Abstract
Histone deacetylase inhibitors (HDIs) induce cell cycle arrest, differentiation, or apoptosis in numerous cancer cell types both in vivo and in vitro. These dramatic effects are the result of a specific reprogramming of gene expression. However, the mechanism by which these agents activate the transcription of some genes, such as p21WAF1, but repress others, such as cyclin D1, is currently unknown. We have been studying the human SRC gene as a model for HDI-mediated transcriptional repression. We found previously that both the tissue-specific and housekeeping SRC promoters were equally repressed by HDIs. Here we show that, despite an overt dissimilarity, both SRC promoters do share similar core promoter elements and transcription is TAF1 dependent. Detailed analysis of the SRC promoters suggested that both core and proximal promoter elements were responsible for HDI-mediated repression. This was confirmed in a series of promoter-swapping experiments with the HDI-inducible, TAF1-independent p21WAF1 promoter. Remarkably, all the SRC-p21WAF1 chimeric promoter constructs were not only repressed by HDIs but also dependent on TAF1. Together these experiments suggest that the overall promoter architecture, rather than discrete response elements, is responsible for HDI-mediated repression, and they implicate core promoter elements in particular as potential mediators of this response.
In order for an individual cell to properly execute its gene expression program, the appropriate combinations of factors must be present at specific regulatory regions of the genome at the correct time. In large part, the diversity of the promoters of individual genes directs these requirements; therefore, eukaryotic promoter structure and function have been an area of intense study. RNA polymerase II-dependent promoters can be broken down into two basic components. The core promoter recruits the general transcriptional apparatus and supports basal transcription, while the proximal promoter recruits transcriptional activators, which are necessary for appropriately activated transcription (26). In vitro transcription reconstitution assays have provided an important framework for the specific mechanistic roles the general transcription factors (GTFs) play at core promoters (32, 36). For example, an important step in the assembly of a preinitiation complex at the promoter is the binding of the GTF TFIID (7). TFIID is comprised of the TATA binding protein (TBP) and 10 to 12 TBP-associated factors (TAFs) (53). For core promoters that contain TATA elements, the TBP component of TFIID is important for TFIID binding and, hence, supports basal transcription (26). Another common functional element, termed the initiator (Inr), can also be found in eukaryotic core promoters (39). For this class of core promoter, the largest TFIID component, TAF1 (formerly referred to as TAFII250), has been implicated in recruiting TFIID by binding the Inr in conjunction with TAF2 (formerly referred to as TAFII150) (6). Interestingly, TAF1 offers additional functions at promoters such as acetyltransferase, E1 ubiquitin activation, E2 ubiquitin conjugation, and two Ser/Thr kinase activities, in addition to two bromodomains, which play a role in binding to acetylated histones (52). The precise targets of these activities are not known, but the importance of TAF1 is highlighted by findings that the transcription of 18 and 30% of cellular genes in hamster and Saccharomyces cerevisiae, respectively, is absolutely dependent on its function (18, 31). Furthermore, TAF1 inactivation in Drosophila melanogaster is lethal (51).
A major obstacle that must be overcome for transcription to occur is a repressive chromatin structure. The regulation of chromatin dynamics is achieved in large part through the acetylation and deacetylation of the N-terminal tails on nucleosomal histone proteins (23). Histone acetylation is catalyzed by histone acetyltransferase enzymes and strongly correlates with transcriptional activity (15, 33). Indeed, transcriptional activators, in addition to influencing the thermodynamics and kinetics of preinitiation complex assembly, also recruit coactivator complexes which contain histone acetyltransferase enzymes, such as p300/CBP and PCAF (42). Corepressor complexes, conversely, contain histone deacetylase (HDAC) activities and are recruited to promoters by transcriptional repressors (33). Although histones are the best-defined substrates for these activities, other proteins involved in transcription, such as p53, EKLF, GATA-1, TFIIE, and TFIIF, are also acetylated and deacetylated by these enzymes (42). Uncoupling of the fine balance between acetylation and deacetylation is thought to occur upon treatment of cells with HDAC inhibitors (HDIs) such as trichostatin A (TSA) and butyrate. Indeed, treatment with HDIs leads to the accumulation of hyperacetylated nuclear histones (50). Current excitement surrounding these agents stems from their ability to elicit G1/S arrest, differentiation, and/or apoptosis of transformed cells in culture and animal models (28). It would be expected that treatment with these agents would cause a general induction of many cellular genes. However, the present view is that HDIs reprogram gene expression and only affect a very specific subset of genes (27). For example, the most well characterized response to HDI treatment is the p53-independent transcriptional induction of the WAF1 gene, which encodes the cell cycle inhibitor p21WAF1 (19, 30). Induction of WAF1 has been demonstrated as essential for the growth inhibitory effects of these agents (2). However, HDIs can also directly repress genes such as cyclin D1 (25) and c-Myc (16, 41). The repression of these growth-promoting genes offers further explanation for the anticancer effectiveness of HDIs. At the present time, it is largely unknown how HDIs repress gene expression. Recent studies, however, have suggested that the mechanism of HDI-mediated gene expression modulation is direct and that changes in the promoters' chromatin structure, resulting from disrupted acetylation or deacetylation dynamics, are secondary effects (22, 29).
We have recently demonstrated that c-Src mRNA and protein expression are inhibited by treatment of a diverse array of cancer cell lines with HDIs (24). Activation and/or overexpression of the c-Src tyrosine kinase has been a consistent finding in colon and other cancers (3), and it has been shown that this activation is at the level of SRC transcription in a subset of human colon cancer cell lines (9). c-Src is the human homologue of the transforming v-Src oncogene encoded in the Rous sarcoma virus genome. SRC transcription is controlled by two disparate promoters separated by approximately 1 kb (5). Despite their apparent dissimilarity, both promoters are directly inhibited following treatment with TSA and butyrate. These observations, coupled with the lack of reports describing the mechanism of gene repression by HDIs, prompted an investigation into the mechanism of SRC inhibition by these agents. A fundamental regulatory similarity was observed for these two promoters in that they both contain Inr elements in their core regions and are TAF1 dependent. Interestingly, the effects of TSA and butyrate on the SRC1A promoter were blocked in cells harboring a temperature-sensitive TAF1 mutant, suggesting that TAF1 could play a role in the repression of transcription mediated by HDIs. The generation of chimeric promoters demonstrated that proximal and core promoter elements from both SRC promoters could independently confer HDI-mediated repression on the WAF1 promoter. Further analysis of these chimeric promoters showed that they were TAF1 dependent even though WAF1 is normally TAF1 independent. In summary, these findings represent the first, potentially functional link between promoter architecture, TAF1 dependence, and HDI-mediated transcriptional repression.
MATERIALS AND METHOD
Cell lines and tissue culture
The HT29 and SW480 human colon adenocarcinoma cell lines were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (FCS). Human HepG2 hepatocellular carcinoma cells were obtained from the American Type Culture Collection and propagated in DMEM-F-12 with 10% FCS. BHK-21 hamster cells and their derivative, tsBN462, were obtained from T. Sekiguchi (Kyushu University, Fukuoka, Japan) and grown in DMEM with 10% FCS. All cells were maintained at 37°C and 5% CO2, except for tsBN462 cells, which were maintained at 33°C and 5% CO2.
Plasmid-based constructs
The hepatocyte nucear factor 1α (HNF-1α)/GAL4 and HNF-1β/GAL4 fusion constructs, as well as plasmid pSG424, were obtained from R. O'Brien (Vanderbilt University, Nashville, Tenn.) (43). GAL4/Sp1 and GAL4/Sp3 fusions, as well as plasmid pM, were obtained from T. Sakai (Kyoto Univeristy, Kyoto, Japan) (30). GAL4/VP16 was obtained from D. Anderson (Saskatchewan Cancer Agency). Plasmid pCMV-hTAFII250 was a gift of R. Tjian (Howard Hughes Medical Institute). The −145 SRC1α-CAT, −145 SRC1α-CAT HNFmut, 0.2 SRC1A-CAT, 0.38 SRC1A-CAT, and 0.38 SRC1A-CAT GC1/GA2mut reporters (where CAT is chloramphenicol acetyltransferase), as well as the SRC1A TC tract deletion constructs have been described previously (4, 5, 34).
