Abstract
Parameters that regulate or affect the cell cycle or the DNA repair choice between non-homologous end-joining and homology-directed repair (HDR) are excellent targets to enhance therapeutic gene targeting. Here, we have evaluated the impact of five cell-cycle modulating drugs on targeted genome engineering mediated by DNA double-strand break (DSB)-inducing nucleases, such as zinc-finger nucleases (ZFNs). For a side-by-side comparison, we have established four reporter cell lines by integrating a mutated EGFP gene into either three transformed human cell lines or primary umbilical cord–derived mesenchymal stromal cells (UC-MSCs). After treatment with different cytostatic drugs, cells were transduced with adeno-associated virus (AAV) vectors that encode a nuclease or a repair donor to rescue EGFP expression through DSB-induced HDR. We show that transient cell-cycle arrest increased AAV transduction and AAV-mediated HDR up to six-fold in human cell lines and ten-fold in UC-MSCs, respectively. Targeted gene correction was observed in up to 34% of transduced cells. Both the absolute and the relative gene-targeting frequencies were dependent on the cell type, the cytostatic drug, the vector dose, and the nuclease. Treatment of cells with the cyclin-dependent kinase inhibitor indirubin-3′-monoxime was especially promising as this compound combined high stimulatory effects with minimal cytotoxicity. In conclusion, indirubin-3′-monoxime significantly improved AAV transduction and the efficiency of AAV/ZFN-mediated gene targeting and may thus represent a promising compound to enhance DSB-mediated genome engineering in human stem cells, such as UC-MSCs, which hold great promise for future clinical applications.
Rahman and colleagues evaluate the impact of five cell cycle-modulating drugs on targeted genome engineering mediated by zinc-finger nucleases (ZFNs). They show that treatment of cells with the cyclin-dependent kinase inhibitor indirubin-3′-monoxime is especially promising as this compound significantly improves AAV transduction and the efficiency of AAV/ZFN-mediated gene targeting in vitro.
Introduction
The precise manipulation of complex genomes has been a hallmark of studying gene function in vivo for more than two decades (Capecchi, 1989). Gene targeting, the underlying technology, is based on the homologous recombination (HR) pathway and describes the targeted transfer of genetic information from an exogenous donor DNA to the host genome. Meanwhile, this technology has been increasingly employed in biotechnology, and therapeutic applications are tangible, as epitomized by the introduction of targeted alterations in the genome of multi- and pluripotent human stem cells (Chamberlain et al., 2004; Hockemeyer et al., 2009; Zou et al., 2009; Benabdallah et al., 2010). Two major platforms have been described to overcome the low HR frequency in such cells: a viral vector platform based on adeno-associated virus (AAV) (Chamberlain et al., 2004; Mitsui et al., 2009; Khan et al., 2010) and DNA double-strand break (DSB)-inducing agents, such as meganucleases (Silva et al., 2011; Takeuchi et al., 2011), zinc-finger nucleases (ZFNs) (Urnov et al., 2010; Carroll, 2011; Rahman et al., 2011), and transcriptional activator-like effector (TALE) nucleases (Bogdanove and Voytas, 2011; Mussolino and Cathomen, 2012).
Early studies involving the natural homing endonuclease I-SceI, which recognizes an 18-bp target site, demonstrated that the creation of a DSB in the target gene stimulates recombination with an exogenous donor DNA up to 10,000-fold (Rouet et al., 1994; Choulika et al., 1995). The enhancing effect of DSBs is mainly due to activation of the cellular DNA damage response. In the presence of high concentrations of an appropriately designed donor molecule, sequence information from the donor DNA can be transferred to the target locus by HR-based homology-directed repair (HDR). When employing designer ZFNs, gene conversion frequencies of up to 50% were reported in human cell lines in the absence of selection (Lombardo et al., 2007; Maeder et al., 2008). In combination with AAV donor vectors, gene-targeting frequencies of up to 65% in transduced human cell lines were reported (Miller et al., 2003; Porteus et al., 2003; Gellhaus et al., 2010; Hirsch et al., 2010 ; Händel et al., 2012), suggesting that a combination of the two platforms leads to synergistic effects.
Gene targeting in human stem cells has remained challenging. Although ZFN-mediated targeted integration has been achieved in up to 40% of a mesenchymal stromal cell (MSC) population (Benabdallah et al., 2010), such high frequencies have remained unmatched in other therapeutically relevant human stem cells, including embryonic stem or induced pluripotent stem cells (Mitsui et al., 2009; Zou et al., 2009). Hence, gene targeting in human stem cells has been dependent on the co-integration of a selection marker to enrich for targeting events (Hockemeyer et al., 2009; Mitsui et al., 2009; Zou et al., 2009; Khan et al., 2010). Importantly, MSCs, which bear differentiation potential to multiple lineages, such as bone, neurons, and cardiac tissues, have been effectively transduced with AAV serotype 2 vectors (Kumar et al., 2004; Chng et al., 2007). In concordance with the differential availability of the cellular DNA repair factors that mediate HDR or nonhomologous end-joining (NHEJ) throughout the cell cycle, AAV-mediated gene targeting has been shown to be dependent on the cell-cycle phase (Trobridge et al., 2005). Consequently, it has been speculated that transient cell-cycle arrest can affect the efficiency of ZFN- and AAV-mediated genome editing. A few studies have reported that a transient arrest in the G2 phase using the microtubule inhibitors vinblastine or nocodazole augment ZFN-mediated HDR up to seven-fold (Urnov et al., 2005; Potts et al., 2006; Maeder et al., 2008; Olsen et al., 2010). However, these compounds are highly toxic to cells and therefore not useful in a therapeutic context.
The present study was designed to assess the effect of the cell cycle on DSB-induced gene targeting and to identify compounds that stimulate HDR activity with minimal cytotoxicity. We identified the cyclin-dependent kinase (CDK) inhibitor indirubin-3′-monoxime as a promising candidate to increase AAV transduction and AAV/ZFN-mediated gene targeting by transiently arresting the cell cycle. Because indirubin-3′-monoxime is a well-tolerated anti-cancer drug (Eisenbrand et al., 2004), it may present a promising compound to enhance DSB-dependent genome engineering in future clinical applications.
Materials and Methods
Plasmids and vectors
Plasmid pRK5.mCherry was generated by subcloning the mCherry cassette from pRSET-BmCherry (gift of Dr. Roger Y. Tsien) into vector pRK5 (Alwin et al., 2005). All other plasmids have been described before: plasmids encoding AAV vector in Gellhaus et al. (2010); ZFN expression plasmids in Alwin et al. (2005) and Szczepek et al. (2007); and control vector pRK5.LHA-SceIΔ in Szczepek et al. (2007). Maps and sequences of all plasmids are available upon request.
Cell-cycle profiling
HeLa, HT-1080, and U-2 OS cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Biochrom AG) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (Invitrogen). Umbilical cord–derived mesenchymal stromal cells (UC-MSCs) were isolated as previously described (Hatlapatka et al., 2011) and cultured in α-MEM Glutamax (Invitrogen) supplemented with 10% FCS selected for MSC (Stem Cell Technologies), 100 IU/ml penicillin, and 10 mg/ml streptomycin (PAA Laboratories GmbH). The polyclonal target cells HeLa.696, HT1080.696, U2OS.696, and UC-MSC.696 were generated by lentiviral transduction as previously described (Cornu and Cathomen, 2007; Gellhaus et al., 2010). The phenotype of UC-MSC was evaluated by flow cytometric analysis of surface antigens typical for MSC (CD105, CD73, CD90) and absence of hematopoietic stem cell markers (CD45, HLA-DR, CD14, CD34, CD19). All antibodies had been purchased from Becton Dickinson and staining procedure was performed as previously described (Hatlapatka et al., 2011). For cell-cycle profiling, cells were kept in serum-free conditions for 24 h before 200,000 cells were seeded per well of a 6-well plate. Four hours later, cells were incubated for 4 h (vinblastine) or 24 h (hydroxyurea, indirubin-3′-monoxime, L-mimosine or etoposide) with the indicated components (Sigma-Aldrich). Then, cells were fixed overnight in ethanol 24, 48, or 72 h after treatment. Before staining with propidium iodide solution (3.8 mM sodium citrate, 25 μg/ml of PI, 0.2 mg/ml of RNase A), low molecular weight DNA was released by incubating samples in DNA extraction buffer (200 mM Na2HPO4, 100 mM citric acid pH 7.8). At least 10,000 cells were assessed by flow cytometry (FACSCalibur, BD Biosciences), and cell cycle profile was evaluated by plotting FL3-W versus FL3-A (see Supplementary Fig. S1 for details, available online at www.liebertonline.com/hum). The number of cells containing a fragmented genome due to apoptosis was quantified by determining the percentage of subG1 events (see Supplementary Fig. S2 for details).
