Abstract
MicroRNAs (miRNAs) function as 21–24 nucleotide guide RNAs that use partial base-pairing to recognize target messenger RNAs and repress their expression. As a large fraction of protein-coding genes are under miRNA control, production of the appropriate level of specific miRNAs at the right time and in the right place is integral to most gene regulatory pathways. MiRNA biogenesis initiates with transcription, followed by multiple processing steps to produce the mature miRNA. Every step of miRNA production is subject to regulation and disruption of these control mechanisms has been linked to numerous human diseases, where the balance between the expression of miRNAs and their targets becomes distorted. Here we review the basic steps of miRNA biogenesis and describe the various factors that control miRNA transcription, processing and stability in animal cells. The tremendous effort put into producing the appropriate type and level of specific miRNAs underscores the critical role of these small RNAs in gene regulation.
Keywords: miRNA, Dicer, Drosha, Argonaute, mirtron
Introduction
In the past decade, the study of non-coding RNAs (ncRNAs) has taken center stage with the recognition that these small molecules function in virtually all cellular pathways. In multicellular organisms, the three major classes of small ncRNAs are microRNAs (miRNAs), small interfering (siRNAs), and Piwi-interacting RNAs (piRNAs), which all bind to complementary sequences within target RNAs to generally down-regulate gene expression (Aalto and Pasquinelli, 2012). A distinguishing feature of these post-transcriptional gene regulators is their unique biogenesis pathways. This review focuses on the biogenesis of miRNAs and the factors that regulate this pathway in animals.
The first miRNAs, lin-4 and let-7, were discovered through genetic screens in Caenorhabditis elegans as genes important for developmental timing (Lee et al., 1993, Reinhart et al., 2000). Since their discovery in C. elegans, miRNAs have been found across animal and plant species. In many cases the expression patterns and recognition of target mRNAs by specific miRNAs are conserved as well. Since miRNAs can target mRNAs through imperfect binding, each miRNA has many targets, giving miRNAs the potential to regulate over half of the human genome (Bartel, 2009, Rigoutsos, 2009). Thus, miRNAs control many different biological pathways, and interference with miRNA function can lead to a number of human diseases, including neurological disorders, cancer and cardiovascular complications (Bussing et al., 2008, Sayed and Abdellatif, 2011).
MiRNA biogenesis begins in the nucleus with transcription, usually by RNA polymerase II, to create the long primary miRNA (pri-miRNA) housing a hairpin that contains the mature sequence (Figure 1) (Kim et al., 2009, Winter et al., 2009). The hairpin is excised by the Microprocessor that includes Drosha, an RNase III enzyme, and its cofactor DGCR8 (also known as Pasha), producing the 60–70 nucleotide precursor miRNA (pre-mRNA). The precursor hairpin is exported out of the nucleus by Exportin 5 where another RNase III enzyme, Dicer, processes the precursor into the 21–24 nucleotide duplex miRNA. The strand destined to be the mature sequence is then loaded onto Argonaute, forming the miRNA induced silencing complex (miRISC) along with other proteins. Using imperfect base pairing, miRNAs guide RISC to specific mRNAs to down-regulate their expression by triggering mRNA destabilization or translational repression (Huntzinger and Izaurralde, 2011, Pasquinelli, 2012). MiRNA biogenesis can be regulated at each of these steps through accessory proteins to ensure proper miRNA homeostasis (Table 1). Since small deviations in miRNA levels can perturb the regulation of many target genes, control of miRNA biogenesis is essential for the maintenance of normal cell biology.
Figure 1. General miRNA pathway.
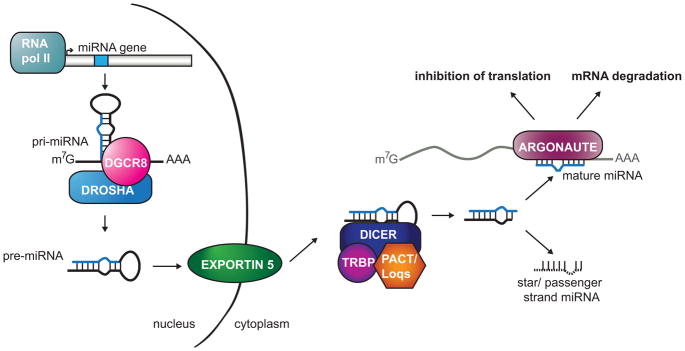
MiRNAs are typically transcribed by RNA polymerase II to produce the primary miRNA (pri-miRNA) transcript. The pri-miRNA is cleaved by the Microprocessor, which includes Drosha and DGCR8, resulting in the precursor miRNA (pre-miRNA) hairpin. The pre-miRNA is exported out of the nucleus by Exportin 5 to be further processed by the Dicer complex. Dicer cleaves the loop, and one strand of the miRNA duplex is loaded onto Argonaute to form the miRNA-Induced Silencing Complex (miRISC); the other strand, called the star or passenger, is degraded. miRISC is then capable of down-regulating gene expression through mRNA decay or translation inhibition. A color version of the figure is available online.
Table 1.
Regulators of miRNA biogenesis
| Factor | Type of gene product | Effect on miRNA biogenesis |
|---|---|---|
| ADAR1/2 | Adenosine deaminase that acts on RNA | Editing leads to reduced processing of pri- or pre-miRNAs |
| p68/DDX5 | DEAD-box RNA helicase | Facilitation of pri-miRNA processing |
| p72/DDX17 | DEAD-box RNA helicase | Facilitation of pri-miRNA processing |
| p53 | Transcription factor; tumor suppressor | Stimulation of pri-miRNA processing; regulation of transcription |
| SMAD proteins | Transcription factors | Stimulation of pri-miRNA processing; regulation of transcription |
| BRCA1 | DNA binding protein; tumor suppressor | Stimulation of pri-miRNA processing; regulation of transcription |
| hnRNP A1 | RNA binding protein | Stimulation or repression of pri-miRNA processing |
| KSRP | RNA binding protein | Stimulation of pri- or pre-miRNA processing |
| TDP-43 | DNA binding protein | Stimulation of pri- or pre-miRNA processing |
| LIN-28 | RNA binding protein | Repression of let-7 pri- or pre-miRNA processing |
| TUT4/Zcchc11 | Terminal RNA uridylyl transferase | Uridylation of let-7 pre-miRNAs, reducing their stability |
| TRBP | dsRNA binding protein | Facilitation of pre-miRNA processing |
| PACT/Loqs | dsRNA binding protein | Facilitation of pre-miRNA processing |
| RBM3 | RNA binding protein | Stimulation of pre-miRNA processing |
| MCPIP1/Zc3h12a | Nuclease | Cleavage of pre-miRNAs |
| Argonaute | RNA binding protein; endonucleolytic activity in some members | Stabilization of mature miRNAs; processing of pre-miR-451 |
| Nibbler | 3′ – 5′ exonuclease | Trimming of mature miRNA 3′ ends |
| XRN-1/2 | 5′ – 3′ exonuclease | Degradation of mature miRNAs |
| GLD-2 | cytoplasmic poly(A) polymerase | 3′ A-addition stabilizes mature miR-122 |
| QKI | RNA binding protein | Stabilization of mature miR-20a |
| miR-709 | Nuclear miRNA | Repression of pri-miR-15/16-1 processing |
| Argonaute with let-7 | miRNA complex | Stimulation of pri-let-7 miRNA processing |
In this review, each step of animal miRNA biogenesis is discussed and several key regulators that control miRNA processing and stability are highlighted. Additionally, miRNAs that mature independently of core biogenesis factors are described. Finally, the new discovery that mature miRNAs can directly regulate the processing of miRNA substrates is presented and exemplifies the surprises still unfolding in this small RNA pathway.
Genomic arrangement of miRNAs
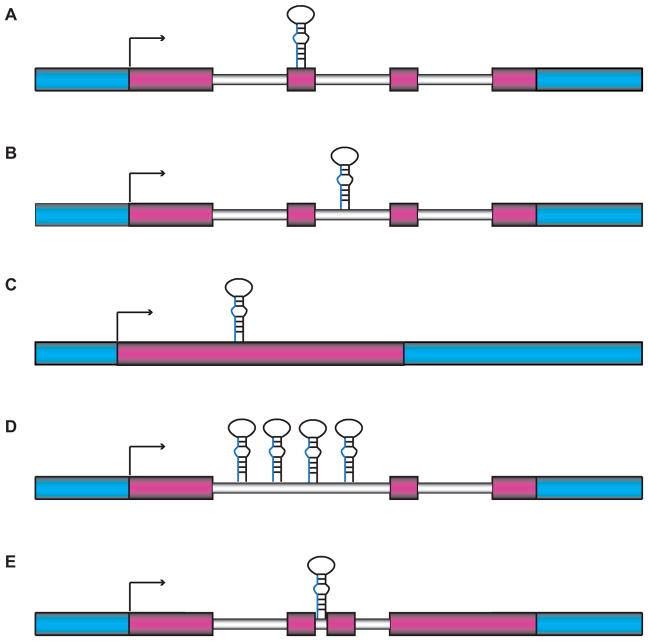
Most multi-cellular organisms encode dozens to hundreds of miRNA genes. Some miRNAs are housed within protein coding genes while others are in noncoding RNA (ncRNA) transcription units (Figure 2a,b) (Rodriguez et al., 2004). In mammalian cells more than 70% of miRNAs are located within an intron, with the majority residing in a protein-coding gene. The remaining miRNAs are located in ncRNA transcription units in approximately equal ratios of exonic and intronic miRNAs. Furthermore, alternative splicing of certain transcripts can cause the miRNA to be either in an intron or exon. Alternatively, in C. elegans only about 15% of miRNAs are under the control of a host gene promoter, with the rest being regulated by independent promoters (Martinez et al., 2008b). About 30% of these miRNA genes overlap protein-coding genes in the antisense direction, leaving over 100 miRNAs with their own promoters (Figure 2c). In some cases, there is a direct correlation in expression between the host gene and miRNA (Baskerville and Bartel, 2005, Rodriguez et al., 2004). However, expression of the host gene and mature miRNA can also be uncoupled by the use of alternative promoters and regulated processing.
Figure 2. MiRNA genomic locations.
MiRNAs can be located within exons (A) or introns (B) of coding or non-coding transcripts. Some miRNA genes are intergenic and not subject to splicing (C). MiRNA clusters consist of multiple miRNAs co-transcribed as part of one primary transcript, which can be intronic (D) or exonic (not shown) as part of a coding RNA or a non-coding RNA. MiRNAs that are excised from introns by splicing instead of Microprocessor activity are called mirtrons (E). In this figure, blue represents non-transcribed DNA, pink represents exons, and gray represents introns. A color version of the figure is available online.
It is quite common for multiple miRNAs to be transcribed as one long transcriptional unit called a “cluster” (Figure 2d) (Lau et al., 2001, Lee et al., 2002). MiRNAs in this genomic arrangement are sometimes, but not always, related and considered members of a family. Typically, a family of miRNAs includes all miRNAs, regardless of genomic location, that share nucleotides 2–7, often called the seed sequence (Bartel, 2009). For example, in humans there are thirteen members of the let-7 family, seven of which reside in clusters with other let-7 miRNAs (Mondol and Pasquinelli, 2012). Since seed pairing is often a primary determinant for target recognition, families of miRNAs have the capacity to regulate common genes. Thus, co-transcription of a family of miRNAs in a cluster provides an efficient means for the regulation of shared targets.
The complexity of miRNA genome arrangements can coordinate biological functions (Rodriguez et al., 2004). For example, the protein coding gene dynamin-3 includes miR-3120 within an intron on the sense strand (Scott et al., 2012). Antisense to this same intron is miR-214, which exhibits an expression pattern similar to that of dynamin-3 and miR-3120, despite being transcribed in the opposite orientation. Together, dynamin-3, miR-3120 and miR-214 all seem to be involved in synaptic vesicle recycling and function in neuronal cells. (Scott et al., 2012)
Transcription of miRNAs
The vast majority of miRNAs are transcribed by RNA polymerase II either as part of host gene transcripts or as independent transcription units. The nascent RNAs, called miRNA primary transcripts, receive 5′ methylated caps and 3′ polyadenylated tails similar to most other Pol II products (Bracht et al., 2004, Cai et al., 2004, Lee et al., 2004). In rare cases, transcription by RNA polymerase III has been shown to occur for miRNAs that are near Alu repeats (Borchert et al., 2006). At least for Pol II transcribed miRNAs, there is ample evidence that processing of the miRNA primary transcript to the precursor form can occur co-transcriptionally (Ballarino et al., 2009, Morlando et al., 2008, Pawlicki and Steitz, 2009). The processing complex, generally called the Microprocessor, has been shown to associate with the miRNA expressing genes in an RNA-dependent manner through chromatin immunoprecipitation (ChIP) analyses and localization studies (Morlando et al., 2008, Pawlicki and Steitz, 2009). In cases where the miRNA is intronic, release of the precursor has been shown to precede the completion of mRNA splicing (Morlando et al., 2008, Kim and Kim, 2007). Additionally, processing of a miRNA from an intron does not seem to reduce pre-mRNA splicing efficiency and, in at least one case, the two catalytic events mutually enhance each other (Kim and Kim, 2007, Pawlicki and Steitz, 2009, Janas et al., 2011).