All site-directed mutagenesis was performed by using the QuickChange protocol (Stratagene). The SRC1AΔGC1/GA2-GAL4-CAT construct was made by introducing a KpnI site into 0.38 SRC1A-CAT, downstream of the GC1 site, by using mutagenic primers 5′-GCTTCTGTGCCCGGTACCCCCACCCCGCCC and 5′-GGGCGGGGTGGGGGTACCGGGCACAGAAGC. The resulting vector was digested with NarI/KpnI, and a synthetic double-stranded DNA cassette created by annealing sense (5′-CGCCCTGGCGTCGGAGTACTGTCCTCCGATCGGCCCGGTAC, GAL4 site underlined) and antisense (5′-CGGGCCGATCGGAGGACAGTACTCCGACGCCAGGG) oligonucleotides was inserted into this site. The GA2 site was then replaced in this construct by removing a BssHII fragment and inserting a double-stranded cassette created by annealing single-stranded sense (5′-CGCGCTTCCTCCTTCCTCCTCCTCCCGGCTGCTCGGAGTACTGTCCTCCGATCGCG, GAL4 recognition sequence underlined) and antisense (5′-CGCGCGCGATCGGAGGACAGTACTCCGAGCAGCCGGGAGGAGGAGGAAGGAGGAAG) oligonucleotides. To create the SRC1αΔHNF-GAL4-CAT reporter, the SrcHNF site was directly changed to a GAL4 site in the −145 SRC1α-CAT vector by using mutagenic primers spanning the HNF site (5′-GCTGGGGGCCCGCCCTGAGCCCCTGGGAATCGGAGTACTGTCCTCCGATCGGCCTTGCAAACAAGTGCGGCCATTTCAC, GAL4 site underlined, and 5′-GTGAATGGCCGCACTTGTTTGCAAGGCCGATCGGAGGACAGTACTCCGATTCCCAGGGCTCAGGGCGGGCCCCCAGC).
All SRC1A 3′ deletion constructs were based on the 0.38 SRC1A promoter which had been inserted into a pCAT3 Basic (Promega) reporter vector with the synthetic intron (HindIII fragment) removed. SRC1A3′ΔSacII-CAT was derived by the digestion of 0.38 SRC1A-CAT with SacII/HindIII, creation of blunt ends, and religation. The remaining SRC1A 3′ promoter deletions were isolated as PCR fragments from 0.38 SRC1A-CAT by using a common forward primer (SRC1A/1α3′ΔFWD, 5′-GGTACCGAGCTCTTACGCGTGC) in conjunction with specific reverse primers (SRC1AΔ+13REV, 5′-CCGCTCAAGCTTCCAGGCCGG; SRC1AΔ-26REV, 3′-AGAAAGCTTGAGAGAGAAAGGG; and SRC1AΔ-60REV, AGGAAGCTTCGGCGGCCCGGG). SRC1α 3′ promoter deletion fragments were isolated from −145 SRC1α-CAT as PCR fragments by using a common forward primer (SRC1A/1α3′ΔFWD) paired with specific reverse primers (SRC1αΔ+99REV, GGTAAGCTTGTGCTAGATGAATGG; SRC1αΔ+41REV, GGGAAGCTTGAGGTGCCACAGC; and SRC1αΔ-20REV, GGCCAAGCTTGTTTGCAAGGC). PCR products were digested with SacI/HindIII and cloned directly into SacI/HindIII-digested pCAT3-Basic.
Vector pWWP-Luc (WAF1-Luc) was a gift of Bert Vogelstein (11). A 2.3-kb HindIII fragment from this construct was subcloned into a HindIII-digested pBlue shuttle vector. The WAF1 promoter was deleted to position −210, relative to the transcription start site, via PstI digestion and religation. To create −210 WAF1-CAT, the truncated WAF1 promoter was removed from pBlue with SacI/SalI and cloned into SacI/SalI-digested pCAT3-Basic. To allow for further truncation of this WAF1 promoter, a SacI site was introduced at the −101 position by using the mutagenic primers 5′-GGGCGGTCCCGGGCGGAGCTCTGGGCCGAGCGAGGGTCCC and 5′-GGGACCCGCGCTCGGCCCAGAGCTCCGCCCGGGACCGCCC. This mutant construct was subsequently digested with SacI and religated to create −101 WAF1-CAT. A −145 SRC1α-CAT variant, harboring a SacII recognition site at position −10, was created by using mutagenic primers 5′-GCAAACAAGTGCGGCCATTTCCGCGGCCCAGGCTGGCTTCTGC and 5′-GCAGAAGCCAGCCTGGGCCGCGGAAATGGCCGCACTTGTTTGC. A similar variant of 0.38 SRC1A-CAT3-Basic, with a SacII recognition site engineered at position −10, was created by using the primers 5′-CGATCTGTCTCTCCCGGCCCGCGGTCCATTCCGGCCTGGGAGC and 5′-GCTCCCAGGCCGGAATGGACCGCGGGCCGGGAGAGACAGATCG. The WAF1 core promoter was amplified from −210 WAF1-CAT via PCR with primers 5′-GGCGCCGCGGTTGTATATCAGG and 5′-TTTCTCCATGGTGGCTTTACC. The chimeric promoter constructs 0.38SRC1A:WAFcore-CAT and −145SRC1α:WAFcore-CAT were derived by digesting the WAF1 PCR product with SacII/NcoI and cloning it into the SacII-engineered, SacII/NcoI-digested SRC1A or SRC1α CAT constructs, respectively. Similarly, EcoRI recognition sites were engineered in the −101 WAF1-CAT and −210 WAF1-CAT vectors by using site-directed mutagenesis with the primers 5′-CGGGCGGGGCGGTTGGAATTCAGGGCCGCGCTGAGC and 5′-GCTCAGCGCGGCCCTGAATTCCAACCGCCCCGCCCG. The SRC1A and SRC1α core promoters were isolated from 0.38 SRC1A-CAT-Basic and −145 SRC1α-CAT via PCR using a common reverse primer, CAT3NcoREV, paired with the appropriate 1A- or 1α-specific forward primer (1AEcoFWD, 5′-CTCCGAATTCTCCCTTTCTCTCTCG; 1αEcoFWD, 5′-GGTTAGAATTCAAGCCAGCCTTGC). The SRC1A core promoter PCR product was digested with EcoRI/NcoI and cloned directly into EcoRI/NcoI-digested −101 WAF1-CAT or −210 WAF1-CAT to create −101WAF1:SRC1Acore-CAT and −210WAF1:SRC1Acore-CAT, respectively. The SRC1α core promoter PCR product was similarly digested and ligated with −210 WAF1-CAT or −101 WAF1-CAT, thus generating −210WAF1:SRC1αcore-CAT and −101WAF1:SRC1αcore-CAT, respectively. In all cases, the promoter regions were isolated from final constructs by restriction digestion and reintroduced into the original parental vector. All promoter cassettes were completely sequenced to verify their integrity.
Transient transfections
All plasmid constructs used in transfection experiments were isolated and purified by using an EndoFree Plasmid Maxi Kit from QIAGEN. SW480, HepG2, and HT29 cells were transfected and treated with 1 μM TSA exactly as previously described (24). For tsBN462 and BHK-21 transfection experiments, cells were transfected at 33°C. Transfections were performed by using Superfect reagent (QIAGEN) with reaction mixtures as previously described (24). For TAF1 cotransfection experiments, mixtures consisted of 1.0 μg of CAT reporter construct, 0.5 μg of vector pCMV-βGal, and 62.5 or 125 ng of vector pCMV-hTAFII250. pBluescript was added to these mixtures to ensure a final DNA mass of 2.0 μg. Following transfection, fresh media was added to the cells, and they were allowed to grow at 33°C for an additional 36 h. Cells were then left untreated or exposed to TSA (1 mM) or sodium butyrate (5 mM) for 18 h at 33 or 39°C before harvesting. Transfected cells were lysed, and levels of CAT expression were subsequently determined with a CAT enzyme-linked immunosorbent assay kit (Roche). The CMV-βGal construct used in these experiments was activated by TSA and butyrate treatment, as well as by the shift to 39°C in tsBN462 cells. This consistent response, therefore, served as a useful internal control and was measured by a colorimetric β-galactosidase assay (13). All CAT levels were corrected to protein concentrations in transfected cell lysates.
Mapping of SRC1A transcription start sites
SRC1A promoter transcription start sites were mapped in HepG2, HT29, and SW480 cells exactly as described previously (4).
EMSAs
Hemagglutinin (HA)-TAF1 (human TAFII250) and FLAG-TAF2 (human TAFII150) were immunoaffinity purified from Sf9 cell lysates and assembled into heterodimers in vitro as described elsewhere (8). Double-stranded, 32P-labeled probes encompassing the SRC1A or SRC1α core promoters were generated. For electrophoretic mobility shift assay (EMSA) reactions, 5 μg of TAF1/2 heterodimer was incubated in a binding buffer (17) at 25°C or 37°C for 20 min. Labeled, double-stranded probes were subsequently added, and the reaction proceeded at the same temperature for an additional 20 min. For competition experiments, double-stranded competitors representing wild-type SRC1A or SRC1α core promoters or SRC core promoters harboring mutations in the Inr elements were added to the initial preincubation reaction with the TAF1/2 heterodimer prior to the addition of 32P-labeled probe. Bound and unbound core promoter probes were fractionated on agarose gels as described previously (17) and visualized via autoradiography.
