Abstract
Background and Aims
The coffee genus (Coffea) comprises 124 species, and is indigenous to the Old World Tropics. Due to its immense economic importance, Coffea has been the focus of numerous genetic diversity studies, but despite this effort it remains insufficiently studied. In this study the genetic diversity and genetic structure of Coffea across Africa and the Indian Ocean islands is investigated.
Methods
Genetic data were produced using 13 polymorphic nuclear microsatellite markers (simple sequence repeats, SSRs), including seven expressed sequence tag-SSRs, and the data were analysed using model- and non-model-based methods. The study includes a total of 728 individuals from 60 species.
Key Results
Across Africa and the Indian Ocean islands Coffea comprises a closely related group of species with an overall pattern of genotypes running from west to east. Genetic structure was identified in accordance with pre-determined geographical regions and phylogenetic groups. There is a good relationship between morpho-taxonomic species delimitations and genetic units. Genetic diversity in African and Indian Ocean Coffea is high in terms of number of alleles detected, and Madagascar appears to represent a place of significant diversification in terms of allelic richness and species diversity.
Conclusions
Cross-species SSR transferability in African and Indian Ocean islands Coffea was very efficient. On the basis of the number of private alleles, diversification in East Africa and the Indian Ocean islands appears to be more recent than in West and West-Central Africa, although this general trend is complicated in Africa by the position of species belonging to lineages connecting the main geographical regions. The general pattern of phylogeography is not in agreement with an overall east to west (Mascarene, Madagascar, East Africa, West Africa) increase in genome size, the high proportion of shared alleles between the four regions or the high numbers of exclusive shared alleles between pairs or triplets of regions.
Keywords: Africa, Coffea, coffee, crop wild relatives (CWRs), genetic diversity, genetic structure, Indian Ocean islands, Madagascar, Mascarenes, microsatellites, Rubiaceae, simple sequence repeats (SSRs)
INTRODUCTION
The coffee genus (Coffea; Rubiaceae) comprises 124 species, and occurs naturally in Africa, Madagascar, the Comoros Islands, Mascarene Islands, Indian subcontinent, south tropical Asia, south-eastern Asia and Australasia (Davis, 2010, 2011; Davis et al., 2006, 2011). There are two main coffee crop species, Coffea arabica and C. canephora, which provide a global commodity second only to oil, accounting for exports worth an estimated US$ 15·4 billion in 2009/10 (International Coffee Organization (ICO), 2012). Given its immense economic importance, Coffea has been the focus of numerous phylogenetic and genetic diversity studies.
Phylogenetic studies based on sequence data from plastid and nuclear regions (Lashermes et al., 1997; Cros et al., 1998; Davis et al., 2007; Maurin et al., 2007; Anthony et al., 2010; Davis et al., 2011; Nowak et al., 2012) have provided a solid framework for understanding the evolutionary history of Coffea. In the most recent phylogenetic study of Coffea, Davis et al. (2011) retrieved and recognized six main well-supported lineages: (1) African ‘Psilanthus’ clade (i.e. short-styled African Coffea); (2) Asian and Australasian ‘Psilanthus’ clade (short-styled non-African Coffea); (3) the Lower Guinea/Congolian (LG/C) clade [species from West and Central Africa, west of the Great Rift Valley, with some species (C. canephora and C. liberica) also occurring in the Upper Guinea Region]; (4) the Upper Guinea (UG) clade (three Upper Guinea endemics); the East-Central Africa (E-CAfr) clade [species straddling the Great Rift Valley but with one species (C. anthonyi) in West and Central Africa]; (5) the Indian Ocean (Madagascar, Comoros and Mascarenes) (IO) clade, which includes the Mascarene (MAS) clade and the Madagascar (MAD) clade; (6) the dry-adapted Madagascan baracoffea alliance; an East African (EA) clade was also consistently retrieved in the above-mentioned phylogenetic analyses but with inconsistent levels of support.
Even though extensive multiple sampling of natural populations in Africa and in Madagascar was carried out between the 1960s and 1980s, detailed analyses of genetic diversity with molecular markers have only included a relatively small sampling of species, with studies largely focused on C. arabica and C. canephora and their proposed close relatives (reviewed by de Kochko et al., 2010). The Cameroon–Gabon–Congo region, East Africa and Madagascar have been posited as the three main centres of diversification (Berthaud, 1986; Charrier, 1978) in the African–West Indian Ocean region, but have been largely ignored in terms of studies. Given the high morphological variation observed in Madagascan and Mascarene Coffea, Leroy (1971) and Charrier (1978) addressed a specific question: should Coffea spp. be considered as such or only as morphologically divergent populations? In this respect Leroy's uncertainty is clearly manifest on herbarium specimens of Coffea spp. collected from Madagascar, which were re-annotated by him with identifications indicating increasingly broad species concepts. Several new and morphologically discrete species were never published by him (Davis and Rakotonasolo, 2001a, b, 2004, 2008), and it seems as if he had a crisis of confidence regarding species delimitation. Similar issues regarding species delimitation were raised by Charrier and Berthaud (1985) for some of the East African species studied by Leroy (1982) and Hamon et al. (1984). There is also a need for a more fine-scale understanding of species relationships, as sequencing studies have so far not provided sufficient resolution in some areas, notably Madagascar (Maurin et al., 2007).
The development of simple sequence repeat (SSR) markers for Coffea (Moncada and McCouch, 2004; Poncet et al., 2007) has provided a key resource for investigating genetic diversity in the genus. Studies include hybridization (Anthony et al., 2000a; Ruas et al., 2003; Tesfaye et al., 2007; Gomez et al., 2010), domestication (Anthony et al., 2002a, b), cultivated genepools (Prakash et al., 2005), breeding (Cubry et al., 2008), and in situ and ex situ conservation (Krishnan et al., 2013a, b). In this study we apply nuclear microsatellite (SSR) markers to assess more precisely genetic structure and diversity of African and Indian Ocean island Coffea. A broad-scale assessment of genetic diversity for Coffea has not previously been undertaken. SSRs may have shortcomings (e.g. high mutation rates and size homoplasy; Estoup et al., 2002) but they display strong advantages compared with other methods (e.g. they are highly polymorphic, co-dominant and relatively easy to use) and have proved successful in cases where sequence data require corroboration (Richard and Thorpe, 2001), have been problematic (Ochieng et al., 2007) or where fine-scale evolutionary insights are sought (Petren et al., 1999). Our focus here is on the species occurring in Africa, Madagascar, the Comoros and the Mascarenes, as these areas include several of the main Coffea lineages (see above and Methods) and a large proportion of the species diversity (60 of the 124 known species). Our study does not include the short-styled lineages of Coffea, i.e. those formerly placed in the genus Psilanthus (20 species), or the baracoffea alliance (nine species), which are notable omissions. However, the evolutionary history of the short-styled lineages and the baracoffea alliance are largely separate from the lineages investigated here, comprising separate African and Africa–Asian–Australasian radiations (Davis et al., 2011), and an isolated monophyletic lineage (Maurin et al., 2007; Davis and Rakotonasolo, 2008; Nowak et al., 2012), respectively. Moreover, the baracoffea alliance and short-styled lineage species are either absent from germplasm collections or DNA is in short supply.
The specific objectives of this study are to: (1) investigate the inter- and intraspecific genetic diversity across the African and the Indian Ocean islands; (2) evaluate genetic structure and its correspondence with species delimitation; (3) investigate areas of high diversification and see whether this corresponds to centres of genetic diversity; and (4) ascertain whether the genetic structure and species relationships generated by SSR data correspond to phylogenetic signals based on data from other sources. The data were analysed using methods more usually applied to population genetics, including non-model- and model-based methods of analyses. The study includes 728 accessions from 60 species.
MATERIAL AND METHODS
Plant material
Africa and the Indian Ocean islands (Madagascar, Comoros and Mascarenes) represent the natural distribution for 112 species of Coffea (Davis et al., 2006). These areas also comprise a large part of the distribution range and several of the major lineages of Coffea (Maurin et al., 2007; Davis et al., 2011; and see Introduction), including the Lower Guinea/Congolian (LG/C) clade, Upper Guinea (UG) clade, East-Central African (E-CAfr) clade, East Africa (EA) clade, and Indian Ocean (IO) clade [including Madagascan (MAD) and Mascarene (MAS) clades]. These species grow in various tropical habitats and from sea level to 2000 m (Leroy, 1968; Charrier, 1978; Davis et al., 2006). There is complete endemicity for Madagascar, Comoros and Mascarenes, and in Africa the vast majority of the species are restricted to one side of the Great Rift Valley, with only a few species straddling this high mountainous area (Stoffelen, 1998; Davis et al., 2006; Maurin et al., 2007). According to Davis (2010, 2011) and Davis et al. (2006, 2010, 2011) species numbers are distributed as follows: Africa (46 species), Madagascar (61 species), Comoros (one species), Mascarene Islands [three species; although a fourth (C. bernardiana) is sometimes recognized], Indian subcontinent and Sri Lanka (six species), south tropical Asia (two species), south-eastern Asia (four species) and Australasia (one species); and for Africa, either side of the Great Rift Valley: West and West-Central Africa (24 species), and East Africa, including Rift Valley (22 species), giving 124 species in total.
The study uses 728 individuals of wild origin, via germplasm collections: 421 individuals, from 51 populations, representing 39 species from Comoros, Mauritius and Madagascar; and 307 individuals from 36 populations representing 21 African species (60 species in total). Leaves from Madagascan and Comorian species, obtained from the Kianjavato Coffee Research Station (KCRS) in Madagascar, were sampled between 2009 and 2010. The African and the Mascarene (Mauritian) species were sampled from the Coffea collection maintained by IRD at the Armeflhor de Bassin Martin Station, Saint-Pierre, Reunion [originating from representatives of the international Coffea collection maintained at Divo's research station, Ivory Coast (Hamon et al., 1998)]. These Coffea germplasm collections, assembled between the 1960s and 1980s, represent exceptional taxonomic and geographical coverage for Africa, Madagascar and the Mascarenes. They were created without taxonomic, commercial or regional bias, and, in most cases, include multiple population representatives. Their accessions have been verified and vouchered by Coffea taxonomists, and the identifications of a large proportion of the KRCS and IRD collections have been corroborated using sequence data (Maurin et al., 2007).
Mature leaves were sampled from one to 14 trees (depending on availability) per species or population, lyophilized and stored until use. Details of the investigated accessions are given in the Appendix, including provenance data; locations are visualized in Fig. 1. Vouchers specimens are maintained at FOFIFA, Antananarivo (TEF), the Antananarivo Herbarium (TAN), the Royal Botanic Gardens, Kew (K), the Natural History Museum, Paris (P), and Missouri Botanical Garden (MO; herbarium abbreviations after Holmgren et al., 1990).
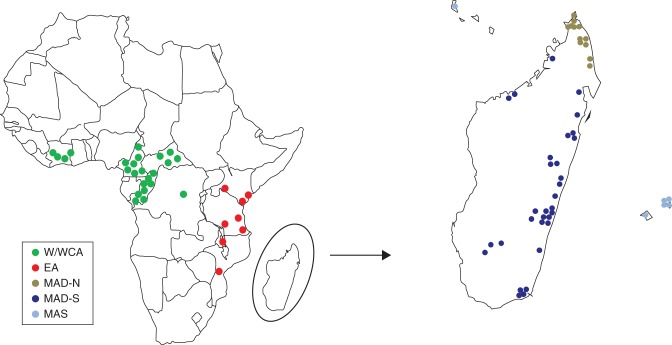
Fig. 1.
Location of populations (see Appendix). W/WCA, West and West-Central Africa; EA, East Africa; MAD-N, Madagascar north; MAD-S, Madagascar south; and MAS, the Mascarenes, and the Comoros Islands.
Assignment of accessions and populations to geographical regions
The accessions were divided into two main areas, four regions and two sub-regions (for Madagascar) according to their origin (provenance). The regions are as follows: (1) West and West-Central Africa (W/WCA), (2) East Africa (EA), (3) Madagascar (MAD) and (4) Mascarenes (including Comoros) (MAS). The sub-regions are (3i) Madagascar north (MAD-N) and (3ii) Madagascar south (MAD-S). For some analyses and parts of the discussion the regions have also been grouped together to form two main areas: (1) and (2) comprise Africa; and (3) and (4) the Indian Ocean islands (IOIs). Division into two regions for Africa is based on established phytogeographical regions (White, 1979, 1983), either side of the Great Rift Valley. The sub-regions for Madagascar are based on the observation that north and south Madagascar house different groups of Coffea spp., which appears, at least in part (A. P. Davis, unpubl. data), to correspond to the major bioclimatic divisions (Cornet, 1974) and vegetation types (Moat and Smith, 2007) of Madagascar (see below). Preliminary analyses indicated that Mauritius and Comoros formed a genetic grouping, and as the latter has only one species (C. humblotiana), it was grouped with the Mascarenes for simplification. These divisions and the accessions assigned to them comprise ample coverage of all the long-styled lineages (i.e. the delimitation of Coffea pre-2011; Davis et al., 2011), except the baracoffea alliance (Davis and Rakotonasolo, 2008), which has not been sampled.
Most of the accessions from Madagascar south (MAD-S) are from the humid evergreen forests of eastern Madagascar, except the following: Coffea perrieri, which is restricted to humid gallery (riverine) forest running through the otherwise dry vegetation of western Madagascar (i.e. Western Dry Forest; Moat and Smith, 2007); and C. sakarahae, which occurs in seasonally dry evergreen–deciduous forest of southern-central Madagascar. Our northern Madagascar (MAD-N) accessions are mostly species restricted to seasonally dry habitats in deciduous–evergreen forests, or seasonally dry evergreen and evergreen–deciduous forest, although C. bonnieri is from humid (evergreen) montane vegetation. Coffea tetragona is a western species but occurs in Sambirano vegetation, which is an extension of the humid forest into western Madagascar, at a mid altitude range (e.g. 500 m and above).
