Abstract
By using selected 2H and 15N labels, we have examined the influence of a central proline residue upon the properties of a defined peptide that spans lipid bilayer membranes by solid-state NMR spectroscopy. For this purpose, GWALP23 (acetyl-GGALW5LALALALALALALW19LAGA-ethanolamide) is a suitable model peptide that employs—for the purpose of interfacial anchoring—only one tryptophan residue on either end of a central alpha-helical core sequence. Because of its systematic behavior in lipid bilayer membranes of differing thickness (see J. Biol. Chem. 285, 31723), we utilize GWALP23 as a well-characterized framework for introducing guest residues within a transmembrane sequence; for example, a central proline yields acetyl-GGALW5LALALAP12ALALALW19LAGA-ethanolamide. We synthesized the GWALP23-P12 with specifically placed 2H and 15N labels for solid-state NMR spectroscopy, and examined the peptide orientation and segmental tilt in oriented DMPC lipid bilayer membranes using combined (2H)-GALA and (15N-1H) high resolution separated local field methods. In DMPC bilayer membranes, the peptide segments N-terminal and C-terminal to the proline are both tilted substantially with respect to the bilayer normal, by about 34° and 29° (± 5°), respectively. While the tilt increases for both segments when proline is present, the range and extent of the individual segment motions are comparable or less than those of the entire GWALP23 peptide in bilayer membranes. In DMPC, the proline induces a kink of about 30° (± 5°), with an apparent helix unwinding or “swivel” angle of about 70°. In DLPC and DOPC, based on 2H NMR data only, the kink angle and swivel angle probability distributions overlap those of DMPC, yet the most probable kink angle appears somewhat smaller than in DMPC. As has been described for GWALP23 itself, the C-terminal helix ends before Ala-21 in the phospholipids DMPC and DLPC, yet remains intact through Ala-21 in DOPC. The dynamics of bilayer-incorporated, membrane-spanning GWALP23 and GWALP23-P12 are less extensive than observed for WALP-family peptides that have more than two interfacial Trp residues.
Keywords: Lipid bilayer, GWALP23 peptide geometry and dynamics, SAMPI4, PISEMA analysis, 15N solid-state NMR, GALA analysis, deuterium solid-state NMR, WALP peptide
The functions of membrane proteins are sometimes reliant on the presence and positioning of specific proline residues (1–5), which may introduce a kink within a transmembrane helical sequence. To examine the consequences of proline on structure and dynamics, it is of interest to study the effects of a proline residue within model transmembrane peptide sequences. Significant unresolved questions pertain not only to an overall kink angle, but also to the peptide dynamics and the orientations of individual segments on either side of the proline with respect to a lipid bilayer normal. Some of these issues have been addressed using a system in which two Trp anchors flank a central transmembrane helical core sequence, WALP19, acetyl-GWW(LA)6LWWA-ethanolamide (6). Nevertheless, some of the system properties are difficult to interpret when too many interfacial tryptophans are present. Multiple Trp anchors tend to complicate the behavior of transmembrane peptides, for example, by inducing significant peptide dynamics and variations in the helix tilt direction (7). Remarkably, the extent of dynamic averaging of NMR resonances is reduced when only two Trps—or one Trp and one Tyr—are present (8). Therefore, with only single Trp anchors near each membrane interface, the transmembrane peptide behavior has been found to be more predictable and systematic with respect to the helix tilt magnitude, direction and lipid bilayer thickness (7–10).
Here we characterize a central proline residue within the context of a transmembrane peptide having only one Trp anchor near each end, using lipid bilayers of varying thicknesses. For this purpose we incorporated proline in GWALP23 (9), resulting in GWALP23-P12 (acetyl-GGALWLALALAP12ALALALWLAGA-ethanolamide) (Figure 1). GWALP23 itself exhibits a well-defined, tilted transmembrane orientation that scales with the lipid bilayer thickness (7, 8). To study the effects of proline incorporation on the behavior of GWALP23, we incorporated 2H-Ala or 15N-labeled Leu and Ala residues at specific positions within the sequence, on both sides of the proline.
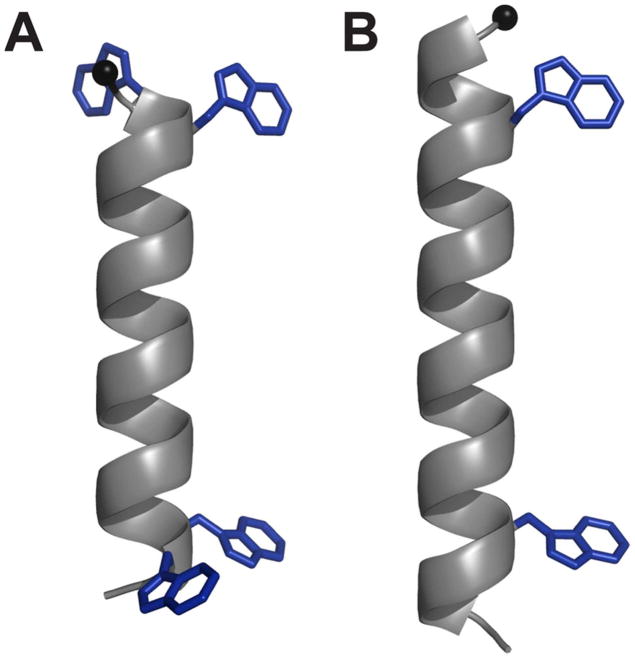
Figure 1.
Representative models of (A) WALP19 with 4 Trp residues, and (B) GWALP23 with only 2 Trp residues. The core helical sequence between the inner tryptophans is identical in the two peptides. The Trp indole side chains are shown in arbitrary orientations. (See also Table 1.)
When considering the helix breaking properties of proline, the N-terminal domain is both expected and observed to be more sensitive (6), because a hydrogen bond is missing between the proline residue and the amino acid four residues earlier in the sequence. The distortion introduced into the transmembrane helix by the proline, which effectively divides the continuous α-helix into two sections, is of interest to characterize, in terms of not only the helix kink angle but also the individual tilt magnitudes and directions for the N- and C-terminal domains. If sufficient data can be obtained, aspects of the segmental dynamics also can be characterized. For this reason, deuterium quadrupolar splitting values from 2H-Ala residues (11) are used together with 15N-1H dipolar coupling and 15N chemical shift values from 15N-Ala and 15N-Leu labeled residues (9, 12, 13), enabling a combined 2H and 15N solid-state NMR analysis of the properties of the GWALP23-P12 helical segments in DMPC.
The combined use of the 15N and 2H based solid-state NMR techniques, analyzed together, provides a robust determination of the peptide properties. Advantages are offered by both the complementary nature of these NMR techniques and the significant number of data points available for each peptide segment. Considering that GWALP23 has a relatively long alpha-helical domain with only one anchoring Trp residue near each end, this peptide is a favorable candidate for such studies. The insertion of Pro at position 12 in the transmembrane core of GWALP23 is expected to preserve sufficiently long α-helical segments for determination of the orientations of the segments on both sides of the proline.
Previous work from our lab revealed that the proline residue in WALP19-P10 interrupts the helix and results in two segments with distinct orientations and a kink angle of about 20° in DOPC bilayers (6). Uncertainties remain, nevertheless, concerning the respective N- and C-terminal segment orientations and dynamics. The presence of only single Trp anchors and the availability of 15N data have enabled us to obtain additional information about GWALP23-P12.
Materials and methods
Fmoc-amino acids and resin were obtained from Novabiochem (San Diego, CA) or Advanced Chemtech (Louisville, KY). Isotope enriched 15N Fmoc-L-amino acids were from Cambridge Isotope Laboratiories (Andover, MA) and Sigma-Aldrich (St. Louis, MO). Deuterium labeled alanine and 2H-depleted water were from Cambridge. This commercial L-alanine-d4 was Fmoc-protected using Fmoc-ON-succinimide (Novabiochem), as described (14). Ethanolamine and trifluoroethanol (TFE) were from Sigma-Aldrich. All lipids (DH-o-PC, DLPC, DMPC, DM-o-PC and DOPC) were acquired from Avanti polar lipids (Alabaster, AL). Other solvents were the highest grade available from EMD (Gibbstown, NJ).
