hence, HuH-7 cells were infected with AxdV-F-GFP and three days later, the transduced AdV DNA was examined using a Southern blot technique (Fig. 2b). The pre-vector DNA (2.7 kb) containing the VA genes clearly shifted to VA-deleted AdV DNA (2.2 kb), and no 2.7-kb band was detected in this assay. The result indicates that the VA RNAs in the pre-vector were efficiently removed during the replication of the pre-vector genome in 293hde12 cells. The same results were obtained when AxdV-4F-GFP was used (data not shown). The copy numbers of these viral genomes were examined using quantitative real-time PCR (qPCR) and VAI- and VAII-specific probes (Fig. 1a). Although the secondary structures of VAI and VAII are very similar, these probes were highly specific for one to the other (Supplementary Fig. S1). The pre-vector genome was present at approximately 1/90 or less, indicating that the purity of the VA-deleted AdV was about 99% or more.
Almost complete removal of contaminated pre-vector
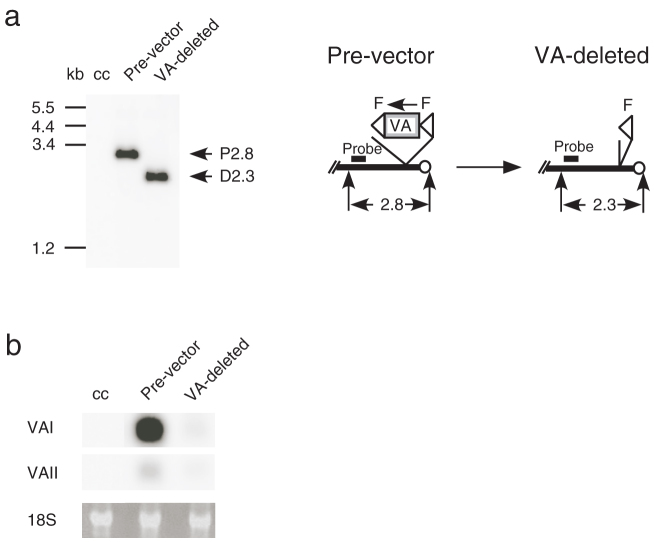
To examine the level of VA RNAs expressed from contaminated pre-vector, the total cellular RNA of HuH-7 cells infected with AxdV-F-GFP was extracted and the expressed VA RNAs were analyzed using a northern blot technique (Fig. 2c). Neither VAI nor VAII were detected. In contrast, the pre-vector AxdV-FVF-GFP expressed considerable amounts of VAI and VAII. The same results were obtained when AxdV-4F-GFP was used (data not shown). The FG AdV AxCAGFP expressing GFP under control of CAG promoter does express similar amount of VA RNAs (data not shown). Expressed VAI and VAII RNA in cells infected with AxdV-4F-GFP or AxdV-F-GFP were also examined using qPCR after reverse transcription (Table 2). Both VAI and VAII were hardly detected, compared with the pre-vector infected in parallel, especially when using AxdV-4F-GFP, though 1% to 3% of the VA RNAs were detected when infected with AxdV-F-GFP. The Table 2 also showed that AxdV-4F-GFP and AxdV-F-GFP preparations expressed VA RNAI only about 1/300 and 1/120 less than FG AdV (0.02/5.97 and 0.05/5.97), respectively. These results correlated well with those of the Southern blot and qPCR analyses described above. Because contamination by the pre-vector of AxdV-4F-GFP was hardly detected using qPCR, an extremely sensitive bioassay was performed; 293 cells were infected with the VA-deleted AdV stock and the possible presences of pre-vector genome and VA RNAs expressed from the pre-vector were examined using Southern and northern blot techniques, respectively. Only the pre-vector genome must replicate efficiently in 293 cells. The amplified pre-vector genome was still not detected in a Southern blot analysis (Fig. 3a), whereas the VA RNAs were slightly detected using a northern blot technique (Fig. 3b). Although the experiments using 293 cells were not quantitative, these results suggested that the amount of VA RNAs expressed from contaminated pre-vector in target cells other than 293 cells was minimal when the second stock was used. These results indicated that the VA-deleted AdVs produced using this production system were almost pure and may be used without consideration of the effect of VA RNAs in most studies. The contaminated circular DNA containing the VA-RNA genes was completely removed by purification of the vector.