RESULT
The SRC core promoters share common elements
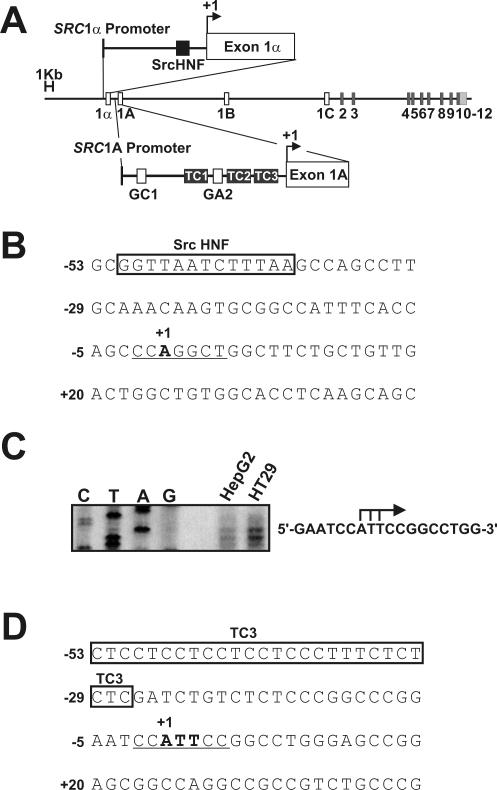
We have demonstrated that SRC transcription is directly repressed following treatment with HDIs, leading to a significant reduction in cellular c-Src mRNA and protein levels in diverse human cancer cell lines (24). A major interest, therefore, became to elucidate the mechanism of this repression. The SRC gene is comprised of 15 exons; exons 1B and 1C are noncoding, and exons 2 to 12 encode the functional c-Src protein (Fig. 1A). The extreme 5′ exons, 1A and 1α, are each associated with their own promoter, which we have termed SRC1A and SRC1α, respectively (4, 5, 34). Alternative SRC promoter usage results in two c-Src transcripts that have identical coding capacities but differ in their extreme 5′ exon composition. Interestingly, our studies of HDI action have shown that both of the SRC promoters are equally and effectively inhibited following HDI treatment (24). Therefore, we hypothesized there must be some commonality in their regulation that could account for this repression. Previous reports from our laboratory, however, have demonstrated that these promoters are quite different. For example, the ubiquitously expressed SRC1A promoter is regulated by two Sp-family binding sites, termed GC1 and GA2, as well as by three perfect polypurine-polypyrimidine tracts (TC1, TC2, and TC3), which bind hnRNP K protein (Fig. 1A) (35). In contrast, the tissue-restricted SRC1α promoter is absolutely dependent on a single HNF binding site that we have shown binds and responds to HNF-1α (Fig. 1A) (5).
FIG. 1.
The SRC core promoters contain Inr elements. (A) Schematic of the human SRC locus on chromosome 20. Proximal promoter elements are shown and defined in the text. (B) Sequence of the SRC1α core promoter regions surrounding the SRC1α transcription start site. The Inr element is underlined. (C) Transcription start site mapping in the SRC1A promoter. S1 protection analysis was performed to determine the initiation sites for transcription in the SRC1A promoter in HepG2 and HT29 cells. The residues mapped to the extreme 5′ termini of exon 1A-containing transcripts are denoted with bent black arrows on the right. (D) Sequence of the SRC1A core promoter regions surrounding the SRC1A transcription start site. The Inr element is underlined.
Because of the apparent dissimilarity in the identity and composition of the proximal regions of the two SRC promoters, the potential for common core elements was assessed. Both SRC promoters lack consensus TATA or CCAAT regulatory motifs but, rather, contain sequences that resemble Inr elements (4, 5). For example, the major transcription initiation site in the SRC1α promoter maps to a CCA(+1)GGCT motif 39 bp downstream from the HNF site in HepG2 and HT29 cells (Fig. 1B) (5). To determine if transcription is initiated from a similar motif in the SRC1A core promoter, we employed an S1-nuclease protection strategy in HepG2 and HT29 cells (Fig. 1C). Three major sites of transcription initiation were mapped to a core CCA(+1)TTC in these cells, 27 bp downstream from the TC3 tract in the SRC1A promoter (Fig. 1D). This SRC1A transcription start site core perfectly matched the Inr consensus sequence of YYA(+1)NTYY (40). These observations led to the hypothesis that both SRC promoters contained functional Inr elements in their core regions. It was further hypothesized that this commonality in their regulation could explain their repression following HDI treatment.
The SRC1A and SRC1α core promoters are Inr driven and TAF1 dependent
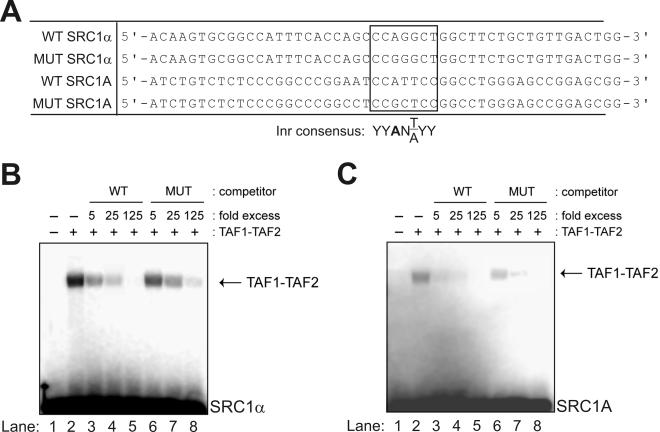
A fundamental property of many promoters with Inr elements is their ability to directly bind a heterodimer of the two largest TFIID components, TAF1 and TAF2 (6). Therefore, to ascertain whether the observed motifs in the SRC1A and SRC1α core promoters were bona fide Inr elements, we assessed their ability to bind a TAF1-TAF2 heterodimer derived from epitope-tagged human recombinant TAF1 and TAF2 proteins. Double-stranded oligonucleotides, representing wild-type and Inr mutant forms of the SRC core promoters, were used as labeled probes and unlabeled competitors in these binding experiments (Fig. 2A). As anticipated, both the wild-type SRC1α and SRC1A core promoters were able to bind recombinant TAF1-TAF2 (Fig. 2B and C, lane 2). This binding was effectively competed away with an excess of unlabeled probe (Fig. 2B and C, lanes 3 to 5). However, unlabeled double-stranded oligonucleotides representing Inr mutant forms of the SRC1α or SRC1A core promoters were less efficient competitors for TAF1-TAF2 binding (Fig. 2B and C, compare lanes 6 through 8 to lanes 3 through 5). These results showed that the SRC1α and SRC1A core promoters bind a TAF1-TAF2 heterodimer and that the Inr core plays a role in this binding.
FIG. 2.
Both SRC core promoters bind a TAF1-TAF2 heterodimer. (A) Oligonucleotides used in EMSAs. Proposed SRC1A and SRC1α Inr elements as well as the consensus sequence for an Inr element are shown. (B) EMSAs were performed using a recombinant human TAF1-TAF2 heterodimer (17) and a 32P-labeled probe representing the SRC1α core promoter. Competitions were performed with excess unlabeled wild-type probe (lanes 3 to 5) or an unlabeled duplex harboring a mutation in the SRC1α Inr core (lanes 6 to 8). (C) EMSAs were performed using a recombinant TAF1-TAF2 heterodimer and a 32P-labeled probe representing the SRC1A core promoter. Competitions were performed using excess unlabeled wild-type probe (lanes 3 to 5) or an unlabeled duplex harboring mutations in the SRC1A Inr core (lanes 6 to 8). WT, wild type; MUT, mutant.
