Abstract
Although Escherichia albertii is an emerging intestinal pathogen, it has been associated only with sporadic human infections. In this study, we determined that a human gastroenteritis outbreak at a restaurant in Japan had E. albertii as the major causative agent.
Keywords: Escherichia albertii, enteric infections, bacteria, outbreak, human gastroenteritis, Japan
Escherichia albertii is an emerging human and bird pathogen that belongs to the attaching and effacing group of pathogens. This group of pathogens forms lesions on intestinal epithelial cell surfaces by the combined action of intimin, an eae gene–encoded outer membrane protein, and type III secretion system effectors (1–4).
Recently, we found that E. albertii represents a substantial proportion of the strains that had previously been identified as eae-positive Escherichia coli, enteropathogenic E. coli or enterohemorrhagic E. coli; 26 of the 179 eae-positive strains analyzed were found to be E. albertii (5). Furthermore, E. albertii is also a potential Shiga toxin 2f (Stx2f)–producing bacterial species (5). However, no E. albertii–associated gastroenteritis outbreak has been reported, which generates doubts regarding the clinical role of this microorganism. In this study, we revisited an outbreak of gastroenteritis that was presumed to have been caused by eae-positive atypical E. coli OUT:HNM (6) to determine if it was actually caused by E. albertii.
The Study
An outbreak of gastroenteritis occurred at the end of May 2011 in Kumamoto, Japan, among persons who attended 1 of 2 parties held in a Japanese restaurant on May 29. We reviewed case records for the 94 persons who attended the parties. A total of 48 persons became ill; 43 of them attended the first party (a total of 86 attended), and 5 attended the second party (a total of 8 attended). The ill participants had not eaten any food in common except for the meals served at the restaurant. The main symptoms of the patients were diarrhea (83%), abdominal pain (69%), fever (44%; mean temperature 37.2°C), and nausea (29%). The mean incubation period was 19 h.
A routine protocol to identify bacteria and viruses (Technical Appendix) was used by our laboratory to examine 54 fecal specimens from 44 party participants and 10 members of the restaurant kitchen staff (7 party participants and all of the kitchen staff were asymptomatic). Atypical E. coli (lactose negative; OUT:HNM) strains harboring the eae gene and E. coli OUT:H18 strains harboring the stx2d and astA (but not eae) genes were isolated from 24 and 3 specimen, respectively; 7 specimens yielded both strains (Table 1). The stx2-positive/eae-negative E. coli strains were found to be serotype O183 (a recently described O serotype) by agglutination testing with O183-specific antiserum (S. Iyoda, M. Ohnishi, unpub. data).
Table 1. Isolates from fecal specimens of party participants during outbreak of gastroenteritis associated with Escherichia albertii, Japan*.
| Isolate | Origin of isolates |
|||||
|---|---|---|---|---|---|---|
| Participants, n = 44 |
Kitchen staff, n = 10 |
|||||
| Symptomatic | Asymptomatic | No information | Symptomatic | Asymptomatic | ||
| E. albertii† | 21 | 1 | 0 | 0 | 2 | |
| E. albertii† and E. coli O183:H18‡ | 7 | 0 | 0 | 0 | 0 | |
| E. coli O183:H18‡ | 3 | 0 | 0 | 0 | 0 | |
| None | 6 | 5 | 1 | 0 | 8 | |
*None, negative for both pathogens. †Initially identified as atypical (lactose negative) E. coli OUT:HNM harboring the intimin (eae) gene. ‡Initially identified as eae negative E. coli OUT:H18 harboring the Shiga toxin 2d and enteroaggregative E.coli heat-stable toxin genes.
All atypical E. coli strains showed identical or nearly identical XbaI-digested DNA banding patterns by pulsed-field gel electrophoresis, and the 10 E. coli O183:H18 strains also exhibited identical patterns (Figure). The source of the infection was most likely the meals served in the restaurant, but a bacteriological examination of the meal or of the ingredients used to prepare the meal was not possible because none of the food was preserved for analysis.
Figure.