Microsatellite analyses
An initial set of 20 SSR markers was selected, mainly on the basis of their chromosome location (distributed on the Coffea linkage groups as defined by Coulibaly et al., 2003; Lefebvre-Pautigny et al., 2010; de Kochko et al., 2010) and on the expected amplicon size for easier reads after multiplexing. DNA was purified using the Qiagen DNeasy Plant Maxi Kit® or the DNeasy Plant Mini Kit® (Quiagen, Valencia, CA, USA) according to the manufacturer's instructions. Quantification was undertaken using a NanoDrop TM 1000 Spectrophotometer (LabTech, Paris, France). PCR conditions and all information on the markers are given in Plechakova et al. (2009). Alleles were detected using fluorescently labelled forward primers and sizes were determined on an Abi Prism 3100 (Applied Bioscience, Foster City, CA, USA) automated sequencer using GeneScan™ –400HD (Applied Bioscience) as an internal lane size standard. Reads were scored using GeneMapper ver. 3·7 (Applied Bioscience). Finally, after eliminating loci that were too difficult to read, or contributed too many missing data to the global set, results are given for 13 of the 20 markers tested (listed in Table 1 with their allele features).
Table 1.
SSRs used in this study with their tandem repeated motif, linkage groups location and total number of alleles (AN,Total); allele size ranges observed in Africa and Indian Ocean islands (IOI), data for C. canephora and C. arabica are given as reference.
| Marker | SSR motif | C. canephora linkage group | AN,Total | Allele size range (bp) |
||
|---|---|---|---|---|---|---|
| IOI | C. canephora (C. arabica) | Africa | ||||
| ES69a | AGG | I | 24 | 151–205 | 172–193 | 151–205 |
| ES12a | CAG | K | 15 | 148–192 | 160–184 | 151–192 |
| ES74a | AGG | A | 12 | 210–240 | 219–225 | 210–240 |
| ES13a | GA | J | 24 | 219–263 | 235–255 | 223–263 |
| ES42a | TTC | B | 12 | 150–177 | 156–177 | 150–180 |
| ES84a | AGA | F | 18 | 176–218 | 185–201 | 185–218 |
| ES90a | AAG | K | 15 | 200–218 | 188–215 | 188–238 |
| C2_At4g35070a | AT | I | 15 | 192–220 | 210–218 | 200–222 |
| A8742b | GT | D | 30 | 124–162 | (110) | 124–160 |
| M804b | GT | C | 23 | 302–332 | (294) | 288–336 |
| A8809b | ATG | B | 14 | 123–162 | (144) | 138–165 |
| M821b | GT | E | 28 | 160–188 | (148) | 156–196 |
| A8856b | GA | C | 38 | 210–292 | (209) | 192–274 |
Markers derived from aC. canephora (Plechakova et al., 2009); bC. arabica (Poncet et al., 2004); cLefebvre-Pautigny et al. (2010) and de Kochko et al. (2010). Codes prefixed with ES and C represent EST-SSRs, and those prefixed by A or M are anonymous genomic sequences obtained from a C. arabica SSR library produced by Rovelli et al. (2000).
General arrangement of genetic variation
To assess the general arrangement of genetic variation, principal component analyses (PCAs) were performed on the individual allelic frequencies matrix, structured into three genotype sets: (1) a global set of 705 genotypes (after excluding genotypes introduced to Reunion with doubtful accession data) accounting for a total of 233 alleles, with genotypes assigned to the four main regions: W/WCA, EA, MAD and MAS; (2) an African set of 284 (198 alleles) genotypes (W/WCA and EA); and (3) an Indian Ocean island (IOI) set of 421 (165 alleles) genotypes (MAD-N, MAD-S and MAS). The PCAs were performed using R software ver. 2·11·1 (Husson et al., 2008; http://cran.r-project.org/). The significance of the first eigenvalues (axes) was tested using the statistical tests of Patterson et al. (2006).
Identification of genetic units
To assess the genetic homogeneity of each main geographical region, the SSR data for Africa and the IOIs were independently submitted to STRUCTURE ver. 2·2 (Pritchard et al., 2000), which is a probabilistic-based clustering method for investigating population structure using multi-locus genotype data. In the case of well-delimited genetic units in a meta-population, K groups corresponding to the K genetic units are expected. Admixed genotypes are identified by their highest probability memberships to a given K-cluster lower than 60 % and graphically represented by a juxtaposition of several colours. Given that neither the African nor the IOI data sets appeared as unique genetic units (Fig. 3), each region and sub-region data set was independently submitted to STRUCTURE. Our predefined populations were either taxonomic units (mostly species; see Appendix) or true populations. A series of K-values from 2 to 25 [depending on the predefined geographical region (see above) and the results obtained] were tested. In all cases, the following parameters were used: no prior information on ancestral populations; admixture model with allele frequencies correlated among populations; each run of 700 000 Markov chain Monte Carlo iterations with 30 000 as burn-in, with five runs for each K-value tested. The determination of the best K-value was considered as the modal value of (ΔK) an ad hoc quantity as proposed by Evanno et al. (2005). We also took into consideration the α parameter, given as output with values close to zero indicating the presence of structure (most individuals belong to one or another population) whereas α > 1 means that most individuals are admixed (Falush et al., 2003). Then, we considered the probability memberships of each genotype. When the predefined populations (Coffea spp. or true populations) did not correspond to well-defined genetic units most genotypes appeared admixed, with the probability membership to one cluster lower than 60 %. Conversely, when the majority of genotypes assigned to one K-cluster attained at least 80 % membership we considered them as part of well-delimited genetic units and verified the correspondence with their taxonomic status (i.e. species and botanical varieties).
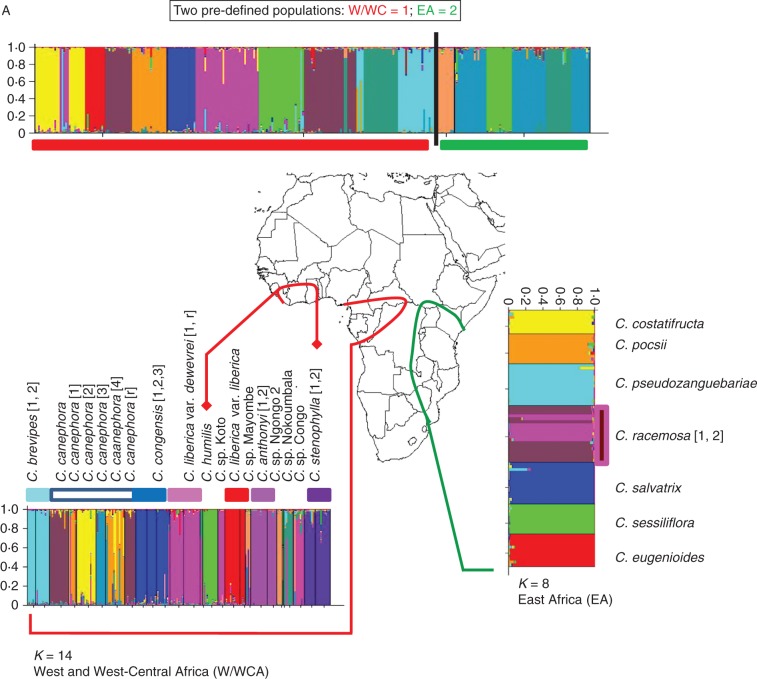
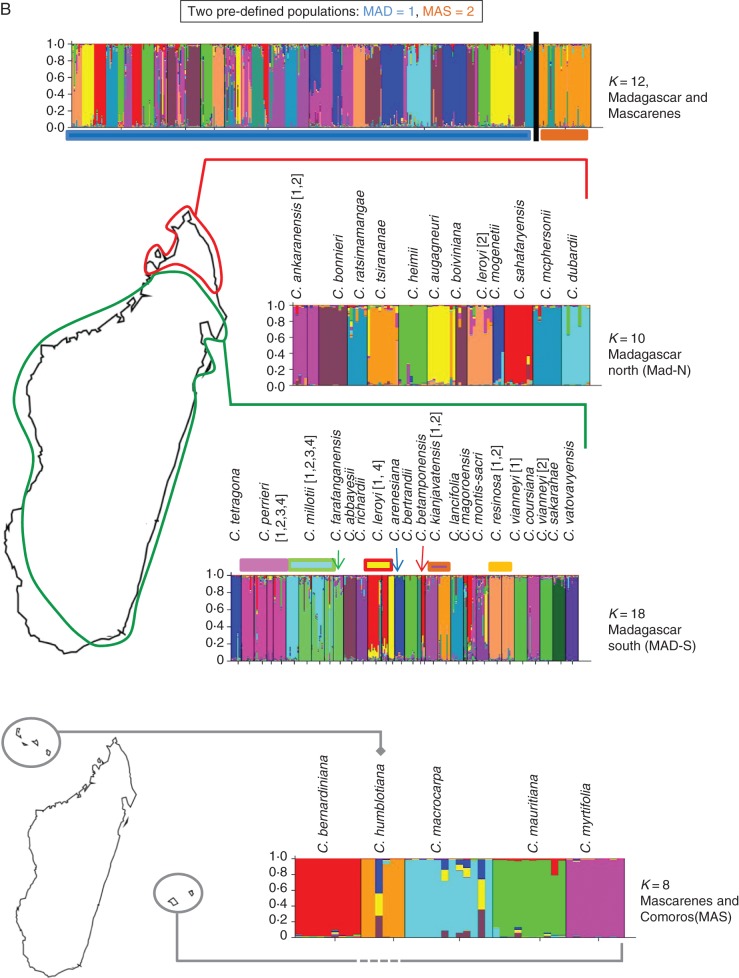
Fig. 3.
Analysis of Coffea genotypes using STRUCTURE superimposed on maps of Africa and Madagascar. Bar-plots representing the genotypes given on the x-axis distributed in pre-defined populations and coloured following their membership to the K-dependent clustering. For each geographical set analysed, the probability membership for each genotype in each K-cluster is given on the y-axis. Individuals with multiple colours have admixed genotypes. For each analysis, the ΔK values were calculated according to Evanno et al. (2005) in order to determine the best K value (see Supplementary Data Fig. S2). Species and population number/code (as specified in the Appendix) are given alongside the relevant bar-plots. (A) Analysis of 307 African Coffea genotypes. The two pre-defined African regions (West and West-Central Africa (W/WCA; 222 genotypes, including 51 genotypes from Upper Guinea centre of endemism) and East Africa (EA; 85 genotypes). (B) Analysis of 421 Indian Ocean Island area Coffea genotypes, by regions and sub-regions. Madagascar north (MAD-N; 104 genotypes); Madagascar south (MAD-S; 272 genotypes); and Mascarenes including the Comoros (MAS; 45 genotypes).
Genetic relationships
The groupings of genotypes into operational units for the analysis of genetic relationships was made on the basis of three processes: (1) analysis of genotypes undertaken using STRUCTURE (Pritchard et al., 2000); (2) the use of predefined taxonomic units (e.g. species) as based on morphology (Davis et al., 2006), and predefined populations, i.e. the accessions as received (but with identification updates via cross-checking with accession data and with reference to voucher specimens); (3) and genome size (as recorded by Cros et al., 1995; Noirot et al., 2003; Razafinarivo et al., 2012): statistically equivalent genome size was used as an additional deciding character for the grouping of specific accessions. Of the initial 87 potential populations surveyed, six were assimilated, leaving 81 for the analysis of genetic relationships (Fig. 4). Six accessions were assimilated, mainly on the basis of the STRUCTURE analysis (i.e. due to near-identical genetic similarity): the two C. canephora populations from Reunion ([r1], [r2]; see Appendix), identified as C. canephora without suffix in Fig. 4; C. sp. ‘Congo’ (OB accession group; Appendix) and C. sp. ‘Ngongo 2’ (OF accession group), which were merged with C. sp. ‘Nkoumbala’ (OI accession group) and C. mayombensis, respectively; C. perrieri ([4] A.730), which was assimilated with C. perrieri ([2] A.305); the two populations of C. mangoroensis ([1] A.401; [2] A.402); and the two populations of C. liberica var. liberica (EA accession group; and [r] LIB accession group). Effectively, this reduces the number of terminals for the analyses of genetic relationships; retention of these accessions in the analyses places them with the assimilated accessions (not shown).
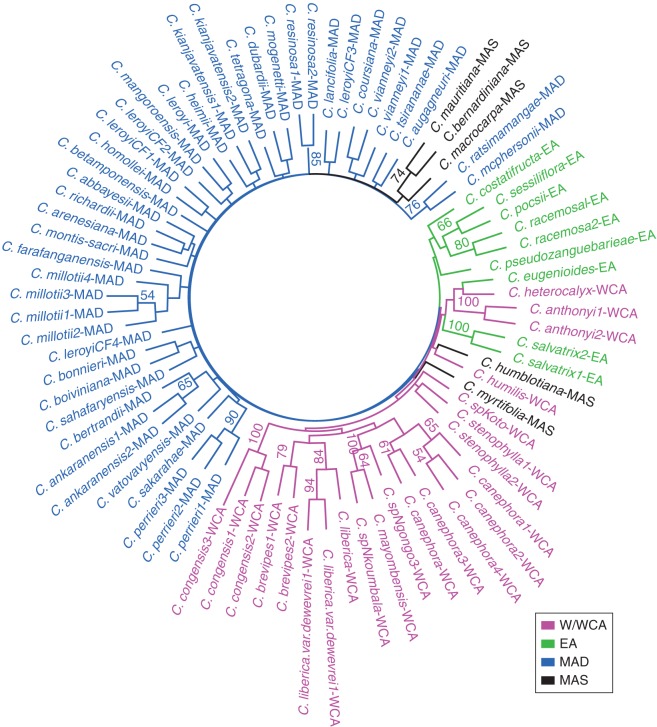
Fig. 4.
Unrooted neighbour-joining trees based on 13 microsatellite markers, using shared allele distance (DA,S) (Chakraborty and Jin, 1993), to show the genetic relationships among 81 African and Indian Ocean Coffea populations. Trees obtained from 500 bootstrap iterations; bootstrap values greater >50 % are shown on branches. W/WCA, West and West-Central Africa; EA, East Africa; MAD, Madagascar; and MAS, Mascarenes including the Comoros. Species/population names according to the Appendix.
