Abstract
Approximate Bayesian computation has become an essential tool for the analysis of complex stochastic models when the likelihood function is numerically unavailable. However, the well-established statistical method of empirical likelihood provides another route to such settings that bypasses simulations from the model and the choices of the approximate Bayesian computation parameters (summary statistics, distance, tolerance), while being convergent in the number of observations. Furthermore, bypassing model simulations may lead to significant time savings in complex models, for instance those found in population genetics. The Bayesian computation with empirical likelihood algorithm we develop in this paper also provides an evaluation of its own performance through an associated effective sample size. The method is illustrated using several examples, including estimation of standard distributions, time series, and population genetics models.
Keywords: autoregressive models, Bayesian statistics, likelihood-free methods, coalescent model
Bayesian statistical inference cannot easily operate when the likelihood function associated with the data is not entirely known, or cannot be computed in a manageable time, as is the case in most population genetic models (1–3). The fundamental reason for this difficulty with population genetics is that the statistical model associated with coalescent data needs to integrate over trees of high complexity. Similar computational issues with the likelihood function often occur in hidden Markov and other dynamic models (4). In those settings, traditional approximation tools based on stochastic simulation (5) are unavailable or unreliable. Indeed, the complexity of the latent structure defining the likelihood makes simulation of such structures too unstable to be trusted. Such settings call for alternative and often cruder approximations. The approximate Bayesian computation (ABC) methodology (1, 6) is a popular solution that bypasses the computation of the likelihood function (surveys in refs. 7 and 8); ref. 9 validates a conditional version of ABC that applies to hierarchical Bayes models in a wide generality.
The fast and polytomous development of the ABC algorithm is indicated by the rising literature in the domain, at both the methodological and the application levels. For instance, a whole new area of population genetic modeling (8, 10) has been explored thanks to the availability of such methods. However, practitioners and theoreticians both show a reluctance in adopting ABC, with some doubt about the validity of the method (11–13). We propose in this paper to supplement the ABC approach with a generic and convergent likelihood approximation called the empirical likelihood that validates this Bayesian computational technique as a convergent inferential method when the number of observations grows to infinity. The empirical likelihood perspective, introduced by ref. 14, is a robust statistical approach that does not require the specification of the likelihood function. However, although it does not appear to have been used before in the settings that now rely on ABC, this data analysis method also is a broadly (albeit not universally) applicable and often fast approach, the approximation of which differs from the one found in ABC algorithms, even though both are rooted in nonparametric statistics. Therefore, this methodology can be used both as a solution per se and as a benchmark against which to test the ABC output in many cases. This paper presents the Bayesian computation via empirical likelihood (BCel) algorithm and illustrates its performances on selected representative examples, comparing the outcome with the true posterior density whenever available, and with an ABC approximation (15) otherwise.
Statistical Methods
ABC Algorithm.
The primary purpose of the ABC algorithm is to approximate simulation from the centerpiece of Bayesian inference, the posterior distribution 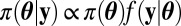 , when it cannot be numerically computed but when the distributions corresponding to both the prior π and the likelihood f can be simulated by manageable computer devices. The original (6) ABC algorithm is as follows: given a sample y of observations from the sample space, a sample of parameters
, when it cannot be numerically computed but when the distributions corresponding to both the prior π and the likelihood f can be simulated by manageable computer devices. The original (6) ABC algorithm is as follows: given a sample y of observations from the sample space, a sample of parameters  is produced by
is produced by
Algorithm 1: ABC sampler.
for i = 1 to M do
repeat
Generate 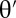 from the prior distribution π(⋅)
from the prior distribution π(⋅)
Generate z from the likelihood 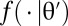
until 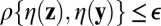
set 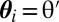 ,
,
end for
The parameters of the ABC algorithm are the summary statistic η, the distance 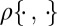 , and the tolerance level
, and the tolerance level 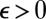 . The basic justification of the ABC approximation is that, when using a sufficient statistic η, the distribution of the
. The basic justification of the ABC approximation is that, when using a sufficient statistic η, the distribution of the 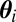 ’s in the output of the algorithm converges to the genuine posterior distribution when ε goes to zero (16).
’s in the output of the algorithm converges to the genuine posterior distribution when ε goes to zero (16).
In practice, however, the statistic η is nonsufficient and at best the approximation then converges to the genuine posterior 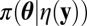 when ɛ goes to zero. This loss of information seems to be a necessary price to pay for the access to computable quantities. Furthermore, as argued below, it can be evaluated against the empirical likelihood approximation when the latter is available. Indeed, this approach does not require an information reduction through the choice of a tolerance zone or of a nonsufficient summary statistic.
when ɛ goes to zero. This loss of information seems to be a necessary price to pay for the access to computable quantities. Furthermore, as argued below, it can be evaluated against the empirical likelihood approximation when the latter is available. Indeed, this approach does not require an information reduction through the choice of a tolerance zone or of a nonsufficient summary statistic.
Empirical Likelihood.
Owen (14) developed empirical likelihood techniques as a robust alternative to classical likelihood approaches. He demonstrated that, for some categories of statistical models, this approach inherited the convergence properties of standard likelihood at a much lower cost in assumptions about the model (as detailed in SI Text). Whereas ABC algorithms do require a fully defined and often complex (hence debatable) statistical model, we argue that one should take advantage of the approximation device provided by the empirical likelihood to overcome most of the calibration difficulties encountered by ABC, at least as a convenient benchmark against which to test ABC solutions.