Recombinant AAV and cell transduction
Recombinant AAV vectors (rAAV) were produced and titrated as previously described (Gellhaus et al., 2010). HeLa, HT-1080, or U-2 OS cells were incubated in serum-free DMEM for 24 h before 50,000 cells per well of a 12-well plate were seeded and treated with cytostatic drugs as described above. To assess AAV transduction, cells were infected with rAAV.donor-REx at 20 gc/cell (HeLa and U-2 OS) or 50 gc/cell (HT-1080). For AAV-based gene targeting, polyclonal target cell lines were infected with rAAV.donor-SceI at 2,000 gc/cell alone or with rAAV.donor at 2,000 gc/cell in combination with I-SceI or ZFN expression vectors at 2000, 500, or 200 gc/cell. For AAV-mediated gene correction in human primary cells, 5×104 UC-MSC.696 were seeded in 24-well plates coated with 0.1% porcine gelatin (Sigma-Aldrich) 6–8 h before transduction with 10,000 gc/cell of AAV-ZFN and 2,000 gc/cell of AAV-donor. The frequencies of gene targeting and transduction were determined 4 or 11 days after transduction by flow cytometry, respectively.
Transfection-based gene targeting
100,000 serum-starved HeLa.696 cells were seeded into each well of a 12-well plate and transfected using polyethylenimine (PEI) as previously described (Söllü et al., 2010). Twenty-five ng of nuclease expression vectors (ZFN or I-SceI) were co-transfected with 1.6 μg of donor DNA (pTR.dGFPiN) and 100 ng of pRK5.mCherry as an internal transfection control. After 24 h, cells were rinsed and incubated with 40 mM hydroxyurea for 24 h or 40 nM vinblastine for 4 h. At 3 and 7 days post-transfection, the percentage of EGFP and mCherry-positive cells was determined by flow cytometry. Relative gene targeting is indicated as EGFP positive cells in the mCherry positive cell fraction.
Western blotting
Western blots were basically performed as described before (Händel et al., 2009). Cells were harvested 72 h post-infection with AAV vectors and resuspended in lysis buffer. Equal amounts of proteins (60 μg) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-P membrane (Millipore). Proteins were detected using antibodies recognizing the HA tag (Novus Biologicals; 1:5,000) and visualized after incubation with an IRDye-800CW–labeled secondary antibody (Li-Cor Bioscience; 1:20,000) using an Odyssey® Infrared Imaging System (Li-Cor Bioscience).
Quantitative PCR
Total DNA from polyclonal cells was isolated 30 days after AAV transduction using the Invisorb Spin Cell Mini Kit (Invitek). For quantitative real-time polymerase chain reaction (PCR), SYBR Green (DyNAmo™ Capillary SYBR Green, Finnzymes) and template concentrations of 100 ng genomic DNA were used. The PCR cycling conditions were as follows: Denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 10 sec, 60°C for 20 sec, and 72°C for 20 sec. Random integration (RI) #538 RIfw (5′-tgcttacatccgttctcgtg) and #539 RIrv (5′-cgcagacccttaaccaggta); target locus (TL) #361 TLfw (5′-gaggagctgttcaccggg) and #237 TLrv (5′-cgtaggtcagggtggtcac); gene targeting (GT) #136 GTfw (5′-caagggcgaggagctgtt) and #559 GTrv (5′-ctcggcgcgggtcttgtag). The samples for detection of gene targeting were pre-amplified using 0.125 U of Taq polymerase (Peqlab Biotechnologie GmbH) using 300 ng of genomic DNA, 300μM of each primer (#361 TLfw and #559 GTrv), 200mM dNTPs, and 1x reaction buffer. Cycling conditions were as follows: denaturation at 94°C for 4 min, followed by 13 cycles of 94°C for 30 s, 60°C for 30 s, 2 min at 72°C, and a final extension of 7 min at 72°C. The product of this first PCR reaction was purified with the Invisorb PCR Purification Kit (Invitek), and 1 out of 20 μl was used as a template in the qualitative PCR (qPCR).
Genotyping
Allele-specific PCR using primer pair #136 GTfw and #559 GTrv was employed to detect gene correction on the genome level, as described previously (Händel et al., 2012). Briefly, cells were harvested 16 days after transduction, and 300 ng of genomic DNA was used for PCR. Control reaction was performed with PTBP2 gene-specific primers (#1274 PTBPfw 5′-tctcattccctatgttcatgc and #1275 PTBPrv 5′-gttcccgcagaatggtgaggtg). PCR products (328 bp and 150 bp, respectively) were visualized by electrophoresis on a 2% agarose gel.
Statistical analysis
All experiments were independently performed at least three times. Error bars represent standard deviation, and statistical significance was determined using a two-sided Student's t-test with unequal variance.
Results
Optimizing transient cell-cycle arrest
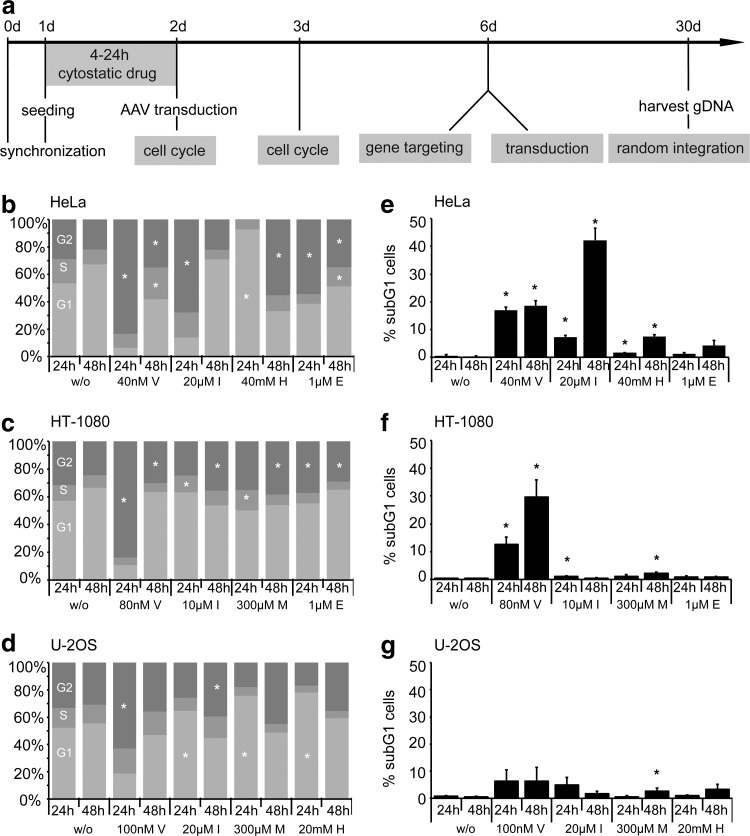
An initial set of experiments was performed to determine the optimal drug concentration to transiently arrest the cell cycle with minimal toxicity in three different human cell lines. To this end, HeLa (aneuploid adenocarcinoma), HT-1080 (quasi-diploid fibrosarcoma), and U-2 OS (hypertriploid osteosarcoma) cells were serum starved for one day prior to incubation with a broad range of concentrations of etoposide, hydroxyurea, indirubin-3′-monoxime, L-mimosine, and vinblastine. Cell-cycle profiles and drug-induced apoptosis were measured at day 1 and 2 after treatment with cytostatics by staining fixed cells with propidium iodide and ensuing analysis of DNA content by flow cytometry (Fig. 1a and Supplementary Fig. S1). The cell cycle profiles in response to optimal concentrations of the chosen chemicals are depicted in Figure 1b–d. The microtubule inhibitor vinblastine induced the strongest G2 arrest in the three tested cell lines. Incubation of cells with the CDK inhibitor indirubin-3′-monoxime affected the cell cycle with different kinetics and induced a significant G2/M arrest in HeLa cells at day 1 after treatment and in HT-1080 and U-2 OS cells after two days. On the other hand, some cytostatic compounds induced significant effects only in a subset of the tested cell lines. For instance, treatment with hydroxyurea induced a robust G1 arrest in HeLa and U-2 OS cells but did not affect HT-1080 cells (not shown).
