Abstract
Members of the myosin-I family of molecular motors are expressed in many eukaryotes where they are involved in a multitude of critical processes. Humans express 8 distinct members of the myosin-I family, making it the second largest family of myosins expressed in humans. Despite the high degree of sequence conservation in the motor and light chain binding domains of these myosins, recent studies have revealed surprising diversity of function and regulation arising from isoform-specific differences in these domains. Here we review the regulation of myosin-I function and localization by the motor and light chain binding domains.
Keywords: myosin-I, mechanochemistry, light chain binding domain, motor domain
Myosin-I Diversity
Myosin-Is are widely expressed, single-headed members of the myosin superfamily that comprise the second largest myosin family in vertebrates (8 isoforms; [1]). Myosin-Is participate in a number of crucial cellular processes related to membrane morphology and trafficking [2–7], pathogen response [5, 8], tension sensing [2, 9–11], regulation of actin dynamics [3, 12–15], and nuclear transcription [16]. For example, in vertebrates, Myo1a plays roles in microvilli structure and dynamics [9, 17], Myo1b in the formation of post-Golgi carriers [7], Myo1c in GLUT4 exocytosis [4, 18, 19] and the trafficking of cholesterol-rich membranes [5], and Myo1e in receptor-mediated endocytosis [20]. Clearly, myosins-Is display impressive functional diversity, raising the question of how such diversity can be achieved from a single motor family.
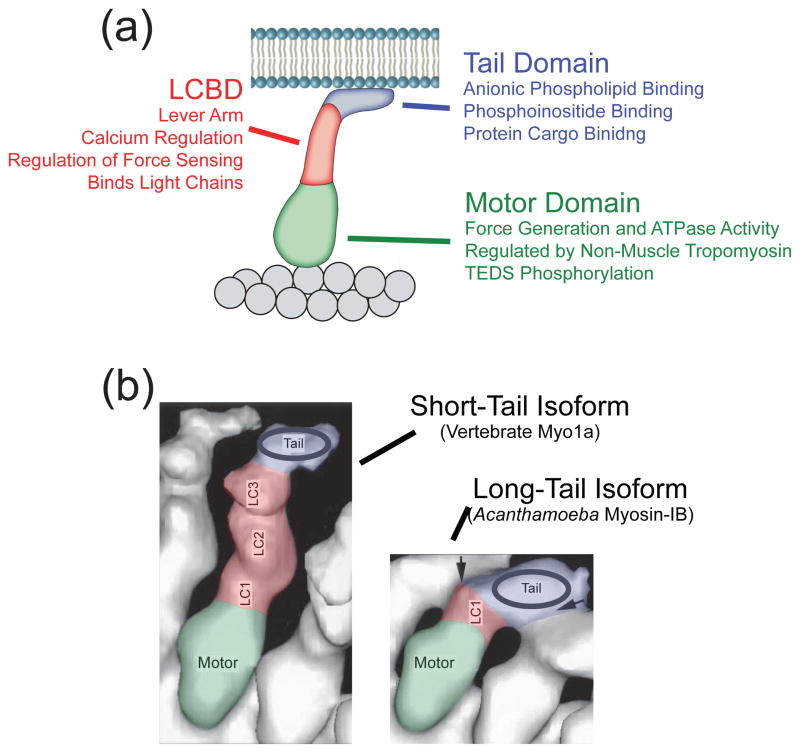
All myosin-I isoforms have a similar structural organization, consisting of a motor domain, light chain binding domain (LCBD), and tail domain (Figure 1). The motor and LCBDs are strongly conserved, while the tail domain shows more sequence divergence [21]. The motor domain binds actin and hydrolyzes ATP. The LCBD, which is composed of IQ motifs that bind light chains, acts as a lever-arm, amplifying the small conformational changes that occur at the nucleotide binding site into larger displacements as measured from the end of the LCBD. Together, these domains power motility and perform mechanical work. The myosin-I tail domain binds phospholipids and accessory proteins, enabling myosin-I to link membranes to the actin cytoskeleton. All characterized tail domains contain a positively-charged lipid-binding region and a pleckstrin homology (PH) domain that mediates phospholipid specificity in some isoforms [22, 23]. Long-tailed myosin-I isoforms also contain an SH3 domain and a proline-rich region that binds actin and regulates actin dynamics (Box 1).
Figure 1.