Assume that the dataset y is composed of n independent replicates 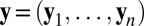 of some random vector Y with density f. Rather than defining the likelihood from the density f as usual, the empirical likelihood method starts by defining parameters of interest θ as functionals of f, for instance as moments of f, and it then profiles a nonparametric likelihood. More precisely, given a set of constraints of the form
of some random vector Y with density f. Rather than defining the likelihood from the density f as usual, the empirical likelihood method starts by defining parameters of interest θ as functionals of f, for instance as moments of f, and it then profiles a nonparametric likelihood. More precisely, given a set of constraints of the form
where the dimension of h sets the number of constraints unequivocally defining θ, the empirical likelihood is defined as
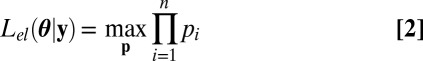 |
for p in the set 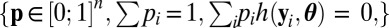 . For instance, in the one-dimensional case when
. For instance, in the one-dimensional case when 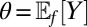 , the empirical likelihood in θ is the maximum of the product
, the empirical likelihood in θ is the maximum of the product  under the constraint
under the constraint 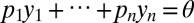 . (Solving 2 is done using the R package “emplik” developed by ref. 17 and based on the Newton–Lagrange algorithm.) When the data are not independent and identically distributed (iid), an underlying iid structure may sometimes be exploited, as illustrated in Dynamic Model. However, this is not always the case, meaning that the empirical likelihood method remains out of reach in some complex cases when ABC can still be implemented. Furthermore, as pointed out in SI Text by a quote from Owen (18), the validation of the approach depends on a choice of the set of constraints that ensures convergence.
. (Solving 2 is done using the R package “emplik” developed by ref. 17 and based on the Newton–Lagrange algorithm.) When the data are not independent and identically distributed (iid), an underlying iid structure may sometimes be exploited, as illustrated in Dynamic Model. However, this is not always the case, meaning that the empirical likelihood method remains out of reach in some complex cases when ABC can still be implemented. Furthermore, as pointed out in SI Text by a quote from Owen (18), the validation of the approach depends on a choice of the set of constraints that ensures convergence.
Whereas the convergence of the empirical likelihood is well-established (SI Text and ref. 18), the Bayesian use of empirical likelihoods has been little examined in the past, apart from a Monte Carlo study in ref. 19, and a probabilistic one in ref. 20.
BCel.
The most natural use of the empirical likelihood approximation is to act as if this representation was an exact likelihood, as in ref. 19. Incorporating this perspective into a basic sampler leads to the following algorithm:
Algorithm 2: Basic BCel sampler.
for i = 1 to M do
Generate θi from the prior distribution π(⋅)
Set the weight 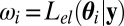
end for
The output of BCel is a sample of size M of parameters with associated weights, which operate as an importance sampling output (5). Thus, the performance of the algorithm can be evaluated through the effective sample size (ESS):
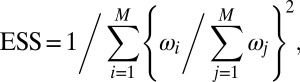 |
which approximates the size of an iid sample with the same variance as the original sample. As shown in ref. 21, this quantity is always between 1 (corresponding to a very poor outcome) and M (corresponding to an iid perfect outcome).
Any algorithm that samples from a posterior distribution (e.g., Markov chain Monte Carlo, population Monte Carlo, sequential Monte Carlo algorithms, ref. 5) may instead use the empirical likelihood as a proxy to the exact likelihood. For instance, to speed up the computation in the population genetics model introduced below, we resorted to the adaptive multiple importance sampling (AMIS, ref. 22), which is easy to parallelize on a multicore computer. Although the original target distribution is 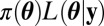 and the AMIS algorithm uses several (multivariate) Student’s t distributions denoted
and the AMIS algorithm uses several (multivariate) Student’s t distributions denoted 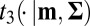 (i.e., with three degrees of freedom, centered at mean m, and with covariance matrix Σ) as an importance sampling distribution, the algorithm can be adapted to the empirical likelihood in a straightforward manner:
(i.e., with three degrees of freedom, centered at mean m, and with covariance matrix Σ) as an importance sampling distribution, the algorithm can be adapted to the empirical likelihood in a straightforward manner:
Algorithm 3: BCel–AMIS sampler.
for i = 1 to M do
Generate 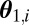 from the prior distribution
from the prior distribution 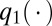
Set 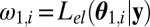
end for
for t = 2 to TM do
Compute weighted mean mt and weighted variance matrix 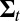 of the
of the 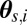 (
(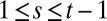 ,
, 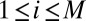 ).
).
Denote 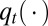 the density of
the density of 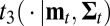 .
.
for i = 1 to M do
Generate 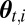 from
from 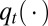 .
.
Set 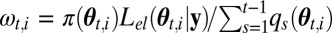
end for
for r = 1 to t − 1 do
for i = 1 to M do
Update the weight of 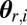 as
as
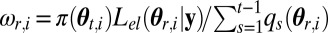
end for
end for
end for
The output is thus a weighted sample 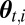 of size
of size 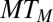 .
.
In contrast with ABC, BCel algorithms do not usually require simulations from the sampling model, given that 2 provides a converging and nonparametric approximation of the likelihood function. This feature thus induces very significative improvements in computing time when the production of pseudodatasets is time-consuming, because solving 1 is usually immediate. This is for instance the case in population genetics and Time Gains in Population Genetic Models provides an illustration of a huge improvement in comparison with ABC in two experiments described below. However, the improvement in speed may vanish in cases when producing an iid structure connected with the constraint 1 requires simulations from the sampling model, as illustrated by a counterexample for point processes in SI Text, BCel, and ABC, then breaking even in terms of computing time. Even though the computing time required by BCel is customarily negligible compared with ABC (or does not induce any extra time as in the point process counterexample), we further caution against opposing both approaches solely based on computing times, because they differ in the approximations they provide to a genuine Bayesian analysis and thus should be used in conjunction.
Using empirical likelihoods means there is no calibration of the many tuning parameters of ABC algorithms; most significantly, the likelihood ratio acts as a natural distance and importance weights produce an implicit and self-defined quantile on the original sample simulated from the prior. Notwithstanding these appealing qualities, BCel still requires calibration, in particular in the choices of the parametrization of the sampling distribution and of the corresponding constraints 1 defining the empirical likelihood. Some examples are discussed below. The BCel – AMIS sampler also implies computing values of the prior density, up to a constant, which may be a hindrance in peculiar cases.
Composite Likelihood in Population Genetics.
ABC was introduced by population geneticists (2, 6, 10) interested in statistical inference about the evolutionary history of species, as no likelihood-based approach existed apart from very rudimentary and hence unrealistic situations. This approach has been used in a number of biological studies (23–25), most of them including model choice. It is therefore crucial to obtain insights into the validity of such studies, particularly when they have economic, biological, or ecological consequences (e.g., ref. 26). This can be achieved in part by running a comparison using BCel. Furthermore, given the major gain in computing time due to the absence of replications of the data, BCel can be applied to more complex biological models.