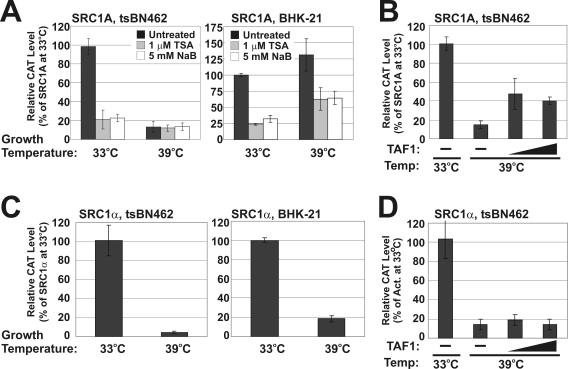
Another property of many promoters that contain Inr elements in their core regions is a dependence on TAF1 for full activity. Commonly used tools for studying promoters' TAF1 dependence are the BHK-21 (baby hamster kidney)-derived tsBN462 and ts13 cell lines, which harbor identical G690D mutations in the TAF1 protein (14). When maintained at 33°C, G690D TAF1 retains wild-type function; therefore, these cells grow normally. However, when the cells are shifted to the restrictive temperature of 39°C, G690D TAF1 acetyltransferase as well as Inr binding activities are impaired, resulting in G1/S arrest (10, 17, 48). Cell cycle arrest has been partly attributed to inhibited TAF1 function, leading to the induction of genes encoding the cell cycle inhibitors p21 and p27 and repression of cyclins D and A1 (37, 38, 45). Therefore, to determine if G690D TAF1 also affected the transcriptional activity of the SRC promoters, we performed transient transfections with SRC1α and SRC1A CAT reporters in tsBN462 cells as well as parental BHK-21 cells. Following a shift in growth temperature from 33 to 39°C, the activity of the SRC1A promoter was decreased in tsBN462 but not BHK-21 cells (Fig. 3A). This decrease in SRC1A activity at 39°C in tsBN462 cells was partially rescued by coexpression of wild-type TAF1 (Fig. 3B). These results showed that the SRC1A promoter is indeed TAF1 dependent. Concurrently, we also felt it prudent to characterize the response of the SRC1A promoter to HDIs in this hamster-derived model system, because the cell lines that we normally use to study SRC transcription are of human origin. Indeed, at 33°C TSA and butyrate had a similar negative effect on SRC1A transcription in tsBN462 and BHK-21 cells (Fig. 3A). Interestingly, however, at 39°C the SRC1A promoter was repressed by TSA and butyrate in BHK-21 cells but not in tsBN462 cells (Fig. 3A). These data therefore hinted at the possibility that compromised TAF1 function was blocking the repressive effects of HDIs on SRC1A in the tsBN462 cell line. When we assessed the TAF1 dependence of the SRC1α promoter in this model system, we were surprised to observe lower activity at 39°C compared to levels at 33°C in both tsBN462 and BHK-21 cells (Fig. 3C). As a result, the decrease in SRC1α activity following a shift of tsBN462 cells from 33 to 39°C was not rescued by wild-type TAF1 coexpression (Fig. 3D). These findings for SRC1α suggested that there was a temperature effect unrelated to TAF1 that was preventing an accurate determination of TAF1 dependence in these cell lines. Nevertheless, the results from the TAF1-TAF2 binding studies strongly suggested that the SRC1α promoter, like the SRC1A promoter, contains a functional Inr element.
FIG. 3.
TAF1 dependence of the SRC promoters. (A) tsBN462 and BHK-21 cells were transfected with a minimal SRC1A promoter reporter, 0.38 SRC1A-CAT (34), and grown at 33°C for 36 h. Following treatment with 1 μM TSA or 5 mM sodium butyrate, transfected cells were grown at 33°C (permissive) or 39°C (restrictive) for 18 h. CAT levels were subsequently determined. (B) tsBN462 cells were transfected with 0.38 SRC1A-CAT with or without CMV-hTAFII250. Following 36 h of growth at 33°C, cells were shifted to 39°C and grown for an additional 18 h. CAT levels were subsequently determined. (C) tsBN462 and BHK-21 cells were transfected with a minimal SRC1α promoter reporter, −145 SRC1α-CAT (5), and grown at 33°C for 36 h. Following treatment with 1 μM TSA or 5 mM sodium butyrate, transfected cells were grown at 33 or 39°C for 18 h. CAT levels were subsequently determined. (D) tsBN462 cells were transfected with −145 SRC1α-CAT as described for panel B. Act, activity.
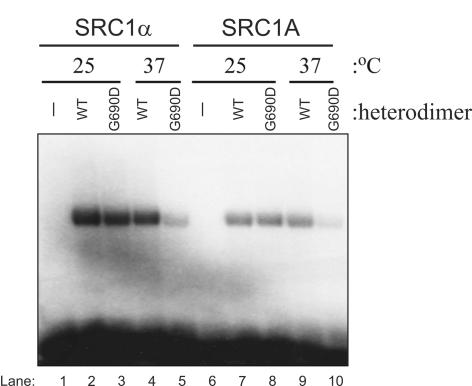
Previous studies have shown that the G690D TAF1 mutation compromises both acetyltransferase activity and Inr binding at the restrictive temperature (10, 17). Therefore, to determine if the decrease in SRC promoter activity following a shift to the restrictive temperature in tsBN462 cells could in part be due to reduced TAF1 core promoter binding, EMSAs were performed by using the SRC1A and SRC1α core promoters with recombinant wild-type or G690D mutant TAF1-TAF2 heterodimers (Fig. 4). These binding assays were performed at either 25°C or 37°C, as these temperatures have previously been characterized as being permissive and restrictive, respectively, for in vitro transcription reactions with ts13 cell lysates (48). We observed strong binding of TAF1-TAF2, containing either wild-type or G690D forms of TAF1, to the SRC1α and SRC1A core promoters at 25°C (Fig. 4, lanes 2, 3, 7, and 8). However, binding of the TAF1-TAF2 dimer containing G690D TAF1 to both SRC core promoters was compromised at 37°C (Fig. 4, lanes 5 and 10). Conversely, the TAF1-TAF2 dimer containing wild-type TAF1 did not display a decrease in binding to the SRC core promoters at this temperature (Fig. 4, lanes 4 and 9). These results support the hypothesis that both SRC promoters contain functional Inr elements and are TAF1 dependent. These data further suggest that the SRC transcriptional defect at 39°C could be due in part to reduced binding of TAF1 to the SRC core promoters.
FIG. 4.
SRC core promoter binding is inhibited for a G690D TAF1-TAF2 heterodimer at 37°C. EMSAs were performed at 25°C or 37°C with wild-type or G690D mutant TAF1 in a TAF1-TAF2 heterodimer and 32P-labeled probes representing the SRC1α or SRC1A core promoters. WT, wild type.
Search for a SRC core promoter HDI response element
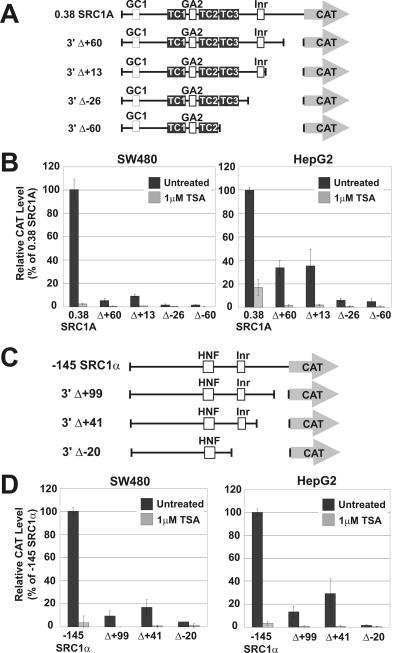
Our characterization of the SRC1A and SRC1α promoters highlighted that they are both Inr driven and TAF1 dependent. Therefore, we next elected to address the role these core promoter elements played in human cancer cell lines, as well as to assess their function in HDI-mediated transcriptional repression by analyzing a series of SRC promoter-CAT constructs harboring deletions in the core promoter regions (Fig. 5A and C). We were surprised to observe that, for both promoters, small deletions from the 3′ end had significant negative effects on their activity, suggesting that important functional elements reside in these fragments (Fig. 5B and D). In addition, 3′ deletion of the SRC1A promoter to position +60 and up to position +13 had a much more pronounced effect in SW480 cells compared to the effect in HepG2 cells (Fig. 5B). Deletions that eliminated the Inr element from the SRC1A promoter (Δ-26 and Δ-60) nearly abolished all detectable transcriptional activity in both these cell lines, lending further evidence to the hypothesis that the SRC1A promoter is Inr driven. However, despite these observations, core promoter deletions were unable to block transcriptional repression in response to TSA treatment. Deletions in the SRC1α core promoter had a very similar effect to deletions in the SRC1A core promoter, and exclusion of the Inr element (Δ-20) severely compromised promoter activity (Fig. 3D). Again, regardless of the effect the core promoter deletions had on SRC1α transcriptional activity, further repression was still consistently observed following treatment with TSA (Fig. 3D). These data therefore showed that there are core elements in addition to the Inr that are crucial for full transcriptional activity of both SRC promoters and that a distinct core promoter element in either SRC promoter is not solely responsible for mediating the effects of HDIs.