Two methods were used to estimate the between-individual genetic distance, both methods being based on the individual allelic frequencies matrix: (1) shared allele distance (DA,S) as defined by Chakraborty and Jin (1993), on the assumption of a stepwise mutation model (SMM); and (2) Rogers' (1972) Euclidian genetic distances. The shared allele distance (DA,S) should retain linearity with increasing time (Chakraborty and Jin, 1993). Rogers' (1972) Euclidian genetic distances were used on the basis that the mutation model (evolutionary model) is unknown, enabling two different approaches to be compared. For the shared allele distance an unrooted neighbour-joining tree (Saitou and Nei, 1987) was constructed; and for non-model genetic distances an unrooted tree was constructed using UPGMA aggregating methods. In both cases the accessions were assigned to the four main regions (W/WCA, EA, MAD, MAS). To assess the degree of statistical support, 500 bootstrap iterations were performed on the data set. All calculations were made in PowerMarker ver. 3·25 (Liu and Muse, 2005). A minimum 50 % majority rule consensus tree was built from the 500 retrieved bootstrap trees, using the CONSENSE package of PHYLIP ver. 3·69 (Felsenstein, 2005). Bootstrap support (BS) values >50 % were added to the relevant branches. BS was designated as poor/low (50–70 %), moderate (71–84 %), and strong (85–100 %). Trees were visualized in SeaView ver. 4·2.5 (Gouy et al., 2010) and drawn using INKSCAPE ver. 0·48·2-1 (http://inkscape.org/).
Regional analysis of genetic variation
To determine the genetic variation for each geographical area, other than their main area of origin (e.g. Africa, Madagascar) the genotypes were again assigned to regions/sub-regions, as outlined above, that is: (1) W/WCA, (2) EA, (3) MAD, (3i) MAD-N and (3ii) MAD-S, and (4) MAS. For some analyses regions were grouped together to form two main areas: (1) and (2) for Africa; and (3) and (4) for the IOIs. Comparative analyses were performed on the different geographical sets, as follows: Africa [(1) and (2)] vs. IOIs [(3i, 3ii) and (4)] (set 1); MAD [(3i) and (3ii)] vs. MAS [(4)] vs. Africa [(1) and (2)] (set 2); (1) W/WCA vs. (2) EA vs. regions within the IOIs [(3i, ii) and (4)] (set 3). For each geographical set, the following genetic parameters were estimated (means and standard deviations provided where relevant): total number of alleles per locus (NA,Total), effective alleles per locus (Ne = 1/(1 – He), shared alleles (As), number of private alleles (Ap) and their proportion relative to the total number of alleles detected in a given set, observed heterozygosity (Ho), expected heterozygosity (He), percentage polymorphic loci at 0·05 (PL) and fixation indices (F). Calculations were performed in GenAlEx6 (Peakall and Smouse, 2006). Deviation from Hardy–Weinberg equilibrium was tested using GENEPOP 4·0.10 web version (Raymond and Rousset, 1995; Rousset, 2008). We also calculated the probability of the presence of null alleles (rb) using MICRO-CHECKER ver. 2·2.3 (Van Oosterhout et al., 2004) and estimated their frequencies at every locus in each species according to Brookfield (1996) as rb = (He – Ho)/(1 + He), where He is the expected heterozygosity under the Hardy–Weinberg hypothesis and Ho the observed heterozygosity. Averaged values over all loci are given in Table 2. These parameters were also estimated for each population and then averaged per area/region/sub-region, giving the mean values of the mean population of each region considered (Table 3). Pairwise Fst (a standardized measure of genetic variance among population) averaged over all loci were calculated based on pairwise differences between ‘populations’ for set 3, and values of significance at 0·05 level were assessed using 1000 permutations (see Table 6). The number of migrants (M; see Tables 6 and 7) were estimated from Nm = [(1/Fst) – 1]/4 and M = 2Nm for diploids. To determine the proportion of genetic variance explained by the differences between and among regions an analysis of molecular variance (AMOVA) was also performed using comparative sets (sets 1–3, see Table 8). Fst, number of migrants (Nm) and AMOVA calculations were performed in ARLEQUIN ver. 3·5 (Excoffier et al., 2005).
Table 2.
Genetic parameters for the different predefined geographical areas/regions/sub-regions
| Area/region/sub-region | Se | PL | AN | AN,e | Ho | He | F | rb | AP/AN,Total | AS (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Mean Africa | 253·2 | 100 | 15·23 | 6·84 | 0·34 | 0·83 | 0·59 | 0·26 | 67/198 | 66·2 |
| s.d. | ±13·34 | – | ±4·76 | ±2·37 | ±0·13 | ±0·08 | ±0·14 | ±0·06 | ||
| Mean IOI | 396·1 | 100 | 12·77 | 4·12 | 0·27 | 0·65 | 0·57 | 0·22 | 35/166 | 78·9 |
| s.d. | ±28·04 | – | ±6·75 | ±3·27 | ±0·13 | ±0·19 | ±0·17 | ±0·08 | ||
| Mean W/WCA | 169·8 | 100 | 13·54 | 6·29 | 0·35 | 0·81 | 0·57 | 0·25 | 44/176* | 75·0 |
| s.d. | ±5·89 | – | ±4·73 | ±2·14 | ±0·13 | ±0·09 | ±0·14 | ±0·06 | ||
| Mean EA | 79·62 | 100 | 8·07 | 4·58 | 0·31 | 0·74 | 0·58 | 0·24 | 11/105* | 89·5 |
| s.d. | ±8·62 | – | ±2·95 | ±1·78 | ±0·18 | ±0·11 | ±0·22 | ±0·09 | ||
| Mean MAD | 352·9 | 100 | 12·0 | 3·99 | 0·28 | 0·63 | 0·52 | 0·20 | 28/156 | 82·0 |
| s.d. | ±27·54 | – | ±7·09 | ±3·38 | ±0·14 | ±0·22 | ±0·20 | ±0·10 | ||
| Mean MAS | 43·15 | 100 | 6·23 | 3·20 | 0·19 | 0·613 | 0·69 | 0·25 | 4/81* | 95·1 |
| s.d. | ±2·23 | – | ±1·58 | ±1·46 | ±0·13 | ±0·19 | ±0·17 | ±0·09 | ||
| Mean MAD-S | 260·7 | 100 | 10·92 | 3·68 | 0·26 | 0·603 | 0·53 | 0·19 | 19/142* | 86·6 |
| s.d. | ±16·86 | – | ±6·75 | ±3·06 | ±0·13 | ±0·22 | ±0·20 | ±0·10 | ||
| Mean MAD-N | 92·23 | 100 | 8·23 | 4·01 | 0·33 | 0·67 | 0·47 | 0·19 | 4/107* | 96·3 |
| s.d. | ±10·70 | – | ±3·29 | ±2·03 | ±0·17 | ±0·20 | ±0·24 | ±0·11 |
Se: effective sample size; AS: proportion of shared alleles. Each following parameter was averaged on the 13 loci analysed. PL: polymorhic loci at 5 %; AN: number of alleles; AN,e: efficient alleles; Ho: observed heterozygosity; He: expected heterozygosity; F: fixation index and rb: null alleles. AP/AN,Total: number of private alleles/total number of alleles. For example, among the 105 alleles in total (AN,Total) found in EA, 11 are private alleles (AP). *Obtained with the subregions of Africa and IOI.
Table 3.
Average genetic parameters for each population (details are given in Supplementary Data Table S1) for the different predefined geographical areas/regions/sub-regions, with mean values and standard deviation
| Area/region/subregion | Se | PL | AN | AN,e | Ho | He | F | rb |
|---|---|---|---|---|---|---|---|---|
| Mean Africa | 9·29 | 0·8 | 2·9 | 2·08 | 0·34 | 0·39 | 0·13 | 0·04 |
| s.d. | ±2·51 | ±0·18 | ±0·84 | ±0·55 | ±0·12 | ±0·13 | ±0·17 | ±0·05 |
| Mean IOI | 8·53 | 0·68 | 2·18 | 1·67 | 0·27 | 0·29 | 0·04 | 0·01 |
| s.d. | ±1·81 | ±0·15 | ±0·45 | ±0·31 | ±0·09 | ±0·09 | ±0·21 | ±0·04 |
| Mean W/WCA | 9·04 | 0·8 | 3·08 | 2·19 | 0·35 | 0·41 | 0·11 | 0·04 |
| s.d. | ±2·55 | ±0·20 | ±0·92 | ±0·59 | ±0·13 | ±0·14 | ±0·16 | ±0·05 |
| Mean EA | 9·95 | 0·8 | 2·42 | 1·77 | 0·3 | 0·35 | 0·18 | 0·04 |
| s.d. | ±2·43 | ±0·12 | ±0·29 | ±0·24 | ±0·1 | ±0·08 | ±0·21 | ±0·04 |
| Mean MAD | 8·57 | 0·69 | 2·22 | 1·71 | 0·28 | 0·3 | 0·02 | 0·01 |
| s.d. | ±1·79 | ±0·14 | ±0·44 | ±0·30 | ±0·08 | ±0·09 | ±0·22 | ±0·04 |
| Mean MAS | 8·23 | 0·58 | 1·86 | 1·38 | 0·16 | 0·2 | 0·15 | 0·02 |
| s.d. | ±2·18 | ±0·20 | ±0·42 | ±0·20 | ±0·08 | ±0·08 | ±0·08 | ±0·02 |
| Mean MAD-S | 8·68 | 0·66 | 2·15 | 1·66 | 0·27 | 0·28 | 0 | 0 |
| s.d. | ±1·76 | ±0·14 | ±0·46 | ±0·31 | ±0·08 | ±0·08 | ±0·23 | ±0·04 |
| Mean MAD-N | 8·21 | 0·79 | 2·45 | 1·89 | 0·33 | 0·38 | 0·1 | 0·03 |
| s.d. | ±3·74 | ±0·11 | ±1·00 | ±0·83 | ±0·15 | ±0·17 | ±0·24 | ±0·22 |
Se: effective sample size; AP/AN,Total: private alleles/total number of alleles; AS: proportion of shared alleles. Each following parameter was averaged over the 13 loci analysed. PL: polymorh loci at 5 %; AN: number of alleles; AN,e: efficient alleles; Ho: observed heterozygosity; He: expected heterozygosity; F: fixation index and rb: null alleles.
Table 6.
Pairwise Fst and migrant estimates
| Regions/sub-regions (set 3) | W/WCA | EA | MAD-N | MAD-S | MAS |
|---|---|---|---|---|---|
| W/WCA | – | 8·6 | 5·6 | 4·6 | 4·6 |
| EA | 0·11 | – | 6·2 | 5·1 | 4 |
| MAD-N | 0·13 | 0·13 | – | 19 | 5·8 |
| MAD-S | 0·17 | 0·17 | 0·055 | – | 4·8 |
| MAS | 0·19 | 0·22 | 0·12 | 0·16 | – |
Fst calculations (below the diagonal) and estimates of the number of migrants (M = 2*Nm) (above the diagonal) were performed on the four regions and sub-regions of Madagascar.
Regions: West and West-Central Africa (W/WCA), East Africa (EA), and Madagascar (MAD) and Mascarenes (MAS); sub-regions: Madagascar north (MAD-N) and Madagascar south (MAD-S).
Table 7.
Number of migrants (M = 2*Nm) estimated between regions/sub-regions considering different combinations
| Regions | M | Min – Max M per locus |
|---|---|---|
| Set 1: Africa/IOIs | 7 | 1·4 (ES13) – 92·6 (M804) |
| Set 2: Africa/MAD/MAS | 4·2 | 1·4 (ES13) – 18·6 (M821) |
| Set 3: W/WCA/EA/MAD-N/MAD-S/MAS | 3·6 | 1·2 (ES13) – 11·6 (M821) |
| IOIs: MAS/MAD-N/MAD-S | 5 | 1·2 (ES13) – 31·6 (M821) |
| Africa: W/WCA/EA | 8·6 | 2·8 (C2_At4g35070) – 38·1 (ES12) |
| Madagascar: MAD-N/MAD-S | 19 | 4·7 (ES84) – 194·2 (ES12) |
Calculations averaged over all loci (M) and range, minimum – maximum values obtained per locus are given, with the corresponding locus name in parentheses.
Areas: Africa and IOIs (Indian Ocean Islands); regions: West and West-Central Africa (W/WCA), East Africa (EA), and Madagascar (MAD) and Mascarenes (MAS); sub-regions: Madagascar north (MAD-N) and Madagascar south (MAD-S).
Table 8.
Genetic variation distribution (AMOVA results) considering the different geographical sets (see material and methods)
| Source of variation | Sum of squares | Variance components | Percentage variation | Statistics | P |
|---|---|---|---|---|---|
| Set 1: Africa vs. IOI | |||||
| Among population | 414·63 | 0·63 | 12 | Fst = 0·116 | 0·01 |
| Within population | 6578·56 | 4·80 | 88 | ||
| Total | |||||
| Set 2: Africa vs. MAD vs. MAS | |||||
| Among population | 532·28 | 0·69 | 13 | Fst = 0·129 | 0·01 |
| Within population | 6460·91 | 4·72 | 87 | ||
| Total | 6993·19 | 5·42 | |||
| Set 3: EA vs. W/WCA vs. MAS vs. MAD-N vs. MAD-S | |||||
| Among population | 722·76 | 0·70 | 13 | Fst = 0·133 | 0·01 |
| Within population | 6270·43 | 4·59 | 87 | ||
| Total | 6993·19 | 5·29 | |||
RESULTS
General arrangement of genetic variation
Based on all the genotypes for which the country of origin was known (705 in total), the PCA revealed an almost continuous genotype distribution, shown as a three-dimensional factorial plot (Fig. 2A). The statistical significance for the first three axes of the PCA, using the analysis method of Patterson et al. (2006), was robust (x1 = 199·3, P < < 0·001; x2 = 131·3, P < < 0·001 and x3 = 127·4, P < < 0·001, respectively). Overlap between the African and IOI genotypes is manifest. Despite this, MAD genotypes were strongly grouped towards axis 1 negative values and African genotypes, especially those from W/WCA, were mainly distributed towards axis 1 positive values and along axis 2. Two African species (C. congensis and C. anthonyi) are separated (Fig. 2A). EA and MAS genotypes occupied an intermediate position between W/WCA and MAD. EA, MAD and MAS genotypes are tightly clustered to one end of the W/WCA genotypes, with the vast majority of the MAS genotypes clustering closely with MAD genotypes. The four regions are not clearly separated, but they have a clustered, non-random structure, with a general pattern of distribution from west (W/WCA) genotypes to east (EA > MAD > MAS). In the Africa-only PCA, the statistical significance (Patterson et al., 2006) for the first three axes was also robust (x1 = 77·09, P < < 0·001; x2 = 66·8, P < < 0·001; and x3 = 59·8, P < < 0·001, respectively). West and West-Central Africa and EA genotypes overlap but if the Upper Guinea species C. humilis and C. stenophylla [Upper Guinea (UG) clade of phylogenetic sequence analyses; e.g. Maurin et al. (2007)] and C. anthonyi and C. eugenioides [the East-Central Africa (E-CAfr) clade; e.g. Davis et al. (2011)] are removed there is an almost clear separation (Fig. 2B). Within the W/WCA genotypes, C. anthonyi is distinctly separated and, although East African genotypes are widely distributed along axis 1, C. pseudozanguebariae is also separated (Fig. 2B). Separate analysis conducted for the IOIs showed three highly significant principal axes (x1 = 111·18, P < < 0·001; x2 = 97·6, P < < 0·001; and x3 = 93·6, P < < 0·001, respectively) and revealed a near-discrete cluster (with slight overlap) for the MAS genotypes; MAD-N clusters within MAD-S (Fig. 2C). MAD-S genotypes have considerably more variation than MAD-N. Coffea humblotiana (from the Comoros) is clustered with the MAD-N genotypes.