Genome wide analyses of human miRNAs have provided a map of their promoter regions and chromatin signatures (Corcoran et al., 2009, Ozsolak et al., 2008, Monteys et al., 2010). These studies indicate that about one-third of intronic miRNAs are transcribed independently of their host gene. Additionally, transcriptional start sites can be located thousands of nucleotides upstream of the miRNA sequence. Many of the promoters contain features typical of Pol II promoters, including TATA boxes and initiator elements (Corcoran et al., 2009, Ozsolak et al., 2008, Monteys et al., 2010, Zhou et al., 2007).
Transcriptional regulation is at least partially responsible for the specific spatial and temporal expression patterns exhibited by many miRNAs. Several genome wide approaches have been taken to match transcription factors to miRNA genes. In mouse embryonic stem cells (ESCs), key pluripotency factors, including Oct4, Sox2, Nanog and Tcf3, are associated with the most abundantly expressed miRNAs in this cell type, as shown by ChIP (Marson et al., 2008). Surprisingly, these same transcription factors also occupy the promoters of some miRNAs that only become de-repressed as ESCs differentiate into a specific cellular lineage. In these cases, though, the Polycomb complex is also present, exerting its silencing function. In another approach, yeast one-hybrid (Y1H) assays were used to screen for transcription factors that associate with miRNA promoters in C. elegans (Martinez et al., 2008a). This genome wide survey revealed multiple cases of feedback loops where transcription factors that regulate specific miRNAs are themselves targets of those miRNAs (Martinez et al., 2008a). Since transcription factors can positively or negatively regulate miRNA expression, feedback loops can either buffer or amplify their own expression, respectively.
The Microprocessor
Most primary miRNA transcripts are processed into their precursor forms in the nucleus by the Microprocessor complex (Lee et al., 2003, Gregory et al., 2004, Han et al., 2004, Landthaler et al., 2004, Denli et al., 2004). Minimally, the Microprocessor consists of the RNase III enzyme Drosha and DGCR8 (DiGeorge syndrome critical region gene 8), also named Pasha (Partner of Drosha) (Gregory et al., 2004, Han et al., 2004, Landthaler et al., 2004, Denli et al., 2004). RNase III enzymes are known to cleave RNA and leave 2-nt overhangs, a characteristic seen on the 3′ end of pre-miRNAs (Lee et al., 2003, Han et al., 2004). Drosha contains two RNase III domains that are each responsible for making a cut to release the hairpin from the primary transcript (Han et al., 2004). Although Drosha harbors a recognizable double-stranded RNA binding domain (dsRBD), it requires DGCR8 with its two dsRBDs to directly bind to the RNA substrate (Yeom et al., 2006). Drosha and DGCR8 co-precipitate with each other as a complex that is sufficient for in vitro processing reactions. Additionally, depletion of either protein leads to the reduction of precursor and mature miRNAs in human cells, Drosophila cells, and C. elegans (Lee et al., 2003, Gregory et al., 2004, Han et al., 2004, Landthaler et al., 2004, Denli et al., 2004). Other proteins associated with the Microprocessor, such as helicases and heterogeneous nuclear ribonucleoproteins (hnRNPs), may help facilitate co-transcriptional processing and be differentially important for processing of specific primary transcripts.
The Microprocessor is responsible for processing most miRNAs in a cell and, thus, the structure of the RNA is more important than its sequence for substrate recognition. Within a primary transcript, the approximately 65 nt precursor sequence typically includes a partially base-paired stem region followed by a terminal loop. Systematic studies altering the structure of model RNA substrates led to the discovery of several important structural components that mark the precursor hairpin to be released by the Microprocessor: double stranded nature of the stem, ssRNA basal segments flanking the stem and a large terminal loop of at least 10 nt (Zeng et al., 2005, Han et al., 2006, Zeng and Cullen, 2005). While there seems to be consensus on the importance of the first two features, the role of the terminal loop has been debated (Zeng et al., 2005, Han et al., 2006, Zeng and Cullen, 2005, Zhang and Zeng, 2010). One study concluded that the Drosha cleavage site is mostly determined by the distance from the single-stranded basal region to the base of the stem and that the loop is not required for processing (Han et al., 2006). However, other reports have indicated a strong influence of the terminal loop over the efficiency of the cleavage reaction (Zeng et al., 2005, Han et al., 2006, Zeng and Cullen, 2005, Zhang and Zeng, 2010). It is likely that in vivo the single stranded flanking sequence, hairpin stem and terminal loop all contribute to the ability of the Microprocessor to accurately detect and efficiently process miRNA precursors from primary substrates.
Primary processing is further regulated by the amount of Microprocessor available to the cell. Microprocessor levels are auto-regulated, as Drosha and DGCR8 post-transcriptionally regulate each other. The Microprocessor binds to a hairpin, which resembles a miRNA precursor, within the 5′ UTR of DGCR8 mRNA and cleaves the mRNA (Han et al., 2009, Kadener et al., 2009, Macias et al., 2012, Triboulet et al., 2009). Thus, depletion of Drosha stabilizes the DGCR8 mRNA leading to increased levels of DGCR8 mRNA and protein. Conversely, depletion of DGCR8 causes a decrease in Drosha protein levels. Mutations in DGCR8 that prevent it from binding to Drosha also reduce Drosha levels. These results indicate that DGCR8 stabilizes Drosha through protein-protein interactions (Han et al., 2009). The auto-regulatory loop of the key Microprocessor proteins ensures homeostasis of miRNA levels and represents another step where miRNA maturation can be controlled.
Modifiers of Microprocessor Activity
The structure of primary miRNAs can be altered through editing of nucleotides within and flanking the hairpin. ADAR (adenosine deaminase that acts on RNA) enzymes are able to convert adenosine to inosine (A-to-I) using dsRNA as a substrate. This conversion causes changes in dsRNA structure and overall stability due to the wobble base pairing of I:U (Bass and Weintraub, 1988, Wagner et al., 1989). Primary miR-22 was the first miRNA shown to be edited in such a way by ADAR1 and ADAR2 in human and mouse brain tissue (Luciano et al., 2004). ADAR proteins have since been shown to act on other specific miRNA processing substrates, resulting in reduced cleavage by Drosha or Dicer (Yang et al., 2006, Kawahara et al., 2007). In some cases of editing, processing is unaffected but the nucleotide change in the seed sequence of the miRNA leads to differential targeting (Blow et al., 2006, Kawahara et al., 2007). The extent of ADAR-mediated editing that affects miRNA processing and target recognition is presently unclear, as the fraction of the modified transcript is often low and errors in high throughput sequencing results raise concerns about the validity of potential deaminase events (Warf et al., 2012, Alon et al., 2012, Vesely et al., 2012, Peng et al., 2012).
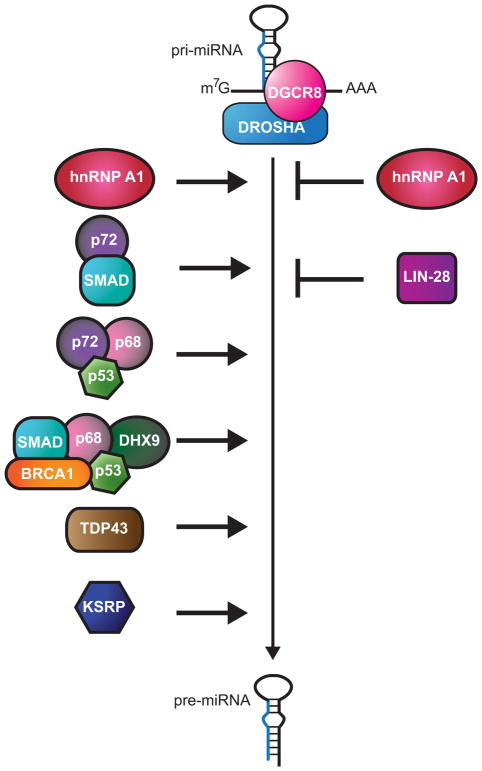
In vivo, Drosha and DGCR8 are associated with cofactors that facilitate the processing of certain miRNAs (Figure 3). While the functional relevance of the approximately 20 different proteins that have been found to associate with Drosha is yet to be determined (Gregory et al., 2004), a few have been characterized as modifiers of Microprocessor activity. The DEAD-box RNA helicases p68 (DDX5) and p72 (DDX17) co-precipitate with Drosha and are important for processing of select miRNAs (Gregory et al., 2004, Fukuda et al., 2007). The subset of miRNAs affected by loss of p68 or p72 is largely, but not entirely, overlapping (Fukuda et al., 2007). In mice, genetic disruption of p68 and p72 results in embryonic and neonatal lethality, respectively (Fukuda et al., 2007). Since these genes are also involved in rRNA processing, the contribution of rRNA and miRNA deficiencies to the lethality is unclear. The exact role of p68/p72 in mediating Drosha cleavage of specific miRNAs is yet to be defined, but their helicase activities could enable favorable structural rearrangements of primary miRNAs or dislodge inhibitory proteins bound to the RNA substrates.
Figure 3. Regulation of Microprocessor activity.
To the left of the vertical arrow are factors that positively regulate processing of pri-miRNAs to pre-miRNAs and to the right are proteins that negatively regulate this step. A color version of the figure is available online.
One function of p68/p72 seems to be to bridge other regulatory factors, including p53 and SMAD proteins, to the Microprocessor to regulate the biogenesis of certain miRNAs. The transcription factor p53 is a tumor suppressor gene that becomes active under stress conditions. It is well known that in response to certain cellular stresses, p53 induces transcription of genes involved in halting the cell cycle, promoting apoptosis and triggering senescence (Lane and Levine, 2010). Recent studies have shown that transcriptional and post-transcriptional regulation of specific miRNAs is another way p53 induces these stress responses. Through its canonical function, p53 binds the promoters of specific miRNA genes, in particular the miR-34 family members, to up-regulate their transcription (Chang et al., 2007, Corney et al., 2007, He et al., 2007, Raver-Shapira et al., 2007, Tazawa et al., 2007). More surprisingly, p53 also binds to the Microprocessor in a p68/p72-dependent manner to stimulate the processing of certain miRNAs (Suzuki et al., 2009). Analysis of miRNA levels showed a correlation between miRNAs that are positively dependent on p72 for processing and miRNAs that are up-regulated after stress occurs. Unexpectedly, many of these miRNAs showed no change in primary transcript levels but an increase in precursor and mature. Through yet to be resolved mechanisms, p53 recruits specific primary miRNAs to the Microprocessor through p68/p72, resulting in enhanced processing. These up-regulated miRNAs, including miR-16-1, miR-143 and miR-145, target cell proliferation and cell cycle proteins including KRAS and CDK6 (cyclin dependent kinase 6). CDK6 is also repressed by miR-34 family miRNAs, which are transcriptionally induced by p53 (Suzuki et al., 2009). Thus, the regulation of miRNA biogenesis at multiple levels reinforces the tumor suppressive function of p53 (Chang et al., 2007, Corney et al., 2007, He et al., 2007, Raver-Shapira et al., 2007, Tazawa et al., 2007).
The SMAD proteins also regulate miRNA biogenesis through two distinct pathways with the help of p68 (Blahna and Hata, 2012). The SMAD proteins are a part of the TGF-β (transforming growth factor β) signal cascade pathway, which regulates growth and development (Conidi et al., 2011). Generally, upon induction of this pathway, SMAD proteins translocate to the nucleus and bind DNA specifically at Smad-binding elements (SBE) to activate or repress transcription with the aid of other transcription factors (Massague et al., 2005). SMAD proteins can regulate transcription of miRNAs by binding to their promoters along with other proteins, including p68 (Warner et al., 2004, Blahna and Hata, 2012, Davis et al., 2010). Several miRNA genes contain SBEs that put their expression under the control of SMADs in a tissue specific manner (Davis et al., 2010, Yao et al., 2010). For example, TGF-β directly induces the expression of miR-216 and miR-217 in glomerular meningeal cells and represses the expression of miR-24 in myoblasts (Kato et al., 2009, Sun et al., 2008). In addition to binding to SBEs in DNA, SMAD proteins are also capable of binding a very similar sequence within the stem region of pri-miRNA transcripts (thus named the RNA Smad-binding element, R-SBE) regulated by TGF-β signaling at the post-transcriptional level. About 20 human miRNAs share this motif and undergo enhanced processing in response to TGF-β signaling (Davis et al., 2008, Davis et al., 2010). Binding of these pri-miRNAs by SMAD proteins seems to stabilize their interaction with Drosha in a p68-dependent manner, facilitating processing to the precursor forms.