FIG. 1.
Cell-cycle profile and toxicity. (a) Experimental setup. Cells were synchronized by serum starvation for one day prior to incubation with cytostatic drugs for 4 h (vinblastine) or 24 h (all others). Cell-cycle profiles were measured 24 h and 48 h later by flow cytometry. For gene targeting, cells were transduced on day two with the AAV vectors, and the extent of gene targeting and transduction was determined by flow cytometry on day six. After 30 days in culture, cells were harvested and genomic DNA extracted for analysis of AAV random integration. (b–d) Cell-cycle profiles of HeLa (b), HT-1080 (c), and U-2 OS (d) cells. At 24 h and 48 h after treatment with cytostatics, cells were fixed, stained with propidium iodide, and analyzed by flow cytometry. The percentage of cells in G1 (light gray), S (gray), and G2/M (dark gray) phase is indicated with a bar graph. Asterisks mark a significant difference (p<0.05) from nontreated cells. (e–g) Cytostatic treatment-induced toxicity in HeLa (e), HT-1080 (f), and U-2 OS (g) cells. At 24 h and 48 h after treatment, cells were fixed, stained with propidium iodide, and analyzed by flow cytometry. The percentage of cells in subG1 phase is indicated. Asterisks mark significant differences (p<0.05) from nontreated cells. E, etoposide; H, hydroxyurea; I, indirubin-3′-monoxime; M, L-mimosine; V, vinblastine; w/o, without cytostatics.
To assess the cytotoxicity associated with the treatment regiment, the extent of apoptosis was determined by quantifying the subG1 fraction (Supplementary Fig. S2). Vinblastine was associated with high toxicity in all cell lines tested (Fig. 1e–g). The anti-cancer drug indirubin-3′-monoxime was described as a potent inducer of caspase-mediated apoptosis in HeLa cells (Shi and Shen, 2008), but the substance revealed no or only mild toxicity in HT-1080 and U-2 OS cells. The other compounds, hydroxyurea, etoposide, and L-mimosine, provoked no or only mild cytotoxicity in all tested cell lines at the concentrations used.
In summary, we have compared five drugs for conditions to induce a transient cell-cycle arrest in the G1 or G2/M phase with the lowest possible toxicity in three different cell lines. The observed differences with regard to the degree of cell-cycle arrest and cytotoxicity is most likely a reflection of the different genetic background and chromosomal alterations of the used cell lines.
Transient cell-cycle arrest augments AAV-mediated gene targeting
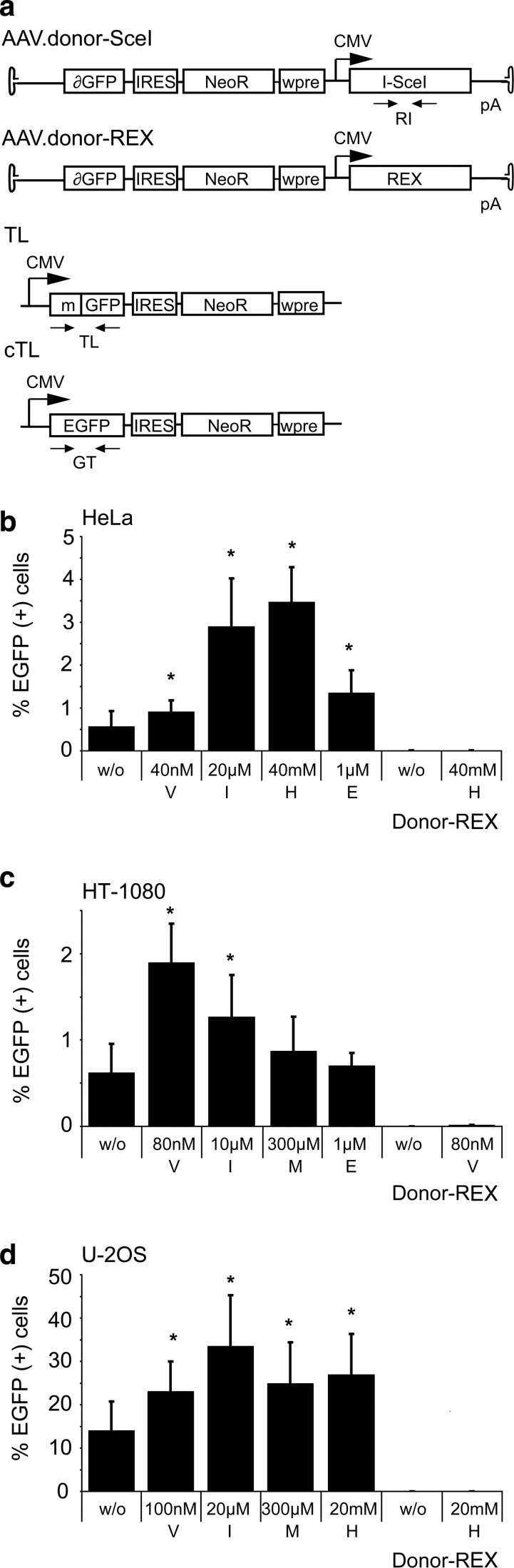
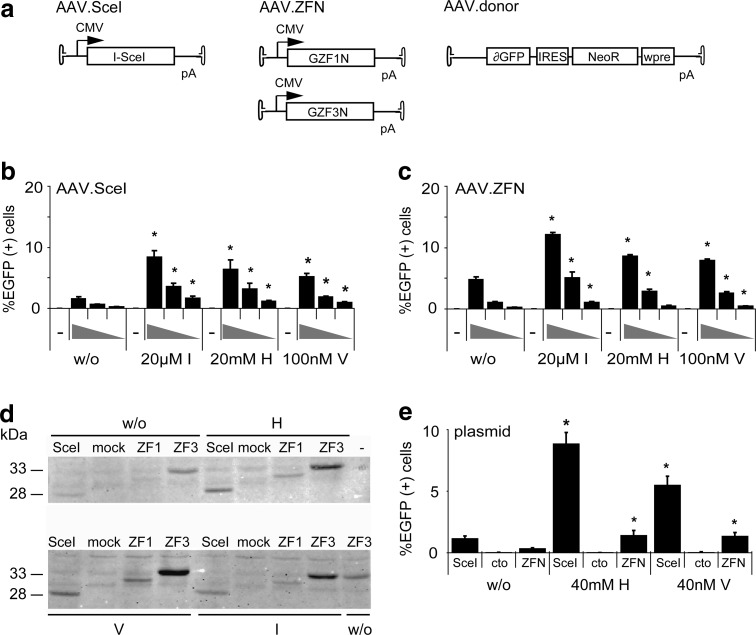
In order to assess quantitatively whether a transient G2 arrest enhances gene targeting after induction of a DSB in the target locus, we employed a previously described enhanced green fluorescent protein (EGFP) rescue assay (Gellhaus et al., 2010). Target cell lines that carry a mutated EGFP target locus were generated by lentiviral transduction and subsequent selection in the presence of neomycin. To ensure that results are comparable between the different cell lines, we worked with polyclonal cell populations, and the parental cells were transduced with less than 0.1 transducing units per cell to make sure that individual cells are unlikely to contain more than one target locus. The EGFP open reading frame in the target locus is disrupted by binding sites for I-SceI and a ZFN. Upon delivery of a donor DNA in the form of an AAV vector, EGFP expression can be rescued by HDR between the target locus and the AAV donor (Fig. 2a). As shown previously, creation of a DSB in the target locus by expression of I-SceI stimulates gene correction significantly (Miller et al., 2003; Porteus et al., 2003; Gellhaus et al., 2010). HeLa, HT-1080, and U-2 OS-based target cell lines were treated with cytostatic drugs and then infected with 2,000 genome copies (gc) of AAV.donor-SceI or AAV.donor-REx per