Abstract
Smad proteins are the most well-characterized intracellular effectors of the transforming growth factor β (TGF-β) signal. The ability of the Smads to act as transcriptional activators via TGF-β-induced recruitment to Smad binding elements (SBE) within the promoters of TGF-β target genes has been firmly established. However, the elucidation of the molecular mechanisms involved in TGF-β-mediated transcriptional repression are only recently being uncovered. The proto-oncogene c-myc is repressed by TGF-β, and this repression is required for the manifestation of the TGF-β cytostatic program in specific cell types. We have shown that Smad3 is required for both TGF-β-induced repression of c-myc and subsequent growth arrest in keratinocytes. The transcriptional repression of c-myc is dependent on direct Smad3 binding to a novel Smad binding site, termed a repressive Smad binding element (RSBE), within the TGF-β inhibitory element (TIE) of the c-myc promoter. The c-myc TIE is a composite element, comprised of an overlapping RSBE and a consensus E2F site, that is capable of binding at least Smad3, Smad4, E2F-4, and p107. The RSBE is distinct from the previously defined SBE and may partially dictate, in conjunction with the promoter context of the overlapping E2F site, whether the Smad3-containing complex actively represses, as opposed to transactivates, the c-myc promoter.
The transforming growth factor β (TGF-β) superfamily of ligands, which includes bone morphogenetic proteins, activins, nodals, and TGF-βs, are multifunctional regulators of cellular differentiation, apoptosis, and proliferation. The tumor-suppressive TGF-β pathway, constituents of which are mutated in a variety of human cancers (8, 16, 52, 55, 65), potently inhibits the proliferation of most normal lymphoid, endothelial, and epithelial cells (46, 52, 56). The initiation of this cytostatic program in certain cell types is dependent upon the ability of TGF-β to repress the transcriptional initiation of the proto-oncogene c-myc (3, 12, 89). Although much is known about the molecular mechanisms involved in TGF-β-mediated gene activation, the question of how the TGF-β pathway actively inhibits the transcription of the subset of genes that it represses, such as c-myc, is less well elucidated.
TGF-β initiates its signal through binding the TGF-β type II serine/threonine (Ser/Thr) receptor kinase, which in turn phosphorylates the type I Ser/Thr receptor, forming an activated receptor complex (85, 86). The activated type I receptor is then able to phosphorylate its intracellular effector substrates, which include the highly homologous receptor Smads, Smad2 and Smad3 (5, 20, 23, 30, 51, 66, 73). These cytoplasmically retained Smads then form heteromeric complexes with the common signaling Smad, Smad4, and translocate to the nucleus (1, 43, 49, 58, 92). Nuclear heteromeric Smad complexes modulate the transcription of TGF-β target genes through physical association with other cofactors as well as direct recruitment to specific, promoter-contained sequence (4, 14, 53, 54, 60, 79, 94).
The Smad binding elements (SBEs) GTCT and its palindrome AGAC within particular promoters are capable of binding the N-terminal Mad homology domain 1 (MH1) of Smad3 (70, 91) and mediate the TGF-β-induced transactivation of various TGF-β target genes, such as c-jun and plasminogen activator inhibitor 1 (PAI-1) (19, 75, 83). The C-terminal MH2 domain of the Smads, tethered to the MH1 domain by a proline-rich linker, contains a transactivation domain and is able to bind the coactivators CBP/p300 as well as other transcription factors, such as proteins of the AP-1 family (37, 47, 59, 62, 63, 69, 83, 90).
Recent studies have begun to elucidate the mechanisms involved in the TGF-β-mediated repression of c-myc and have expanded our understanding of how the TGF-β cytostatic program is initiated. TGF-β upregulates the expression of the cyclin-dependent kinase (CDK) inhibitors p15 and p21, and the elevated CDK inhibitor levels in turn suppress the activity of the G1-phase-specific CDKs 2 and 4 (15, 28, 64). The subsequent hypophosphorylated state of the CDK substrate pocket proteins, pRb, p107, and p130, leads to pocket protein sequestration of the E2F family of transcription factors and, thus, inhibits E2F-1-, -2-, and -3-mediated transactivation of genes required for G1-to-S-phase cell cycle progression (18, 21, 57, 78). In normally cycling cells, hypophosphorylated pocket proteins also associate with E2F-4 and -5, forming complexes that repress the expression of specific genes in early G1 necessary for a properly coordinated cell cycle (18, 21, 78). It has been demonstrated that the TGF-β downregulation of c-Myc expression is required for p15 and p21 transactivation, and these findings highlight the importance of TGF-β-mediated c-myc repression in initiating the TGF-β-induced G1-phase cell cycle arrest program (12, 68, 74, 81).
Although the SBE is known to directly bind Smad3 and mediate TGF-β transactivation of specific genes, a TGF-β inhibitory element (TIE) was first characterized as essential to mediating TGF-β repressive effects in the promoter of the TGF-β-repressed gene stromelysin 1 (39). The identified stromelysin 1 TIE was compared to the promoter sequence of other TGF-β-repressed genes, such as collagenase 1/matrix metalloproteinase 1 (MMP-1), urokinase, and c-myc, and the following nucleotides, in which uppercase letters signify invariance and lowercase letters signify preferred sequence, became known as a consensus TIE: 5′-GNNTTGGtGa-3′ (39). TIEs within the rat stromelysin and rabbit MMP-1 promoters have been shown to bind TGF-β-induced c-Fos, and this mechanism of c-Fos upregulation and subsequent TIE binding has been reported as essential in mediating the transcriptional repression of these promoters by TGF-β (39, 82). Although the recruitment of c-Fos to the putative c-myc TIE (5′-GGCTTGGCGG-3′; nucleotides −84 to −75 relative to the major P2 transcriptional start site) has not been implicated in the TGF-β-mediated repression of c-myc, an overlapping, consensus E2F binding site (5′-GGCGGGAAA-3′; −79 to −71) has been shown to be important for transactivation of the c-myc promoter by E2F and Ets transcription factors (33, 67). Recently, the c-myc TIE has been characterized as essential to TGF-β-mediated repression of c-myc and, importantly, a biotinylated c-myc TIE oligonucleotide was shown to be capable of directly or indirectly precipitating Smad3 and Smad4 (10).
Armed with evidence that targeted deletion of Smad3 results in inhibition of TGF-β-induced repression of c-myc, we set out to define how Smad3, a protein known to be involved in the transactivation of TGF-β target genes through promoter-contained SBE binding, was involved in the inhibition of c-myc transcriptional initiation. Along with two complementary reports that were published during the completion of this study (11, 89), we demonstrate that Smad3 is required for TGF-β-mediated repression of c-myc through a mechanism that involves direct Smad3 binding to the c-myc TIE. In addition, these three studies all demonstrate that the c-myc TIE is unique from the consensus TIE 5′-GNNTTGGtGa-3′ in that it overlaps with a consensus E2F site that is capable of precipitating E2F-4 (11, 89), thus constituting an extended TIE, which heretofore will be simply referred to as the c-myc TIE. It was nicely demonstrated by Chen et al. that Smad3 directly interacts with both E2F-4 or -5 and p107, forming a repressive complex recruited to the c-myc TIE in response to TGF-β regardless of cell cycle stage, and that E2F-4, E2F-5, and p107 were required for TGF-β-mediated c-myc repression (11). Our study is the first to carefully map the Smad3 binding site within the c-myc TIE and demonstrate its distinction from a previously defined SBE. We demonstrate that the “SBE-like” sequences adjacent to and contained within the c-myc TIE are dispensable for both Smad binding and mediation of the repressive response to TGF-β. Thus, the c-myc TIE is a unique composite element that is comprised of both an E2F site and a novel Smad3 binding site, termed a repressive Smad binding element (RSBE). Smad3 can act context specifically as either a transcriptional activator or active repressor, dependent upon the protein cofactors with which it associates and the promoter sequence to which it is recruited: the SBE, as in many documented cases, or the RSBE, as in the case of c-myc, respectively.
MATERIALS AND METHOD
Plasmid construction and mutagenesis
The wild-type c-myc promoter reporter (c-mycDel-5pGL3, or WT c-myc) contains nucleotides −279 to + 337 relative to the c-myc P2 transcriptional start site. This construct was generated by subcloning a SmaI/PvuII fragment excised from the c-myc Del-1 construct generously provided by Bert Vogelstein into the SmaI site of the pGL3-basic luciferase reporter vector (Promega) (29). Correct orientation was confirmed by diagnostic restriction enzyme digestion. Corresponding reporters with 2-bp scanning mutations across the c-myc TIE (M2-M8) were generated with the QuikChange site-directed mutagenesis kit (Stratagene) using the 30-bp oligonucleotides depicted below in Fig. 5A and the aforementioned c-mycDel-5pGL3 as template according to the manufacturer's protocol. Four copies of the sequence encompassing the c-myc TIE from −87 to −72 relative to the P2 transcriptional start site (5′-AGAGGCTTGGCGGGAA-3′) were inserted upstream of a simian virus 40 (SV40) promoter contained within the pGL3-promoter vector (Promega). This construct, 4x WT TIE, was created by subcloning two copies of the annealed, complementary oligonucleotides, 5′-GATCAGAGGCTTGGCGGGAAAGAGGCTTGGCGGGAA-3′ and 5′-GATCTTCCCGCCAAGCCTCTTTCCCGCCAAGCCTCT-3′, into the BglII site of the pGL3-promoter vector. A corresponding reporter, 4x M5/M7 TIE, was constructed that contained 4-bp mutations within the TIE by directionally subcloning a XhoI/BglII-digested, double-strand oligonucleotide (5′-CGGGCTCGAGATCAGAGGCTTATCGATAAAGAGGCTTATCGATAAGATCAGAGGCTTATCGATAAAGAGGCTTATCGATAAGATCTCGGA-3′) into the XhoI/BglII sites of the pGL3-promoter vector. The underlined sequence represents the mutations within the four TIE copies and the only distinctions between the 4x M5/M7 TIE and 4x WT TIE. The correct sequences of all the constructed plasmids were confirmed by sequencing. 3TP-Lux and the PAI-1 promoter construct, p800LUC, have been previously described and were kind gifts from Joan Massagué and David J. Loskutoff, respectively (38, 85).
FIG. 5.