Another DNA binding protein that also recognizes miRNA primary transcripts and regulates their processing is the breast cancer 1 (BRCA1) protein (Kawai and Amano, 2012). BRCA1 is a tumor suppressor that is responsible for repairing DNA damage, maintaining genomic stability, and controlling proper cell growth (Huen et al., 2010). Loss of BRCA1 activity leads to an increased risk of developing breast and ovarian cancer (Barnes and Antoniou, 2012). New studies suggest that some of its tumor suppressive functions may be mediated through its regulation of miRNA biogenesis. By recruiting histone deacetylases, BRCA1 has been shown to repress the transcription of miR-155, a so called oncomiR based on its general cell growth promoting activity (Chang et al., 2011). Conversely, BRCA1 promotes the processing of some miRNAs considered to have tumor suppressive effects, such as let-7, miR-16 and miR-145 (Kawai and Amano, 2012). Overexpression of BRCA1 causes an increase in the mature levels of these miRNAs, with a slight decrease in their primary transcript levels. Additionally, the DNA binding domain of BRCA1 associates with specific miRNA primary transcripts in vivo and in vitro by recognizing the base of their hairpins. BRCA1 interacts with Drosha, p68, Smad3 and p53 in a largely RNA-dependent manner. Another helicase, DHX9, also associates with the pri-miRNAs regulated by BRCA1 and depletion of this factor attenuates BRCA1-induced processing (Kawai and Amano, 2012). What distinguishes specific miRNAs for regulation by BRCA1 and how Drosha and its associated helicases and classic DNA binding proteins might cooperate to enhance processing are unresolved questions.
The terminal loop of some pri-miRNAs is a key feature for regulation by specific RNA binding proteins. For example, hnRNP A1 binds to the terminal loop of specific miRNAs and can enhance or repress cleavage by the Microprocessor (Michlewski et al., 2008, Michlewski and Caceres, 2010, Michlewski et al., 2010). hnRNP A1 has many roles in mRNA metabolism in the nucleus, including alternative splicing, stability, and export of mRNAs (He and Smith, 2009). Given its broad mRNA binding profile, it was surprising that only one miRNA, miR-18a, was found to be directly associated with hnRNP A1 in HeLa cells, using cross-linking immunoprecipitation (CLIP) experiments (Guil and Caceres, 2007). miR-18a is one of six miRNAs in the mir-17-92 cluster, and is the only one regulated by hnRNP A1 (Guil and Caceres, 2007). hnRNP A1 binds to the terminal loop as well as at the bottom of the stem, causing a bulge that improves Microprocessor access for cleavage (Michlewski et al., 2008, Michlewski et al., 2010). Subsequently, hnRNP A1 was shown to also recognize the terminal loop of let-7a-1 (Michlewski and Caceres, 2010). In contrast to its effect on processing of miR-18a, binding of hnRNP A1 to primary let-7a-1 inhibits cleavage by Drosha. At least part of this repressive activity is explained by competition between hnRNP A1 and factors that stimulate Drosha processing, such as KH-type splicing regulatory protein (KSRP), for binding to the terminal loop of let-7a-1 (Michlewski and Caceres, 2010).
Factors that bind to the terminal loop of miRNAs are capable of regulating primary and precursor processing if they are not restricted to a subcellular compartment. For example, KSRP has a mild to moderate effect on the biogenesis of more than thirty different miRNAs by stimulating both Drosha and Dicer activity in the nucleus and cytoplasm, respectively (Trabucchi et al., 2009). KSRP is a multi-functional protein with roles in alternative splicing, mRNA decay and miRNA biogenesis (Briata et al., 2011). KSRP binds preferentially to G-rich regions in the terminal loop of specific pri-miRNAs and enhances processing by stabilizing their interaction with Drosha. Furthermore, KSRP is predicted to remain associated with the precursor form of the miRNAs, enabling it to also stimulate Dicer cleavage in the cytoplasm (Trabucchi et al., 2009). The ability of KSRP to enhance miRNA processing can be regulated in response to DNA damage. Ataxia-telangiectasia mutated (ATM) kinase responds to dsDNA breaks by phosphorylating various protein targets in an effort to arrest cell cycle progression while DNA repair mechanisms ensue (Harper and Elledge, 2007). A newly identified target of ATM is KSRP (Zhang et al., 2011). Activation of ATM by DNA damage results in phosphorylation of KSRP, which stabilizes its interaction with specific miRNAs leading to enhanced processing (Zhang et al., 2011). Consistent with its general anti-oncogenic activity, some of the miRNAs up-regulated by ATM through KSRP, such as let-7 and miR-16, also support tumor suppressive pathways.
Another factor that enhances miRNA maturation in the nucleus and cytoplasm is the Tar DNA-binding protein-43 (TDP-43). It was recently shown that TDP-43 interacts with the Microprocessor and Dicer to facilitate processing of a subset of miRNAs (Buratti et al., 2010, Kawahara and Mieda-Sato, 2012). TDP-43 is important for mRNA processing in almost all steps including transcription, splicing, transport and translation (Lagier-Tourenne et al., 2010). For that reason, it is not surprising that TDP-43 is involved in multiple steps of miRNA processing. TDP-43 was shown to associate with terminal loop sequences in a small set of miRNAs and promote their interaction with Drosha and Dicer complexes (Kawahara and Mieda-Sato, 2012). However, a specific binding motif sequence was not defined. The C-terminus of TDP-43 mediates its association with Drosha and Dicer complexes in a partially RNA-independent manner (Kawahara and Mieda-Sato, 2012). One of the miRNAs regulated by TDP-43, miR-132-3p, was previously shown to be important for dendritic growth (Magill et al., 2010), and over-expression of this miRNA in neuronal cell culture partially restored neurite outgrowth in cells depleted of TDP-43 (Kawahara and Mieda-Sato, 2012). These findings implicate miRNA biogenesis as an important function of TDP-43 in neuronal physiology and raise the possibility that loss of this activity may contribute to some cases of amyotrophic lateral sclerosis (ALS) and front temporal lobar degeneration (FTLD), where TDP-43 accumulates in abnormal cytoplasmic aggregates (Lagier-Tourenne et al., 2010).
The first step of miRNA processing is also subject to negative regulation through factors that bind primary transcripts and prevent their cleavage by the Microprocessor (Figure 3). The well conserved RNA binding protein, LIN-28, regulates maturation of let-7 miRNA by binding to pri-let-7 to prevent Drosha processing in mammals and C. elegans (Newman et al., 2008, Viswanathan et al., 2008, Piskounova et al., 2008, Van Wynsberghe et al., 2011). LIN-28 is expressed in the earliest stages of development and is important for maintaining a stem cell like state, while let-7 promotes cellular differentiation pathways (Bussing et al., 2008, Roush and Slack, 2008, Viswanathan and Daley, 2010). While mature let-7 is typically undetectable in undifferentiated cells, primary let-7 transcripts are expressed at these early stages in development (Thomson et al., 2006, Van Wynsberghe et al., 2011, Viswanathan et al., 2008). As cells move towards more differentiated fates, LIN-28 protein levels fall, releasing the let-7 primary transcripts from processing inhibition. LIN-28 recognizes conserved sequences in two distinct regions of the loop found in most let-7 family members (Nam et al., 2011, Piskounova et al., 2011, Newman et al., 2008, Heo et al., 2009, Lightfoot et al., 2011). In C. elegans, the single lin-28 gene encodes a protein that binds to pri-let-7 co-transcriptionally to prevent processing by the Microprocessor (Van Wynsberghe et al., 2011). The mammalian genome encodes two Lin28 paralogs, LIN28A and LIN28B. The nuclear Lin28B protein binds primary let-7 transcripts and appears to sequester them in nucleoli to prevent access by the Microprocessor (Piskounova et al., 2011). Lin28A, however, is primarily localized in the cytoplasm, where it blocks the Dicer step of processing (Piskounova et al., 2011), which will be discussed further in the section on precursor processing.
Alternative pri-miRNA processing pathways
The recognition that some miRNA precursors mapped precisely to short introns in Drosophila and C. elegans led to the discovery of mirtrons (Figure 2e) (Ruby et al., 2007, Okamura et al., 2007). These miRNAs depend on splicing, instead of Microprocessor activity, for release of the precursor forms (Ruby et al., 2007, Okamura et al., 2007, Martin et al., 2009). Deep sequencing efforts and computational analyses have uncovered representatives in vertebrates, invertebrates and plants (Westholm and Lai, 2011). Although mirtron-derived miRNAs tend to be less abundant than those processed by the canonical pathway, they have been shown to repress the expression of targets with perfect or seed-matched sites (Ruby et al., 2007, Okamura et al., 2007, Martin et al., 2009). Following splicing, mirtrons undergo debranching to resolve the lariat and then fold into hairpin structures resembling precursor miRNAs (Westholm and Lai, 2011). At this point, mirtrons follow the typical miRNA processing pathway and are transported to the cytoplasm by Exportin-5, cleaved by Dicer and the mature strands are loaded onto Argonaute proteins. Some introns give rise to precursor miRNAs that are defined by splicing only on one end and contain nucleotide extensions on either the 5′ or 3′ termini. These “tailed mirtrons” require exonucleases to shave off the extra nucleotides to form structures compatible with Exportin-5 binding and transport to the cytoplasm (Babiarz et al., 2008, Flynt et al., 2010, Okamura et al., 2007, Ruby et al., 2007). The nuclear exosome is responsible for trimming mirtrons with 3′ tails (Flynt et al., 2010), but the activity that removes extra 5′ nucleotides is yet to be determined.
Although introns composed entirely of a miRNA precursor would implicate them as splicing-dependent mirtrons, a recent study showed that this is not always the case. Biogenesis of the human intronic miRNAs, miR-1225 and miR-1228, was found to be independent of splicing, DGCR8, Exportin-5, Dicer and Ago2 (Havens et al., 2012). Surprisingly, these splicing-independent mirtron-like miRNAs, termed simtrons, do require Drosha for generation of the precursors (Havens et al., 2012). The other factors needed to produce the mature miRNAs are yet to be uncovered. This is the first example of Drosha activity without the aid of DGCR8 in vivo and raises the question of whether a different dsRBP couples with this RNase to process certain substrates.
Export out of the nucleus
Precursor miRNAs must be exported out of the nucleus to be further processed by Dicer (Lee et al., 2002). Most nuclear transport receptors export RNA out of the nucleus by using the cofactor Ran-GTP; once in the cytoplasm hydrolysis of GTP to GDP allows the RNA to be released (Lei and Silver, 2002). Exportin 5 (XPO5) binds to pre-miRNAs with high affinity in a Ran-GTP dependent manner in vitro and in vivo (Lund et al., 2004, Yi et al., 2003, Bohnsack et al., 2004). Loss of Exportin 5 leads to a reduction in pre-miRNAs and, therefore, a decline in miRNA function in vivo (Zeng and Cullen, 2004, Yi et al., 2003). Exportin 5 preferentially binds to transport substrates that have short 3′ overhangs, terminal loops of at least 5 nucleotides, and base-paired stems greater then 15 nucleotides, characteristics of most miRNA precursors (Zeng and Cullen, 2004, Lund et al., 2004, Gwizdek et al., 2003, Okada et al., 2009). In addition to its transport role, Exportin 5 also stabilizes pre-miRNAs by preventing degradation by exonucleases (Zeng and Cullen, 2004).
The presence of inactivating mutations in Exportin 5 has been linked to the reduced mature miRNA levels observed in several types of tumors (Melo et al., 2010). Frameshift mutations that cause truncation of Exportin 5 were identified in a considerable fraction of the cancer cell lines and primary tumors examined. The truncated form of Exportin 5 failed to mediate transport of miRNA precursors, thus, prohibiting their access to Dicer processing in the cytoplasm. This defect led to reduced general levels of mature miRNAs, including those that function as tumor suppressors. Reintroduction of functional Exportin 5 into mutant cells supported precursor miRNA export and restoration of mature miRNA levels. Additionally, the presence of wild-type Exportin 5 imparted growth inhibiting properties to these cell lines. Thus, in at least some settings, Exportin 5 functions as a tumor suppressor by facilitating the maturation of miRNAs needed to regulate the expression of cell proliferative genes.
