Abstract
Gene transcription is regulated in response to environmental changes as well as developmental cues. In mammalian cells subjected to stress conditions such as heat shock, transcription of most protein-coding genes decreases, while the transcription of heat shock protein genes increases. Repression involves direct binding to RNA polymerase II (Pol II) of certain non-coding RNAs (ncRNAs) that are upregulated upon heat shock. Another class of ncRNAs is also upregulated and binds to Pol II, but does not inhibit transcription. Incorporation of repressive ncRNAs into pre-initiation complexes prevents transcription initiation, while non-repressive ncRNAs are displaced from Pol II by TFIIF. Here, we present cryo-EM reconstructions of human Pol II in complex with six different ncRNAs from mouse and human. Our structures show that both repressive and non-repressive ncRNAs bind to a conserved binding site within the cleft of Pol II. The site, also shared with a previously characterized yeast aptamer, is close to the active center and thus in an ideal position to regulate transcription. Importantly, additional RNA elements extend flexibly beyond the docking site. We propose that the differences concerning the repressive activity of the ncRNA analyzed must be due to the distinct character of these more unstructured, flexible segments of the RNA that emanate from the cleft.
Keywords: transcription regulation, RNA polymerase II, electron microscopy, non-coding RNA, structure
INTRODUCTION
Initiation of transcription is one of the major control points for the regulation of gene expression. The assembly of the pre-initiation complex (PIC), consisting of the core enzyme RNA polymerase II (Pol II) and general transcription factors, is precisely regulated through the interplay of numerous protein factors1 as well as non-coding RNAs (ncRNAs)2. The general transcription factors aid Pol II in selecting the transcription start site and in transitioning from initiation to elongation states. Typically, transcription factors involved in gene-specific regulation have a modular structure that includes a DNA binding domain and an activation domain that binds to the general transcription machinery, typically TFIID or mediator complex1.
Non-coding RNAs that regulate transcription by binding directly to Pol II have been identified in the mouse and human systems. In both organisms, the ncRNAs are transcribed from short interspersed elements (SINEs) by RNA polymerase III3 in response to cellular stresses. The mouse genome contains two major SINEs, named B2 and B1, whose transcription levels rapidly increase upon heat shock. Both B2 and B1 RNA bind to Pol II with high affinity and specificity, and compete with each other for binding to Pol II. However, only B2 RNA represses transcription. B2 RNA contains distinct Pol II binding and transcriptional repression domains, a molecular organization that resembles that of protein regulators of Pol II transcription8. Upon docking of B2 RNA onto Pol II, the ncRNA/Pol II complex assembles into stable PICs and renders them inactive by preventing Pol II from establishing contacts with the promoter DNA, thereby inhibiting the formation of closed complexes9. Notably, B2 RNA has to bind before assembly of the complete PIC in order to inhibit transcription, as it is not able to invade closed complexes once Pol II has engaged the DNA9.
The cellular levels of the second major mouse SINE ncRNA, B1, are also increased upon heat shock. Although the affinities of B1 and B2 RNA for Pol II are similar, B1 RNA cannot prevent inhibition by B2 RNA in vitro. This can be explained by the fact that the general transcription factor TFIIF decreases the stability of B1 RNA/Pol II complexes and thereby facilitates their dissociation10. The biological function of B1 RNA still remains unclear6.
The human genome contains a single predominant SINE, termed Alu. The Alu RNA transcript is unrelated in primary sequence to B2 RNA and does not share any similarities in overall secondary structure. However, Alu RNA functions in the same way as B2 RNA: it is transcribed by RNA polymerase III upon heat shock, associates with Pol II, and acts as a transcriptional repressor11. Alu RNA consists of two repeats, the folding pattern of which is similar to that of B1 RNA12. While both repeats can bind to Pol II independently, only one of them, termed Alu-RA, is able to repress transcription11. The other repeat, termed scAlu, has been identified as a shortened transcript of Alu RNA in human cells and is thought to be similar in its fold and function to B1 RNA. Interestingly, a repression domain from human Alu RNA can be fused to the Pol II binding domain of mouse B1 RNA to yield a functional repressor, confirming the modular organization of the ncRNAs11. Like protein transcription factors, the ncRNAs contain distinct domains for binding tightly to Pol II, and repression domains that are structurally and functionally independent from the interaction domain.