Table 2. Expression levels of VAI and VAII RNAs measured using qPCR.
| VA RNAs | |||||
|---|---|---|---|---|---|
| I | II | ||||
| AdVs | Type | Copies ×108 | Ratio (%) | Copies ×108 | Ratio (%) |
| AxdV-4FVF-GFP | pre-vector | 4.47 ± 0.81 | 100 | 1.36 ± 0.15 | 100 |
| AxdV-4F-GFP | VA-deleted | 0.02 ± 0.00 | <1 | ND | <1 |
| AxdV-FVF–GFP | pre-vector | 1.81 ± 0.06 | 100 | 1.83 ± 0.32 | 100 |
| AxdV-F-GFP | VA-deleted | 0.05 ± 0.00 | 2.9 | 0.03 ± 0.00 | 1.4 |
| AxCAGFP | control FG AdV | 5.97 ± 0.83 | 4.17 ± 0.16 | ||
HuH-7 cells were infected with 50 μL of conventional stock of the pre-vector and the second stock of VA-deleted AdV. The method for preparation of the second stock is described in the Method section.
Figure 3. Viral genome and expressed VA-RNA of contaminated pre-vector replicating in 293 cells.
(a) Removal of the FVF fragment from the genome of the replicating pre-vector. Total DNAs of 293 cells infected with 15 μL of AxdV-4F-GFP stock were digested with EcoRV and the DNA fragment containing FVF and F was detected using a Southern blot technique. Other presentations are the same as in Fig. 1. (b) Detection of the expressed VA RNAs. Total RNAs of the same infected 293 cells were analyzed using a northern blot technique. 18 S, 18 S ribosomal RNA.
To examine whether the VA-deleted AdV expresses a gene product as efficiently as FG AdV, HeLa cells were infected with VA-deleted AdVs and pre-vectors expressing GFP at the same transduction unit value and the resulting fluorescence was measured (Table 3). The VA-deleted AdVs showed similar expression levels to those of their pre-vectors.
Table 3. GFP expression in target cells infected with VA-deleted AdVs.
| Ratio | |||
|---|---|---|---|
| AdV | Type | fluorescence | expressed mRNA |
| AxdV-4FVF-GFP | pre-vector | 1 | 1 |
| AxdV-4F-GFP | VA-deleted | 0.97 ± 0.15 | 0.81 ± 0.16 |
| AxdV-FVF-GFP | pre-vector | 1 | 1 |
| AxdV-F-GFP | VA-deleted | 1.14 ± 0.31 | 1.18 ± 0.20 |
HeLa cells were infected with pre-vector and VA-deleted AdV at a transduction MOI of 10. Three days later, GFP fluorescence was measured using a fluorometer and the GFP mRNA was quantified using reverse transcriptase and qPCR.
Discussion
The efficient removal of VA RNA genes from the pre-virus genomes is crucial for this strategy. We here showed that 293hde12 cell line that highly and constitutively express FLP recombinase was sufficient for this purpose. The cell lines constitutively expressing Cre have been used for production of helper-dependent AdVs, which contain a large DNA up to about 30kb. The viral packaging region of the helper virus must be excised out from the replicating viral genome but the excision efficiency is less than that described here13,14 and hence the contaminated helper virus must be removed by the buoyant centrifugation using cesium chloride. Probably such incomplete excision might result from the toxicity of highly expressed Cre in 293 cells15,16,17,18, while no toxicity of FLP has so far been reported. Therefore, 293hde12 cells played the most important role in this production system.
Furthermore, we demonstrated in this work that the AdVs containing two FRT sequences at the VA RNA region in the viral genome, AxdV-F-GFP etc., were viable and their titers were comparable to FG AdV. When a foreign DNA sequence is inserted into the AdV genome, even when the sequence is short such as recombination targets, the AdVs are often not viable or their titers are very low, probably because the small insertion influences on the replication of the AdV genome. For this reason only three positions, i.e. as the substitutions of E1 and E3 genes and the insertion upstream of E4 genes, are generally used. Hence, we first constructed AxdV-4F-GFP because we knew that the AdVs containing foreign DNA at the E4 insertion position was very stable shown by our early works1,19. Then, we tried the original site of VA RNA genes and demonstrated that the FRTs can successfully be inserted at the VA RNA region causing only slight reduction of the AdV titers.