Fig. 2.
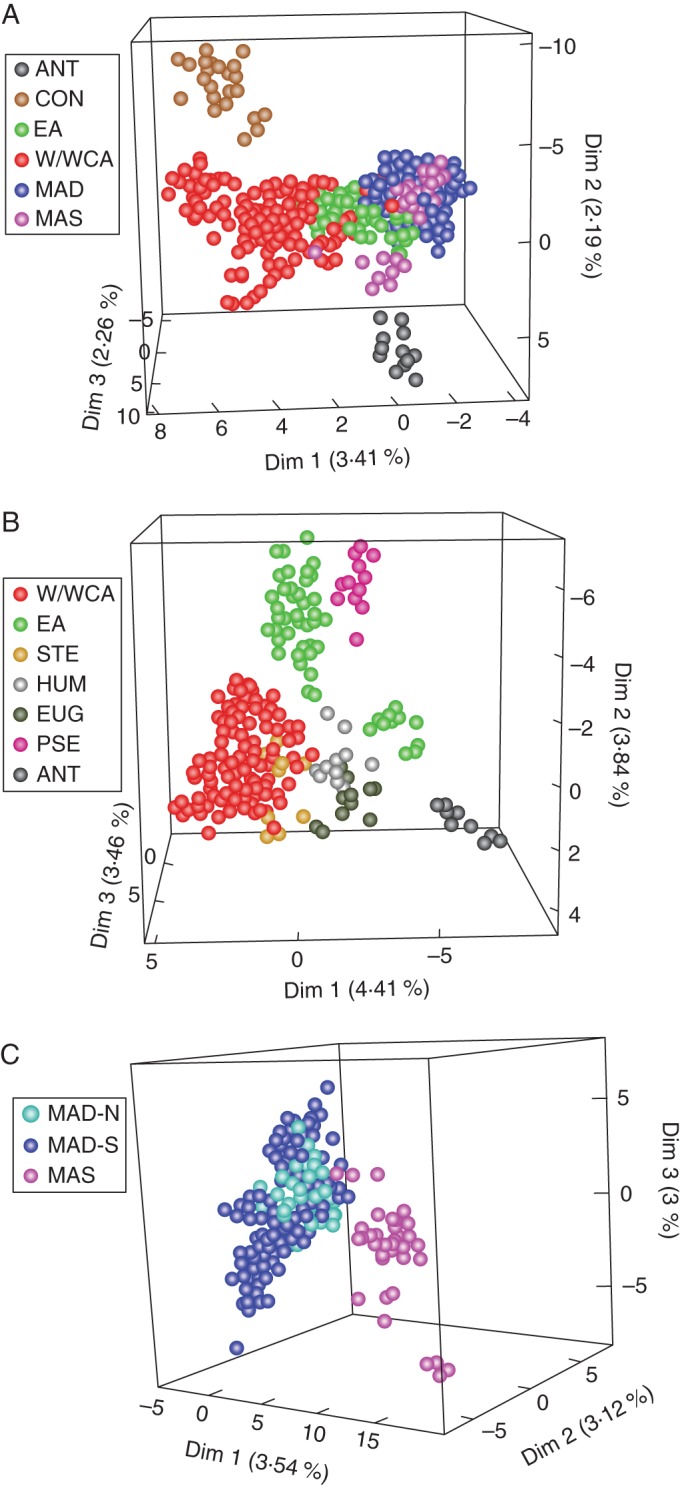
PCAs of Coffea genotypes; distribution in three-dimensional factorial plan, based on individual allele frequencies. (A) PCA of African and Indian Ocean Island Coffea genotypes (705 genotypes and 233 alleles). W/WCA, West and West-Central Africa, including C. congensis (CON) and C. anthonyi (ANT); EA, East Africa; MAD, Madagascar; and MAS, Macarenes including the Comoros. (B) PCA of African Coffea genotypes (284 genotypes and 198 alleles). W/WCA, West and West-Central Africa, including C. stenophylla (STE), C. humilis (HUM) and C. anthonyi (ANT); and EA, East Africa, including C. eugenioides (EUG), and C. pseudozanguebariae (PSE). (C) PCA of Indian Ocean Island Coffea genotypes (421 genotypes and 165 alleles). MAD-N, Madagascar north; MAD-S, Madagacar south; and MAS Macarenes including the Comoros.
There is a good general geographical structuring in the PCA analyses, although the paucity of separated genotypes and populations (i.e. those from more than one locality, see Appendix) justified population genetic methods to assess Coffea genetic diversity more fully.
Identification of genetic units
Based on the best-fitting K-values for our data (K = 11 for Africa and K = 12 for IOIs), as revealed by STRUCTURE, none of the geographical areas or regions (corresponding to predefined populations or species) appeared to constitute genetically homogeneous ‘meta-populations’ (Fig. 3). When the STRUCTURE analysis was performed independently on regions and sub-regions, even if some genotypes appeared as admixtures, it was possible to define sets of genotypes belonging to only one K-cluster, and these corresponded (with few exceptions; see below) to pre-determined taxonomic units, i.e. species (Fig. 3). For instance in W/WCA all species are well structured, although some populations of C. canephora appeared considerably differentiated (Fig. 3A). In EA the situation was clear: each predefined taxonomic unit (species) corresponded to only one structured group, with the exception of C. racemosa, which is split into two clusters.
For the Mascarene genotypes, genetic groupings corresponded to pre-determined taxonomic units (i.e. species), although two genotypes of C. humblotiana and C. macrocarpa appeared as admixtures (Fig. 3B). In MAD, the situation appears more complex, especially in the eastern humid forest region (MAD-S), the area of Madagascar with the highest Coffea species richness (i.e. species number), although most species were retrieved as well-delimited genetic units (Fig. 3B). Some populations of species such as C. millotii (formerly C. dolichophylla; [3]; Appendix), C. leroyi and the two accessions of C. kianjavatensis (A.602 [1], A.213 [2]; Appendix) also appeared well differentiated. Coffea leroyi occurs in two different groupings, although A.317 ([cf. 3]) is only represented by one genotype, and the other (A.956 [cf. 2]) is from MAD-N and is unlikely to be closely related to this otherwise more southerly occurring species. Conversely, different populations for a given species appeared as separate genetic unit entities, i.e. two C. millotii populations (A.222 [2], A.206 [3]; Appendix) and two C. kianjavatensis populations (A.602 [1], A.213 [2]; see Appendix). Some were in complete admixture, e.g. C. mangoroensis.
Genetic relationships
Analyses of genetic distance based on individual allelic frequencies matrices for 81 populations produced trees of genetic relationships for Coffea across Africa and the IOIs. UPGMA of Euclidian genetic distances (Rogers, 1972) and neighbour-joining of shared allele distance (DA,S) (Chakraborty and Jin, 1993) produced similar results, although the latter performed slightly better, as judged by: greater agreement with previous hypotheses of relationships based on sequence studies (see Introduction and Methods), and the results produced by our PCA and STRUCTURE analysis, and slightly improved bootstrap support values. Specific differences include: (1) the two populations of C. stenophylla come together at the base of W/WCA Africa (not grouped in the UPGMA); and (2) there is better resolution for W/WCA Africa. There are some minor differences but these are on short branch lengths. On this basis we decided to describe the results based on the neighbour-joining of shared allele distance (DA,S); the UPGMA tree is provided as Supplementary Data Fig. S1.
Moderate to strong BS values (>70 %) were produced for some branches on the neighbour-joining tree, but generally support was zero to negligible across the study group (Fig. 4). All populations belonging to the same species fall into their respective groups, mostly with BS, apart from C. leroyi (MAD). Aside from well-supported groups at the species level, other groupings receiving around 50 % BS or higher are species pairs: including C. pocsii and C. sessiliflora (BS = 66 %) (EA); C. mauritiana and C. bernardiniana (BS = 74 %) (MAS); C. mcphersonii and C. ratsimamangae (BS = 76 %) (MAD)' and C. sp. ‘Ngongo 3’ (including C. sp. ‘Ngongo 2’), C. sp. ‘Nkoumbala’ and C. mayombensis (BS = 100 %) (W/WCA).
Based on reasonable branch lengths, but zero or negligible BS, the following can be stated. African and IOI species are separated into two clusters (Fig. 4). In EA, the low-altitude species (C. costatifructa, C. sessiliflora, C. pocsii and C. racemosa) group together, and are separated from the higher altitude species C. salvatrix. In W/WCA, C. anthonyi, C. heterocalyx and C. eugenioides are grouped despite their distribution across Africa. Populations of C. canephora are grouped together and nested within other W/WCA species. In MAD, many relationships are represented by groups with short branch lengths and zero to negligible BS (<50 %).
Regional analysis of genetic variation
Regarding the genetic diversity among the geographical regions/sub-regions, globally there was a higher total number of alleles per locus (NA) in Africa than in the IOIs (15·2 versus 12·8). More similar values were obtained by comparing W/WCA and MAD (13·5 and 12, Table 4) knowing that sample size for W/WCA was less than half that for MAD but included the highly polymorphic C. canephora (contributing seven alleles). In MAD, NA ranged from 8·2 (MAD-N) to 10·9 (MAD-S). Heterozygote deficiency (Ho) was detected for all loci, both for the IOIs and for the African regions. For each sub-region and all loci, testing for Hardy–Weinberg equilibrium (where H1= heterozygote deficit) gave a P value of 0·0000 and a standard error of 0·0000. Hence, Hardy–Weinberg equilibrium was rejected and a heterozygote deficit was observed. Averaged over all loci, Ho was less than half He. He decreased stepwise from west to east: W/WCA (0·82), EA (0·74), MAD (0·63), MAS (0·61). High fixation indices (F) were at least partly explained by the presence of a high proportion of null alleles. At the population level (Table 3) the same genetic parameters averaged for each of the populations for the area/region/sub-region are not significantly different. This means that the main components of variation are between the pooled populations for each area/region/sub-region.
Table 4.
Pairwise AP and AS per sub-region, calculated in pairwise comparisons
| W/WCA | /EA | /MAS | /MAD | ||
|---|---|---|---|---|---|
| AP | W/WCA | – | 93/176 (52·8) | 106/176 (60·2) | 62/176 (35·2) |
| EA | 22/105 (20·9) | – | 53/105 (50·5) | 24/105 (22·8) | |
| MAS | 11/81 (13·6) | 29/81 (35·8) | – | 10/81 (12·3) | |
| MAD | 42/156 (26·9) | 75/156 (48) | 85/156 (54·5) | – | |
| AS | W/WCA | – | 83/198 (41·9) | 70/198 (35·3) | 114/218 (52·3) |
| EA | – | 52/134 (38·8) | 81/180 (45) | ||
| MAS | – | 71/166 (42·8) | |||
| AS/total | all | 47/233 (20·2) | |||
The first line indicates, for example, W/WCA AP/AN compared with EA, MAS and MAD. Among the 176 W/WCA alleles, 93 are private alleles relative to EA, 106 are relative to MAS and 62 to MAD. Percentages are given in parentheses. For AS, the proportion is relative to the total alleles exhibited by the pair of sub-regions compared. For example, the pair (W/WCA)/EA shared 83 alleles of a total of 198. The second section gives the proportion of shared alleles by all sub-regions.
The proportion of shared alleles (AS) within each region (Table 2) is high: 66·2 % for Africa vs. 78·9 % for IOI; and by region: W/WCA (75 %), EA (89·5 %), MAD (82 %) and MAS (95·1 %). Conversely, the proportion (AP/NA,Total) of private alleles (AP) within each region and sub-region is much lower and varies from 3·7 % (4/107) for MAD-N to 25 % (44/176) for W/WCA (Table 2). From west to east AP decreases stepwise (apart from EA): W/WCA (44), EA (11), MAD (28), MAS (4). Exclusive AS values (AS,ex) between region pairs or triplets are found (Table 5). They are unequal in numbers but represented by numerous loci and not only rare alleles (frequency <0·05). For instance, the 27 AS,ex between W/WCA and MAD covered 10 loci but none is frequent in the two regions compared, whereas the 12 AS,ex between W/WCA and EA included three frequent alleles in the two regions, represented at all seven loci.
Table 5.