Our current structural understanding of ncRNA-mediated transcriptional repression derives from a model based on the crystal structure of a complex of yeast Pol II (yPol II) with the central core of the synthetic RNA aptamer FC14, which was selected based on its ability to bind to Pol II. The crystal structure shows the RNA bound in the central cleft of Pol II, indicating the location of a potential binding site for ncRNAs. To gain further insights into the molecular mechanisms underlying transcriptional regulation by naturally occurring ncRNAs, we have used cryo-electron microscopy (cryo-EM) and single particle three-dimensional image reconstruction to determine structures of human Pol II in complex with repressive and non-repressive mouse and human ncRNAs. We found that naturally occurring ncRNAs bind to a similar binding site in hPol II as the synthetic FC RNA in yPol II and that this site is shared between repressive and non-repressive ncRNAs.
RESULTS
Cryo-EM reconstructions of hPol II in complex with ncRNAs
To identify the binding site for ncRNAs on Pol II and to study the differences between repressive and non-repressive ncRNAs, we analyzed complexes of hPol II and different repressive and non-repressive ncRNAs from mouse (B2, B2 [81-131] (a minimally repressive fragment of B2 RNA), and B1 RNA) and human (Alu, Alu-RA and scAlu RNA) by cryo-EM and three-dimensional image reconstruction. Human Pol II was purified from HeLa cell nuclei, incubated with a three-fold molar excess of each in vitro-transcribed ncRNA, and visualized by cryo-EM. We used a low-pass filtered model of apo hPol II, previously obtained using cryo-negative stain15, as the initial reference for Euler angle assignment15. Reconstructions were generated and refined from each data set in iterative steps of projection matching using EMAN216. The final refinement and CTF correction were done using FREALIGN. Our data sets contained between 15,814 (for the B2/hPol II sample) and 31,423 particles (for the B2 [81-131]/hPolII sample) and they all refined to a resolution of about 25 Å, according to the 0.5 cut-off in the Fourier shell correlation curve.
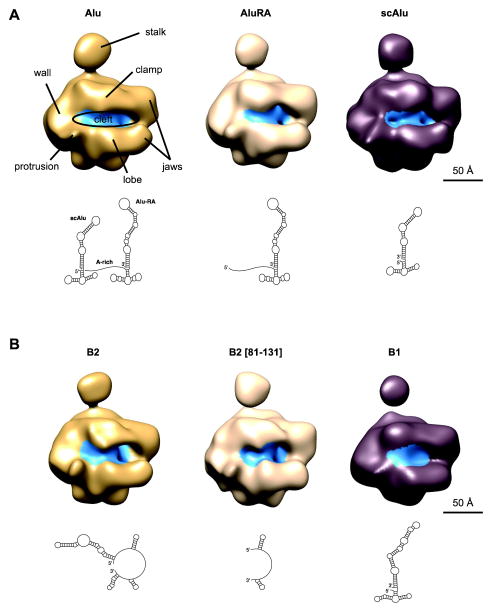
Overall, our ncRNA/Pol II structures (Figure 1) are similar to each other and to the model of apo Pol II determined by cryo-negative staining EM15. Comparison of all six cryo-EM structures shows that the clamp is in a slightly different conformation in each model; in addition, the conformation and density of the stalk also varies between the different models. Importantly, in all the hPol II/ncRNA reconstructions we observe additional density within the cleft of Pol II when compared to the previous cryo-negative stain apo hPol II structures15 (Fig 1, blue label). This location in the enzyme is near the active site and thus in the region that accommodates the DNA in an actively transcribing complex.
Figure 1. Cryo-EM maps of hPol II in complex with non-coding RNAs from mouse and human.
(A) Cryo-EM maps of Pol II in complex with the human non-coding RNAs Alu, Alu-RA and scAlu RNA. The secondary structures of ncRNAs as derived by biochemical methods are depicted below each structure. Pol II domains are labeled in the cryo-EM map of hPol II in complex with full-length Alu RNA. Regions of the volume occupied by ncRNAs are colored in blue.
(B) Cryo-EM maps of hPol II in complex with the mouse non-coding RNAs B2, B2 [81-131], and B1 RNA. The secondary structures of ncRNAs as derived by biochemical methods are depicted below each structure.