MicroRNA biogenesis can be regulated by titration of Exportin 5 activity. Saturation of this transporter by the expression of artificial short hairpins can impair endogenous precursor miRNA transport and, thus, maturation, leading to toxicity in mice (Grimm et al., 2006). A recently discovered role for Exportin 5 in transporting Dicer mRNA to the cytoplasm points to a surprising additional link between this transport factor and miRNA maturation. Down-regulation of Exportin 5 resulted in decreased Dicer protein through a post-transcriptional mechanism that was associated with nuclear accumulation of Dicer mRNA (Bennasser et al., 2011). Dicer mRNA was shown to interact with Exportin 5 in vivo and in vitro, although the binding element remains to be defined (Bennasser et al., 2011). While the dependence of both miRNA precursors and Dicer mRNA on Exportin 5 provides a mechanism for balancing levels of substrate and enzyme, it also presents a regulatory point for exploitation. The potential for viral hijacking of this pathway was demonstrated by showing that expression of the Adenoviral VA RNA I, a known substrate of Exportin 5 (Gwizdek et al., 2001), resulted in decreased Dicer protein levels and this provided an advantage for Adenoviral replication in HeLa cells (Bennasser et al., 2011).
The role of Dicer in miRNA maturation
Once exported out of the nucleus, miRNAs must be processed further by the RNase III enzyme, Dicer, in order to reach their mature forms of 21–24 nucleotides. Originally, Dicer was linked to small RNAs by the demonstration that it is responsible for cleavage of long dsRNA into 22 nucleotide small interfering RNAs (siRNAs) in the RNAi pathway (Bernstein et al., 2001). Shortly thereafter, Dicer was shown to be the enzyme that converts cellular miRNA precursors into the mature forms in Drosophila and C. elegans (Ketting et al., 2001, Hutvagner et al., 2001, Grishok et al., 2001, Knight and Bass, 2001). Most animal and plant species express at least one Dicer protein, which is characterized by the presence of helicase, PAZ, dsRNA binding, and two RNase III domains. The PAZ domain recognizes the 2-nt overhang at the base of the stem of precursor miRNAs (MacRae and Doudna, 2007). In most metazoans, Dicer typically measures from the monophosphorylated 5′ end of the precursor to position the RNase III domains for intramolecular dimerization and cleavage approximately 21–24 nt away (Park et al., 2011). A recent electron microscopy (EM) reconstitution of human Dicer revealed that the helicase domain is in the vicinity of the nuclease domains and may contribute to recognition of the loop region in miRNA precursors (Lau et al., 2012). Once precursor miRNAs are properly loaded into Dicer, cleavage can occur, where each RNase domain cleaves one of the strands, releasing the loop (MacRae and Doudna, 2007).
Regulation of Dicer activity
Regulation of the precursor to mature step can occur in a variety of ways. Specific precursor miRNAs can be detained in the nucleus to prevent maturation in certain cell types (Lee et al., 2008). Precursor maturation can also be regulated globally by affecting Dicer levels and activity. Human Dicer mRNA is itself a target of miRNA repression via three conserved let-7 binding sites within its coding sequence (Forman et al., 2008). Additionally, Dicer protein negatively regulates its own catalytic activity through its helicase domain. Mutation or deletion of the helicase domain in human Dicer causes enhanced cleavage efficiency, but does not affect binding affinity to miRNAs (Ma et al., 2008). This auto-inhibitory effect may be modulated by binding of Dicer cofactors to the helicase domain.
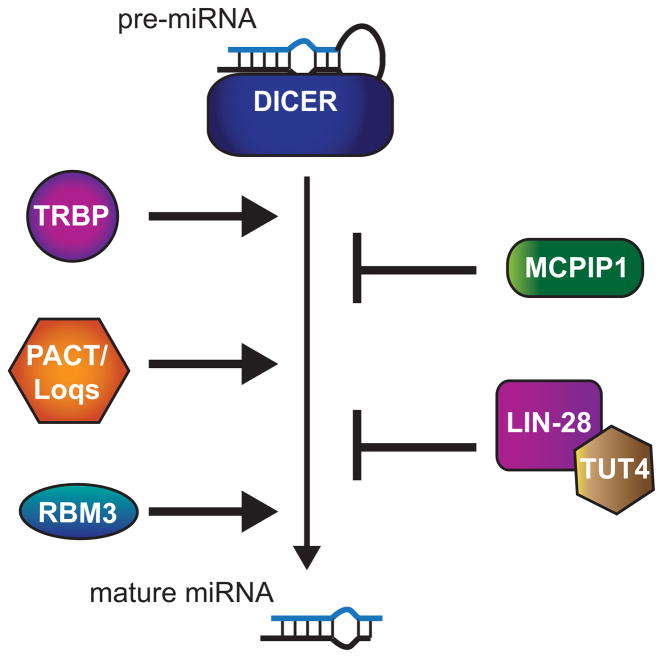
Dicer activity can be altered through many protein interactions (Figure 4). One Dicer interactor that increases cleavage efficiency is the HIV-1 TAR RNA-binding protein (TRBP). While Dicer is capable of cleaving precursor miRNAs into their mature forms without any cofactors, TRBP enhances this reaction in mammalian cells (Ma et al., 2008, Chakravarthy et al., 2010, Koscianska et al., 2011). TRBP has three RNA binding domains, but its association with Dicer is RNA-independent and direct through Dicer’s helicase domain (Haase et al., 2005, Lee et al., 2006, Ma et al., 2008). The exact role TRBP plays in precursor cleavage remains under debate. Some studies have found that the TRBP and Dicer interaction stabilizes Dicer protein (Chendrimada et al., 2005, Koscianska et al., 2011), while others found no effect on Dicer levels when TRBP is lost (Haase et al., 2005, Lee et al., 2006). Depletion of TRBP causes a decrease in mature miRNAs, and, in some studies, a decrease in precursor levels (Koscianska et al., 2011, Chakravarthy et al., 2010, Haase et al., 2005). This is likely due to a combined loss of Dicer stabilization by TRBP and absence of the stimulatory effect on Dicer activity. Inactivating mutations in TRBP have been found to be associated with several types of sporadic and hereditary cancers (Melo et al., 2009). In these instances Dicer protein is destabilized as well, explaining the general down-regulation of miRNA levels in tumors deficient in TRBP.
Figure 4. Regulation of Dicer activity.
To the left of the vertical arrow are proteins that positively regulate processing of pre-miRNAs to mature miRNAs and to the right are proteins that negatively regulate this step. A color version of the figure is available online.
The dsRNA binding proteins, protein activator of PKR (PACT) in mammals and Loquacious (Loqs) in Drosophila, also associate with Dicer and facilitate processing of miRNA precursors (Lee et al., 2006, Forstemann et al., 2005, Jiang et al., 2005, Saito et al., 2005). PACT and TRBP associate directly with each other as well as with Dicer in the cytoplasm (Lee et al., 2006). Depletion of PACT or TRBP in mammalian cells results in reduced mature miRNA levels, indicating that the two factors are not entirely redundant (Haase et al., 2005, Lee et al., 2006, Koscianska et al., 2011, Chakravarthy et al., 2010). It has been suggested that these Dicer cofactors help synchronize the dual cleavage by each of the RNase III domains (Koscianska et al., 2011).
Recently, another RNA binding protein, RBM3, was found to stimulate the processing of pre-miRNAs. RBM3 is an mRNA chaperone that enhances translation and is up-regulated under stress conditions including hypoxia and hypothermia (Danno et al., 1997, Williams et al., 2005, Dresios et al., 2005, Smart et al., 2007). Microarray analysis of neuronal cell lines subjected to RNAi against RBM3 showed a significant change in expression of over 150 miRNAs (Pilotte et al., 2011). The vast majority of miRNAs were reduced, and overexpression of RBM3 showed the opposite effect on mature miRNA accumulation. Since miRNA primary levels were unaffected, but precursor levels rose after knockdown of RBM3, the Dicer processing step appears to be regulated by this factor. Consistent with this idea, RBM3 was shown to bind miRNA precursors in vivo and in vitro and gradient fractionation studies suggest that RBM3 promotes association of precursor miRNAs with the Dicer complex. One output of induced RBM3 expression under stress conditions seems to be up-regulation of miRNAs, as hypothermia resulted in increased mature levels of several miRNAs in a RBM3 dependent manner (Pilotte et al., 2011). Thus, RBM3 may be an important regulator of miRNA biogenesis under specific conditions. Further analysis needs to be done to explore the role of this factor, such as determining if RBM3 helps recruit precursor miRNAs to the Dicer complex in the cell or prevents inhibitory factors from associating with precursor miRNAs.
Global repression of miRNA precursor conversion to mature can also be induced by certain conditions (Figure 4). The monocyte chemoattractant protein 1-induced protein 1 (MCPIP1), also known as Zinc-finger CCCH-type containing 12A (Zc3h12a), was recently shown to suppress miRNA biogenesis by degrading miRNA precursors (Suzuki et al., 2011). MCPIP1 is a nuclease that is induced during the inflammatory response in macrophages to limit the expression of several cytokines and chemokines by promoting mRNA decay (Cifuentes et al., 2010b). In addition to specific mRNAs, miRNA precursors in general appear to be targets of MCPIP1’s endonucleolytic activity (Suzuki et al., 2011). MCPIP1 catalyzes cleavage within the loop region of miRNA precursors, which antagonizes Dicer processing and results in rapid degradation of the cleaved fragments. Thus, induction of the inflammatory response or other conditions where MCPIP1 levels are elevated, such as in certain cancers, can dampen miRNA regulatory pathways in general through the pruning of miRNA precursor levels.
In contrast to general regulation of miRNA precursor processing, LIN28 is exceptionally selective in its miRNA targets. As mentioned earlier LIN28 represses both the Drosha and Dicer processing steps by recognizing specific loop sequences in let-7 miRNAs across species (Newman et al., 2008, Viswanathan et al., 2008, Piskounova et al., 2011, Van Wynsberghe et al., 2011, Heo et al., 2009, Heo et al., 2008, Lehrbach et al., 2009, Rybak et al., 2008). In mammalian cells, LIN28A regulates Dicer processing of pre-let-7 in the cytoplasm with the help of Tut4 (Rybak et al., 2008, Heo et al., 2008, Piskounova et al., 2011, Hagan et al., 2009). Tut4 is a terminal RNA uridylyl transferase that adds uridines to the 3′ ends of RNAs, marking them for degradation in some cases (Martin and Keller, 2007). While LIN28A alone can block Dicer processing, it also functions to recruit Tut4 to let-7 precursor miRNAs (Heo et al., 2008, Heo et al., 2009, Hagan et al., 2009, Lehrbach et al., 2009, Piskounova et al., 2011). The addition of uridines to pre-let-7 miRNAs makes them susceptible to rapid degradation by a yet to be discovered nuclease.
The ability of LIN28 proteins to regulate let-7 biogenesis at multiple steps underscores the opposing roles of these proteins and miRNAs: LIN28 promotes cellular pluripotency while let-7 typically supports differentiated fates (Viswanathan and Daley, 2010). Fitting with these functions, LIN28 is highly expressed in stem cells and then is down-regulated as cells undergo differentiation. Reactivation of LIN28 has been observed in several types of tumors and this correlates with decreased let-7 levels (Viswanathan et al., 2009, West et al., 2009). Remarkably, reintroduction of let-7 into transformed cells can halt oncogenic cell growth (Viswanathan et al., 2009). Thus, proper regulation of the LIN28 and let-7 nexus is essential for normal cellular growth and development.
The role of Argonaute in miRNA maturation
Processing of miRNA precursors in vivo can be linked to deposition of the mature miRNA products into miRISC through a miRNA loading complex, consisting of Dicer, TRBP and Argonaute (Gregory et al., 2005, Maniataki and Mourelatos, 2005, Liu et al., 2012). While the catalytic activity of Argonaute is not required for processing of most miRNAs, its presence in the complex enables uptake of the mature product, freeing Dicer for more precursor substrates. In some cases, Argonaute exerts its endonucleolytic cleavage activity on miRNA precursors that contain well-paired stem regions. In mammalian cells, Ago2 has been shown to cleave the 3′ arm of the hairpin, producing an Ago2-cleaved precursor miRNA (ac-pre-miRNA), prior to Dicer processing (Diederichs and Haber, 2007). It is hypothesized that this intermediate step may play a role in determining mature miRNA strand selection.