Peptide synthesis was performed on an Applied Biosystems 433A peptide synthesizer (Life Technologies, Foster City, CA), using modified Fmoc chemistry with extended coupling times and additional deprotection steps. L-Ala-d4 or 15N-amino acids were incorporated at specific residues during peptide synthesis. Using a dual labeling strategy, deuterated alanine was incorporated at 100% abundance and at lower 50% abundance in two different sequence positions, in the same peptide (15). During peptide synthesis, a five-fold excess of Fmoc-amino acid was used for unlabeled residues and for 15N-Leu and 15N-Ala. Two-fold excess Fmoc-L-Ala-d4 followed by a second coupling with five-fold excess unlabeled Fmoc-L-Ala was used for deuterated alanines (16). Protecting groups, N-acetyl and C-ethanolamide, served to mask the charges at the ends of the peptides. The C-ethanolamide group was introduced during cleavage of a peptide from Wang resin, using 20% ethanolamine in dichloromethane, for 48 hr under constant agitation at 22 °C (17). The reaction was quenched using deionized water, upon which the peptide precipitated. Precipitated peptide was centrifuged to form a pellet, which was lyophilized twice from 1 ml of acetonitrile/water (1:1, v/v). The purity of peptides was evaluated via reversed-phase HPLC using a 4.6 × 50 mm Zorbax SB-C8 column packed with 3.5 μm octyl-silica (Agilent Technologies, Santa Clara, CA), operated at 1 mL/min using a methanol/water gradient from 85% to 99% methanol over five min. The peptide identity was confirmed using MALDI mass spectrometry.
Samples for NMR spectroscopy were prepared using peptides denoted in Table 1. Mechanically oriented samples for deuterium NMR spectroscopy were prepared with a 1:40 peptide:lipid ratio using DLPC, DMPC or DOPC. Magnetically oriented samples for high resolution separated local field (SLF) experiments (SAMPI4) (18) were prepared using a ratio of 1:80 peptide to long-chain lipid, DMPC for deuterium studies or DM-o-PC for 15N based studies, in addition to short-chain lipid DH-o-PC present to enable bicelle formation. The ether-linked lipids DM-o-PC and DH-o-PC, chosen for some experiments based on their superior chemical stability, show similar behavior to the corresponding ester lipids DMPC and dihexanoyl-PC (19, 20).
Table 1.
Sequences of peptidesa
| WALP19-P10 | acetyl-GWWLALALAPALALALWWA-ethanolamide |
| GWALP23-P12 | acetyl-GGALWLALALAPALALALWLAGA-ethanolamide |
| A16GWALP23-P12 | acetyl-GGALWLALALAPALAAALWLAGA-ethanolamide |
WALP19-P10 is included for reference. Labels were incorporated for 2H and 15N NMR experiments. Deuterium labels were introduced in two alanines per GWALP23-P12 peptide, using partial (p) and full (f) deuteration at the following positions: 7f and 9p, 13p and 15f, 17f and 21p; and 15p and 16f (in the mutant A16GWALP23-P12 sequence). Two GWALP23-P12 peptides were synthesized with uniform 15N enrichment, covering residues 13–17, or residues 6–11 together with 13–18.
[NB: please use a fixed-width font for the sequences in Table 1, and align them with respect to the central “P” as shown.]
For preparation of mechanically oriented samples, a peptide-lipid mixture was made from 2 μmol peptide in a TFE stock solution, and 80 μmol lipid in chloroform. Solvents were evaporated under a stream of N2, followed by drying under vacuum (10−3 torr) for at least 24 hr. The peptide-lipid mixture was dissolved in 95% methanol, 5% water, and was distributed evenly over ~40 glass slides (4.8 × 23 × 0.07 mm; Marienfeld, Lauda-Königshofen, Germany). The slides with the mixture were placed under vacuum for at least 48 hr, after which the peptide-lipid films were hydrated using deuterium depleted water to a hydration level of 45% (w/w). The hydrated slides were stacked under gentle pressure and then sealed in a glass cuvette. To allow for formation and alignment of lipid bilayers, the samples were incubated for at least 48 hr at 42 °C. To verify the orientation of mechanically aligned bilayers, solid-state 31P NMR spectra (1H decoupled) were recorded at 50 °C using a Bruker Avance 300 Spectrometer (Bruker Instruments, Billerica, MA).
Bicelle samples, which contain lipid bilayer discs that orient in a magnetic field, were made using 0.76 μmole 2H- or 15N-labeled peptide, 61 μmole DM-o-PC (or DMPC) and 19 μmole DH-o-PC, giving a value of 3.2 for the q ratio of long to short lipid. Peptide in TFE and DM-o-PC in chloroform were mixed together in appropriate amounts. Solvents were evaporated under N2 (g) flow and samples subsequently were dried under vacuum (10−3 torr) for at least 36 hr. The DMPC-peptide films were hydrated using 125 μl deuterium-depleted water at 45 °C, resulting in formation of multilamellar vesicles, which were further mixed with DH-o-PC suspended in 50 μl deuterium depleted water. To promote bicelle formation, a series of freeze/thaw steps were performed until a clear solution with low viscosity at low temperature (~4 °C) was obtained. (An increase of temperature results in an increased sample viscosity up to the point of ~45 °C, upon which the viscosity lowers again.) Samples were transferred to 5 mm glass vials (New Era Enterprises, Vineland, NJ) and sealed in preparation for NMR experiments.
Deuterium NMR spectra were recorded at 46 MHz using a Bruker Avance 300 Spectrometer, employing a quadrupolar echo sequence with full phase cycling (21), a 4.5 μs 90° pulse time, 90 ms interpulse delay and 125 μs echo delay. The sample temperature was 50 °C for glass slides and 42 °C for bicelles. The 2H spectra were processed using a line broadening of 200 Hz. Deuterium quadrupolar splitting values were assigned to individual labeled alanine methyl groups based on the distances between corresponding peak maxima and the relative peak intensities associated with the respective isotopic abundances used for particular residues in the peptide sequence.
For 15N-enriched peptides, 15N chemical shifts and 15N-1H dipolar coupling values were recorded using 500 MHz Bruker Avance and Varian Inova spectrometers and established pulse sequences (13, 18, 22–24). Solid-state NMR high resolution separated local field SAMPI4 experiments (which are complementary to PISEMA) (18) were performed using a 1 ms CP contact time, RF field strengths of approximately 50 kHz; 54 t1 points were acquired using 8.0 ms of acquisition time in the direct (t2) dimension and a 7.5 s recycle delay. The sample temperature was 42 °C.
Combinations of 2H quadrupolar splitting magnitudes, along with 15N chemical shift and 15N-1H dipolar coupling values, were used to calculate the orientations of the different peptide segments in DMPC. We performed these calculations both with a semi-static model and with a more dynamic model that incorporates Gaussian distributions for the tilt and direction of tilt (25).
The analysis using semi-static peptide dynamics involves a principal order parameter Szz to estimate overall peptide motion with respect to an apparent average peptide orientation. These calculations are based on the GALA analysis, equations [1] and [3], as previously described (11, 25, 26). We incorporate also 15N-1H dipolar coupling and 15N chemical shift values obtained from SAMPI4 spectra, using equations [2], [4] and [5], to determine a best fit to the experimental data (27). These calculations are performed using τ, ρ and Szz as variable parameters to fit the data for the isotope-labeled residues based on ideal α-helix geometry.
| [Equation 1] |
| [Equation 2] |
| [Equation 3] |
| [Equation 4] |
The general equation for the chemical shift is:
| [Equation 5] |
In Equation 5, the σii are the chemical shift tensor components and Hii the corresponding projections of the applied magnetic field. The chemical shift tensor components (σ11, σ22, σ33) were (64, 77, 224) ppm, as reported earlier for model dipeptides (28) and small proteins (29). Chemical shift values were converted from ppm to Hz, and a principal order parameter Szz was applied as described (30). The dependence of 15N chemical shift on θ has been described (12, 13, 30).