This production system described here may allow the practical application of VA-deleted AdVs as a possible alternative to current FG AdVs, since the transduction titer was only one order lower than that of FG AdVs and, in fact, we have already produced more than twenty VA-deleted AdVs expressing various genes without any difficulty. Because VA RNAs may disturb experimental results that are obtained using FG AdVs to some extent, VA-deleted AdVs are likely to be preferable for many types of basic research using adenovirus vectors. When examining the influences of VA RNAs on previously obtained results, the pre-vector can be used as an ideal control for VA-deleted AdVs. Although the target genes of VA RNAs are not clear at present, Aparicio et al.5 reported that the expressions of many genes including TIA-1, a splicing and translation regulator, are downregulated by mivaRNAs, which are the processed products of VA RNAs, and they proposed that TIA-1 may be one such target of mivaRNAs. We compared the expression of TIA-1 using a VA-deleted AdV and its pre-vector and observed that TIA-1 expression was slightly downregulated but was not statistically significant in our assay system. Because they did not use a VA-deleted vector but rather E1-containing viruses that efficiently replicate in infected cells and probably express much more VA RNAs. Therefore, their results are not necessarily contradictory and may be explained by the quantitative differences of expressed VA RNAs. We found other genes that were downregulated by VA RNAs (YK, unpublished results). Studies on the influences of VA RNAs on immune responses in vivo are also underway. So far, we found that the immune response caused by VA RNA was lower than that by aberrantly expressed viral pIX protein20 but quantitative analyses are needed to clarify this issue. Notably, VA-deleted AdVs are probably superior to the current FG AdV for researches involving AdVs expressing siRNA and miRNA, since VA RNAs expressed by FG AdVs may compete with them and disturb the effects of these RNAs3. In fact, an shRNA expressed using VA-deleted AdV was more effective than that using current FG AdV (manuscript in preparation). These small-RNA technologies are extensively used in various research fields including signal transduction, cell differentiation and iPS study. Because more than three hundred papers have been published so far using FG AdVs expressing siRNA or miRNA based on our PubMed searching, VA-deleted AdV may be valuable in these fields.
The production level may be sufficient to apply VA-deleted AdVs to gene therapy as a safer alternative to existing practices. To what extent VA-deleted AdVs reduce immune responses in gene therapy of humans remain to be elucidated. Another possible contribution to gene therapy using AdVs is that, VA-deleted AdVs may considerably reduce contamination with replication-competent AdVs (RCA), one of the problems in the production of gene-therapy grade AdVs, because even if VA-deleted RCA is generated, only very low levels of replication would occur in human cells. Moreover, as proposed by Carnero et al.4, a VA-deleted replication-competent adenovirus produced using this method could be used as an oncolytic virus that specifically replicate in cells with inactive PKR, such as tumor cells with activated ras or Epstein-Barr virus.
The same strategy described here could also be applied to studies of other DNA viruses, such as herpes viruses, because drug-resistant cell lines that highly express viral genes essential for the viral life cycle are often difficult to obtain because of their slight toxicity to cell growth. Moreover, using this strategy and recombinase-expressing cells, two or more genes could be deleted simultaneously using mutant loxP 227221 and FRT mutants22,23 that exclusively recombine two identical mutant recombinase-targets. In conclusion, this strategy may accelerate studies using adenovirus vectors and may contribute to gene therapy.
Methods
Cells and virus titration
Human cell lines of 29324, HeLa and HuH-7 are derived from the embryonic kidney, the cervical carcinoma and the hepatocellular carcinoma, respectively. These cells were cultured in DMEM supplemented with 10% fetal calf serum (FCS). The 293 cells constitutively express adenoviral E1 genes and support the replication of E1-substituted AdVs. 293hde12 is a 293 cell line possessing the hFLPe gene8, an improved version of the FLPe gene9, in which the codon usage has been changed to that used in humans and which produces more FLPe enzyme. 293hde12 cells were cultured in DMEM supplemented with 10% FCS plus geneticin (0.75 mg/mL). After infection with AdVs, the cells were maintained in DMEM supplemented with 5% FCS without geneticin.
All the AdVs were titrated using the method of transduction titer known as the relative vector titer (copies/mL)12. In this titration method, the copy number of viral genomes successfully transduced into the infected target cells, such as HeLa or HuH-7 cells, are measured using real-time PCR. The transduction titer method can be used not only for VA-deleted AdV, but also for FG AdVs, including the pre-vectors. This method enabled us to compare the various titers, since the transduction titer is not influenced by the growth rate of 293 cells, even if an expressed gene product (such as Cre or dsRed) is slightly deleterious to 293 cells12. When the gene product is not deleterious, the titer obtained using this method corresponds to about one fifth of that using either a 50% tissue culture infectious dose (TCID50/mL)25,26 or a plaque assay; the reason for this difference is probably because the transduction efficiency of 293 cells is much higher than that of other cells. The sequences of TaqMan probes for the titration (named AdV-110,12) are derived from Ad5 pIX gene: forward primer, 5′-TGTGATGGGCTCCAGCATT-3′; probe, 5′-ATGGTCGCCCCGTCCTGCC-3′; reverse primer, 5′-TCGTAGGTCAAGGTAGTAGAGTTTGC-3′. A recommended protocol of the titration is available as Supplementary data of reference 12.