The main caveat when using the empirical likelihood in such settings is selecting a constraint 1 on the parameter of interest: In phylogeography, parameters like divergence dates, effective population sizes, mutation rates, etc., cannot be expressed as moments of the sampling distribution at a given locus. In particular, the data are not iid. However, when considering microsatellite loci with the stepwise mutation model (27) and evolutionary scenarios composed of divergence, we can derive the pairwise composite scores whose zero is the pairwise maximum likelihood estimator (MLE). Composite likelihoods have been proved consistent for estimating recombination rates, introducing an approximation of the dependency structure between nearby loci (28–31). (Also, ref. 32 gives composite likelihoods used in a likelihood-free setting.)
More specifically, we are approximating the intralocus likelihood by a product over all pairs of genes in the sample at a given locus. Assuming that 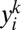 denotes the allele of the ith gene in the sample at the kth locus, and that ϕ is the vector of parameters, then the so-called pairwise likelihood of the data at the kth locus, namely
denotes the allele of the ith gene in the sample at the kth locus, and that ϕ is the vector of parameters, then the so-called pairwise likelihood of the data at the kth locus, namely 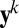 , is defined by
, is defined by
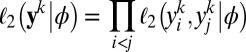 |
and the corresponding pairwise score function is 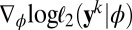 . Pairwise score equations
. Pairwise score equations
provide a constraint 1 in every way comparable to the score equations that give the maximum likelihood estimator and which is quite powerful for empirical likelihood derivations (ref. 18), pp 48–50). Hence the empirical likelihood of the full dataset 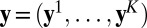 given ϕ is computed with 2 under the (multidimensional) constraint that
given ϕ is computed with 2 under the (multidimensional) constraint that
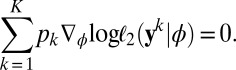 |
When the effective population size is identical over all populations of the demographic scenario, the time axis may be scaled so that coalescence of two genes in Kingman’s genealogy occurs with rate 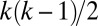 if there are k lineages. In this modified scale, mutations at a given locus arise with rate
if there are k lineages. In this modified scale, mutations at a given locus arise with rate 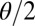 along the gene genealogy. Our mutation model is the simple stepwise mutation model of ref. 27; i.e., the number of repeats of the mutated gene increases or decreases by 1 unit with equal probability. Given two microsatellite allelic states x1 and x2, their pairwise likelihood
along the gene genealogy. Our mutation model is the simple stepwise mutation model of ref. 27; i.e., the number of repeats of the mutated gene increases or decreases by 1 unit with equal probability. Given two microsatellite allelic states x1 and x2, their pairwise likelihood 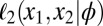 depends only on the difference of the states
depends only on the difference of the states 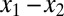 . If both genes belong to individuals that lie in the same deme, then (SI Text and ref. 33)
. If both genes belong to individuals that lie in the same deme, then (SI Text and ref. 33)
where 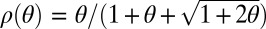 . If the two genes belong to individuals from demes having diverged at time τ, then (33)
. If the two genes belong to individuals from demes having diverged at time τ, then (33)
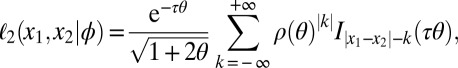 |
where 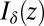 denotes the δth-order modified Bessel function of the first kind evaluated at z. Computing the pairwise scores, i.e., partial derivatives of
denotes the δth-order modified Bessel function of the first kind evaluated at z. Computing the pairwise scores, i.e., partial derivatives of 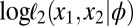 from those equations, is straightforward by virtue of recurrence properties of the Bessel functions. Algorithm BCel is therefore directly available in this setting, and furthermore at a cost much lower than the one associated with ABC algorithms (Table S1).
from those equations, is straightforward by virtue of recurrence properties of the Bessel functions. Algorithm BCel is therefore directly available in this setting, and furthermore at a cost much lower than the one associated with ABC algorithms (Table S1).
Results
Normal Distribution.
Starting with the benchmark of a normal distribution with known variance (equal to 1), we can check that the empirical likelihood allows for a proper recovery of the true posterior distribution on the mean. Fig. S1 shows that a constraint 1 based on the mean works well, as do the two constraints on mean and second central moment, 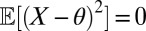 (Fig. S2). On the other hand, using the first three central moments in the empirical likelihood may degrade the fit (three cases in Fig. S3). Whereas this poor fit is not signaled by the ESS (which is often larger than in Figs. S1 and S2, because of the growing disconnection between the approximation and the true likelihood and hence a more uniform range of the weights), a parallel run of the method with different collections of constraints does detect the discrepancy. This illustrates the variability of the empirical likelihood approximation, as well as its sensitivity to the choice of defining constraints. Although a drawback of the method, this variability can be tested and evaluated by comparing outcomes, due to often limited computing costs. This toy experiment also supports the generic recommendation (18) to keep the number of constraints and parameters equal.
(Fig. S2). On the other hand, using the first three central moments in the empirical likelihood may degrade the fit (three cases in Fig. S3). Whereas this poor fit is not signaled by the ESS (which is often larger than in Figs. S1 and S2, because of the growing disconnection between the approximation and the true likelihood and hence a more uniform range of the weights), a parallel run of the method with different collections of constraints does detect the discrepancy. This illustrates the variability of the empirical likelihood approximation, as well as its sensitivity to the choice of defining constraints. Although a drawback of the method, this variability can be tested and evaluated by comparing outcomes, due to often limited computing costs. This toy experiment also supports the generic recommendation (18) to keep the number of constraints and parameters equal.
Quantile Distributions.