Comparison with a cryo-EM reconstruction of apo hPol II
To assert the presence of the ncRNAs at the cleft of hPol II we decided to make a more detailed comparison between the ncRNA-bound structures and apo hPol II. To that aim we pursued the structural characterization of nucleotide-free apo hPol II (see Experimental procedures) by cryo-EM and three-dimensional image reconstruction. Our initial 2D reference-free analysis suggested that the clamp of hPol II was adopting a broad range of conformations (Supplementary Figure 2B). In agreement with this indication, the reconstruction showed limited density for the clamp region, indicative of multiple conformations for this region of the complex, as previously also described for the cryo-negative stain study of apo hPol II. In an attempt to sort out different conformational states in three dimensions we implemented an automated multi-model approach19. The data set, containing a total of 36,863 particles, was sorted into three distinct conformations to account for some of the conformational flexibility of the sample (Supplementary figure 2D). One of the models refined more poorly and probably incorporated misshapen complexes, false particles or a mixture of conformations still unsorted. The other two reconstructions were an improvement with respect to the initial, unsorted model and recover a density for the clamp that approximately accommodates the mass of this segment.
Given the variability in clamp position for the apo hPol II structures, it is not surprising that the relatively small differences we observed among hPol II/ncRNA complexes also concern the clamp and stalk, and confirm, one more time, that these structural elements are highly mobile. We tried to sort our data sets of the ncRNA-bound samples using the same multi-model approach, but did not obtain clearly distinct conformations. This result indicates that the binding of ncRNAs restricts the movement of the clamp. However, flexibility is still limiting the resolution of our structures. In the case of the ncRNA-bound reconstructions, conformational heterogeneity, contributing to resolution-limiting errors in alignment, may be majorly attributed by the ncRNA itself.
For comparison of apo Pol II with the ncRNA-bound structures, we used conformation 3 of the former, for which the position of the clamp is better defined. Once again, the comparison shows that binding of ncRNAs has a stabilizing effect on the clamp of Pol II, as evidenced by the larger volume this domain occupies in our ncRNA-bound structures. This result indicates that these domains are in a more stable conformation and therefore not averaged out due to their flexibility. The stabilization of the clamp and stalk domains is likely due to the direct interaction of the ncRNAs with the clamp, restricting the movement of the latter and, as a result, of the stalk as well, which is directly engaged with the clamp region.
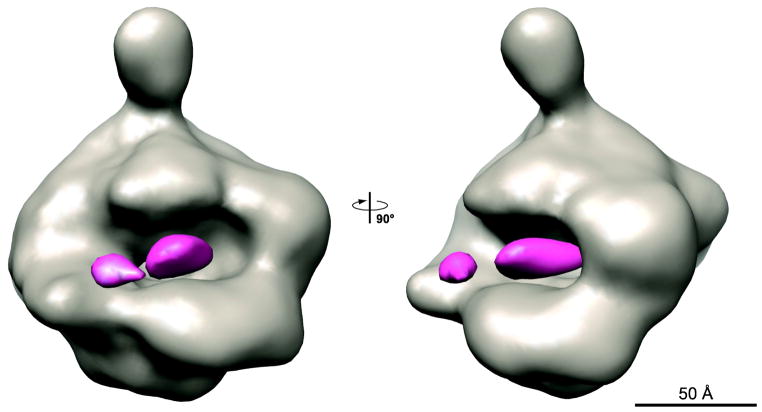
With these new cryo-EM structures of the apo hPol II, we were in a position to quantitatively confirm if the ncRNA-bound structures contain additional density in the cleft of hPol II. To that aim we calculated difference maps between each of the ncRNA-bound Pol II structures and that of apo Pol II in conformation 3. Similar results were obtained for all of them. Figure 2 shows the difference map obtained using the Alu RNA-bound Pol II. The extra density defines the cleft as the main Pol II-binding domain for the ncRNAs. The additional density, adjacent to the so-called wall region of Pol II, has a similar position in all the ncRNA-bound structures at the present resolution (data not shown). We attribute this density to ncRNAs bound in the cleft of hPol II. Importantly, however, this density cannot account for the length of any of the ncRNA constructs and indicates that large regions in the ncRNAs are flexible and in variable positions with respect to the hPol II structure. In spite of the stabilizing effect of the ncRNAs on the conformation of the hPol II complex, the conformational heterogeneity of the ncRNA itself must be playing an important part in limiting the resolution of our cryo-EM reconstructions.
Figure 2. Difference map between the hPol II/Alu RNA complex and apo hPol II in two orthogonal views.
The cryo-EM map of apo hPol II (model 3, see supplementary Figure 2) is shown in grey, the difference density is colored in pink.
Comparison of the cryo-EM structures of ncRNA-bound hPol II with the synthetic FC RNA aptamer-yPol II crystal structure
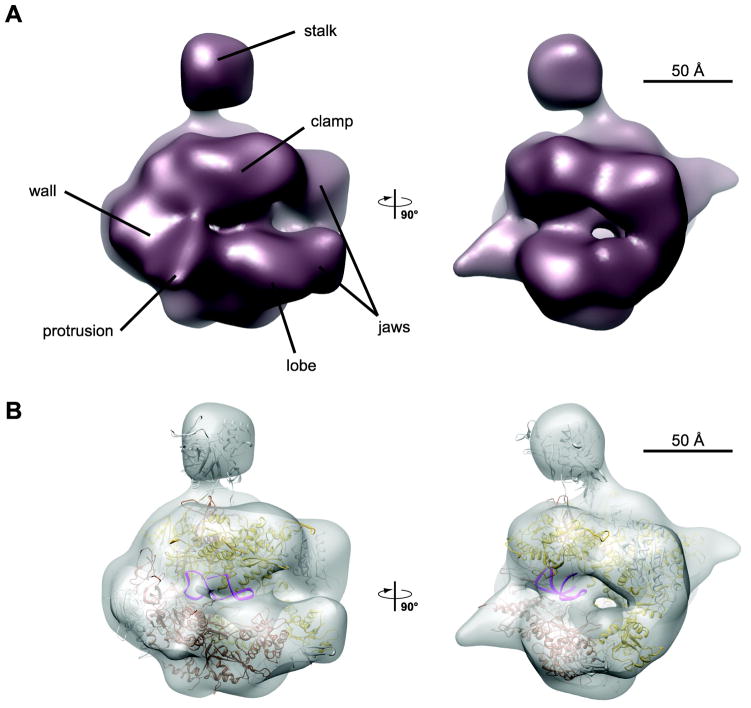
To compare our cryo-EM maps with the crystal structure of yPol II bound to the core of the synthetic RNA aptamer FC14 (PDB code 2B63), we docked the atomic model into our maps of hPol II in complex with the different ncRNAs. The overall fit was similar across all maps; as an example, Figure 3 shows the crystal structure docked into the map of hPol II in complex with scAlu RNA, the human non-repressive ncRNA. The docking reveals a good fit between the two structures, and shows that the additional density we observe in the cryo-EM maps corresponds to the position of the FC RNA aptamer core in the yeast crystal structure. This docking illustrates that the extra density could be accounted for by an RNA molecule of similar length to that used in the crystallographic studies, which comprised 33 nucleotides. As expected, this is considerably shorter than the actual lengths of the ncRNAs used in this study, which range between 51 nucleotides, for the minimal repressive fragment of B2 RNA (termed B2 [81-131]), to the 281-nucleotide long full-length Alu RNA. This result indicates that the major portion of the ncRNAs is disordered or not stably bound to Pol II and therefore not visible in our cryo-EM maps, which are generated by averaging a large number of molecules.
Figure 3. Cryo-EM map of hPol II in complex with scAlu RNA.
(A) Final reconstruction of the hPol II/scAlu complex at 25Å resolution in two orthogonal views.
(B) Rigid body docking of the yeast Pol II/FC* RNA complex (PDB code 2B63) into the cryo-EM density map.
RNA elements beyond the docking site are flexible
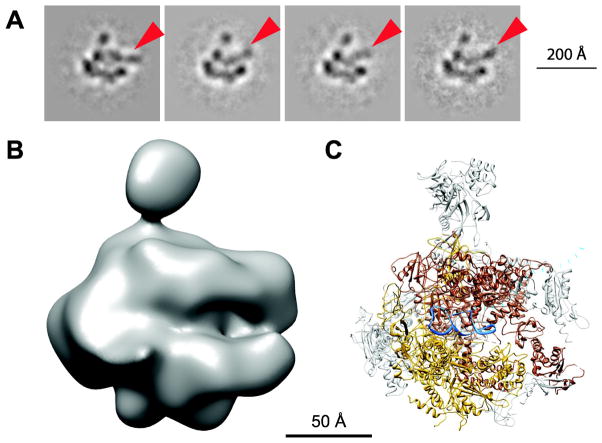
Docking of the FC RNA/yPol II crystal structure into our cryo-EM maps revealed that the additional density visible in the cleft can account for ~33 nucleotides, representing only a small portion of the ncRNAs used in this study. To determine if additional parts of the RNA can be detected in our data sets, we subjected them to multivariate statistical analysis and 2D reference-free classification. In the case of Alu RNA we observed additional density, protruding from the cleft, that is in slightly different positions in different class averages (Figure 4). This data set, which was collected using a CCD camera, had higher contrast and likely facilitated the visualization of very flexible elements in class averages with small number of particle images. Since our three-dimensional structure represents an average over all molecules, this density is not visible in our 3D maps. Thus, a number of related evidence lead us to conclude that RNA elements that extend beyond the major docking site in the cleft are not stably bound to hPol II: (1) the visualization of flexible RNA regions in different conformations as they extend from the cleft, (2) the fact that the density assigned to ncRNA within the average 3D cryo-EM reconstruction only accommodates a small segment of the ncRNAs bound to hPol II, and (3) the limit in resolution of the structures due to conformational heterogeneity of the ncRNA.