Plasmids
The pVA41da plasmid contains a DNA fragment covering the entire VAI and VAII from nt position 10576–11034 (Fig. 1a) of adenovirus type 5. A splicing acceptor site upstream of the pTp gene was disrupted, as described in the text. The region of functional VAI and VAII was excised as a HindIII-XbaI fragment and inserted at a SwaI site of pUFwF27; the resulting plasmid and DNA fragment containing intact VAI and VAII flanked two FRTs were named pUFVA41daF and FVF fragment, respectively.
Vector construction
All the AdVs described here were constructed using the cosmid cassette pAxcwit2 containing the full-length AdV genome28. The pre-vector cassette pAxdV-4FVF-w (w refers to the SwaI cloning site at the E1 region, see below) possesses the AdV genome in which VAI and VAII genes are disrupted by 15-nt and 17-nt deletions in their B-box sequences, respectively (Fig. 1b). The AdV genome contains the FVF fragment at the SnaBI site (nt position 35770) located in the E4 region at 165-nt downstream from the right end of the Ad5 genome10. A GFP-expressing unit under the control of the EF1α promoter29 was inserted into the SwaI cloning site at the authentic E1 substitution region to obtain the pre-vector cosmid pAxdV-4FVF-GFP. The EF1α promoter in the left orientation was adopted to express the GFP because the use of the promoter in this manner greatly reduces the immune response of AdV, compared with the CAG and CMV promoters20,30. The pre-vector AxdV-4FVF-NCre and VA-deleted AdV AxdV-4F-NCre possess identical structures to AxdV-4FVF-GFP and AxdV-4F-GFP, respectively, except that the GFP gene is replaced by a Cre recombinase gene tagged with a nuclear localization signal31. The other pre-vector cosmid cassette pAxdV-FVF-4c lacks the VAI and VAII genes because of a large deletion from nt 10635 to nt 11012 and instead carries the FVF fragment. Note that this cassette contains two cloning sites, the SwaI site in the E1 region and the ClaI site in the E4 region (as described above), and is useful for the simultaneous expression of two genes. The pre-vector cosmid pAxdV-FVF-GFP was obtained by inserting the GFP expression unit described above at the SwaI site. Also, another pre-vector cosmid pAxdV-FVF-4Che contains the Cherry marker gene (Clontech) under the control of an SRα promoter32 at this ClaI site. Then, the pre-vector AxdV-FVF-GFP, the VA-deleted vector AxdV-F-GFP, the pre-vector AxdV-FVF-4SRChe, and the VA-deleted AdV, AxdV-F-4SRChe were obtained using these pre-vector cosmid cassettes. The procedure to produce VA-deleted AdVs was as follows: the pre-vector genome in the cosmid cassette described above was excised with PacI and transfected into 293 cells. The obtained pre-vector was then amplified twice using 293 cells and was used to infect 293hde12 cells at 10 copies/cell of transduction multiplicity of infection (MOI) to obtain VA-deleted AdVs. Then, This VA-deleted AdVs were used to infect 293hde12 cells at 10 copies/cell. The resultant viral stock is called the second stock in the Result section.
Quantitative Real-time PCR
The RNA expressions of VAI and VAII were quantified using the primers and probes described in Fig. 1a. The sequences of the GFP primers were as follows: forward primer, CTACAACAGCCACAACGTCTATATCA; probe, CGACAAGCAGAAGAACGGCATCAAGG; reverse primer, ATGTTGTTGCGGATCTTGAAG. The total RNA of the infected cells was extracted, and the amounts of the expressed target RNA and 18S-rRNA (correction standard) were quantified using reverse-transcription and real-time PCR (Applied Biosystems Prism 7000); the ratio of the target RNA to 18S-rRNA was then calculated. The linear correlation between the amount of infected vector and the Ct values was confirmed (Supplementary Fig. S2). To quantify the DNA amount of VA RNAs and the AdV genome, the infected total cell DNA was prepared from cells using a previously described method33,34 or a DNA preparation kit (Macherey-Nagel, through TaKaRa Bio). Quantitative PCR was performed to detect the AdV genome using a probe for the pIX gene described above10,12. The amount of chromosomal DNA was simultaneously measured to correct the Ct values of the viral genome per cell, and the corrected Ct was shown throughout. The probes were derived from the sequence of the human β-actin gene for HeLa and HuH710. The qPCR reaction was performed according to the manufacturer's protocol: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min (Applied BioSystems).