FIG. 5.
Role of core promoter elements in TSA-mediated SRC repression. Various SRC1A deletion constructs, depicted in panel A, were transfected in HepG2 and SW480 cells and analyzed for their response to 1 μM TSA (B). Various SRC1α deletion constructs, depicted in panel C, were transfected in HepG2 and SW480 cells and analyzed for their response to 1 μM TSA (D).
Search for a SRC proximal promoter HDI response element
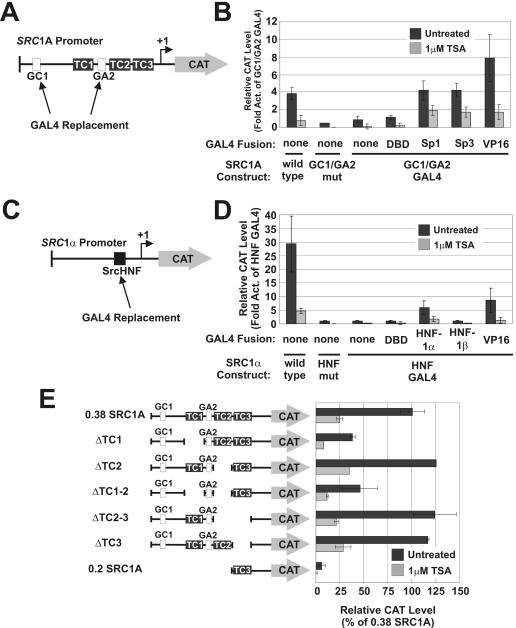
Since we could not identify a discrete core element in either SRC promoter that was responsible for the repressive effects of HDIs, we next studied the role of proximal promoter elements in mediating this effect. Previous studies with the WAF1 promoter have implicated Sp-family binding sites in transcriptional activation following HDI treatment (19, 30). Therefore, we chose to assess the role of the SRC1A GC1 and GA2 elements in HDI-mediated repression. To this end, the GC1 and GA2 sites were replaced in a 0.38 SRC1A promoter reporter construct with binding sites for the GAL4 yeast transcription factor (Fig. 6A), and transfections in HepG2 liver carcinoma cells were performed (Fig. 6B). GAL4 replacement impaired SRC1A activity, but following cotransfection with GAL4-Sp1 or GAL4-Sp3 fusions, SRC1A activity was restored to levels of the wild-type promoter (Fig. 6B). When these transfected cells were exposed to TSA, SRC1A activity was consistently repressed regardless of whether the promoter construct was analyzed alone or if an Sp1- or Sp3-GAL4 fusion was employed for cotransfection (Fig. 6B). Strikingly, when the SRC1A GC1/GA2-GAL4 promoter was transactivated by GAL4 fused to the strong viral VP16 activator, repression was still observed following TSA treatment (Fig. 6B). These findings suggested that the repressive effects of TSA were not specific for Sp1 or Sp3 binding to GC1 or GA2. Therefore, TSA was not inhibiting SRC1A activity through the Sp-family binding sites. A similar approach was taken to study the HNF-1α binding site in the SRC1α promoter (Fig. 6C). We observed that a GAL4-HNF-1α fusion, but not a GAL4-HNF-1β fusion, was able to moderately transactivate the SRC1α HNF-GAL4 promoter (Fig. 6D). However, regardless of whether the SRC1α HNF-GAL4 construct was transactivated by GAL4-HNF-1α or GAL4-VP16, it was still significantly repressed by TSA (Fig. 6D). These findings suggested that the HNF site in the SRC1α promoter was not responsible for mediating the repressive effect of HDIs.
FIG. 6.
Role of proximal promoter elements in TSA-mediated SRC repression. (A) The GC1 and GA2 sites were both mutated or replaced in the SRC1A promoter with GAL4 binding sites. (B) The SRC1AΔGC1/GA2-GAL4 promoter was subsequently transactivated with various GAL4 transcription factor fusions, and the response to treatment with 1 μM TSA was assessed in HepG2 cells. (C) The SrcHNF site was mutated or replaced in the SRC1α promoter with a GAL4 binding site. (D) The SRC1αΔHNF-GAL4 promoter was subsequently transactivated with various GAL4 transcription factor fusions, and the response to treatment with 1 μM TSA was assessed in HepG2 cells. (E) Various SRC1A constructs harboring deletions in upstream activation sequences were evaluated for their response to 1 μM TSA in HepG2 cells. Act, activity; mut, mutant.
We next sought to determine if an element(s) other than GC1 or GA2 was responsible for mediating the inhibitory effects of TSA on the SRC1A promoter. A number of 5′ and TC1, TC2, and TC3 SRC1A internal promoter deletions based on the 0.38 SRC1A-CAT promoter construct have previously been analyzed in transfection experiments (4, 34). Regardless of the deletion generated, all of these constructs were still further repressed following treatment with TSA (Fig. 6E). We systematically deleted the SRC1A promoter in its entirety in the present study (Fig. 5A and B and 6E) and were unable to observe any significant change in its response to TSA. These findings therefore, led to the conclusion that a single distinct element was not responsible for HDI-mediated SRC transcriptional repression. This conclusion suggested that multiple promoter elements, or overall promoter architecture, could be responsible for SRC repression by HDIs.
SRC1α and SRC1A proximal and core promoter elements can independently confer HDI-mediated repression on the WAF1 promoter
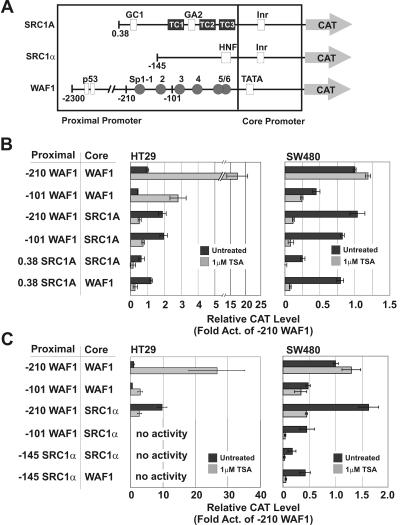
Our data have shown that the SRC promoters are both Inr driven and TAF1 dependent and have hinted at a role for TAF1 in SRC repression by HDIs. However, a single promoter element responsible for this repression was not observed. Of significance, therefore, is the observation that TAF1 dependence does not map to a single promoter element; rather, proximal and core promoter elements from a TAF1-dependent promoter can each confer TAF1 dependence on a normally TAF1-independent promoter (49). In light of this, an important question was whether the TAF1 dependence of the SRC promoters was responsible for their repression by HDIs. We therefore took a stringent approach and asked whether proximal and core promoter elements from the SRC promoters could independently confer HDI-mediated repression on a TAF1-independent, heterologous promoter normally activated by HDIs. To this end, we generated a series of chimeric SRC:WAF1 and WAF1:SRC promoter CAT reporters and analyzed their responses to HDIs following transfection in HT29 and SW480 colon cancer cells (Fig. 7). Previous studies have shown that the WAF1 promoter contains a consensus TATA box and a crucial Sp-family binding element, Sp1-3, which mediates its activation following HDI treatment (Fig. 7A) (19, 30). In agreement with these studies, two WAF1 promoter CAT constructs, −210 WAF1-CAT and −101 WAF1-CAT, were significantly induced following TSA treatment in transfected HT29 cells (Fig. 7B). Conversely, in SW480 cells, constitutively strong WAF1 promoter activity was observed, which was not induced following TSA treatment (Fig. 7B). However, when the WAF1 core promoter was replaced in the −210 WAF1 or −101 WAF1 constructs with either the SRC1A (Fig. 7B) or SRC1α (Fig. 7C) core promoters, repression was observed following TSA treatment in both HT29 and SW480 cells. Similarly, when the WAF1 proximal promoter was replaced in these constructs with the SRC1A (Fig. 7B) or SRC1α (Fig. 7C) proximal promoter, repression was observed following TSA treatment in both cell lines. The −145 SRC1α, −101 WAF1:SRC1α, and −145 SRC1α:WAF1 reporters had undetectable activities in HT29 cells. Taken together, these results demonstrated that proximal and core promoter elements from both the SRC1A and SRC1α promoters independently conferred HDI-mediated repression on the heterologous WAF1 promoter.
FIG. 7.