The quadrupolar splitting, equation [1], depends upon the static coupling constant QCC, which is defined in equation [3] and has a value ~1/3 of 168 kHz, namely 56 kHz, for C-D3 groups (6, 11). Other parameters include the orientation angle θ for the alanine Cα-Cβ bond with respect to the bilayer normal, and the macroscopic sample orientation angle β for the bilayer normal with respect to the external magnetic field. The peptide geometry is reflected in angle ε// between peptide helix axis and the Cα-Cβ bond vector for deuterium labeled alanine residues (11). Angle θ depends on the peptide geometry (ε//) and the orientation of the peptide’s helix axis (τ and ρ). While earlier analysis (11) involved searching for appropriate values of ε//, current procedure fixes ε// at a known appropriate value of 59.4° and varies Szz to estimate the dynamics and obtain a best fit.
The dipolar coupling values are dependent on a coupling constant of 10.22 kHz (assuming the NH bond length of 1.06 Å in equation 4) (31, 32), and the angle θ which in turn is dependent on the τ, ρ, and ε// values. For an α-helical peptide, defined by angles (Φ, Ψ, Ω) = (−65, −40, 180) (33), the ε// angle is 14° between the peptide helix axis and the N-H bond vector for 15N labeled residues. In the combined analysis, the 2H methyl quadrupolar couplings, 15N-1H dipolar couplings and 15N chemical shifts were given equal weights; to account for peak dispersion and poor signal-to-noise, a relative weighting factor of 0.5 was applied to the observed and calculated backbone Cα-2H quadrupolar couplings.
Using these equations and parameters, probable peptide segment orientations were calculated by considering the best fit (lowest RMSD between calculated values and NMR observables) for a range of peptide orientations. Helix rotation ρ is referenced in relation to Cα of the first amino acid (Gly1) of the peptide (11). To calculate a kink angle (κ) between two α-helical segments, one considers both the tilt (τ) and rotation (ρ) differences between the segments N- and C-terminal to the proline; equation 6 (15).
| [Equation 6] |
We have also performed calculations that consider more complex peptide dynamics, in which στ and σρ describe the widths of Gaussian distributions for the peptide tilt and rotation (25). In this analysis, a principal order parameter Szz is set to 0.88 to estimate isotropic fluctuations (25), and further anisotropic variations in τ and ρ are then described by fitting στ and σρ (25). A best-fit RMSD to observed dipolar and quadrupolar couplings is based on the parameters τ, ρ, στ and σρ, following ref. (25). Whereas the semi-static analysis employs three parameters, the Gaussian analysis involves a four-parameter fit.
Results
The synthetic GWALP23-P12 peptides gave the predicted molecular mass and isotope enrichment values (Fig S1 of the Supporting Information), and were at least 95% pure by reversed-phase HPLC (Fig S2 of the Supporting Information).
Peptides were incorporated into mechanically or magnetically aligned lipid bilayers. Oriented glass-plate samples were prepared using peptides containing specific deuterated alanine residues. To confirm mechanical alignment of the bilayers, 31P NMR spectra were recorded (Fig S3 of the Supporting Information). From the 2H NMR spectra for samples aligned with β = 90° (Figures 2–3), or with β = 0° (Figures S4–S5 of the Supporting Information), unique quadrupolar splitting magnitudes were observed for each labeled alanine in the sequence. Resonances were assigned to the corresponding alanine residues based on relative signal intensities, bearing in mind the isotopic abundance present for each of the labeled residues (Table 1). The observed quadrupolar splitting magnitudes (reported in Table 2 for the β = 0° orientation) were reduced by a factor of two, as expected, when a sample was turned from β = 0° to β = 90°.
Figure 2.
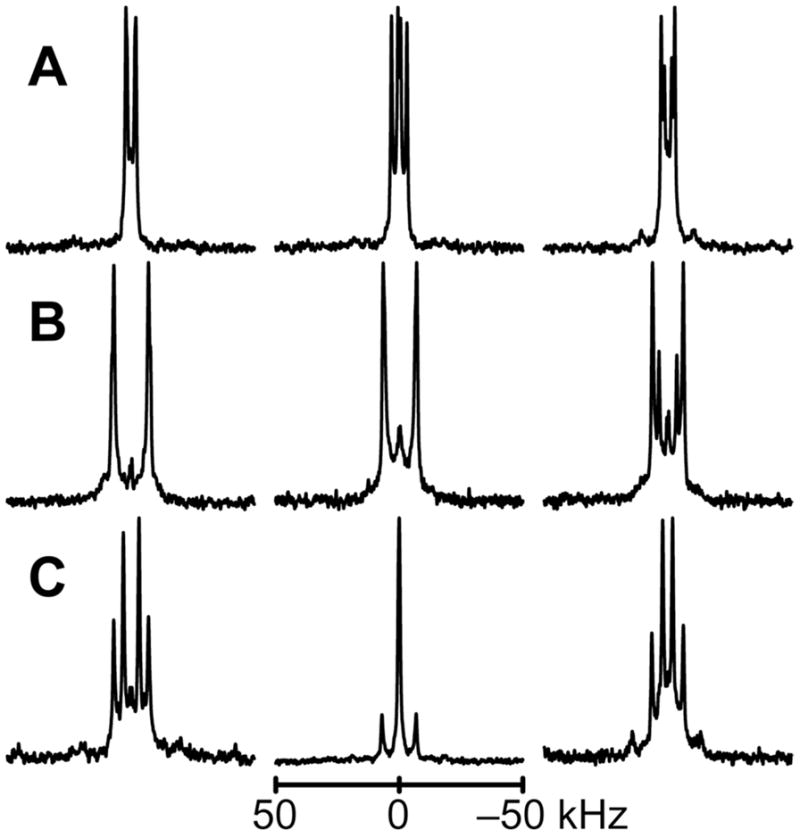
2H NMR spectra for labeled Ala methyl groups C-terminal to the proline in GWALP23-P12, in oriented bilayers of DLPC, DMPC and DOPC. Sample orientation is β = 90°. Deuterium isotope levels in the labeled alanines are: A. 100% Ala17, 50% Ala21. B. 100% Ala15, 50% Ala13. C. 100% Ala16, 50% Ala15.
Figure 3.
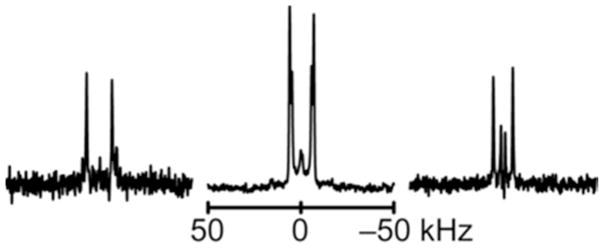
2H NMR spectra for labeled Ala methyl groups N-terminal to the proline in GWALP23-P12, in oriented bilayers of (left to right) DLPC, DMPC and DOPC. Sample orientation is β = 90°. The deuterium isotope abundance is 100% in Ala7 and 50% in Ala9.
Table 2.
Observed 2H quadrupolar splitting magnitudes (kHz) for labeled alanines in GWALP23-P12a
| Ala | DLPC | DMPC | DOPC |
|---|---|---|---|
| 7 | 27.6 | 26.8 | 21.2 |
| 9 | 36.9 | 20.8 | 4.4 |
| 13 | 28.5 | 25.4 | 14.3 |
| 15 | 28.5 | 27.6 | 25.0 |
| 15b | 28.0 | 28.0 | 25.9 |
| 16 | 12.3 | 0.5c | 8.1 |
| 17 | 8.2 | 12.2 | 6.0 |
| 21 | 7.2c | 2.2c | 10.2 |
| 9d | 66d | ||
| 21d | 89d | 70d | 53d |
Values in kHz for the β = 0° sample orientation. Values represent the CD3 side chain except where noted.
Values for A15 when A16 replaces L16.
Value not used for tilt analysis, due to helix fraying.
Backbone Cα deuteron.