Distribution of alleles
| 1/2 | 1/3 | 1/4 | 2/3 | 3/4 | 1/2/3 | 1/2/4 | 1/3/4 | 2/3/4 | 1/2/3/4 | |
|---|---|---|---|---|---|---|---|---|---|---|
| AS,ex | 12 | 27 | 5 | 7 | 3 | 23 | 1 | 17 | 4 | 47 |
| LOC | 7 | 10 | 5 | 5 | 3 | 12 | 1 | 8 | 4 | 13 |
| AS,freq | 3 | 0 | 1 | 1 | 0 | 4 | 0 | 1 | 2 | 20 |
AS,ex: number of exclusive shared alleles between region pairs and triplets (1, W/WCA; 2, EA; 3, MAD; 4, MAS). LOC: the number of loci involved. AS,freq: the number of frequent alleles (frequency ≥0·05) shared in the comparison.
Regions: West and West-Central Africa (W/WCA), East Africa (EA), and Madagascar (MAD) and Mascarenes (MAS).
In pairwise comparisons (Table 4), W/WCA exhibited the highest proportion of AP (from 35·2 to 60·2 %), whereas the lowest values were for MAS (13·6–35·8 %). The proportion AS varied from 35·3 % for W/WCA vs. MAS to 52·3 % for W/WCA vs. MAD. However, the proportion of AS by all regions is 20·2 %, of which 42·5 % are frequent (>0·05) in all regions.
Pairwise Fst values for regions and sub-regions were all significant (P = 0·0000, Table 6). The strongest value was obtained between an African and IOIs region (0·22 for MAS vs. EA) and the lowest for the MAD sub-regions (0·055 for MAD-N vs. MAD-S). The MAS appeared strongly differentiated from Africa and MAD but the lowest value was for MAD-N (0·12). Consequently, averaged over all loci, the number of migrants was of the same magnitude between MAS and MAD (MAD-S = 4·8 and MAD-N = 5·8) and between Africa and IOI sub-regions (4 to 6·2), compared with that found among regions in Africa (EA vs. W/WCA = 8·6) or in MAD (19) (Table 6). However, considerable differences in the number of migrants exchanged were obtained depending on the locus considered, for example from 4·7 (ES84) to 194·2 (ES12) between MAD-N and MAD-S and as much as 92·6 (M804) between Africa and the IOIs (Table 7). Whatever the geographical set considered, similar AMOVA results were obtained (Table 8). In summary, the greatest part of the variation is explained by within-region/sub-region (87–88 %) rather than among-region/sub-region (12–13 %) differences, and especially for the regions (W/WCA, EA, MAD and MAS). These results infer intra-regional differentiation, i.e. radiations within the regions/sub-regions, rather than across Africa and the IOIs in general, in agreement with the Fst results.
DISCUSSION
Application of SSR markers in multiple species analyses
SSRs display strong advantages compared with other methods (e.g. they are highly polymorphic, co-dominant and easy to use), but there are also several constraints to their use [e.g. high mutation rates, and size homoplasy (size constraints; see below); Estoup et al. (2002)]. Traditionally, SSRs are widely used for population genetics (Guajardo et al., 2010; Born et al., 2011; Legrand et al., 2011) and genetic mapping (Burrell et al., 2011). Multilocus SSR studies have also been used to investigate species delimitation (Richard and Thorpe, 2001), phylogenetic relationships within genera (Orsini et al., 2004; Cubry et al., 2008), and even phylogenetic relationships across genera (Petren et al., 1999; Ritz et al., 2000; Ochieng et al., 2007). Despite the apparent success of SSRs in studies above the species level, investigation of genetic structure, diversity and relationships across species and species groups (e.g. genera) has not received much attention. The slow take-up of the SSR approach at higher levels of taxonomic hierarchy may be due to concerns over transferability of SSRs across a large range of species, and issues regarding detectable orthology (i.e. inherited from a shared ancestor) and other causes of homoplasy. To address these points, we discuss below some specific features of SSRs.
Cross-species transferability and SSR homoplasy
Given the large number of species used in this study (60 in total), no prior selection was made as to the potential polymorphism that they can reveal, as monomorphic markers for one species could be polymorphic for another. The aim of this strategy is to limit the bias of the genetic variation by over-estimation, due to the removal of the least variable markers (Chikhi, 2008). In our study, the level of polymorphism depended on the locus used; a minimum of 12 alleles for ES42 and ES74 and a maximum of 38 alleles for A8856 were detected. Similarly, no initial selection on their species-discriminating power was made, in order to avoid, or at least limit, possible bias. Ellegren et al. (1997) reported that microsatellite markers tested on one species produced shorter repeats in another species (ascertainment bias or size constraint). Here, nine SSR markers were developed using C. canephora. Seven and five of them produced shorter repeats in species originating from IOIs and Africa, respectively. However, six of the seven, and all five of five, also gave a larger allele size range, respectively (Table 1). Finally, orthology was assumed after the analysis of amplification patterns, length (same or similar range) and number of amplicons produced (two maximum for diploids at unique loci). Of course, the constraints for the evolution of microsatellites in anonymous non-coding genomic regions is not the same as for expressed sequence tag (EST) microsatellites and this resulted in a higher number of alleles for nuclear SSRs than for EST-SSRs. However, the constraint should be similar whatever the species considered and there is no reason to believe that this was not the case in our study. Therefore, if the constraints are different depending on the nature of the SSR (nuclear or EST), they will be similar irrespective of the species studied. In fact, of 47 shared alleles (20 % of the total number of alleles), 24 belong to anonymous loci and 23 to ESTs, and so this observation would not be in agreement with a hypothesis that constraints on allele size range are responsible for the percentage of shared alleles.
Regardless of previously defined phylogenetic relationships (e.g. Maurin et al., 2007) and using pre-defined PCR conditions for amplification (Plechakova et al., 2009), only 13–14 % of 452 and 341 initial genotypes from Africa and the IOIs, respectively, gave too much missing data with the 20 loci tested. These values were 50 % lower (6·2–6·9 %) for the 13 loci and 728 genotypes retained. In the case of the study presented here, the high mutation rate of SSRs is expected to be an advantage to assess Coffea genetic structuring and to estimate genetic relationships between genotypes, and among populations and species. In this study, 21 of 233 alleles differed by one base from initial forecasts based on the repeat unit length. Assuming that homoplasy can be buffered by the large number of genotypes analysed, as shown by Ochieng et al. (2007), we inferred a low level of homoplasy, and used a non-evolutionary model-based approach (PCA), model-based population genetics methods (i.e. the software STRUCTURE) and a shared allele distance analysis based on an SMM.
Cross-species SSR transferability within African and IOI species was efficient. Previous cross-species amplification among African Coffea spp., and a small number of Madagascan (MAD) species, has already proved efficient (Combes et al., 2000; Bhat et al., 2004; Poncet et al., 2004, 2007; Aggarwal et al., 2007; Hendre et al., 2008, Krishnan et al., 2013a, b). Our results now firmly demonstrate the potential of SSRs to study genetic structure and diversity and estimate genetic relationships in Coffea, concurring with reports on SSR transferability across distantly related plant species (Shepherd et al., 2006; Ochieng et al., 2007; Plechakova et al., 2009).
General arrangement of genetic variation
A multivariate approach using PCA demonstrated that across Africa and the IOIs, Coffea can be considered as a large, closely related group of species, with few clearly defined genetic clusters (Fig. 2A). Despite the paucity of clear-cut clusters, the PCA revealed structure in accordance with the main geographical regions (as above) and an overall pattern of genotypes running from west (W/WCA) to east (EA > MAD > MAS). No overlap between W/WCA and MAD species was observed (Fig. 2A), and this result is in agreement with the failure (or very low rates) of hybridization between African and Madagascan species, as demonstrated by low numbers of stunted F1 interspecific hybrids, and progeny with low fertility (Charrier, 1978).
In the separate PCA of the African area, the Upper Guinea species C. humilis and C. stenophylla (UG clade of phylogenetic sequences analysis; Maurin et al., 2007; Anthony et al., 2010; Davis et al., 2011; Nowak et al., 2012) are nested within W/WCA and are intermediate between W/WCA and EA, respectively (Fig. 2A). This position is consistent with the sequencing studies mentioned above and our unrooted radial tree based on shared allele distance (Fig. 4). Likewise, the near-clustering of C. anthonyi and C. eugenioides is also consistent with sequencing studies (the EC-Afr clade: Maurin et al., 2007; Davis et al., 2011; Nowak et al., 2012), and our neighbour-joining tree (Fig. 4).
The near-discrete cluster for the MAS genotypes (Fig. 2C) is also congruent with sequencing studies, which show that the Mascarene species form a well-supported clade (Maurin et al., 2007; Davis et al., 2011). The nested position (not indicated in Fig. 2C) of C. humblotiana (from the Comoros) within MAD-N is in agreement with the sequencing results of Maurin et al. (2007).
Identification of genetic units
Our results justified the use of the software STRUCTURE (Pritchard et al., 2000) to test for congruence between genetic and taxonomic units. Previously, Orsini et al. (2004) demonstrated that clusters/groups produced by STRUCTURE corresponded with pre-defined species in Drosophila. In our study we revealed a good relationship between species delimitations (and botanical varieties for C. liberica) and genetic units based on SSR data. Thus, in Coffea there is good correspondence between genetic structure and morphology (Davis et al., 2006), genome size (Noirot et al., 2003; Razafinarivo et al., 2012), seed chemistry (Dussert et al., 2008) and leaf chemistry (Campa et al., 2012).
The SSR data reveal some cases that are not consistent with anticipated genetic structure. The situation for our accessions of C. canephora (Appendix), which show admixture, is likely to reflect the complex natural variation of this species and/or the numerous exchanges that have occurred during the last century or so since this species became widely cultivated all over the world (Cheney, 1925). The genotypes surveyed reveal similar patterns to that identified in hybrids between the Congolese and Guinean groups, as defined by Gomez et al. (2009). In East Africa, each pre-defined taxonomic unit (i.e. species) corresponded to only one structured group, with the exception of C. racemosa. This species is split into two clusters: one corresponding to genotypes obtained from the Brazilian collection and the other from the collection constituted in Côte d'Ivoire and recently transferred in the field at the Biological Resources Centre, Saint Pierre, Reunion, France (Appendix). Coffea racemosa has quite a considerable geographical range, occurring in Mozambique, Zimbabwe and Kwa-Zulu-Natal (Davis et al., 2006), and it is likely that the original origin of these genotypes is different. Another exception is C. stenophylla. In a STRUCTURE analysis of W/WCA genotypes (not shown) the two populations of C. stenophylla were well differentiated (suggesting that the populations are isolated geographically and gene flow between them is limited), as they share few alleles (see below). However, in the second round (after removal of C. canephora accessions from Reunion) the improved K-value did not differentiate the two populations, and this was also the same for accessions of C. brevipes, C. anthonyi and C. congensis. Two genotypes from the accession of C. humblotiana and C. macrocarpa appeared as admixtures, although we think that this is probably be due to a labelling mistake or other sampling error.
Genetic relationships
Our tree of genetic relationships (Fig. 4) based on shared allele distance (DA,S) is congruent with phylogenetic studies based on plastid and internal transcribed spacer (ITS) sequencing (Maurin et al., 2007; Davis et al., 2011; see Introduction) and other data (see below), although BS for most groupings above the level of species is absent or negligible. There is a clear separation between Africa and the IOIs, as retrieved on the basis of ITS and plastid sequencing (Davis et al., 2011) and Ty1-copia LTR-retrotransposon data (Hamon et al., 2011), although this does not agree with the paraphyly of African and IOIs species identified using low-copy nuclear marker sequencing (Nowak et al., 2012). The Mascarene species (except C. myrtifolia) formed a separate clade close to the Madagascan (MAD) species group, a relationship consistent with Maurin et al. (2007) and Davis et al. (2011). The position of C. myrtifolia cannot be readily explained and further sampling of this species is required. The grouping of W/WCA species is consistent with the Lower Guinean/Congolian (LG/C) clade of Maurin et al. (2007), Anthony et al. (2010; as Guineo-Congolian) and Davis et al. (2011); and the grouping of East African low-altitude species (C. costatifructa, C. sessiliflora, C. pocsii and C. racemosa) is also congruent with the aforementioned sequencing studies. The close relationship between C. anthonyi, C. eugenioides and C. heterocalyx is consistent with the East-Central African (EC-Afr) clade revealed by sequence data (Maurin et al., 2007; Anthony et al., 2010; Davis et al., 2011) and leaf chemistry (Campa et al., 2012). The position of the EC-Afr clade with EA species and one of the Upper Guinea species (C. humilis) is consistent with plastid sequence data (Cros et al., 1998; Maurin et al., 2007; Davis et al., 2011), but is in disagreement with low-copy nuclear region sequencing (Nowak et al., 2012), which places it with Lower Guinea/Congolian species (i.e. our W/WCA).
The Upper Guinea endemics (C. stenophylla, C. humilis, C. togoensis) formed a well-supported clade for Maurin et al. (2007) and Davis et al. (2011). In our analysis, C. stenophylla and C. humilis do not fall in the same clade, although this is not supported by bootstrap values (Fig. 4). One feature of the Upper Guinea (UG) clade is that C. humilis has the biggest genome in Coffea (representing the uppermost size limit for W/WCA African species) and C. stenophylla has the smallest for W/WCA and is within the range of genome sizes for EA (1·76 pg and 1·28 pg/2C estimated from fresh leaves, respectively; Noirot et al., 2003). In addition, C. stenophylla is found in drier habitats, compared with C. humilis, and has black fruits (like many East African species), whereas C. humilis occurs in wetter habitats and has red fruits (as in the majority of W/WCA species). Further work is needed to assess fully the evolutionary origin of, and relationships within, the Upper Guinea clade, and their relationships with other Coffea spp.