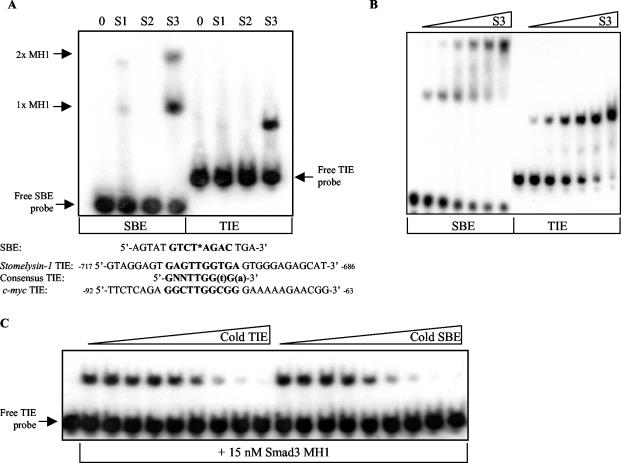
The c-myc TIE contains a novel Smad binding site, or RSBE, maximally comprised of 5′-TTGGCGGGAA-3′. (A) DNA cold competitor EMSAs similar to that described in the legend for Fig. 4C were performed to map the Smad3 binding sequence. A 15 nM concentration of recombinant Smad3 MH1 domain was incubated with radiolabeled, wild-type c-myc TIE probe and a 1.365 μM concentration of the indicated unlabeled TIE competitor. Complexes of Smad3 MH1 bound to the radiolabeled probe (S3 MH1) and unbound, free probe are indicated by arrows. The sequences for the wild-type (WT) c-myc TIE and various, scanning TIE mutants (M1 to M10 and M“SBE” or MS TIE) are listed below. The mutated nucleotides are underlined, and the sequence with homology to a consensus TIE is depicted in bold. The top panel demonstrates an equal amount of employed cold competitor DNA. The shown EMSA is representative of five similar experiments. (B) The underlined guanines 5′-GGCGG-3′ (−79 to −75 relative to the P2 transcriptional start site) within the c-myc TIE were shown to be important in mediating Smad3 binding by methylation interference assay and are boxed. The 30-bp, wild-type c-myc TIE oligonucleotide was radiolabeled on the 5′ end of the sense strand of DNA, methylated with DMS, and then incubated with recombinant Smad3 MH1 domain. The methylated DNA-protein mix was separated by PAGE, and free, or unbound, probe as well as protein-shifted probe was isolated by electroelution. The free (outer two lanes) and Smad3 MH1-bound (middle lane) probes were then piperidine cleaved and separated by denaturing PAGE. Nucleotide −86 relative to the c-myc P2 transcriptional start site is shown at the bottom and −62 is at the top. (C) The binding affinity of recombinant Smad3 MH1 domain (S3) to wild-type c-myc TIE (WT TIE) probe compared to M“SBE” TIE probe was assessed by EMSA. M“SBE” TIE represents a c-myc TIE sequence in which the putative SBE sequence, 5′-GGCT-3′ (−84 to −81) was mutated to TTAA. Threefold serial dilutions of recombinant Smad3 MH1, from 1.668 to 2.29 mM, were incubated with WT TIE and M“SBE” TIE probes as indicated. The presented EMSA is representative of four independent experiments. (D) A 300-μg aliquot of whole-cell lysate (WCL) obtained from sonicated HaCaT cells treated with vehicle or TGF-β for 1 h was incubated with equimolar amounts of biotinylated double-strand oligonucleotides (B-DNA). The SBE sequence used is listed in Fig. 4A, and the composition of the wild-type and mutated c-myc TIE oligonucleotides (WT, MS, and M3 to M6) is depicted in panel A. DNA-bound protein complexes were isolated by subsequent incubation with streptavidin-linked agarose beads, centrifugation, and washing. DNA-bound protein was then separated by sodium dodecyl sulfate-PAGE and analyzed by Western blotting for Smad3 and Smad4. (E) The DNAP experiment shown is exactly the same as that depicted in panel D with the exception that 300 μg of nuclear extract (NE) was used in place of whole-cell lysate. The shown DNAPs are representative of three independent experiments.
Cell culture and isolation
HaCaT cells, spontaneously immortalized human keratinocytes, were kindly provided by Nobert E. Fusenig and maintained in α-MEM (Life Technologies) supplemented with 10% fetal bovine serum (FBS). Two stable transfectant HaCaT cell lines, HaCaT neomycin-resistant control (HaCaT control) and HaCaT Smad3D407E/clone 3.9 (HaCaT Smad3 dominant negative [S3 DN]), were a generous gift of Mitsuyasu Kato (26). These HaCaT lines were first cultured in MCDB 153 medium (Sigma) supplemented with 10 ng of human epidermal growth factor (Roche)/ml, 10% dialyzed FBS, and 300 mg of G418 (Life Technologies)/ml and then weaned and maintained in α-MEM (Life Technologies) plus 10% FBS and 300 mg of G418/ml. Primary mouse keratinocytes were isolated from 1- to 3-day postnatal littermates birthed by Smad3ex1/+ heterozygotes of a 50:50 C57BL6:129/Sv mixed-strain background (14). Trunk skin was removed, washed three times in phosphate-buffered saline (PBS) with antibiotics (kanamycin, amphotericin, penicillin, and streptomycin [KAPS]), and digested in 0.25% trypsin and KAPS overnight at 4°C followed by 20 min at 37°C. Trypsin was then neutralized with trypsin neutralization buffer (Clonetics), and the trunk skin was removed. The epidermis was carefully peeled from the dermis in a 10-cm2 culture dish containing low-CaCl2 keratinocyte growth medium (KGM; Clonetics), and the dermis was discarded. A scalpel was used to gently scrape the surface of the epidermis that was previously adjacent to the dermis to remove viable keratinocytes. After the rest of the epidermal sheet was discarded, the keratinocytes were cultured in the aforementioned low-CaCl2 KGM that included CaCl2-free keratinocyte basal medium (KBM), 5 μg of insulin/ml, 0.5 μg of hydrocortisone/ml, 30 μg of bovine pituitary extract/ml, and 30 μM CaCl2 (all from Clonetics); 10 ng of murine epidermal growth factor (Roche)/ml, penicillin, and streptomycin. BALB/c murine keratinocytes (BALB/MK) were a generous gift from Harold L. Moses (13). The BALB/MK cells were passaged in CaCl2-free KBM (Clonetics) supplemented with 10% dialyzed fetal calf serum, 40 μM CaCl2, 4 ng of murine epidermal growth factor (Roche)/ml, penicillin, and streptomycin. Crude colon epithelial cell preps were isolated from 3- to 5-month-old Smad3 wild-type or homozygous null mice (14). Colons were excised, split longitudinally, and rinsed three times in PBS, and the lumen was scraped with a scalpel to remove the epithelium. Epithelial cells were collected in PBS, gently pelleted, and lysed in buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 50 mM NaF, 0.5% NP-40, 1 mM dithiothreitol (DTT), and a mixture of protease and phosphatase inhibitors (10 mM β-glycero-phosphate, 0.5 mM NaF, 0.5 mM sodium orthovanadate, 0.1 mM sodium molybdate, 5 μg of antipain/ml, 5 μg of aprotinin/ml, 5 μg of leupeptin/ml, 0.5 μg of pepstatin/ml, and 5 μg of soybean trypsin inhibitor/ml). Western blotting of the colon epithelium extracts was performed with anti-c-Myc (sc-42; Santa Cruz Biotechnology) and anti-actin (Sigma).
[3H]thymidine incorporation assay
Primary murine keratinocytes were isolated as described previously from wild-type and Smad3ex1/ex1 homozygous null littermates. A total of 20,000 cells/well were seeded in 12-well plates and cultured under normal conditions. The cells were treated with 100 pM TGF-β or vehicle for 30 h and labeled with 5 μCi of [3H]thymidine for the last 5 h of treatment. Labeled cells were then fixed in 10% trichloroacetic acid and lysed with 0.2 N NaOH, and the DNA-incorporated [3H]thymidine was measured by a scintillation counter.
RPA
Control and Smad3 DN HaCaT cells were seeded at 3 × 106 cells/10-cm2 plate and grown for 24 h under the described serum conditions. The cells were then treated with 100 pM TGF-β for the indicated times. Total RNA was isolated with RNeasy (Qiagen) according to the manufacturer's protocol. Five micrograms of RNA harvested from each time point was assayed according to the manufacturer's protocol with a RiboQuant MultiProbe RNase protection assay (RPA) kit (PharMingen) using 5 × 105 cpm/sample of the riboprobes generated with the in vitro transcription kit (PharMingen) and a custom template set (PharMingen) that included PAI-1, c-myc, L32, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
For the rest of the RPAs, antisense riboprobes were made from a murine β-actin riboprobe template supplied with the Ambion RPAII kit as well as a PCR product of the murine c-myc cDNA that incorporated the T7 promoter sequence. Purified PCR primers used for the generation of the c-myc template were 5′-GAATTAATACGACTCAGTATAGGAGGGTTTTGTTTTTGTTTTTTGTTTTTTCCAGAGTTTCGAAGCTGTTC-3′ and 5′-CAACCGCAAGTGCTCCAGCC-3′. The β-actin and gel-purified c-myc templates were transcribed with T7 polymerase, and 105 cpm of both the resulting riboprobes was used per sample following an additional gel purification. Primary murine keratinocytes were isolated and cultured as described previously and treated with the indicated concentration of TGF-β or vehicle. BALB/MK cells were cultured as described above and either were treated with 400 nM TGF-β or vehicle or were infected with the indicated adenoviruses. Total mRNA was isolated with RNeasy (Qiagen), and RPAs were performed with the RPAII kit (Ambion) as specified by the manufacturer. The intensity of the protected mRNAs was quantified by densitometry of phosphor images created by exposing the dried gels through which the RPA samples were resolved to storage phosphor screens (Molecular Dynamics). The phosphor images were analyzed with ImageQuant software (Molecular Dynamics) and a PhosphorImager 445 SI densitometer (Molecular Dynamics).
Production of Smad2, Smad3, and Smad4 recombinant adenoviruses was completed in collaboration with GlaxoSmithKline using an Escherichia coli-based Tn7 transposition strategy. The recombinant genomes were purified and transfected into HEK293 cells. All recombinant adenoviruses were purified on CsCl gradients and dialyzed.