Quantile distributions are defined by a closed-form quantile function 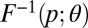 , and generally have no closed form for the density function. They are of great interest because of their flexibility and the ease with which they can be simulated by a simple inversion of the uniform distribution. A range of methods, including ABC approaches (10), has been proposed for estimation (SI Text). We focus here on the four-parameter g-and-k distribution, defined by its quantile function, denoted
, and generally have no closed form for the density function. They are of great interest because of their flexibility and the ease with which they can be simulated by a simple inversion of the uniform distribution. A range of methods, including ABC approaches (10), has been proposed for estimation (SI Text). We focus here on the four-parameter g-and-k distribution, defined by its quantile function, denoted 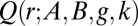 and equal to
and equal to
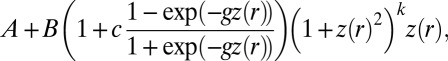 |
where 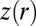 is the rth standard normal quantile; the parameters
is the rth standard normal quantile; the parameters 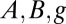 , and k represent location, scale, skewness, and kurtosis, respectively, and c measures the overall asymmetry (34, 35). We evaluated the BCel algorithm for estimating this distribution using two values of
, and k represent location, scale, skewness, and kurtosis, respectively, and c measures the overall asymmetry (34, 35). We evaluated the BCel algorithm for estimating this distribution using two values of 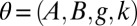 , two sets of priors, and various combinations of n, M, and p, where p is the number of percentiles used as constraints (details in SI Text).
, two sets of priors, and various combinations of n, M, and p, where p is the number of percentiles used as constraints (details in SI Text).
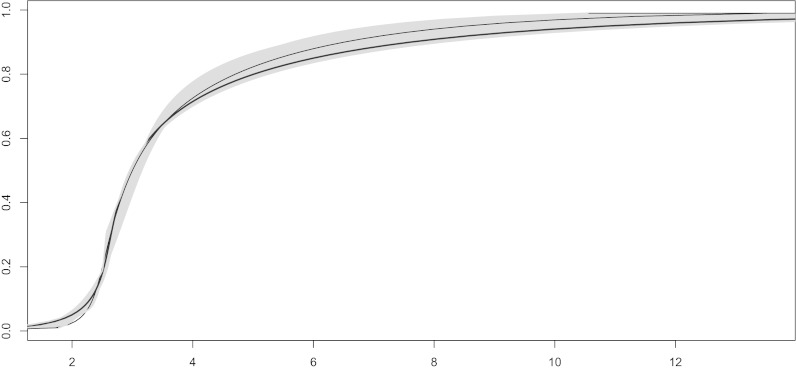
Fig. 1 illustrates the true and fitted curves and a 95% credible region for the case with 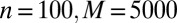 , and
, and 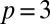 . The corresponding posterior means (SDs) for the parameters
. The corresponding posterior means (SDs) for the parameters 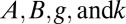 were 3.08(0.14), 1.12(0.23), 1.79(0.25), and 0.41(0.12), respectively. The choice of sample size and number of constraints did not substantively affect the accuracy of parameter estimates, but the precision was noticeably improved for the larger sample size (Figs. S4–S6).
were 3.08(0.14), 1.12(0.23), 1.79(0.25), and 0.41(0.12), respectively. The choice of sample size and number of constraints did not substantively affect the accuracy of parameter estimates, but the precision was noticeably improved for the larger sample size (Figs. S4–S6).
Fig. 1.
True (black) and fitted (brown) cdf functions with a pointwise 95% credible (shaded gray) region centered on the fitted cumulative distribution function for a dataset of 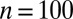 observations from the g-and-k distribution, based on
observations from the g-and-k distribution, based on 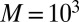 simulations of BCel.
simulations of BCel.
The accuracy and precision of the estimates were broadly comparable with the results obtained by ref. 36 for the same distribution. Based on the whole experiment, the parameters A and B were well estimated in all cases, whereas the estimates of g and k were poorer for smaller values of n and M. For small n the estimates were more subject to the vagaries of sampling variation, whereas for small M they were subject to the influence of a smaller number of very large importance weights. However, given the speed of BCel compared with competing ABC algorithms, it is feasible to use even larger values of M than considered in this experiment, because there is no requirement to simulate new datasets at each iteration. Moreover, this experiment is based on the very basic case of sampling from the prior; the results would be further improved by using an analog of BCel–AMIS or alternative approaches similar to those proposed by ref. 37 for ABC.
Dynamic Models.
In dynamic models, the difficulty with empirical likelihood stems from the dependence in the data 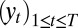 . However, these models can be represented as transforms of unobserved iid sequences
. However, these models can be represented as transforms of unobserved iid sequences 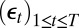 . The recovery of a converging empirical likelihood representation thus requires the reconstitution of the
. The recovery of a converging empirical likelihood representation thus requires the reconstitution of the 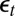 ’s as transforms of the data y and of the parameter θ. Independence between the
’s as transforms of the data y and of the parameter θ. Independence between the 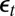 ’s is then at least as important as moment conditions. (This implies equivalent computing times for ABC and BCel.)
’s is then at least as important as moment conditions. (This implies equivalent computing times for ABC and BCel.)
For instance, consider a simple dynamic model, namely the autoregressive conditionally heteroskedastic 1 [Arch(1)] model:
with a uniform prior over the simplex, i.e., 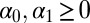 ,
, 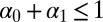 . Whereas this model can be handled by other means, because the likelihood function is available we will compare here the behavior of ABC and BCel algorithms.
. Whereas this model can be handled by other means, because the likelihood function is available we will compare here the behavior of ABC and BCel algorithms.