At the species level almost all pre-determined species replicates clustered together, with good levels of BS (Fig. 4), apart from C. leroyi (Madagasar) and C. stenophylla (Upper Guinea). These exceptions are discussed in the section ‘Identification of genetic units’. Strong bootstrap support (100) for the grouping of C. sp. ‘Ngongo 3’ (including C. sp. ‘Ngongo 2’), C. sp. ‘Nkoumbala’ and C. mayombensis (BS = 100) and morphological examination of voucher specimens and geographical location suggests that these accessions are all referable to a single species, i.e. C. mayombensis. Aside from well-supported groups at the species level, other groupings receiving 50 % BS or higher are three species pairs: C. pocsii and C. sessiliflora (East African lowlands); C. mauritiana and C. bernardiniana (Mascarenes); and C. mcphersonii and C. ratsimamangae (northern Madagascar). The close relationship between C. pocsii and C. sessiliflora has been identified on the basis of ITS and plastid sequencing studies (Maurin et al., 2007) but the others have not. The alliance of C. mcphersonii and C. ratsimamangae is entirely consistent with morphology and geographical distribution (Davis and Rakotonasolo, 2001a). The grouping of C. mauritiana and C. bernardiniana is noteworthy because the latter is considered conspecific with C. macrocapra (Leroy, 1989), presumably on the basis of general leaf and fruit morphology. The genome sizes of C. mauritiana and C. bernardiniana are different [1·23 and 0·96 pg, respectively; 1·17 pg for C. macrocarpa (Razafinarivo et al., 2012)], and the fruits mature at different times (end of July and early September, respectively).
Regional analysis of genetic variation
The genetic structuring and differentiation for W/WCA is better than all the other three regions (EA, MAD, MAS), as shown by the higher number of private alleles (Ap) and lower percentage of shared alleles (AS). W/WCA also possesses the highest He. If we consider that the mutation rate for a given SSR would be similar for both the source and the tested species (Harr et al., 1998) and that the number of mutations should increase with the time of divergence, we could infer that the speciation in EA, MAD and MAS was later than that in W/WCA or that the selective pressure in W/WCA was stronger than in the other regions, although both of these suggestions are highly speculative. For the four regions, W/WCA, EA, MAD and MAS, He is higher than Ho, meaning that there is a strong differentiation among the regions. We know that this is congruent with the fact that each region includes more than one true species (i.e. there are species radiations in these areas). Inter-regional differentiation and regional unity is also supported by the numbers of AP and AS obtained for each of the four regions (Table 2).
For each sub-region and all loci the Hardy–Weinberg equilibrium was rejected and a heterozygote deficit was observed. This deficit was partly the result of the presence of null alleles, intra- regional/sub-regional differentiation and/or inbreeding, although the role of each of these factors cannot be determined.
General discussion
Our study has shown that SSR data and methods more usually applied to population genetics (including model- and non-model-based approaches) can be used successfully across a large species complex to obtain an overview of genetic structuring and diversity. We have shown that the vast majority of African and IOI Coffea spp. examined possess significant genetic structure, corresponding to well-defined genetic units, i.e. they represent a tangible and practical means of expressing genetic cohesion, in good agreement with other data, including morphology, distribution, bioclimate and secondary chemistry (leaf and seed). The correspondence between morphotaxonomic units and genetic structure in Coffea is significant, as it shows that species provide robust units for in situ and ex situ conservation. Genetic structure and diversity corresponds to geography and diversification (i.e. species richness), which in the case of our study were regions and sub-regions [W/WCA, EA, MAD (MAD-N, MAD-S), MAS], pre-defined on the basis of biotic and abiotic criteria. Genetic relationships retrieved with SSRs were highly congruent with phylogenetic analyses based on plastid and nuclear sequencing, although bootstrap support for relationships above the level of species was generally lacking.
That speciation in East Africa (EA), Madagascar (MAD) and the Mascarenes (MAS) could have occurred later in West and West-Central Africa (W/WCA) is not in agreement with the distribution of genome size in Coffea. All species of Coffea so far examined, except C. arabica (an allotetraploid), are diploid (Bouharmont, 1963; Louarn, 1972) and have the same number of chromosomes, but with differences in genome size. Cros et al. (1995) and Noirot et al. (2003) found that there is a global increase in species genome size from East Africa (1·21 pg/2C) to West and West and West-Central Africa (1·52 pg/2C). The Madagascan and Mascarene species of Coffea (Razafinarivo et al., 2012) have a similar average genome size (1·19 pg/2C) to species from East Africa, with the smallest genome sizes being found more frequently in Mauritius, Comoros and north Madagascar. In angiosperm groups there is a general trend for genome size to increase with time (Soltis et al., 2003), although decreases in DNA content are often reported (e.g. Wendel and Cronn, 2002; Price et al., 2005; Johnston et al., 2005).
Our study shows SSRs have considerable potential for examining the genetic diversity of Coffea. An improved understanding of genetic relationships for the Madagascan and Mauritian species groups is still urgently required, and this might be achieved by using an improved and up-scaled SSR approach. It will also be necessary to examine those lineages not yet sampled, in particular the baracoffea alliance and short-styled lineages (previously included in Psilanthus), although the constraint for these groups is the lack of effective sampling, particularly as their species occur in remote and diffuse localities.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was undertaken in collaboration with the National Center of Applied Research and Rural Development (Foibe Fikarohana ampiharina amin'ny Fampandrosoana ny eny Ambanivohitra: FOFIFA), in Madagascar, through the RubiComp network. We thank the FOFIFA staff at the Kianjavato Coffee Research Station (KCRS), Madagascar, and the Ueshima Coffee Corporation (UCC), Japan, for allowing us to access the KCRS collections. We thank Fanchon Mora for her technical assistance and Muriel Gros-Balthazard for kindly providing personal R script. N.J.R holds a PhD fellowship from IRD. We also thank the various germplasm collections (see Appendix) for the use of original material, either directly sourced (i.e. KCRS) or via the Centre de Ressources Biologiques Coffea, Reunion; and the herbaria at FOFIFA, Antananarivo (TEF), Antananarivo Herbarium (TAN), Royal Botanic Gardens, Kew (K), the Natural History Museum, Paris (P), and Missouri Botanical Garden (MO; herbarium abbreviations after Holmgren et al., 1990), for allowing us access to their collections. We gratefully acknowledge three anonymous reviewers for their constructive comments and careful review of an earlier version of this contribution. This study was partially supported by the Conseil Régional de la Réunion and the Préfecture de la Région Réunion (FEADER project).
APPENDIX
Accession information for African and Indian Ocean Island Coffea taxa investigated in this study
APPENDIX.
| Species name [population number/code] | No. of individuals | Voucher/herbarium code | Country of origin | Geographical region/sub-region | Forest type | Germplasm collection source |
|---|---|---|---|---|---|---|
| C. abbayesii J.-F.Leroy | 10 | A.601 (K, P, TAN) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. ankaranensis A.P.Davis & Rakotonas. [1] | 4 | A.525 (K, TEF, TAN) | Madagascar | MAD-N | Deciduous–evergreen forest | KCRS |
| C. ankaranensis A.P.Davis & Rakotonas. [2] | 5 | A.808 (TEF) | Madagascar | MAD-N | Deciduous–evergreen forest | KCRS |
| C. anthonyi Stoff. & F.Anthony [1] | 12 | OD54, 55, 61-65, 68-72 (K) | Cameroon [LG/C] | W/WCA | Evergreen forest | BRC |
| C. anthonyi Stoff. & F.Anthony [2] | 7 | OE52-54, 56, 57, 21/12, 22/3 (K) | Congo [LG/C] | W/WCA | Evergreen forest | BRC |
| C. arenesiana J.-F.Leroy | 8 | A.403 (K, P) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. augagneuri Dubard | 10 | A.966 (TEF) | Madagascar | MAD-N | Deciduous–evergreen forest | KCRS |
| C. bernardiniana J.-F.Leroy | 9 | coll. anon. (K) | Mauritius | MAS | Evergreen forest (including drier open-canopy, and dwarf canopy forest) | BRC |
| C. bertrandii A.Chev. | 11 | A.5 (K, P, TAN) | Madagascar | MAD-S | Deciduous–evergreen forest (transitional humid–dry forest) | KCRS |
| C. betamponensis Portères & J.-F.Leroy | 3 | A.573 (TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. boiviniana (Baill.)Drake | 4 | A.980 (K, P) | Madagascar | MAD-N | Evergreen forest (seasonally dry) | KCRS |
| C. bonnieri Dubard | 10 | A.535 (K, MO, P) | Madagascar | MAD-N | Evergreen forest | KCRS |
| C. brevipes Hiern | 6 | JA52-54, 56, 62, 66 (K) | Cameroon [LG/C] | W/WCA | Evergreen forest | BRC |
| C. brevipes Hiern | 10 | JB53, 56, 57, 62, 64-66, 68-70 (K) | Cameroon [LG/C] | W/WCA | Evergreen forest | BRC |
| C. canephora Pierre ex A.Froehner [1] | 6 | BA53, 55, 58, 59, 38/5, 38/6 (K) | Ivory Coast [UG] | W/WCA | Evergreen forest | BRC |
| C. canephora Pierre ex A.Froehner [2] | 14 | BB53-57, 60, 62, 64, 66-70, 39/10 (K) | CAR. [LG/C] | W/WCA | Evergreen forest | BRC |
| C. canephora Pierre ex A.Froehner [3] | 7 | BC51, 53,57, 58, 60, 62, 40/5 (K) | CAR [LG/C] | W/WCA | Evergreen forest | BRC |
| C. canephora Pierre ex A.Froehner [4] | 14 | BD54-57, 59, 60, 62-69 (K) | Cameroon [LG/C] | W/WCA | Evergreen forest | BRC |
| C. canephora Pierre ex A.Froehner [r1] | 14 | BBL42/16, 42/17, 42/21, 42/22, 43/7, 43/8, 43/12, 43/13, 43/17, 43/18, 43/22, 43/23, 44/10, 44/11 (K) | Not known | W/WCA | Evergreen forest | BB |
| C. canephora Pierre ex A.Froehner [r2] | 8 | CAN-DAF1-4, 7, 8, 11, 12 (K) | Not known | W/WCA | Evergreen forest | DAF |
| C. congensis A.Froehner [1] | 8 | CA51, 52, 54, 56, 58, 59, 61, 69 (K) | CAR [LG/C] | W/WCA | Evergreen forest (riverine species) | BRC |
| C. congensis A.Froehner [2] | 7 | CB51, 52, 56, 58, 61, 65, 66 (K) | Cameroon [LG/C] | W/WCA | Evergreen forest (riverine species) | BRC |
| C. congensis A.Froehner [3] | 10 | CC52-54, 56, 61, 65, 67, 68, 70, 73 (K) | Congo [LG/C] | W/WCA | Evergreen forest (riverine species) | BRC |
| C. costatifructa Bridson | 8 | OH54, 59-62, 64, 08128 (K) | Tanzania | EA | Deciduous–evergreen forest | BRC |
| C. coursiana J.-F.Leroy | 10 | A.570 (K, TAN, TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. dubardii Jum. | 10 | A.969 (K, P, TAN) | Madagascar | MAD-N | Evergreen-deciduous forest | KCRS |
| C. eugenioides S.Moore | 11 | DA54, 56, 58-60, 68, 71, 74, 75, 77, 78 (D) | Kenya | EA | Evergreen forest | BRC |
| C. farafanganensis J.-F.Leroy | 8 | A.208 (P, TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. heimii J.-F.Leroy | 10 | A.516 (K, TEF) | Madagascar | MAD-N | Evergreen–deciduous forest | KCRS |
| C. heterocalyx Stoff. | 2 | JC 65 (K) | DRC [LG/C] | W/WCA | Evergreen forest | BRC |
| C. homollei J.-F.Leroy | 3 | A.945 (TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. humblotiana Baill. | 6 | A.230 (K, MO, TAN) | Comoros | MAS | Evergreen forest | KCRS |
| C. humilis A.Chev. | 11 | G52, 56-59, 63, 67-69, 72, 46/26 (K) | Ivory Coast [UG] | W/WCA | Evergreen forest | BRC |
| C. kianjavatensis J.-F.Leroy [1] | 10 | A.213 (K, P, TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. kianjavatensis J.-F.Leroy [2] | 10 | A.602 (K, MO) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. lancifolia A.Chev. var. auriculata J.-F.Leroy | 10 | A.320 (K, P) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. leroyi A.P.Davis [1] | 10 | A.315 (K, MO, P, TAN) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. leroyi A.P.Davis [2] | 5 | A.310 (K, P, MO, TAN) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. leroyi A.P.Davis cf. [cf. 1] | 5 | A.227 (K, P) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. leroyi A.P.Davis cf. (C. costei ined.) [cf. 2] | 9 | A.956 (K, P, TAN, TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. leroyi A.P.Davis cf. (C. daphnoides ined.) [cf. 3] | 1 | A.317 (K, P, TAN) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. liberica Bull. ex Hiern var. liberica [1] | 11 | EA51, 52, 61-64, 66, 67, 69, 70, 44/23 (K) | Ivory Coast [UG] | W/WCA | Evergreen forest | BRC |
| C. liberica Bull. ex Hiern var. liberica [r] | 4 | LIB-1, 2, Tr1, Tr2 (K) | Ivory Coast [UG] | W/WCA | Evergreen forest | BRC |