Southern blotting analysis
HuH-7 cells in a 6-cm dish were infected with VA-deleted AdV and its pre-vector, and three days later the total DNA was prepared from the dish. Before alkaline treatment, the agarose gel was exposed to 0.1-N HCl for partial depurination, causing the DNA fragmentation of several hundred base pairs to obtain the complete transfer to the membrane33. The DNA was then transferred to the nylon membrane Hybond-N (Amersham GE) using the capillary-transfer method. Specific DNA was detected using a DIG DNA Labeling and Detection Kit (Roche Diagnostics). The probe DNA fragment derived from the viral genome was labeled with digoxigenin-UTP, and specific DNA was detected using the chemiluminescence of CDP-Star (Roche Diagnostics); the bands were visualized using LAS-4000 (Fuji Film).
Northern blotting analysis
The cells indicated in the figure legends were infected with pre-vector and VA-deleted AdV, respectively, at a transduction MOI of 10 copies/mL. The total RNA of the infected cells was extracted, and 30 μg per lane was electrophoresed in the agarose gel with tris-acetate-EDTA buffer. The RNA was transferred to the nylon membrane Hybond-N+ (Amersham GE) using the capillary-transfer method. Specific RNA was detected using a DIGDNA Labeling and Detection Kit (Roche Diagnostics).
Author Contributions
A.M. performed the experiments and contributed to the writing of the manuscript. Z.P., M.S., H.F., Y.O. and S.K. performed the experiments. I.S. discussed the data and wrote the manuscript. Y.K. designed the strategies and performed the experiments. All the authors discussed the results and commented on the manuscript.
Supplementary Material
supplementary info
Acknowledgments
We thank Ms M. Terashima for excellent technical support and Ms E. Kondo for secretarial assistance. This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology to Y.K. and S.K. and the Ministry of Health, Labour and welfare ; by Research on the innovative development and the practical application of new drugs for hepatitis B to I.S.
References
- Miyake S. et al. Efficient generation of recombinant adenoviruses using adenovirus DNA-terminal protein complex and a cosmid bearing the full-length virus genome. Proc. Natl. Acad. Sci. U S A 93, 1320–1324 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton M. & Graham F. L. Site-specific recombination mediated by an adenovirus vector expressing the Cre recombinase protein: a molecular switch for control of gene expression. J. Virol. 69, 4600–4606 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. & Cullen B. R. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 78, 12868–12876 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnero E., Sutherland J. D. & Fortes P. Adenovirus and miRNAs. Biochim. Biophys. Acta 1809, 660–667 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O. et al. Adenovirus VA RNA-derived miRNAs target cellular genes involved in cell growth, gene expression and DNA repair. Nucleic Acids Res 38, 750–763 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat R. A., Domer P. H. & Thimmappaya B. Structural requirements of adenovirus VAI RNA for its translation enhancement function. Mol. Cell Biol. 5, 187–196 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machitani M. et al. Development of an adenovirus vector lacking the expression of virus-associated RNAs. J. Control Release 154, 285–289 (2011). [DOI] [PubMed] [Google Scholar]
- Takata Y., Kondo S., Goda N., Kanegae Y. & Saito I. Comparison of efficiency between FLPe and Cre for recombinase-mediated cassette exchange in vitro and in adenovirus vector production. Genes Cells 16, 765–777 (2011). [DOI] [PubMed] [Google Scholar]
- Buchholz F., Angrand P. O. & Stewart A. F. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16, 657–662 (1998). [DOI] [PubMed] [Google Scholar]
- Kanegae Y. et al. High-level expression by tissue/cancer-specific promoter with strict specificity using a single-adenoviral vector. Nucleic Acids Res. 39, e7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Takata Y., Nakano M., Saito I. & Kanegae Y. Activities of various FLP recombinases expressed by adenovirus vectors in mammalian cells. J. Mol. Biol. 390, 221–230 (2009). [DOI] [PubMed] [Google Scholar]