Myosin-I structural domains. (a) Cartoon showing the interaction of the domains of myosin-I with actin and the plasma membrane. Each of the domains plays an important role in the regulation and control of myosin-I function. The regulatory effects of each domain are listed. (b) Three dimensional cryo- electron microcopy reconstructions of a short-tailed (Myo1a [94]) and a long-tailed (Acanthamoeba myosin-IB [95]) myosin-I isoform (colored) bound to actin (white). The colors correspond to the domains shown in (a). Individual light chains (LC1, 2, 3) are indicated. Note that different isoforms have different numbers of light chains. Also, note the additional density in the tail domain of the long-tailed isoform.
Box 1. Myosin-I Tail Domain.
The myosin-I tail domain has a crucial role in controlling myosin-I subcellular localization and function (for review, see [79]). It is divided into three tail-homology (TH) subdomains based on sequence motifs that have been termed TH1, TH2, and TH3. Long-tailed isoforms contain all three subdomains, while short-tailed isoforms contain only TH1.
TH1
A key feature of myosin-I isoforms is their ability to bind membranes directly via the interaction of positively-charged regions in TH1 with anionic phospholipids. All characterized myosin-I isoforms can nonspecifically bind phospholipids via delocalized electrostatic interactions [80–82]. Some isoforms bind with high affinity to poly-phosphoinositides [6, 22, 23, 83] through a pleckstrin homology domain (PH) in TH1. The PH domain is necessary for the proper membrane localization of Myo1b [84], Myo1c [22], Myo1f, and Myo1g [11, 85]; however, phosphoinositide binding does not appear to be necessary for membrane association of Myo1a [82], Myo1e [81], or Acanthamoeba myosin-IC [86].
Several TH1 binding proteins have been identified in vertebrate isoforms. These include sucrase–isomaltase [87], NF-kappa-B-essential modulator [88], NEPH-1 [89], PHR1 [90], RalA [19], and Rictor [15]. New experiments have also suggested that the TH1 region of vertebrate Myo1c binds directly to G-actin, but not F-actin [13].
TH2
The TH2 region in lower eukaryotes is enriched in glycine, proline, and either alanine or glutamine residues, and it has been shown in some isoforms to be an ATP-insensitive actin-filament binding site [80]. This secondary actin-binding site allows myosin-I to act as an actin cross-linking protein. Interestingly, a Dictyostelium myosin-I isoform was shown to bind to microtubules via a TH1–TH2 interaction [91]. The TH2 region in vertebrate isoforms contains a short stretch of proline and lysine or glutamine dipeptide repeats that has been shown to be important for recruitment of myosin-I to endocytic structures [3, 20]
TH3
The TH3 region is a src homology-3 (SH3) domain that lies at the C-terminus of the protein or within the TH2 domain. Several proteins have been found to bind TH3 directly. Notably, Xu et al. discovered the first protein to bind this region [92], named CARMIL for its ability to function as a capping protein, Arp2/3, and myosin-I linker [93]. Subsequent experiments have identified additional binding partners including dynamin, verprolin, synaptojanin, and possibly N-WASP and verprolin family members WIP and WIRE [3, 12, 20]. Clearly, these proteins point to an important role for long-tailed myosins in regulation of actin dynamics and endocytosis.
Much research has focused on determining how the different domains control isoform-specific subcellular localization and mechanical function. The tail domains have the most sequence divergence between isoforms, so it is not surprising that they play crucial roles in the differential targeting of the isoforms. It has been surprising to learn that the motor and LCBDs are also important in the control and adaptability of myosin-I function. These domains not only have the remarkable ability to change the myosin’s motile activity and power output in response to mechanical loads, but they also have roles in directing myosin-I subcellular localization. This review will focus on the features of the motor and LCBDs that enable them to control myosin-I function.
