Abstract
Transgenic mosquitoes generated by transposable elements (TE) often poorly express transgenes due to position effects. To avoid these effects, the ΦC31 site-directed recombination system was used to insert transgenes into a locus favorable for gene expression in Aedes aegypti. We describe phenotypes of mariner Mos1 TE and ΦC31 transgenic mosquitoes expressing the EGFP reporter in midguts of bloodfed females. Mosquitoes of nine TE-generated lines (estimated transformation frequency (TF): 9.3%) clearly expressed the eye-specific selection marker but only 2/9 lines robustly expressed the EGFP reporter. The piggyBac TE-generated ΦC31 docking strain attP26 supported recombination with attB site containing donors at an estimated TF of 1.7–4.9%. Using a codon-optimized ΦC31 integrase mutant instead of the ‘wild-type’ enzyme did not affect TF. Site-directed recombination of line attP26 with an attB-containing donor expressing EGFP from the Ae. aegypti carboxypeptidase promoter produced one transgenic line with bloodfed females expressing the reporter in midgut tissue. Docking strain attP26 also supported robust expression of Flock House virus B2 from the Ae. aegypti poly-ubiquitin promoter. Our data confirm that eye-specific selection marker expression alone is not a reliable indicator for robust gene-of-interest expression in Ae. aegypti and that the ΦC31 system can ensure predictable transgene expression in this mosquito species.
Keywords: Aedes aegypti, transposable element, ΦC31, reporter gene, transgenic
Introduction
Genetically-modified mosquitoes are used to characterize gene regulation or innate immunity (Kokoza et al., 2000; 2001; Shin et al., 2003; Adelman et al. 2008; Papathanos et al., 2009; Khoo et al., 2010) and for developing vector-based, disease control strategies (Ito et al., 2003; Franz et al., 2006, 2009; Alphey et al., 2008; Catteruccia et al., 2009; Olson and Franz, 2009; Mathur et al. 2010)). The phenotype of genetically modified mosquitoes is determined by several factors including the regulatory elements that drive transgene expression, the choice and design of the expressed effectors, the transgene insertion site within the genomic DNA and associated fitness costs. The availability of the full-length genome sequence of Ae. aegypti allows for the improvement and optimization of transgene design. Researchers can now choose among strong, tissue-specific promoters that allow transgene expression in midgut, fat body, germline tissue, or salivary glands (Edwards, et al., 2000; Kokoza et al., 2000; Moreira et al., 2000; Adelman et al., 2007; Mathur et al., 2010). In addition, two non-tissue specific, constitutive promoters of ubiquitin-encoding genes have been described (Anderson et al., 2010). Fitness costs in mosquitoes vary among different insertions and most likely depend on the transgene design and integration site in the vector genome (Cateruccia et al., 2003; Marrelli et al., 2006; Amenya et al., 2010).
Class II transposable elements (TE) such as piggyBac or mariner Mos1 are used routinely for mosquito germline transformation. However, these TEs integrate quasi-randomly into the host genome because they recognize a short genome motif as a target site (TTAA or TA, respectively; [Jacobson et al., 1986; Wang and Fraser, 1993; O’Brochta et al., 2003]), and these sites are frequent and dispersed throughout the genome. One potential strategy to overcome transgene position effects is the use of chromatin domain insulators such as scs and scs’ derived from of the Drosophila gypsy TE or the chicken β-globin 5’HS4 element (Kuhn and Geyer, 2003). Both insulators have been used successfully in Drosophila but so far not in mosquitoes (Sarkar et al., 2006). Another approach exploits the site-directed recombination system of bacteriophage ΦC31, in which two short recognition sequences, the phage attachment site attP and the bacterial attachment site, attB, recombine unidirectionally in the presence of the ΦC31 integrase (Thorpe and Smith, 1998), to place a transgene in a characterized region of the genome. A large exogenous circular DNA molecule carrying the attB recognition site can be integrated in the host genome at an attP recognition site-containing locus (Groth et al., 2000, 2004; Nimmo et al., 2006; Labbe et al., 2010). Although functional in Drosophila and mosquitoes, ΦC31-based recombination has not yet been widely used in mosquitoes (Nimmo et al., 2006; Amenya et al., 2010; Labbe et al., 2010; Meredith et al., 2011). In contrast to ΦC31, other recombination systems such as Cre/Lox of bacteriophage P1 or FLP/FRT of yeast have not yet shown efficient recombination-mediated insertion of exogenous DNA into mosquitoes (Morris et al., 1991; Jasinskiene et al., 2003; Nimmo et al., 2006).
The aim of this study was to validate the ΦC31 system as a powerful tool to facilitate predictable transgene expression in a target tissue of Ae. aegypti. Here we compare phenotypes of transgenic mosquitoes generated by either TE insertion or ΦC31 recombination that express enhanced green fluorescent protein (EGFP) in the female midgut. EGFP expression and transgene integration patterns are described for the transgenic lines. We also show that the ΦC31 system facilitates predictable, robust gene expression of an immunity related gene under control of a constitutive promoter. Furthermore, we evaluate the effects of ΦC31 integrase codon optimization on recombination efficiency.
Results
Generation and characterization of mariner Mos1 transformed mosquitoes expressing a reporter gene in midguts of bloodfed females
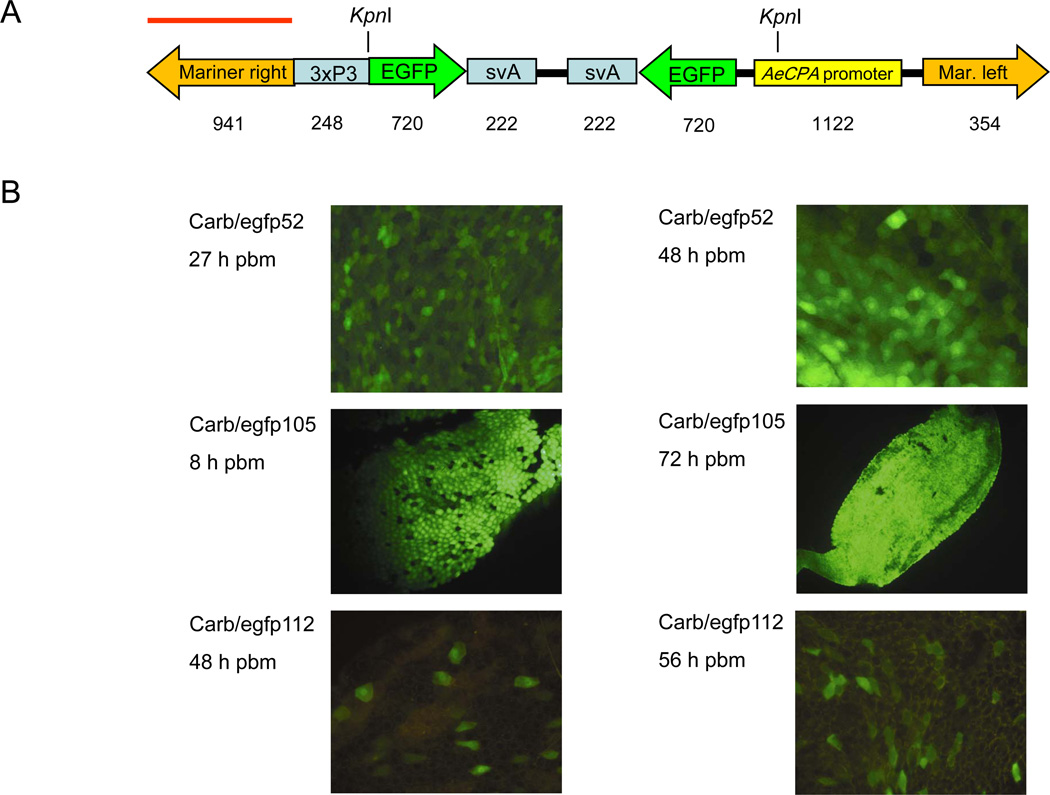
The mariner Mos1TE contains an EGFP expression cassette under control of the bloodmeal-inducible, midgut-specific Ae. aegypti carboxypeptidase A (AeCPA) promoter and an eye-specific screenable marker using the same fluorescent protein (Fig. 1A). We co-injected this donor plasmid along with a helper plasmid expressing the Mos1 transposase into 1140 Higgs White Eye (HWE) strain preblastoderm embryos (Table 1). When 193 G0 mosquitoes were out-crossed to HWE and G1 progeny larvae were screened for eye-specific expression, we found EGFP positive larvae in nine crosses, Carb/egfp52, 54, 86, 105, 112, 157, 169, 188, and 191. This results in an estimated transformation frequency (TF) of 9.3% with the assumption that the average fertility rate for each male or female individual was 0.5 (Table 1; Adelman et al., 2002). All nine crosses resulted in stable mosquito lines clearly expressing the EGFP selection marker in their eyes. However, ~90–95% of all transgenic (EGFP-positive) larvae in lines Carb/egfp86 and 105 developed into males. The sex-specific inheritance was observed in subsequent generations of Carb/egfp105 until G6. From G6 to G19 the ratio of transgenic males to females shifted gradually to 57.7% (males) : 42.3% (females) by G19 (n=324). The ratio between transgenic males and females was balanced (~50:50) from G2 onwards in all other families. Outcrossing of transgenic heterozygous G1 mosquitoes of the nine lines to HWE resulted in progeny of which approximately 50% expressed EGFP in the eyes. Female transgenic G2 mosquitoes of all nine lines were analyzed visually for gene-of-interest expression. Based on the reported expression profile directed by the AeCPA promoter (Edwards et al., 2000), EGFP expression in midgut tissue was monitored between 4–96 h post-bloodmeal (pbm). EGFP expression became visible in midguts of females that received a bloodmeal in three lines, Carb/egfp52, 105, and 112 (Fig. 1B; Table 2). In all other lines, reporter gene expression was never observed in the midgut. Carb/egfp105 exhibited the strongest level of EGFP expression among the positive mosquito lines. Reporter gene expression was apparent during the entire observation period, and at 24 h pbm, the entire midgut epithelium was expressing EGFP. EGFP expression was found to be slightly weaker in Carb/egfp52 than in Carb/egfp105, and started to become visible at 8 h pbm. Carb/egfp112 consistently showed only weak reporter gene expression in midgut tissue throughout the time course with only a few isolated cells of the epithelium expressing EGFP after 8 h pbm.
Figure 1.
Midgut-specific reporter gene expression in mariner Mos1 transformed Aedes aegypti. (A). Diagram of the mariner Mos1 TE based transgene pMos-carb/EGFP/svA. Locations of KpnI sites and probe (red bar) for Southern analysis are indicated. Numbers below the diagram indicate DNA fragment sizes in base pairs (bp). (B). EGFP expression in female midguts of the pMos-carb/EGFP/svA transformed lines Carb/egfp52, 105, and 112 at 8–72 h post-bloodmeal (pbm). EGFP was viewed under a fluorescent microscope equipped with EGFP specific filter sets.
Table 1.
Transformation data of Aedes aegypti lines (HWE strain) using mariner Mos1 or piggyBac TEs as insertion vectors or ΦC31 for site-directed recombination
| Mos1-Carb/egfp | attP26-attB, ‘WT’ ΦC31 |
attP26-attB, ‘mut.’ ΦC31 |
piggyBac- attP site |
attP26- attB/Carb/egfp |
|
|---|---|---|---|---|---|
| No. of embryos injected | 1140 | 297 | 557 | 800 | 1340 |
| No. of G0 survivors | 193 | 89 | 181 | 82 | 120 |
| No. of G1 families (f) or pools (p)1 | 116f | 13f, 3p | 22f, 5p | 41f | 27p |
| No. of G2 transgenic families or pools | 9f | 1f, 1p | 2f, 2p | 2f | 1p |
| estimated TF or RF2 | 9.3 % | 4.5 % | 4.4 % | 4.9 % | 1.7 % |
family: 1 transgenic male × 20 HWE females or 4–5 transgenic males × 40–50 HWE females; pool sizes ranged from 4 transgenic females × 4 HWE males to 19 transgenic females × 16 HWE males.
estimated transformation or recombination frequency: number of independent integration events divided by number of surviving G0 mosquitoes × 0.5 (estimated average fertility rate per individual).
Table 2.
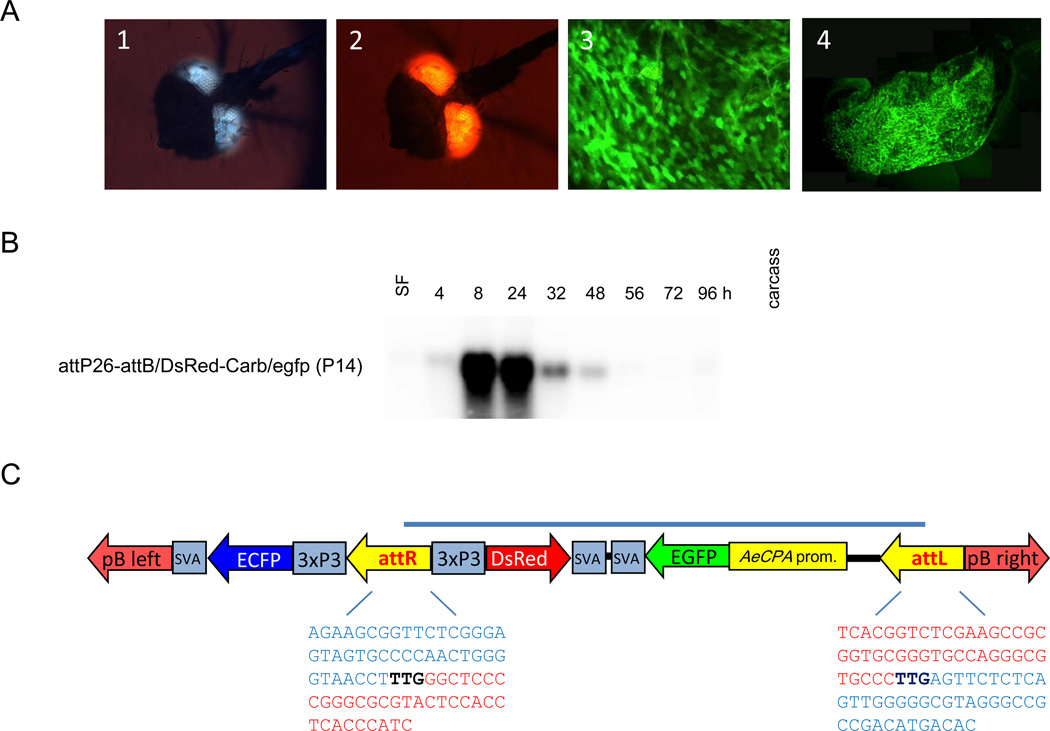
EGFP expression patterns in midgut epithelium of bloodfed Mos1-Carb/egfp and attP26-attB/DsRed-Carb/egfp (P14) mosquitoes
| family | 4 h pbm |
8 h pbm |
24 h pbm |
32 h pbm |
48 h pbm |
56 h pbm |
72 h pbm |
96 h pbm |
|---|---|---|---|---|---|---|---|---|
| Carb/egfp52 | − | + | ++ | +++ | +++ | ++ | + | + |
| 54 | − | − | − | − | − | − | − | − |
| 86 | − | − | − | − | − | − | − | − |
| 105 | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 112 | − | − | + | + | + | + | − | − |
| 157 | − | − | − | − | − | − | − | − |
| 169 | − | − | − | − | − | − | − | − |
| 188 | − | − | − | − | − | − | − | − |
| 191 | − | − | − | − | − | − | − | − |
| P14 | − | + | ++ | +++ | +++ | +++ | + | − |
− = no midgut-specific EGFP expression; + weak EGFP expression; ++ intermediate EGFP expression; +++ very strong EGFP expression.
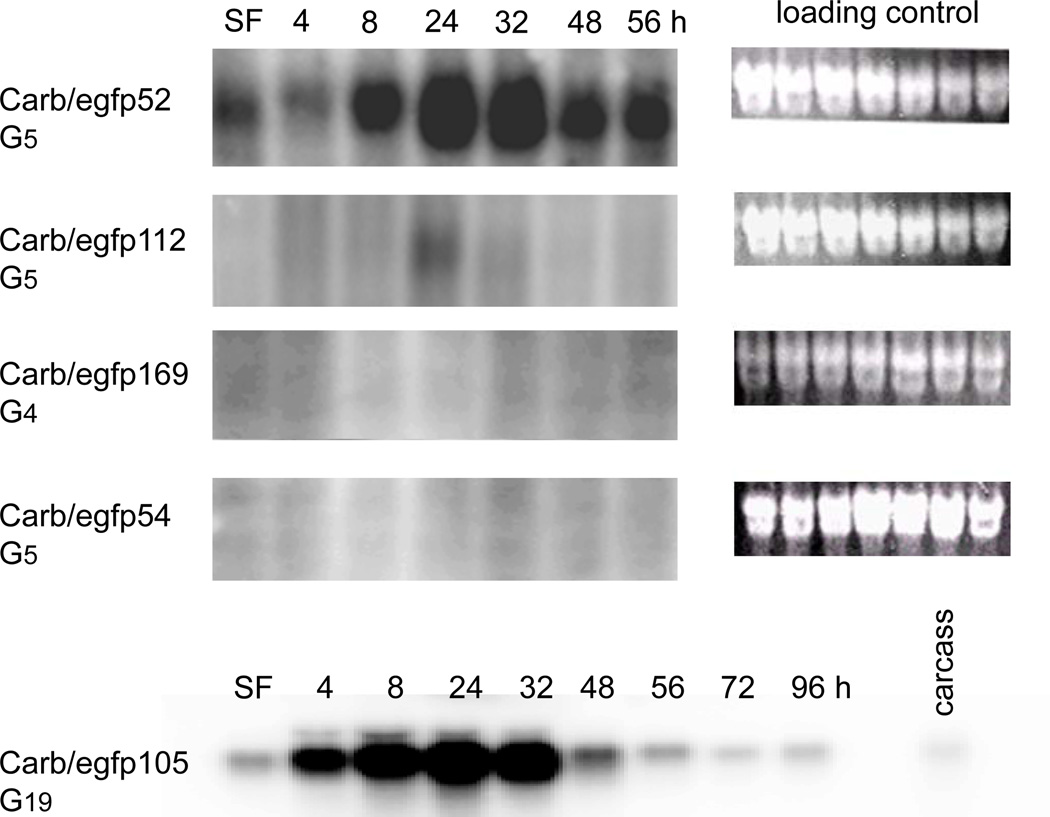
EGFP transcription profiles in midguts of G4-G5 Carb/egfp52, 54, 112, 157, 169, 188, 191, and G19 Carb/egfp105 females were evaluated by Northern blot analysis to confirm visual reporter gene expression patterns in midgut tissue of the transgenic lines (Fig. 2). EGFP mRNAs were abundant in the female midguts of Carb/egfp52 and105, but were low in Carb/egfp112. No signals were seen with RNA prepared from midguts of Carb/egfp 54, 157, 169, 188, and 191 mosquitoes. Midgut-specific EGFP expression was evaluated for line Carb/egfp105 G19 females, since almost all transgenic individuals of this line were males in earlier generations. Carb/egfp52 and 105 females showed strongest transcriptional activity for EGFP in midgut tissue at 24 and 32 h pbm, respectively. EGFP transcripts were abundant until 56 h pbm in midguts of line Carb/egfp52 and 48 h pbm in line Carb/egfp105, although the latter showed far stronger marker gene expression in situ.
Figure 2.
Northern blot analyses of EGFP transcription in female midguts of the mariner Mos1 transformed lines (G4–5) Carb/egfp52, 112, 169, 54, and (G19) 105. Samples were prepared from midguts harvested after sugarfeeding (SF) or 4–96 h pbm. The 10 carcasses of Carb/egfp105 mosquitoes were without heads. The hybridization pattern of Carb/egfp157, 188, and 191 (not shown) resembled those of Carb/egfp54 and 169. Total RNA was extracted from pools of 25 midguts per time point. Blots were hybridized with random primed 32P-dCTP- labeled DNA probes or in vitro-transcribed 32P-dUTP-labeled probes corresponding to the EGFP gene at 48°C. Ethidium bromide stained agarose gels are shown as loading controls.
Southern blot analyses of the transgene integration patterns using KpnI digested genomic DNA of Carb/egfp52, 54, 86, 105, and 112 females hybridized to a probe comprising the ~940 bp right arm of the Mos1 TE showed signals corresponding to DNA fragments >1000 bp in length (Fig. 3A). Multiple TE integration events were detected in Carb/egfp105, single insertions in Carb/egfp52, 86, and 112, and no discrete signals were evident in Carb/egfp54. Thus, the stronger reporter gene expression levels in line Carb/egfp105 as compared to Carb/egfp52 could be caused by a dose effect due to high copy number integrations of the transgene.
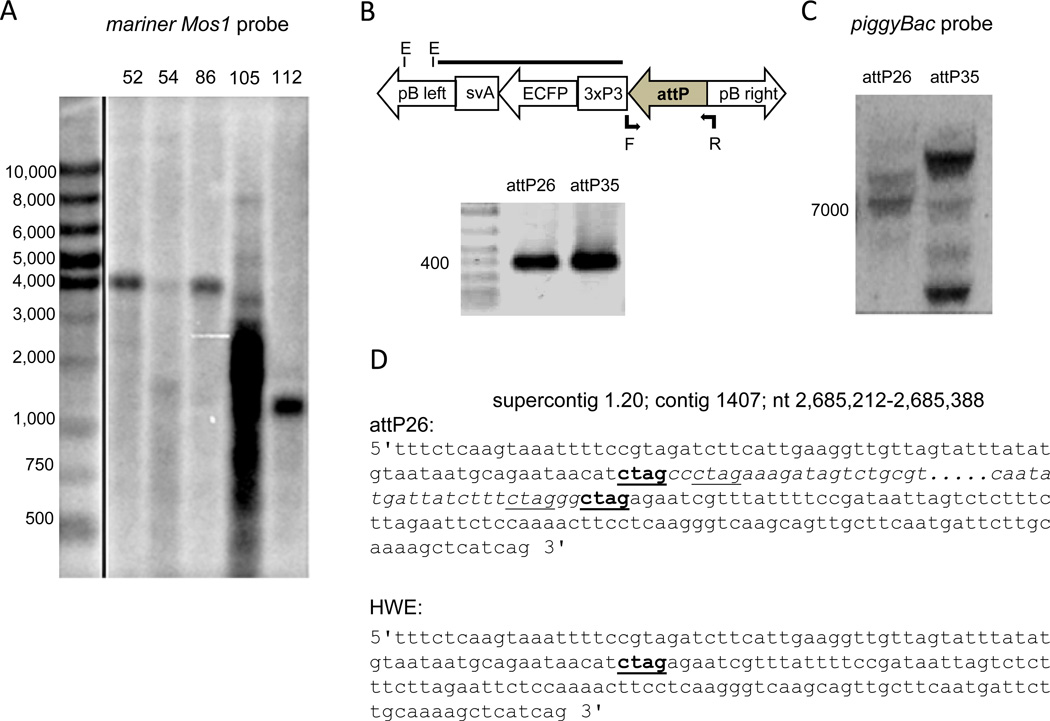
Figure 3.
Genotypic analysis of transgenic mosquito lines. (A). Southern blot analysis of mariner Mos1 transformed lines Carb/egfp52, 54, 86, 105, and 112. Total DNA was extracted from 10 females per line, followed by digestion with KpnI. Blots were hybridized with a random-primed 32P-dCTP-labeled DNA probe corresponding to the right arm of the mariner Mos1 TE at 48°C. (B). Detection of the attP site-specific recombination site in transgenic mosquitoes of lines attP26 and attP35. Total DNA of five females was extracted and used as template for gene amplification using primers corresponding to the 171 bp attP site and 220 bp flanking cloning sites of plasmid Bac[3xP3-ECFPfa]attP shown above as a diagram (Nimmo et al., 2006). (C). Southern blot analysis of piggyBac transformed lines attP26 and attP35. Total DNA was extracted from 3 females per line, followed by digestion with EcoRV. Blots were hybridized with a random primed 32P-dCTP labeled DNA probe corresponding to the left arm/3xP3/ECFP encoding region of the piggyBac TE at 48°C. (D). Partial sequence (nt position 2,685,212–2,685,388) of the Ae. aegypti genomic DNA of supercontig 1.2, contig 1407. Highlighted in blue: partial sequence of piggyBac left arm; highlighted in green: partial sequence of piggyBac right arm. The CTAG sequence motif is underlined and/or in bold.
Generation and characterization of piggyBac TE transformed mosquitoes containing the attP site for ΦC31-mediated recombination
Eight hundred embryos were co-injected with pBac[3xP3-ECFPfa]attP (Fig. 3B; Nimmo et al., 2006) and the piggyBac helper plasmid to generate a HWE-based docking strain containing the attP landing site. The survival rate was 10%, and after out-crossing to the HWE recipient strain, 33 G1 families and eight female pools were established. Progeny of two families, attP26 and attP35, clearly expressed the enhanced cyan fluorescent protein (ECFP) marker in their eyes. Presence of the attP site in the genome of the lines was confirmed by gene amplification using primers complementary to the 171 bp attP site and flanking cloning sites (Fig. 3B). Both attP site-containing docking lines were intercrossed until G14. Thereafter lines attP26 and attP35 were made homozygous. Southern blot analysis revealed two and four possible integration events in lines attP26 and attP35, respectively (Fig. 3C). Differences in signal intensity among the various hybridizing DNA fragments in the Southern blots could be due to independent assortments of the different insertions into the mosquito genome. It is likely that not every individual inherited each copy of the transgene in a homozygous state, resulting in potentially weaker hybridization signals.
We were unable to obtain DNA-hybridization patterns with higher resolution for line attP26 despite several attempts. This prompted us to perform Genome Walking on genomic DNA of this line to confirm the number of transgene integration events. Genome Walking revealed one TE integration event for line attP26 within supercontig 1.20 (contig 1407) at nt position 2,685,285 or 2,685,289. The piggyBac TE is inserted within the first intron of a gene, AAEL001004, whose function is unknown. This was the only integration site identified in repeated experiments using multiple restriction endonuclease-digested genomic DNA libraries in the Genome Walking procedure. The piggyBac integration pattern is unusual because it did not integrate at a TTAA motif in the host genome. Instead, insertion occurred at a 5’CTAG3’ sequence motif (nt position 2,685,285–2,685,289) and upon integration the sequence motif was duplicated (Fig. 3D). This was confirmed in five independent gene amplification, cDNA cloning, and sequencing experiments. Curiously, both inverted terminal repeat (ITR) sequences of piggyBac contain the CTAG motif as well (Fig. 3D).
Comparing recombination efficiencies of ‘wild-type’ or codon-optimized ΦC31 integrases
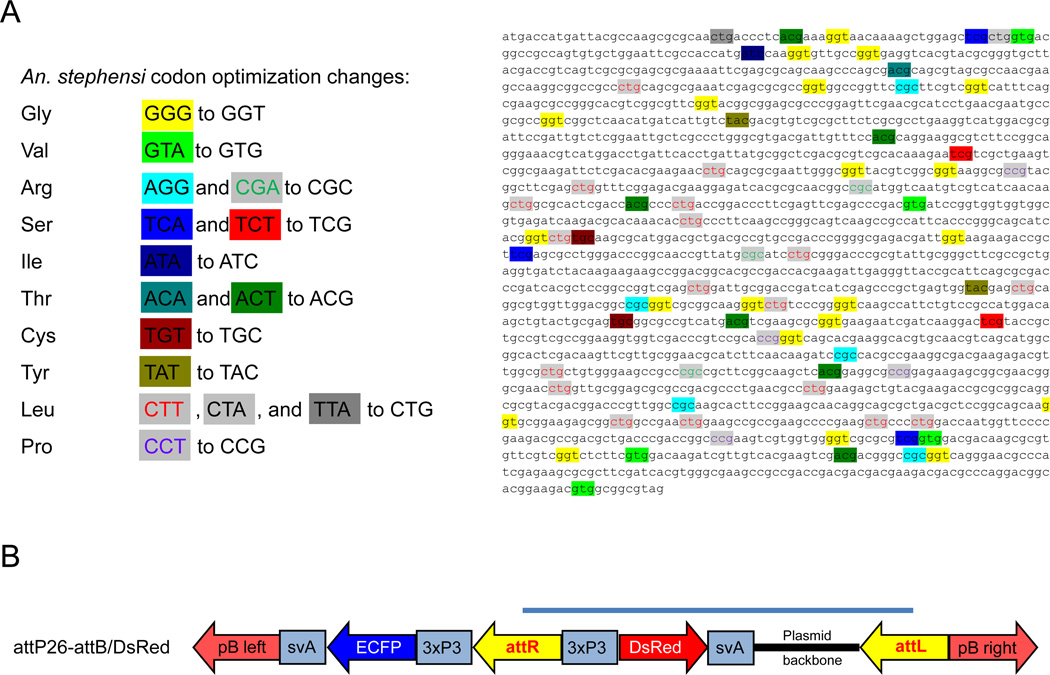
The ΦC31 integrase mutant P3 (Keravala et al., 2009) was codon-optimized for use in the mosquito Anopheles stephensi using the Codon Usage Database (http://www.kazusa.or.jp/codon/; Nakamura et al., 2000). Base triplet sequences coding for certain amino acids were mutated as shown in Fig. 4A. Only two of these 10 mutations, AGG or CGA to CGC and ACA or ACT to TCG do not occur at highest frequencies in the Ae. aegypti genome. This prompted us to use the An. stephensi optimized ΦC31 mutant for the experiments here. Females from the attP26 docking strain were transformed with the donor plasmid pattB-DsRed2 (Nimmo et al., 2006) using mRNA in vitro transcribed from either ‘wild-type’ or codon-optimized ΦC31 integrase cDNA (Fig. 4B). Thirty percent (30%) of the 297 attP26 embryos injected with pattB-DsRed2 and ‘wild-type’ integrase mRNA survived (Table 1). The survivors were grouped in three female pools (4–17 females crossed with 4–15 HWE males per pool) and 13 male pools (4–5 males crossed with 40 HWE females per family). Offspring that showed site-directed recombination with the donor plasmid based on eye-specific ECFP and DsRed expression were observed in one male and one female pool. A total of 557 attP26 embryos were injected with the codon-optimized ΦC31 mRNA. Again, ~30% of the injected eggs survived and resulting adults were combined in five female pools (9–19 females crossed with 9–16 HWE males per pool) and 22 male pools (4–5 males crossed with 50 HWE females per family). Two female and two male pools produced recombinant offspring. Thus, 12.5% and 15% of the pools containing mosquitoes transformed with the ‘wild-type’ and codon-optimized ΦC31 integrases respectively, had recombination events with the donor plasmid (Fig. 4B). Intensity of DsRed eye marker expression was very similar in all six attP26-attB/DsRed recombinant lines as would be expected after donor integration into the same attP site containing docking strain. Furthermore, the estimated TF were essentially the same for the transgenic lines generated with codon-optimized or ‘wild-type’ integrases, ranging from 4.4% to 4.5% (Table 1). This indicates that the codon-optimized ΦC31 integrase did not promote higher recombination rates in Ae. aegypti than the ‘wild-type’ enzyme in these experiments. For the following ΦC31 site-directed recombination experiments we used the ‘wild-type’ integrase.
Figure 4.
Codon optimization of the ΦC31 integrase and ΦC31-mediated recombination of line attP26 with the attB/DsRed2 donor plasmid. (A). Codon optimization was performed on the DNA sequence of ΦC31 integrase P3 mutant (Keravala et al., 2009), which contained an additional 33 amino acid sequence upstream of the ‘wild-type’ ΦC31 start codon. The DNA sequence of the P3 mutant was codon optimized according to the most frequent codon usage in the An. stephensi genome, which is similar to that of Ae. aegypti. (B). Diagram of the attB/DsRed2 donor plasmid integration into the genome of docking strain attP26 following recombination between attB and attP in presence of ΦC31(‘wild-type’) or codon optimized ΦC31 P3 mutant integrases, respectively. As a consequence of recombination, attP and attB sites are converted into attL and attR. Grey bar indicates the part of the transgene that originates from the donor plasmid.
Generation and characterization of ΦC31 recombinants expressing a reporter gene in midguts of bloodfed females and a viral suppressor of RNA interference
We co-injected 1340 embryos of the homozygous attP26 line with the donor plasmid pattB-carb/EGFP/svA and ΦC31 integrase mRNA (Table 1). One hundred and twenty individuals survived and 22 male and five female pools were established. Each male pool contained three males and each female pool was made up of as many as 15 females originating from the injected embryos. One male pool, P14, contained mosquitoes that expressed strongly both ECFP and DsRed in their eyes when viewed under the respective fluorescent filter sets, indicating recombination between the attP and attB sites of the docking strain and the donor, respectively. Two experiments in which 725 and 1130 embryos of the attP35 docking strain were injected with the same donor plasmid did not result in any recombination-mediated integration (data not shown). This supports the conclusion that the attP landing sites in line attP35 are integrated at loci that do not promote sufficient recombination using the ΦC31 system.