- Pei Z., Kondo S., Kanegae Y. & Saito I. Copy number of adenoviral vector genome transduced into target cells can be measured using quantitative PCR: application to vector titration. Biochem. Biophys. Res. Commun. 417, 945–950 (2012). [DOI] [PubMed] [Google Scholar]
- Ng P., Evelegh C., Cummings D. & Graham F. L. Cre levels limit packaging signal excision efficiency in the Cre/loxP helper-dependent adenoviral vector system. J. Virol. 76, 4181–4189 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P., Parks R. J. & Graham F. L. Preparation of helper-dependent adenoviral vectors. Methods Mol. Med. 69, 371–388 (2002). [DOI] [PubMed] [Google Scholar]
- Loonstra A. et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. U S A 98, 9209–9214 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A., Brandon E. P., Kootstra N., Gage F. H. & Verma I. M. Delivery of the Cre recombinase by a self-deleting lentiviral vector: efficient gene targeting in vivo. Proc. Natl. Acad. Sci. U S A 98, 11450–11455 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D. P. & Livingston D. M. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell 8, 233–243 (2001). [DOI] [PubMed] [Google Scholar]
- Baba Y., Nakano M., Yamada Y., Saito I. & Kanegae Y. Practical range of effective dose for Cre recombinase-expressing recombinant adenovirus without cell toxicity in mammalian cells. Microbiol. Immunol. 49, 559–570 (2005). [DOI] [PubMed] [Google Scholar]
- Saito I., Oya Y., Yamamoto K., Yuasa T. & Shimojo H. Construction of nondefective adenovirus type 5 bearing a 2.8-kilobase hepatitis B virus DNA near the right end of its genome. J. Virol. 54, 711–719 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M. et al. Expression of pIX gene induced by transgene promoter: possible cause of host immune response in first-generation adenoviral vectors. Hum. Gene Ther. 18, 925–936 (2007). [DOI] [PubMed] [Google Scholar]
- Lee G. & Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216, 55–65 (1998). [DOI] [PubMed] [Google Scholar]
- Schlake T. & Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33, 12746–12751 (1994). [DOI] [PubMed] [Google Scholar]
- Nakano M., Ishimura M., Chiba J., Kanegae Y. & Saito I. DNA substrates influence the recombination efficiency mediated by FLP recombinase expressed in mammalian cells. Microbiol. Immunol. 45, 657–665 (2001). [DOI] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C. & Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36, 59–74 (1977). [DOI] [PubMed] [Google Scholar]
- Precious B. & Russell W. C. Growth, purification and titration of adenoviruses. In Virology: a practical approach. (ed. Mahy B. W. J.) 128–152 (IRL press, Oxford; 1991). [Google Scholar]
- Kanegae Y., Makimura M. & Saito I. A simple and efficient method for purification of infectious recombinant adenovirus. Jpn J. Med. Sci. Biol. 47, 157–166 (1994). [DOI] [PubMed] [Google Scholar]
- Nakano M. et al. Efficient gene activation in cultured mammalian cells mediated by FLP recombinase-expressing recombinant adenovirus. Nucleic Acids Res. 29, E40 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H., Terashima M., Koshikawa M., Kanegae Y. & Saito I. Possible mechanism of adenovirus generation from a cloned viral genome tagged with nucleotides at its ends. Microbiol. Immunol. 50, 643–654 (2006). [DOI] [PubMed] [Google Scholar]
- Kim D. W., Uetsuki T., Kaziro Y., Yamaguchi N. & Sugano S. Use of the human elongation factor 1 alpha promoter as a versatile and efficient expression system. Gene 91, 217–223 (1990). [DOI] [PubMed] [Google Scholar]
- Chiyo T. et al. Conditional gene expression in hepatitis C virus transgenic mice without induction of severe liver injury using a non-inflammatory Cre-expressing adenovirus. Virus Res. 160, 89–97 (2011). [DOI] [PubMed] [Google Scholar]
- Kanegae Y. et al. Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 23, 3816–3821 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe Y. et al. SR alpha promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell Biol. 8, 466–472 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I., Groves R., Giulotto E., Rolfe M. & Stark G. R. Evolution and stability of chromosomal DNA coamplified with the CAD gene. Mol. Cell Biol. 9, 2445–2452 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M. et al. Production of viral vectors using recombinase-mediated cassette exchange. Nucleic Acids Res. 33, e76 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary info