Abstract
A challenging goal in neuroscience is to be able to read out, or decode, mental content from brain activity. Recent functional magnetic resonance imaging (fMRI) studies have decoded orientation1,2, position3, and object category4,5 from activity in visual cortex. However, these studies typically used relatively simple stimuli (e.g. gratings) or images drawn from fixed categories (e.g. faces, houses), and decoding was based on prior measurements of brain activity evoked by those same stimuli or categories. To overcome these limitations, we develop a decoding method based on quantitative receptive field models that characterize the relationship between visual stimuli and fMRI activity in early visual areas. These models describe the tuning of individual voxels for space, orientation, and spatial frequency, and are estimated directly from responses evoked by natural images. We show that these receptive field models make it possible to identify, from a large set of completely novel natural images, which specific image was seen by an observer. Identification is not a mere consequence of the retinotopic organization of visual areas; simpler receptive field models that describe only spatial tuning yield much poorer identification performance. Our results suggest that it may soon be possible to reconstruct a picture of a person’s visual experience from brain activity measurements alone.
Keywords: decoding, brain reading, receptive field, Gabor wavelets, visual cortex, fMRI
Imagine a general brain-reading device that could reconstruct a picture of a person’s visual experience at any moment in time6. This general visual decoder would have great scientific and practical utility. For example, we could use the decoder to investigate differences in perception across people, to study covert mental processes such as attention, and perhaps even to access the visual content of purely mental phenomena such as dreams and imagery. The decoder would also serve as a useful benchmark of our understanding of how the brain represents sensory information.
How do we build a general visual decoder? We consider as a first step the problem of image identification3,7,8. This problem is analogous to the classic “pick a card, any card” magic trick: We begin with a large, arbitrary set of images. The observer picks an image from the set and views it while brain activity is measured. Is it possible to use the measured brain activity to identify which specific image was seen?
To ensure that a solution to the image identification problem will be applicable to general visual decoding, we introduce two challenging requirements6. First, it must be possible to identify novel images. Conventional classification-based decoding methods can be used to identify images if brain activity evoked by those images has been measured previously, but they cannot be used to identify novel images (see Supplementary Discussion). Second, it must be possible to identify natural images. Natural images have complex statistical structure9 and are much more difficult to parameterize than simple artificial stimuli such as gratings or pre-segmented objects. Because neural processing of visual stimuli is nonlinear, a decoder that can identify simple stimuli may fail when confronted with complex natural images.
Our experiment consisted of two stages (Fig. 1). In the first stage, model estimation, fMRI data were recorded from visual areas V1, V2, and V3 while each subject viewed 1,750 natural images. These data were used to estimate a quantitative receptive field model10 for each voxel (Fig. 2). The model was based on a Gabor wavelet pyramid11,13 and described tuning along the dimensions of space3,14,19, orientation1,2,20, and spatial frequency21,22. (See Supplementary Discussion for a comparison of our receptive field analysis to those of previous studies.)
Fig. 1. Schematic of experiment.
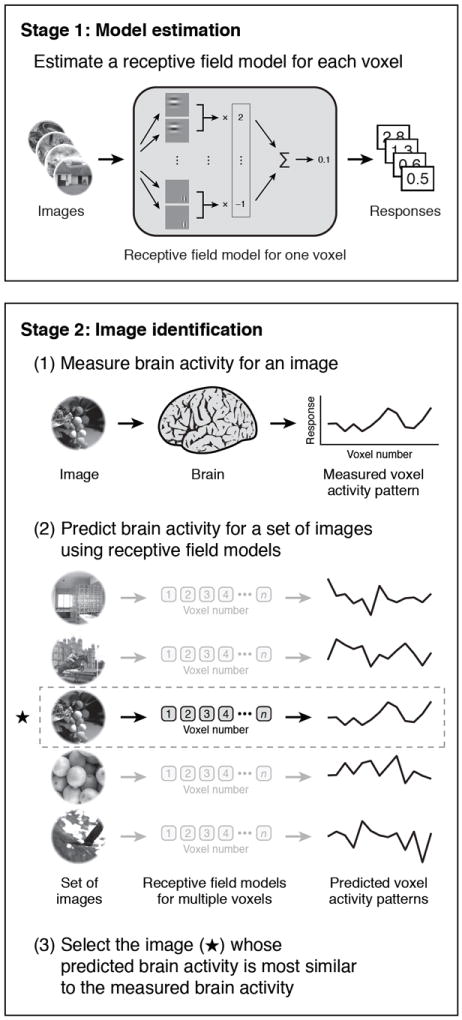
The experiment consisted of two stages. In the first stage, model estimation, fMRI data were recorded while each subject viewed a large collection of natural images. These data were used to estimate a quantitative receptive field model10 for each voxel. The model was based on a Gabor wavelet pyramid11,13 and described tuning along the dimensions of space3,14,19, orientation1,2,20, and spatial frequency21,22. In the second stage, image identification, fMRI data were recorded while each subject viewed a collection of novel natural images. For each measurement of brain activity, we attempted to identify which specific image had been seen. This was accomplished by using the estimated receptive field models to predict brain activity for a set of potential images and then selecting the image whose predicted activity most closely matches the measured activity.
Fig. 2. Receptive field model for a representative voxel.
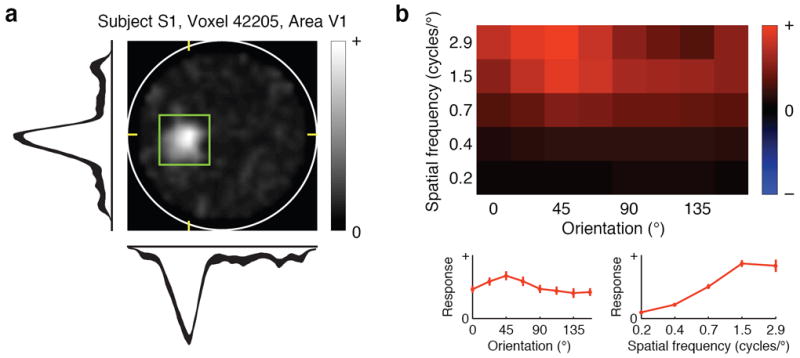
a, Spatial envelope. The intensity of each pixel indicates the sensitivity of the receptive field (RF) to that location. The white circle delineates the bounds of the stimulus (20° × 20°) and the green square delineates the estimated RF location. Horizontal and vertical slices through the spatial envelope are shown below and to the left. These intersect the peak of the spatial envelope, as indicated by yellow tick marks. The thickness of each slice profile indicates ± 1 s.e.m. This RF is located in the left hemifield, just below the horizontal meridian. b, Orientation and spatial frequency tuning curves. The top matrix depicts the joint orientation and spatial frequency tuning of the RF, and the bottom two plots give the marginal orientation and spatial frequency tuning curves. Error bars indicate ± 1 s.e.m. This RF has broadband orientation tuning and high-pass spatial frequency tuning. For additional RF examples and population summaries of RF properties, see Supplementary Figs. 9 11.
In the second stage, image identification, fMRI data were recorded while each subject viewed 120 novel natural images. This yielded 120 distinct voxel activity patterns for each subject. For each voxel activity pattern we attempted to identify which image had been seen. To do this, the receptive field models estimated in the first stage of the experiment were used to predict the voxel activity pattern that would be evoked by each of the 120 images. The image whose predicted voxel activity pattern was most correlated (Pearson’s r) with the measured voxel activity pattern was selected.
Identification performance for one subject is illustrated in Fig. 3. For this subject 92% (110/120) of the images were identified correctly (subject S1), while chance performance is just 0.8% (1/120). For a second subject 72% (86/120) of the images were identified correctly (subject S2). These high performance levels demonstrate the validity of our decoding approach, and indicate that our receptive field models accurately characterize the selectivity of individual voxels to natural images.
Fig. 3. Identification performance.
