Abstract
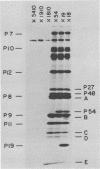
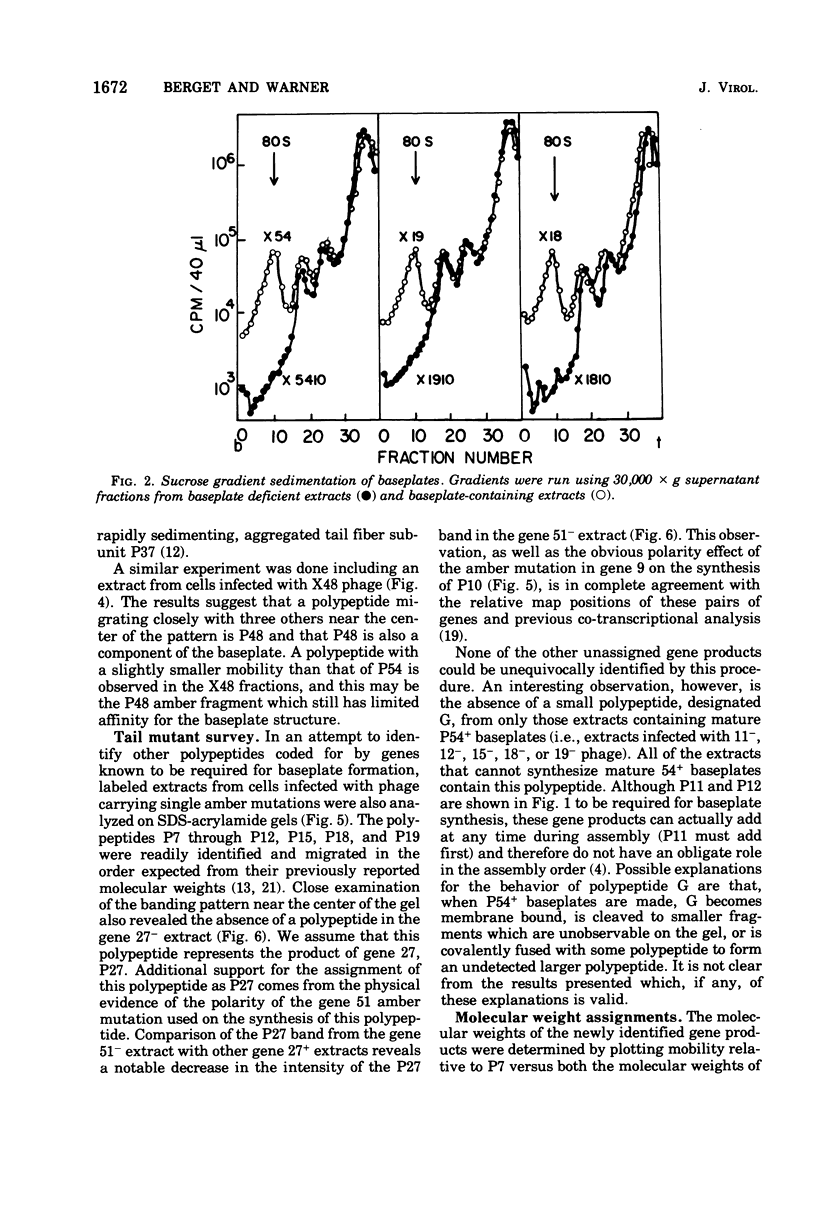
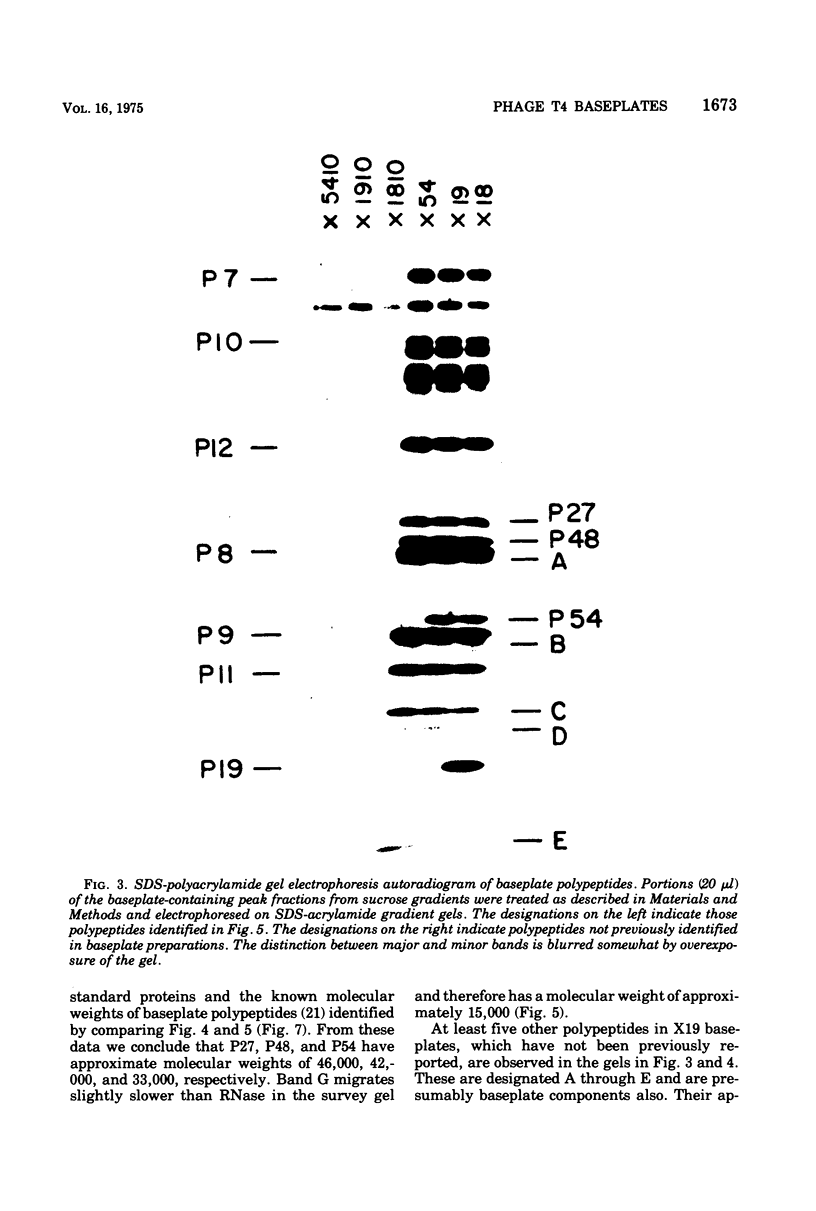
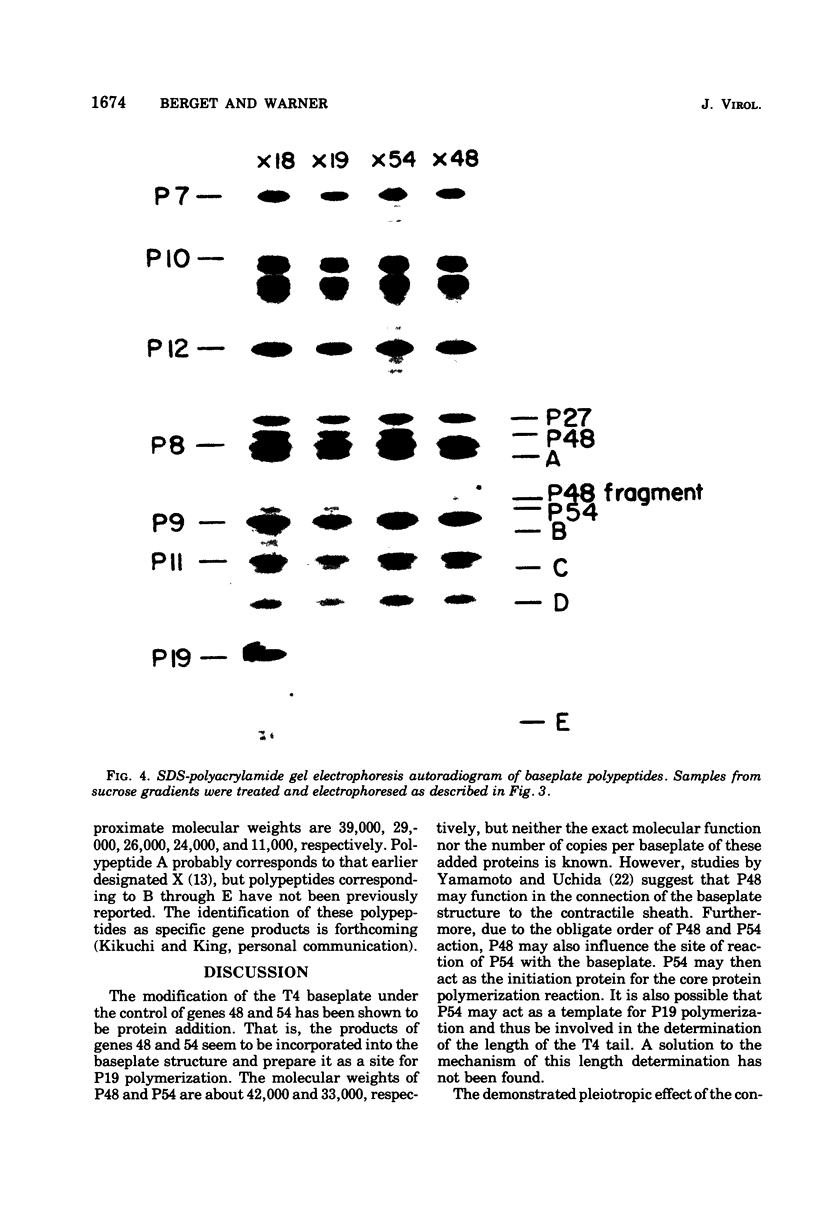
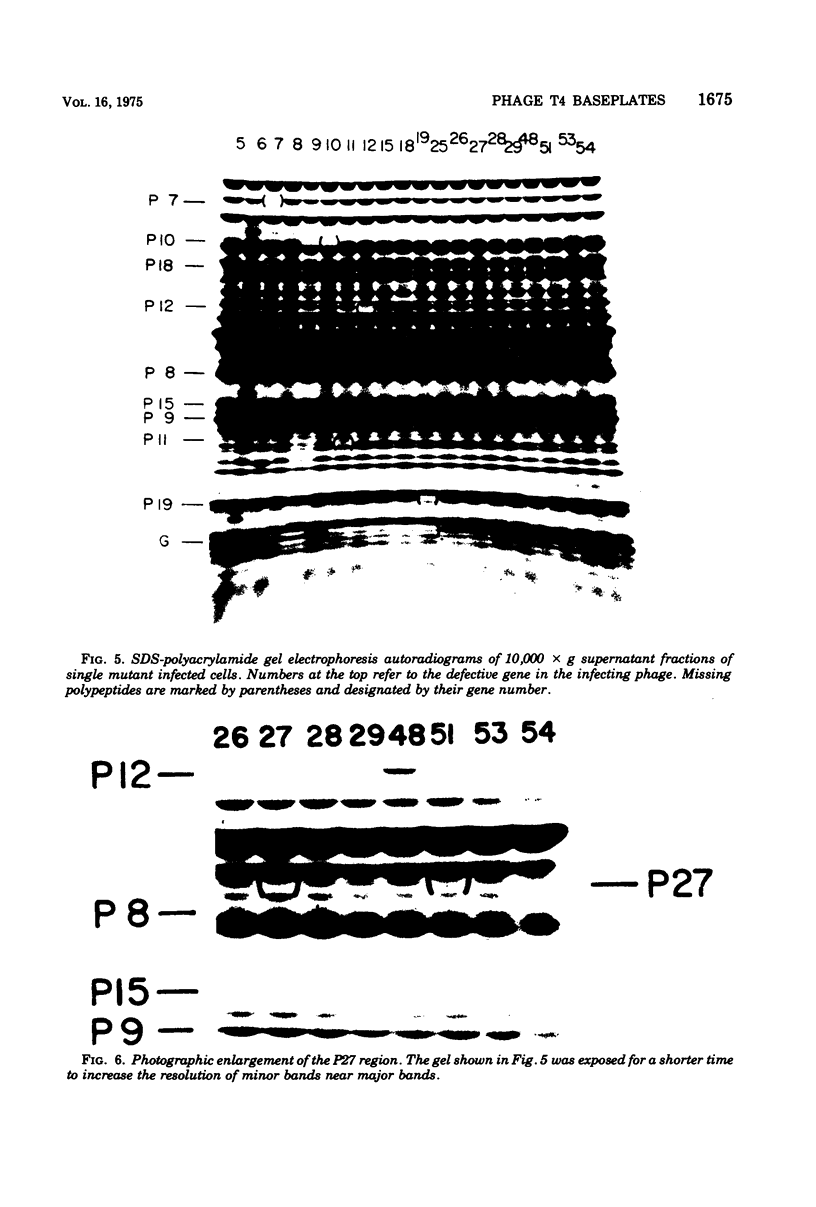
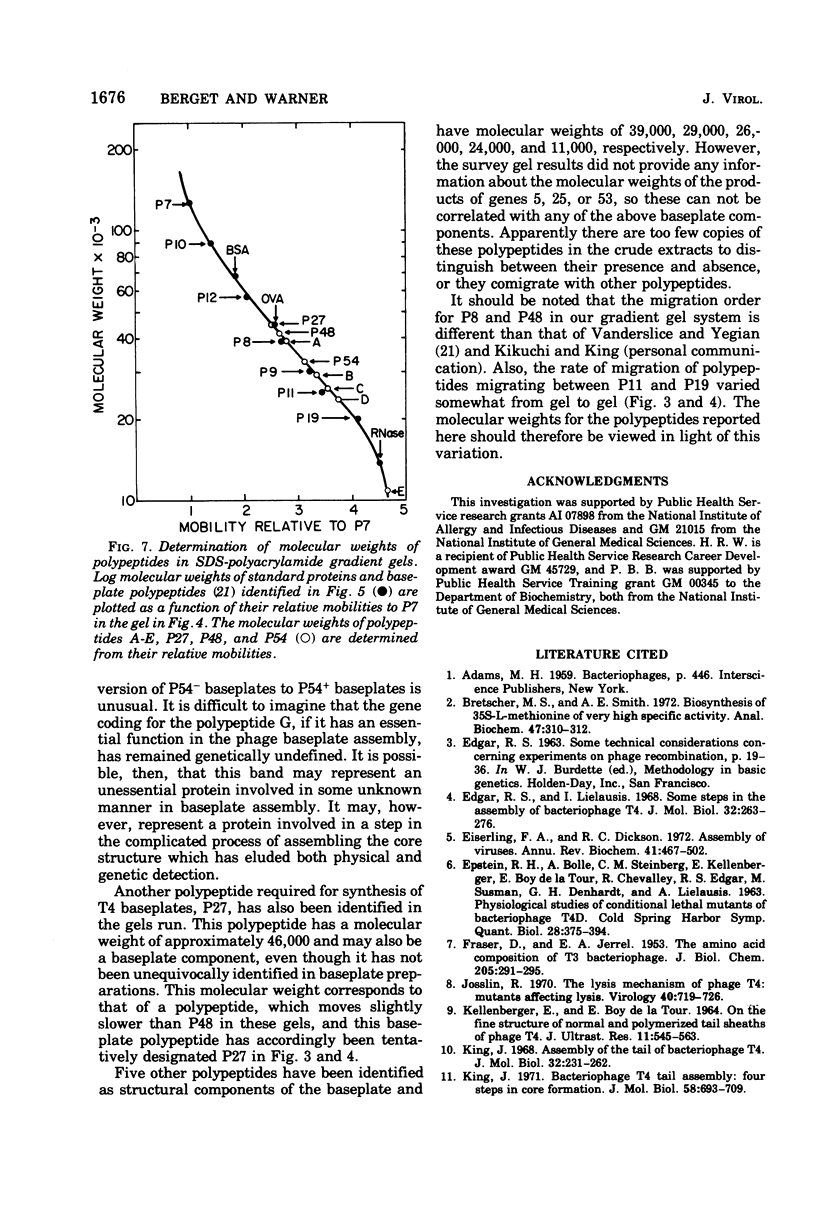
The involvement of two bacteriophage T4 gene products in the initiation of T4 tail tube and sheath polymerization on mature baseplates has been studied by radioautography of acrylamide gels of various partially completed tail structures. The products of genes 48 and 54 (P48[the nomenclature P48 refers to the protein product of bacteriophage T4 gene 48] and P54), which are known to be required for the synthesis of mature baseplates, have been shown to be structural components of the baseplate. These gene products have molecular weights of 42,000 and 33,000, respectively. The addition of P54 to the baseplate not only permits the polymerization of the core protein, P19, onto the baseplate, but also caused