cell. Treatment with hydroxyurea and indirubin-3′-monoxime–enhanced AAV-based gene targeting in HeLa cells by a factor of 5–6, while vinblastine and etoposide had significant but more modest effects (Fig. 2b). In HT-1080 cells, treatment with vinblastine and indirubin-3′-monoxime revealed the strongest effects with a 2- to 3-fold stimulation of EGFP rescue (Fig. 2c). While AAV-based gene targeting was able to rescue EGFP expression in about 0.6% of infected HeLa and HT-1080 cells, U-2 OS cells were much more susceptible to gene correction with AAV vectors (Fig. 2d). In the absence of cytostatics, 14% of cells were EGFP-positive upon infection with AAV.donor-SceI. Treatment of cells with indirubin-3′-monoxime could stimulate gene correction more than 2-fold to 34% of corrected cells. Of note, after infection with the control vector AAV.donor-REx, the number of EGFP-positive cells was not significantly above background for all target cell lines, confirming that efficient AAV-mediated gene targeting is dependent on the creation of a DSB in the EGFP target locus.
FIG. 2.
Effect of cytostatic drugs on DNA double-strand break-stimulated gene targeting. (a) Setup of DSB-mediated gene correction. The target locus (TL) contains a disrupted EGFP (mGFP) gene with binding sites for I-SceI and zinc-finger nucleases (ZFNs). After infection with vector AAV.donor-SceI, the I-SceI–induced DSB stimulates homologous recombination with the AAV donor to restore EGFP (cTL). AAV.donor-REx expresses DsRed-Express and served as a control vector. Small arrows indicate primer binding sites for qPCR analysis (Fig. 4). (b–d) Rescue of EGFP expression in HeLa (b), HT-1080 (c), and U-2 OS cells (d). A total of 50,000 cells were treated with cytostatic drugs, as indicated, before infection with AAV.donor-SceI (2,000 gc/cell). The extent of gene correction was measured by counting EGFP-positive cells by flow cytometry four days after transduction. Graphs indicate average and standard deviation of three independent experiments. Asterisks mark significant differences (p<0.05) from nontreated cells. IRES, internal ribosome entry site; NeoR, neomycin resistance gene; wpre, woodchuck hepatitis post-transcriptional regulatory element; cTL, corrected target locus; E, etoposide; H, hydroxyurea; I, indirubin-3′-monoxime; M, L-mimosine; V, vinblastine; w/o, without cytostatics.
In conclusion, transient cell-cycle arrest induced by particular cytostatics enhanced AAV-mediated gene correction in a cell-type–specific manner. Only the CDK-inhibitor indirubin-3′-monoxime showed at least a 2-fold stimulatory effect in all three tested cell lines.
Cytostatic drugs augment AAV transduction
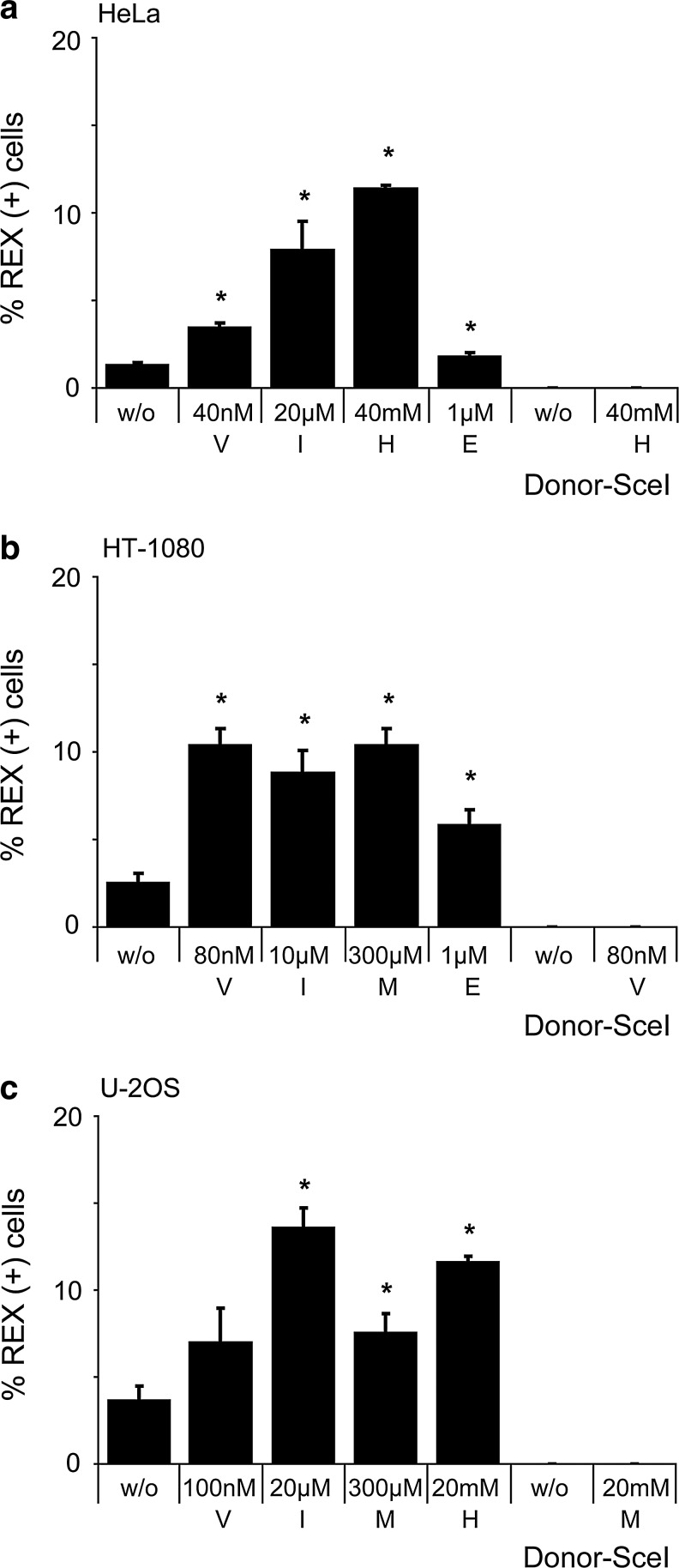
Reagents that induce DNA damage have been described to enhance the transduction of AAV in cultured cells and this effect has been mainly attributed to their properties of activating the DNA damage response (Alexander et al., 1994; Russell et al., 1995). Using the donor vector containing the REx expression cassette, we compared the effects of the different drugs on AAV transduction in the three different cell lines. Upon infection with 20 or 50 gc/cell of AAV.donor-REx, 1–4% of cells were scored REx-positive by flow cytometry four days later (Fig. 3). Preincubation of cells with the examined cytostatic drugs increased AAV transduction significantly between 1.5 to 8.3-fold, depending on the cell line and the specific drug used. However, only indirubin-3′-monoxime and vinblastine showed a stimulatory effect in all three cell lines. Infection with AAV.donor-SceI served as a negative control. These results show that the beneficial effects of the cytostatic drugs on AAV transduction parallels the increase in AAV-mediated gene targeting (Fig. 2), suggesting that transduction is a rate-limiting step in AAV-mediated gene targeting.