EMSAs
Electrophoretic mobility shift assays (EMSAs) were performed with untagged, purified recombinant protein. Recombinant Smad3 MH1, residues 1 to 145, as well as the corresponding homologous domains of Smad1 and Smad2 were a generous gift of Yigong Shi (70). Recombinant E2F-4 (residues 11 to 86) and DP-1 (residues 105 to 200) DNA binding domain (DBD) proteins were a generous gift of Ning Zheng (93). Briefly, glutathione S-transferase (GST)-fusion proteins were overexpressed in E. coli and isolated by using a glutathione-Sepharose column. The protein fragments of interest were released from the GST tag bound to the glutathione-Sepharose column by specific protease digestion and purified by cation-exchange and gel-filtration chromatography. The indicated DNA probes were generated by end labeling 200 ng of polyacrylamide gel electrophoresis (PAGE)-purified, double-stranded oligonucleotide with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (6,000 Ci/mmol; NEN) and further purified by nondenaturing PAGE. The recombinant protein-DNA binding assays were performed essentially as described elsewhere (40, 70). The indicated amounts of protein were incubated with 20,000 to 50,000 cpm (less than 20 ng/ml) of the indicated DNA probe in binding buffer (25 mM Tris [pH 7.5], 80 mM NaCl, 35 mM KCl, 5 mM MgCl2, 10% glycerol, 2% NP-40, 1 mM DTT) and 20 μg of the nonspecific competitor poly(dI-dC) · poly(dI-dC) (Amersham Pharmacia Biotechnology)/ml for 30 min at 4°C. For the cold competition EMSAs, the indicated double-strand oligonucleotides were purified by PAGE and quantified, and the indicated amount of unlabeled oligonucleotide was added to the binding assay. The sequences of the employed probe and cold competitor oligonucleotides are listed below in Fig. 4A and 5A. EMSA samples were then separated on 5% polyacrylamide (75:1 acrylamide to bis-acrylamide) gels at 4°C in Tris-borate running buffer that lacked EDTA, dried, and visualized by autoradiography.
FIG. 4.
The MH1 domain of Smad3 specifically and directly binds to the c-myc TIE with a relative affinity similar to that of the SBE. (A) A 15 nM concentration of recombinant Smad1 (S1), Smad2 (S2), or Smad3 (S3) MH1 domain was individually incubated with radiolabeled SBE and c-myc TIE-containing probes, and an EMSA was performed as described in Materials and Methods. Two Smad3 MH1 complexes shifted the 16-bp SBE probe due to the incorporated palindromic double site, as depicted in bold. One molecule of the MH1 domain bound to one molecule of SBE is represented by 1x MH1, whereas two molecules of the MH1 domain bound to one molecule of DNA are represented by 2x MH1. The employed 30-bp c-myc TIE probe, equivalent to nucleotides of the c-myc promoter −92 to −63 relative to the P2 transcriptional start site, is depicted below. Sequence encompassing the first identified TIE within the rat stromelysin promoter as well as the composition of the subsequently proposed “consensus” TIE are listed below. Nucleotides in bold represent invariance, whereas lowercase letters signify preferred nucleotides. (B) The relative affinities of the Smad3 MH1-SBE and Smad3-TIE interactions were determined by EMSA, in which increasing concentrations of Smad3 MH1 recombinant protein (S3) were incubated with a fixed amount of radiolabeled SBE or c-myc TIE probe. Threefold serial dilutions of protein, from 556 to 2.29 nM, were used as indicated. The free or unbound probes are the lowest bands depicted within their respective lanes. (C) The similarity of the relative affinities of Smad3 MH1 for SBE and c-myc TIE probes was confirmed by a DNA cold competitor EMSA as described in Materials and Methods. Fifteen nanomolar Smad3 MH1 domain recombinant protein was incubated with radiolabeled c-myc TIE and increasing amounts of cold competitor DNA. The EMSA samples incubated with unlabeled c-myc TIE oligonucleotide are shown on the left, whereas those with unlabeled SBE are on the right. Threefold dilutions of DNA, from 700 to 0.320 nM, were used as indicated. All the data depicted are representative of at least three independent experiments.
Methylation interference
Methylation interference assays were performed as previously described (90). Probe for the methylation interference assay was generated in a similar fashion as that for EMSA probes with the following exceptions: the “top” or sense, single-strand oligonucleotide of the c-myc TIE was 5′-end labeled, and after heat inactivation of T4 polynucleotide kinase and PAGE purification was subsequently annealed with the unlabeled “bottom” strand. The annealed, double-strand probe was then PAGE purified, and specific activity was assessed. Next, 106 cpm of purified probe was methylated by incubation with 4 μl of dimethyl sulfate (DMS) in 600 μl of DMS reaction buffer (50 mM sodium cacodylate [pH 8.0] and 1 mM EDTA [pH 8.0]) for 5 m at 25°C. The methylation reaction was stopped with the addition of 160 μl of DMS stop buffer (1.5 mM sodium acetate [pH 7.0] and 1 M β-mercaptoethanol), 20 μg of tRNA, and 1.2 ml of ethanol (EtOH). Following EtOH precipitation and subsequent resuspension in 10 mM Tris (pH 8.0), the methylated probe was used in an EMSA as described above, with the exception that 125,000 cpm of probe was incubated in EMSA binding buffer containing 100 μg of poly(dI-dC) · poly(dI-dC)/ml with 225 μM recombinant Smad3 MH1. The undried EMSA gel, through which both the binding sample and free probe were resolved, was briefly exposed to film to visualize the protein-DNA and unbound DNA complexes. These complexes were excised from the gel, electroeluted, EtOH precipitated, and resuspended in 100 μl of 1 M piperidine. Samples were then heated for 30 min at 90°C, and piperidine was subsequently removed by lyophilization and resuspension in distilled H2O three times. After an additional lyophilization, the isolated samples were resuspended in formamide loading buffer, heated for 2 min at 95°C, placed on ice, and subsequently separated on a 12% polyacrylamide-urea sequencing gel. The resolved, piperidine-cleaved products were visualized by autoradiography.
DNAP
HaCaT cells were seeded at 5 × 106 cells/10-cm2 plate and cultured for 24 h. Cells were treated with 100 pM TGF-β or vehicle for 1 h, washed twice with PBS, and collected in PBS with a cell lifter. After the cells were pelleted at 735 × g and resuspended in DNA affinity precipitation (DNAP) buffer (25 mM Tris [pH 7.5], 80 mM NaCl, 35 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, and 0.1% IPEGAL CA-630 with the previously described mixture of phosphatase and protease inhibitors and the addition of 0.5 mM NaF), the cells were disrupted for 10 s three times at setting 2 on a Heat Systems ultrasonic processor XL and rotated for 30 min at 4°C. The extracted whole-cell lysates were cleared by centrifugation at 10,000 × g for 20 min at 4°C, and protein concentrations were assessed. Nuclear extract was derived from cells treated identically as described above, except that cells were lysed in hypotonic lysis (HL) buffer (10 mM HEPES [pH 7.5], 10 mM KCl, 3 mM MgCl2, 0.05% NP-40, 1 mM EDTA [pH 8.0], 1 mM DTT, and the described mixture of phosphatase and protease inhibitors with the addition of 0.5 mM NaF), and the nuclei were pelleted, washed in HL buffer, and finally resuspended in DNAP buffer including 320 nM NaCl. After rotational incubation, sonication, and dilution of the NaCl concentration to 80 mM, the nuclear extract was processed identically to the whole-cell lysate. A 300-μg aliquot of cell extract was then incubated by rotation with equimolar amounts, 2.675 μg of the 16-bp SBE and 5 μg of the 30-bp TIEs, of the indicated PAGE-purified, biotinylated double-strand oligonucleotides for 16 h at 4°C. The sequences of the employed biotinylated oligonucleotides are depicted below in Fig. 4A and 5A. Ten microliters of prewashed streptavidin-beaded agarose (Pierce) was added to each sample and rotated for an additional hour at 4°C. Streptavidin-agarose-DNA-protein complexes were pelleted by centrifugation at 1,310 × g for 10 min, washed three times with DNAP buffer, and analyzed by sodium dodecyl sulfate-PAGE and subsequent immunoblotting. Western blotting was performed with anti-Smad3 (51-1500; Zymed Laboratories), anti-Smad4 (sc-7966; Santa Cruz), anti-E2F-4 (sc-1082; Santa Cruz), and anti-p107 (sc-318; Santa Cruz).
Luciferase reporter assays
A total of 0.15 × 106 HaCaT cells/well were seeded into six-well plates and grown under normal culture conditions for 24 h. Cells were transfected with a standard DEAE-dextran transfection protocol essentially as described previously (15, 85). Three micrograms of total DNA including 1.5 μg of the indicated reporter construct (see Fig. 6), 0.125 μg of a cytomegalovirus β-galactosidase (CMV-β-Gal) expression vector, and 1.375 μg of filler vector were transfected into the cells of each well in triplicate for each condition shown. At 16 h posttransfection, 100 pM TGF-β or vehicle and fresh medium was added to each well, and 24 h later luciferase activity was assessed from harvested total cell lysates. The determined luciferase activity in each sample was normalized to measured levels of corresponding β-Gal expression to account for differences in transfection efficiencies.
FIG. 6.
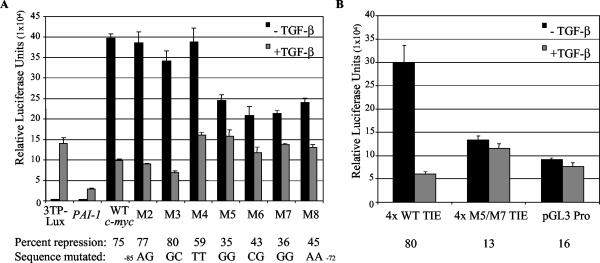
The RSBE characterized by Smad-DNA interaction studies within the c-myc TIE is required and sufficient for TGF-β-mediated transcriptional repression. (A) HaCaT cells were transiently transfected with equal amounts of the indicated promoter-driven luciferase reporters and cultured in the absence and presence of 100 pM TGF-β for 24 h. Cells were then harvested, and levels of luciferase activity were measured by a luminometer. 3TP-lux and PAI-1 are promoter reporters that are transactivated by TGF-β in part through recruitment of Smad3 to contained SBEs. Nucleotides −279 to +337 relative to the P2 transcriptional start site of the c-myc promoter were subcloned into a reporter vector, and mutations were created (M2 to M8) corresponding to those utilized in the characterization of the protein-DNA interaction. The sequence that was mutated within the reporter constructs (−85 to −72) is listed below. The percent of TGF-β-mediated repression was calculated as the measured luciferase activity from exponentially growing cells (−TGF-β) (control) divided by the difference of the measured activity of TGF-β-treated cells (+TGF-β) from that of the control, and this value is listed below the indicated reporter construct. (B) HaCaTs were transiently transfected as for panel A with luciferase reporter constructs containing four copies of sequence (−87 to −72) encompassing the wild-type c-myc TIE (4x WT TIE) or corresponding M5/M7 mutant TIE (4x M5/M7 TIE), subcloned upstream of an SV40 promoter-driven reporter vector. The empty vector, pGL3 Pro, was also transfected to serve as a negative control. Percent repression was calculated as for panel A and is listed below the indicated construct. The presented data are representative of three independent experiments.