Control of Myosin-I Unloaded Kinetics by the Motor Domain
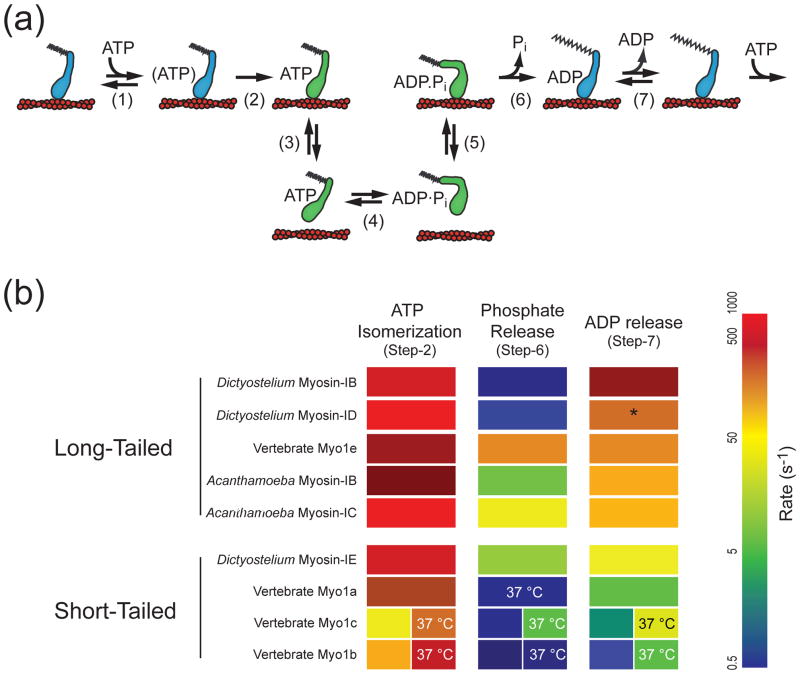
All characterized members of the myosin-I family follow a similar ATPase pathway in which myosin generates force via structural changes associated with nucleotide binding and release (Figure 2). Characterized myosin-I isoforms have ATPase rates that are limited by the rate of phosphate release and unloaded velocities that are limited by the rate of ADP release [24–35]. As such, myosin-Is are low duty ratio motors (i.e. they spend a majority of their biochemical cycle detached from actin) in the absence of force. Despite these kinetic similarities, the rates and equilibrium constants of the transitions in the ATPase cycle vary substantially between isoforms, resulting in differences in actin gliding rates and power outputs [36]. Sorting the characterized myosin-I isoforms by the rate of ADP release reveals that long-tailed myosin-I isoforms have faster ADP release rates than short-tailed isoforms (Figure 2b). Isoforms can be further divided into categories that correspond to the classes set forth by Bloemink and Geeves [37] of fast movers (similar to skeletal muscle myosin-II), efficient force holders (similar to smooth muscle myosin-II), and strain sensors (similar to Myo1b). Dictyostelium myosin-IB and ID isoforms have slow ATPase rates compared to the other long-tailed isoforms, but their rates of ADP release (and thus motility rates) are faster than the other isoforms [32, 33]. This leads to a low duty ratio that is optimized for fast movement, highlighting that sliding velocity and steady-state ATPase rates are not necessarily correlated. The other long-tailed myosins have higher duty ratios (though still < 0.5) and slower motility rates, suggesting that are efficient force holders). The short-tailed isoforms are slow movers with lower duty ratios, suggesting tension sensing roles. These classifications are important for understanding myosin-I isoforms, but as discussed below, it is not possible to extrapolate tension-sensing capabilities solely from an analysis of the kinetics.
Figure 2.
Diversity of unloaded kinetics in the myosin-I family. (a) ATPase mechanochemical cycle followed by all myosin isoforms. Blue denotes force-bearing states while green denotes weak-binding or detached states. While all myosin follow the same pathway, the rate and equilibrium constants vary between isoforms. During the course of the mechanochemical cycle, one ATP is consumed to produce mechanical work. (b) The rates of critical transitions in the myosin-I ATPase cycle are tuned for the isoform specific function [24–26, 28, 29, 32–34, 48, 57]. All rates are reported at 20–25 °C unless otherwise noted. The myosin isoforms were ordered by ADP release rate (fastest to slowest), and the rates of the ATP isomerization (step-2), phosphate release (step-6), and ADP release (step-6) are depicted by colors on a logarithmic scale. The asterisk denotes a rate that was assumed based on the motility rate rather than a direct measurement. For all of the myosin-I isoforms, phosphate release is the rate-limiting step of the ATPase cycle, meaning that all of the isoforms are low duty ratio motors (i.e. they spend a majority of their biochemical cycle detached from actin). Interestingly, the rate of phosphate release appears to be uncorrelated with the tail length. The rate of the isomerization following ATP binding (i.e. the maximal rate of ATP binding at saturating ATP concentrations) is fast for all of the isoforms. The rate of ADP release, the transition that sets the motility rate, is quite diverse within the myosin-I family (varying from >200 s−1 for Dictyostelium myosin-IB to 1.6 s−1. Note that while the rate of ADP release is faster for long-tailed isoforms than for short-tailed isoforms, the rate of phosphate release varies greatly and does not necessarily correlate with the tail length.