First, a natural empirical likelihood representation is based on the reconstituted 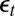 ’s, defined as
’s, defined as 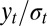 when the
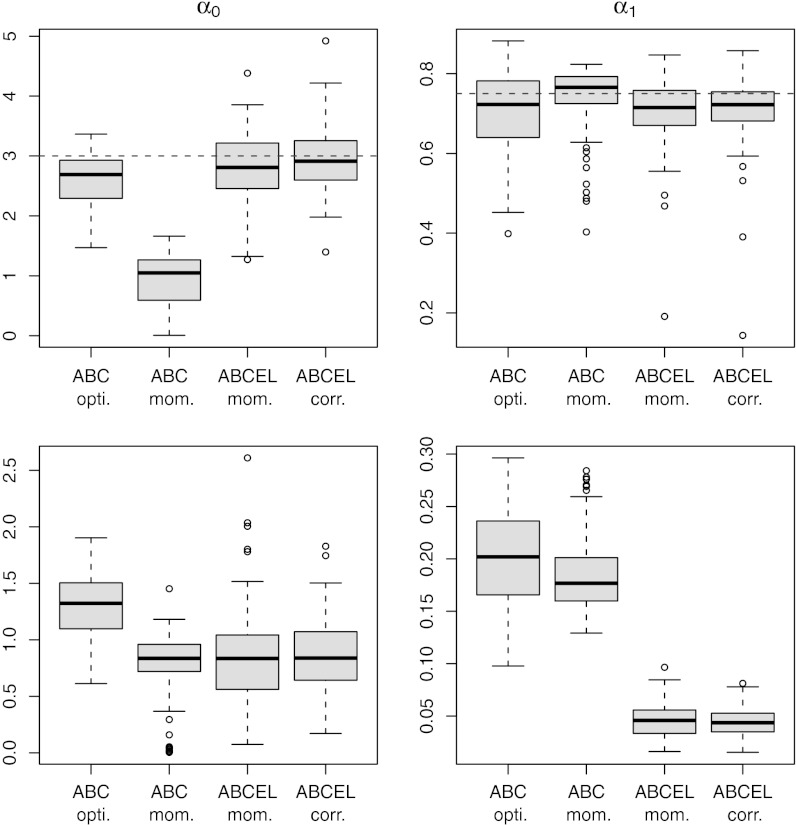
when the 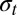 ’s are derived recursively. Fig. 2 shows the result of estimating both parameters
’s are derived recursively. Fig. 2 shows the result of estimating both parameters 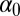 and
and 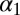 when Algorithm ABC uses as summary statistics either the least-square estimates of the parameters [derived from the series
when Algorithm ABC uses as summary statistics either the least-square estimates of the parameters [derived from the series 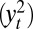 ], which we label “optimal ABC” in connection with ref. 38, or the mean of the series
], which we label “optimal ABC” in connection with ref. 38, or the mean of the series 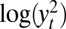 supplemented by the first two autocorrelations of the series
supplemented by the first two autocorrelations of the series 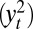 . The constraints in the empirical likelihood are either based on the first three moments of the reconstituted
. The constraints in the empirical likelihood are either based on the first three moments of the reconstituted 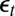 ’s or on the variance of those
’s or on the variance of those 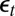 ’s complemented by both the correlations between the
’s complemented by both the correlations between the 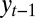 ’s and the
’s and the 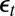 ’s and between the
’s and between the 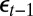 ’s and the
’s and the 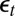 ’s. As seen from this experiment, BCel does as well as the optimal ABC for the estimation of the parameters, but further brings a reduction in the variability of those estimates, thanks to the importance weights.
’s. As seen from this experiment, BCel does as well as the optimal ABC for the estimation of the parameters, but further brings a reduction in the variability of those estimates, thanks to the importance weights.
Fig. 2.
(Upper) Comparison of ABC evaluations of posterior expectations, with true values given in dashed lines. (Lower) Posterior variances of the parameters 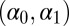 of the ARCH (1) model with 100 observations. The first two columns correspond to two choices of summary statistics for the ABC algorithm (least-squares estimates and mean of the
of the ARCH (1) model with 100 observations. The first two columns correspond to two choices of summary statistics for the ABC algorithm (least-squares estimates and mean of the 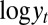 ’s plus autocorrelations of order 2 and 3, respectively). The last two columns correspond to two sets of constraints for the BCel alternative (first three moments and second moment plus autocorrelation of order 1 plus correlation with previous observation for the reconstituted
’s plus autocorrelations of order 2 and 3, respectively). The last two columns correspond to two sets of constraints for the BCel alternative (first three moments and second moment plus autocorrelation of order 1 plus correlation with previous observation for the reconstituted 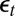 ’s). All experiments are based on the same reference table of 104 simulations, with the tolerance ε chosen as the 1% quantile of the distances.
’s). All experiments are based on the same reference table of 104 simulations, with the tolerance ε chosen as the 1% quantile of the distances.
A much more complex dynamic model is the generalized ARCH (GARCH)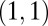 model of ref. 39 that can be formalized as the observation of
model of ref. 39 that can be formalized as the observation of 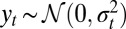 when
when
under the constraints 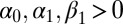 and
and 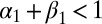 , that is,
, that is, 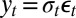 . Given the constraints on the parameters, a natural priority is to choose an exponential distribution on
. Given the constraints on the parameters, a natural priority is to choose an exponential distribution on 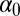 , for instance an exponential
, for instance an exponential 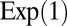 distribution, and a Dirichlet
distribution, and a Dirichlet 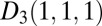 on
on 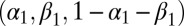 . An ABC approach requires the choice of summary statistics, which are necessarily nonsufficient because the model is a state-space model. Following ref. 38, we use the MLE as summary statistics, relying on the R function garch for its derivation despite its lack of stability. Because ref. 40 derived natural score constraints for the empirical likelihood associated with this model, we used their constraints to build our BCel algorithm. Fig. 3 provides a comparison of both approaches with the MLE. It shows in particular that the ABC algorithm is unable to produce acceptable inference in this case, even in the most favorable case when it is initialized at a satisfactory maximum likelihood estimate (as shown by the bottom row). The BCel algorithm is performing better, even though it fails to catch the correct range of
. An ABC approach requires the choice of summary statistics, which are necessarily nonsufficient because the model is a state-space model. Following ref. 38, we use the MLE as summary statistics, relying on the R function garch for its derivation despite its lack of stability. Because ref. 40 derived natural score constraints for the empirical likelihood associated with this model, we used their constraints to build our BCel algorithm. Fig. 3 provides a comparison of both approaches with the MLE. It shows in particular that the ABC algorithm is unable to produce acceptable inference in this case, even in the most favorable case when it is initialized at a satisfactory maximum likelihood estimate (as shown by the bottom row). The BCel algorithm is performing better, even though it fails to catch the correct range of 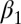 .
.
Fig. 3.