FIG. 3.
Effect of cytostatic drugs on AAV transduction. Transduction frequency in HeLa (a), HT-1080 (b), and U-2 OS (c) cells. After treatment with cytostatic drugs, 50,000 cells were transduced with AAV.donor-REx at 20 gc/cell (HeLa, U-2 OS) or 50 gc/cell (HT-1080) and assessed by flow cytometry four days later. Graphs represent average percentage of REx-positive cells with standard deviation, and asterisks mark significant (p<0.05) differences in transduction frequency of treated vs. nontreated cells. Control infections were performed with AAV.donor-SceI. E, etoposide; H, hydroxyurea; I, indirubin-3′-monoxime; M, L-mimosine; V, vinblastine; w/o, without cytostatics.
Targeting ratio is cell-type dependent
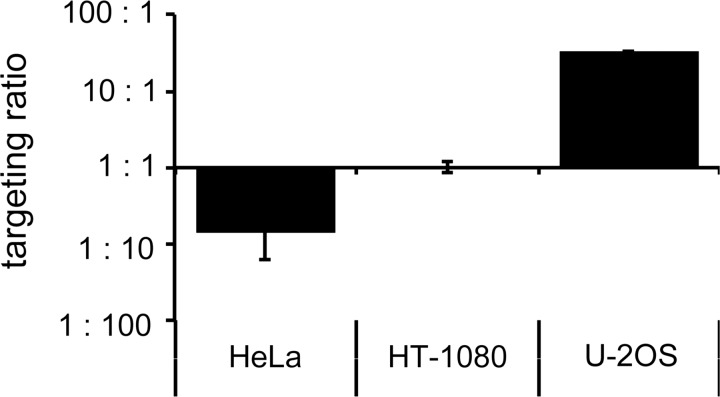
The cellular proteins mediating HDR are mainly available in the G2 phase of the cell cycle, while NHEJ factors are more prominent in G1 (Shrivastav et al., 2008). Changing the cell-cycle profile during AAV infection could therefore affect the fate of the donor DNA in terms of whether it will be used as a template for HDR-mediated gene targeting or for NHEJ-mediated illegitimate recombination, respectively. To answer this question, the gene targeting frequency of cells transduced with AAV.donor-SceI was offset against the random integration frequency of the vector, as determined by flow cytometry and quantitative PCR on extracted genomic DNA. The targeting ratio was defined here as the quotient between the gene targeting frequency and the illegitimate integration frequency. Similar to the diverse responses to treatment with cytostatic drugs in terms of cell-cycle profile, the different cell lines dealt differently with the exogenous vector DNA. While in HeLa cells the AAV vector was ∼7 times more likely to integrate into the host genome than to serve as a template for HDR, the ratio was about 1:1 in HT-1080 cells, and 33:1 in favor of gene targeting in U-2 OS cells (Fig. 4). However, none of the cytostatic drugs had a significant effect on the targeting ratio in any of the tested cell lines (Supplementary Fig. S3). In brief, while the targeting ratio itself was highly cell-type dependent, probably reflecting the cells' distinct ability to cope with DSBs and exogenous DNA, treatment of cells with cytostatic drugs did not significantly affect the balance between gene targeting and random integration.
FIG. 4.
Targeting ratio. I-SceI-induced AAV-mediated gene targeting was performed as described in Figure 1. Quantitative PCR was performed on genomic DNA extracted 30 days post transduction with primer pairs (see Fig. 1a) to quantify the number of randomly integrated AAV vector genomes (RI) or target locus (TL) as an internal control. The targeting ratio is indicated as the quotient between the number of gene-targeting events (percent EGFP-positive cells at day six) and RI events (I-SceI copies per TL at day 30). Graph represents average targeting ratio with standard deviation.
Transient cell-cycle arrest augments zinc-finger nuclease-mediated gene targeting in cell lines
While I-SceI has been widely used as a benchmark in the genome engineering field, DSB-enhanced gene targeting in a therapeutic context is only possible when employing customizable designer nucleases, such as zinc-finger nucleases (ZFNs). Here we used previously characterized ZFNs with heterodimeric FokI domains that recognize a target site in the EGFP reporter (Szczepek et al., 2007). To directly compare I-SceI with ZFN-induced gene targeting in an AAV context, the respective nucleases were expressed from separate vectors and independently from donor administration (Fig. 5a). As expected, transduction of the U-2 OS target cell line with increasing amounts of AAV vector revealed that both I-SceI (Fig. 5b) and ZFN-mediated gene targeting (Fig. 5c) was strictly dose-dependent. Depending on the individual cytostatic and the vector dose, a transient cell-cycle arrest improved AAV-mediated gene targeting up to 5.3-fold. Also, there was no considerable difference between cell-cycle–dependent stimulation of AAV.SceI and AAV.ZFN-induced gene targeting, suggesting that these cytostatic drugs can be used to enhance any kind of DSB-mediated gene targeting.
FIG. 5.
AAV-mediated gene targeting with zinc-finger nucleases. (a) Experimental setup. Target cell lines contain a disrupted EGFP (mGFP) gene with binding sites for ZFN and I-SceI. A DSB to stimulate gene targeting between target locus and AAV.donor was created by transduction with either two AAV vectors encoding ZFN subunits GZF1N and GZF3N or with an I-SceI AAV expression vector, respectively. (b, c) AAV-mediated gene targeting in U-2 OS target cells induced by I-SceI (b) or ZFN expression (c). After pretreatment with cytostatic drugs, cells were transduced with various AAV vector amounts (AAV.donor 2,000 gc/cell in all samples; AAV.ZFN or AAV.SceI 2,000, 500, or 200 gc/cell, respectively) and assessed after four days by flow cytometry. Graphs represent average percentage of EGFP-positive cells with standard deviation and asterisks mark significant (p<0.05) differences between treated and nontreated cells. (d) Expression analysis. U-2 OS cells were transduced with AAV.ZFN or AAV.SceI (2,000 gc/cell). Cells were harvested after 72 h and I-SceI, GZF1N (ZF1), and GZF3N (ZF3) expression levels detected using an HA tag-specific antibody. Mock transduced cells served as a control. (e) Plasmid-based gene targeting in HeLa target cells. Cells were transfected with nuclease expression plasmids, donor plasmid, and pRK5.mCherry as a transfection marker. After 8 h, transfected cells were treated with cytostatic drugs and evaluated by flow cytometry three days later. Graphs indicate percentage of EGFP-positive cells with standard deviation, normalized for transfection efficiency. Asterisks mark significant (p<0.05) differences in gene correction in treated vs. untreated (w/o) cells. Transfection with a nonfunctional nuclease is indicated as “cto.” H, hydroxyurea; I, indirubin-3′-monoxime; V, vinblastine; w/o, without cytostatics.