| C. liberica Bull. ex Hiern var. dewevrei (De Wild. & T. Durand) Lebrun [1] | 10 | EB51-53, 56, 58, 60, 63, 64, 68, 69 (K) | CAR [LG/C] | W/WCA | Evergreen forest | BRC |
| C. liberica Bull. ex Hiern var. dewevrei (De Wild. & T. Durand) Lebrun [r] | 12 | DAF1-1, 1-2, 2-1, 2-2, 3-1, 3-2, 4-1, 4-2, 5-1, 5-2, 6-1, 6-2 (K) | CAR [LG/C] | W/WCA | Evergreen forest | BRC |
| C. macrocarpa A.Rich. | 12 | coll. anon. (K) | Mauritius | MAS | Evergreen forest (including drier open-canopy, and dwarf canopy forest) | BRC |
| C. mangoroensis Portères [1] | 7 | A.401 (K) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. mangoroensis Portères [2] | 3 | A.402 (P) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. mauritiana Lam. | 10 | coll. anon. (K) | Reunion | MAS | Evergreen forest (including dwarf and ‘high-altitude’ cloud forest) | BRC |
| C. mayombensis A.Chev. | 4 | OC51, 55, 56, 37/1 (K) | Congo [LG/C] | W/WCA | Evergreen forest | BRC |
| C. mcphersonii A.P.Davis & Rakotonas. | 10 | A.977 (K, P, MO, TAN) | Madagascar | MAD-N | Evergreen (seasonally dry) or evergreen–deciduous forest | KCRS |
| C. millotii J.-F.Leroy [1] | 10 | A.721 (TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. millotii J.-F.Leroy [2] | 10 | A.222 (TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. millotiiJ.-F.Leroy (C. dolichophylla J.-F.Leroy) [3] | 10 | A.206 (P) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. millotii J.-F.Leroy (C. ambodirianensis Portères) [4] | 7 | A.572 (K, TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. mogenetti Dubard cf. | 4 | A.975 (TEF) | Madagascar | MAD-N | Evergreen (seasonally dry) or evergreen–deciduous forest | KCRS |
| C. montis-sacri A.P.Davis | 10 | A.321 (K, TAN) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. myrtifolia (A. Rich. ex DC.) J.-F.Leroy | 8 | coll. anon. (K) | Mauritius | MAS | Evergreen forest (sub-humid, including drier low-canopy evergreen forest) | BRC |
| C. perrieri Jum. & H.Perrier [1] | 10 | A.12 (TEF) | Madagascar | MAD-S | Gallery forest (mostly evergreen) | KCRS |
| C. perrieri Jum. & H.Perrier [2] | 10 | A305 (TEF) | Madagascar | MAD-S | Gallery forest (mostly evergreen) | KCRS |
| C. perrieri Jum. & H.Perrier [3] | 5 | A.732 (TEF) | Madagascar | MAD-S | Gallery forest (mostly evergreen) | KCRS |
| C. perrieri Jum. & H.Perrier [4] | 10 | A.730 (TEF) | Madagascar | MAD-S | Gallery forest (mostly evergreen) | KCRS |
| C. pocsii Bridson | 10 | PB52, 57-59, 61, 65, 68, 78, 08163, 08170 (K) | Tanzania | EA | Evergreen forest (seasonally dry) | BRC |
| C. pseudozanguebariae Bridson | 14 | H52-55, 58-61, 63, 65, 66, 68-70 (K) | Kenya | EA | Evergreen forest (seasonally dry) or evergreen–deciduous forest | BRC |
| C. racemosa Lour. [1] | 6 | IA51, 52, 55, 56, 61, 62 (K) | Mozambique | EA | Deciduous–evergreen or evergreen forest (seasonally dry) | BRA |
| C. racemosa Lour. [2] | 10 | IB52, 54, 55, 57-62, 11/6 (K) | Mozambique | EA | Deciduous–evergreen or evergreen forest (seasonally dry) | BRC |
| C. ratsimamangae A.P.Davis & Rakotonas. | 7 | A.528 (P) | Madagascar | MAD-N | Deciduous–evergreen forest | KCRS |
| C. resinosa(Hook.f.)Radlk. [1] | 10 | A.71 (TEF) | Madagascar | MAD-S | Littoral forest (evergreen) | KCRS |
| C. resinosa(Hook.f.)Radlk. [2] | 10 | A.8 (P, TEF) | Madagascar | MAD-S | Littoral forest (evergreen) | KCRS |
| C. richardii J.-F.Leroy | 9 | A.575 (TEF) | Madagascar | MAD-S | Littoral forest (evergreen) | KCRS |
| C. sahafaryensis J.-F. Leroy | 10 | A.978 (P) | Madagascar | MAD-N | Littoral forest (evergreen–deciduous) | KCRS |
| C. sakarahae J.-F. Leroy | 10 | A.304 (P, TEF) | Madagascar | MAD-S | Evergreen–deciduous forest | KCRS |
| C. salvatrix Swynn. & Philipson [1] | 3 | LA51, 56, 60 (K) | Tanzania | EA | Evergreen forest | BRA |
| C. salvatrix Swynn. & Philipson [2] | 11 | LB51-53, 57, 61-63, 66-69 (K) | Tanzania | EA | Evergreen forest | BRC |
| C. sessiliflora Bridson | 10 | PA55-58, 60, 63-65, 67, 70 (K) | Kenya | EA | Evergreen forest (seasonally dry) | BRC |
| C. sp. ‘Congo’ | 8 | OB56, 58, 60, 61, 65, 66, 36/3, 36/9 (K) | Congo [LG/C] | W/WCA | Evergreen forest | BRC |
| C. sp. ‘Koto’ | 6 | EC51-53, 57, 66, 67 (K) | Cameroon [LG/C] | W/WCA | Evergreen forest | BRC |
| C. sp. ‘Ngongo 2’ | 4 | OF52, 60, 63, 64 (K) | Congo [LG/C] | W/WCA | Evergreen forest | BRC |
| C. sp. ‘Ngongo 3’ | 1 | OG65 (K) | Congo [LG/C] | W/WCA | Evergreen forest | BRC |
| C. sp. ‘Nkoumbala’ | 8 | OI52, 55, 60, 65-68, 71 (K) | Cameroon [LG/C] | W/WCA | Evergreen forest | BRC |
| C. stenophylla G.Don. [1] | 8 | FA51, 54, 56, 59, 62, 63, 66, 68 (K) | Ivory Coast [UG] | W/WCA | Evergreen forest | BRC |
| C. stenophylla G.Don. [2] | 11 | FB52-55, 57-61, 64, 33/12, 33/13 (K) | Ivory Coast [UG] | W/WCA | Evergreen forest | BRC |
| C. tetragona Jum. & H.Perrier | 8 | A.252 (K, MO, TAN) | Madagascar | MAD-S | Evergreen (seasonally dry; Sambirano*) | KCRS |
| C. tsirananae J.-F.Leroy | 11 | A.515 (TEF) | Madagascar | MAD-N | Deciduous–evergreen forest | KCRS |
| C. vatovavyensis J.-F.Leroy | 10 | A.830 (K, TAN) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. vianneyi J.-F.Leroy [1] | 10 | A.20 (K, P) | Madagascar | MAD-S | Evergreen forest | KCRS |
| C. vianneyi J.-F.Leroy [1] | 10 | A.946 (TEF) | Madagascar | MAD-S | Evergreen forest | KCRS |
Abbreviations – Countries: Central African Republic (CAR); Democratic Republic of Congo (DRC). Centres of endemism for West Africa (after White 1979, 1983): Lower Guinea-Congolian (LG/C); Upper Guinea (UG). Regions: West and West-Central Africa (W/WCA), East Africa (EA), Madagascar (MAD) and Mascarenes (MAS); subregions: Madagascar north (MAD-N), and Madagascar south (MAD-S). Origin of germplasm material: Bois-Blanc Reunion (BB); unknown germplasm collection from Brazil (BRA); Centre de Ressources Biologiques Coffea, Saint Pierre, Reunion (BRC); Direction de l'Agriculture et de la Forêt (DAF) collection, Reunion; Kianjavato Coffee Research Station, Madagascar (KCRS). Herbarium abbreviations after Holmgren et al. (1990).
* Sambirano vegetation represents a specific humid forest type in western Madagascar, which shares species occurrences and phylogeographical associations with the humid forest of eastern Madagascar, but mixed at lower altitudes with those from the west.
LITERATURE CITED
- Aggarwal KR, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, Singh L. Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theoretical and Applied Genetics. 2007;114:359–372. doi: 10.1007/s00122-006-0440-x. [DOI] [PubMed] [Google Scholar]
- Anthony F, Combes M-C, Astorga C, Bertrand B, Graziosi G, Lashermes P. The origin of cultivated Coffea arabica L. varieties revealed by AFLP and SSR markers. Theoretical and Applied Genetics. 2002a;104:894–900. doi: 10.1007/s00122-001-0798-8. [DOI] [PubMed] [Google Scholar]
- Anthony F, Quiros Q, Topart P, Bertrand B, Lashermes P. Detection by simple sequence repeat markers of introgression from Coffea canephora in Coffea arabica cultivars. Plant Breeding. 2002b;121:542–544. [Google Scholar]
- Anthony F, Diniz LEC, Combes M-C, Lashermes P. Adaptive radiation in Coffea subgenus Coffea L. (Rubiaceae) in Africa and Madagascar. Plant Systematics and Evolution. 2010;285:51–64. [Google Scholar]
- Bhat P, Krishnakuma V, Hendre PS, Rajendrakumar P, Varshney RK, Aggarwalrk RK. Identification and characterization of expressed sequence tags-derived simple sequence repeats markers from robusta coffee variety ‘C×R’ (an interspecific hybrid of Coffea canephora×Coffea congensis) Molecular Ecology Notes. 2004;5:80–83. [Google Scholar]
- Berthaud J. ORSTOM, Série TDM 188. Paris: ORSTOM; 1986. Les ressources génétiques pour l'amélioration des caféiers Africains diploïdes. [Google Scholar]
- Born C, Alvarez N, McKey D, et al. Insights into the biogeographical history of the Lower Guinea Forest Domain: evidence for the role of refugia in the intraspecific differentiation of Aucoumea klaineana. Molecular Ecology. 2011;20:131–142. doi: 10.1111/j.1365-294X.2010.04919.x. [DOI] [PubMed] [Google Scholar]
- Bouharmont J. Somatic chromosomes of some Coffea species. Euphytica. 1963;12:254–257. [Google Scholar]
- Brookfield JFY. A simple new method for estimating null allele frequency from heterozygote deficiency. Moleculary Ecology. 1996;5:453–455. doi: 10.1111/j.1365-294x.1996.tb00336.x. [DOI] [PubMed] [Google Scholar]
- Burrell AM, No E-G, Pepper AE. Discovery of nuclear and plastid microsatellites, and other key genomic information, in the rare endemic plant (Caulanthus amplexicaulis var. barbarae) using minimal 454 pyrosequencing. Conservation Genetics Resources. 2011;3:753–755. [Google Scholar]
- Campa C, Mondolot L, Rakotondravao A, et al. A survey of mangiferin and hydroxycinnamic acid ester accumulation in coffee (Coffea L.) leaves: biological implications and uses. Annals of Botany. 2012;110:595–613. doi: 10.1093/aob/mcs119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Jin L. Determination of relatedness between individuals by DNA fingerprinting. Human Biology. 1993;65:875–895. [PubMed] [Google Scholar]
- Charrier A. La structure génétique des caféiers spontanés de la région malgache (Mascarocoffea). Leurs relations avec les caféiers d'origine africaine (Eucoffea) France: PhD Thesis, Université Paris-Sud Orsay; 1978. [Google Scholar]
- Charrier A, Berthaud J. Botanical classification of coffee. In: Clifford MN, Willson KC, editors. Coffee botany, biochemistry and production of beans and beverage. London: Croom Helm; 1985. pp. 13–47. [Google Scholar]
- Cheney RH. Coffee. A monograph of the economic species of the genus Coffea L. New York: New York University Press; 1925. [Google Scholar]
- Chikhi L. How accurate can genetic data be? Heredity. 2008;101:471–472. doi: 10.1038/hdy.2008.106. [DOI] [PubMed] [Google Scholar]
- Combes MC, Andrzejewski S, Anthony F, et al. Characterization of microsatellite loci in Coffea arabica and related coffee species. Molecular Ecology. 2000;9:1171–1193. doi: 10.1046/j.1365-294x.2000.00954-5.x. [DOI] [PubMed] [Google Scholar]
- Cornet A. Essai de cartographie bioclimatique à Madagascar. Vol. 55. Paris: ORSTOM (Office de la Recherche Scientifique et Technnique Outre-Mer), notice explicative; 1974. pp. 1–28. ISBN 2-7099-0339-3. [Google Scholar]
- Coulibaly I, Revol B, Noirot M, et al. AFLP and SSR polymorphism in a Coffea interspecific backcross progency [(C. heterocalyx) × C. canephora] Theoretical and Applied Genetics. 2003;107:1171–1193. doi: 10.1007/s00122-003-1355-4. [DOI] [PubMed] [Google Scholar]
- Cros I, Combes MC, Chabrillange N, Desangles AM, Hamon S, et al. Nuclear DNA content in the subgenus Coffea (Rubiaceae): inter and intra specific variation in African species. Canadian Journal of Botany. 1995;73:14–20. [Google Scholar]
- Cros J, Combes MC, Trouslot P, et al. Phylogenetic analysis of chloroplast DNA variation in Coffea L. Molecular Phylogenetics and Evolution. 1998;9:109–117. doi: 10.1006/mpev.1997.0453. [DOI] [PubMed] [Google Scholar]
- Cubry P, Musoli P, Legnate H, et al. Diversity in coffee assessed with SSR markers: structure of the genus Coffea and perspectives for breeding. Genome. 2008;51:50–63. doi: 10.1139/g07-096. [DOI] [PubMed] [Google Scholar]
- Davis AP. Six species of Psilanthus transferred to Coffea (Coffeeae, Rubiaceae) Phytotaxa. 2010;10:41–45. [Google Scholar]
- Davis AP. Psilanthus mannii, the type species of Psilanthus, transferred to Coffea. Nordic Journal of Botany. 2011;29:471–472. [Google Scholar]
- Davis AP, Rakotonasolo F. Three new species of Coffea L. (Rubiaceae) from NE Madagascar. Adansonia, Sér 3. 2001a;23:137–146. [Google Scholar]
- Davis AP, Rakotonasolo F. Two new species of Coffea L. (Rubiaceae) from northern Madagascar. Adansonia, Sér 3. 2001b;23:337–345. [Google Scholar]
- Davis AP, Rakotonasolo F. New species of Coffea L. (Rubiaceae) from Madagascar. Botanical Journal of the Linnean Society. 2004;142:111–118. [Google Scholar]
- Davis AP, Rakotonasolo F. A taxonomic revision of the baracoffea alliance: nine remarkable Coffea species from western Madagascar. Botanical Journal of the Linnean Society. 2008;158:355–390. [Google Scholar]
- Davis AP, Govaerts R, Bridson DM, Stoffelen P. An annotated taxonomic conspectus of the genus Coffea (Rubiaceae) Botanical Journal of the Linnean Society. 2006;142:465–512. [Google Scholar]
- Davis AP, Chester M, Maurin O, Fay M. Searching for the relatives of Coffea (Rubiaceae, Ixoroideae): the circumscription and phylogeny of Coffeeae based on plastid sequence data and morphology. American Journal of Botany. 2007;94:313–329. doi: 10.3732/ajb.94.3.313. [DOI] [PubMed] [Google Scholar]
- Davis AP, Rakotonasolo F, DeBlock P. Coffea toshii sp. nov. (Rubiaceae) from Madagascar. Nordic Journal of Botany. 2010;28:134–136. [Google Scholar]
- Davis AP, Tosh J, Ruch N, Fay M. Growing coffee: Psilanthus (Rubiaceae) subsumed on the basis of molecular and morphological data; implications for the size, morphology, distribution and evolutionary history of Coffea. Botanical Journal of the Linnean Society. 2011;167:357–377. [Google Scholar]