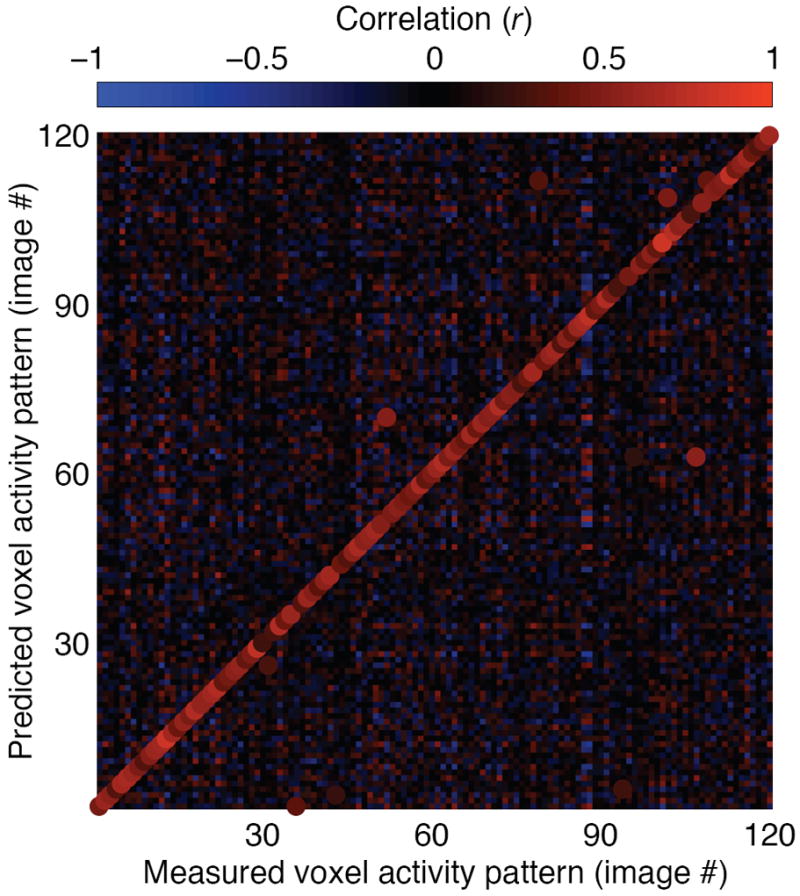
In the image identification stage of the experiment, fMRI data were recorded while each subject viewed 120 novel natural images that had not been used to estimate the receptive field models. For each of the 120 measured voxel activity patterns we attempted to identify which image had been seen. This figure illustrates identification performance for one subject (S1). The color at the mth column and nth row represents the correlation between the measured voxel activity pattern for the mth image and the predicted voxel activity pattern for the nth image. The highest correlation in each column is designated by an enlarged dot of the appropriate color, and indicates the image selected by the identification algorithm. For this subject 92% (110/120) of the images were identified correctly.
A general visual decoder would be especially useful if it could operate on brain activity evoked by a single perceptual event. However, because fMRI data are noisy the results reported above were obtained using voxel activity patterns averaged across 13 repeated trials. We therefore attempted identification using voxel activity patterns from single trials. Single-trial performance was 51% (834/1620) and 32% (516/1620) for subjects S1 and S2, respectively (Fig. 4a); once again, chance performance is just 0.8% (13.5/1620). These results suggest that it may be feasible to decode the content of perceptual experiences in real-time7,23.
Fig. 4. Factors that impact identification performance.
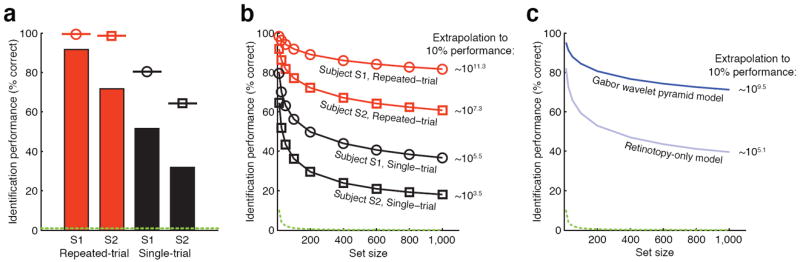
a, Summary of identification performance. The bars indicate empirical performance for a set size of 120 images, the marker above each bar indicates the estimated noise ceiling (i.e. the theoretical maximum performance given the level of noise in the data), and the dashed green line indicates chance performance. The noise ceiling estimates suggest that the difference in performance across subjects is due to intrinsic differences in the level of noise. b, Scaling of identification performance with set size. The x-axis indicates set size, the y-axis indicates identification performance, and the number to the right of each line gives the estimated set size at which performance declines to 10% correct. In all cases performance scaled very well with set size. c, Retinotopy-only model versus Gabor wavelet pyramid model. Identification was attempted using an alternative retinotopy-only model that captures only the location and size of each voxel’s receptive field. This model performed substantially worse than the Gabor wavelet pyramid model, indicating that spatial tuning alone is insufficient to achieve optimal identification performance. (Results reflect repeated-trial performance averaged across subjects; see Supplementary Fig. 5 for detailed results.)