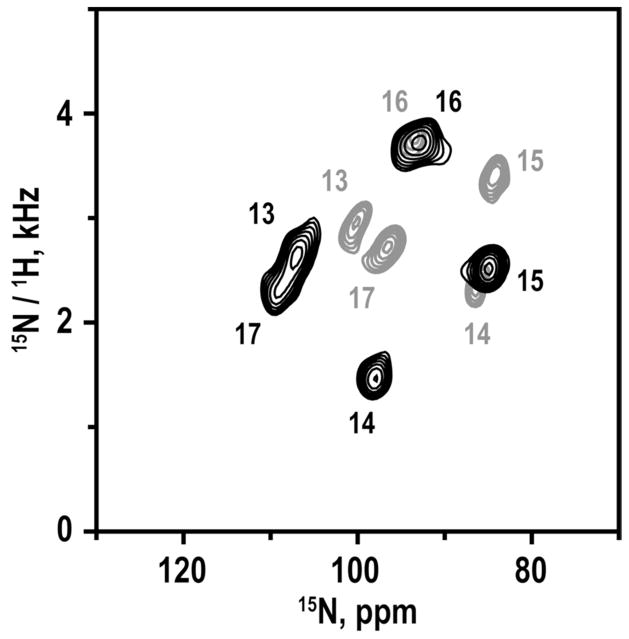
Bicelle samples for magnetic alignment were prepared using 15N-labeled Leu and Ala in positions 13–17 (five residues) C-terminal to the proline, or in positions 5–11 and 13–18 (six residues on either side of the proline). The SAMPI4 spectrum for residues 13–17 was compared to that for the same residues in GWALP23. These spectra, each showing only five signals, can be assigned in unique fashion (Figure 4) for a right-handed α-helix, because the signals from residues 13 and 17 must be near each other on a helical wheel, and the helix sense is known. The spectral overlay (Figure 4) also reveals a larger spread of the five assigned peaks when proline-12 is present. For example, the 1H- 15N dipolar couplings for residues 13–17 range from 1.3–3.5 kHz when P12 is present (Table 3) but only from 2.4–3.8 kHz for GWALP23 without the proline (30). The shift of the centroid of the pattern of five peaks (Figure 4) suggests a somewhat different tilt for the C-terminal helical segment of GWALP23 when P12 is present. The expanded size (“diameter”) of the pattern may indicate somewhat reduced motional averaging of the NMR observables when P12 is present. Further analysis of the segmental orientation and dynamics is presented below.
Figure 4.
Overlay of SAMPI4 spectra for 15N-labeled residues 13–17 in GWALP23 (gray) and GWALP23-P12 (black), with the peak assignments indicated.
Table 3.
Dipolar couplings and 15N chemical shift values for 15N-1H groups in GWALP23-P12a
| Residue | 15N-1H dipolar coupling | 15N chemical shift |
|---|---|---|
| 6 | 0.7 kHz | 111.0 ppm |
| 7 | 1.5 | 89.8 |
| 8 | 2.8 | 95.6 |
| 9 | 1.5 | 118.5 |
| 10 | 0.3 | 103.0 |
| 11b | 1.2b | 96.2b |
| 13 | 2.4 | 107.5 |
| 14 | 1.3 | 98.5 |
| 15 | 2.3 | 85.5 |
| 16 | 3.5 | 94.0 |
| 17 | 2.1 | 109.3 |
| 18 | 1.7 | 88.5 |
Values are listed for bicelle samples, corresponding to a β = 90° sample orientation.
Values for residue 11 were excluded from the analysis of the N-terminal segment because the helix is broken at this point.
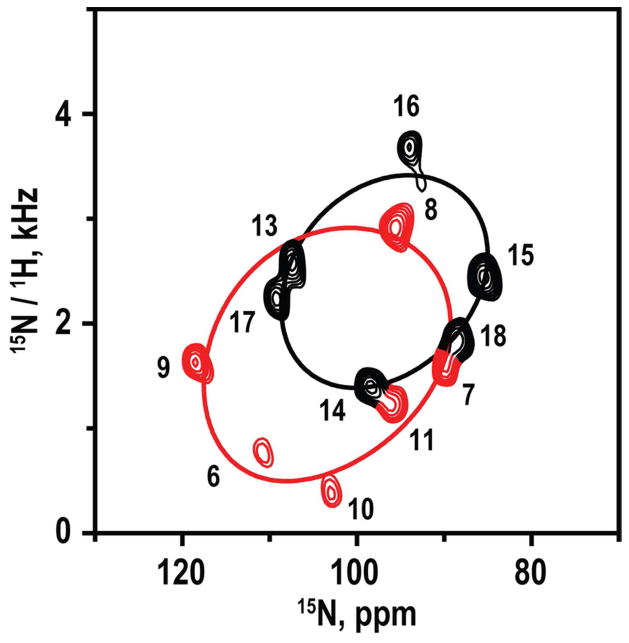
The full SAMPI4 spectrum (Figure 5) shows a striking pattern that indicates directly the presence of two helical wheels with different tilt angles in the DMPC/DHPC bicelles. With the peaks for residues 13–17 assigned, the remaining peaks for residue 18 and residues 6–11 were assigned by considering a difference spectrum, the right-handed α-helix geometry, the absence of a signal for proline-12, and a distortion from helical geometry at residue 11, adjacent to the proline (Figure 5). The assigned resonances on the PISA wheels yield directions of tilt (see below), for the segments C- and N-terminal to proline, that are consistent with the tilt directions deduced independently from GALA analysis of deuterated alanines alone. Dipolar coupling and 15N chemical shift values were recorded (Table 3) for the labeled residues based upon the assigned PISA wheel peaks. (As noted in Table 3, the values for residue 11 were excluded from the analysis because the helix is broken at that location. Helix distortion also has been observed for the residue preceding Pro in WALP19-P10 (6).)
Figure 5.
SAMPI4 spectrum for 15N-labeled residues 6–11 and 13–18 GWALP23-P12 in magnetically oriented DM-o-PC/DH-o-PC bicelle bilayers. The spectrum was recorded from a single sample. Peaks were assigned by comparison with the spectrum for residue 13–17 (Figure 4) and are colored red for residues N-terminal to the proline. Helical (“PISA”) wheel patterns are superimposed to represent the orientations of the N-terminal and C-terminal segments (Table 4).
When not only 2H quadrupolar coupling magnitudes but also 15N chemical shifts and 15N-1H dipolar coupling values are available—namely for the case of DMPC bilayers (in combination with DM-o-PC/DH-o-PC bicelle samples)—the segment orientations can be determined using a combined analysis involving all available data (Table 4). For the respective temperatures of ~50 °C for bilayers and ~42 °C for bicelles, it has been determined that the peptide principal order parameter is similar between the two systems (30). The analysis is based on calculation models in which the theoretical quadrupolar and dipolar waves, together with the distribution of 15N chemical shifts, most closely approximate the values found experimentally, as determined by a root mean squared deviation. We determined average apparent peptide segment orientations in DMPC using both semi-static and Gaussian dynamic approximations (see Table 4). The results indicate a tilt of the N-terminal segment of about 30° for a semi-static model, or a closely similar value of 34° for a Gaussian analysis, with a corresponding στ of only 19°. The tilt angle observed for the C-terminal segment is 22° for a semi-static model, or about 29° with a στ of notably 0° for a converged Gaussian dynamic analysis. The direction of tilt ρo for the N-terminal segment is about 263° for either dynamic model (relative to Gly1 Cα), while the semi-static and Gaussian analyses also agree for the direction of C-terminal segment tilt and yield a value of about 330° (Table 4). The GWALP23 helix itself exhibits ρo of about 312° (when proline is absent), so the respective segmental tilt changes direction are about −50° and +20°, giving a proline-induced difference in the direction of segment tilt |Δρo| of about 70°. The Gaussian analysis indicates substantial σρ values of about 34° for the N-terminal segment and about 48° for the C-terminal segment. We note that the larger σρ value for the C-terminal together with the larger στ value for the N-terminal may suggest the presence of some coupled dynamics (see Discussion). The distributions of results are evident in the calculated dipolar wave plots (Figure 6) and in RMSD plots for (τo, ρo) and (στ, σρ) (Figures 7–8). Once again, the observed segmental tilt parameters should be compared to the reported (τo, ρo) of (9°, 312°) for GWALP23 in DMPC, in the absence of the proline (7). The central proline increases the magnitude of tilt for both segments, with respect to the DMPC bilayer normal, while exerting opposite effects on the N- and C-terminal segment tilt directions.