The increase in AAV transduction after treatment of cells with particular cytostatic drugs implied that the observed augmentation in gene targeting (Fig. 2) was exclusively due to enhanced transduction efficiency (Fig. 3). Consistent with this implication was the observation that AAV-mediated I-SceI and ZFN expression was enhanced by treating cells with hydroxyurea, vinblastine, and indirubin-3′-monoxime (Fig. 5d). To demonstrate that transient cell-cycle arrest has a direct stimulatory effect on gene targeting and not only on AAV transduction, the HeLa target cell line was transfected with the AAV vector–encoding plasmid DNA. As expected, expression of either I-SceI or the ZFN improved gene correction at the target locus significantly and resulted in 1.2% or 0.3% of EGFP-positive cells, respectively (Fig. 5e). A further significant increase (up to 7.6-fold) in gene targeting was observed upon treatment of cells with either hydroxyurea or vinblastine. These results verify that transient cell-cycle arrest is a general means of improving DSB-mediated gene targeting.
Indirubin-3′-monoxime enhances AAV/ZFN-mediated gene targeting in human mesenchymal stromal cells
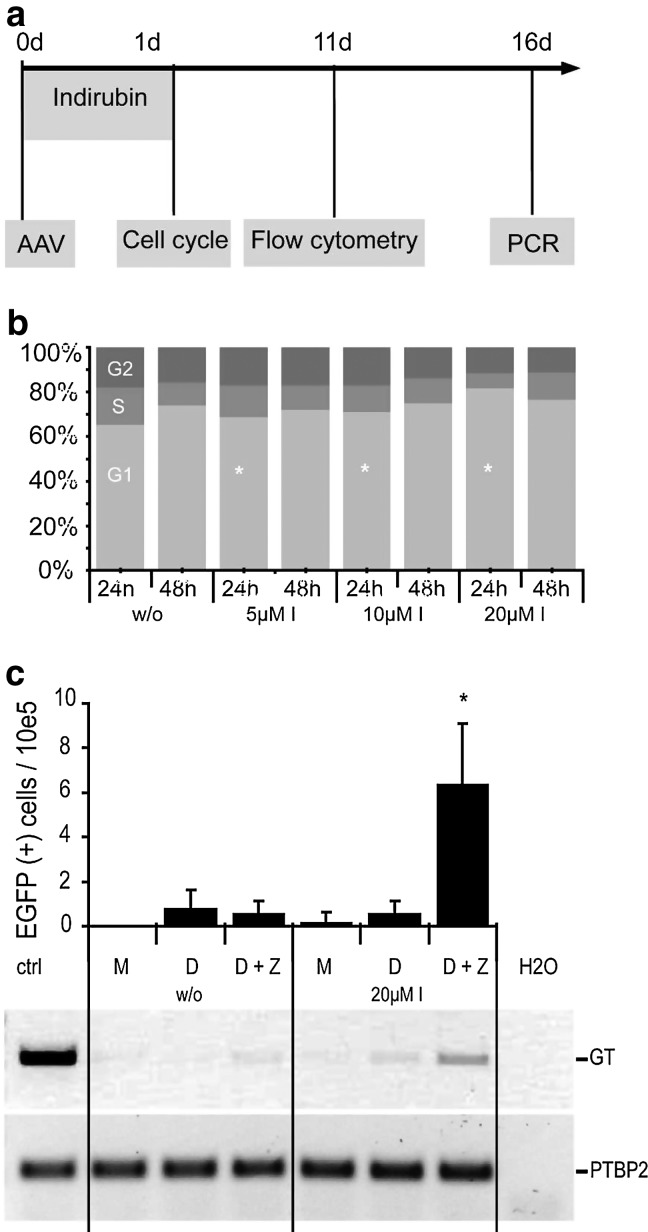
To demonstrate that transient cell-cycle arrest improves gene targeting also in human stem cells, the defective EGFP reporter was introduced into umbilical cord–derived mesenchymal stromal cells (UC-MSCs). These primary cells were transduced with the same AAV vectors, previous to treatment with cytostatic drugs (Fig. 6a). Notably, UC-MSCs did not exhibit cell cycle changes in response to any of the drugs tested before (not shown) with the exception of indirubin-3′-monoxime, which induced a significant, dose-dependent G1 arrest (Fig. 6b). At the highest concentration, treatment with indirubin-3′-monoxime led to a 10-fold increase in EGFP-positive cells compared to untreated cells, as measured by flow cytometry 11 days post-transduction (Fig. 6c). PCR-based genotyping, which was used to verify gene correction on the genome level (Händel et al., 2012), confirmed the stimulatory effect of indirubin-3′-monoxime on AAV/ZFN-mediated gene targeting. Of note, treatment with the cytostatic drug did not lead to profound cell toxicity (Supplementary Fig. S4) and did not alter MSC properties. After initial cell-cycle arrest, the cells regained their high proliferative capacity and displayed typical morphology as well as characteristic surface markers (Supplementary Fig. S5).
FIG. 6.
ZFN-mediated gene targeting in human mesenchymal stromal cells. (a) Experimental setup. UC-MSC.696 cells were seeded in a 24-well plate (50,000 cells/well) 6 h before transduction with 10,000 gc/cell of AAV-ZFN and 2,000 gc/cell of AAV-donor and then treated with indirubin-3′-monoxime. (b) Cell-cycle analysis of UC-MSC.696 cells after treatment with indirubin-3′-monoxime. At 24 h and 48 h after treatment with cytostatics, cells were fixed, stained with propidium iodide, and analyzed by flow cytometry. The percentage of cells in G1 (light gray), S (gray), and G2/M (dark gray) phase is indicated as a bar graph. Asterisks mark a significant difference (p<0.05) from nontreated cells. (c) Gene correction in UC-MSC. The graph shows the numbers of EGFP-positive cells per 100,000 cells measured by flow cytometry. Asterisk indicates a significant increase (p<0.05) over nontreated cells. To verify the presence of faithfully corrected EGFP gene on the genome level, allele-specific PCR was performed. A PCR encompassing a fraction of the PTBP2 gene was used as a control (N=5). I, indirubin-3′-monoxime; M, mock; D, donor; D+Z, donor and ZFN; GT, gene targeting; PTBP2, locus-encoding polypyrimidine-binding protein 2.
Discussion
Targeted genome engineering, such as gene correction or the integration of a therapeutic transgene cassette into a “neutral site” in the human genome, will be safer and more efficient if NHEJ-mediated random integration of the donor DNA can be suppressed and HR-mediated gene targeting enhanced. Conventional gene targeting, e.g., the generation of gene knockouts in murine ES cells, likely depends on the occurrence of a natural DSB at the target locus to mediate HR between the targeting vector and the gene of interest. The intentional creation of a DSB by a designer nuclease, on the other hand, has been shown to significantly enhance HR-based genome engineering if sufficient amounts of appropriately designed donor molecules are available (Lombardo et al., 2007). Since illegitimate integration is only mildly (if at all) increased by the created DSB (Miller et al., 2003; Gellhaus et al., 2010), the net balance between the two repair pathways strongly shifts toward HR. One way to shift the balance further is the ectopic expression of HR factors (Yanez and Porter, 1999) or, vice versa, the suppression of factors involved in NHEJ (Fattah et al., 2008) or sister chromatid HR (Potts et al., 2006). For instance, the genetic knockout of one Ku70 allele, a gene encoding a protein that forms an active complex with Ku86 and protein kinase catalytic subunit (DNA-PKcs) at DSBs, increased AAV-mediated gene targeting 6-fold (Fattah et al., 2008). Whereas this effect could be further enhanced by siRNA-mediated knockdown of the second Ku70 allele (Fattah et al., 2008), knockdown of DNA-PKcs did not have an effect on AAV-mediated gene targeting (Vasileva et al., 2006). On the other hand, although it has been shown that Rad51 and Rad54 are essential for both ZFN and AAV-mediated gene targeting (Vasileva et al., 2006; Bozas et al., 2009), human cells stably over-expressing RAD51 revealed only a two-fold increase in conventional, plasmid-based gene targeting (Yanez and Porter, 1999). On the whole, it remains open to discussion whether a genetic intervention to shift the balance in AAV or ZFN-mediated gene targeting from NHEJ to HR is meaningful in a therapeutic context.