The ATPase activities and cellular localization of myosin-I isoforms from lower eukaryotes are regulated by phosphorylation of a residue near the actin-binding interface called the TEDS site [38]. In most myosins, this site is occupied by a negatively charged aspartate or glutamate residue, whereas in lower eukaryote myosin-Is (and vertebrate myosin-VI [38]), this site contains either a phosphorylatable threonine or serine residue. The TEDS residue is phosphorylated by p21-activated kinase family members [38], which in Acanthamoeba, results in activation of the actin-activated ATPase activity by increasing the rate of phosphate release [28]. Phosphorylation of the TEDS site does not appear to affect any other rate constants of the ATPase cycle, although it may increase the actin affinity in some myosin-I isoforms [33]. In both the phosphorylated and dephosphorylated states, phosphate release remains the rate-limiting step for the ATPase cycle and thus the myosin remains a low duty-ratio motor.
Myosin-Is from higher eukaryotes do not require TEDS phosphorylation since they have an aspartate or glutamate at this site, highlighting the importance of the negative charge at this region for activity. Substitution of the TEDS residue in Acanthamoeba and Dictyostelium myosin-Is [39] with a negatively charged residue that mimics phosphorylation activates the ATPase activity.
Control of Motor Activity and Power Output
The motile activities of myosin-I isoforms are modulated by force, i.e., myosin-I motors dynamically adjust their sliding rates to forces opposing or assisting the movement of the LCBD. This force-dependent effect on contraction was first observed in muscle tissue in the early 1900s. Even those not familiar with the details of muscle physiology intuitively know that muscles contract faster when working against low loads than when moving heavy objects, and that you burn more calories per unit time doing cardiovascular exercises (i.e., aerobic exercises) than strength training. The slowing of muscle contraction with force is due to force slowing the rates of transitions in the ATPase cycle. These force-dependent effects are an inherent property of all myosins. New studies show that the magnitude and mechanisms of force sensing vary substantially between myosin-I isoforms [34, 40–42]. This point is significant for understanding the molecular roles of myosin-Is since the expected mechanical properties of a tension-sensing anchor and a transporter (two possible roles for myosin-Is) are quite different; whereas a tension-sensing anchor would be expected to have a low power output that is non-zero over a narrow range of forces, a transporter would be expected to be able to perform work over a range of forces.
The most dramatic effect of force on myosin behavior is seen with the vertebrate Myo1b isoform [40]. Myo1b is expressed in the liver, testes, lungs, kidneys, brain, heart, and intestines [43]. The MYO1B gene transcript is alternatively spliced in the region encoding the LCBD, resulting in isoforms that contain either 6 (Myo1ba), 5 (Myo1bb), or 4 (Myo1bc) IQ motifs that bind calmodulin with various affinities (i.e. they have different lever arm lengths). Myo1b is enriched at the plasma membrane and it has been associated with trafficking of endocytic vesicles and the formation of post-Golgi carriers [7].
The force dependence of Myo1ba was recently probed [41] using single molecule techniques [44]. Specifically, optical tweezers were used to measure the kinetics of actomyosin detachment as a function of force. Strikingly, the rate of actomyosin detachment (which is directly proportional to the motility rate) is strongly sensitive to forces that oppose the working stroke (Figure 3b). The effect of force on a reaction rate can be modeled using the Bell equation [45]:
| Eq. 1 |
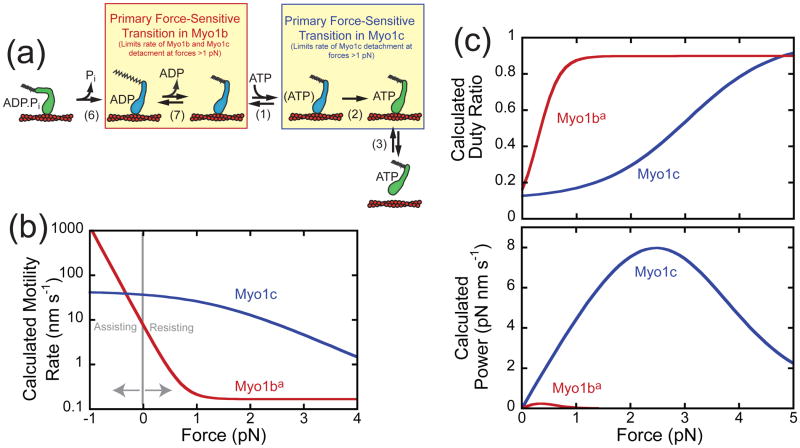
where k0 is the reaction rate in the absence of force, F is the force on the molecule, kB is Boltzman’s constant, T is the temperature, and d is the distance to the transition state, also known as the distance parameter. A larger distance parameter corresponds to a more force-sensitive transition. The actomyosin detachment rate of Myo1ba is extraordinarily force-sensitive with a distance parameter of 18 nm, which is far larger than the distance parameter measured for any other myosin [46, 47] (Fig. 3). At < 0.5 pN of force, the rate of ADP release slows to the point that it becomes the rate-limiting transition in the ATPase cycle and Myo1ba becomes a high duty-ratio motor [40, 41]. With just 1 pN of force, the average attachment lifetime exceeds 50 s, meaning that the myosin remains attached to actin for an extended period of time. Moreover, the power output of Myo1bais low and non-zero over a very small range of forces. Taken together, these results clearly demonstrate that Myo1b has not evolved to play a role in rapid transport processes, but rather it acts as a tension-sensing anchor protein. Therefore the application of force provides a mechanism for regulating the anchoring activities of Myo1ba, which may be consistent with its proposed role in pulling membranes to create post-Golgi carriers [7].