Table 4.
Calculated orientations in DMPC of segments N- and C-terminal to proline in GWALP23-P12, using Gaussian dynamics and semi-static calculation modelsa
| Segment | Model | τ0 | στ | ρ0 | σρ | Szz | RMSD | nb |
|---|---|---|---|---|---|---|---|---|
| N-terminal | Gaussian | 35° | 18° | 263° | 36° | 0.88c | 1.1 kHz | 13 (2+1+5+5) |
| semi-static | 30° | n.a.d | 263° | n.a.d | 0.64 | 1.1 | 13 (2+1+5+5) | |
| C-terminal | Gaussian | 29° | 0° | 329° | 48° | 0.88c | 0.9 | 15 (3+0+6+6) |
| semi-static | 22° | n.a.d | 332° | n.a.d | 0.72 | 1.0 | 15 (3+0+6+6) |
The Gaussian model for the dynamics uses a fixed principal order parameter Szz (25), representing the dynamic extent of (mis)alignment (angle α) between the molecular z-axis and its average orientation, characterized by the time average Szz = 〈3 cos2α − 1〉/2 (34). Within this context, further motions can be characterized by the widths στ and σρ of Gaussian distributions about the average values of tilt magnitude τ0 and tilt direction ρ0 (25). An alternative semi-static analysis, using three parameters instead of four, determines the best fit (lowest RMSD) as a function of τ0, ρ0 and Szz. Weighting factors for the 2H methyl and backbone quadrupolar couplings, 1H-15N dipolar couplings and 15N chemical shifts were 1.0, 0.5, 1.0 and 1.0, respectively.
Number of data points in the format: total (2H methyl, 2H backbone, 1H-15N dipolar, 15N chemical shift).
Fixed value.
Not applicable.
Figure 6.
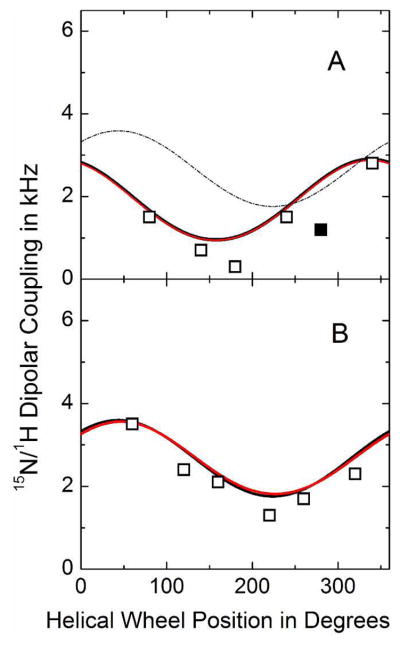
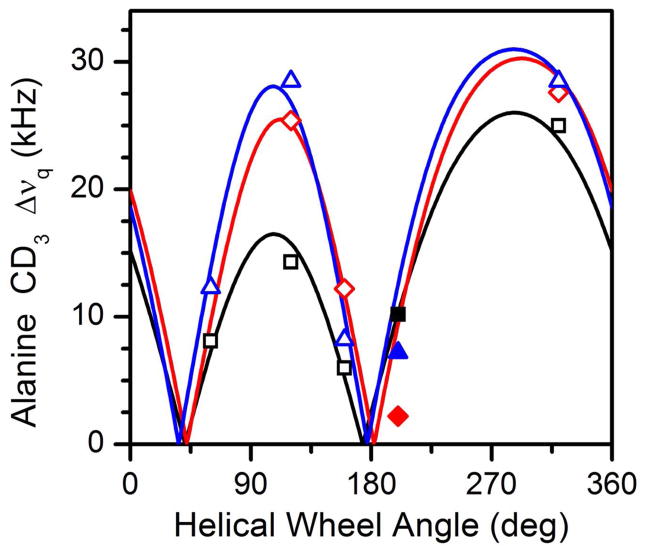
Calculated dipolar wave plots for the N-terminal (A) and C-terminal (B) segments of GWALP23-P12 in oriented DMPC bilayers. Wave functions based on semi-static (red) or Gaussian (black) calculations essentially overlap. Data points indicated by open symbols have been used in the calculations, whereas the solid symbol represents a measurement for A11 that was excluded from the analysis (because the helix is broken prior to P12). For comparison, the fit to the C-terminal (B) is included also in the form of the thin dashed curve in panel A.
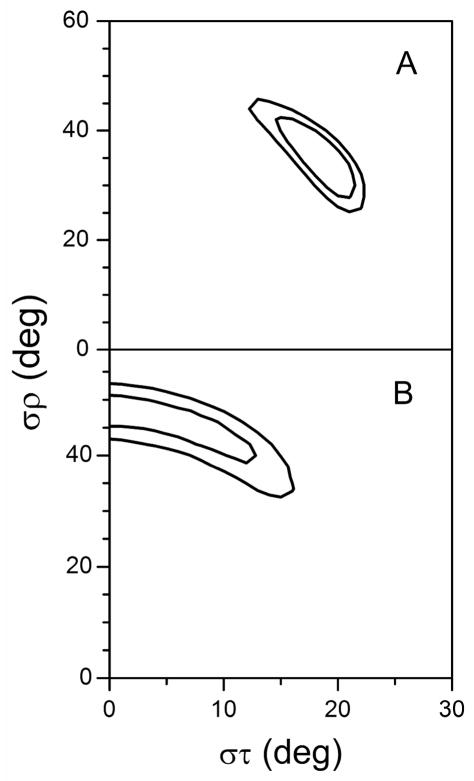
Figure 7.
Gaussian dynamics. RMSD (στ, σρ) graphs for the Gaussian dynamics analysis of the N-terminal (A) and C-terminal (B) helical segments of GWALP23-P12 in oriented DMPC bilayers. Contours are drawn at 1.3, 1.4 and 1.5 kHz (A), or 0.9 and 1.0 kHz (B).
Figure 8.
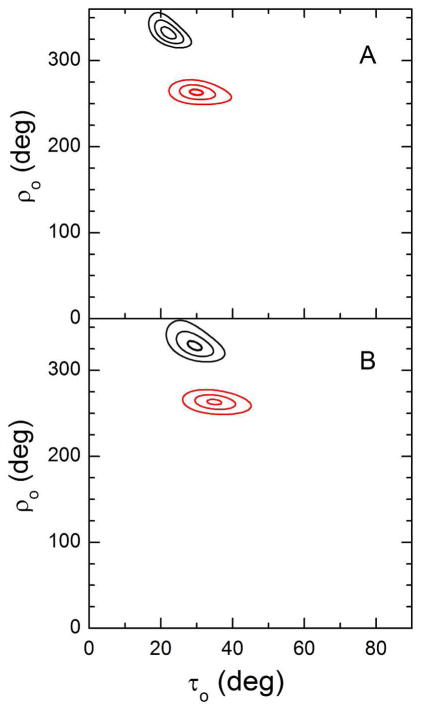
Segmental tilt. RMSD plots for the tilt of the N-terminal (red) and C-terminal (black) helical segments of GWALP23-P12 in oriented DMPC bilayers. A. RMSD plotted as a function of τ and ρ values for the optimum Szz in semi-static calculations. B. RMSD plotted as a function of τ and ρ values for the optimum values of στ and σρ in Gaussian calculations (see Figure 7 and Table 4). The RMSD contour levels are 1, 2 and 3 kHz.