The availability of NHEJ and HR repair factors is strongly cell-type dependent. For example, we and others have shown previously that DSB-induced gene targeting works particularly well in U-2 OS cells (Händel et al., 2009; Gellhaus et al., 2010; Chen et al., 2011). Moreover, the balance between the pathways changes during progression through the cell cycle (Shrivastav et al., 2008). Even though NHEJ seems to be active throughout the whole cell cycle, the DSB repair shifts from NHEJ toward HR as the cell progresses from G1 to S/G2, as for instance, evidenced by a lower activity of the NHEJ factor DNA-PKcs (Lee et al., 1997) and increased expression of the HR factors RAD51 and RAD52 (Chen et al., 1997) in the S phase. Hence, the cell-cycle status is an important factor in HR-based genome engineering. The balance between HDR and NHEJ can be shifted toward HDR by chemical or genetic inhibition of NHEJ, which led to up to 10-fold increase of AAV HDR-based gene targeting (Paulk et al., 2012). A few compounds used in chemotherapy regiments have been described to enhance ZFN-mediated gene targeting, such as vinblastine (Urnov et al., 2005; Potts et al., 2006; Maeder et al., 2008) and nocodazole (Olsen et al., 2010). Our results extend this observation to AAV/ZFN-mediated gene targeting and confirm that the stimulatory potential on gene targeting is conveyed by transiently arresting the cells in G1 or G2. However, microtubule inhibitors are rather cytotoxic, as substantiated by this study, and their application in a therapeutic context is not advisable. Here, we have identified indirubin-3′-monoxime as a robust enhancer of AAV/ZFN-mediated gene targeting in three different cell lines and in primary human MSC. The anti-cancer drug indirubin has been characterized as a potent CDK-inhibitor (Hoessel et al., 1999), and more active derivatives, such as indirubin-3′-monoxime, have been shown to induce apoptosis in a number of cancer cells, including HeLa (Shi and Shen, 2008), and to inhibit the growth of tumors in clinical studies and in animal models without major side effects (Eisenbrand et al., 2004). Of note, while the stimulatory potential of the other cytostatic drugs to enhance AAV/ZFN-mediated gene correction was limited to certain cell types, treatment with indirubin-3′-monoxime showed a significant activity in all cells tested here.
It has been known for a while that substances that induce DNA damage, such as cis-platinum, hydroxyurea, camptothecin, or etoposide increase AAV transduction in cultured cells (Alexander et al., 1994; Russell et al., 1995). This effect has been attributed to activation of the cellular DNA damage response, which in turn is important to process the vector genome to generate a transcription-competent template (Cervelli et al., 2008). The development of capsid variants optimized for cell targeting and/or trafficking (Büning et al., 2008; Li et al., 2008), as well as the use of compounds that do not directly damage the DNA, such as proteasome inhibitors (Jennings et al., 2005; Monahan et al., 2010), have further improved AAV transduction efficiencies both in cultured cells (Ellis et al., 2012) and in vivo (Paulk et al., 2012). The effect might be linked to the nuclear trafficking of the viral particles. In combination with sodium butyrate, a histone deacetylase inhibitor, ZFN-mediated gene targeting could be increased up to six-fold in a human cell line (Ellis et al., 2012). These drugs are FDA-approved, which might facilitate integration into clinical protocols for rAAV-based gene therapy; however, cytotoxic side effects on relevant primary target cells need to be investigated. Here we show that AAV transduction can also be augmented with a CDK inhibitor that arrests the cell cycle without obvious cytotoxicity. The fact that the stimulatory effect of indirubin-3′-monoxime on AAV transduction parallels the increase in AAV-mediated gene targeting identifies transduction as another rate-limiting step in AAV-mediated gene targeting. Interestingly, depending on the cell type, indirubin-mediated cell-cycle arrest occurred in the G1 or G2 phase of the cell cycle, yet in both instances we found an increase in gene targeting. This suggests that the transient cell-cycle arrest had a dual effect in AAV/ZFN-mediated gene targeting: (i) an increase in AAV transduction that leads to an augmented nuclease expression and (ii) a longer transition time through the cell cycle that may facilitate gene targeting.
In conclusion, we have identified and characterized the application of small molecule drugs for improving AAV/ZFN-mediated gene targeting by transiently slowing transition through the cell cycle. In particular, incubation of cells with indirubin-3′-monoxime, a well-tolerated CDK inhibitor with minimal toxicity in normal cells, augmented the gene-targeting frequency up to 10-fold to correct a mutated reporter gene in up to 34% of infected human cells. Given the recent success in AAV-mediated gene targeting in other multipotent (Chamberlain et al., 2004; Jang et al., 2011) or pluripotent stem cells (Mitsui et al., 2009; Khan et al., 2010; Asuri et al., 2012), respectively, as well as in vivo (Papadopoulos et al., 1997; Paulk et al., 2012), these findings present a promising strategy to enhance AAV/ZFN-mediated genome engineering for potential clinical applications.
Supplementary Material
Acknowledgments
We thank Eva Guhl (Charité Medical School, Berlin) for technical assistance, Roger Y. Tsien (University of California, San Diego) for plasmid pRSET-BmCherry, Ralf Hass (Hannover Medical School, Hannover) for providing human umbilical cords, Cornelia Kasper (Leibniz University, Hannover) for UC-MSC isolation protocol, Marcin Majka and Danuta Jarocha (Jagiellonian University, Cracow) for supportive work regarding MSC, and Heimo Riedel (West Virginia University, Morgantown) and Eva Händel (Hannover Medical School, Hannover) for critical discussions. This work was supported by grants from the German Research Foundation (SFB 738 to T.C. and R.J.; SPP1230–Ca311/2 to T.C.; Cluster of Excellence REBIRTH to D.C.), the European Commission's 7th Framework Programme (PERSIST-222878 to T.C.), the Niedersächsische Krebsgesellschaft (D.C.), and the Kind-Philipp-Stiftung für Leukämieforschung (K.P.)
Author Disclosure Statement
The authors declare no potential conflict of interest.