Recombinant P14 females expressed EGFP in their midguts between 8–72 h pbm (Table 2, Fig. 5A). The strongest reporter gene expression was observed at 32–56 h pbm, which was similar to the expression pattern of Carb/egfp52. In addition, P14 females, similar to Carb/egfp52, exhibited weaker EGFP expression in midgut tissue than line Carb/egfp105. Northern blot analysis showed abundant EGFP mRNA in P14 midgut tissue at 8–24 h pbm (Fig. 5B). From 24 h pbm onwards, EGFP mRNA levels declined steeply and were only weakly detectable at 48 h pbm. Thus, P14 recombinant females showed slightly weaker transcriptional activity of the EGFP gene in midgut tissue than the TE transformed lines Carb/egfp52 and Carb/egfp105 (Fig. 2). Gene amplification analysis using primers surrounding the recombination sites in P14 and subsequent DNA sequencing of the amplicons showed that the attP site of docking strain attP26 and the attB site of the donor plasmid had been recombined to become attL and attR sites (Fig. 5C). The ΦC31 recombinants showed relatively balanced sex ratios similar to G19 Carb/egfp105 mosquitoes: 46.1 % females : 53.9% males (n=1786).
Figure 5.
ΦC31-mediated recombination of docking strain attP26 with the attB/DsRed2-carb/EGFP/svA donor plasmid. (A). Eye-specific marker gene expression in line P14. (1) ECFP expression originates from the selection marker of line attP26; (2) DsRed expression originates from the donor plasmid. (3) Midgut-specific EGFP expression represents the gene-of-interest of the donor and was visible at 48 h post-bloodmeal (pbm) in individual epithelial cells of (4) the entire midgut containing a bloodmeal. (B). Northern blot analysis for the detection of EGFP transcripts in female midguts of attP26-attB/DsRed-carb/egfp (P14) mosquitoes after sugarfeeding (SF) or 4–96 h pbm. Total RNA was extracted from 25 midguts per time point or 10 headless carcasses. Blots were hybridized with a random primed 32P-dCTP labeled DNA probe corresponding to the EGFP gene at 48°C. (C). Diagram of the donor plasmid integration pattern following recombination between the attP site of line attP26 and the attB site of the donor. attP and attB sites are converted into attL and attR consisting of DNA sequence originating from attP (blue) and attB (red). In bold and black: crossover recognition motif. attL and attR sequences were confirmed by gene amplification and DNA sequencing of the amplicons. Grey bar indicates the part of the transgene that originates from the donor plasmid.
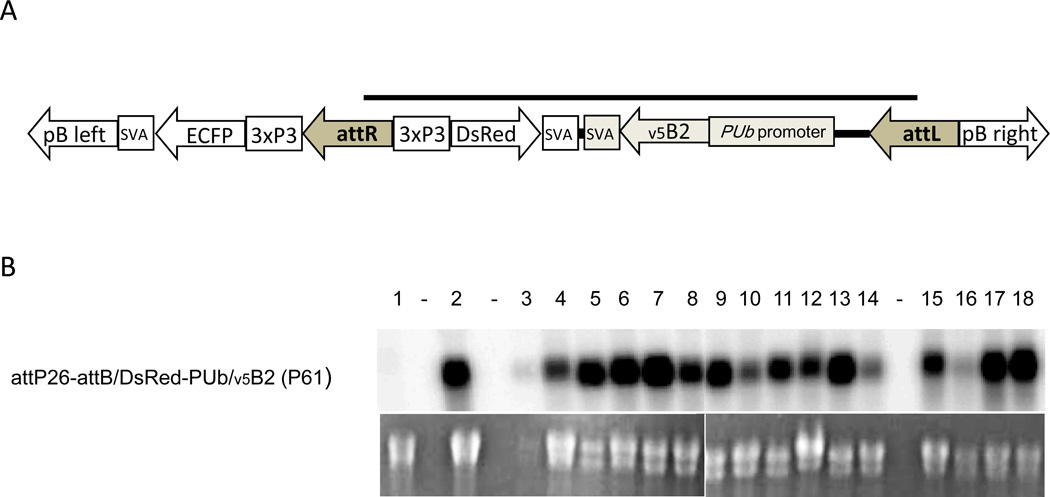
In another experiment we generated attP26 based ΦC31 recombinants expressing the Flock House virus B2 protein, a viral suppressor of RNA interference, from the non-tissue specific, constitutive Ae. aegypti poly-ubiquitin (PUb) promoter (Fig. 6A; [Li et al., 2002; Anderson et al., 2010]). We obtained one pool, P61, in which larvae expressed both fluorescent eye markers, ECFP and DsRed originating from docking strain attP26 and the donor plasmid, respectively. Northern blot analysis showed the presence of v5B2 transcripts in larvae, males, sugarfed, and bloodfed females of P61 mosquitoes (Fig. 6B). Thus, docking strain attP26 repeatedly facilitated predictable, robust gene-of-interest expression following recombination with two different donor plasmids. The v5B2 expressing P61 mosquitoes will be further characterized and extensively analyzed in vector competence studies using a range of arboviruses. This work will be presented elsewhere.
Figure 6.
ΦC31-mediated recombination of line attP26 with the attB/DsRed2-PUb/v5B2/ svA donor plasmid to constitutively express the Flock House virus B2 suppressor of RNA interference in Ae. aegypti. (A) Diagram of the donor plasmid integration pattern following recombination between the attP site of line attP26 and the attB site of the donor. Grey bar indicates the part of the transgene that originates from the donor plasmid. (B) Northern blot showing v5B2 expression over time in various tissues of recombinant P61 mosquitoes. Lanes: (1) docking strain attP26 (negative control); (2) P61 larvae; (3) P61 females - midguts, 1 day post-emergence (pe); (4) carcasses, 1 day pe; (5) midguts, 3 days pe; (6) carcasses, 3 days pe; (7) midguts, 5 days pe; (8) carcasses, 5 days pe; (9) midguts, 7 days pe; (10) carcasses, 7 days pe; (11) midguts of bloodfed females, 7 days post-bloodmeal (pbm); (12) carcasses of bloodfed females, 7 days pbm; (13) midguts of bloodfed females, 14 days pbm; (14) carcasses of bloodfed females, 14 days pbm; (15) P61 males - 1 day pe; (16) 3 days pe; (17) 5 day pe; (18) 7 days pe.
Discussion
We show here that although several transformed lines of Ae. aegypti can be routinely obtained from a single experiment when using mariner Mos1 as an insertion vector, only a fraction of them express the transgene in a manner consistent with the expression profile characteristic of the endogenous gene from which the regulatory DNA was derived. Only two of nine marinerMos1 generated transgenic lines, Carb/egfp52 and Carb/egfp105, expressed strongly the EGFP reporter gene in female midguts after induction by a bloodmeal. In Carb/egfp105 mosquitoes, the TE appeared to be integrated into the genome in high copy numbers, which resulted in a smear pattern in Southern blot analysis. A mass integration event into the genome of Ae. aegypti has been described before for the piggyBac TE but not for mariner Mos1 (Adelman et al., 2004). Whereas the piggyBac mass integration resulted in a highly unstable transgene expression pattern, the gene-of-interest was stably expressed in line Carb/egfp105 for at least 19 generations. The quasi-random integration patterns of mariner Mos1 and piggyBac often lead to integrations at loci that exhibit position effects (Nimmo et al., 2006; Labbe et al., 2010). These position effects are caused in part by integration in heterochromatic regions of the genome or nearby repressors and/or enhancers of endogenous genes (Wallrath and Elgin, 1995; Sarkar et al., 2006; Anderson et al., 2010). However, expression of the eye-specific selection marker appears to be relatively robust despite position effects, since the marker was clearly visible among all nine transformants. Thus, the selection marker cannot be seen as a reliable indicator to predict gene-of-interest expression levels in transformed Ae. aegypti.
We tested the ΦC31 site-directed recombination system as a strategy to overcome transgene position effects. An essential component of this system is an attP site-containing docking strain, which has the landing site integrated at a region of the genome that is permissive for strong gene-of-interest expression. Of two such lines established, attP26 and attP35, ΦC31-mediated recombinants could only be recovered from the former. This is notable because the Southern analysis showed at least four integrations of the docking site DNA in attP35. Apparently, none of these integrations was in a locus favorable for site-directed recombination. In contrast, at least one of the two sites in attP26 accepted integrase-mediated insertions at rates detectable with the number of embryos we injected. Comparisons of EGFP expression in Carb/egfp52 and 105 with attP26-attB/DsRed-Carb/egfp (P14) recombinants showed slightly weaker gene-of-interest expression levels for the latter. Position of the attP site within the mosquito genome and differences in insert copy number could account for these observations. Nevertheless, ΦC31-mediated recombinants based on line attP26 repeatedly facilitated robust gene-of-interest expression. This was shown by the abundance of v5B2 mRNA among samples of attP26-attB/DsRed-PUb/v5B2 (P61) mosquitoes, which express the viral suppressor of RNA interference, B2, from the constitutive PUb promoter. Taken together this study supports the argument that any attP docking strain generated for the ΦC31 system needs to be evaluated empirically for gene-of-interest expression after recombination. Similar to the observation with TE generated transformants, selection marker expression alone is not a reliable indicator for the gene-of-interest expression potential of ΦC31 docking strains.