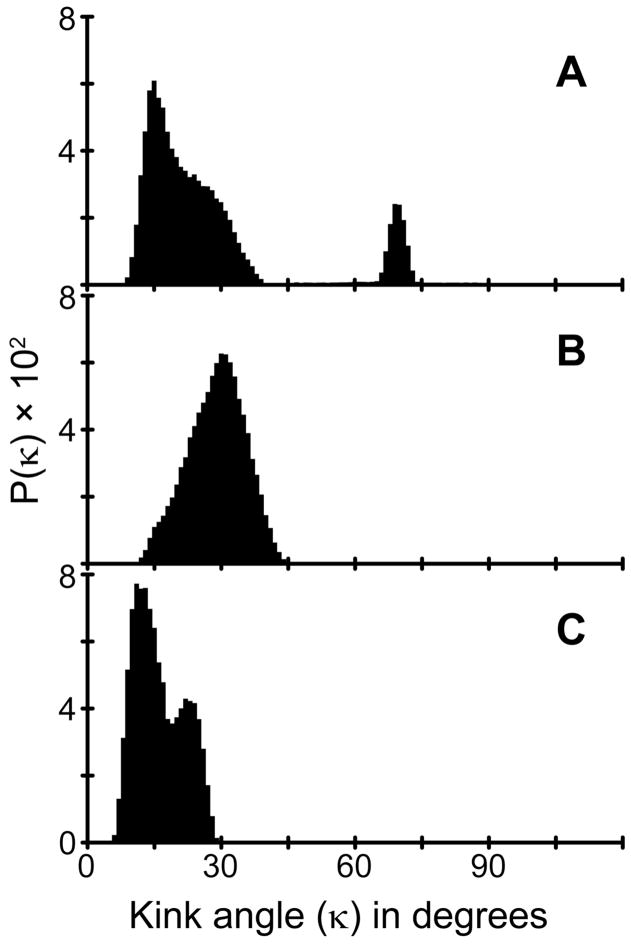
Using the orientations determined for the segments on each side of the proline residue, it is feasible to calculate a kink (κ) angle introduced into the peptide as described in equation [6]. From the segment orientations shown in Figure 8, we calculated a probability distribution for the κ value for GWALP23-P12 in DMPC. The most probable κ value is about 30° (± 5° (Figure 9B). As noted, the helix unwinding or “swivel” angle |Δρo| is 65°–70° in DMPC, regardless of the method employed for the dynamic analysis.
Figure 9.
Distributions of apparent kink angles, κ (degrees), calculated from rmsd (τ ,ρ) analysis of the N- and C-terminal segments of GWALP23-P12 in (A) DLPC, (B) DMPC and (C) DOPC. Regions with rmsd of <3 kHz (C-terminal) or <2 kHz (N-terminal) were binned at a τ of 0.5° and a ρ of 3°.
The segment analysis in lipids other than DMPC is less straightforward because 15N data are not available. To increase somewhat the size of the 2H data sets for the C-terminal in DLPC and DOPC, we included a variation in which a substitution (L16A) could provide an additional data point. This type of substitution often has only a small effect on the helical properties of the peptide segment C-terminal to Pro, and therefore is likely to fit the GALA wave and provide valuable additional quadrupolar splitting values (6). As previously, also in this study we incorporated a control label (Ala-15) adjacent to the L16A substitution. The 2H NMR spectra indicate that the control residue 15 changed little (Table 2) when the L16A substitution was introduced. Because the segment N-terminal to proline can be more sensitive to mutations (6), mutations were not introduced in this region. Additional data points for both segments emerged fortuitously when the quadrupolar splittings were observed from the backbone Cα deuterons of residues 9 and 21 (Table 2). In lipids where the C-terminal segment remained helical through resides 21 (see below), both the backbone and side-chain 2H-signals for Ala-21 could be employed in the analysis.
With the smaller numbers of data points, a Gaussian dynamic analysis of segmental tilt in DLPC or DOPC is not feasible at this time, as it leads to a high degeneracy of solutions. Nevertheless, we have estimated the allowed distributions of apparent tilt magnitudes τo and directions ρo for the segments in DLPC and DOPC (Figure S6 of the Supporting Information), based upon available 2H quadrupolar splitting magnitudes and using (assumed) order parameters that are close to those deduced in DMPC. The semi-static estimates suggest modest segment adjustments in DLPC or DOPC. For the C-terminal segment, ρo remains close to 330° in all three lipids, while τo in DLPC or DOPC may decrease slightly (Figure S6) from the 22° observed by semi-static analysis in DMPC (Figure 8A). For the N-terminal segment, with the caveat of limited data, the currently allowed ranges of τo and ρo are larger in DLPC and DOPC (Figure S6). (For the kink analysis in DOPC, we considered only values within the major distribution of τo between 0°–45° and ρo between 180°–360° (Figure S6).) When the segmental data are considered together, the resulting probability plots (Figure 9) suggest that the kink angle (κ) distributions for DLPC and DOPC overlap that for DMPC, although the most probable κ may be somewhat smaller in DLPC or DOPC than in DMPC. Figure S7 (see Supporting Information) illustrates the swivel angle distributions for GWALP23-P12 in the three lipids. As is the case for DMPC, also in DLPC and DOPC the most probable swivel angle magnitude is about 70° (Δρ is −70° when expressed as (ρN – ρC)), although several other choices exhibit non-zero probability under the current data limitations (Figure S7).
We also tested whether the C-terminal segment remains helical through a deuterated Ala-21 residue. In parallel studies it had been determined that the GWALP23 helix ends at or near Trp-19, namely in the region between residues 19 and 21 (7). We find a similar result for the C-terminal segment of GWALP23-P12 in DLPC and DMPC, although the segment nevertheless does remain helical for residues 13–21 in the thicker DOPC bilayer membranes. As a consequence, the quadrupolar splitting obtained for Ala-21 also could be used for the GALA analysis in DOPC but not in DLPC or DMPC. In Figure 10 we show the fitted semi-static GALA waves for the C-terminal segment in each of the lipid membranes. While the A21 data point (solid symbol) fits on a GALA curve for DOPC, the deviations of A21 from the best-fit quadrupolar waves exceed 5 kHz in both DLPC and DMPC. The lipid bilayer thickness influences the helicity of the C-terminal segment.
Figure 10.
Quadrupolar waves from semi-static GALA analysis of the C-terminal segment of GWALP23-P12 in lipid bilayer membranes of DOPC (black), DMPC (red), and DLPC (blue). The solid symbols indicate data points for A21, which resides outside of the core region defined by W5 and W19. A21 remains part of the core helix in DOPC, but helix distortion moves the A21 quadrupolar splitting more than 5 kHz below the best-fit curves for DLPC and DMPC.
Discussion
The biological occurrence of transmembrane proline-containing peptides is sufficiently common that it is important to understand the implications of the presence of a proline residue for the structure, dynamics and positioning of a transmembrane sequence. The combined analysis of deuterium (2H) and 15N NMR spectral data, reported here, enabled robust determination of a proline-induced kink angle in a designed model peptide in DMPC. We find that introduction of proline into a previously “ideal” GWALP23 α-helix introduces a moderately large kink angle which changes relatively little with the lipid bilayer thickness. In the DLPC, DMPC and DOPC lipids tested, the peptide segments on either side of the proline both adopt significant tilt angles with respect to the lipid membrane normal. Indeed, both segments increase their tilt angles, in comparison with the previously reported whole-peptide tilt of GWALP23 itself in each of the lipid bilayer membranes (7).
Remarkably, regardless of the host lipid identity, the segmental directions of tilt for the C-terminal of GWALP23-P12 remain the same in DLPC, DMPC and DOPC (Figure 8 and Figure S6). Compared to the reference GWALP23, the introduction of Pro-12 alters the C-terminal tilt direction by about 20°. In DMPC, the N-terminal tilt direction changes oppositely by ~50°, leading to a swivel or approximate helix unwinding angle of close to −70° = (ρN – ρC). With a caveat due to the data limitations, it appears likely that the swivel angle of about −70° is preserved in DLPC and DOPC (Figure S7 of the Supporting Information).
The use of combined 1H- 15N high resolution SLF and 2H GALA based solid-state NMR methodologies is advantageous for calculating the average apparent orientations of the segments before and after proline in GWALP23-P12 in lipid bilayers (bicelles) of DMPC (DM-o-PC). At the respective experimental temperatures, the peptide (segment) principal order parameters are about the same in the bilayers at 50 °C and in the bicelles at 42 °C. The combined data sets allow us to deduce a kink angle of ~30° in DMPC. We note that both segments increase their tilt, from that of about 9° for GWALP23 itself, with respect to the DMPC bilayer normal, when P12 is introduced (see Table 4).