RESULT
Smad3 is required for TGF-β-induced repression of c-myc mRNA
We have previously shown that Smad3 is integral for TGF-β-mediated growth inhibition in primary fibroblast and lymphoid cells (14). As the TGF-β cytostatic effect is particularly potent in epithelial cells, the requirement of Smad3 in TGF-β-mediated growth inhibition was assessed in primary keratinocytes by [3H]thymidine incorporation. As seen in Fig. 1A, TGF-β inhibited proliferation of wild-type epithelial cells by 85.6% compared to only 32% in Smad3ex1/ex1 homozygous null cells. Stemming from the fact that c-myc plays an important role in the TGF-β antiproliferative program in epithelial cells and from the observation that targeted deletion of Smad3 leads to elevated levels of c-Myc in primary colon epithelium (Fig. 1B), we next sought to determine if Smad3 was involved in the reported TGF-β-induced repression of c-myc transcriptional initiation (61). In wild-type primary murine keratinocytes, the level of c-myc RNA decreased by 2.3-fold within 1 h of TGF-β treatment, whereas there was only a 1.4-fold decrease in cells harboring a targeted deletion of Smad3 (Fig. 2A and B). In a similar experiment, the level of c-myc mRNA in response to TGF-β was assessed in control and Smad3 DN-overexpressing keratinocyte cell lines (HaCaTs). In the control cell line, TGF-β repressed c-myc mRNA levels within 2 h and maintained this repression for up to 8 h, compared to no observed repression in the Smad3 DN cell line (Fig. 2C). In these cells, the level of PAI-1 mRNA in the absence and presence of TGF-β was assayed to serve as a positive control. PAI-1 mRNA levels gradually increased over the measured time course of TGF-β treatment in the control cell line, and this increase was inhibited in the Smad3 DN cell line (Fig. 2C). These results indicate that Smad3 is involved not only in the TGF-β-mediated transcriptional activation of PAI-1 but also the TGF-β-induced repression of c-myc.
FIG. 1.
Targeted deletion of Smad3 results in elimination of TGF-β growth inhibitory effects and elevated levels of c-Myc. (A) The rate of DNA synthesis of primary keratinocytes derived from wild-type (+/+) and Smad3ex1/ex1 homozygous null (−/−) murine keratinocytes treated with vehicle or 100 pM TGF-β was measured by [3H]thymidine incorporation as described in Materials and Methods. The difference of the measured amount of incorporated [3H]thymidine of cells treated with vehicle minus that of TGF-β-treated cells was divided by the vehicle control counts per minute and plotted as the percent growth inhibition. The presented data are representative of values obtained from three individual lines of each genotype. (B) Murine colon epithelium was isolated from wild-type (+/+) and Smad3ex1/ex1 homozygous null (−/−) 3- to 5-month-old littermates, and Western analysis for c-Myc and actin, as a loading control, of 30 μg of protein from the epithelium lysate are depicted. Brackets indicate two individual sets of wild-type and Smad3ex1/ex1 homozygous null littermates (litter A and litter B).
FIG. 2.
TGF-β-mediated repression of c-myc transcriptional initiation is reduced in the absence of functional Smad3. (A) BALB/MK cells or primary keratinocytes derived from wild-type and Smad3ex1/ex1 homozygous null littermates were treated with 400 nM TGF-β or vehicle for 1 h. RPAs of 10 μg of BALB/MK and 15 μg of primary keratinocyte total RNA were performed as described in Materials and Methods. The depicted RPA is representative of three independent experiments. (B) Densitometric analysis of a phosphorimager scan of the RPA depicted in panel A was used to quantify mRNA levels. Values for c-myc band intensities were normalized to β-actin levels, which were quantified with a lighter phosphorimage exposure to obtain more accurate assessments of the relatively high β-actin intensities. The repression was calculated by dividing the quantified, normalized value of c-myc in the absence of TGF-β (control) by corresponding levels in the presence of TGF-β. (C) Total RNA was isolated from control HaCaT and S3 DN HaCaT cells treated with 100 pM TGF-β for 0, 2, 4, and 8 h, as indicated. RPAs of 5 μg of total RNA per condition were performed as described in Materials and Methods to determine mRNA levels of c-myc, PAI-1, L32, and GAPDH. PAI-1 levels were assessed as a positive control, as this gene is known to be transactivated by TGF-β through a mechanism that is partially dependent on Smad3 recruitment to promoter-contained SBEs. L32 and GAPDH mRNA levels served as loading control indicators. The depicted RPA is representative of three independent experiments.
Smad overexpression is sufficient for repression of c-myc mRNA
Although it was demonstrated that Smad3 played a role in the TGF-β-mediated repression of c-myc, we next sought to determine if Smad overexpression was sufficient for repression of c-myc mRNA. The adenovirus-mediated overexpression of Smad2, Smad3, Smad4, Smad2 and -4, and Smad3 and -4 in exponentially growing BALB/MK cells led to 1.3-, 1.9-, 1.8-, 1.3-, and 2.9-fold repression of c-myc mRNA, respectively, compared to a vector control level set at 1 (Fig. 3). Overexpression of both Smad3 and -4 resulted in a repression of c-myc levels comparable to those in uninfected cells treated with TGF-β (Fig. 3A). Adenovirus infection with the control virus did not alter the basal levels of c-myc transcript.
FIG. 3.
Smad3 and Smad4 cooverexpression represses endogenous c-myc expression. (A) BALB/MK cells either were treated with 400 nM TGF-β or vehicle for 60 min or were infected with recombinant adenoviruses (Ad) carrying empty CMV vector (V) or vector containing cDNA for Smad2 (2), Smad3 (3), and/or Smad4 (4) at a total multiplicity of infection of 100 for 12 h. RPA of 15 μg of total RNA isolated from these cells is shown. Results are representative of three experiments. Immunoblot analysis of whole-cell lysates from the infected cells with anti-Smad1/2/3 (sc-7960; Santa Cruz) is shown in the middle panel. The blot was reprobed with anti-Smad4 (sc-7966; Santa Cruz), and these results are depicted in the lower panel. (B) Densitometric analysis of a phosphorimager scan of the RPA in panel A was completed, and repression of normalized, quantified values of band intensities was calculated as described in the legend for Fig. 2B and graphed, with the exception that the value derived from empty vector adenovirus-infected cells was used as the control level.
Recombinant Smad3 MH1 domain specifically and directly binds to the c-myc TGF-β TIE with similar relative affinity as a consensus SBE
After establishing that Smad3 plays a critical role in c-myc repression and with the knowledge that Smad3 and -4 are recruited to the c-myc TIE (10), we next determined if Smad3 directly binds to the c-myc TIE or if Smads are indirectly recruited to the TIE through another DNA binding factor. Recombinant preparations of the MH1 domain of Smad3 (residues 1 to 145) and the homologous domains of Smad1 and -2 were incubated with radiolabeled SBE and c-myc TIE probes (the sequences of which can be seen in Fig. 4A). Smad3 MH1 bound directly to the SBE sequence GTCT and its palindrome AGAC with a higher affinity than an equimolar amount of Smad1 or Smad2, as reported previously (70). Importantly, Smad3 also specifically and directly bound the c-myc TIE (Fig. 4A). Since the MH1 domain of Smad3 directly bound to both a consensus SBE and the c-myc TIE, we compared the relative binding affinity of Smad3 to the TIE and Smad3 to the SBE in two experimental contexts. We incubated increasing concentrations of Smad3 MH1 with radiolabeled SBE or c-myc TIE (Fig. 4B), as well as a constant amount of Smad3 and radiolabeled TIE with increasing concentrations of unlabeled TIE or SBE (Fig. 4C). As shown in Fig. 4B and C, similar relative binding affinities of Smad3 MH1 to the SBE and to the TIE were revealed. The dissociation constant of Smad3 MH1 and the employed SBE has been reported to be 1.14 × 10−7 M (70). Either one or two molecules of Smad3 MH1 were found to bind to the palindromic SBE, whereas only one Smad3 MH1-c-myc TIE complex was observed (Fig. 4B and C).
The Smad binding site within the c-myc TIE, maximally comprised of 5′-TTGGCGGGAA-3′, is distinct from a consensus SBE
Since recombinant Smad3 MH1 bound to both a consensus SBE and the c-myc TIE with similar relative affinities yet exerted opposite transcriptional effects, we carried out a series of experiments to map the precise DNA sequence within the TIE to which Smad3 was found to directly interact. In the first set of experiments, 50 nM Smad3 MH1 was incubated with a constant amount of radiolabeled wild-type c-myc TIE and an excess, equimolar amount of cold c-myc TIE competitor oligonucleotides that contained 2-bp scanning mutations as indicated in Fig. 5A. The mutations made in the TIE corresponding to M5, M6, M7 and, to a lesser extent, M4 and M8 inhibited the ability of these oligonucleotides to compete with the radiolabeled, wild-type TIE for Smad3 MH1 binding (Fig. 5A). The sequence of the c-myc promoter spanning mutations M4 to M8 was 5′-TTGGCGGGAA-3′ (−81 to −72 relative to the P2 transcriptional start site) and did not include the adjacent SBE-like sequences 5′-CAGA-3′ (−88 to −85) or 5′-GGCT-3′ (−84 to −81). Secondly, we performed methylation interference assays to pinpoint the guanine nucleotides within the TIE that form direct contacts with Smad3 MH1. Consistent with the results of the EMSA cold competition analysis, methylation of only three specific guanines within the c-myc TIE contained in the scanning mutations M5 and M7 (designated by underlining in 5′-TTGGCGGGAA-3′) interfered with Smad3 MH1 binding (Fig. 5B). Importantly, the mutation of the SBE-like sequence 5′-GGCT-3′ within the c-myc TIE, designated MS or M“SBE” TIE, did not reduce the binding affinity of Smad3 MH1 for this element (Fig. 5C). However, radiolabeled probe in which a 2-bp substitution was made, designated M6 in Fig. 5A, was completely unable to bind up to 450 nM recombinant Smad3 MH1 (data not shown). Taken together, these data clearly indicate that any SBE-like sequence within the c-myc TIE sequence is in fact not sufficient for nor significantly contributing to Smad binding.