Figure 3.
Myo1b and Myo1c are tuned for different mechanochemical functions. (a) Kinetic scheme showing that the primary force-sensitive transition in Myo1b (ADP release, step-7) is different from the primary force-sensitive transition in Myo1c (ATP isomerization, step-2). (b) The calculated motility rates of Myo1ba and Myo1c based on the values for the working stroke, detachment rate, and distance parameters determined using the optical trap [34, 41, 42]. Whereas the velocity of Myo1ba is highly sensitive to forces <1 pN, the velocity of Myo1c is only sensitive to forces >1 pN. Moreover, the force sensitivity of Myo1ba is far greater than the force sensitivity of Myo1c. (c) The calculated duty ratio and power output of Myo1ba and Myo1c as a function of force based on the values of parameters measured using an optical trap [34, 40]. Myo1ba becomes a high duty ratio motor with < 0.5 pN of force, while Myo1c remains a low duty ratio motor until ~2.5 pN of force. While Myo1ba undergoes a transformation to a high duty ratio motor over a narrow range of forces, Myo1c undergoes the transformation over a broader range of forces. Note that the power output of Myo1ba is very low and non-zero only over a narrow range of forces, consistent with the expected properties of a tension-sensitive anchor, while the power output of Myo1c is higher over a broad range of forces, consistent with the expected properties of a transporter. Taken together, these data suggest that despite their similar kinetics, the molecular roles of Myo1ba and Myo1c are likely quite different.
The response of the widely expressed vertebrate myosin-I isoform, Myo1c, to force is substantially different from Myo1b [34]. This result was surprising since Myo1b and Myo1c have similar unloaded ATPase kinetics [26, 48]. Myo1c has been associated with several important cellular processes, including endocytosis [14], exocytosis [5] (including insulin-stimulated GLUT4 translocation to the cell membrane) [18, 19], membrane ruffling [49], transcription of DNA in the nucleus [16], and mechanosensation in sensory hair cells [30, 50, 51]. Like Myo1b, it is a low duty ratio motor (i.e. phosphate release limits the overall ATPase cycle) with slow motility that is limited by the rate of ADP release in the absence of force (Figure 2b; [30, 48, 50]). In contrast to Myo1b, single-molecule measurements revealed that actin-attachment durations of Myo1c are largely independent of forces < 1 pN and that force-sensitive detachment only occurs at forces >1 pN (Figure 3b). The distance parameter for Myo1c (d=5.2 nm) is larger than that of other myosins [46, 47], but still lower than that of Myo1b. Also, Myo1c does not become a high duty ratio motor until the load exceeds ~2.5 pN. Plotting the average power output of Myo1c as a function of force (Figure 3c) shows that Myo1c is able to generate power over a range of forces, more similar to smooth muscle myosin-II than Myo1b. Based on these results, it appears that Myo1c has evolved to be a slow transporter, capable of generating power over a range of loads, and not a tension-sensitive anchor like Myo1b[18]. Interestingly, the primary force-sensitive transition in Myo1c is the isomerization following ATP binding, not ADP release as in Myo1b.