The quality of the combined analysis, using data from macroscopically oriented DMPC bilayers (on glass slides) together with data from magnetically oriented DM-o-PC bicelles, suggests that not only segment motions but also the tilt parameters vary only slightly between the bilayer and bicelle samples. Furthermore, when the bicelle-derived 1H- 15N SAMPI4 data (Figure 4; Table 3) are analyzed alone, in the absence of the bilayer-derived 2H quadrupolar splittings, the resulting apparent τ0 value of 29° for the C-terminal segment is identical to that deduced from the combined bilayer and bicelle data (Table 4). The deduced ρ0 values also are nearly identical from bicelles to bilayers, giving in each case about 330° ± 2° for the C-terminal segment. Similarly, for the N-terminal segment, the deduced τ0 remains nearly constant at 35°–36° (Table 4), regardless of whether only the 15N data or the full data sets are analyzed, while in this case the apparent ρ0 increases slightly, from 263° (for the full data sets) to 273° (15N data only, in bicelles). To summarize, the SAMPI4 results from bicelle samples and the GALA results from bilayer samples agree closely for the tilt magnitude and direction for both of the helical segments, preceding and following the proline, in DMPC.
In addition to calculation of the peptide tilt and rotation, simple visual observation of helical wheel representations with respect to the obtained SAMPI4 spectra provides a striking confirmation of the kink introduced by the proline. This can be recognized easily, as there are two distinct sets of resonances that fit to separate helical wheel patterns in the two-dimensional SAMPI4 spectrum (Figure 5).
The semi-static and Gaussian treatments of the dynamics of GWALP23-P12 in DMPC agree quite well. Considering the semi-static model, the peptide orientation consists of a 30° tilted N-terminal segment connected to a 22° tilted C-terminal segment via a 30° kink angle. The kink angle between the segments also reflects the −69° difference in rotation for the two segments, 263° and 332° respectively. Considering the dynamic Gaussian model, we calculate a 35° tilted N-terminus connected to the 29° tilted C-terminus, with −66° swivel angle (difference in rotation) and a 34° kink angle. Overall, the agreement is quite good between the two models for the analysis, as they give similar kink angles and nearly identical swivel angles. The magnitude of the 30–34° kink angle is somewhat larger than the value of 19° determined previously for WALP19 in DOPC (6). The modest difference in the kink angle could reflect the different lipid environment and/or the different numbers of Trp anchor residues, as well as their positions relative to the proline ring. While the “inner” Trps 3 and 17 of WALP19 have the same radial and longitudinal separation as Trps 5 and 19 of GWALP23, the “outer” Trps of WALP19 are absent in GWALP23.
The respective στ and σρ values from the Gaussian analyses (Table 4) suggest slightly more rotational averaging for the C-terminal segment, together with a larger value of στ for the N-terminal segment. We note that the connecting proline should couple some of the segment motions, such that the C-terminal and N-terminal dynamics should not be completely independent. In this vein, rotation of the C-terminal helix could lead to tilt variation (στ motion) of the N-terminal helix, and vice versa. The extent of the helix wobble, nevertheless, can be larger than the change in an apparent τ value, because not all projections of wobbling motions will influence the angle between a helix axis and the membrane normal. The semi-static analysis by itself suggests furthermore a somewhat lower principal order parameter of 0.64 for the N-terminal segment (Table 4). The relative N-terminal mobility stands in accord with the location of the missing hydrogen bond—before proline—and with previous observations that the segment preceding proline is more susceptible to helix unwinding (3, 6). Internal helix motions also would likely not be coupled through the proline.
The proline connection furthermore might decrease each segment’s preferred range and extent of motion. Indeed, the larger sizes of the PISA wheels for both of the individual segments (Figures 4–5), compared to the PISA wheel of GWALP23 itself (Figure 4), suggest smaller extents of dynamic averaging for both the C-terminal and the N-terminal when proline-12 is present in GWALP23.
In comparison to previous studies on the proline-containing WALP19-P10 in DOPC bilayers, we find for GWALP23-P10 in DMPC a modestly larger κ angle, 30°–34° versus 19°, along with significantly larger τ angles for each of the proline flanking segments, namely 30–35° and 22–29° versus 7° and 12°, respectively (6). The greater hydrophobic length of GWALP23-P12 in the thinner DMPC bilayer probably accounts for the larger segmental tilt values, compared to WALP19-P10 in DOPC, while the influence on the proline-induced kink angle κ is somewhat less dramatic.
In addition to the studies of GWALP23-P12 in DMPC bilayers, we have performed 2H NMR experiments using thicker and thinner lipid bilayer systems. Although fewer data points are available than in DMPC, we nevertheless estimate the allowed ranges of segmental tilt (Figure S6) and the kink and swivel probabilities (Figure 9 and Figure S7) in DLPC and DOPC. The results suggest modest yet non-systematic changes in the kink angle with bilayer thickness, as the most probable κ value, based on available data, may be slightly less in DLPC and DOPC than in DMPC. Because the C-terminal helix is intact through A21 in DOPC, but not in DMPC or DLPC, partial unwinding may represent a portion of the helix adaptation to the lipids. The entire segment preceding proline is furthermore susceptible to helix unwinding (3, 6), presumably to varying extents depending upon the immediate environment.
Concerning the biological relevance, it is of note that an array of biologically occurring proline-containing transmembrane helices display a wide range of proline-induced kink angles from 0°–70° (3). The present results demonstrate the use of 1H- 15N SAMPI4 and 2H GALA solid-state NMR methods to determine the relative orientations of transmembrane helical segments on either side of a localized distortion. The methods should prove relevant for investigating α-helical transmembrane sequences containing a helix-breaking residue such as proline or glycine, also known for its potential helix-breaking properties. In this context, one notes the advantages conferred by the joint application of 2H and 15N-1H NMR methods, and furthermore the combined use of aligned lipid bicelle and lipid bilayer samples.
In summary, the full SAMPI4 spectrum (Figure 5) illustrates directly the kink in GWALP23-P12. The combined analysis of deuterium (2H) and 1H- 15N NMR observables has enabled robust determination of the proline-induced kink angle in the designed peptide GWALP23-P12 in DMPC. For proteins, the proline influence on domain orientation and dynamics could have decisive consequences, especially considering that structure and function are tightly intertwined. As a result, it is well acknowledged that cellular functions could be influenced critically by the presence or absence of specific transmembrane proline residues.
Supplementary Material
Acknowledgments
We thank James Hinton and Marvin Leister for many helpful discussions.
Abbreviations
- DH-o-PC
ether analogue of 1,2-Dihexanoylphosphatidylcholine
- DLPC
1,2-Dilauroylphosphatidylcholine
- DMPC
1,2-Dimyristoylphosphatidylcholine
- DM-o-PC
ether analogue of 1,2-Dimyristoylphosphatidylcholine
- DOPC
1,2-Dioleoylphosphatidylcholine
- Fmoc
fluorenylmethoxycarbonyl
- GALA
Geometric Analysis of Labeled Alanines
- HPLC
high-performance liquid chromatography
- MALDI
matrix-assisted laser-desorption ionization
- NMR
nuclear magnetic resonance
- PISEMA
Polarization Inversion with Spin Exchange at Magic Angle
- TEA
triethylamine
- TFE
trifluoroethanol
- TM
transmembrane
- GWALP23
acetyl-GGALWLALALALALALALWLAGA-ethanolamide
Footnotes
This work was supported in part by NSF grant MCB 0841227 and by the Arkansas Biosciences Institute. The peptide facility was supported by NIH grants RR31154 and RR16460. The 2H NMR facility in Fayetteville AR was supported by NIH grants RR31154 and GM103450. The 15N NMR facility in La Jolla CA was supported by the Biotechnology Research Center for NMR Molecular Imaging of Proteins at the University of California, San Diego, which is supported by NIH grant P41EB002031.