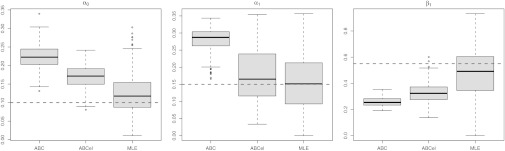
Comparison of evaluations of posterior expectations, with true values in dashed lines, of the parameters 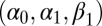 of the GARCH (1) model with 250 observations. The first row corresponds to an optimal ABC algorithm (using the MLE as summary statistic and with the tolerance ε chosen as the 5% quantile of the distances), the second row corresponds to the BCel algorithm based on the constraints derived in ref. 40, and the third row corresponds to the MLE derived by the R procedure GARCH initialized at the true parameter value.
of the GARCH (1) model with 250 observations. The first row corresponds to an optimal ABC algorithm (using the MLE as summary statistic and with the tolerance ε chosen as the 5% quantile of the distances), the second row corresponds to the BCel algorithm based on the constraints derived in ref. 40, and the third row corresponds to the MLE derived by the R procedure GARCH initialized at the true parameter value.
Another type of non-iid model relying on the superposition of an unknown number of gamma point processes and processed in ref. 41 through a (non-Bayesian) alternative to ABC is discussed in SI Text as an additional illustration of the possibilities of the empirical likelihood perspective for complex models, offering a free benchmark for evaluating the ABC outcome. Fig. S7 shows a clear improvement brought by BCel over the corresponding ABC outcome.
Population Genetics.
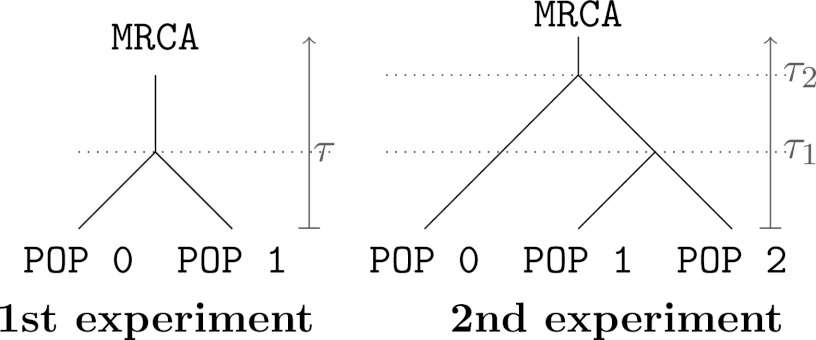
We compare our proposal with the reliable ABC-based estimates given by ref. 3. We set up two toy experiments that are designed to defeat ABC, using pseudoobserved data. The two evolutionary scenarios are given in Fig. 4. In all experiments, we only consider microsatellite loci and assume that the effective population size is identical over all populations of the scenario.
Fig. 4.
Evolutionary scenarios of the two experiments in population genetics.
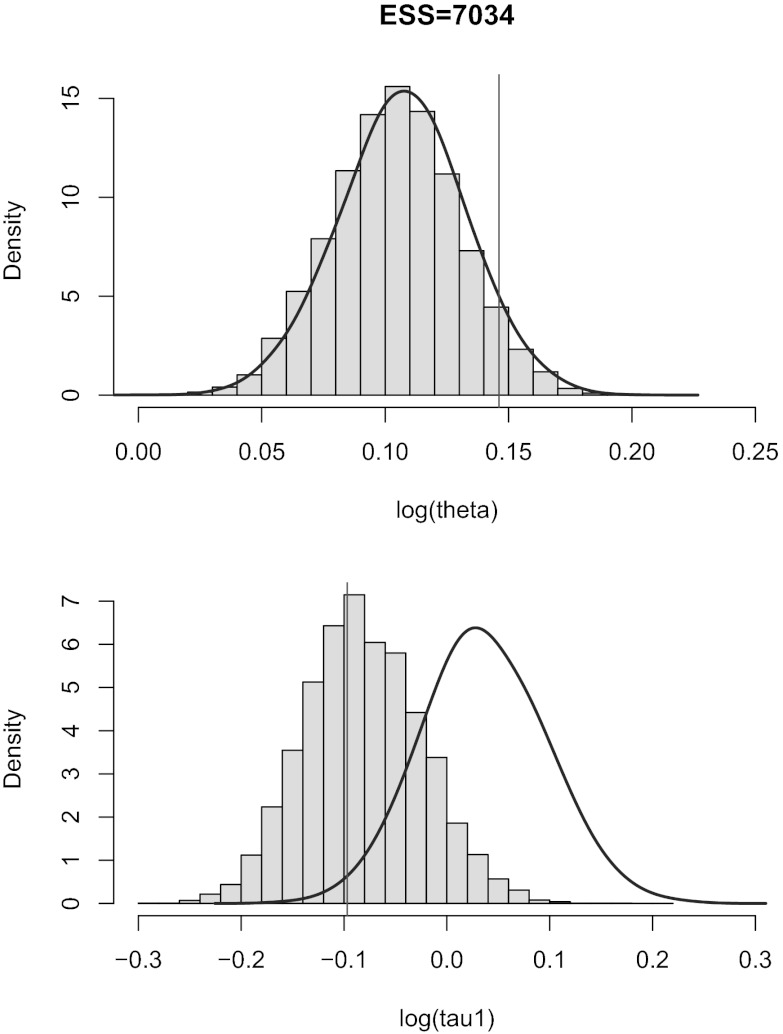
In the first experiment, we consider two populations which diverged at time τ in the past (Fig. 4, Left). Our pseudoobserved datasets are made of 30 diploid individuals per population genotyped at 100 independent loci. We compare the marginal posterior distributions of the unknown parameters θ and τ computed with the ABC method [using the Do It Yourself ABC (DIYABC) software of ref. 42] and with the BCel–AMIS sampler. In this case, results are improved when the θ component of the composite scores, namely 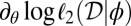 , is restricted to the sum over all pairs of genes lying in the same population. Otherwise, as can be checked via a quick simulation experiment, BCel systematically underestimates θ. Fig. 5 shows the typical discrepancy between both results: ABC and BCel agree on the mutation rate θ, but the BCel estimation of τ is more accurate (Table 1).