To determine if endogenous Smad proteins were recruited to the same sequence of the TIE as defined above, DNAP binding studies were performed with lysate from cells treated in the absence and presence of TGF-β and biotinylated double-strand oligonucleotides. As shown in Fig. 5D, elevated levels of Smad3 and Smad4 were precipitated by biotinylated SBE and wild-type TIE oligonucleotides from whole-cell lysate derived from cells treated with TGF-β for 1 h, demonstrating TGF-β-inducible binding of the Smads to the two distinct DNA sequences (Fig. 5D). Mutations of the biotinylated TIE oligonucleotides represented by M4, M5, and M6 resulted in a loss of Smad3 and Smad4 precipitation, whereas the MS and M3 mutations had no effect on endogenous Smad binding. An identical experiment is depicted in Fig. 5E, with the exception that nuclear extract was employed instead of whole-cell lysate. Although the amount of Smad3 that was precipitated by the TIE was comparable to that bound to the SBE, the amount of Smad4 associated with the TIE was less than that precipitated by the SBE, possibly indicating a difference in the stoichiometry of Smad complexes bound to the two DNA sequences (Fig. 5D and E). Taken together, these data demonstrate that recombinant Smad3 MH1 and endogenous Smads bind a sequence (5′-TTGGCGGGAA-3′) within the c-myc promoter that is distinct from a consensus SBE (5′-GTCT-3′ or its palindrome, 5′-AGAC-3′). Thus, the c-myc TIE contains a novel Smad binding site, or an RSBE.
The RSBE is both required and sufficient for TGF-β-mediated repression of the c-myc promoter
After mapping the RSBE within the c-myc TIE, we set out to determine if the sequence that mediates Smad binding is also required functionally in mediating TGF-β-induced repression of the c-myc promoter. Luciferase reporter constructs were created in which the nucleotides −279 to + 337 relative to the P2 transcriptional start site were subcloned into the pGL3-basic vector (c-mycDel-5pGL3, or WT c-myc), and the scanning mutations M2-M8 (Fig. 5A) were incorporated in seven corresponding constructs. The wild-type and M2 to M8 luciferase reporters, in addition to two reporters known to be activated in a TGF-β-inducible manner partially dependent on promoter contained SBEs, 3TP-Lux and p800LUC PAI-1 (19, 38, 75, 85), were transfected into exponentially growing HaCaT cells and treated in the presence and absence of 100 pM TGF-β. TGF-β treatment resulted in the transactivation of 3TP-Lux and the PAI-1 promoter reporters as expected, yet inhibited the activity of the WT c-myc reporter by 75% (Fig. 6A). The directed mutations within the TIE designated by M4, M5, M6, M7, and M8 diminished the ability of TGF-β to repress the corresponding reporters (59, 35, 43, 36, and 45% repression, respectively), whereas the transcriptional activity of the reporters harboring mutations M2 and M3 were repressed by percentages similar to that demonstrated by the WT construct (Fig. 6A).
To determine if the RSBE within the c-myc TIE was sufficient to mediate TGF-β-induced repression, four copies of the c-myc promoter sequence encompassing the TIE (−87 to −72 relative to the P2 transcriptional start site) were cloned into an SV40 promoter-driven luciferase vector. The transcriptional activity of this construct, 4x WT TIE, was repressed by 80% in the presence of TGF-β, whereas the level of TGF-β-mediated repression of a corresponding reporter with 4-bp mutations corresponding to M5 and M7, 4x M5/M7 TIE, was comparable to that of the vector alone (Fig. 6B).
Interestingly, the reporters with scanning mutations M5 to M8, as well as the 4x M5/M7 TIE construct, displayed considerably lower levels of basal transcriptional activity compared to their wild-type counterparts (Fig. 6A and B). This suggested that the sequence contained within the scanning mutants M5 to M8 (5′-GGCGGGAA-3′; −79 to −72) not only comprise at least part of the RSBE, but also part of an element responsible for mediating basal transactivation responsiveness to factors within the medium. Cells were transfected with 4x WT TIE, 4x M5/M7 TIE, and the vector pGL3 Pro and serum starved, and transcriptional activity was measured in response to 10% FBS. The luciferase activity measured from cell lysate containing the wild-type vector exhibited a 1.9-fold increase in response to serum, whereas the corresponding 4x M5/M7 TIE and vector-containing lysates displayed only 0.26- and 0.41-fold increases, respectively (data not shown).
The c-myc TIE is a composite RSBE and E2F binding site
Sequence overlapping the c-myc TIE (5′-GGCGGGAAA-3′; −79 to −71) constitutes a consensus E2F binding site (33, 67), and thus this site may mediate transactivating effects of E2F-1, -2, and -3 and/or repressive effects of E2F-4 and -5. Thus, it was hypothesized that TGF-β-mediated repression of c-myc may involve the displacement of an activating E2F from this site by a Smad3-containing complex and/or the TGF-β-induced recruitment of a complex containing Smad3 that functions in conjunction with repressive E2Fs. Although E2Fs are able to bind E2F sites as homodimers, E2F/DP heterodimers exhibit higher affinities for DNA binding, increased levels of transactivation ability, and enhanced pocket protein binding (6, 31, 34, 42). EMSAs revealed that recombinant DBDs of E2F-1 and E2F-4 homodimers and also E2F-4/DP-1 and E2F-4/DP-2 heterodimers were able to bind wild-type TIE probe, yet not the employed SBE probe (data not shown). A similar experiment as that depicted in Fig. 5A was performed to map the E2F binding domain with the exception that a 150 nM equimolar mixture of the DBDs of E2F-4 and DP-1 was employed instead of recombinant Smad3 MH1. As shown in Fig. 7A, mutations in the unlabeled oligonucleotides corresponding to M5 to M8 and to a lesser extent M4 and M9 eliminated competition for E2F-4/DP-1 heterodimer binding from the labeled wild-type c-myc TIE probe. These data confirm that the putative E2F binding site is capable of direct association with E2F-4/DP-1 heterodimers and that this site closely overlaps with the defined RSBE. To confirm these in vitro binding results, DNAPs were performed as previously described. Biotinylated, double-strand wild-type c-myc TIE, MS, M3, and to a lesser extent M4 oligonucleotides were able to precipitate endogenous E2F-4 in the presence and absence of TGF-β, whereas mutations corresponding to M5 and M6 inhibited this binding (Fig. 7B). There were no detectable levels of E2F-1, -2, and -3 precipitated in this assay (data not shown). As E2F-4 is known to exert its repressive effects on most promoters in concert with the pocket proteins (pRb, p107, and/or p130) through direct protein-protein association, the immunoblots generated from the DNAP assays were probed with antibody against these proteins. p107 exhibited the same binding pattern as E2F-4, whereas pRb and p130 precipitation was not detected (Fig. 7B and data not shown).
FIG. 7.
The extended c-myc TIE is comprised of a closely overlapping RSBE and E2F site. (A) An E2F-4/DP-1 heterodimer binding site within the c-myc TIE was mapped as described in the legend for Fig. 5A with the exception that 150 nM of an equimolar mixture of recombinant E2F-4/DP-1 DBDs was employed in place of Smad3 MH1. The extended c-myc TIE is depicted below, with the sequence with homology to a consensus TIE in bold, the RSBE in italics, and the E2F site underlined. (B) A DNAP was performed with whole-cell lysate as described in the legend for Fig. 5A, and immunoblots were probed for p107 and E2F-4. The data presented are representative of three independent experiments.
DISCUSSION
The cell cycle progresses in a tightly coordinated fashion in which growth-promoting factors are counterbalanced by antiproliferative pathways. The proto-oncogene c-myc is a transcription factor involved in the induction of various genes necessary for cell cycle progression, such as cyclin D2, and it has been shown that the ability of c-Myc to induce S phase is dependent on E2F-2 and -3 (22, 27, 45). Upregulated levels of c-Myc through dysregulation of its promoter by the abnormal presence or absence of transactivating factors or through gene amplification has been implicated in the initiation and/or progression of a variety of human cancers (7, 9, 24, 25, 32, 71, 72). In contrast, the TGF-β signaling pathway has potent antiproliferative effects that most likely contribute to its ability to act in a tumor-suppressive capacity. The TGF-β-mediated repression of c-myc is clearly an important event in the manifestation of this pathway's cytostatic program, in that c-myc repression is essential for subsequent transactivation of the CDK inhibitors p15 and p21, and c-Myc overexpression has been shown to counteract these growth inhibitory effects in various epithelial cells (3, 12, 68, 74, 81, 89). Thus, the elucidation of the molecular mechanisms involved in TGF-β-mediated repression of c-myc in normal cells may provide insight into how cancerous cells become refractory to the TGF-β antiproliferative signal.
We and others have provided data that demonstrate that the Smads play an essential role in TGF-β-mediated repression of c-myc. TGF-β induces the recruitment of Smad3 and Smad4 to an element within the c-myc promoter similar to the previously identified stromelysin TIE through a direct interaction with the MH1 domain of Smad3 (10, 11, 39, 89). Although Smad3 is known to interact with corepressor molecules, such as c-Ski, SnoN, TGIF, mSin3A, and SNIP1 (2, 41, 48, 50, 76, 77, 84, 88), these interactions have only been implicated in the subsequent inhibition of preceding TGF-β-mediated transactivation of target genes rather than a process of TGF-β-induced active repression, such as in the case of c-myc. Thus, our findings, in conjunction with those in two complementary reports (11, 89), currently represent the only characterized instance of Smad involvement in the active repression of a gene repressed by TGF-β through a mechanism that requires the direct interaction of Smad3 with specific promoter-contained sequence.