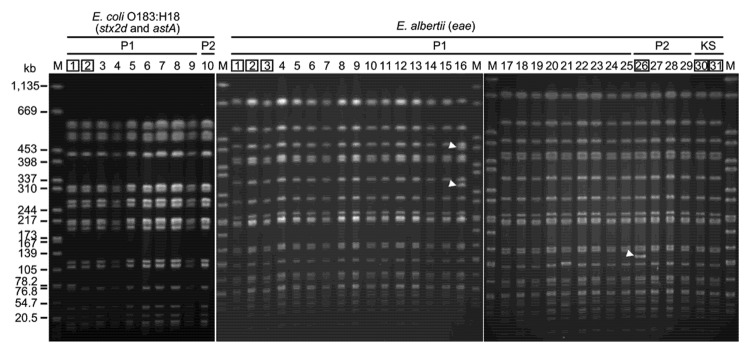
XbaI-digested pulsed-field gel electrophoresis profiles of isolates from fecal specimens collected from patients during an outbreak of human gastroenteritis associated with Escherichia albertii, Japan. Extra bands observed in 2 E. albertii isolates are indicated by arrowheads (only 1 or 2 band differences). The 2 E. coli O183:H18 and 6 E. albertii isolates indicated by numbers in boxes were subjected to multilocus sequence analysis (Technical Appendix). stx2d, Shiga toxin 2d gene; astA, enteroaggregative E. coli heat-stable toxin gene; eae, intimin gene. Lane M, Salmonella enterica serovar Braenderup strain H9812 (used as a DNA size standard); lanes P1, Party 1; lanes P2, Party 2; lanes KS, kitchen staff.
The lactose-negative/eae-positive features of the OUT:HNM strains suggested that these strains might be E. albertii. We examined additional biochemical properties of these strains and found that they exhibited the E. albertii–specific features described (4,5). These features include nonmotility, inability to ferment xylose and lactose, and inability to produce β-d-glucuronidase. The E. coli O183:H18 strains demonstrated common phenotypic and biochemical properties of E. coli (7).
To determine whether the E. albertii–like OUT:HNM strains were E. albertii, we randomly selected 6 strains and determined their phylogeny by multilocus sequence analysis as described (5) (Technical Appendix Table). Results indicated that although the E. coli O183:H18 strain analyzed in parallel belongs to E. coli sensu stricto, the E. albertii–like OUT:HNM strains belong to the E. albertii lineage; all 6 strains showed identical sequences (Technical Appendix Figure).
We further examined the intimin subtype by sequencing the eae gene, the chromosome integration site of the locus of enterocyte effacement encoding the eae gene and a set of type III secretion system genes, and the presence and subtype of the cdtB gene as described (5). Results showed that the E. albertii strains had intimin σ, which is rarely identified in enteropathogenic E. coli or enterohemorrhagic E. coli; the locus of enterocyte effacement was integrated into the pheU tRNA gene; and the cdtB gene of the II/III/V subtype group was present. These features are consistent with recently described genetic features of E. albertii (5).
We divided the party participants into 4 groups according to strain isolation patterns and statistically assessed the association of strain isolation patterns with incidence of clinical symptoms (Table 2). The results indicated that persons infected with only E. albertii or persons infected with E. albertii and E. coli O183:H18 had diarrhea and abdominal pain more frequently than did uninfected persons (p<0.05) and that the incidence of asymptomatic carriers was lower among persons infected only with E. albertii.
Table 2. Clinical symptoms of party participants during outbreak of gastroenteritis associated with Escherichia albertii, by pathogen identified, Japan*.
| Symptom | E. albertii, n = 21† | E. albertii and E. coli O183:H18, n = 7 | E. coli O183:H18, n = 3 | None,‡ n =11§ |
|---|---|---|---|---|
| Diarrhea | 17 (81)¶ | 7 (100)§ | 1 (33) | 4 (36) |
| Abdominal pain | 16 (76)¶ | 6 (86)§ | 2 (67) | 3 (27) |
| Nausea | 5 (24) | 5 (71)§ | 0 | 1 (9) |
| Fever | 8 (38) | 4 (57) | 2 (67) | 4 (36) |
| None | 1 (5)¶ | 0 | 0 | 5 (45) |
*Values are no. (%). None, negative for both pathogens. †One symptomatic person was excluded because no clinical record was available. ‡Negative for both pathogens. §One person was excluded because no clinical record was available. ¶A 2-tailed Fisher exact test (p<0.05) showed significant differences between the groups from which E. albertii or E. coli O183:H18 was isolated and the groups from which they were not isolated.
Nucleotide sequences obtained in this study have been deposited in the DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank database. Accession numbers and other information on sequence analyses are shown in the Technical Appendix.