, is restricted to the sum over all pairs of genes lying in the same population. Otherwise, as can be checked via a quick simulation experiment, BCel systematically underestimates θ. Fig. 5 shows the typical discrepancy between both results: ABC and BCel agree on the mutation rate θ, but the BCel estimation of τ is more accurate (Table 1).
Fig. 5.
Comparison of the original ABC (Curve) with the histogram of the simulation from the BCel–AMIS sampler in the case of the population genetics model given in scenario A, based on uniform priors on 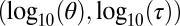 on
on 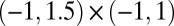 and 104 particles.
and 104 particles.
Table 1.
Comparison of the original ABC and ABCel on 100 Monte Carlo replicates
| Root-mean-square error of posterior mean |
Median absolute deviation of posterior median |
Coverage of the credible interval with probability 0.8 |
||||
| Experiment | ABC | ABCel | ABC | ABCel | ABC | ABCel |
| First experiment | ||||||
| θ | 0.0971 | 0.0949 | 0.071 | 0.059 | 0.68 | 0.81 |
| τ | 0.315 | 0.117 | 0.272 | 0.077 | 1.0 | 0.80 |
| Second experiment | ||||||
| θ | 0.0593 | 0.0794 | 0.0484 | 0.0528 | 0.79 | 0.76 |
| τ1 | 0.472 | 0.483 | 0.320 | 0.280 | 0.88 | 0.76 |
| τ2 | 29.6 | 4.76 | 4.13 | 3.36 | 0.89 | 0.79 |
We use two point estimates of the parameters (i) posterior mean and (ii) posterior median, and measured the error between the estimation and the “true” value used to generate the observation with (i) the root-mean-square error in the case of the posterior mean and (ii) the median absolute deviation in the case of the posterior median. We also compare credible intervals (with probability 0.8) through the proportion of Monte Carlo replicates in which the true value falls into this interval.
In the second experiment, we consider three populations (Fig. 4, Right): the last two populations diverged at time 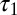 and their common ancestral population diverged from the first population at time
and their common ancestral population diverged from the first population at time 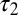 . The sample comprises 30 diploid individuals per population genotyped at 100 independent loci. In contrast with the first experiment, all components of the composite scores are computed here by summing over all pairs of genes whatever the population to which they belong. The results given in Table 1 show that ABC and BCel mainly agree on both parameters θ and
. The sample comprises 30 diploid individuals per population genotyped at 100 independent loci. In contrast with the first experiment, all components of the composite scores are computed here by summing over all pairs of genes whatever the population to which they belong. The results given in Table 1 show that ABC and BCel mainly agree on both parameters θ and 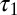 , but BCel is slightly more accurate than ABC on
, but BCel is slightly more accurate than ABC on 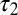 .
.
Table S1 gives a comparison of the computing times for both algorithms, showing the difference of magnitudes between them. This is due to the simulation of the simulated datasets for ABC: Although this difference should not be overinterpreted, it signals a potential for self-assessment and testing that is missing for ABC methods.
Discussion
Compared with ABC methods, the (often) significant time savings provided by BCel due to the lack of pseudosample simulation may open wider ranges for processing models involving complex likelihoods. For instance, in population genetics, ABC is severely hindered by the time spent simulating a dataset when modeling isolation by distance in a continuously distributed population, or when studying a large set of SNP markers even on quite simple evolution scenarios. Moreover, when the dataset is composed of large sets of markers, the summary statistics proposed in ABC (in DIYABC, these are averages of some quantitative statistics over all loci) ignores some (statistical) information, whereas BCel manages to recover most of it, more specifically to estimate divergence on large datasets. Improvements in accuracy of estimation and computational efficiency are also possible in other contexts as illustrated in the range of examples given above.
Even when BCel requires the same computing time as ABC, it uses the outcome in a very different perspective and provides a benchmark likelihood that helps in evaluating the pertinence of the ABC approximation, as illustrated in the gamma point process of SI.
We acknowledge a caveat of the empirical likelihood: it requires a careful choice of the constraint 1. Those pivotal quantities have to be connected to the parameter in an identifying way, which may require complex manipulations as in the gamma process case or may even be impossible. However, repeated experimentation is often available, as illustrated by the normal example and the population genetic experiments (where we computed the composite score on both a restricted set of pairs and all pairs of genes). Checking for the accuracy of the approximation means that a constraint in BCel should be tested on simulated datasets in controlled experiments where the true parameters are known, although much less than in ABC runs. Then we can test coverage of credibility intervals, and measure the error of various point estimates based on the output of the scheme.
Supplementary Material
Acknowledgments
P.P. and C.P.R. thank Jean-Marie Cornuet for his help and availability. C.R.P. is grateful to Patrice Bertail, Chris Drovandi, Brunero Liseo, and Art Owen for useful discussions. Comments and suggestions from the Editor and Reviewers greatly contributed to improve both the presentation and scope of the paper. P.P. and C.P.R. were partly supported by the Agence Nationale de la Recherche through the 2009–2012 Project Emile.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208827110/-/DCSupplemental.