SRC proximal and core promoter elements can confer TSA-mediated repression on the WAF1 promoter. Wild-type SRC1A and WAF1 promoter constructs depicted in panel A, as well as various promoter chimeras, were assessed for their response to 1 μM TSA following transfection in HT29 and SW480 cells (B). Wild-type SRC1α and WAF1 promoter constructs depicted in panel A, as well as various promoter chimeras, were assessed for their response to 1 μM TSA following transfection in HT29 and SW480 cells (C). Act, activity.
SRC1α and SRC1A proximal and core promoter elements can independently confer TAF1 dependence on the WAF1 promoter
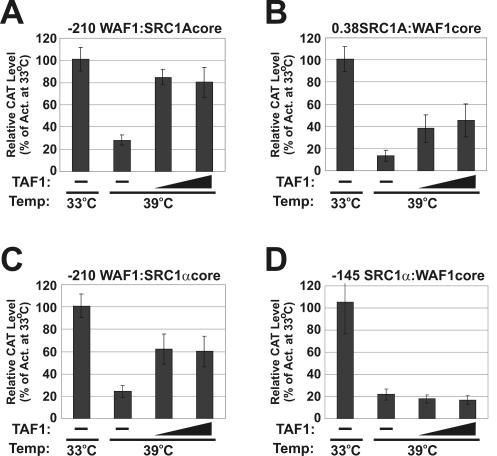
The addition of SRC proximal or core promoter elements to the HDI-activated WAF1 promoter produced a promoter that was repressed by HDIs. To test if these SRC proximal and core promoter elements also independently conferred TAF1 dependence on the WAF1 promoter, we analyzed the activities of the chimeric SRC:WAF1 constructs in tsBN462 cells (Fig. 8). We found that all WAF1 constructs had extremely high activity in tsBN462 and BHK-21 cells at both 33 and 39°C (data not shown), thus preventing an accurate analysis of activity. However, previous studies with ts13 cells stably expressing a WAF1-CAT construct showed that WAF1 transcription is TAF1 independent and even slightly induced following a shift from 33 to 39°C in these cells (37). In contrast, we observed a strong decrease in promoter activity for both the −210 WAF1:SRC1A and 0.38 SRC1A:WAF1 chimeras following a shift from 33 to 39°C in tsBN462 cells (Fig. 8A and B). The decrease in activity of these chimeras at 39°C was rescued partially or in full by coexpression of wild-type TAF1. These findings confirmed that the SRC1A proximal and core promoter elements independently conferred TAF1 dependence on the WAF1 promoter. Similarly, the −210 WAF1:SRC1α chimera showed diminished activity following a shift from 33 to 39°C (Fig. 8C). However, in contrast to our previous results with the SRC1α promoter alone (Fig. 3C), the activity of the −210 WAF1:SRC1α chimera was elevated by coexpression of wild-type TAF1 at 39°C. Rescue of the transcriptional block at 39°C by TAF1 was not observed for the −145 SRC1α:WAF1 chimera (Fig. 8D). These results, therefore, lend further support to the hypothesis that the SRC1α core promoter is indeed TAF1 dependent and that the unusual, temperature-sensitive property of SRC1α is mediated by the proximal promoter. Furthermore, similar to the results attained for the SRC1A promoter, the TAF1 dependence of the SRC1α core promoter was conferred on the heterologous TAF1-independent WAF1 promoter.
FIG. 8.
SRC upstream activation sequences and core promoter elements can confer TAF1 dependence on the WAF1 promoter. (A) A chimeric −210 WAF1:SRC1A core construct was transfected alone or in combination with increasing amounts of a wild-type TAF1 expression vector into tsBN462 cells. Cells were grown at 33°C for 36 h and then maintained at 33°C or shifted to 39°C for an additional 18 h. CAT levels were subsequently determined. (B) A chimeric 0.38 SRC1A:WAF1 core construct was analyzed as described for panel A. (C) A chimeric −210 WAF1:SRC1α core construct was analyzed as described for panel A. (D) A chimeric −145 SRC1α:WAF1 core construct was analyzed as described for panel A. Act, activity.
In summary, these results have shown that both SRC promoters' proximal and core elements could each confer TAF1 dependence and HDI-mediated repression on the TAF1-independent, HDI-induced WAF1 promoter. This observation explains the inability of promoter deletion and GAL4 replacement strategies to identify single, discrete HDI response elements, because precise elements in mammalian promoters that dictate TAF1 dependence have not been clearly defined. These results, therefore, suggest that the inherent TAF1 dependence of the SRC promoters could explain their repression in response to HDIs. This possibility is supported by the observed block of HDI-mediated SRC1A repression by the G690D TAF1 mutant at the restrictive temperature.
DISCUSSION
HDIs have been described as exciting agents with impressive anticancer potential (47). Indeed, these agents effectively reverse the transformed phenotype of various tumor cell lines in vivo and in vitro by inhibiting proliferation and inducing differentiation and/or apoptosis (28). Such anticancer properties of HDIs have been hypothesized to result from the highly selective changes in gene expression that ensue following treatment. Indeed, the most well-described cellular response to treatment with these agents is the p53-independent activation of the potent cell cycle inhibitor p21WAF1 (19, 30). Because of these previous findings, several classes of HDIs are currently being analyzed for their antitumor effectiveness in various phases of human clinical trials (47).
Given the potential clinical importance of these agents, it is surprising that relatively few studies have focused on the mechanisms by which HDIs modulate gene expression. HDIs are generally thought to exert their effects at the level of chromatin. Indeed, treatment with HDIs in vivo has been shown to lead to the accumulation of hyperacetylated nuclear histone proteins in both tumor and normal tissues (50). However, models that rely purely on a general relaxation of chromatin to explain gene induction fail to account for the hyperacetylation of nonhistone proteins that could occur following HDI treatment, including transcription factors such as p53, EKLF, and GATA-1, as well as the GTFs TFIIE and TFIIF (42). Furthermore, these models offer very little explanation for the mechanism of gene repression by HDIs, and global gene expression studies have shown that just as many genes are repressed as are activated by these agents (27). Clearly, a model of global histone hyperacetylation could not also account for the observation that the expression of only a very select subset of genes is altered in response to HDIs (27, 46). Indeed, the most comprehensive studies into the mechanism of gene activation by these agents have involved the WAF1 promoter, and these concluded that specific Sp-family binding sites are responsible for transcriptional induction (19, 30). More recently, the HDI apicidin was shown to require phosphatidylinositol-3-kinase, via protein kinase C-ɛ signaling, to elicit activation of WAF1 transcription (22). As has been shown previously, this apicidin-mediated WAF1 activation was associated with histone H3 hyperacetylation (22). Intriguingly, treatment with a phosphatidylinositol-3-kinase inhibitor markedly attenuated WAF1 induction by apicidin but did not affect histone H3 hyperacetylation at the WAF1 promoter (22). Therefore, this mechanistic study has suggested that histone hyperacetylation is a separate and/or secondary effect and may not play a direct role in the induction of genes such as WAF1 following HDI treatment. This hypothesis is supported by another recent study of the mechanism of TSA-mediated repression of the mouse mammary tumor virus (MMTV) promoter (29). Similar to findings from the study of WAF1 induction by apicidin, results of this study showed that changes in the well-characterized MMTV promoter nucleosome structure following TSA treatment were a secondary effect and not the cause of the observed MMTV repression (29). Rather, this study concluded that nonhistone proteins essential for basal transcription initiation steps were altered by HDI treatment (29).
A previous study discovered that HDIs were able to effectively inhibit c-Src expression in a wide variety of human cancer cell lines by directly repressing SRC transcription (24). The equal repression of both SRC promoters by HDIs suggested to us that a common mechanism might exist. In the present study we were able to demonstrate that the SRC1A and SRC1α core promoters were similar in that they were both Inr driven and TAF1 dependent in the tsBN462 system. Interestingly, we noted that the repressive effects of HDIs on the SRC1A promoter were completely blocked in tsBN462 cells grown at the restrictive temperature of 39°C. Although the activity of the SRC1A promoter was significantly repressed at 39°C, CAT reporter levels were still safely within a dynamic range. Therefore, we felt this finding was potentially significant because it was the only time we had noted a complete absence of SRC1A repression in any transfection experiments following HDI treatment. Interestingly, this TAF1 dependence could be conferred on the heterologous, TAF1-independent WAF1 promoter by SRC proximal or core promoter elements. Most remarkable was the observation that HDI-mediated repression was also conferred upon the WAF1 promoter by SRC proximal or core promoter elements.