Argonaute also allows a specific miRNA to bypass Dicer processing. In mammalian cells, Ago2 is the only Argonaute capable of endonuclease activity (Liu et al., 2004, Rivas et al., 2005), making it a candidate for catalyzing the maturation of a miRNA unaffected by Dicer loss. While depletion of Dicer in most species results in diminished mature miRNA levels, miR-451 continues to be produced in human, mouse and zebrafish cells. This puzzle was solved with the discovery that Ago2 cleaves the 3′ stem and an unknown nuclease trims the 5′ half of pre-miR-451 to produce the 23 nt mature miRNA (Cifuentes et al., 2010a, Cheloufi et al., 2010, Yang et al., 2010). Pre-miR-451 is structurally different from canonical precursors as it has a 42-nt hairpin with a 17-nt stem, while Dicer is thought to require at least 19-nt in the stem for proper cleavage (Siolas et al., 2005). The unique secondary structure of the hairpin is likely what determines if processing occurs through Ago2, as artificial pre-miRNAs that imitate the same structure and length are also cleaved efficiently by Ago2 (Yang et al., 2012, Cifuentes et al., 2010a, Yang et al., 2010). It is yet to be determined if any other endogenous miRNAs are capable of being processed in this same Dicer-independent manner.
After Dicer produces a final 21–24-nt sized miRNA duplex, one strand is loaded onto an Argonaute protein, forming a miRNA-induced silencing complex (miRISC). The mature miRNA serves as a guide to recruit Argonaute and associated cofactors to target mRNAs, resulting in mRNA destabilization and translational repression (Huntzinger and Izaurralde, 2011, Pasquinelli, 2012). There are three categories of Argonaute proteins including the Ago, Piwi and worm specific subfamilies (Hock and Meister, 2008, Tolia and Joshua-Tor, 2007). Members of the Ago subfamily function in siRNA and miRNA pathways, while the Piwi subfamily interacts with piRNAs to regulate gene expression primarily in the germ-line of animals. Argonaute proteins are present across animal and plant species, however their number and roles vary from species to species. The yeast Schizosaccharomyces pombe has only one Argonaute protein, while C. elegans has a total of 27 with specific members dedicated to miRNA (i.e. Argonaute Like Genes 1 and 2, ALG-1/2) or RNAi (i.e. RNAi Defective 1, RDE-1) pathways. Drosophila melanogaster has 5 Argonautes, but miRNAs primarily load onto Ago1. The mouse and human genomes encode 8 Argonautes with Agos 1–4 forming the Ago subfamily. MiRNAs and siRNAs seem to load indiscriminately onto these four Argonautes, although Ago2 is the only catalytically active one capable of cleaving mRNAs that pair perfectly with the loaded small RNA.
Since Dicer cleavage produces a dsRNA product, the duplex must be unwound so that one of the strands can enter Argonaute to be functionally active. The single stranded mature miRNA loaded onto miRISC is considered the “guide,” while the other “passenger” strand is degraded. The thermodynamic stability of the miRNA duplex largely determines which strand will be incorporated into miRISC. Most miRNA duplexes are destabilized through imperfect base pairing, causing gaps, bulges, and weak interactions. The strand that is less stably paired at its 5′ terminus is more likely to be selected for Argonaute loading (Khvorova et al., 2003, Schwarz et al., 2003). It was originally thought that the unincorporated passenger strand, also known as the star (*) strand, is subject to rapid degradation. However, there are cases where the star strands are loaded onto Argonaute and can direct repression of target mRNAs (Okamura et al., 2008). In fact the abundance of guide versus star strand miRNAs has been observed to reverse under some conditions or in different developmental stages (Shin, 2008). Thus, factors besides the thermodynamic stability of base pairing can also influence the accumulation of mature miRNA species.
Recent structural studies have elucidated the architecture of Argonaute proteins loaded with small RNA guides (Elkayam et al., 2012, Nakanishi et al., 2012, Schirle and MacRae, 2012). Argonaute proteins have three domains important for miRNA loading: the PAZ, MID and PIWI domain. The guide RNA threads through the protein with the 3′ end in contact with the PAZ domain and the 5′ monophosphate on the first nucleotide stabilized by a highly basic pocket in the MID domain. The central region of the RNA is located in the PIWI domain, which in some Argonaute proteins contains catalytically active residues that mediate cleavage of target mRNAs that pair perfectly with the guide. In animals, miRNAs generally lack sufficient complementarity to induce target mRNA cleavage and instead Argonaute interacts with cofactors that promote deadenylation and degradation of the mRNA or interfere with its translation (Hock and Meister, 2008, Huntzinger and Izaurralde, 2011, Pasquinelli, 2012, Tolia and Joshua-Tor, 2007).
After loading onto Argonaute, some miRNAs undergo further processing. In Drosophila, the 3′ to 5′ exonuclease Nibbler trims the 3′ nucleotides from a large fraction of miRNAs (Han et al., 2011, Liu et al., 2011). Previously, size heterogeneity in some miRNAs, which often ranges from 21–24 nucleotides, was attributed to imprecise cleavage by Dicer or Drosha. The discovery that Nibbler removes 1–3 nucleotides from the 3′ ends of mature miRNAs associated with Argonaute points to a new source of the differently sized isoforms. Depletion of Nibbler activity reduced miRNA targeting efficiency, suggesting an important role for miRNA re-sizing (Han et al., 2011). Flies lacking Nibbler exhibited developmental defects and lethality, although it is yet to be demonstrated that these phenotypes are attributable to reduced miRNA function (Han et al., 2011, Liu et al., 2011). Heterogeneity in miRNA sizes has been noted across species and homologs of Nibbler are present in humans (EXD3) and worms (MUT-7), raising the possibility that 3′ end trimming may be a general step in the biogenesis of many miRNAs.
MiRNA stability
In addition to its key role as an effector of miRNA targeting, Argonaute also regulates the stability of associated miRNAs. Argonaute appears to be the limiting factor for miRNA biogenesis as over-expression of any of the four Argonaute proteins in human cells leads to a global increase in mature miRNAs (Diederichs and Haber, 2007). Likewise, knockdown of Ago2 by RNAi causes a significant decrease in miRNA levels. Similarly the Argonaute protein that functions with miRNAs in C. elegans, ALG-1, is important for miRNA stability, as well as precursor processing through an unclear mechanism (Grishok et al., 2001). In C. elegans, association of miRNAs with Argonaute proteins has been shown to protect them from exonucleolytic degradation by XRN-1 and XRN-2 (Figure 5a) (Chatterjee and Grosshans, 2009). These 5′ to 3′ exonucleases have been found to degrade miRNAs in a sequence-independent manner in vitro and in vivo, and RNAi depletion of xrn-1 or xrn-2 leads to an increase in mature miRNA levels in C. elegans. Interestingly, miRNAs engaged with mRNA targets are immune to XRN-1/2 nuclease activity in vitro. However, when targets are unavailable these exonucleases facilitate miRNA unloading from Argonaute, enabling degradation to occur (Chatterjee and Grosshans, 2009). This target-mediated miRNA protection (TMMP) has been shown to function in vivo, as the levels of lowly expressed mature miRNAs can be increased through introduction of reporters with complementary binding sites (Chatterjee et al., 2011). Therefore, the stability of miRNAs may be at least partially dependent on the number of target sites in a cell at any given time in C. elegans.
Figure 5. Effect of target interactions on miRNA stability.
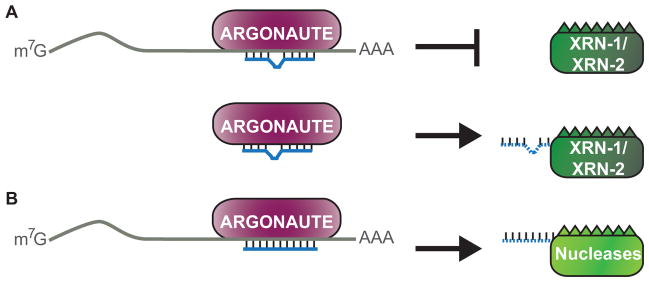
(A) In C. elegans, mRNA target interactions stabilize the association of miRNAs with Argonaute, which protects the miRNA from exonucleolytic degradation by XRN-1/2. When targets are unavailable miRNAs dislodge from Argonaute and undergo degradation by XRN-1/2. (B) In Drosophila and mammalian cells, nearly perfect complementarity of miRNAs to their targets triggers degradation of the miRNAs by unknown nucleases. A color version of the figure is available online.
It is yet to be determined if TMMP and homologs of XRN-1/2 control miRNA stability in other organisms. In fact, to the contrary, there is evidence that pairing of miRNAs to target sequences in Drosophila and mammalian cells can lead to reduced mature miRNA levels (Figure 5b) (Cazalla et al., 2010, Ameres et al., 2010, Baccarini et al., 2011, Libri et al., 2012). So far, two examples of viruses expressing RNAs that induce miRNA destabilization through pairing interactions have been documented. Partial base-pairing between the host cell miR-27a and the U-rich RNA, HSUR 1, encoded by Herpesvirus saimiri results in degradation of this miRNA during infection (Cazalla et al., 2010). This same miRNA is also targeted for destabilization by the m169 transcript expressed by murine cytomegalovirus (Libri et al., 2012). In both cases, miR-27a forms a partial duplex, typical of animal miRNA-target interactions, with the viral RNAs and this leads to down-regulation of the miRNA through unknown mechanisms. It is also unclear why pairing of miR-27a to these viral RNAs, but not its numerous other targets, destabilizes the miRNA. Since this effect can be transferred to other miRNAs designed to base pair to the viral RNAs, HSUR 1 and m169 must harbor features that recruit destabilizing factors to the captured miRNAs.
It appears that the degree of complementarity between the miRNA and target also influences miRNA stability, with perfect base pairing increasing miRNA decay rates (Figure 5b) (Ameres et al., 2010, Baccarini et al., 2011). In Drosophila, extensive base pairing has been shown to trigger the addition of multiple 3′ nucleotides and trimming of the miRNA (Ameres et al., 2010). In human cells a single uridine was most commonly added to miR-223 when matched to a perfect or near perfect target site, which correlated with its degradation (Baccarini et al., 2011). Adenosines and uridines are preferentially added to miRNAs involved in extensive base pairing interactions, though it is presently unknown which enzyme is responsible for these additions (Ameres et al., 2010, Wyman et al., 2011). Since near perfect complementarity is needed for efficient miRNA degradation through this pathway, the pervasiveness of this mechanism is questionable as most endogenous targets include mismatches and bulges.
The addition of non-templated nucleotides to the 3′ end of a miRNA does not always lead to degradation. The cytoplasmic poly(A) polymerase GLD-2 catalyzes the addition of a single adenosine to the 3′ end of miR-122 (Katoh et al., 2009). The Dicer product of pre-miR-122 processing is a 22 nt mature miRNA. When GLD-2 is present, miR-122 can be found in a 23 nt form through the addition of a single adenosine. In mice lacking GLD-2, levels of this 23 nt RNA were strongly reduced and this correlated with diminished overall levels of mature miR-122. Thus, GLD-2 is thought to adenylate mature miR-122, which prevents exonucleases from shortening the miRNA and, thus, reducing its half life. While GLD-2 has been shown to also adenylate 7SL RNA, there is no evidence for GLD-2 involvement in the stabilization of any other miRNAs (Katoh et al., 2009). It is likely there are additional modifications that can affect miRNA stability, although many may be miRNA specific and, thus, are yet to be discovered.
Another example of a factor that regulates the levels of a specific miRNA is the Signal Transduction and Activation of RNA (STAR) RNA binding protein Quaking (QKI). QKI was found to bind and stabilize miR-20a in human cells (Chen et al., 2012). miR-20a is expressed from a cluster of miRNAs, but only the level of mature miR-20a is influenced by QKI. QKI functions as a tumor suppressor of glioblastoma multiforme (GBM), at least partly through its regulation of miR-20a stability. In the context of GBM, TGFβ̃R2 can be oncogenic when not properly regulated through miR-20a expression. In the absence of QKI, levels of mature miR-20a drop which causes release of the direct target TGFβ̃R2 from repression (Chen et al., 2012). The mysteries of how QKI specifically regulates miR-20a and how this interaction stabilizes the miRNA are yet to be solved.
miRNA-guided regulation of biogenesis
Recent studies have shown that mature miRNAs can regulate miRNA biogenesis directly by interacting with primary miRNAs and controlling the Drosha processing step. The first example of this was observed in mouse cells where miR-709 was shown to negatively regulate biogenesis of miR-15a/16-1 to control the apoptosis pathway (Figure 6a) (Tang et al., 2012). Although most mature miRNAs reside primarily in the cytoplasm, miR-709 is enriched in the nucleus of mouse liver cells. One function of nuclear miR-709 seems to be to bind a complementary site in the primary transcript for the clustered miR-15a and miR-16-1 miRNAs and repress Drosha processing. Interestingly, serum depletion results in redistribution of miR-709 to the cytoplasm and consequently increased levels of miR-15a/16-1. Elevation of miR-15a/16-1 then promotes the apoptosis program by targeting the anti-apoptotic gene Bcl-2 (Tang et al., 2012). This work demonstrates that miRNAs can directly regulate the processing of other miRNAs and raises many new questions regarding the mechanism and control over nuclear localization of miR-709.