References
- 1.Tavaré S, Balding DJ, Griffiths RC, Donnelly P. Inferring coalescence times from DNA sequence data. Genetics. 1997;145(2):505–518. doi: 10.1093/genetics/145.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162(4):2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornuet JM, et al. Inferring population history with DIY ABC: A user-friendly approach to approximate Bayesian computation. Bioinformatics. 2008;24(23):2713–2719. doi: 10.1093/bioinformatics/btn514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cappé O, Moulines E, Rydén T. Hidden Markov Models. New York: Springer; 2004. [Google Scholar]
- 5.Robert C, Casella G. Monte Carlo Statistical Methods. 2nd Ed. New York: Springer; 2004. [Google Scholar]
- 6.Pritchard JK, Seielstad MT, Perez-Lezaun A, Feldman MW. Population growth of human Y chromosomes: A study of Y chromosome microsatellites. Mol Biol Evol. 1999;16(12):1791–1798. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- 7.Beaumont M. Approximate Bayesian computation in evolution and ecology. Annu Rev Ecol Evol Syst. 2010;41:379–406. [Google Scholar]
- 8.Lopes JS, Beaumont MA. ABC: A useful Bayesian tool for the analysis of population data. Infect Genet Evol. 2010;10(6):826–833. doi: 10.1016/j.meegid.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Bazin E, Dawson KJ, Beaumont MA. Likelihood-free inference of population structure and local adaptation in a Bayesian hierarchical model. Genetics. 2010;185(2):587–602. doi: 10.1534/genetics.109.112391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marjoram P, Molitor J, Plagnol V, Tavaré S. Markov chain Monte Carlo without likelihoods. Proc Natl Acad Sci USA. 2003;100(26):15324–15328. doi: 10.1073/pnas.0306899100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Templeton AR. Coherent and incoherent inference in phylogeography and human evolution. Proc Natl Acad Sci USA. 2010;107(14):6376–6381. doi: 10.1073/pnas.0910647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger JO, Fienberg SE, Raftery AE, Robert CP. Incoherent phylogeographic inference. Proc Natl Acad Sci USA. 2010;107(41):E157–author reply E158. doi: 10.1073/pnas.1008762107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robert CP, Cornuet JM, Marin JM, Pillai NS. Lack of confidence in approximate Bayesian computation model choice. Proc Natl Acad Sci USA. 2011;108(37):15112–15117. doi: 10.1073/pnas.1102900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owen AB. Empirical likelihood ratio confidence intervals for a single functional. Biometrika. 1988;75(2):237–249. [Google Scholar]
- 15.Marin J, Pudlo P, Robert C, Ryder R. Approximate Bayesian computational methods. Stat Comput. 2012;22(6):1167–1180. [Google Scholar]
- 16.Biau G, Cérou F, Guyader A. 2012. New Insights into Approximate Bayesian Computation. Tech Rep HAL 00721164. Available at http://hal.inria.fr/hal-00721164/en.
- 17.Zhou M. 2012. emplik: Empirical likelihood ratio for censored/truncated data. R package version 0.9-8-2.
- 18.Owen AB. Empirical Likelihood. London: Chapman & Hall; 2001. [Google Scholar]
- 19.Lazar NA. Bayesian empirical likelihood. Biometrika. 2003;90:319–326. [Google Scholar]
- 20.Schennach SM. Bayesian exponentially tilted empirical likelihood. Biometrika. 2005;92(1):31–46. doi: 10.1093/biomet/asaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J. Monte Carlo Strategies in Scientific Computing. New York: Springer; 2001. [Google Scholar]
- 22.Cornuet JM, Marin JM, Mira A, Robert C. Adaptive multiple importance sampling. Scand J Stat. 2012;39(4):798–812. [Google Scholar]
- 23.Estoup A, Beaumont M, Sennedot F, Moritz C, Cornuet JM. Genetic analysis of complex demographic scenarios: Spatially expanding populations of the cane toad, Bufo marinus. Evolution. 2004;58(9):2021–2036. doi: 10.1111/j.0014-3820.2004.tb00487.x. [DOI] [PubMed] [Google Scholar]
- 24.Estoup A, Clegg SM. Bayesian inferences on the recent island colonization history by the bird Zosterops lateralis lateralis. Mol Ecol. 2003;12(3):657–674. doi: 10.1046/j.1365-294x.2003.01761.x. [DOI] [PubMed] [Google Scholar]
- 25.Fagundes NJ, et al. Statistical evaluation of alternative models of human evolution. Proc Natl Acad Sci USA. 2007;104(45):17614–17619. doi: 10.1073/pnas.0708280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombaert E, et al. Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PLoS ONE. 2010;5(3):e9743. doi: 10.1371/journal.pone.0009743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohta H, Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet Res. 1973;22(2):201–204. doi: 10.1017/s0016672300012994. and reprinted in (2007) 89(5–6):367–370. [DOI] [PubMed] [Google Scholar]
- 28.Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159(4):1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160(2):765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160(3):1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fearnhead P. Consistency of estimators of the population-scaled recombination rate. Theor Popul Biol. 2003;64(1):67–79. doi: 10.1016/s0040-5809(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 32.Barthelmé S, Chopin N. 2012. Expectation-propagation for likelihood-free inference. Tech Rep arXiv:1107.5959v2. Available at http://arxiv.org/pdf/1107.5959.
- 33.Wilson IJ, Balding DJ. Genealogical inference from microsatellite data. Genetics. 1998;150(1):499–510. doi: 10.1093/genetics/150.1.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haynes M, MacGillivray H, Mengersen K. Robustness of ranking and selection rules using generalized g-and k distributions. J Stat Plann Inference. 1997;65(1):45–66. [Google Scholar]
- 35.Gilchrist W. Statistical Modelling with Quantile Functions. London: Chapman & Hall; 2000. [Google Scholar]
- 36.Allingham D, King R, Mengersen K. Bayesian estimation of quantile distributions. Stat Comput. 2009;19(2):189–201. [Google Scholar]
- 37.Drovandi C, Pettitt A. Likelihood-free Bayesian estimation of multivariate quantile distributions. Comput Stat Data Anal. 2011;55(9):2541–2556. [Google Scholar]
- 38.Fearnhead P, Prangle D. Semi-automatic approximate Bayesian computation. J R Stat Soc, B. 2012;74(3):419–474. [Google Scholar]
- 39.Bollerslev T. Generalized autoregressive conditional heteroskedasticity. J Econom. 1986;31(3):307–327. [Google Scholar]
- 40.Chan N, Ling S. Empirical likelihood for GARCH models. Econometric Theory. 2006;22(3):403–428. [Google Scholar]
- 41.Cox D, Kartsonaki C. The fitting of complex parametric models. Biometrika. 2012;99(3):741–747. [Google Scholar]
- 42.Cornuet JM, Ravigné V, Estoup A. Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0) BMC Bioinf. 2010;11:401. doi: 10.1186/1471-2105-11-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.