It was proposed that short-tailed myosin-I isoforms are tension sensors while long-tailed isoforms are built for fast motility [37]. This assumption has yet to be tested rigorously; however, the diversity of tension sensing behaviors, even within the short-tailed family members, indicate that mechanical experiments are needed to draw conclusions about the mechanochemistry of different myosin-I isoforms and their respective molecular roles in the cell.
LCBD Control of Motor Activity
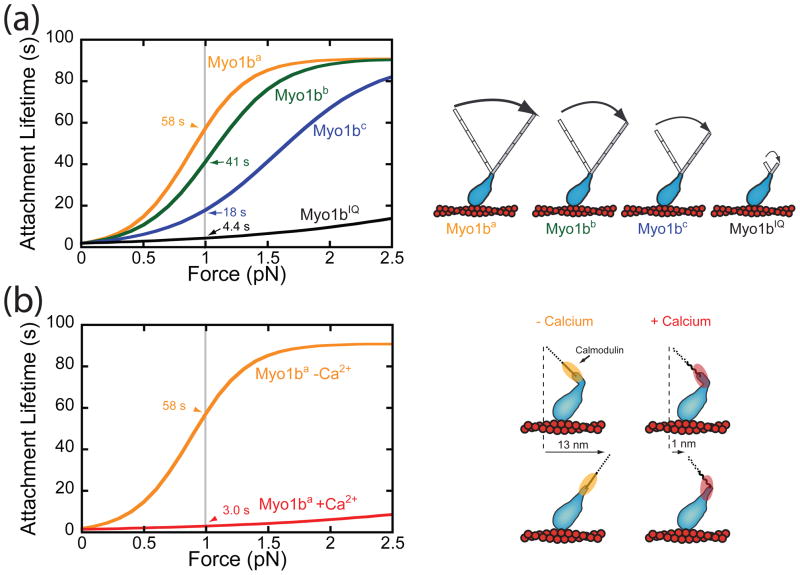
The LCBD is composed of IQ motifs, α-helical sequences of ~23 amino acids that bind calmodulin and calmodulin-like proteins. Calmodulin binding to IQ motifs likely provides mechanical stiffness to the LCBD, allowing it to act as the motor’s lever arm [52]. The size of the working stroke and the motility rate are linearly related to the LCBD length [53]. The force-sensitive kinetic transitions of some myosins are coupled to lever arm rotation [40, 46, 47, 54], so myosins with longer lever arms are expected to be more force sensitive (as Archimedes said, “Give me a lever long enough and a fulcrum on which to place it, and I shall move the world”). This idea was tested directly with Myo1b splice isoforms [41]. As described above, the MYO1B gene transcript is alternatively spliced in the LCBD, resulting in isoforms with identical motor and tail domains but different length LCBDs. Single-molecule techniques were used to examine the force sensitivity of the splice isoforms and these experiments showed that force affects the lifetime of myosin-Is with longer LCBDs more strongly. In fact, there is a linear relationship between LCBD length and the distance parameter, with the force sensitivity of Myo1ba (6 IQ) > Myo1bb (5 IQ) >Myo1bc (4 IQ) (Figure 4a). These results show that alternative-splicing of the LCBD provides a mechanism for controlling force sensitivity. This is important because expression of different Myo1b isoforms could lead to different anchoring behaviors and power outputs for different force regimes. It is not currently known how changes in splicing are related to the molecular role of the myosin in vivo; however, the alternatively-spliced MYO1B transcripts have been shown to be differentially expressed in rat tissues [43, 49], and changes in the presence of transcripts were shown to accompany the epithelial to mesenchymal transitions in human cells [55]. It is possible that different isoforms with different anchoring thresholds may be expressed in response to mechanical changes in the intracellular or extracellular environments.
Figure 4.
Control of force sensitivity by the LCBD (a) The calculated attachment lifetime as a function of force for Myo1b splice isoforms containing either 1 (an artificial construct), 4, 5, or 6 IQ motifs based on the values of parameters measured using an optical trap [41]. The attachment lifetime of the different isoforms increases as a function of force for all of the isoforms examined. The extent of the effect of force on the lifetime varies between the different isoforms with the greatest force sensitivity in the myosin with the longest LCBD. The numbers next to the grey line show the attachment lifetime of the different isoforms at 1 pN of force. (b) (left) The calculated attachment lifetime of Myo1bain the presence and absence of 100 μM calcium as a function of force based on the values of parameters measured using an optical trap [42]. As can be seen, the addition of calcium causes a dramatic decrease in the attachment duration at 1 pN.. (right) Model showing the effects of calcium on Myo1b lever arm rotation. In the absence of calcium, conformational changes in the motor domain cause the LCBD to generate a 13 nm working stroke. In the presence of calcium, the effective length of the lever arm and the force sensitivity of Myo1b are both reduced due to structural changes in the LCBD domain.