Careful analysis of the sequence within the c-myc promoter that mediates direct Smad3 binding, as well as the repressive effects of TGF-β on the transcriptional initiation of c-myc, reveals a novel Smad3 binding site that is distinct from an SBE and has been termed an RSBE. Although it has been reported that an SBE-like sequence, 5′-GGCT-3′ (−84 to −81), mediates Smad3 binding to the c-myc promoter (11), we have found that this sequence is dispensable for both Smad3 binding and TGF-β-mediated repression of c-myc reporter activity (Fig. 5 and 6). This apparent discrepancy can be explained by the inclusion of the 3′ T (−80) to the SBE-like sequence mutated in the previously published report, in which mutation of the TIE sequence 5′-GGCTT-3′ (−84 to −80) to 5′-TTAAA-3′ was shown to inhibit endogenous Smad3 and Smad4 precipitation by a 30-bp biotinylated oligonucleotide (11). We have demonstrated that mutation of only the 4 nucleotides that comprise the SBE-like sequence has no effect on binding to recombinant Smad3 MH1 or on precipitation of endogenous Smad3- and Smad4-containing complexes. The T 3′ to this sequence (−80), however, seems to mark the 5′ start of the relevant Smad binding site, as the mutation corresponding to M4, in which the underlined sequence of 5′-GGCTTGGCGGGAA-3′ (−81 to −80) was altered to CG, markedly inhibits Smad binding. Our EMSA and DNAP binding studies in conjunction with the methylation interference analysis (Fig. 5) clearly defined an RSBE that is maximally comprised of 5′-TTGGCGGGAA-3′ (−81 to −72).
The c-myc TIE was initially identified by its homology with the stromelysin TIE, yet the c-myc element is distinguished from the previously characterized consensus TIE (5′-GNNTTGGtGa-3′) by the presence of an overlapping E2F site (39). The sequence 5′-GGCGGGAAA-3′ (−79 to −72) is a consensus E2F binding site that was reported to be capable of binding E2F protein preparations affinity purified from adenovirus-infected HeLa cell extract (33). As mentioned above, scanning mutations made across this site in c-myc luciferase reporters resulted in a drop in basal transcriptional activity as well as inhibition of serum-induced transactivation (Fig. 6). Furthermore, it has been reported that separate E2F-1 and Ets-1 overexpression resulted in transactivation of a c-myc promoter reporter that was dependent upon this E2F binding site (33), although, in contradiction, adenovirus-mediated E2F-1 expression and regulated overexpression of E2F-1, -2, and -3 was shown to not be sufficient for transactivation of the endogenous c-myc promoter (17, 80). Given the overlapping Smad3 and E2F binding sites, we hypothesized that TGF-β may induce the displacement of an activating E2F-1, -2, or -3, or an Ets-containing complex by a Smad3-containing complex and that this competition may contribute to TGF-β-mediated repression of c-myc. However, we were not able to detect any of these E2Fs or Ets 1/2 in our DNAP assays, and thus we could not confirm the validity of this hypothesis. The identity of the factor(s) responsible for conferring serum responsiveness, possibly E2F or Ets transcription factors, will have to be revealed through a more sensitive assay, such as chromatin immunoprecipitation (78).
Another possibility we considered, not mutually exclusive with the scenario described above, was that a repressive E2F, E2F-4, and/or -5 may work in conjunction with Smad3 in TGF-β-induced repression of c-myc transcriptional initiation. We confirmed that recombinant E2F-4 DBD homodimers and E2F-4/DP-1 heterodimers directly bind the consensus E2F site overlapping the RSBE (Fig. 7A and data not shown), and that this sequence is responsible for precipitating endogenous E2F-4 and p107 (Fig. 7B). Although the binding of Smad3 and E2F-4 to the composite RSBE/E2F site extensively overlaps, there does appear to be a distinction in DNA recognition when the proteins bind independently from one another. Methylation of the underlined guanines in the sequence 5′TTGGCGGGAAA3′ (−81 to −72) was shown to interfere with binding of affinity-purified E2F (33), whereas we have shown that methylation of the guanines in bold interfered with Smad3 MH1 binding (Fig. 5B). It has been previously demonstrated that the Smad3 MH2 domain binds E2F-4 and p107, allowing the formation of a multimeric protein complex that is recruited to the c-myc TIE in response to TGF-β, and that E2F-4 and p107 are functionally required for TGF-β-mediated repression of c-myc (11). Although our study does not conflict with the functional significance of this previously published study, the mechanistic model of how a repressive Smad3, E2F-4, and p107-containing repressive complex binds to the c-myc TIE needs to be modified to account for the binding of Smad3 to the embedded RSBE we have defined, rather than to the adjacent “SBE-like” sequence reported.
Given our thorough mapping of both the MH1 domain of Smad3 and the E2F-4 DBD to a closely overlapping DNA sequence, and also the previously reported demonstration that Smad3, E2F-4, and the wild-type c-myc TIE are able to form a tertiary complex (11), we propose that in an endogenous context the MH1 domain of Smad3 binds to the RSBE and the E2F-4 DBD contacts the overlapping E2F site of the c-myc TIE simultaneously in response to TGF-β. We were not able to demonstrate simultaneous binding of the recombinant Smad3 MH1 fragment with E2F-4 DBD homodimers or E2F-4/DP-1 DBD heterodimers to an EMSA probe encompassing the c-myc TIE (data not shown). However, it is possible that in the correct context full-length Smad3 and full-length E2F-4 will prove to bind simultaneously to the composite RSBE/E2F site, considering that the MH2 domain of Smad3 directly contacts a domain of E2F-4 contained within residues 1 to 168 (11) and adjacent to its DBD (amino acids 11 to 86). The protein-protein interface between the Smad3 MH2 domain and E2F-4 residues 1 to 168 may result in a continuous DNA-binding interface comprised of portions of the Smad3 MH1 domain and the E2F-4 DBD capable of TGF-β-induced binding to the composite RSBE/E2F site within the c-myc promoter. We were able to demonstrate that both full-length Smad3 fused to GST and full-length E2F-4 recombinant proteins are capable of binding a wild-type TIE EMSA probe and that the revealed protein-DNA interactions were entirely inhibited with the mutation of two nucleotides (as designated by the M6 TIE listed in Fig. 5A) within the composite RSBE/E2F site (data not shown). However, the incubation of the two full-length proteins with the wild-type TIE EMSA probe did not reveal a complex that necessarily suggested both proteins bound to one DNA molecule simultaneously. It is possible that such a tertiary complex can only form with appropriate posttranslational modifications of the full-length proteins, as occurs in the endogenous context of TGF-β signaling. Towards this end, we performed similar experiments as described above with a mutant Smad3-GST fusion protein that was hoped to mimic the serine phosphorylations that occur upon type I receptor activation (i.e., the two most carboxyl serines were mutated to aspartic acid residues), yet similar results as that with the wild-type protein were obtained (data not shown). It remains possible that recombinant full-length Smad3 in which the appropriate serines are phosphorylated as opposed to “pseudophosphorylated” may yield a specific conformation that facilitates both E2F-4 and DNA binding. The generation of phosphorylated Smad3 might be achieved by an in vitro kinase assay with activated type I receptor or, more specifically, by an expressed protein ligation strategy (87). Alternatively, or perhaps in addition to required posttranslational modifications, full-length Smad3 and E2F-4 may only be able to bind to the c-myc TIE in the presence of additional proteins, e.g., Smad4, DP, and/or pocket protein family members, in a multimeric complex.
The established ability of Smad3 and Smad4 to act as effectors of TGF-β-induced transactivation of various genes through direct interaction of the Smad3 MH1 domain with promoter-contained SBEs raises the question of how these same proteins can mediate the repression of a gene such as c-myc. Firstly, context-specific cofactors to which the Smads bind participate in determining whether a Smad-containing complex will transactivate, inhibit preceding transactivation, or actively repress the promoters to which it is recruited. Cofactor binding preference is partially determined by cell type, environment of extracellular signals to which the cell is exposed, cofactor subcellular localization, and surrounding promoter-contained cofactor binding elements. For an example of the latter case, one can compare the synergistic transactivation of Smad3 and AP-1 family members bound to their respective sites on the c-jun promoter to the transcriptional effects of Smad3, E2F-4, and p107 bound to the composite RSBE/E2F site on the c-myc promoter (83). It remains to be determined if the Smad-binding cofactors that are involved in inhibiting preceding TGF-β-induced transactivation, such as c-Ski, SnoN, TGIF, mSin3A, and SNIP1, also contribute to Smad-mediated active repression of transcription. In addition, different Smad DNA binding sites may also participate in determining the functional fate of the Smads in terms of their effects on transcription and cofactor binding preference. Our study defined the novel RSBE to which Smad3 directly binds and is required for TGF-β-mediated repression of c-myc. As mentioned, this RSBE is distinct from an SBE, which mediates TGF-β-induced transactivation of genes such as PAI-1 and c-jun, although the Smad3 MH1 domain directly binds to both DNA elements with similar relative affinity (Fig. 3B and C) (19, 38, 75, 83). It is thus possible that the different Smad3-binding DNA elements, the c-myc RSBE and the consensus SBE, induce distinct conformations of Smad3 that exhibit differential binding affinities for specific cofactors (44).
The c-myc extended TIE is a composite element comprised of a newly defined a RSBE and E2F site that is essential to TGF-β-mediated repression of c-myc. It remains to be demonstrated if similar elements and a corresponding mode of transcriptional repression that occurs in the case of the c-myc promoter are involved in the regulation of other genes that are actively repressed by TGF-β. It will be interesting to determine if other putative TIEs that lack an overlapping E2F site, traditionally defined by the sequence 5′-GNNTTGGtGa-3′, contain an embedded RSBE and mediate Smad3 binding-dependent transcriptional repression of the gene's promoters in which these sites exist, such as the rat stromelysin promoter (39). However, it is possible that Smad3-dependent transcriptional repression of certain TGF-β target genes will prove to require a composite RSBE/E2F site. CDK-activating tyrosine phosphatase cdc25A is an example of another gene that may be transcriptionally repressed by TGF-β in a mechanistically similar manner as c-myc. TGF-β-mediated repression of cdc25A contributes to the TGF-β cytostatic program (36). In keratinocytes, this induced repression has been shown to be a delayed event, subsequent to CDK inhibitor transactivation, that is dependent on a promoter-contained E2F site (5′-TTTGGCGCCAA-3′; −62 to −52) (35). This site is comparable to the RSBE/E2F site defined in the c-myc promoter (5′-TTTCCCGCCAA-3′; −71 to −81) and, considering that TGF-β rapidly inhibits cdc25A mRNA in mammary epithelial cells (36), it is possible that Smad3 recruitment to this site is required for TGF-β-mediated repression of cdc25A in breast epithelium.