We have so far demonstrated identification of a single image drawn from a set of 120 images, but a general visual decoder should be able to handle much larger sets of images. To investigate this issue we measured identification performance for various set sizes up to 1,000 images (Fig. 4b). As set size increased 10-fold from 100 to 1,000, performance only declined slightly, from 92% to 82% (subject S1, repeated-trial). Extrapolation of these measurements (see Supplementary Methods) suggests that performance for this subject would remain above 10% even up to a set size of ~1011.3 images. This is more than 100 times larger than the number of images currently indexed by Google (~108.9 images; source: http://www.google.com/whatsnew/, June 4, 2007).
Early visual areas are organized retinotopically, and voxels are known to reflect this organization14,16,18. Could our results be a mere consequence of retinotopy? To answer this question we attempted identification using an alternative model that captures the location and size of each voxel’s receptive field but discards orientation and spatial frequency information (Fig. 4c). Performance for this retinotopy-only model declined to 10% correct at a set size of just ~105.1 images, whereas performance for the Gabor wavelet pyramid model did not decline to 10% correct until ~109.5 images were included in the set (repeated-trial, performance extrapolated and averaged across subjects). This result indicates that spatial tuning alone does not yield optimal identification performance; identification improves substantially when orientation and spatial frequency tuning are included in the model.
To further investigate the impact of orientation and spatial frequency tuning, we measured identification performance after imposing constraints on the orientation and spatial frequency tuning of the Gabor wavelet pyramid model (Supplementary Fig. 8). The results indicate that both orientation and spatial frequency tuning contribute to identification performance, but that the latter makes the larger contribution. This is consistent with recent studies demonstrating that voxels have only slight orientation bias1,2. We also find that voxel-to-voxel variation in orientation and spatial frequency tuning contributes to identification performance. This reinforces the growing realization in the fMRI community that information may be present in fine-grained patterns of voxel activity6.
To be practical our identification algorithm must perform well even when brain activity is measured long after estimation of the receptive field models. To assess performance over time2,4,6,23 we attempted identification for a set of 120 novel natural images that were seen approximately two months after the initial experiment. In this case 82% (99/120) of the images were identified correctly (chance performance 0.8%; subject S1, repeated-trial). We also evaluated identification performance for a set of 12 novel natural images that were seen more than a year after the initial experiment. In this case 100% (12/12) of the images were identified correctly (chance performance 8%; subject S1, repeated-trial). These results demonstrate that the stimulus-related information that can be decoded from voxel activity remains largely stable over time.
Why does identification sometimes fail? Inspection revealed that identification errors tended to occur when the selected image was visually similar to the correct image. This suggests that noise in measured voxel activity patterns causes the identification algorithm to confuse images that have similar features.
Functional MRI signals have modest spatial resolution and reflect hemodynamic activity that is only indirectly coupled to neural activity24,25. Despite these limitations we have shown that fMRI signals can be used to achieve remarkable levels of identification performance. This indicates that fMRI signals contain a considerable amount of stimulus-related information4 and that this information can be successfully decoded in practice.
Identification of novel natural images brings us close to achieving a general visual decoder. The final step will require devising a way to reconstruct the image seen by the observer, instead of selecting the image from a known set. Stanley and co-workers26 reconstructed natural movies by modeling the luminance of individual image pixels as a linear function of single-unit activity in cat LGN. This approach assumes a linear relationship between luminance and the activity of the recorded units, but this condition does not hold in fMRI27,28.
An alternative approach to reconstruction is to incorporate receptive field models into a statistical inference framework. In such a framework, receptive field models are used to infer the most likely image given a measured activity pattern. This model-based approach has a long history in both theoretical and experimental neuroscience29,30. Recently, Thirion and co-workers3 used it to reconstruct spatial maps of contrast from fMRI activity in human visual cortex. The success of the approach depends critically on how well the receptive field models predict brain activity. The present study demonstrates that our receptive field models have sufficient predictive power to enable identification of novel natural images, even for the case of extremely large sets of images. We are therefore optimistic that the model-based approach will make possible the reconstruction of natural images from human brain activity.