Figure 6. Regulation of pri-miRNA processing by mature miRNAs.
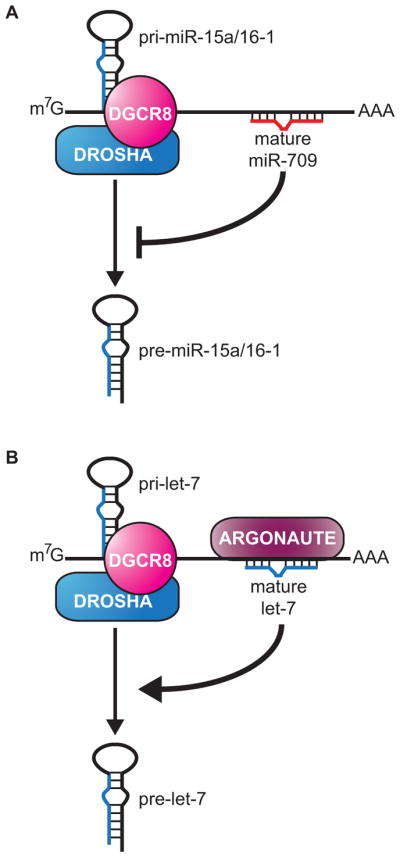
(A) In mouse cells, mature miR-709 binds to pri-miR-15a/16-1 to negatively regulate processing of the primary transcript to its precursor form. (B) In C. elegans, Argonaute (ALG-1) and mature let-7 bind to pri-let-7, positively regulating its processing to pre-let-7. A color version of the figure is available online.
The let-7 miRNA is capable of directly regulating its own processing through a novel auto-regulatory loop in C. elegans (Figure 6b). Analysis of sequences bound by the Argonaute dedicated to miRNA function in worms, ALG-1, revealed an interaction site in the let-7 primary transcript (Zisoulis et al., 2012, Zisoulis et al., 2010). Within this ALG-1 bound region is a conserved let-7 complementary sequence that is necessary for targeting by the Argonaute complex. Both let-7 and ALG-1 are detectable in nuclear fractions and association with pri-let-7 was shown to occur in this compartment (Zisoulis et al., 2012). Surprisingly, disruption of ALG-1 binding to the let-7 primary transcript resulted in reduced processing, implying a new role for Argonaute in positively regulating miRNA biogenesis. Since Argonaute was also found to interact with let-7 primary transcripts in human cells, auto-regulation of let-7 biogenesis may operate across species. This surprising new role for let-7 and Argonaute demonstrates that miRISC has targets beyond protein-coding mRNAs and adds processing regulation to its list of post-transcriptional functions. The mechanistic details and generality of direct processing regulation by miRNAs are yet to be determined.
Concluding remarks
Since the establishment of miRNAs as a new class of regulatory RNA a little over ten years ago, an outstanding question has been how these tiny RNAs are produced. The general factors involved in miRNA synthesis and processing have been elucidated. However, exceptions to the canonical miRNA biogenesis pathway continue to emerge. The fact that some miRNAs bypass core processing factors is an important consideration when interpreting phenotypes that result from inactivation of such proteins. Biogenesis of miRNAs is subject to complex regulation at both the transcriptional and post-transcriptional levels. Unexpectedly, several classic DNA binding proteins, such as p53, SMAD and BRCA1, appear to be directly involved in controlling the processing of specific miRNAs. Likewise, RNA binding proteins with established roles in regulating translation or stability of mRNAs have also been found to control the biogenesis of certain miRNAs. In some cases, such as the recognition of let-7 by LIN-28, the specificity determinants for particular miRNAs have been uncovered. In many others, though, it is yet to be determined how specific miRNAs are chosen for regulated processing or stability by particular factors. Additionally, for most instances of regulated miRNA biogenesis the detailed mechanisms are yet to be resolved. The recent discovery that miRNAs can recruit regulatory factors to enhance or repress the processing of other miRNA transcripts not only presents a new mechanism for controlling the expression of specific miRNAs but also reveals a novel function for the miRNA effector complex. The multitude of factors and pathways involved in miRNA production and maintenance indicates that these small RNAs are subject to just as much regulation as they exert over other genes.
Acknowledgments
We thank members of the Pasquinelli lab for critical reading of the manuscript.
Footnotes
Declaration of interest
The authors report no declarations of interest.
References
- AALTO AP, PASQUINELLI AE. Small non-coding RNAs mount a silent revolution in gene expression. Current opinion in cell biology. 2012;24:333–40. doi: 10.1016/j.ceb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALON S, MOR E, VIGNEAULT F, CHURCH GM, LOCATELLI F, GALEANO F, GALLO A, SHOMRON N, EISENBERG E. Systematic identification of edited microRNAs in the human brain. Genome research. 2012;22:1533–40. doi: 10.1101/gr.131573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMERES SL, HORWICH MD, HUNG JH, XU J, GHILDIYAL M, WENG Z, ZAMORE PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–9. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BABIARZ JE, RUBY JG, WANG Y, BARTEL DP, BLELLOCH R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes & development. 2008;22:2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BACCARINI A, CHAUHAN H, GARDNER TJ, JAYAPRAKASH AD, SACHIDANANDAM R, BROWN BD. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Current biology: CB. 2011;21:369–76. doi: 10.1016/j.cub.2011.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALLARINO M, PAGANO F, GIRARDI E, MORLANDO M, CACCHIARELLI D, MARCHIONI M, PROUDFOOT NJ, BOZZONI I. Coupled RNA processing and transcription of intergenic primary microRNAs. Molecular and cellular biology. 2009;29:5632–8. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARNES DR, ANTONIOU AC. Unravelling modifiers of breast and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers: update on genetic modifiers. Journal of internal medicine. 2012;271:331–43. doi: 10.1111/j.1365-2796.2011.02502.x. [DOI] [PubMed] [Google Scholar]
- BARTEL DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASKERVILLE S, BARTEL DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–7. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASS BL, WEINTRAUB H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–98. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- BENNASSER Y, CHABLE-BESSIA C, TRIBOULET R, GIBBINGS D, GWIZDEK C, DARGEMONT C, KREMER EJ, VOINNET O, BENKIRANE M. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nature structural & molecular biology. 2011;18:323–7. doi: 10.1038/nsmb.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERNSTEIN E, CAUDY AA, HAMMOND SM, HANNON GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- BLAHNA MT, HATA A. Smad-mediated regulation of microRNA biosynthesis. FEBS letters. 2012;586:1906–12. doi: 10.1016/j.febslet.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLOW MJ, GROCOCK RJ, VAN DONGEN S, ENRIGHT AJ, DICKS E, FUTREAL PA, WOOSTER R, STRATTON MR. RNA editing of human microRNAs. Genome biology. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOHNSACK MT, CZAPLINSKI K, GORLICH D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BORCHERT GM, LANIER W, DAVIDSON BL. RNA polymerase III transcribes human microRNAs. Nature structural & molecular biology. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- BRACHT J, HUNTER S, EACHUS R, WEEKS P, PASQUINELLI AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–94. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRIATA P, CHEN CY, GIOVARELLI M, PASERO M, TRABUCCHI M, RAMOS A, GHERZI R. KSRP, many functions for a single protein. Frontiers in bioscience: a journal and virtual library. 2011;16:1787–96. doi: 10.2741/3821. [DOI] [PubMed] [Google Scholar]
- BURATTI E, DE CONTI L, STUANI C, ROMANO M, BARALLE M, BARALLE F. Nuclear factor TDP-43 can affect selected microRNA levels. The FEBS journal. 2010;277:2268–81. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- BUSSING I, SLACK FJ, GROSSHANS H. let-7 microRNAs in development, stem cells and cancer. Trends in molecular medicine. 2008;14:400–9. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- CAI X, HAGEDORN CH, CULLEN BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAZALLA D, YARIO T, STEITZ JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–6. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAKRAVARTHY S, STERNBERG SH, KELLENBERGER CA, DOUDNA JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. Journal of molecular biology. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANG S, WANG RH, AKAGI K, KIM KA, MARTIN BK, CAVALLONE L, HAINES DC, BASIK M, MAI P, POGGI E, ISAACS C, LOOI LM, MUN KS, GREENE MH, BYERS SW, TEO SH, DENG CX, SHARAN SK. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nature medicine. 2011;17:1275–82. doi: 10.1038/nm.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANG TC, WENTZEL EA, KENT OA, RAMACHANDRAN K, MULLENDORE M, LEE KH, FELDMANN G, YAMAKUCHI M, FERLITO M, LOWENSTEIN CJ, ARKING DE, BEER MA, MAITRA A, MENDELL JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Molecular cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHATTERJEE S, FASLER M, BUSSING I, GROSSHANS H. Target-mediated protection of endogenous microRNAs in C. elegans. Developmental cell. 2011;20:388–96. doi: 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- CHATTERJEE S, GROSSHANS H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–9. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- CHELOUFI S, DOS SANTOS CO, CHONG MM, HANNON GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN AJ, PAIK JH, ZHANG H, SHUKLA SA, MORTENSEN R, HU J, YING H, HU B, HURT J, FARNY N, DONG C, XIAO Y, WANG YA, SILVER PA, CHIN L, VASUDEVAN S, DEPINHO RA. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes & development. 2012;26:1459–72. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHENDRIMADA TP, GREGORY RI, KUMARASWAMY E, NORMAN J, COOCH N, NISHIKURA K, SHIEKHATTAR R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIFUENTES D, XUE H, TAYLOR DW, PATNODE H, MISHIMA Y, CHELOUFI S, MA E, MANE S, HANNON GJ, LAWSON ND, WOLFE SA, GIRALDEZ AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010a;328:1694–8. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIFUENTES RA, CRUZ-TAPIAS P, ROJAS-VILLARRAGA A, ANAYA JM. ZC3H12A (MCPIP1): molecular characteristics and clinical implications. Clinica chimica acta; international journal of clinical chemistry. 2010b;411:1862–8. doi: 10.1016/j.cca.2010.08.033. [DOI] [PubMed] [Google Scholar]
- CONIDI A, CAZZOLA S, BEETS K, CODDENS K, COLLART C, CORNELIS F, COX L, JOKE D, DOBREVA MP, DRIES R, ESGUERRA C, FRANCIS A, IBRAHIMI A, KROES R, LESAGE F, MAAS E, MOYA I, PEREIRA PN, STAPPERS E, STRYJEWSKA A, VAN DEN BERGHE V, VERMEIRE L, VERSTAPPEN G, SEUNTJENS E, UMANS L, ZWIJSEN A, HUYLEBROECK D. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFbeta/BMP signaling in vivo. Cytokine & growth factor reviews. 2011;22:287–300. doi: 10.1016/j.cytogfr.2011.11.006. [DOI] [PubMed] [Google Scholar]
- CORCORAN DL, PANDIT KV, GORDON B, BHATTACHARJEE A, KAMINSKI N, BENOS PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PloS one. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CORNEY DC, FLESKEN-NIKITIN A, GODWIN AK, WANG W, NIKITIN AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer research. 2007;67:8433–8. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- DANNO S, NISHIYAMA H, HIGASHITSUJI H, YOKOI H, XUE JH, ITOH K, MATSUDA T, FUJITA J. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochemical and biophysical research communications. 1997;236:804–7. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- DAVIS BN, HILYARD AC, LAGNA G, HATA A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS BN, HILYARD AC, NGUYEN PH, LAGNA G, HATA A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Molecular cell. 2010;39:373–84. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENLI AM, TOPS BB, PLASTERK RH, KETTING RF, HANNON GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- DIEDERICHS S, HABER DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- DRESIOS J, ASCHRAFI A, OWENS GC, VANDERKLISH PW, EDELMAN GM, MAURO VP. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1865–70. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELKAYAM E, KUHN CD, TOCILJ A, HAASE AD, GREENE EM, HANNON GJ, JOSHUA-TOR L. The Structure of Human Argonaute-2 in Complex with miR-20a. Cell. 2012;150:100–10. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLYNT AS, GREIMANN JC, CHUNG WJ, LIMA CD, LAI EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Molecular cell. 2010;38:900–7. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FORMAN JJ, LEGESSE-MILLER A, COLLER HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14879–84. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FORSTEMANN K, TOMARI Y, DU T, VAGIN VV, DENLI AM, BRATU DP, KLATTENHOFF C, THEURKAUF WE, ZAMORE PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS biology. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUKUDA T, YAMAGATA K, FUJIYAMA S, MATSUMOTO T, KOSHIDA I, YOSHIMURA K, MIHARA M, NAITOU M, ENDOH H, NAKAMURA T, AKIMOTO C, YAMAMOTO Y, KATAGIRI T, FOULDS C, TAKEZAWA S, KITAGAWA H, TAKEYAMA K, O’MALLEY BW, KATO S. DEAD-box RNAhelicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nature cell biology. 2007;9:604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- GREGORY RI, CHENDRIMADA TP, COOCH N, SHIEKHATTAR R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- GREGORY RI, YAN KP, AMUTHAN G, CHENDRIMADA T, DORATOTAJ B, COOCH N, SHIEKHATTAR R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- GRIMM D, STREETZ KL, JOPLING CL, STORM TA, PANDEY K, DAVIS CR, MARION P, SALAZAR F, KAY MA. Fatality in mice due to over saturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- GRISHOK A, PASQUINELLI AE, CONTE D, LI N, PARRISH S, HA I, BAILLIE DL, FIRE A, RUVKUN G, MELLO CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- GUIL S, CACERES JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nature structural & molecular biology. 2007;14:591–6. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- GWIZDEK C, BERTRAND E, DARGEMONT C, LEFEBVRE JC, BLANCHARD JM, SINGER RH, DOGLIO A. Terminal minihelix, a novel RNA motif that directs polymerase III transcripts to the cell cytoplasm. Terminal minihelix and RNA export. The Journal of biological chemistry. 2001;276:25910–8. doi: 10.1074/jbc.M100493200. [DOI] [PubMed] [Google Scholar]