References
- Alexander I.E. Russell D.W. Miller A.D. DNA-damaging agents greatly increase the transduction of nondividing cells by adeno-associated virus vectors. J. Virol. 1994;68:8282–8287. doi: 10.1128/jvi.68.12.8282-8287.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwin S. Gere M.B. Guhl E, et al. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- Asuri P. Bartel M.A. Vazin T., et al. Directed evolution of adeno-associated virus for enhanced gene delivery and gene targeting in human pluripotent stem cells. Mol. Ther. 2012;20:329–338. doi: 10.1038/mt.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdallah B.F. Allard E. Yao S., et al. Targeted gene addition to human mesenchymal stromal cells as a cell-based plasma-soluble protein delivery platform. Cytotherapy. 2010;12:394–399. doi: 10.3109/14653240903583803. [DOI] [PubMed] [Google Scholar]
- Bogdanove A.J. Voytas D.F. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Bozas A. Beumer K.J. Trautman J.K. Carroll D. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics. 2009;182:641–651. doi: 10.1534/genetics.109.101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büning H. Perabo L. Coutelle O., et al. Recent developments in adeno-associated virus vector technology. J. Gene Med. 2008;10:717–733. doi: 10.1002/jgm.1205. [DOI] [PubMed] [Google Scholar]
- Capecchi M.R. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervelli T. Palacios J.A. Zentilin L., et al. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J. Cell Sci. 2008;121:349–357. doi: 10.1242/jcs.003632. [DOI] [PubMed] [Google Scholar]
- Chamberlain J.R. Schwarze U. Wang P.R., et al. Gene targeting in stem cells from individuals with osteogenesis imperfecta. Science. 2004;303:1198–1201. doi: 10.1126/science.1088757. [DOI] [PubMed] [Google Scholar]
- Chen F. Nastasi A. Shen Z., et al. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutat. Res. 1997;384:205–211. doi: 10.1016/s0921-8777(97)00020-7. [DOI] [PubMed] [Google Scholar]
- Chen F. Pruett-Miller S.M. Huang Y., et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng K. Larsen S.R. Zhou S., et al. Specific adeno-associated virus serotypes facilitate efficient gene transfer into human and non-human primate mesenchymal stromal cells. J. Gene Med. 2007;9:22–32. doi: 10.1002/jgm.990. [DOI] [PubMed] [Google Scholar]
- Choulika A. Perrin A. Dujon B. Nicolas J.F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu T.I. Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- Eisenbrand G. Hippe F. Jakobs S. Muehlbeyer S. Molecular mechanisms of indirubin and its derivatives: novel anticancer molecules with their origin in traditional Chinese phytomedicine. J. Cancer Res. Clin. Oncol. 2004;130:627–635. doi: 10.1007/s00432-004-0579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis B.L. Hirsch M.L. Porter S.N., et al. Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Gene Ther. 2012 doi: 10.1038/gt.2011.211. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah F.J. Lichter N.F. Fattah K.R., et al. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8703–8708. doi: 10.1073/pnas.0712060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellhaus K. Cornu T.I. Heilbronn R. Cathomen T. Fate of recombinant adeno-associated viral vector genomes during DNA double-strand break-induced gene targeting in human cells. Hum. Gene Ther. 2010;21:543–553. doi: 10.1089/hum.2009.167. [DOI] [PubMed] [Google Scholar]
- Händel E.M. Alwin S. Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol. Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händel E.M. Gellhaus K. Khan K., et al. Versatile and efficient genome editing in human cells by combining zinc-finger nucleases with adeno-associated viral vectors. Hum. Gene Ther. 2012;23:321–329. doi: 10.1089/hum.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatlapatka T. Moretti P. Lavrentieva A., et al. Optimization of culture conditions for the expansion of umbilical cord-derived mesenchymal stem or stromal cell-like cells using xeno-free culture conditions. Tissue Eng. Part C Methods. 2011;17:485–493. doi: 10.1089/ten.TEC.2010.0406. [DOI] [PubMed] [Google Scholar]
- Hirsch M.L. Green L. Porteus M.H. Samulski R.J. Self-complementary AAV mediates gene targeting and enhances endonuclease delivery for double-strand break repair. Gene Ther. 2010;17:1175–1180. doi: 10.1038/gt.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D. Soldner F. Beard C., et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoessel R. Leclerc S. Endicott J.A., et al. Indirubin, the active constituent of a Chinese antileukaemia medicine, inhibits cyclin-dependent kinases. Nat. Cell Biol. 1999;1:60–67. doi: 10.1038/9035. [DOI] [PubMed] [Google Scholar]
- Jang J.H. Koerber J.T. Kim J.S., et al. An evolved adeno-associated viral variant enhances gene delivery and gene targeting in neural stem cells. Mol. Ther. 2011;19:667–675. doi: 10.1038/mt.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings K. Miyamae T. Traister R., et al. Proteasome inhibition enhances AAV-mediated transgene expression in human synoviocytes in vitro and in vivo. Mol. Ther. 2005;11:600–607. doi: 10.1016/j.ymthe.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Khan I.F. Hirata R.K. Wang P.R., et al. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 2010;18:1192–1199. doi: 10.1038/mt.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Mahendra G. Nagy T.R. Ponnazhagan S. Osteogenic differentiation of recombinant adeno-associated virus 2-transduced murine mesenchymal stem cells and development of an immunocompetent mouse model for ex vivo osteoporosis gene therapy. Hum. Gene Ther. 2004;15:1197–1206. doi: 10.1089/hum.2004.15.1197. [DOI] [PubMed] [Google Scholar]
- Lee S.E. Mitchell R.A. Cheng A. Hendrickson E.A. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell Biol. 1997;17:1425–1433. doi: 10.1128/mcb.17.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Asokan A. Wu Z., et al. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol. Ther. 2008;16:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- Lombardo A. Genovese P. Beausejour C.M., et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Maeder M.L. Thibodeau-Beganny S. Osiak A., et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.G. Petek L.M. Russell D.W. Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol. Cell Biol. 2003;23:3550–3557. doi: 10.1128/MCB.23.10.3550-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K. Suzuki K. Aizawa E., et al. Gene targeting in human pluripotent stem cells with adeno-associated virus vectors. Biochem. Biophys. Res. Commun. 2009;388:711–717. doi: 10.1016/j.bbrc.2009.08.075. [DOI] [PubMed] [Google Scholar]
- Monahan P.E. Lothrop C.D. Sun J., et al. Proteasome inhibitors enhance gene delivery by AAV virus vectors expressing large genomes in hemophilia mouse and dog models: a strategy for broad clinical application. Mol. Ther. 2010;18:1907–1916. doi: 10.1038/mt.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C. Cathomen T. TALE nucleases: tailored genome engineering made easy. Curr. Opin. Biotechnol. 2012;23:644–650. doi: 10.1016/j.copbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Olsen P.A. Gelazauskaite M. Randol M. Krauss S. Analysis of illegitimate genomic integration mediated by zinc-finger nucleases: implications for specificity of targeted gene correction. BMC Mol. Biol. 2010;11:35. doi: 10.1186/1471-2199-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos V. Amri H. Li H., et al. Targeted disruption of the peripheral-type benzodiazepine receptor gene inhibits steroidogenesis in the R2C Leydig tumor cell line. J. Biol. Chem. 1997;272:32129–32135. doi: 10.1074/jbc.272.51.32129. [DOI] [PubMed] [Google Scholar]
- Paulk N.K. Loza L.M. Finegold M.J. Grompe M. AAV-mediated gene targeting is significantly enhanced by transient inhibition of nonhomologous end joining or the proteasome in vivo. Hum. Gene Ther. 2012;23:658–665. doi: 10.1089/hum.2012.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus M.H. Cathomen T. Weitzman M.D. Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol. Cell Biol. 2003;23:3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P.R. Porteus M.H. Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S.H. Maeder M.L. Joung J.K. Cathomen T. Zinc-finger nucleases for somatic gene therapy: the next frontier. Hum. Gene Ther. 2011;22:925–933. doi: 10.1089/hum.2011.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P. Smih F. Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D.W. Alexander I.E. Miller A.D. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5719–5723. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. Shen H.M. Critical role of Bid and Bax in indirubin-3′-monoxime-induced apoptosis in human cancer cells. Biochem. Pharmacol. 2008;75:1729–1742. doi: 10.1016/j.bcp.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Shrivastav M. De Haro L.P. Nickoloff J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Silva G. Poirot L. Galetto R., et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllü C. Pars K. Cornu T.I., et al. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. 2010;38:8269–8276. doi: 10.1093/nar/gkq720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M. Brondani V. Buchel J., et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Takeuchi R. Lambert A.R. Mak A.N., et al. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13077–13082. doi: 10.1073/pnas.1107719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobridge G. Hirata R.K. Russell D.W. Gene targeting by adeno-associated virus vectors is cell-cycle dependent. Hum. Gene Ther. 2005;16:522–526. doi: 10.1089/hum.2005.16.522. [DOI] [PubMed] [Google Scholar]
- Urnov F.D. Miller J.C. Lee Y.L., et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Urnov F.D. Rebar E.J. Holmes M.C., et al. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Vasileva A. Linden R.M. Jessberger R. Homologous recombination is required for AAV-mediated gene targeting. Nucleic Acids Res. 2006;34:3345–3360. doi: 10.1093/nar/gkl455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez R.J. Porter A.C. Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther. 1999;6:1282–1290. doi: 10.1038/sj.gt.3300945. [DOI] [PubMed] [Google Scholar]
- Zou J. Maeder M.L. Mali P., et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.