Acknowledgments
This work was supported by National Institutes of Health grants CA75368 and CA83770 to X.-F. Wang. J.P.F. and D.S.W. were supported by predoctoral fellowships from the Department of Defense Breast Cancer Research Program, DAMD17-98-1-8067 and DAMD17-00-1-0229, respectively. N.T.L was supported by a National Science Foundation predoctoral fellowship.
We thank Bert Vogelstein, Joan Massagué, David J. Loskutoff, Mitsuyasu Kato, Harold L. Moses, and Ning Zheng for providing reagents. We are also grateful to Suzanne D. Neill and John E. Bisi of GlaxoSmithKline for generating the Smad-expressing adenovirus constructs and Allan Balmain and Sheelagh Frame for advice on primary keratinocyte isolation. Joseph R. Nevins, Paloma M. Giangrande, and Ester P. Black were a source of helpful information regarding E2F function.
REFERENCE
- 1.Abdollah, S., M. Macias-Silva, T. Tsukazaki, H. Hayashi, L. Attisano, and J. L. Wrana. 1997. TβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 272:27678-27685. [DOI] [PubMed] [Google Scholar]
- 2.Akiyoshi, S., H. Inoue, J. Hanai, K. Kusanagi, N. Nemoto, K. Miyazono, and M. Kawabata. 1999. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. J. Biol. Chem. 274:35269-35277. [DOI] [PubMed] [Google Scholar]
- 3.Alexandrow, M. G., M. Kawabata, M. Aakre, and H. L. Moses. 1995. Overexpression of the c-Myc oncoprotein blocks the growth-inhibitory response but is required for the mitogenic effects of transforming growth factor beta 1. Proc. Natl. Acad. Sci. USA 92:3239-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attisano, L., and J. L. Wrana. 2000. Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 12:235-243. [DOI] [PubMed] [Google Scholar]
- 5.Baker, J. C., and R. M. Harland. 1997. From receptor to nucleus: the Smad pathway. Curr. Opin. Genet. Dev. 7:467-473. [DOI] [PubMed] [Google Scholar]
- 6.Bandara, L. R., V. M. Buck, M. Zamanian, L. H. Johnston, and N. B. La Thangue. 1993. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J. 12:4317-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieche, I., I. Laurendeau, S. Tozlu, M. Olivi, D. Vidaud, R. Lidereau, and M. Vidaud. 1999. Quantitation of MYC gene expression in sporadic breast tumors with a real-time reverse transcription-PCR assay. Cancer Res. 59:2759-2765. [PubMed] [Google Scholar]
- 8.Blobe, G. C., W. P. Schiemann, and H. F. Lodish. 2000. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 342:1350-1358. [DOI] [PubMed] [Google Scholar]
- 9.Bonilla, M., M. Ramirez, J. Lopez-Cueto, and P. Gariglio. 1988. In vivo amplification and rearrangement of c-myc oncogene in human breast tumors. J. Natl. Cancer Inst. 80:665-671. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C. R., Y. Kang, and J. Massague. 2001. Defective repression of c-myc in breast cancer cells: a loss at the core of the transforming growth factor beta growth arrest program. Proc. Natl. Acad. Sci. USA 98:992-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, C. R., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 110:19-32. [DOI] [PubMed] [Google Scholar]
- 12.Claassen, G. F., and S. R. Hann. 2000. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta-induced cell-cycle arrest. Proc. Natl. Acad. Sci. USA 97:9498-9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffey, R. J., Jr., C. C. Bascom, N. J. Sipes, R. Graves-Deal, B. E. Weissman, and H. L. Moses. 1988. Selective inhibition of growth-related gene expression in murine keratinocytes by transforming growth factor beta. Mol. Cell. Biol. 8:3088-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datto, M. B., J. P. Frederick, L. Pan, A. J. Borton, Y. Zhuang, and X. F. Wang. 1999. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol. Cell. Biol. 19:2495-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datto, M. B., Y. Li, J. F. Panus, D. J. Howe, Y. Xiong, and X. F. Wang. 1995. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc. Natl. Acad. Sci. USA 92:5545-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Caestecker, M. P., E. Piek, and A. B. Roberts. 2000. Role of transforming growth factor-beta signaling in cancer. J. Natl. Cancer Inst. 92:1388-1402. [DOI] [PubMed] [Google Scholar]
- 17.DeGregori, J., T. Kowalik, and J. R. Nevins. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 15:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J. M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derynck, R., and Y. Zhang. 1996. Intracellular signalling: the mad way to do it. Curr. Biol. 6:1226-1229. [DOI] [PubMed] [Google Scholar]
- 21.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 22.Eisenman, R. N. 2001. Deconstructing myc. Genes Dev. 15:2023-2030. [DOI] [PubMed] [Google Scholar]
- 23.Eppert, K., S. W. Scherer, H. Ozcelik, R. Pirone, P. Hoodless, H. Kim, L. C. Tsui, B. Bapat, S. Gallinger, I. L. Andrulis, G. H. Thomsen, J. L. Wrana, and L. Attisano. 1996. MADR2 maps to 18q21 and encodes a TGFβ-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell 86:543-552. [DOI] [PubMed] [Google Scholar]
- 24.Erisman, M. D., P. G. Rothberg, R. E. Diehl, C. C. Morse, J. M. Spandorfer, and S. M. Astrin. 1985. Deregulation of c-myc gene expression in human colon carcinoma is not accompanied by amplification or rearrangement of the gene. Mol. Cell. Biol. 5:1969-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erisman, M. D., J. K. Scott, R. A. Watt, and S. M. Astrin. 1988. The c-myc protein is constitutively expressed at elevated levels in colorectal carcinoma cell lines. Oncogene 2:367-378. [PubMed] [Google Scholar]
- 26.Goto, D., K. Yagi, H. Inoue, I. Iwamoto, M. Kawabata, K. Miyazono, and M. Kato. 1998. A single missense mutant of Smad3 inhibits activation of both Smad2 and Smad3, and has a dominant negative effect on TGF-beta signals. FEBS Lett. 430:201-204. [DOI] [PubMed] [Google Scholar]
- 27.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell. Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 28.Hannon, G. J., and D. Beach. 1994. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 371:257-261. [DOI] [PubMed] [Google Scholar]
- 29.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 30.Heldin, C. H., K. Miyazono, and P. ten Dijke. 1997. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 390:465-471. [DOI] [PubMed] [Google Scholar]
- 31.Helin, K., C. L. Wu, A. R. Fattaey, J. A. Lees, B. D. Dynlacht, C. Ngwu, and E. Harlow. 1993. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7:1850-1861. [DOI] [PubMed] [Google Scholar]
- 32.Henriksson, M., and B. Luscher. 1996. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv. Cancer Res. 68:109-182. [DOI] [PubMed] [Google Scholar]
- 33.Hiebert, S. W., M. Lipp, and J. R. Nevins. 1989. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc. Natl. Acad. Sci. USA 86:3594-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber, H. E., G. Edwards, P. J. Goodhart, D. R. Patrick, P. S. Huang, M. Ivey-Hoyle, S. F. Barnett, A. Oliff, and D. C. Heimbrook. 1993. Transcription factor E2F binds DNA as a heterodimer. Proc. Natl. Acad. Sci. USA 90:3525-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iavarone, A., and J. Massague. 1999. E2F and histone deacetylase mediate transforming growth factor β repression of cdc25A during keratinocyte cell cycle arrest. Mol. Cell. Biol. 19:916-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iavarone, A., and J. Massague. 1997. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-beta in cells lacking the CDK inhibitor p15. Nature 387:417-422. [DOI] [PubMed] [Google Scholar]
- 37.Janknecht, R., N. J. Wells, and T. Hunter. 1998. TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev. 12:2114-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keeton, M. R., S. A. Curriden, A. J. van Zonneveld, and D. J. Loskutoff. 1991. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor beta. J. Biol. Chem. 266:23048-23052. [PubMed] [Google Scholar]
- 39.Kerr, L. D., D. B. Miller, and L. M. Matrisian. 1990. TGF-beta 1 inhibition of transin/stromelysin gene expression is mediated through a Fos binding sequence. Cell 61:267-278. [DOI] [PubMed] [Google Scholar]
- 40.Kim, J., K. Johnson, H. J. Chen, S. Carroll, and A. Laughon. 1997. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature 388:304-308. [DOI] [PubMed] [Google Scholar]
- 41.Kim, R. H., D. Wang, M. Tsang, J. Martin, C. Huff, M. P. de Caestecker, W. T. Parks, X. Meng, R. J. Lechleider, T. Wang, and A. B. Roberts. 2000. A novel smad nuclear interacting protein, SNIP1, suppresses p300-dependent TGF-beta signal transduction. Genes Dev. 14:1605-1616. [PMC free article] [PubMed] [Google Scholar]
- 42.Krek, W., D. M. Livingston, and S. Shirodkar. 1993. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science 262:1557-1560. [DOI] [PubMed] [Google Scholar]
- 43.Lagna, G., A. Hata, A. Hemmati-Brivanlou, and J. Massague. 1996. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature 383:832-836. [DOI] [PubMed] [Google Scholar]
- 44.Lefstin, J. A., and K. R. Yamamoto. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392:885-888. [DOI] [PubMed] [Google Scholar]
- 45.Leone, G., R. Sears, E. Huang, R. Rempel, F. Nuckolls, C. H. Park, P. Giangrande, L. Wu, H. I. Saavedra, S. J. Field, M. A. Thompson, H. Yang, Y. Fujiwara, M. E. Greenberg, S. Orkin, C. Smith, and J. R. Nevins. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8:105-113. [DOI] [PubMed] [Google Scholar]
- 46.Letterio, J. J., and A. B. Roberts. 1997. TGF-beta: a critical modulator of immune cell function. Clin. Immunol. Immunopathol. 84:244-250. [DOI] [PubMed] [Google Scholar]