It could still be argued, however, that the repression of the SRC promoters we have observed is the result of local changes in chromatin structure. We suggest, rather, that SRC repression is more likely direct and results from acetylation of nonhistone factors associated with the SRC promoters. For example, we utilized episomal templates for this study, which would be expected to possess different nucleosomal structures than the endogenous gene. Despite these likely differences in chromatin structure, we still consistently observed repression of these constructs, which matched the repression we have observed for the endogenous gene (24). In addition, we have shown by chromatin immunoprecipitation assays that both the endogenous SRC1A and SRC1α promoters were associated with acetylated histone H3 in untreated HT29 and SW480 cells and that this acetylation pattern did not change with TSA treatment (S. Dehm and K. Bonham, unpublished observations). Finally, in this study we created many SRC promoter deletion constructs which would certainly have altered nucleosomal structures compared with the wild-type constructs, but these deletions did not change the response of these reporters to HDIs. Therefore, these observations strongly suggest that SRC repression is independent of chromatin structure, a finding which parallels mechanistic studies of WAF1 induction (22) and MMTV repression (29) in response to HDIs.
A previous study has addressed the mechanism of gene repression by the HDI butyrate by using the cyclin D1 promoter as a model (25). This report identified an 11-bp butyrate response element that could confer weak butyrate-mediated repression when placed upstream of a thymidine kinase promoter. However, the repression mediated by the cyclin D1 butyrate response element was only twofold, and the basal activities of the promoter constructs in untreated cells were not included in this report (25). We have observed that the basal activities of reporter constructs in untreated cells could significantly affect the fold induction or repression mediated by HDIs. We chose, therefore, to take a qualitative approach and catalogue responses to HDIs as activated, repressed, or unaffected. A need for such caution is supported by the observation that the elements in the WAF1 promoter that are considered to be important for induction in response to HDIs are those that impair promoter activity most significantly when mutated and analyzed in transfection experiments (19, 30). A more recent study of the transcriptional inhibition of the Hmga2 gene following TSA treatment has suggested that repression results from a decrease in Sp1 and Sp3 binding to the proximal promoter (12). However, again, basal activities determined for the constructs used in the transfection experiments were not included in this study; rather, the conclusions in this report relied strongly on calculated changes in fold induction or fold repression (12).
In the present study, we could not identify a single HDI response element in either SRC promoter. This result therefore suggests that multiple elements or an overall architecture is important for the SRC promoters' response to these agents. Furthermore, our results suggest an interesting potential link between SRC promoter TAF1 dependence and SRC promoter repression by HDIs. The importance of this relationship was strengthened by studies with SRC:WAF1 and WAF1:SRC chimeras, which demonstrated that conferring TAF1 dependence on the WAF1 promoter also rendered it repressible in response to TSA treatment. Interestingly, other promoters share these features. The cyclin D1 and cyclin A promoters, much like the SRC1α and SRC1A promoters, are also Inr driven, significantly inhibited in tsBN462 cells following a shift to the restrictive temperature, and repressed in response to HDI treatment (20, 44, 45, 49). Conversely, promoters of genes that are activated in response to HDI treatment, such as WAF1, C-FOS, and CMV, contain TATA elements and are unaffected, or even slightly induced, in tsBN462 cells following a shift to the restrictive temperature (1, 37, 49). These findings suggest that there could be a more general, potentially functional, link between Inr-driven, TAF1-dependent promoters and HDI-mediated repression. Indeed, treatment of various cells with HDIs elicits a similar response of G1/S arrest and/or apoptosis as the shift of tsBN462 or ts13 cells from 33 to 39°C. Some fundamental questions therefore arise, such as what precisely determines TAF1 dependence and what determines HDI-mediated repression. These questions are especially intriguing considering the ability of SRC upstream activation sequences to convert the TATA-containing WAF1 core promoter into a TAF1-dependent, HDI-repressed promoter. Therefore, it will be of considerable future importance to dissect the complex architectural features of these promoters that dictate both TAF1 dependence and HDI-mediated repression. Currently, we are investigating these features, as well as the apparent generality of the link between TAF1 dependence and HDI-mediated repression.
While our results do not clearly establish whether TAF1 plays a direct or functional role in HDI-mediated repression, they do point to a key association between these two properties, which are common to both SRC promoters. Therefore, we can at the present time only speculate that TAF1 could serve some mechanistic role in repression mediated by HDIs. For example, TAF1 could acetylate an unidentified factor at either of the SRC promoters; this modification would, we propose, negatively influence SRC transcription. This acetylation would be balanced by one or more specific HDAC activities associated with the SRC promoters. Upon treatment with HDIs, the balance would shift towards acetylation of this putative factor, resulting in the SRC transcriptional repression we have observed in various cell lines. This model would also explain the apparent block of HDI-mediated SRC repression in tsBN462 cells grown at 39°C. Conversely, HDI treatment could result in prevention of TAF1 binding to the SRC core promoters or in TAF1 exclusion from the TFIID complex, thus accounting for SRC transcriptional repression. This model suggests that the apparent block of HDI-mediated SRC repression in tsBN462 cells grown at 39°C would be due to the fact that G690D TAF1 core promoter binding has already been abolished. With these models in mind, however, it is also quite possible that TAF1 could be indirectly involved in HDI-mediated SRC repression. Further experimental investigation is essential to test these models and accurately conclude if the association between TAF1 dependence and HDI-mediated repression is functional.
In summary, this is the first report describing a potential link between promoter TAF1 dependence and HDI-mediated transcriptional repression. If TAF1 plays a direct role, then substrates of TAF1 acetyltransferase activity that could mediate this repression have not yet been described. TAF1 has very weak activity towards histone proteins (52) but has been shown to acetylate TFIIEβ and the RAP74 subunit of TFIIF in vitro (21). The effect these modifications have on the function of these general transcription factors is not known. In order to clarify the role of TAF1 acetyltransferase activity in core promoter recognition and the repressive effects of HDIs, critical acetylated TAF1 substrates will have to be identified specifically in the context of HDI-repressed promoters.
Acknowledgments
We thank Bill Roesler for critical reading of the manuscript.
This work was supported by a Canadian Institutes of Health Research operating grant to K.B. and research project grant RPG-98-201-CCG from the American Cancer Society to E.W. S.D. was funded in part by a Natural Sciences and Engineering Research Council of Canada Ph.D. studentship. T.H. was funded in part by National Research Service Award GM7750 from NIGMS.