- GWIZDEK C, OSSAREH-NAZARI B, BROWNAWELL AM, DOGLIO A, BERTRAND E, MACARA IG, DARGEMONT C. Exportin-5 mediates nuclear export of minihelix-containing RNAs. The Journal of biological chemistry. 2003;278:5505–8. doi: 10.1074/jbc.C200668200. [DOI] [PubMed] [Google Scholar]
- HAASE AD, JASKIEWICZ L, ZHANG H, LAINE S, SACK R, GATIGNOL A, FILIPOWICZ W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO reports. 2005;6:961–7. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAGAN JP, PISKOUNOVA E, GREGORY RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature structural & molecular biology. 2009;16:1021–5. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN BW, HUNG JH, WENG Z, ZAMORE PD, AMERES SL. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Current biology: CB. 2011;21:1878–87. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN J, LEE Y, YEOM KH, KIM YK, JIN H, KIM VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes & development. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN J, LEE Y, YEOM KH, NAM JW, HEO I, RHEE JK, SOHN SY, CHO Y, ZHANG BT, KIM VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- HAN J, PEDERSEN JS, KWON SC, BELAIR CD, KIM YK, YEOM KH, YANG WY, HAUSSLER D, BLELLOCH R, KIM VN. Posttranscriptional cross regulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARPER JW, ELLEDGE SJ. The DNA damage response: ten years after. Molecular cell. 2007;28:739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- HAVENS MA, REICH AA, DUELLI DM, HASTINGS ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic acids research. 2012;40:4626–40. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE L, HE X, LIM LP, DE STANCHINA E, XUAN Z, LIANG Y, XUE W, ZENDER L, MAGNUS J, RIDZON D, JACKSON AL, LINSLEY PS, CHEN C, LOWE SW, CLEARY MA, HANNON GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE Y, SMITH R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cellular and molecular life sciences: CMLS. 2009;66:1239–56. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEO I, JOO C, CHO J, HA M, HAN J, KIM VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Molecular cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- HEO I, JOO C, KIM YK, HA M, YOON MJ, CHO J, YEOM KH, HAN J, KIM VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- HOCK J, MEISTER G. The Argonaute protein family. Genome biology. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUEN MS, SY SM, CHEN J. BRCA1 and its toolbox for the maintenance of genome integrity. Nature reviews Molecular cell biology. 2010;11:138–48. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNTZINGER E, IZAURRALDE E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews Genetics. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- HUTVAGNER G, MCLACHLAN J, PASQUINELLI AE, BALINT E, TUSCHL T, ZAMORE PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- JANAS MM, KHALED M, SCHUBERT S, BERNSTEIN JG, GOLAN D, VEGUILLA RA, FISHER DE, SHOMRON N, LEVY C, NOVINA CD. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS genetics. 2011;7:e1002330. doi: 10.1371/journal.pgen.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG F, YE X, LIU X, FINCHER L, MCKEARIN D, LIU Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes & development. 2005;19:1674–9. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KADENER S, RODRIGUEZ J, ABRUZZI KC, KHODOR YL, SUGINO K, MARR MT, 2ND, NELSON S, ROSBASH M. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15:537–45. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATO M, PUTTA S, WANG M, YUAN H, LANTING L, NAIR I, GUNN A, NAKAGAWA Y, SHIMANO H, TODOROV I, ROSSI JJ, NATARAJAN R. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nature cell biology. 2009;11:881–9. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATOH T, SAKAGUCHI Y, MIYAUCHI K, SUZUKI T, KASHIWABARA S, BABA T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & development. 2009;23:433–8. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWAHARA Y, MIEDA-SATO A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3347–52. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWAHARA Y, ZINSHTEYN B, CHENDRIMADA TP, SHIEKHATTAR R, NISHIKURA K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO reports. 2007;8:763–9. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWAI S, AMANO A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. The Journal of cell biology. 2012;197:201–8. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KETTING RF, FISCHER SE, BERNSTEIN E, SIJEN T, HANNON GJ, PLASTERK RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & development. 2001;15:2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHVOROVA A, REYNOLDS A, JAYASENA SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- KIM VN, HAN J, SIOMI MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- KIM YK, KIM VN. Processing of intronic microRNAs. The EMBO journal. 2007;26:775–83. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KNIGHT SW, BASS BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–71. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOSCIANSKA E, STAREGA-ROSLAN J, KRZYZOSIAK WJ. The role of Dicer protein partners in the processing of microRNA precursors. PloS one. 2011;6:e28548. doi: 10.1371/journal.pone.0028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAGIER-TOURENNE C, POLYMENIDOU M, CLEVELAND DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Human molecular genetics. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANDTHALER M, YALCIN A, TUSCHL T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Current biology: CB. 2004;14:2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- LANE D, LEVINE A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harbor perspectives in biology. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAU NC, LIM LP, WEINSTEIN EG, BARTEL DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- LAU PW, GUILEY KZ, DE N, POTTER CS, CARRAGHER B, MACRAE IJ. The molecular architecture of human Dicer. Nature structural & molecular biology. 2012;19:436–40. doi: 10.1038/nsmb.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE EJ, BAEK M, GUSEV Y, BRACKETT DJ, NUOVO GJ, SCHMITTGEN TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE RC, FEINBAUM RL, AMBROS V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- LEE Y, AHN C, HAN J, CHOI H, KIM J, YIM J, LEE J, PROVOST P, RADMARK O, KIM S, KIM VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- LEE Y, HUR I, PARK SY, KIM YK, SUH MR, KIM VN. The role of PACT in the RNA silencing pathway. The EMBO journal. 2006;25:522–32. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE Y, JEON K, LEE JT, KIM S, KIM VN. MicroRNA maturation: stepwise processing and subcellular localization. The EMBO journal. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE Y, KIM M, HAN J, YEOM KH, LEE S, BAEK SH, KIM VN. MicroRNA genes are transcribed by RNA polymerase II. The EMBO journal. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEHRBACH NJ, ARMISEN J, LIGHTFOOT HL, MURFITT KJ, BUGAUT A, BALASUBRAMANIAN S, MISKA EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nature structural & molecular biology. 2009;16:1016–20. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEI EP, SILVER PA. Protein and RNA export from the nucleus. Developmental cell. 2002;2:261–72. doi: 10.1016/s1534-5807(02)00134-x. [DOI] [PubMed] [Google Scholar]
- LIBRI V, HELWAK A, MIESEN P, SANTHAKUMAR D, BORGER JG, KUDLA G, GREY F, TOLLERVEY D, BUCK AH. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:279–84. doi: 10.1073/pnas.1114204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIGHTFOOT HL, BUGAUT A, ARMISEN J, LEHRBACH NJ, MISKA EA, BALASUBRAMANIAN S. A LIN28-dependent structural change in pre-let-7g directly inhibits dicer processing. Biochemistry. 2011;50:7514–21. doi: 10.1021/bi200851d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU J, CARMELL MA, RIVAS FV, MARSDEN CG, THOMSON JM, SONG JJ, HAMMOND SM, JOSHUA-TOR L, HANNON GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- LIU N, ABE M, SABIN LR, HENDRIKS GJ, NAQVI AS, YU Z, CHERRY S, BONINI NM. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Current biology: CB. 2011;21:1888–93. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU X, JIN DY, MCMANUS MT, MOURELATOS Z. Precursor microRNA-programmed silencing complex assembly pathways in mammals. Molecular cell. 2012;46:507–17. doi: 10.1016/j.molcel.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUCIANO DJ, MIRSKY H, VENDETTI NJ, MAAS S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–7. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUND E, GUTTINGER S, CALADO A, DAHLBERG JE, KUTAY U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- MA E, MACRAE IJ, KIRSCH JF, DOUDNA JA. Autoinhibition of human dicer by its internal helicase domain. Journal of molecular biology. 2008;380:237–43. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACIAS S, PLASS M, STAJUDA A, MICHLEWSKI G, EYRAS E, CACERES JF. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nature structural & molecular biology. 2012;19:760–6. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACRAE IJ, DOUDNA JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Current opinion in structural biology. 2007;17:138–45. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- MAGILL ST, CAMBRONNE XA, LUIKART BW, LIOY DT, LEIGHTON BH, WESTBROOK GL, MANDEL G, GOODMAN RH. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20382–7. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANIATAKI E, MOURELATOS Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & development. 2005;19:2979–90. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARSON A, LEVINE SS, COLE MF, FRAMPTON GM, BRAMBRINK T, JOHNSTONE S, GUENTHER MG, JOHNSTON WK, WERNIG M, NEWMAN J, CALABRESE JM, DENNIS LM, VOLKERT TL, GUPTA S, LOVE J, HANNETT N, SHARP PA, BARTEL DP, JAENISCH R, YOUNG RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–33. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN G, KELLER W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–49. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN R, SMIBERT P, YALCIN A, TYLER DM, SCHAFER U, TUSCHL T, LAI EC. A Drosophila pasha mutant distinguishes the canonical microRNA and mirtron pathways. Molecular and cellular biology. 2009;29:861–70. doi: 10.1128/MCB.01524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTINEZ NJ, OW MC, BARRASA MI, HAMMELL M, SEQUERRA R, DOUCETTE-STAMM L, ROTH FP, AMBROS VR, WALHOUT AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes & development. 2008a;22:2535–49. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTINEZ NJ, OW MC, REECE-HOYES JS, BARRASA MI, AMBROS VR, WALHOUT AJ. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome research. 2008b;18:2005–15. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASSAGUE J, SEOANE J, WOTTON D. Smad transcription factors. Genes & development. 2005;19:2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- MELO SA, MOUTINHO C, ROPERO S, CALIN GA, ROSSI S, SPIZZO R, FERNANDEZ AF, DAVALOS V, VILLANUEVA A, MONTOYA G, YAMAMOTO H, SCHWARTZ S, JR, ESTELLER M. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer cell. 2010;18:303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- MELO SA, ROPERO S, MOUTINHO C, AALTONEN LA, YAMAMOTO H, CALIN GA, ROSSI S, FERNANDEZ AF, CARNEIRO F, OLIVEIRA C, FERREIRA B, LIU CG, VILLANUEVA A, CAPELLA G, SCHWARTZ S, JR, SHIEKHATTAR R, ESTELLER M. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nature genetics. 2009;41:365–70. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- MICHLEWSKI G, CACERES JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nature structural & molecular biology. 2010;17:1011–8. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MICHLEWSKI G, GUIL S, CACERES JF. Stimulation of pri-miR-18a Processing by hnRNP A1. Advances in experimental medicine and biology. 2010;700:28–35. doi: 10.1007/978-1-4419-7823-3_3. [DOI] [PubMed] [Google Scholar]
- MICHLEWSKI G, GUIL S, SEMPLE CA, CACERES JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Molecular cell. 2008;32:383–93. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONDOL V, PASQUINELLI AE. Let’s make it happen: the role of let-7 microRNA in development. Current topics in developmental biology. 2012;99:1–30. doi: 10.1016/B978-0-12-387038-4.00001-X. [DOI] [PubMed] [Google Scholar]