Figure 4. Flexibility of the termini of Alu RNA.
(A) Reference-free class averages of the cryo-EM model. RNA protruding from the cleft is marked with arrowheads.
(B) View of the final hPol II/Alu RNA model in the same orientation.
(C) View of the yPol II/FC* RNA crystal structure. Rpb1 is shown in brown, Rpb2 in gold and FC* RNA in blue. RNA protruding from the cleft modeled to fit the 2D class averages is indicated by a dotted blue line.
Cross-linking studies have shown that the general transcription factor TFIIF, which facilitates the dissociation of non-repressive ncRNAs from Pol II, binds to the lobe domain of Pol II. TFIIF is therefore located in an ideal position to interact with flexible RNA elements extending from the cleft. Thus TFIIF may be able to distinguish between repressive and non-repressive ncRNAs based on the specific RNA domains that extend from a common binding site in the cleft of the polymerase.
DISCUSSION
A conserved ncRNA docking site in the cleft of Pol II
Here we describe the cryo-EM reconstructions of human Pol II in complex with both human (Alu and Alu-RA RNA) and mouse (B2 and B2 [81-131] RNA) repressive ncRNAs, as well as their non-repressive counterparts (human scAlu RNA and mouse B1 RNA). Our cryo-EM reconstructions show that the main binding site for ncRNAs is located in the cleft of Pol II, close to the wall region, overlapping with the binding site for nucleic acids in the elongation complex. The binding site is the same for repressive and non-repressive ncRNAs, and it is conserved between mouse and human.
Previous studies have shown that the different naturally occurring ncRNAs compete for binding to Pol II10. The colocalization of the extra density we see for all the ncRNA-bound Pol II structures thus agrees with the competitive binding of these ncRNAs seen in biochemical assays and supports the idea of a common, high-affinity binding site that is shared across species and, more intriguingly, between repressive and non-repressive ncRNAs.
In addition, a poly-guanosine RNA oligonucleotide as well as the synthetic RNA aptamer FC have also been shown to compete with naturally occurring ncRNAs for binding to Pol II. It has been suggested that this competition is due to overlapping binding sites for these ncRNAs, but the possibility that binding of ncRNAs causes a conformational change in Pol II that prevents binding of other ncRNAs to a different docking site through an allosteric mechanism could not be ruled out. Our study shows that naturally occurring ncRNAs from mouse and human indeed bind to the same docking site in the cleft of hPol II. Furthermore, comparison with the crystal structure of yPol II in complex with the central part of FC RNA shows that this synthetic RNA aptamer binds to the same overlapping site as well. The docking site is located within the cleft of Pol II and close to the active center, in an ideal position to regulate transcription. The high-affinity binding site we identified is conserved across species from yeast to human, and we anticipate that other ncRNAs might regulate transcription by binding to this docking site as well.
Comparison with the crystal structure of yPol II/FC* RNA
The crystal structure of yPol II in complex with FC* RNA, the minimal fragment of FC RNA required for binding to Pol II, shows the RNA aptamer bound in the cleft of Pol II, above the bridge helix. The RNA forms two stem-loops that are oriented with their loops facing opposite ends of the cleft. The resolution of our cryo-EM maps is too low to trace the secondary structure of the ncRNAs. However, the domain architecture of the ncRNAs used in this study has been delineated by biochemical methods. B2 RNA, the repressive ncRNA in mouse, has been characterized in great detail, identifying the minimal repressive fragment (B2 [81-131]) as well as the minimal Pol II binding domain (B2 [99-131]). The minimal binding domain contains 32 nucleotides and is of almost the same length as the minimal binding domain of the FC RNA aptamer, which comprises 33 nucleotides. According to an RNase footprinting analysis, the 3′ end of B2 [99-131] RNA forms a short stem loop8, similar to the 5′ stem loop of FC* RNA, which consists of four base pairs and a five-nucleotide long loop. The 5′ end of B2 [99-131] RNA, however, is single-stranded and it seems unlikely that it is able to form a stem loop similar to the hairpin found at the 3′ end of FC* RNA observed in the crystal structure. For the different Alu RNAs and B1 RNA, the location of the Pol II binding domain has not been determined. However, they are overall very similar in their secondary structure: B1 RNA contains a core consisting of two short (5 and 6 base pairs) stem loops, reminiscent of the two stem loops of the Pol II-binding domain of FC RNA (Figure 1 and Figure 3). Interestingly, this region is also reminiscent of an equivalent segment in Alu-RA that neither binds to Pol II, nor repress transcription11. This core is connected to a third, very long stem loop that contains a number of unpaired bulges. In Alu-RA this region has been shown to be sufficient for binding and repression11. Both arms of full-length Alu RNA (scAlu and Alu-RA RNA) are predicted to form secondary structures that are very similar to that of B1 RNA (Figure 1). Comparison of the FC* RNA structure with the secondary structure models of mouse and human ncRNAs shows that the minimal binding domains are of similar length, in good agreement with the additional volume we observe in our cryo-EM maps. However, comparison of their secondary structure reveals no common motif for docking to Pol II’s high affinity RNA binding site, with the exception of a short stem loop that is found in all ncRNAs at either the 3′ or 5′ end.