the disappearance of a polypeptide of molecular weight about 15,000 from the supernatant fraction of infected cells. Another gene product, P27, has been identified in the crude extracts of infected cells. This gene product, which is required for the synthesis of baseplate structures, has the same mobility as one of the unidentified structural polypeptides of the baseplate and is therefore probably also a baseplate component.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bretscher M. S., Smith A. E. Biosynthesis of 35 S-L-methionine of very high specific activity. Anal Biochem. 1972 May;47(1):310–312. doi: 10.1016/0003-2697(72)90308-9. [DOI] [PubMed] [Google Scholar]
- Edgar R. S., Lielausis I. Some steps in the assembly of bacteriophage T4. J Mol Biol. 1968 Mar 14;32(2):263–276. doi: 10.1016/0022-2836(68)90008-9. [DOI] [PubMed] [Google Scholar]
- Eiserling F. A., Dickson R. C. Assembly of viruses. Annu Rev Biochem. 1972;41:467–502. doi: 10.1146/annurev.bi.41.070172.002343. [DOI] [PubMed] [Google Scholar]
- FRASER D., JERREL E. A. The amino acid composition of T3 bacteriophage. J Biol Chem. 1953 Nov;205(1):291–295. [PubMed] [Google Scholar]
- Josslin R. The lysis mechanism of phage T4: mutants affecting lysis. Virology. 1970 Mar;40(3):719–726. doi: 10.1016/0042-6822(70)90216-3. [DOI] [PubMed] [Google Scholar]
- KELLENBERGER E., DELATOUR E. B. ON THE FINE STRUCTURE OF NORMAL AND "POLYMERIZED" TAIL SHEATH OF PHAGE T4. J Ultrastruct Res. 1964 Dec;11:545–563. doi: 10.1016/s0022-5320(64)80081-2. [DOI] [PubMed] [Google Scholar]
- King J. Assembly of the tail of bacteriophage T4. J Mol Biol. 1968 Mar 14;32(2):231–262. doi: 10.1016/0022-2836(68)90007-7. [DOI] [PubMed] [Google Scholar]
- King J. Bacteriophage T4 tail assembly: four steps in core formation. J Mol Biol. 1971 Jun 28;58(3):693–709. doi: 10.1016/0022-2836(71)90034-9. [DOI] [PubMed] [Google Scholar]