Conclusions
In this gastroenteritis outbreak, E. albertii or stx2-positive E. coli O183:H18 was isolated from 24 ill patients; both strains were isolated from 7 patients. Thus, although the responsible meal or food was not identified, it was most likely contaminated with these 2 microorganisms. The contribution or involvement of E. coli O183:H18 in this outbreak is unknown because there were 3 patients from whom only E. coli O183:H18 was isolated and because there were no differences in clinical symptoms between persons infected with E. coli O183:H18 and persons not infected (Tables 1, 2). In contrast, E. albertii was isolated from a larger number of patients, and many fecal specimens yielded only E. albertii (Table 1).
The proportion of persons who had clinical symptoms was also higher for E. albertii–positive party participants than for uninfected persons (Table 2). Therefore, it is plausible that E. albertii was the major causative pathogen of this outbreak. This information indicates that E. albertii can cause gastroenteritis outbreaks among humans (5).
More attention should be given to sporadic cases and outbreak cases caused by this emerging pathogen. It may also be informative to revisit past outbreak cases caused by eae-positive atypical E. coli if pathogens were recorded as being nonmotile, unable to ferment lactose and xylose, and unable to produce β-d-glucuronidase.
Protocol used to identify bacteria and viruses in fecal specimens obtained during a human gastroenteritis outbreak associated with Escherichia albertii, Japan.
Acknowledgments
We thank Sunao Iyoda and Makoto Ohnishi for sharing their unpublished results of E. coli serotyping and Keigo Ekinaga, Haruki Tokunaga, and Ryuusei Higashi for providing materials and epidemiologic information.
This study was supported by a grant-in-aid for scientific research from MEXT Japan (Wakate-B, 23790480) to T.O. and a grant from the Yakult Foundation.
Biography
Dr Ooka is an assistant professor in the Department of Infectious Diseases, Faculty of Medicine, University of Miyazaki, Miyazaki, Japan. His research interests include bacterial genomics and pathogenicity.
Footnotes
Suggested citation for this article: Ooka T, Tokuoka E, Furukawa M, Nagamura T, Ogura Y, Arisawa K, et al. Human gastroenteritis outbreak associated with Escherichia albertii, Japan. Emerg Infect Dis [Internet]. 2013 Jan [date cited]. http://dx.doi.org/10.3201/eid1901.120646
References
- 1.Albert MJ, Alam K, Islam M, Montanaro J, Rahman AS, Haider K, et al. Hafnia alvei, a probable cause of diarrhea in humans. Infect Immun. 1991;59:1507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert MJ, Faruque SM, Ansaruzzaman M, Islam MM, Haider K, Alam K, et al. Sharing of virulence-associated properties at the phenotypic and genetic levels between enteropathogenic Escherichia coli and Hafnia alvei. J Med Microbiol. 1992;37:310–4. 10.1099/00222615-37-5-310 [DOI] [PubMed] [Google Scholar]
- 3.Huys G, Cnockaert M, Janda JM, Swings J. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int J Syst Evol Microbiol. 2003;53:807–10. 10.1099/ijs.0.02475-0 [DOI] [PubMed] [Google Scholar]
- 4.Oaks JL, Besser TE, Walk ST, Gordon DM, Beckmen KB, Burek KA, et al. Escherichia albertii in wild and domestic birds. Emerg Infect Dis. 2010;16:638–46. 10.3201/eid1604.090695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ooka T, Seto K, Kawano K, Kobayashi H, Etoh Y, Ichihara S, et al. Clinical significance of Escherichia albertii. Emerg Infect Dis. 2012;18:488–92. 10.3201/eid1803.111401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokuoka E, Furukawa M, Nagamura T, Harada S, Ekinaga K, Tokunaga H, et al. Food poisoning outbreak due to atypical EPEC OUT:HNM, May 2011—Kumamoto. Infectious Agents Surveillance Report. 2012;33:8–9. [Google Scholar]
- 7.Nataro JP, Bopp CA, Fields PI, Kaper JB, Strockbine NA. Escherichia, Shigella, and Salmonella. In: Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Manual of clinical microbiology, 9th ed. Washington (DC): American Society for Microbiology Press; 2007. p. 670–87. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protocol used to identify bacteria and viruses in fecal specimens obtained during a human gastroenteritis outbreak associated with Escherichia albertii, Japan.