Flexibility of inhibitory ncRNA domains
In addition to a Pol II-binding domain, repressive ncRNAs contain an inhibitory domain, which by itself is not sufficient to repress transcription. The repressive domain of B2 [81-131] RNA, comprising nucleotides 81-98, is predicted to form a stem loop, whereas the repressive domains of Alu RNA have been mapped to a bulge in the long stem loop extending from the base, and the A-rich tail that connects its two arms. In our cryo-EM maps, none of the repressive domains are visible, suggesting that they are not stably bound to Pol II, which would cause them to be averaged out in our reconstructions. However, given that we have identified Pol II’s high-affinity binding site for ncRNAs, we can deduce that the repressive domain of B2 RNA, as well as the long stem loops of B1 and Alu RNAs, are likely located close to the DNA downstream cleft. The flexible portions of each ncRNA that extend beyond the docking site are therefore positioned closely to the known interaction sites of Pol II with TFIIF, the general transcription factor that specifically destabilizes interactions between Pol II and non-repressive ncRNAs. The flexible RNA elements are therefore in an ideal position to be detected by TFIIF, which might then trigger the displacement of non-repressive ncRNAs. Given the similarities between all the analyzed ncRNAs in their binding to the cleft of the polymerase, which very likely reflect the common, high-affinity binding site also shared with the yeast aptamer, differences concerning their repressive activity must be due to the distinct character of the more unstructured, flexible segments of the RNA that emanate from the cleft.
EXPERIMENTAL PROCEDURES
Purification of hPol II and ncRNAs
Human Pol II was purified as described previously15. In short, frozen nuclear pellet from ~90 l of HeLa cell culture was ground using a cheese grater, and slowly dissolved in buffer A (50 mM Tris pH 7.9, 5 mM MgCl2, 0.5 mM EDTA, 25% glycerol, 2 mM DTT, 1 mM NaMBS, 1 mM AEBSF, complete EDTA-free Protease Inhibitor Cocktail Tablets [Roche]). The mixture was sonicated for 2 min under rapid stirring before ammonium sulfate was added to a final concentration of 0.3 M. The resulting gel was sonicated until viscosity was greatly reduced, and then centrifuged at 40,000 rpm in a Ti45 rotor for 90 min at 4 °C. The supernatant was removed and the conductivity adjusted to 0.1 M ammonium sulfate by slowly adding buffer A. Pol II was precipitated using a 42% ammonium sulfate cut, and collected by centrifugation at 30,000 rpm in a Ti45 rotor for 30 min. The pellets were resuspended in buffer B (50 mM Tris pH 7.9, 0.1 mM EDTA, 25% glycerol, 2 mM DTT, 0.1 mM PMSF) to a final concentration of 0.15 M ammonium sulfate, and loaded onto a DEAE52 ion exchange column. The column was washed with 3 column volumes of buffer C (50 mM Tris pH 7.9, 150 mM ammonium sulfate, 0.1 mM EDTA, 25% glycerol, 2 mM DTT, 0.1 mM PMSF), and protein was eluted with buffer D (50 mM Tris pH 7.9, 400 mM ammonium sulfate, 0.1 mM EDTA, 25% glycerol, 2 mM DTT, 0.1 mM PMSF). Pol II containing fractions were pooled and dialyzed against buffer E (25 mM HEPES pH 7.9, 200 mM ammonium sulfate, 0.2 mM EDTA, 10% glycerol, 2 mM DTT, 0.1 mM PMSF). Pol II was further immunopurified on a protein G affinity column containing 8WG16 antibody (NeoClone). The dialyzed sampled was incubated with the beads overnight and subsequently washed 3x with buffer F (25 mM HEPES pH 7.9, 0.2 mM EDTA, 10% glycerol, 500 mM ammonium sulfate, 0.05% NP-40, 2 mM DTT, 0.1 mM PMSF) and 2x with buffer G (25 mM HEPES pH 7.9, 0.2 mM EDTA, 10% glycerol, 200 mM ammonium sulfate, 0.05% NP-40, 2 mM DTT, 0.1 mM PMSF). Pol II was eluted four times in buffer F containing the tri-heptapeptide repeat of the CTD, 3A6.