- Dussert S, Laffargue A, de Kochko A, Joët T. Effectiveness of the fatty acid and sterol composition of seeds for the chemotaxonomy of Coffea subgenus Coffea. Phytochemistry. 2008;69:2950–2960. doi: 10.1016/j.phytochem.2008.09.021. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Moore S, Robinson N, Byrne K, Ward W, Sheldon BC. Microsatellite evolution – a reciprocal study of repeat lengths at homologous loci in cattle and sheep. Molecular Biology and Evolution. 1997;14:854–860. doi: 10.1093/oxfordjournals.molbev.a025826. [DOI] [PubMed] [Google Scholar]
- Estoup A, Jarne P, Cornuet JM. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Molecular Ecology. 2002;11:1591–1604. doi: 10.1046/j.1365-294x.2002.01576.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. ARLEQUIN ver. 3·0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Seattle, USA: Distributed by the author. Department of Genome Sciences, University of Washington; 2005. PHYLIP (Phylogeny Inference Package) version 3·6. [Google Scholar]
- Gomez C, Dussert S, Hamon P, Hamon S, de Kochko A, Poncet V. Current genetic differentiation of Coffea canephora Pierre ex A. Froehn in the Guineo-Congolian African zone: cumulative impact of ancient climatic changes and recent human activities. BMC Evolutionary Biology. 2009;9:167. doi: 10.1186/1471-2148-9-167. http://dx.doi.org/10.1186/1471-2148-9-167 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez C, Batti A, Le Pierrès D, et al. Favourable habitats for Coffea inter-specific hybridization in central New Caledonia: combined genetic and spatial analyses. Journal of Applied Ecology. 2010;114:2731–2744. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Guajardo JCR, Schnabel A, Ennos R, Preuss S, Otero-Arnaiz A, Stone G. Landscape genetics of the key African acacia species Senegalia mellifera (Vahl) – the importance of the Kenyan Rift Valley. Molecular Ecology. 2010;19:5126–5139. doi: 10.1111/j.1365-294X.2010.04833.x. [DOI] [PubMed] [Google Scholar]
- Hamon P, Duroy PO, Dubreuil-Tranchant C, et al. Two novel Ty1-copia retrotransposons isolated from coffee trees can effectively reveal evolutionary relationships in the Coffea genus (Rubiaceae) Molecular Genetics and Genomics. 2011;285:447–460. doi: 10.1007/s00438-011-0617-0. [DOI] [PubMed] [Google Scholar]
- Hamon S, Anthony F, Le Pierrès D. La variabilité génétique des caféiers spontanés de la section Mozambicoffea Chev. A. I) Précisions sur deux espèces affines: Coffea pseudozanguebariae Bridson et C. sp. A Bridson. Bulletin du Museum National d'Histoire naturelle, Section B Adansonia. 1984;2:207–223. [Google Scholar]
- Hamon S, Dussert S, Deu M, et al. Effects of quantitative and qualitative principal component score strategies on the structure of coffee, rice, rubber tree and sorghum core collections. Genetics Selection Evolution. 1998;30(suppl. 1):237–258. [Google Scholar]
- Harr B, Weiss S, David JR, Brem G, Schlötterer C. A microsatellite-based multilocus phylogeny of the Drosophila melanogaster species complex. Current Biology. 1998;8:1183–1186. doi: 10.1016/s0960-9822(07)00490-3. [DOI] [PubMed] [Google Scholar]
- Hendre PS, Phanindranath R, Annapurna V, Lalremruata A, Aggarwal K. Development of new genomic microsatellite markers from robusta coffee (Coffea canephora Pierre ex A. Froehner) showing broad cross-species transferability and utility in genetic studies. BMC Plant Biology. 2008;8:51. doi: 10.1186/1471-2229-8-51. http://dx.doi.org/10.1186/1471-2229-8-51 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren PK, Holmgren NH, Barnett LC. Regnum Vegetabile. 8th edn. New York: New York Botanical Garden; 1990. Index herbariorum. Part 1: the herbaria of the world. [Google Scholar]
- Husson F, Josse J, Lê S. FactoMineR: an R package for multivariate analysis. Journal of Statistical Software. 2008;25:1–18. [Google Scholar]
- International Coffee Organization (ICO) World Coffee Trade. 2012 http://www.ico.org/trade_e.asp?section=About_Coffee. (accessed March 2012) [Google Scholar]
- Johnston JS, Pepper AE, Hall AE, Z, et al. Evolution of genome size in Brassicaceae. Annals of Botany. 2005;95:229–235. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kochko A, Akaffou S, Andrade AC, et al. Advances in Coffea genomics. Advances in Botanical Research. 2010;53:23–63. [Google Scholar]
- Krishnan S, Ranker TA, Davis AP, Rakotomalala J-J. The study of genetic diversity patterns of Coffea commersoniana, an endangered coffee species from Madagascar: a model for conservation of other littoral forest species. Tree Genetics and Genomes. 2013a in press. http://dx.doi.org/10.1007/s11295-012-0545-0 . [Google Scholar]
- Krishnan S, Ranker TA, Davis AP, Rakotomalala J-J. An assessment of the genetic integrity of ex situ germplasm collections of three endangered species of Coffea from Madagascar: implications for the management of field germplasm collections. Genetic Resources and Crop Evolution. 2013b in press. http://dx.doi.org/10.1007/s10722-012-9898-3 . [Google Scholar]
- Lashermes P, Combes MC, Trouslot P, Charrier A. Phylogenetic relationships of coffee-tree species (Coffea L.) as inferred from ITS sequences of nuclear ribosomal DNA. Theoretical and Applied Genetics. 1997;94:947–955. [Google Scholar]
- Lefebvre-Pautigny F, Wu F, Philippot M, et al. High resolution synteny maps allowing direct comparisons between the coffee and tomato genomes. Tree Genetics and Genomes. 2010;6:565–577. [Google Scholar]
- Legrand D, Vautrin D, Lachaise D, Cariou ML. Microsatellite variation suggests a recent fine-scale population structure of Drosophila sechellia, a species endemic of the Seychelles archipelago. Genetica. 2011;139:909–919. doi: 10.1007/s10709-011-9595-8. [DOI] [PubMed] [Google Scholar]
- Leroy J-F. Paysages et forêts autour de Diègo-Suarez. Science et Nature. 1968;88:2–8. [Google Scholar]
- Leroy J-F. Réflexions sur l'évolution naturelle et l'évolution artificielle des ressources génétiques végétales; le cas des Coffea. Bulletin du Jardin Botanique National de Belgique. 1971;41:53–67. [Google Scholar]
- Leroy J-F. L'origine Kenyane du genre Coffea L. et la radiation des espèces à Madagascar. Association Scientifique Internationale du Café (ASIC) 10th Colloque. 1982:413–420. [Google Scholar]
- Leroy J-F. Coffea L. In: Bosser J, Cadet T, Guého J, Marais W, editors. Flore des Mascareignes La Réunion, Maurice, Rodrigues. 107. Caprifoliacées à 108bis. Valérianacées. Surrey: Royal Botanic Gardens, Kew; 1989. pp. 93–99. [Google Scholar]
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Louarn J. Introduction à l'étude génétique des Mascarocoffea: nouvelles déterminations de leurs nombres chromosomiques. Café Cacao Thé. 1972;26:312–316. [Google Scholar]
- Maurin O, Davis AP, Chester M, Mvungi EF, Jaufeerally-Fakim Y, Fay MF. Towards a phylogeny for Coffea (Rubiaceae): identifying well-supported lineages based on nuclear and plastid DNA sequences. Annals of Botany. 2007;100:1565–1583. doi: 10.1093/aob/mcm257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncada P, McCouch S. Simple sequence repeat diversity in diploid and tetraploid Coffea species. Genome. 2004;47:501–509. doi: 10.1139/g03-129. [DOI] [PubMed] [Google Scholar]
- Moat J, Smith P. Atlas of the vegetation of Madagascar. Kew, London: Royal Botanic Gardens; 2007. [Google Scholar]
- Noirot M, Poncet V, Barre P, Hamon P, Hamon S, de Kochko A. Genome size variations in diploid African Coffea species. Annals of Botany. 2003;92:709–714. doi: 10.1093/aob/mcg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MD, Davis AP, Yoder AD. Sequence data from new plastid and nuclear COSII regions resolves early diverging lineages in Coffea (Rubiaceae) Systematic Botany. 2012;37:995–1005. [Google Scholar]
- Ochieng JW, Steane DA, Ladiges PY, Baverstock PR, Henry RJ, Shepherd M. Microsatellites retain phylogenetic signals across genera in eucalypts (Myrtaceae) Genetics and Molecular Biology. 2007;30:1125–1134. [Google Scholar]
- Orsini L, Huttunen S, Schlötterer C. A multilocus microsatellite phylogeny of the Drosophila virilis group. Heredity. 2004;93:161–165. doi: 10.1038/sj.hdy.6800487. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. Plos Genetics. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. http://dx.doi.org/10.1371/journal.pgen.0020190 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petren KB, Grant R, Grant PR. A phylogeny of Darwin's finches based on microsatellite DNA length variation. Proceedings of the Royal Society of London, B. 1999;266:321–329. [Google Scholar]
- Plechakova O, Tranchant-Dubreuil C, Benedet F, et al. MoccaDB – an integrative database for functional, comparative and diversity studies in the Rubiaceae family. BMC Plant Biology. 2009;9:123. doi: 10.1186/1471-2229-9-123. http://dx.doi.org/10.1186/1471-2229-9-123 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet V, Hamon P, Minier J, Carasco C, Hamon S, Noirot M. SSR cross-amplification and variation within coffee trees (Coffea spp.) Genome. 2004;47:1071–1081. doi: 10.1139/g04-064. [DOI] [PubMed] [Google Scholar]
- Poncet V, Dufour M, Hamon P, Hamon S, de Kochko A, Leroy T. Development of genomic microsatellite markers in Coffea canephora and their transferability to other coffee species. Genome. 2007;50:1156–1161. doi: 10.1139/G07-073. [DOI] [PubMed] [Google Scholar]
- Prakash NS, Combes M-C, Dussert S, Naveen S, Lashermes P. Analysis of genetic diversity in Indian robusta genepool (Coffea canephora) in comparison with a representative core collection using SSRs and AFLPs. Genetic Resources and Crop Evolution. 2005;52:333–343. [Google Scholar]
- Price HJ, Dillon SL, Hodnett G, Rooney W, Ross L, Johnston JS. Genome evolution in the genus Sorghum (Poaceae) Annals of Botany. 2005;95:219–227. doi: 10.1093/aob/mci015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens P, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1·2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Razafinarivo NJ, Rakotomalala J-J, Brown SC, et al. Geographical gradients in the genome size variation of wild coffee trees (Coffea) native to Africa and Indian Ocean islands. Tree Genetics and Genomes. 2012;8:1345–1358. [Google Scholar]
- Richard M, Thorpe RS. Can microsatellites be used to infer phylogenies? Evidence from population affinities of the western Canary Island lizard (Gallotia galloti) Molecular Phylogenetics and Evolution. 2001;20:351–360. doi: 10.1006/mpev.2001.0981. [DOI] [PubMed] [Google Scholar]
- Ritz LR, Glowatzki-Mullis ML, MacHugh DE, Gaillard C. Phylogenetic analysis of the tribe Bovini using microsatellites. Animal Genetics. 2000;31:178–185. doi: 10.1046/j.1365-2052.2000.00621.x. [DOI] [PubMed] [Google Scholar]
- Rogers JS. Measures of genetic similarity and genetic distances. Studies in Genetics, University of Texas Publication. 1972;7213:145–153. [Google Scholar]
- Rovelli P, Mettulio R, Anthony F, Anzueto F, Lashermes P, Graziosi G. Microsatellites in Coffea arabica L. In: Sera T, Soccol CR, Pandey A, Roussos S, editors. Coffee biotechnology and quality. Dordrecht: Kluwer Academic Publishers. 2000. pp. 123–133. [Google Scholar]
- Rousset F. GENEPOP'007: a complete reimplementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Ruas PM, Ruas CF, Rampim L, Carvalho VP, Ruas EA, Sera T. Genetic relationship in Coffea species and parentage determination of interspecific hybrids using ISSR (inter-simple sequence repeat) markers. Genetics and Molecular Biology. 2003;26:319–327. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shepherd M, Kasem S, Lee D, Henry R. Construction of microsatellite genetic linkage maps for Corymbia. Silvae Genetica. 2006;55:228–238. [Google Scholar]
- Soltis DE, Soltis PS, Bennett MD, Leitch IJ. Evolution of genome size in the angiosperms. American Journal of Botany. 2003;90:1596–1603. doi: 10.3732/ajb.90.11.1596. [DOI] [PubMed] [Google Scholar]
- Stoffelen P. PhD Thesis, Katholieke Universiteit, Leuven: Belgium; 1998. Coffea and Psilanthus in Tropical Africa: a systematic and palynological study, including a revision of the West and Central African Species. [Google Scholar]
- Tesfaye K, Borsch T, Govers K, Bekele E. Characterization of Coffea chloroplast microsatellites and evidence for the recent divergence of C. arabica and C. eugenioides chloroplast genomes. Genome. 2007;50:1112–1129. doi: 10.1139/G07-088. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Advances in Agronomy. 2002;78:139–186. [Google Scholar]
- White F. The Guineo-Congolian Region and its relationships to other phytochoria. Bulletin du Jardin Botanique National de Belgique. 1979;49:11–55. [Google Scholar]
- White F. The vegetation of Africa. A descriptive memoir to accompany the Unesco/AETFAT/UNSO vegetation map of Africa. Paris: Unesco; 1983. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.