REFERENCE
- 1.Archer, S., S. Meng, J. Wu, J. Johnson, R. Tang, and R. Hodin. 1998. Butyrate inhibits colon carcinoma cell growth through two distinct pathways. Surgery 124:248-253. [PubMed] [Google Scholar]
- 2.Archer, S. Y., S. Meng, A. Shei, and R. A. Hodin. 1998. p21WAF1 is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc. Natl. Acad. Sci. USA 95:6791-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biscardi, J. S., D. A. Tice, and S. J. Parsons. 1999. c-Src, receptor tyrosine kinases, and human cancer. Adv. Cancer Res. 76:61-119. [DOI] [PubMed] [Google Scholar]
- 4.Bonham, K., and D. J. Fujita. 1993. Organization and analysis of the promoter region and 5′ noncoding exons of the human c-src proto-oncogene. Oncogene 8:1973-1981. [PubMed] [Google Scholar]
- 5.Bonham, K., S. A. Ritchie, S. M. Dehm, K. Snyder, and F. M. Boyd. 2000. An alternative, human SRC promoter and its regulation by hepatic nuclear factor-1α. J. Biol. Chem. 275:37604-37611. [DOI] [PubMed] [Google Scholar]
- 6.Chalkley, G. E., and C. P. Verrijzer. 1999. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 18:4835-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, B. S., and M. Hampsey. 2002. Transcription activation: unveiling the essential nature of TFIID. Curr. Biol. 12:R620-R622. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. L., and R. Tjian. 1996. Reconstitution of TATA-binding protein-associated factor/TATA-binding protein complexes for in vitro transcription. Methods Enzymol. 273:208-217. [DOI] [PubMed] [Google Scholar]
- 9.Dehm, S., M. A. Senger, and K. Bonham. 2001. SRC transcriptional activation in a subset of human colon cancer cell lines. FEBS Lett. 487:367-371. [DOI] [PubMed] [Google Scholar]
- 10.Dunphy, E. L., T. Johnson, S. S. Auerbach, and E. H. Wang. 2000. Requirement for TAFII250 acetyltransferase activity in cell cycle progression. Mol. Cell. Biol. 20:1134-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson, M., P. A. Henry, and R. A. Currie. 2003. Histone deacetylase inhibition is associated with transcriptional repression of the Hmga2 gene. Nucleic Acids Res. 31:3123-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall, C. V., P. E. Jacob, G. M. Ringold, and F. Lee. 1983. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J. Mol. Appl. Genet. 2:101-109. [PubMed] [Google Scholar]
- 14.Hayashida, T., T. Sekiguchi, E. Noguchi, H. Sunamoto, T. Ohba, and T. Nishimoto. 1994. The CCG1/TAFII250 gene is mutated in thermosensitive G1 mutants of the BHK21 cell line derived from golden hamster. Gene 141:267-270. [DOI] [PubMed] [Google Scholar]
- 15.Hebbes, T. R., A. W. Thorne, and C. Crane-Robinson. 1988. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heruth, D. P., G. W. Zirnstein, J. F. Bradley, and P. G. Rothberg. 1993. Sodium butyrate causes an increase in the block to transcriptional elongation in the c-myc gene in SW837 rectal carcinoma cells. J. Biol. Chem. 268:20466-20472. [PubMed] [Google Scholar]
- 17.Hilton, T. L., and E. H. Wang. 2003. Transcription factor IID recruitment and Sp1 activation. Dual function of TAF1 in cyclin D1 transcription. J. Biol. Chem. 278:12992-13002. [DOI] [PubMed] [Google Scholar]
- 18.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 19.Huang, L., Y. Sowa, T. Sakai, and A. B. Pardee. 2000. Activation of the p21WAF1/CIP1 promoter independent of p53 by the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) through the Sp1 sites. Oncogene 19:5712-5719. [DOI] [PubMed] [Google Scholar]
- 20.Iacomino, G., M. F. Tecce, C. Grimaldi, M. Tosto, and G. L. Russo. 2001. Transcriptional response of a human colon adenocarcinoma cell line to sodium butyrate. Biochem. Biophys. Res. Commun. 285:1280-1289. [DOI] [PubMed] [Google Scholar]
- 21.Imhof, A., X. J. Yang, V. V. Ogryzko, Y. Nakatani, A. P. Wolffe, and H. Ge. 1997. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 7:689-692. [DOI] [PubMed] [Google Scholar]
- 22.Kim, Y. K., J. W. Han, Y. N. Woo, J. K. Chun, J. Y. Yoo, E. J. Cho, S. Hong, H. Y. Lee, Y. W. Lee, and H. W. Lee. 2003. Expression of p21(WAF1/Cip1) through Sp1 sites by histone deacetylase inhibitor apicidin requires PI 3-kinase-PKC epsilon signaling pathway. Oncogene 22:6023-6031. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 24.Kostyniuk, C. L., S. M. Dehm, D. Batten, and K. Bonham. 2002. The ubiquitous and tissue specific promoters of the human SRC gene are repressed by inhibitors of histone deacetylases. Oncogene 21:6340-6347. [DOI] [PubMed] [Google Scholar]
- 25.Lallemand, F., D. Courilleau, M. Sabbah, G. Redeuilh, and J. Mester. 1996. Direct inhibition of the expression of cyclin D1 gene by sodium butyrate. Biochem. Biophys. Res. Commun. 229:163-169. [DOI] [PubMed] [Google Scholar]
- 26.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 27.Mariadason, J. M., G. A. Corner, and L. H. Augenlicht. 2000. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 60:4561-4572. [PubMed] [Google Scholar]
- 28.Marks, P. A., V. M. Richon, and R. A. Rifkind. 2000. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl. Cancer Inst. 92:1210-1216. [DOI] [PubMed] [Google Scholar]
- 29.Mulholland, N. M., E. Soeth, and C. L. Smith. 2003. Inhibition of MMTV transcription by HDAC inhibitors occurs independent of changes in chromatin remodeling and increased histone acetylation. Oncogene 22:4807-4818. [DOI] [PubMed] [Google Scholar]
- 30.Nakano, K., T. Mizuno, Y. Sowa, T. Orita, T. Yoshino, Y. Okuyama, T. Fujita, N. Ohtani-Fujita, Y. Matsukawa, T. Tokino, H. Yamagishi, T. Oka, H. Nomura, and T. Sakai. 1997. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 272:22199-22206. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien, T., and R. Tjian. 2000. Different functional domains of TAFII250 modulate expression of distinct subsets of mammalian genes. Proc. Natl. Acad. Sci. USA 97:2456-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 33.Pazin, M. J., and J. T. Kadonaga. 1997. What's up and down with histone deacetylation and transcription? Cell 89:325-328. [DOI] [PubMed] [Google Scholar]
- 34.Ritchie, S., F. M. Boyd, J. Wong, and K. Bonham. 2000. Transcription of the human c-Src promoter is dependent on Sp1, a novel pyrimidine binding factor SPy, and can be inhibited by triplex-forming oligonucleotides. J. Biol. Chem. 275:847-854. [DOI] [PubMed] [Google Scholar]
- 35.Ritchie, S. A., M. K. Pasha, D. J. Batten, R. K. Sharma, D. J. Olson, A. R. Ross, and K. Bonham. 2003. Identification of the SRC pyrimidine-binding protein (SPy) as hnRNP K: implications in the regulation of SRC1A transcription. Nucleic Acids Res. 31:1502-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 37.Rushton, J. J., R. A. Steinman, and P. D. Robbins. 1997. Differential regulation of transcription of p21 and cyclin D1 conferred by TAF(II)250. Cell Growth Differ. 8:1099-1104. [PubMed] [Google Scholar]
- 38.Sekiguchi, T., E. Noguchi, T. Hayashida, T. Nakashima, H. Toyoshima, T. Nishimoto, and T. Hunter. 1996. D-type cyclin expression is decreased and p21 and p27 CDK inhibitor expression is increased when tsBN462 CCG1/TAFII250 mutant cells arrest in G1 at the restrictive temperature. Genes Cells 1:687-705. [DOI] [PubMed] [Google Scholar]
- 39.Smale, S. T. 2001. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 15:2503-2508. [DOI] [PubMed] [Google Scholar]
- 40.Smale, S. T., A. Jain, J. Kaufmann, K. H. Emami, K. Lo, and I. P. Garraway. 1998. The initiator element: a paradigm for core promoter heterogeneity within metazoan protein-coding genes. Cold Spring Harbor Symp. Quant. Biol. 63:21-31. [DOI] [PubMed] [Google Scholar]
- 41.Souleimani, A., and C. Asselin. 1993. Regulation of c-myc expression by sodium butyrate in the colon carcinoma cell line Caco-2. FEBS Lett. 326:45-50. [DOI] [PubMed] [Google Scholar]
- 42.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Streeper, R. S., C. A. Svitek, J. K. Goldman, and R. M. O'Brien. 2000. Differential role of hepatocyte nuclear factor-1 in the regulation of glucose-6-phosphatase catalytic subunit gene transcription by cAMP in liver- and kidney-derived cell lines. J. Biol. Chem. 275:12108-12118. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, T., H. Yokozaki, H. Kuniyasu, K. Hayashi, K. Naka, S. Ono, T. Ishikawa, E. Tahara, and W. Yasui. 2000. Effect of trichostatin A on cell growth and expression of cell cycle- and apoptosis-related molecules in human gastric and oral carcinoma cell lines. Int. J. Cancer 88:992-997. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki-Yagawa, Y., M. Guermah, and R. G. Roeder. 1997. The ts13 mutation in the TAFII250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol. Cell. Biol. 17:3284-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Lint, C., S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 47.Vigushin, D. M., and R. C. Coombes. 2002. Histone deacetylase inhibitors in cancer treatment. Anticancer Drugs 13:1-13. [DOI] [PubMed] [Google Scholar]
- 48.Wang, E. H., and R. Tjian. 1994. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science 263:811-814. [DOI] [PubMed] [Google Scholar]
- 49.Wang, E. H., S. Zou, and R. Tjian. 1997. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 11:2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warrell, R. P., Jr., L. Z. He, V. Richon, E. Calleja, and P. P. Pandolfi. 1998. Therapeutic targeting of transcription in acute promyelocytic leukemia by use of an inhibitor of histone deacetylase. J. Natl. Cancer Inst. 90:1621-1625. [DOI] [PubMed] [Google Scholar]
- 51.Wassarman, D. A., N. Aoyagi, L. A. Pile, and E. M. Schlag. 2000. TAF250 is required for multiple developmental events in Drosophila. Proc. Natl. Acad. Sci. USA 97:1154-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wassarman, D. A., and F. Sauer. 2001. TAF(II)250: a transcription toolbox. J. Cell Sci. 114:2895-2902. [DOI] [PubMed] [Google Scholar]
- 53.Woychik, N. A., and M. Hampsey. 2002. The RNA polymerase II machinery: structure illuminates function. Cell 108:453-463. [DOI] [PubMed] [Google Scholar]