Calcium regulates the activity of some vertebrate myosin-I isoforms via interaction with LCBD-bound light chains, such as calmodulin. Calmodulin is a ubiquitously expressed calcium-binding protein that binds to IQ motifs, stiffening the LCBD. Calcium binds directly to calmodulin and affects the actin-activated ATPase activities of the some myosin-I motors [30, 42, 50, 56, 57]. For example, micromolar calcium concentrations increase the unloaded cycling kinetics of Myo1b [42, 56], increasing rates of phosphate release, ADP release, and ATP binding 2–3 fold while not affecting the rate of ATP hydrolysis. The Myo1b duty ratio in the absence of load is largely unaffected by calcium, while motility is completely inhibited in vitro (see below). Calcium affects vertebrate Myo1c differently, as it increases the rate of ADP release and decreases the rate of ATP hydrolysis, resulting in ATP hydrolysis becoming the rate-limiting step for the ATPase cycle [50]. As such, the effective duty ratio of Myo1c is reduced in the presence of calcium. Nevertheless, like Myo1b, calcium inhibits actin gliding in vitro. Calcium also reduces the steady-state ATPase activity of vertebrate Myo1e due to catalytic auto-inhibition by the tail domain [58]. Thus, the effects of calcium on myosin-I unloaded kinetics appear to be quite diverse.
Aside from modulating unloaded kinetics, calcium also affects the mechanical properties of some myosin-I isoforms. As stated above, the kinetic rate constants that control the Myo1b ATPase cycle are increased with calcium, while its ability to power actin gliding in vitro are inhibited [56]. This seemingly contradictory result suggests that calcium uncouples myosin ATPase kinetics from LCBD rotation. In Myo1b, calcium binding to the calmodulin closest to the motor domain causes a conformational change in the calmodulin, altering the ability of the LCBD to act as a lever arm. Recent experiments with Myo1b showed a reduction in the size of the working stroke in the presence of 9 μM free calcium [42]. Moreover, the duration of actin-binding events in the absence of force was shortened in the presence of calcium, consistent with the kinetic experiments showing acceleration of Myo1b kinetics with calcium. Calcium also causes a dramatic reduction in force sensitivity of Myo1b (Figure 4b). In the absence of calcium, 1 pN of resisting force leads to an average attachment lifetime of > 50 s for Myo1ba, while in the presence of calcium, the average attachment lifetime at the same force is only 3 s. Thus calcium regulation of tension sensing provides a potential mechanism for regulating long-lived binding events. For example, if Myo1b is pulling a membrane tether or tethering a vesicle, it is possible that fluctuations in calcium near the cell periphery during a calcium transient could cause the release of the tethered membrane.
The mechanical response of Myo1c to calcium appears to be quite different from Myo1b. In particular, calcium has been reported to cause an increase in the size of the Myo1c working stroke [30]. The cellular significance of this result is not clear, since as cellular calcium concentrations increase, free calmodulin concentrations decrease to a point where motility would be expected to be inhibited [59]. Nevertheless, the mechanism for these isoform differences is not clear, and further studies are necessary to determine how calcium affects other myosin-I isoforms. It is worth noting that in some myosin-I isoforms, the ADP release rate is affected by the free magnesium concentration due to changes of nucleotide affinity at the active site, such that motility rates may be affected by changes in intracellular magnesium concentration [33].
Motor Domain and LCBD Control of Localization
Given the presumed role of the myosin-I tail as the cargo-binding region, it was surprising to learn that the proper subcellular localization of some myosin-I isoforms requires not only the tail domain, but also the motor domain [60]. In fact, actin helps to concentrate myosin-I to specific regions within the cell while tropomyosin decorated actin filaments appear to exclude myosin-I binding in vivo [60, 61] (Figure 5). Recent work has shown that in yeast, myosin-I is preferentially localized to the cell cortex where there is no tropomyosin, while myosin-V localizes to tropomyosin containing actin cables and myosin-II localizes to the stress fibers and actin cables [61]. In fact, in vitro experiments have shown that yeast myosin-V (Myo2p) is a processive motor only on tropomyosin-decorated actin filaments [62]. Thus, the interaction of myosin with tropomyosin decorated actin appears to depend on the specific myosin isoform. This result is important since it demonstrates how myosins (all of which bind actin) can be segregated to particular actin pools in the cell based on the motor domain.