Several reports indicate that the use of the ΦC31 system instead of a TE-based transformation system leads to substantially higher TF in Drosophila and mosquitoes (Groth et al., 2004; Nimmo et al., 2006). This study does not support these observations. Estimated transformation frequencies were similar or higher when using TEs instead of ΦC31. One reason for this could be that the Mos1 or piggyBac transposases in these experiments were generated from a helper plasmid whereas the ΦC31 integrase was supplied as an in vitro-transcribed mRNA. Attempts to supply the integrase as a plasmid under control of the hsp70 promoter did not result in successful recombination events when co-injecting the attB/DsRed-carb/EGFP/svA donor plasmid into embryos (A.W.E. Franz, M.R. Smith, K.E. Olson, unpublished results). We attempted to increase recombination efficiency and specificity for the ΦC31 system by optimizing the integrase coding sequence for expression in mosquitoes (Grosjean and Fiers, 1982). Since the natural environment for ΦC31 integrase expression is the bacterium Streptomyces ambofaciens, the ΦC31 recombinase-encoding sequence is optimized for translation by the tRNA spectrum of the bacterium, which differs from that present in mosquitoes. Codon-optimization has been shown previously to boost gene expression in a specific organism by enabling the cell to use its major tRNAs for translation (Fletcher et al., 2007; Liu, 2009). Codon-optimization was performed with the P3 mutant of ΦC31, which arose from an unexpected recombination event (Keravala et al., 2009). Mutant P3 contained nine engineered amino acid substitutions and an additional 33 amino acid sequence at the N-terminal portion of the protein. As a result, mutant P3 in human cells showed a 2.3-fold higher integration efficiency and a 7-fold higher specificity than the ‘wild-type’ integrase (Keravala et al., 2009). When comparing the estimated recombination frequencies of the mosquito codon-optimized P3 mutant with that of the ‘wild-type’ integrase in Ae. aegypti, we did not observe a significant difference between the two enzyme variants. However, the codon-optimized P3 mutant has been shown to be functional for mosquito transgenesis, even though it might not substantially increase the efficiency of the ΦC31 system in mosquitoes as we expected. More experiments need to be conducted to confirm this observation.
In early intercrossed generations (up to G6) mosquitoes of line Carb/egfp105 exhibited a distorted sex ratio with ~ 95% of the insects being males. This sex ratio bias declined in subsequent generations and among G19 mosquitoes the ratio was relatively balanced. Sex ratio distortions among transgenic mosquitoes have been reported previously (Irvin et al., 2004). The most likely explanation for this observation is that it is a consequence of the genetics of sex-determination in Ae. aegypti. Sex in these mosquitoes is determined by a single autosomal gene on chromosome 1 with maleness being the dominant allele (McClelland, 1962). Males represent heterozygotes (Mm) at the sex locus, while females represent the homozygous, recessive (mm) condition. Integration of the transgene close to the M locus would link it to the male-determining factor. Meiotic recombination in males during inbreeding would move the transgene to the m-bearing chromosome and eventually increase the number of females carrying the transgene. Consistent with this hypothesis, we did not observe high mortalities among larvae, pupae or females of line Carb/egfp105 in early or late generations that might be indicative of a male-drive phenomenon. Despite repeated efforts we were unable to physically map the (multiple) TE integration sites in line Carb/egfp105, which would have been important to confirm linkage to the M locus.
Physical mapping of the piggyBac integration in line attP26 revealed that the TE did not integrate at a TTAA site in the genome as would be expected, but rather used the CTAG motif, which was duplicated upon TE integration. This unexpected result was confirmed by gene amplification using genome-specific primers in combination with primers complementary to the piggyBac left and right arms. The genomic sequence of HWE contains the CTAG motif at nt position 2,685,285, whereas the Ae. aegypti (strain: Liverpool) genome sequence in VectorBase shows the presence of a CTAA motif at the identical locus. The genomic sequences flanking the insertion site match the first intron of the predicted gene, AAEL001004. Almost all piggyBac integration patterns reported to date show insertion at the typical TTAA motif, which appears to be absent in the nearby region of genomic DNA in line attP26 where the TE was integrated. Since there is only one example in the literature for an atypical piggyBac integration at an ATAA site instead of a TTAA site, it is difficult to speculate about our observation (Elick et al., 1996). However, it seems likely that the piggyBac transposase was able to utilize the genomic CTAG motif for TE integration.
This work shows potential advantages and disadvantages when using TE-based transformation or ΦC31-mediated recombination to generate gene-of-interest expressing Ae. aegypti. TE based transformation using a transposase expressing helper plasmid is technically simple and appears to result in higher TF compared to ΦC31-mediated recombination. However, position effects and variable insert copy number often lead to a range of different transgenic phenotypes that need to be analyzed before the optimal phenotype can be chosen. Using the ΦC31 system requires more effort in terms of preparing and supplying the integrase source. However, our study shows that we have identified an attP site-containing docking strain, attP26, that allows strong gene-of-interest expression following ΦC31 mediated recombination with a donor. Furthermore, line attP26 is based on the well characterized, arbovirus competent HWE recipient strain (Franz et al., 2006; Sanchez-Vargas et al., 2009; Khoo et al., 2010). This enables us to generate and use ΦC31 recombinants for studies aimed at investigating mosquito-arbovirus interactions.
Experimental procedures
Construction of the donor plasmids
The EGFP expression cassette was constructed by inserting a cDNA fragment containing the EGFP coding sequence and the SV40A transcription termination signal (originating from pEGFP, Clontech, CA) into the NotI/XbaI digested pSLfa1180fa shuttle vector (Horn et al., 2000). The 1120 bp AeCPA promoter cDNA was inserted into the SacI and NotI digested pSLfa1180fa-EGFP/svA. Finally, the AeCPA/EGFP/svA cassette was transferred from pSLfa1180fa into pMos1 or pattB-DsRed2 using the AscI sites. An additional cloning site containing FseI, AscI, and HindIII restriction endonuclease sites was inserted into the SpeI and NotI sites of the original ΦC31 donor plasmid pattB-DsRed2 (Nimmo et al., 2006).
mariner Mos1-based germline transformation of Ae. aegypti and establishment of transgenic mosquito lines
The eye-pigment-deficient Higgs White Eye strain (HWE), a variant of the Ae. aegypti Rexville D strain was used for germline transformation (Wendell et al., 2000). Mosquitoes were reared at 28°C, 82% humidity under a 12 h darkness / 12 h light regime. Adults were maintained on sucrose. Germline transformations using the mariner MosI TE were performed as previously described (Jasinskiene et al., 1998, 2007; Franz et al. 2006; Khoo et al., 2010). Each surviving G0 male was outcrossed to 20 female HWE. Five female G0 mosquitoes were pooled and outcrossed to one HWE male. To produce offspring, females received bloodmeals from mice according to Institutional Animal Care and Use Committee (IACUC) protocol 09-1365A. Progeny larvae of these crosses (G1) were screened for EGFP expression in their eyes using a fluorescence microscope (Olympus SZX12 Zoom) equipped with an EGFP-specific filter set. Transgenic G1 mosquitoes were outcrossed to the HWE recipient strain and their progeny (G2) were analyzed for gene-of-interest expression.
Generation of attP site containing docking strains and ΦC31 based site-directed recombination
We used plasmid pBac[3xP3-ECFPfa]attP encoding the left and right arms of the piggyBac TE, the attP recombination site and the eye tissue-specific ECFP selection marker to generate ΦC31 docking strains (Nimmo et al., 2006). Pre-blastoderm embryos of the HWE strain were co-injected with piggyBac plasmid DNA and a helper plasmid encoding the transposase under control of the Drosophila hsp70 heat shock promoter. Transgenic mosquitoes were identified and lines established as described above. The attP recombination site was detected in the transgenic mosquito lines by PCR using primers attP FWD: 5’caaatgtgttctgtgatgacctg3’ and attP REV: 5’ctcccttgctactgacattatgg3’. Eventually, attP site containing lines were made homozygous by establishing sibling crosses between transgenic G14 individuals over two generations.