Methods
Stimuli
The stimuli consisted of sequences of natural photos. Photos were obtained from a commercial digital library (Corel Stock Photo Libraries from Corel Corporation, Ontario, Canada), the Berkeley Segmentation Dataset (http://www.eecs.berkeley.edu/Research/Projects/CS/vision/grouping/segbench/), and the authors’ personal collections. The content of the photos included animals, buildings, food, humans, indoor scenes, manmade objects, outdoor scenes, and textures. Photos were converted to grayscale, downsampled so that the smaller of the two image dimensions was 500 px, linearly transformed so that the 1/10th and 99 9/10th percentiles of the original pixel values were mapped to the minimum (0) and maximum (255) pixel values, cropped to the central 500 px × 500 px, masked with a circle, and placed on a gray background (Supplementary Fig. 1a). The luminance of the background was set to the mean luminance across photos, and the outer edge of each photo (10% of the radius of the circular mask) was linearly blended into the background.
The size of the photos was 20° × 20° (500 px × 500 px). A central white square served as the fixation point, and its size was 0.2° × 0.2° (4 px × 4 px). Photos were presented in successive 4-s trials; in each trial, a photo was presented for 1 s and the gray background was presented for 3 s. Each 1-s presentation consisted of a photo being flashed ON-OFF-ON-OFF-ON where ON corresponds to presentation of the photo for 200 ms and OFF corresponds to presentation of the gray background for 200 ms (Supplementary Fig. 1b). The flashing technique increased the signal-to-noise ratio of voxel responses relative to that achieved by presenting each photo continuously for 1 s (data not shown).
Visual stimuli were delivered using the VisuaStim goggles system (Resonance Technology, Northridge, CA). The display resolution was 800 × 600 at 60 Hz. A PowerBook G4 computer (Apple Computer, Cupertino, CA) controlled stimulus presentation using software written in MATLAB 5.2.1 (The Mathworks, Natick, MA) and Psychophysics Toolbox 2.53 (http://psychtoolbox.org).
MRI parameters
The experimental protocol was approved by the UC-Berkeley Committee for the Protection of Human Subjects. MRI data were collected at the Brain Imaging Center at UC-Berkeley using a 4 T INOVA MR scanner (Varian, Inc., Palo Alto, CA) and a quadrature transmit/receive surface coil (Midwest RF, LLC, Hartland, WI). Data were acquired using coronal slices that covered occipital cortex: 18 slices, slice thickness 2.25 mm, slice gap 0.25 mm, field-of-view 128 × 128 mm2. (In one scan session, a slice gap of 0.5 mm was used.) For functional data, a T2*-weighted, single-shot, slice-interleaved, gradient-echo EPI pulse sequence was used: matrix size 64 × 64, TR 1 s, TE 28 ms, flip angle 20°. The nominal spatial resolution of the functional data was 2 × 2 × 2.5 mm3. For anatomical data, a T1-weighted gradient-echo multislice sequence was used: matrix size 256 × 256, TR 0.2 s, TE 5 ms, flip angle 40°.
Data collection
Data for the model estimation and image identification stages of the experiment were collected in the same scan sessions. Two subjects were used: S1 (author T.N., age 33) and S2 (author K.N.K., age 25). Subjects were healthy and had normal or corrected-to-normal vision.
Five scan sessions of data were collected from each subject. Each scan session consisted of five model estimation runs and two image identification runs. Model estimation runs (11 min each) were used for the model estimation stage of the experiment. Each model estimation run consisted of 70 distinct images presented 2 times each. Image identification runs (12 min each) were used for the image identification stage of the experiment. Each image identification run consisted of 12 distinct images presented 13 times each. Images were randomly selected for each run and were mutually exclusive across runs. The total number of distinct images used in the model estimation and image identification runs was 1,750 and 120, respectively. (For additional details on experimental design, see Supplementary Methods.)
Three additional scan sessions of data were collected from subject S1. Two of these were held ~2 months after the main experiment, and consisted of five image identification runs each. The third was held ~14 months after the main experiment, and consisted of one image identification run. The images used in these additional scan sessions were randomly selected and were distinct from the images used in the main experiment.
Data preprocessing
Functional brain volumes were reconstructed and then coregistered to correct differences in head positioning within and across scan sessions. Next, voxel-specific response timecourses were estimated and deconvolved from the time-series data. This produced, for each voxel, an estimate of the amplitude of the response (a single value) to each image used in the model estimation and image identification runs. Finally, voxels were assigned to visual areas based on retinotopic mapping data17 collected in separate scan sessions. (Details on these procedures are given in Supplementary Methods.)