- King J., Laemmli U. K. Bacteriophage T4 tail assembly: structural proteins and their genetic identification. J Mol Biol. 1973 Apr 5;75(2):315–337. doi: 10.1016/0022-2836(73)90024-7. [DOI] [PubMed] [Google Scholar]
- King J., Mykolajewycz N. Bacteriophage T4 tail assembly: proteins of the sheath, core and baseplate. J Mol Biol. 1973 Apr 5;75(2):339–358. doi: 10.1016/0022-2836(73)90025-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Meezan E., Wood W. B. The sequence of gene product interaction in bacteriophage T4 tail core assembly. J Mol Biol. 1971 Jun 28;58(3):685–692. doi: 10.1016/0022-2836(71)90033-7. [DOI] [PubMed] [Google Scholar]
- Smith F. L., Haselkorn R. Proteins associated with ribosomes in T4-infected E. coli. Cold Spring Harb Symp Quant Biol. 1969;34:91–94. doi: 10.1101/sqb.1969.034.01.014. [DOI] [PubMed] [Google Scholar]
- Snustad D. P. Dominance interactions in Escherichia coli cells mixedly infected with bacteriophage T4D wild-type and amber mutants and their possible implications as to type of gene-product function: catalytic vs. stoichiometric. Virology. 1968 Aug;35(4):550–563. doi: 10.1016/0042-6822(68)90285-7. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Crasemann J. M., Yegian C., Stahl M. M., Nakata A. Co-transcribed cistrons in bacteriophage T4. Genetics. 1970 Feb;64(2):157–170. doi: 10.1093/genetics/64.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Bacteriophage T7. Science. 1972 Apr 28;176(4033):367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Vanderslice R. W., Yegian C. D. The identification of late bacteriophage T4 proteins on sodium dodecyl sulfate polyacrylamide gels. Virology. 1974 Jul;60(1):265–275. doi: 10.1016/0042-6822(74)90384-5. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Uchida H. Organization and function of bacteriophage T4 tail. I. Isolation of heat-sensitive T4 tail mutants. Virology. 1973 Mar;52(1):234–245. doi: 10.1016/0042-6822(73)90412-1. [DOI] [PubMed] [Google Scholar]