For determination of the apo structure, hPol II was additionally purified on a heparin column prior to immunopurification. The protein was dialyzed into buffer H (50 mM Tris pH 7.9, 200 mM ammonium sulfate, 0.2 mM EDTA, 20% glycerol, 0.2 mM PMSF, 2 mM DTT) and then loaded onto the heparin column, and eluted with a gradient to 700 mM ammonium sulfate over 14 column volumes. Pol II containing fractions were pooled and dialyzed two times against 2 l buffer H for 1 h each before immunopurification.
Non-coding RNAs were synthesized in vitro by T7 RNA polymerase and gel purified as previously described.
EM sample preparation and data collection
Non-coding RNAs were heated at 95 °C for 1 min and then cooled on ice prior to complex formation with Pol II. Pol II was mixed with a 3x molar excess of each ncRNA, diluted to a final concentration of 60 nM in transcription buffer (10 mM Tris pH 7.9, 10 mM HEPES pH 8.0, 4 mM MgCl2, 50 mM KCl, 0.05% NP-40, 1 mM DTT, 0.1% trehalose), and incubated on ice for 30 min. 4 μl sample were placed onto 400 mesh copper grids covered with a holey carbon film or C-flat grids (Protochips Inc.) that had a thin carbon film floated on top. Grids were glow-discharged for 45 sec in an Edwards carbon evaporator right before use. The samples were incubated on the grids in the incubation chamber of an FEI Vitrobot at 6 °C at a humidity of 100% for 20 sec before being blotted for 6 sec at an offset of -2 mm. They were then plunge-frozen in liquid ethane and transferred into liquid nitrogen for storage. Data for ncRNA/hPol II reconstructions were acquired on film using a Tecnai F20 TWIN transmission electron microscope operated at 200 kV at a nominal magnification of 50,000×. Images were recorded under low-dose conditions (20e−/Å2) with a defocus range from -2 to -5 μm on Kodak SO-163 plate films. Micrographs were digitized using a Nikon Super CoolScan 8000 with a 12.7 μm raster step, resulting in a pixel size of 2.54 Å. Data for the apo hPol II reconstruction as well as an additional data set for the Alu RNA/hPol II complex were acquired on a Gatan 4Kx4K CCD camera using a Tecnai F20 TWIN transmission electron microscope operated at 120 kV at a magnification of 80,000× (1.5 Å per pixel) under low-dose conditions (20e−/Å2) using the MSI-T application of the Leginon data collection software24.
Image processing and volume rendering
For the data sets collected on film, particles were picked semi-automatically using the program boxer from the EMAN software package16. The contrast transfer function was estimated using CTFFIND325. Images were extracted using batchboxer at a window size of 147 × 147 pixels, and normalized using SPIDER26. Iterative projection matching was performed using libraries from the SPARX and EMAN2 image processing packages and a model of cryo-negative stained Pol II15, low-pass filtered to 60 Å, was used as a starting model. Angular increments for projection matching started at 25 degrees and were reduced stepwise to 2 degrees. Refinement and full CTF correction were performed using FREALIGN27. The resolution was estimated based on the Fourier shell correlation of 0.5.
For the data sets collected with the CCD camera using Leginon, particle picking was done using DoG picker28 within the Appion image-processing environment29. Particles were extracted at a window size of 128 pixels at 3.01 Å per pixel. Subsequent data processing was done as described for the data collected on film.
The apo Pol II data set was sorted into three different conformations using an automated multi-model approach as previously described19. In short, Euler angles were assigned to all images using Imagic’s multi reference alignment by comparison with forward projections of a template volume at a uniform angular sampling of 17 degrees. For each Euler angle, three subclass averages were computed, and subclasses with more than 30 images per subclass were clustered into homogeneous groups. Based on these clusters, three reconstructions were computed, which were then used as starting models for multi-model projection matching in EMAN2, followed by final refinement in FREALIGN as described for the ncRNA-bound structures.