For site-directed recombination the coding sequence of the ΦC31 integrase was transcribed in vitro from plasmid pET11C31poly(A) (Nimmo et al., 2006) using the mMessage mMachine T7 Ultra Kit (Applied Biosystems). mRNA was purified using the MegaClear Kit (Applied Biosystems) followed by ethanol precipitation. Integrase mRNA and attB-DsRed2 donor plasmids were co-injected into homozygous embryos of the attP site containing docking strains at final concentrations of 960 ng/µl and 450 ng/µl, respectively.
Mutagenesis of the ΦC31 integrase gene
The 1903 bp coding sequence of the ‘wild-type’ ΦC31 integrase gene was codon-optimized for An. stephensi by commercial cDNA synthesis (Epoch Biolabs, Missouri City, TX) following the algorithm shown in Fig. 4A. Another 339 bp fragment representing the 5’ portion of the cDNA of ΦC31 mutant P3 (Keravala et al., 2009) was similarly codon-optimized. Using restriction sites NdeI and BstBI, a 243 bp fragment of the codon-optimized ‘wild-type’ ΦC31 was then replaced with the 339 bp fragment to generate codon-optimized mutant P3.
Northern blot analysis
Total RNA was extracted from pools of 25 midguts of bloodfed and sugarfed females using TRizol reagent (Invitrogen, Carlsberg, CA). Approximately 15 µg of each RNA sample was electrophoresed in a 1.2 % agarose gel containing 1× MOPS and 0.66 M formaldehyde and blotted overnight onto a positively charged nylon membrane (Ambion). Using the T7 Maxiscript Kit (Ambion), a ~540 nt antisense RNA probe was transcribed in vitro from a plasmid containing the EGFP coding sequence. The probe was labeled with 20 mCi of [alpha-32P]UTP/ml] with an activity of 800 Ci/mmol. Alternatively, 32P-dCTP labeled cDNA probes (3000 ci/mmol) containing a 500 bp fragment of the EGFP gene or a 368 bp fragment including the coding sequence of the Flock House virus B2 gene and the v5 epitope tag of Simian virus 5 were generated using the DECAprime II Kit (Ambion). Blots were hybridized at 58°C for ~16 h each with 20 µl 32P-labeled probe in Ultrahyb hybridization buffer (Ambion). Hybridized blots then were washed twice for 20 min with 2× SSC, 0.1% SDS and twice for 20 min with 0.2× SSC, 0.1% SDS at 65°C. Phosphor storage screens were exposed to the hybridized blots for 20 min up to 3 days and then scanned on a Storm Phosphorimager (Molecular Dynamics Inc.).
Total DNA extraction and Southern blot analysis
For detection of the mariner Mos1 TE, total DNA was extracted from pools of 10 whole-body female mosquitoes using the procedure described byFranz et al. (2006). Extracted DNAs were digested overnight with KpnI. DNAs were precipitated in 100 % ethanol and the resulting pellets resuspended in water. Seven to 15 µg digested DNA were electrophoresed in a 0.8 % agarose gel. The gel was denatured, neutralized and blotted onto a positively-charged nylon membrane using to standard procedures (Sambrook et al., 2001). A DNA probe complementary to the right arm of the mariner Mos1 TE was generated by random-prime labeling with 20 mCi of [alpha-32P]CTP/ml (3000 Ci/mmol activity) using the Megaprime labeling kit (Amersham). Blots were hybridized with the probe at 45 or 48°C and washed at 50 or 55°C before being exposed to phosphor storage screens and scanned as described above. To detect the piggyBac TE in docking strains attP26 and 35, total DNA was extracted from three females of each line and digested overnight with EcoRV. The 2,200 bp [alpha-32P]CTP labeled probe for hybridization was complementary to the left arm/3×P3/ECFP encoding region of the piggyBac TE. Hybridizations were carried out overnight at 48°C.
Genome walking and genotypic characterization of transgenic mosquitoes by gene amplification
Genome walking was performed to physically map the transgene integration locus in docking strain attP26. According to the protocol of the Genome Walker Universal Kit (BD Biosciences Palo Alto, CA), total mosquito DNA was digested with DraI, EcoRV, PvuII, or StuI, followed by ligation to an adaptor molecule provided with the kit. For detection of the piggyBac TE integration site by PCR the following piggyBac specific primers were used in combination with the adaptor primers of the kit: PBleftarm REV: 5’gactcacgcggtcgttatagttcaaaatcagtg3’, PBleftarm nested REV: 5’gacaagcacgcctcagccgagctccaagcggcgac3’, PBrightarm FWD: 5’acgtacttactgtacttactgcccctct3’, PBrightarm nested FWD: 5’gtcgagagcataatattgatatgtgccaaagttg3’. Gene amplifications were performed with Advantage 2 polymerase (Clontech, CA). Resulting amplicons were cloned into the pCR4-TOPO TA cloning vector (Invitrogen, Carlsbad, CA) and then sequenced.
Transgene integration in line attP26 was confirmed from genomic DNA by gene amplification using the primer combinations contig1.2FWD: 5’ttccgtagatcttcattgaaggttgttag3’, PBleftarmREV and PBrightarmFWD, contig1.2REV: 5’ctgcaaatctttccagagattgttccag3’. To amplify the corresponding genomic DNA region from HWE mosquitoes, primer combinations contig1.2FWD and contig1.2REV were used. Site-directed recombination of the pattB-DsRed2-AeCPA/EGFP/svA donor plasmid with the attP site of line attP26 was confirmed using primer combinations attlFWD: 5’gaggtcgacgatgtaggtcac3’, attlREV: 5’accttttctcccttgctactgac3’ and attrFWD: 5’tcaaactaaggcggagtgg3’, attrREV: gatgggtgaggtggagtacg3’ (Nimmo et al., 2006).
Acknowledgements
We thank Cynthia Meredith for her assistance in mosquito maintenance and insectary management. This work was supported by RO1 grant AI073298-01 of NIH-NIAID (A.W.E.F.) and a grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative (A.A.J.).
References
- Adelman ZN, Jasinskiene N, James AA. Development and applications of transgenesis in the yellow fever mosquito, Aedes aegypti. Mol Biochem Parasitol. 2002;121:1–10. doi: 10.1016/s0166-6851(02)00028-2. [DOI] [PubMed] [Google Scholar]
- Adelman ZN, Jasinskiene N, Vally KJM, Peek C, Travanty EA, Olson KE, Brown SE, Stephens JL, Knudson DL, Coates CJ, James AA. Formation and loss of large, unstable tandem arrays of the piggyBac transposable element in the yellow fever mosquito, Aedes aegypti. Transgenic Res. 2004;13:411–425. doi: 10.1007/s11248-004-6067-2. [DOI] [PubMed] [Google Scholar]
- Adelman ZN, Jasinskiene N, Onal S, Juhn J, Ashikyan A, Salampessy M, MacCauley T, James AA. nanos gene control DNA mediates developmentally regulated transposition in the yellow fever mosquito Aedes aegypti. P Natl Acad Sci USA. 2007;104:9970–9975. doi: 10.1073/pnas.0701515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman ZN, Anderson MA, Morazzani EM, Myles KM. A transgenic sensor strain for monitoring the RNAi pathway in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2008;38:705–713. doi: 10.1016/j.ibmb.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alphey L, Nimmo D, O’Connell S, Alphey N. Insect population suppression using engineered insects. Adv Exp Med Biol. 2008;627:93–103. doi: 10.1007/978-0-387-78225-6_8. [DOI] [PubMed] [Google Scholar]
- Amenya DA, Bonizzoni M, Isaacs AT, Jasinskiene N, Chen H, Marinotti O, Yan G, James AA. Comparative fitness assessment of Anopheles stephensi transgenic lines receptive to site-specific integration. Insect Mol Biol. 2010;19:263–269. doi: 10.1111/j.1365-2583.2009.00986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MAE, Gross TL, Myles KM, Adelman ZN. Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Ae. aegypti. Insect Mol Biol. 2010;19:441–449. doi: 10.1111/j.1365-2583.2010.01005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catteruccia F, Godfray HC, Crisanti A. Impact of genetic manipulation on the fitness of Anopheles stephensi mosquitoes. Science. 2003;299:1225–1257. doi: 10.1126/science.1081453. [DOI] [PubMed] [Google Scholar]
- Catteruccia F, Crisanti A, Wimmer EA. Transgenic technologies to induce sterility. Malaria J. 2009;16:8. doi: 10.1186/1475-2875-8-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MJ, Moskalyk LA, Donelly-Doman M, Vlaskova M, Noriega FG, Walker VK, Jacobs-Lorena M. Characterization of a carboxypeptidase A gene from the mosquito, Aedes aegypti. Insect Mol Biol. 2000;9:33–38. doi: 10.1046/j.1365-2583.2000.00159.x. [DOI] [PubMed] [Google Scholar]