Model estimation
A receptive field model was estimated for each voxel based on its responses to the images used in the model estimation runs. The model was based on a Gabor wavelet pyramid11,13. In the model each image is represented by a set of Gabor wavelets differing in size, position, orientation, spatial frequency, and phase (Supplementary Fig. 2). The predicted response is a linear function of the contrast energy contained in quadrature wavelet pairs (Supplementary Fig. 3). Because contrast energy is a nonlinear quantity, this is a linearized model10. The model was able to characterize responses of voxels in visual areas V1, V2, and V3 (Supplementary Table 1) but it did a poor job of characterizing responses in higher visual areas such as V4.
Alternative receptive field models were also used, including the retinotopy-only model and several constrained versions of the Gabor wavelet pyramid model. Details on these models and model estimation procedures are given in Supplementary Methods.
Image identification
Voxel activity patterns were constructed from voxel responses evoked by the images used in the image identification runs. For each voxel activity pattern, the estimated receptive field models were used to identify which specific image had been seen. The identification algorithm is described in the main text. See Supplementary Fig. 4 and Supplementary Methods for details concerning voxel selection, performance for different set sizes, and noise ceiling estimation. See Supplementary Discussion for a comparison of identification to the decoding problems of classification and reconstruction.
Supplementary Material
Acknowledgments
This work was supported by an NDSEG fellowship (K.N.K.), NIH, and UC-Berkeley intramural funds. We thank B. Inglis for MRI assistance, K. Hansen for retinotopic mapping assistance, D. Woods and X. Kang for acquisition of whole-brain anatomical data, and A. Rokem for assistance with scanner operation. We also thank C. Baker, M. D’Esposito, R. Ivry, A. Landau, M. Merolle, F. Theunissen, and the anonymous referees for comments on the manuscript. Finally, we thank S. Nishimoto, R. Redfern, K. Schreiber, B. Willmore, and B. Yu for their help in various aspects of this research.
Footnotes
Author Contributions K.N.K. designed and conducted the experiment and was first author on the paper. K.N.K. and T.N. analyzed the data. R.J.P. provided mathematical ideas and assistance. J.L.G. provided guidance on all aspects of the project. All authors discussed the results and commented on the manuscript.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Haynes JD, Rees G. Predicting the orientation of invisible stimuli from activity in human primary visual cortex. Nature Neurosci. 2005;8:686–691. doi: 10.1038/nn1445. [DOI] [PubMed] [Google Scholar]
- 2.Kamitani Y, Tong F. Decoding the visual and subjective contents of the human brain. Nature Neurosci. 2005;8:679–685. doi: 10.1038/nn1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thirion B, et al. Inverse retinotopy: Inferring the visual content of images from brain activation patterns. Neuroimage. 2006;33:1104–1116. doi: 10.1016/j.neuroimage.2006.06.062. [DOI] [PubMed] [Google Scholar]
- 4.Cox DD, Savoy RL. Functional magnetic resonance imaging (fMRI) “brain reading”: Detecting and classifying distributed patterns of fMRI activity in human visual cortex. Neuroimage. 2003;19:261–270. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 5.Haxby JV, et al. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 2001;293:2425–2430. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- 6.Haynes JD, Rees G. Decoding mental states from brain activity in humans. Nature Rev Neurosci. 2006;7:523–534. doi: 10.1038/nrn1931. [DOI] [PubMed] [Google Scholar]
- 7.Hung CP, Kreiman G, Poggio T, DiCarlo JJ. Fast readout of object identity from macaque inferior temporal cortex. Science. 2005;310:863–866. doi: 10.1126/science.1117593. [DOI] [PubMed] [Google Scholar]
- 8.Tsao DY, Freiwald WA, Tootell RB, Livingstone MS. A cortical region consisting entirely of face-selective cells. Science. 2006;311:670–674. doi: 10.1126/science.1119983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simoncelli EP, Olshausen BA. Natural image statistics and neural representation. Annu Rev Neurosci. 2001;24:1193–1216. doi: 10.1146/annurev.neuro.24.1.1193. [DOI] [PubMed] [Google Scholar]