The volumes representing apo Pol II and Pol II in complex with ncRNAs were filtered to 25Å resolution, and a B-factor of −3,000 was applied using bfactor. The density threshold was set manually. The cryo-EM density maps and crystal structures were rendered with UCSF Chimera30.
To exclude noise beyond the envelope of Pol II, both the ncRNA-bound and apo Pol II cryo-EM structures were multiplied with a mask that was slightly bigger than the volume of the Alu RNA/Pol II complex before subtracting the apo Pol II volume from the volumes of ncRNA-bound structures. Difference maps were calculated using the program diffmap developed in the laboratory of Nikolaus Grigorieff.
The crystal structure of yeast Pol II in complex with the synthetic RNA aptamer FC (PDB code 2B63) was docked into the cryo-EM maps using colores31 from the SITUS program package (situs.biomachina.org).
Supplementary Material
Highlights.
ncRNAs that bind to Pol II are key regulators of transcription upon heat shock
Repressive and non-repressive ncRNAs share a binding site within the cleft of Pol II
ncRNA elements not stably bound to Pol II extend flexibly beyond the cleft
The character of the ncRNA flexible segments may determine repressive activity
ncRNA binding site by the active center of Pol II is conserved from yeast to human
Acknowledgments
We thank M. Jinek for comments on the manuscript. S.K. acknowledges a fellowship from the Boehringer Ingelheim Fonds. The work was supported by NIGMS grants GM63072 (E.N.) and GM068414 (J.A.G.). E.N. is a Howard Hughes Medical Institute investigator.
Footnotes
Accession numbers
Maps will be deposited in the Electron Microscopy Data Bank (EMDB) upon manuscript acceptance.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–78. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 2.Kugel JF, Goodrich JA. Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 2012;37:144–51. doi: 10.1016/j.tibs.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kramerov DA, Tillib SV, Lekakh IV, Ryskov AP, Georgiev GP. Biosynthesis and cytoplasmic distribution of small poly(A)-containing B2 RNA. Biochim Biophys Acta. 1985;824:85–98. doi: 10.1016/0167-4781(85)90084-3. [DOI] [PubMed] [Google Scholar]
- 4.Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–72. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 5.Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–65. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–21. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 7.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–9. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 8.Espinoza CA, Goodrich JA, Kugel JF. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA. 2007;13:583–96. doi: 10.1261/rna.310307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci U S A. 2009;106:5569–74. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner SD, Kugel JF, Goodrich JA. TFIIF facilitates dissociation of RNA polymerase II from noncoding RNAs that lack a repression domain. Mol Cell Biol. 2010;30:91–7. doi: 10.1128/MCB.01115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Labuda D, Sinnett D, Richer C, Deragon JM, Striker G. Evolution of mouse B1 repeats: 7SL RNA folding pattern conserved. J Mol Evol. 1991;32:405–14. doi: 10.1007/BF02101280. [DOI] [PubMed] [Google Scholar]
- 13.Matera AG, Hellmann U, Schmid CW. A transpositionally and transcriptionally competent Alu subfamily. Mol Cell Biol. 1990;10:5424–32. doi: 10.1128/mcb.10.10.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kettenberger H, Eisenfuhr A, Brueckner F, Theis M, Famulok M, Cramer P. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat Struct Mol Biol. 2006;13:44–8. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- 15.Kostek SA, Grob P, De Carlo S, Lipscomb JS, Garczarek F, Nogales E. Molecular architecture and conformational flexibility of human RNA polymerase II. Structure. 2006;14:1691–700. doi: 10.1016/j.str.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 17.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–82. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 18.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–65. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 19.Shatsky M, Hall RJ, Nogales E, Malik J, Brenner SE. Automated multi-model reconstruction from single-particle electron microscopy data. J Struct Biol. 2010;170:98–108. doi: 10.1016/j.jsb.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armache KJ, Kettenberger H, Cramer P. Architecture of initiation-competent 12- subunit RNA polymerase II. Proc Natl Acad Sci U S A. 2003;100:6964–8. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bushnell DA, Kornberg RD. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc Natl Acad Sci U S A. 2003;100:6969–73. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 2010;29:706–16. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fishburn J, Hahn S. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Mol Cell Biol. 2012;32:12–25. doi: 10.1128/MCB.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 26.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 27.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–25. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–13. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.