Supporting Information Available. Mass spectra, HPLC chromatogram, 31P NMR spectra and additional 2H NMR spectra; to confirm peptide identity and purity, and the alignment of lipids and peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Von Heijne G. Proline kinks in transmembrane alpha-helices. J Mol Biol. 1991;218:499–503. doi: 10.1016/0022-2836(91)90695-3. [DOI] [PubMed] [Google Scholar]
- 2.Ulmschneider MB, Sansom MS. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 3.Cordes FS, Bright JN, Sansom MSP. Proline-Induced Distortions of Transmembrane Helices. J Mol Biol. 2002;323:951–960. doi: 10.1016/s0022-2836(02)01006-9. [DOI] [PubMed] [Google Scholar]
- 4.Glukhov E, Shulga YV, Epand RF, Dicu AO, Topham MK, Deber CM, Epand RM. Membrane interactions of the hydrophobic segment of diacylglycerol kinase epsilon. Biochim Biophys Acta. 2007;1768:2549–2558. doi: 10.1016/j.bbamem.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Decaffmeyer M, Shulga YV, Dicu AO, Thomas A, Truant R, Topham MK, Brasseur R, Epand RM. Determination of the topology of the hydrophobic segment of mammalian diacylglycerol kinase epsilon in a cell membrane and its relationship to predictions from modeling. J Mol Biol. 2008;383:797–809. doi: 10.1016/j.jmb.2008.08.076. [DOI] [PubMed] [Google Scholar]
- 6.Thomas R, Vostrikov VV, Greathouse DV, Koeppe RE., 2nd Influence of proline upon the folding and geometry of the WALP19 transmembrane peptide. Biochemistry. 2009;48:11883–11891. doi: 10.1021/bi9016395. [DOI] [PubMed] [Google Scholar]
- 7.Vostrikov VV, Daily AE, Greathouse DV, Koeppe RE., 2nd Charged or aromatic anchor residue dependence of transmembrane peptide tilt. J Biol Chem. 2010;285:31723–31730. doi: 10.1074/jbc.M110.152470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gleason NJ, Vostrikov VV, Greathouse DV, Grant CV, Opella SJ, Koeppe RE., II Tyrosine replacing tryptophan as an anchor in GWALP peptides. Biochemistry. 2012;51:2044–2053. doi: 10.1021/bi201732e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vostrikov VV, Grant CV, Daily AE, Opella SJ, Koeppe RE., 2nd Comparison of “Polarization inversion with spin exchange at magic angle” and “geometric analysis of labeled alanines” methods for transmembrane helix alignment. J Am Chem Soc. 2008;130:12584–12585. doi: 10.1021/ja803734k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vostrikov VV, Hall BA, Greathouse DV, Koeppe RE, 2nd, Sansom MS. Changes in transmembrane helix alignment by arginine residues revealed by solid-state NMR experiments and coarse-grained MD simulations. J Am Chem Soc. 2010;132:5803–5811. doi: 10.1021/ja100598e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Wel PCA, Strandberg E, Killian JA, Koeppe RE., II Geometry and intrinsic tilt of a tryptophan-anchored transmembrane alpha-helix determined by 2H NMR. Biophys J. 2002;83:1479–1488. doi: 10.1016/S0006-3495(02)73918-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. Imaging membrane protein helical wheels. J Magn Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 13.Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J Magn Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ten Kortenaar PBW, Dijk BG, Peeters JM, Raagen BJ, Adams PJ, Tesser GI. Rapid and efficient method for the preparation of Fmoc-amino acids starting from 9-fluorenylmethanol. Int J Pept Prot Res. 1986;27:398–400. [Google Scholar]
- 15.Daily AE, Greathouse DV, van der Wel PCA, Koeppe RE., II Helical distortion in tryptophan and lysine anchored membrane-spanning alpha helices as a function of hydrophobic mismatch: A solid-state deuterium NMR investigation using the GALA method. Biophys J. 2008;94:480–491. doi: 10.1529/biophysj.106.097543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greathouse DV, Koeppe RE, II, Providence LL, Shobana S, Andersen OS. Design and characterization of gramicidin channels. Meth Enzymol. 1999;294:525–550. doi: 10.1016/s0076-6879(99)94031-4. [DOI] [PubMed] [Google Scholar]
- 17.Greathouse DV, Goforth Robyn L, Crawford Toria, van der Wel Patrick, Killian J Antoinette. Optimized aminolysis conditions for cleavage of N-protected peptides from solid-phase resins. J Pept Res. 2001;57:519–527. doi: 10.1034/j.1399-3011.2001.00849.x. [DOI] [PubMed] [Google Scholar]
- 18.Nevzorov AA, Opella SJ. Selective averaging for high-resolution solid-state NMR spectroscopy of aligned samples. J Magn Reson. 2007;185:59–70. doi: 10.1016/j.jmr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Cavagnero S, Dyson HJ, Wright PE. Improved low pH bicelle system for orienting macromolecules over a wide temperature range. J Biomol NMR. 1999;13:387–391. doi: 10.1023/a:1008360022444. [DOI] [PubMed] [Google Scholar]
- 20.Aussenac F, Lavigne B, Dufourc EJ. Toward bicelle stability with ether-linked phospholipids: Temperature, composition, and hydration diagrams by H-2 and P-31 solid-state NMR. Langmuir. 2005;21:7129–7135. doi: 10.1021/la050243a. [DOI] [PubMed] [Google Scholar]
- 21.Davis JH, Jeffrey KR, Valic MI, Bloom M, Higgs TP. Quadrupolar echo deuteron magnetic resonance spectroscopy in ordered hydrocarbon chains. Chem Phys Lett. 1976;42:390–394. [Google Scholar]
- 22.Wu CH, Ramamoorthy A, Opella SJ. High-resolution heteronuclear dipolar solid-state NMR spectroscopy. J Magn Reson Ser A. 1994;109:270–272. [Google Scholar]
- 23.Nevzorov AA, Opella SJ. A “Magic Sandwich” pulse sequence with reduced offset dependence for high-resolution separated local field spectroscopy. J Magn Reson. 2003;164:182–186. doi: 10.1016/s1090-7807(03)00240-4. [DOI] [PubMed] [Google Scholar]
- 24.Cook GA, Opella SJ. NMR studies of p7 protein from hepatitis C virus. Eur Biophys J Biophys Lett. 2010;39:1097–1104. doi: 10.1007/s00249-009-0533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strandberg E, Esteban-Martin S, Saldago J, Ulrich AS. Orientation and dynamics of peptides in membranes calculated from 2H-NMR data. Biophys J. 2009;96:3223–3232. doi: 10.1016/j.bpj.2009.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strandberg E, Özdirekcan S, Rijkers DTS, van der Wel PCA, Koeppe RE, II, Liskamp RMJ, Killian JA. Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by 2H solid-state NMR. Biophys J. 2004;86:3709–3721. doi: 10.1529/biophysj.103.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nevzorov AA, Mesleh MF, Opella SJ. Structure determination of aligned samples of membrane proteins by NMR spectroscopy. Magn Reson Chem. 2004;42:162–171. doi: 10.1002/mrc.1320. [DOI] [PubMed] [Google Scholar]
- 28.Saito H, Ando I, Ramamoorthy A. Chemical shift tensor - The heart of NMR: Insights into biological aspects of proteins. Progr in NMR Spectr. 2010;57:181–228. doi: 10.1016/j.pnmrs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bechinger B, Resende JM, Aisenbrey C. The structural and topological analysis of membrane-associated polypeptides by oriented solid-state NMR spectroscopy: Established concepts and novel developments. Biophys Chem. 2011;153:115–125. doi: 10.1016/j.bpc.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Vostrikov VV, Grant CV, Opella SJ, Koeppe RE., II On the combined analysis of 2H and 15N/1H solid-state NMR data for determination of transmembrane peptide orientation and dynamics. Biophys J. 2011;101:2939–2947. doi: 10.1016/j.bpj.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ketchem RR, Lee KC, Huo S, Cross TA. Macromolecular structural elucidation with solid-state NMR-derived orientational constraints. J Biomol NMR. 1996;8:1–14. doi: 10.1007/BF00198135. [DOI] [PubMed] [Google Scholar]
- 32.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page RC, Kim S, Cross TA. Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure. 2008;16:787–797. doi: 10.1016/j.str.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pulay P, Scherer EM, van der Wel PCA, Koeppe RE. Importance of tensor asymmetry for the analysis of H-2 NMR spectra from deuterated aromatic rings. J Am Chem Soc. 2005;127:17488–17493. doi: 10.1021/ja054935x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.