- MONTEYS AM, SPENGLER RM, WAN J, TECEDOR L, LENNOX KA, XING Y, DAVIDSON BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORLANDO M, BALLARINO M, GROMAK N, PAGANO F, BOZZONI I, PROUDFOOT NJ. Primary microRNA transcripts are processed co-transcriptionally. Nature structural & molecular biology. 2008;15:902–9. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAKANISHI K, WEINBERG DE, BARTEL DP, PATEL DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–74. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAM Y, CHEN C, GREGORY RI, CHOU JJ, SLIZ P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–91. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEWMAN MA, THOMSON JM, HAMMOND SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKADA C, YAMASHITA E, LEE SJ, SHIBATA S, KATAHIRA J, NAKAGAWA A, YONEDA Y, TSUKIHARA T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–9. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- OKAMURA K, HAGEN JW, DUAN H, TYLER DM, LAI EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKAMURA K, PHILLIPS MD, TYLER DM, DUAN H, CHOU YT, LAI EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nature structural & molecular biology. 2008;15:354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OZSOLAK F, POLING LL, WANG Z, LIU H, LIU XS, ROEDER RG, ZHANG X, SONG JS, FISHER DE. Chromatin structure analyses identify miRNA promoters. Genes & development. 2008;22:3172–83. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK JE, HEO I, TIAN Y, SIMANSHU DK, CHANG H, JEE D, PATEL DJ, KIM VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–5. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASQUINELLI AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nature reviews Genetics. 2012;13:271–82. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- PAWLICKI JM, STEITZ JA. Subnuclear compartmentalization of transiently expressed polyadenylated pri-microRNAs: processing at transcription sites or accumulation in SC35 foci. Cell cycle. 2009;8:345–56. doi: 10.4161/cc.8.3.7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PENG Z, CHENG Y, TAN BC, KANG L, TIAN Z, ZHU Y, ZHANG W, LIANG Y, HU X, TAN X, GUO J, DONG Z, BAO L, WANG J. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nature biotechnology. 2012;30:253–60. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- PILOTTE J, DUPONT-VERSTEEGDEN EE, VANDERKLISH PW. Wide spread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3. PloS one. 2011;6:e28446. doi: 10.1371/journal.pone.0028446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PISKOUNOVA E, POLYTARCHOU C, THORNTON JE, LAPIERRE RJ, POTHOULAKIS C, HAGAN JP, ILIOPOULOS D, GREGORY RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–79. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PISKOUNOVA E, VISWANATHAN SR, JANAS M, LAPIERRE RJ, DALEY GQ, SLIZ P, GREGORY RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. The Journal of biological chemistry. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- RAVER-SHAPIRA N, MARCIANO E, MEIRI E, SPECTOR Y, ROSENFELD N, MOSKOVITS N, BENTWICH Z, OREN M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Molecular cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- REINHART BJ, SLACK FJ, BASSON M, PASQUINELLI AE, BETTINGER JC, ROUGVIE AE, HORVITZ HR, RUVKUN G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- RIGOUTSOS I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer research. 2009;69:3245–8. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- RIVAS FV, TOLIA NH, SONG JJ, ARAGON JP, LIU J, HANNON GJ, JOSHUA-TOR L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature structural & molecular biology. 2005;12:340–9. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- RODRIGUEZ A, GRIFFITHS-JONES S, ASHURST JL, BRADLEY A. Identification of mammalian microRNA host genes and transcription units. Genome research. 2004;14:1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROUSH S, SLACK FJ. The let-7 family of microRNAs. Trends in cell biology. 2008;18:505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- RUBY JG, JAN CH, BARTEL DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RYBAK A, FUCHS H, SMIRNOVA L, BRANDT C, POHL EE, NITSCH R, WULCZYN FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature cell biology. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- SAITO K, ISHIZUKA A, SIOMI H, SIOMI MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS biology. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAYED D, ABDELLATIF M. MicroRNAs in development and disease. Physiological reviews. 2011;91:827–87. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- SCHIRLE NT, MACRAE IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–40. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHWARZ DS, HUTVAGNER G, DU T, XU Z, ARONIN N, ZAMORE PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- SCOTT H, HOWARTH J, LEE YB, WONG LF, BANTOUNAS I, PHYLACTOU L, VERKADE P, UNEY JB. MiR-3120 is a mirror microRNA that targets heat shock cognate protein 70 and auxilin messenger RNAs and regulates clathrin vesicle uncoating. The Journal of biological chemistry. 2012;287:14726–33. doi: 10.1074/jbc.M111.326041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIN C. Cleavage of the star strand facilitates assembly of some microRNAs into Ago2-containing silencing complexes in mammals. Molecules and cells. 2008;26:308–13. [PubMed] [Google Scholar]
- SIOLAS D, LERNER C, BURCHARD J, GE W, LINSLEY PS, PADDISON PJ, HANNON GJ, CLEARY MA. Synthetic shRNAs as potent RNAi triggers. Nature biotechnology. 2005;23:227–31. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- SMART F, ASCHRAFI A, ATKINS A, OWENS GC, PILOTTE J, CUNNINGHAM BA, VANDERKLISH PW. Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. Journal of neurochemistry. 2007;101:1367–79. doi: 10.1111/j.1471-4159.2007.04521.x. [DOI] [PubMed] [Google Scholar]
- SUN BS, DONG QZ, YE QH, SUN HJ, JIA HL, ZHU XQ, LIU DY, CHEN J, XUE Q, ZHOU HJ, REN N, QIN LX. Lentiviral-mediated miRNA against osteopontin suppresses tumor growth and metastasis of human hepatocellular carcinoma. Hepatology. 2008;48:1834–42. doi: 10.1002/hep.22531. [DOI] [PubMed] [Google Scholar]
- SUZUKI HI, ARASE M, MATSUYAMA H, CHOI YL, UENO T, MANO H, SUGIMOTO K, MIYAZONO K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Molecular cell. 2011;44:424–36. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- SUZUKI HI, YAMAGATA K, SUGIMOTO K, IWAMOTO T, KATO S, MIYAZONO K. Modulation of microRNA processing by p53. Nature. 2009;460:529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- TANG R, LI L, ZHU D, HOU D, CAO T, GU H, ZHANG J, CHEN J, ZHANG CY, ZEN K. Mouse miRNA-709 directly regulates miRNA-15a/16–1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell research. 2012;22:504–15. doi: 10.1038/cr.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAZAWA H, TSUCHIYA N, IZUMIYA M, NAKAGAMA H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15472–7. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THOMSON JM, NEWMAN M, PARKER JS, MORIN-KENSICKI EM, WRIGHT T, HAMMOND SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & development. 2006;20:2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOLIA NH, JOSHUA-TOR L. Slicer and the argonautes. Nature chemical biology. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- TRABUCCHI M, BRIATA P, GARCIA-MAYORAL M, HAASE AD, FILIPOWICZ W, RAMOS A, GHERZI R, ROSENFELD MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRIBOULET R, CHANG HM, LAPIERRE RJ, GREGORY RI. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–11. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN WYNSBERGHE PM, KAI ZS, MASSIRER KB, BURTON VH, YEO GW, PASQUINELLI AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nature structural & molecular biology. 2011;18:302–8. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VESELY C, TAUBER S, SEDLAZECK FJ, VON HAESELER A, JANTSCH MF. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome research. 2012;22:1468–76. doi: 10.1101/gr.133025.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VISWANATHAN SR, DALEY GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–9. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- VISWANATHAN SR, DALEY GQ, GREGORY RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VISWANATHAN SR, POWERS JT, EINHORN W, HOSHIDA Y, NG TL, TOFFANIN S, O’SULLIVAN M, LU J, PHILLIPS LA, LOCKHART VL, SHAH SP, TANWAR PS, MERMEL CH, BEROUKHIM R, AZAM M, TEIXEIRA J, MEYERSON M, HUGHES TP, LLOVET JM, RADICH J, MULLIGHAN CG, GOLUB TR, SORENSEN PH, DALEY GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nature genetics. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAGNER RW, SMITH JE, COOPERMAN BS, NISHIKURA K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:2647–51. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARF MB, SHEPHERD BA, JOHNSON WE, BASS BL. Effects of ADARs on small RNA processing pathways in C. elegans. Genome research. 2012;22:1488–98. doi: 10.1101/gr.134841.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARNER DR, BHATTACHERJEE V, YIN X, SINGH S, MUKHOPADHYAY P, PISANO MM, GREENE RM. Functional interaction between Smad, CREB binding protein, and p68 RNA helicase. Biochemical and biophysical research communications. 2004;324:70–6. doi: 10.1016/j.bbrc.2004.09.017. [DOI] [PubMed] [Google Scholar]
- WEST JA, VISWANATHAN SR, YABUUCHI A, CUNNIFF K, TAKEUCHI A, PARK IH, SERO JE, ZHU H, PEREZ-ATAYDE A, FRAZIER AL, SURANI MA, DALEY GQ. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–13. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WESTHOLM JO, LAI EC. Mirtrons: microRNA biogenesis via splicing. Biochimie. 2011;93:1897–904. doi: 10.1016/j.biochi.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS DR, EPPERSON LE, LI W, HUGHES MA, TAYLOR R, ROGERS J, MARTIN SL, COSSINS AR, GRACEY AY. Seasonally hibernating phenotype assessed through transcript screening. Physiological genomics. 2005;24:13–22. doi: 10.1152/physiolgenomics.00301.2004. [DOI] [PubMed] [Google Scholar]
- WINTER J, JUNG S, KELLER S, GREGORY RI, DIEDERICHS S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature cell biology. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- WYMAN SK, KNOUF EC, PARKIN RK, FRITZ BR, LIN DW, DENNIS LM, KROUSE MA, WEBSTER PJ, TEWARI M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome research. 2011;21:1450–61. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG JS, MAURIN T, ROBINE N, RASMUSSEN KD, JEFFREY KL, CHANDWANI R, PAPAPETROU EP, SADELAIN M, O’CARROLL D, LAI EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15163–8. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG N, CAO Y, HAN P, ZHU X, SUN L, LI G. Tools for investigation of the RNA endonuclease activity of mammalian Argonaute2 protein. Analytical chemistry. 2012;84:2492–7. doi: 10.1021/ac2032854. [DOI] [PubMed] [Google Scholar]
- YANG W, CHENDRIMADA TP, WANG Q, HIGUCHI M, SEEBURG PH, SHIEKHATTAR R, NISHIKURA K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nature structural & molecular biology. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAO G, YIN M, LIAN J, TIAN H, LIU L, LI X, SUN F. MicroRNA-224 is involved in transforming growth factor-beta-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Molecular endocrinology. 2010;24:540–51. doi: 10.1210/me.2009-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YEOM KH, LEE Y, HAN J, SUH MR, KIM VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic acids research. 2006;34:4622–9. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YI R, QIN Y, MACARA IG, CULLEN BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & development. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZENG Y, CULLEN BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic acids research. 2004;32:4776–85. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZENG Y, CULLEN BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. The Journal of biological chemistry. 2005;280:27595–603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- ZENG Y, YI R, CULLEN BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. The EMBO journal. 2005;24:138–48. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG X, WAN G, BERGER FG, HE X, LU X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Molecular cell. 2011;41:371–83. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG X, ZENG Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic acids research. 2010;38:7689–97. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU X, RUAN J, WANG G, ZHANG W. Characterization and identification of microRNA core promotersin four model species. PLoS computational biology. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZISOULIS DG, KAI ZS, CHANG RK, PASQUINELLI AE. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012;486:541–4. doi: 10.1038/nature11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZISOULIS DG, LOVCI MT, WILBERT ML, HUTT KR, LIANG TY, PASQUINELLI AE, YEO GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nature structural & molecular biology. 2010;17:173–9. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]