- 47.Liberati, N. T., M. B. Datto, J. P. Frederick, X. Shen, C. Wong, E. M. Rougier-Chapman, and X. F. Wang. 1999. Smads bind directly to the Jun family of AP-1 transcription factors. Proc. Natl. Acad. Sci. USA 96:4844-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberati, N. T., M. Moniwa, A. J. Borton, J. R. Davie, and X. F. Wang. 2001. An essential role for Mad homology domain 1 in the association of Smad3 with histone deacetylase activity. J. Biol. Chem. 276:22595-22603. [DOI] [PubMed] [Google Scholar]
- 49.Liu, F., C. Pouponnot, and J. Massague. 1997. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes Dev. 11:3157-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo, K., S. L. Stroschein, W. Wang, D. Chen, E. Martens, S. Zhou, and Q. Zhou. 1999. The Ski oncoprotein interacts with the Smad proteins to repress TGFβ signaling. Genes Dev. 13:2196-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macias-Silva, M., S. Abdollah, P. A. Hoodless, R. Pirone, L. Attisano, and J. L. Wrana. 1996. MADR2 is a substrate of the TGFβ receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell 87:1215-1224. [DOI] [PubMed] [Google Scholar]
- 52.Massagué, J., S. W. Blain, and R. S. Lo. 2000. TGFβ signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 53.Massague, J., and D. Wotton. 2000. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyazono, K. 2000. TGF-beta signaling by Smad proteins. Cytokine Growth Factor Rev. 11:15-22. [DOI] [PubMed] [Google Scholar]
- 55.Miyazono, K. 2000. TGF-beta/SMAD signaling and its involvement in tumor progression. Biol. Pharm. Bull. 23:1125-1130. [DOI] [PubMed] [Google Scholar]
- 56.Moses, H. L. 1992. TGF-beta regulation of epithelial cell proliferation. Mol. Rep. Dev. 32:179-184. [DOI] [PubMed] [Google Scholar]
- 57.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakao, A., T. Imamura, S. Souchelnytskyi, M. Kawabata, A. Ishisaki, E. Oeda, K. Tamaki, J. Hanai, C. H. Heldin, K. Miyazono, and P. ten Dijke. 1997. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 16:5353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishihara, A., J. I. Hanai, N. Okamoto, J. Yanagisawa, S. Kato, K. Miyazono, and M. Kawabata. 1998. Role of p300, a transcriptional coactivator, in signalling of TGF-beta. Genes Cells 3:613-623. [DOI] [PubMed] [Google Scholar]
- 60.Padgett, R. W. 1999. Intracellular signaling: fleshing out the TGFβ pathway. Curr. Biol. 9:R408-R411. [DOI] [PubMed] [Google Scholar]
- 61.Pietenpol, J. A., J. T. Holt, R. W. Stein, and H. L. Moses. 1990. Transforming growth factor beta 1 suppression of c-myc gene transcription: role in inhibition of keratinocyte proliferation. Proc. Natl. Acad. Sci. USA 87:3758-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pouponnot, C., L. Jayaraman, and J. Massague. 1998. Physical and functional interaction of SMADs and p300/CBP. J. Biol. Chem. 273:22865-22868. [DOI] [PubMed] [Google Scholar]
- 63.Qing, J., Y. Zhang, and R. Derynck. 2000. Structural and functional characterization of the transforming growth factor-beta-induced Smad3/c-Jun transcriptional cooperativity. J. Biol. Chem. 275:38802-38812. [DOI] [PubMed] [Google Scholar]
- 64.Reynisdottir, I., K. Polyak, A. Iavarone, and J. Massague. 1995. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 9:1831-1845. [DOI] [PubMed] [Google Scholar]
- 65.Rich, J., A. Borton, and X. Wang. 2001. Transforming growth factor-beta signaling in cancer. Microsc. Res. Tech. 52:363-373. [DOI] [PubMed] [Google Scholar]
- 66.Roberts, A. B. 1999. TGF-beta signaling from receptors to the nucleus. Microbes Infect. 1:1265-1273. [DOI] [PubMed] [Google Scholar]
- 67.Roussel, M. F., J. N. Davis, J. L. Cleveland, J. Ghysdael, and S. W. Hiebert. 1994. Dual control of myc expression through a single DNA binding site targeted by ets family proteins and E2F-1. Oncogene 9:405-415. [PubMed] [Google Scholar]
- 68.Seoane, J., C. Pouponnot, P. Staller, M. Schader, M. Eilers, and J. Massague. 2001. TGFβ influences Myc, Miz-1, and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3:400-408. [DOI] [PubMed] [Google Scholar]
- 69.Shen, X., P. P. Hu, N. T. Liberati, M. B. Datto, J. P. Frederick, and X. F. Wang. 1998. TGF-beta-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol. Biol. Cell 9:3309-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi, Y., Y. F. Wang, L. Jayaraman, H. Yang, J. Massague, and N. P. Pavletich. 1998. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell 94:585-594. [DOI] [PubMed] [Google Scholar]
- 71.Sikora, K., S. Chan, G. Evan, H. Gabra, N. Markham, J. Stewart, and J. Watson. 1987. c-myc oncogene expression in colorectal cancer. Cancer 59:1289-1295. [DOI] [PubMed] [Google Scholar]
- 72.Smith, D. R., T. Myint, and H. S. Goh. 1993. Over-expression of the c-myc proto-oncogene in colorectal carcinoma. Br. J. Cancer 68:407-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Souchelnytskyi, S., K. Tamaki, U. Engstrom, C. Wernstedt, P. ten Dijke, and C. H. Heldin. 1997. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-beta signaling. J. Biol. Chem. 272:28107-28115. [DOI] [PubMed] [Google Scholar]
- 74.Staller, P., K. Peukert, A. Kiermaier, J. Seoane, J. Lukas, H. Karsunky, T. Moroy, J. Bartek, J. Massague, F. Hanel, and M. Eilers. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 3:392-399. [DOI] [PubMed] [Google Scholar]
- 75.Stroschein, S. L., W. Wang, and K. Luo. 1999. Cooperative binding of Smad proteins to two adjacent DNA elements in the plasminogen activator inhibitor-1 promoter mediates transforming growth factor beta-induced smad-dependent transcriptional activation. J. Biol. Chem. 274:9431-9441. [DOI] [PubMed] [Google Scholar]
- 76.Sun, Y., X. Liu, E. N. Eaton, W. S. Lane, H. F. Lodish, and R. A. Weinberg. 1999. Interaction of the Ski oncoprotein with Smad3 regulates TGF-beta signaling. Mol. Cell 4:499-509. [DOI] [PubMed] [Google Scholar]
- 77.Sun, Y., X. Liu, E. Ng-Eaton, H. F. Lodish, and R. A. Weinberg. 1999. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proc. Natl. Acad. Sci. USA 96:12442-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 79.ten Dijke, P., K. Miyazono, and C. H. Heldin. 2000. Signaling inputs converge on nuclear effectors in TGF-beta signaling. Trends Biochem. Sci. 25:64-70. [DOI] [PubMed] [Google Scholar]
- 80.Vigo, E., H. Muller, E. Prosperini, G. Hateboer, P. Cartwright, M. C. Moroni, and K. Helin. 1999. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol. Cell. Biol. 19:6379-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Warner, B. J., S. W. Blain, J. Seoane, and J. Massague. 1999. Myc downregulation by transforming growth factor beta required for activation of the p15Ink4b G1 arrest pathway. Mol. Cell. Biol. 19:5913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White, L. A., T. I. Mitchell, and C. E. Brinckerhoff. 2000. Transforming growth factor beta inhibitory element in the rabbit matrix metalloproteinase-1 (collagenase-1) gene functions as a repressor of constitutive transcription. Biochim. Biophys. Acta 1490:259-268. [DOI] [PubMed] [Google Scholar]
- 83.Wong, C., E. M. Rougier-Chapman, J. P. Frederick, M. B. Datto, N. T. Liberati, J. M. Li, and X. F. Wang. 1999. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor beta. Mol. Cell. Biol. 19:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wotton, D., R. S. Lo, S. Lee, and J. Massague. 1999. A Smad transcriptional corepressor. Cell 97:29-39. [DOI] [PubMed] [Google Scholar]
- 85.Wrana, J. L., L. Attisano, J. Carcamo, A. Zentella, J. Doody, M. Laiho, X. F. Wang, and J. Massague. 1992. TGF beta signals through a heteromeric protein kinase receptor complex. Cell 71:1003-1014. [DOI] [PubMed] [Google Scholar]
- 86.Wrana, J. L., L. Attisano, R. Wieser, F. Ventura, and J. Massague. 1994. Mechanism of activation of the TGF-beta receptor. Nature 370:341-347. [DOI] [PubMed] [Google Scholar]
- 87.Wu, J. W., M. Hu, J. Chai, J. Seoane, M. Huse, C. Li, D. J. Rigotti, S. Kyin, T. W. Muir, R. Fairman, J. Massague, and Y. Shi. 2001. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol. Cell 8:1277-1289. [DOI] [PubMed] [Google Scholar]
- 88.Xu, W., K. Angelis, D. Danielpour, M. M. Haddad, O. Bischof, J. Campisi, E. Stavnezer, and E. E. Medrano. 2000. Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type beta transforming growth factor. Proc. Natl. Acad. Sci. USA 97:5924-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yagi, K., M. Furuhashi, H. Aoki, D. Goto, H. Kuwano, K. Sugamura, K. Miyazono, and M. Kato. 2002. c-myc is a downstream target of the Smad pathway. J. Biol. Chem. 277:854-861. [DOI] [PubMed] [Google Scholar]
- 90.Yingling, J. M., M. B. Datto, C. Wong, J. P. Frederick, N. T. Liberati, and X. F. Wang. 1997. Tumor suppressor Smad4 is a transforming growth factor β-inducible DNA binding protein. Mol. Cell. Biol. 17:7019-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zawel, L., J. L. Dai, P. Buckhaults, S. Zhou, K. W. Kinzler, B. Vogelstein, and S. E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1:611-617. [DOI] [PubMed] [Google Scholar]
- 92.Zhang, Y., T. Musci, and R. Derynck. 1997. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr. Biol. 7:270-276. [DOI] [PubMed] [Google Scholar]
- 93.Zheng, N., E. Fraenkel, C. O. Pabo, and N. P. Pavletich. 1999. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13:666-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zimmerman, C. M., and R. W. Padgett. 2000. Transforming growth factor beta signaling mediators and modulators. Gene 249:17-30. [DOI] [PubMed] [Google Scholar]