- Elick TA, Bauser CA, Principe NM, Fraser MJ. PCR analysis of insertion site specificity, transcription, and structural uniformity of the lepidopteran transposable element piggyBac (IFP2) in the TN-368 cell genome. Genetica. 1996;97:127–139. doi: 10.1007/BF00054620. [DOI] [PubMed] [Google Scholar]
- Fletcher SP, Muto M, Mayfield SP. Optimization of recombinant protein expression in the chloroplasts of green algae. Adv Exp Med Biol. 2007;616:90–98. doi: 10.1007/978-0-387-75532-8_8. [DOI] [PubMed] [Google Scholar]
- Franz AWE, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, Olson KE. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. P Natl Acad Sci USA. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz AWE, Sanchez-Vargas I, Piper J, Smith MR, Khoo CCH, James AA, Olson KE. Stability and loss of a virus resistance phenotype over time in transgenic mosquitoes harbouring an antiviral effector gene. Insect Mol Biol. 2009;18:661–672. doi: 10.1111/j.1365-2583.2009.00908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982;18:199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage ΦC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn C, Jaunich B, Wimmer EA. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev Genes Evol. 2000;210:623–629. doi: 10.1007/s004270000111. [DOI] [PubMed] [Google Scholar]
- Irvin N, Hoddle MS, O’Brochta DA, Carey B, Atkinson PW. Assessing fitness costs for transgenic Aedes aegypti expressing GFP marker and transposase genes. Proc Natl Acad Sci USA. 2004;101:891–896. doi: 10.1073/pnas.0305511101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-Lorena M. Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature. 2002;417:452–455. doi: 10.1038/417452a. [DOI] [PubMed] [Google Scholar]
- Jacobson JW, Medhora MM, Hartl DL. Molecular structure of a somatically unstable transposable element in Drosophila. Proc Natl Acad Sci USA. 1986;83:8684–8688. doi: 10.1073/pnas.83.22.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinskiene N, Coates CJ, Benedict MQ, Cornel AJ, Salazar Rafferty C, James AA, Collins FH. Stable transformation of the yellow fever mosquito, Aedes aegypti, with the Hermes element from the housefly. P Natl Acad Sci USA. 1998;95:3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinskiene N, Coates CJ, Ashikyan A, James AA. High efficiency, site-specific excision of a marker gene by the phage P1 cre-loxP system in the yellow fever mosquito, Aedes aegypti. Nucleic Acids Res. 2003;31:147. doi: 10.1093/nar/gng148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinskiene N, Juhn J, James AA. Microinjection of A. aegypti embryos to obtain transgenic mosquitoes. J Visual Exp. 2007;219 doi: 10.3791/219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keravala A, Lee S, Thyagarajan B, Olivares EC, Gabrovsky VE, Woodard LE, Calos MP. Mutational Derivatives of ΦC31 integrase with increased efficiency and specificity. Mol Therapy. 2009;17:112–120. doi: 10.1038/mt.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo CCH, Sanchez-Vargas I, Piper J, Olson KE, Franz AWE. The RNAi pathway affects midgut infection- and escape barriers for Sindbis virus in Aedes aegypti. BMC Microbiology. 2010;10:130. doi: 10.1186/1471-2180-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoza V, Ahmed A, Cho W-L, Jasinskiene N, James AA, Raikhel AS. Engineering blood meal-activated systemic immunity in the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci USA. 2000;97:9144–9149. doi: 10.1073/pnas.160258197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoza VA, Martin D, Mienaltowski MJ, Ahmed A, Morton CM, Raikhel AS. Transcriptional regulation of the mosquito vitellogenin gene via a blood meal-triggered cascade. Gene. 2001;274:47–65. doi: 10.1016/s0378-1119(01)00602-3. [DOI] [PubMed] [Google Scholar]
- Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol. 2003;15:15259–15265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- Labbe GMC, Nimmo DD, Alphey L. piggybac- and ΦC31-mediated genetic transformation of the Asian tiger mosquito, Ae. albopictus (Skuse) PLoS Negl Trop Dis. 2010;4(8):e788. doi: 10.1371/journal.pntd.0000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Liu D. Design of gene constructs for transgenic maize. Methods Mol Biol. 2009;526:3–20. doi: 10.1007/978-1-59745-494-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrelli MT, Moreira CK, Kelly D, Alphey L, Jacobs-Lorena M. Mosquito transgenesis: what is the fitness cost? Trends Parasitol. 2006;22:197–202. doi: 10.1016/j.pt.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Mathur G, Sanchez-Vargas I, Alvarez D, Olson KE, Marinotti O, James AA. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2010;19:753–763. doi: 10.1111/j.1365-2583.2010.01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland GAH. Sex-linkage in Aedes aegypti. Trans R Soc Trop Med Hyg. 1962;56:4. [Google Scholar]
- Meredith JM, Basu S, Nimmo DD, Larget-Thiery I, Warr EL, Underhill A, McArthur CC, Carter V, Hurd H, Bourgouin C, Eggleston P. Site-specific integration and expression of an anti-malarial gene in transgenic Anopheles gambiae significantly reduces Plasmodium infection. PLoS One. 2011;6:e14587. doi: 10.1371/journal.pone.0014587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira L, Edwards MJ, Adhami F, Jasinskiene N, James AA, Jacobs-Lorena M. Robust gut-specific gene expression in transgenic Aedes aegypti mosquitoes. P Natl Acad Sci USA. 2000;97:10895–10898. doi: 10.1073/pnas.97.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AC, Schaub TL, James AA. FLP-mediated recombination in the vector mosquito, Aedes aegypti. Nucl Acids Res. 1991;19:5895–5900. doi: 10.1093/nar/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases: status for the year 2000. Nucl Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo DD, Alphey L, Meredith JM, Eggleston P. High efficiency site-specific genetic engineering of the mosquito genome. Insect Mol Biol. 2006;15:129–136. doi: 10.1111/j.1365-2583.2006.00615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brochta DA, Sethuraman N, Wilson R, Hice RH, Pinkerton AC, Levesque CS, Bideshi DK, Jasinskiene N, Coates CJ, James AA, Lehane MJ, Atkinson PW. Gene vector and transposable element behavior in mosquitoes. J Exp Biol. 2003;206:3823–3834. doi: 10.1242/jeb.00638. [DOI] [PubMed] [Google Scholar]
- Olson KE, Franz AWE. Controlling dengue virus transmission in the field with genetically modified mosquitoes. In: Clark JM, Bloomquist J, Kawada H, editors. Advances in Human Vector Control. USA: Oxford University Press; 2009. pp. 123–141. [Google Scholar]
- Papathanos PA, Windbichler N, Menchinelli M, Burt A, Crisanti A. The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: a versatile tool for genetic strategies. BMC Molecular Biology. 2009;10:65. doi: 10.1186/1471-2199-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, MacCallum P, Russell D. Molecular Cloning: A Laboratory Manual. 3 Edition. Cold Spring Harbor Laboratory Press; NY: 2001. p. 2344. [Google Scholar]
- Sanchez-Vargas I, Scott JC, Poole BK, Franz AWE, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathogen. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Atapattu A, Belikoff EJ, Heinrich JC, Li X, Horn C, Wimmer EA, Scott MJ. Insulated piggyBac vectors for insect transgenesis. BMC Biotechnol. 2006;6:27. doi: 10.1186/1472-6750-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SW, Kokoza VA, Raikhel AS. Transgenesis and reverse genetics of mosquito innate immunity. J Exp Biol. 2003;206:3835–3843. doi: 10.1242/jeb.00640. [DOI] [PubMed] [Google Scholar]
- Thorpe HM, Smith MCM. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath LL, Elgin CR. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Develop. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- Wang HH, Fraser MJ. TTAA serves as a target site for TFP3 lepidopteran insertions in both nuclear polyhedrosis virus and Trichoplusia ni genomes. Insect Mol Biol. 1993;1:109–116. doi: 10.1111/j.1365-2583.1993.tb00111.x. [DOI] [PubMed] [Google Scholar]
- Wendell MD, Wilson TG, Higgs S, Black WC., IV Chemical and gamma-ray mutagenesis of the white gene in Aedes aegypti. Insect Mol Biol. 2000;9:119–125. doi: 10.1046/j.1365-2583.2000.00166.x. [DOI] [PubMed] [Google Scholar]