- 10.Wu MC, David SV, Gallant JL. Complete functional characterization of sensory neurons by system identification. Annu Rev Neurosci. 2006;29:477–505. doi: 10.1146/annurev.neuro.29.051605.113024. [DOI] [PubMed] [Google Scholar]
- 11.Daugman JG. Uncertainty relation for resolution in space, spatial frequency, and orientation optimized by two-dimensional visual cortical filters. J Opt Soc Am A. 1985;2:1160–1169. doi: 10.1364/josaa.2.001160. [DOI] [PubMed] [Google Scholar]
- 12.Jones JP, Palmer LA. An evaluation of the two-dimensional Gabor filter model of simple receptive fields in cat striate cortex. J Neurophysiol. 1987;58:1233–1258. doi: 10.1152/jn.1987.58.6.1233. [DOI] [PubMed] [Google Scholar]
- 13.Lee TS. Image representation using 2D Gabor wavelets. IEEE Trans Pattern Anal. 1996;18:959–971. [Google Scholar]
- 14.DeYoe EA, et al. Mapping striate and extrastriate visual areas in human cerebral cortex. Proc Natl Acad Sci USA. 1996;93:2382–2386. doi: 10.1073/pnas.93.6.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumoulin SO, Wandell BA. Population receptive field estimates in human visual cortex. Neuroimage. 2008;39:647–660. doi: 10.1016/j.neuroimage.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engel SA, et al. fMRI of human visual cortex. Nature. 1994;369:525. doi: 10.1038/369525a0. [DOI] [PubMed] [Google Scholar]
- 17.Hansen KA, David SV, Gallant JL. Parametric reverse correlation reveals spatial linearity of retinotopic human V1 BOLD response. Neuroimage. 2004;23:233–241. doi: 10.1016/j.neuroimage.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Sereno MI, et al. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 1995;268:889–893. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- 19.Smith AT, Singh KD, Williams AL, Greenlee MW. Estimating receptive field size from fMRI data in human striate and extrastriate visual cortex. Cereb Cortex. 2001;11:1182–1190. doi: 10.1093/cercor/11.12.1182. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki Y, et al. The radial bias: A different slant on visual orientation sensitivity in human and nonhuman primates. Neuron. 2006;51:661–670. doi: 10.1016/j.neuron.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 21.Olman CA, Ugurbil K, Schrater P, Kersten D. BOLD fMRI and psychophysical measurements of contrast response to broadband images. Vision Res. 2004;44:669–683. doi: 10.1016/j.visres.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Singh KD, Smith AT, Greenlee MW. Spatiotemporal frequency and direction sensitivities of human visual areas measured using fMRI. Neuroimage. 2000;12:550–564. doi: 10.1006/nimg.2000.0642. [DOI] [PubMed] [Google Scholar]
- 23.Haynes JD, Rees G. Predicting the stream of consciousness from activity in human visual cortex. Curr Biol. 2005;15:1301–1307. doi: 10.1016/j.cub.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 24.Heeger DJ, Ress D. What does fMRI tell us about neuronal activity? Nature Rev Neurosci. 2002;3:142–151. doi: 10.1038/nrn730. [DOI] [PubMed] [Google Scholar]
- 25.Logothetis NK, Wandell BA. Interpreting the BOLD signal. Annu Rev Physiol. 2004;66:735–769. doi: 10.1146/annurev.physiol.66.082602.092845. [DOI] [PubMed] [Google Scholar]
- 26.Stanley GB, Li FF, Dan Y. Reconstruction of natural scenes from ensemble responses in the lateral geniculate nucleus. J Neurosci. 1999;19:8036–8042. doi: 10.1523/JNEUROSCI.19-18-08036.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes JD, Lotto RB, Rees G. Responses of human visual cortex to uniform surfaces. Proc Natl Acad Sci USA. 2004;101:4286–4291. doi: 10.1073/pnas.0307948101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rainer G, Augath M, Trinath T, Logothetis NK. Nonmonotonic noise tuning of BOLD fMRI signal to natural images in the visual cortex of the anesthetized monkey. Curr Biol. 2001;11:846–854. doi: 10.1016/s0960-9822(01)00242-1. [DOI] [PubMed] [Google Scholar]
- 29.Salinas E, Abbott LF. Vector reconstruction from firing rates. J Comput Neurosci. 1994;1:89–107. doi: 10.1007/BF00962720. [DOI] [PubMed] [Google Scholar]
- 30.Zhang K, Ginzburg I, McNaughton BL, Sejnowski TJ. Interpreting neuronal population activity by reconstruction: Unified framework with application to hippocampal place cells. J Neurophysiol. 1998;79:1017–1044. doi: 10.1152/jn.1998.79.2.1017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