Figure 5.
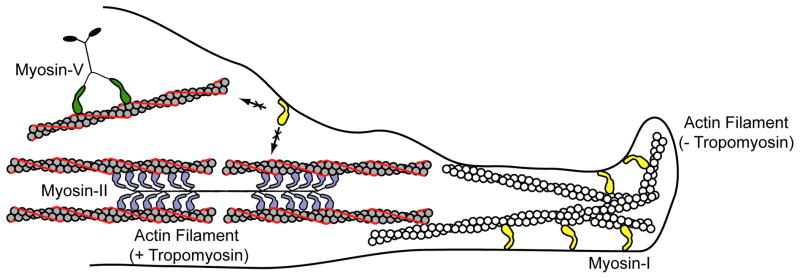
Cartoon showing how tropomyosin causes segregation of different myosins to different actin populations. Myosin-I is unable to interact with tropomyosin decorated actin filaments, so it localizes to the dynamic, tropomyosin-free actin filaments at the cell periphery. Tropomyosin increases the binding of myosin-V, causing myosin-V to segregate to tropomyosin-decorated actin cables. Myosin-II forms filamentous structures that localize to tropomyosin decorated actin filaments in stress fibers and actin cables. Tropomyosin thus provides a mechanism for regulating the cellular localization of different myosins. This regulation is likely mediated by kinetic tuning of the myosin-I motor domain.
Interestingly, the ability of tropomyosin to regulate myosin-I appears to depend on the tropomyosin isoform. Indirect in vivo evidence suggests that the high molecular weight tropomyosin isoform, Tm3, excludes myosin-I binding, whereas the low molecular weight isoform, Tm5, has little effect on myosin-I localization [63]. Moreover, while nonmuscle Xenopus tropomyosin Tm4 stimulates the ATPase activity of cardiac muscle myosin-II, it inhibits the activity of myosin-I in vitro [64]. These results suggest that different myosin isoforms are kinetically tuned to respond to different tropomyosins. It was recently suggested that a long surface-loop of myosin-I (loop 4) located at the actin-binding site might sterically clash with tropomyosin, preventing strong-binding to actin [65]. This suggestion was tested by experiments in which loop 4 of myosin-I was exchanged with the shorter loop 4 from Dictyostelium myosin-II. This chimera could interact with tropomyosin decorated actin in the in vitro motility assay (although it did not localize to tropomyosin decorated filaments in vivo) [66]. Despite these studies, the mechanism by which tropomyosin regulates myosin-I activity is still not clear. It is possible that tropomyosin affects the kinetic transition that regulates the ability of myosin to bind strongly the actin [67]. Moreover, different tropomyosin isoforms appear to lie in different azimuthal positions along the actin filament depending on both the tropomyosin and actin isoform, providing another possible explanation for why myosin-I may interact differently with different tropomyosins. Interestingly, the TEDS site is in close proximity to both loop 4 and the tropomyosin binding interface on actin. Similar to TEDS phosphorylation [68], tropomyosin regulates the weak to strong transition in striated muscle. It is intriguing to speculate that these mechanisms are linked, wherein the TEDS loop might be sterically regulated by tropomyosin.
There have been some cell biology experiments showing that TEDS phosphorylation is important in vivo [39, 69]. It has been shown that TEDS phosphorylation is necessary for proper cellular localization of myosin-I in Schizosaccharomyces pombe [39] and Candida albicans [70], but not in Dictyostelium [71]. When active, yeast myosin-I has been associated with both stimulating Arp2/3-mediated polymerization of cortical actin patches and regulating fluid-phase endocytosis. Moreover, TEDS phosphorylation appears to be necessary for regulating membrane internalization during endocytosis [39, 72]. Future studies are needed to understand these mechanisms and how they regulate cellular localization better.
Interestingly, the LCBD domain may also play a role in directing the cellular localization of myosin-I. The LCBD appears to play a role in the subcellular localization of Myo1c in some cell types [73], indicating that light chains may have a role in myosin-I targeting. Myosin-I isoforms from lower eukaryotes co-purify with calmodulin-like proteins (e.g., [74, 75]) rather than calmodulin. Additionally, vertebrate Myo1c has been shown to bind calcium-binding protein 1 (CaBP1) and calcium- and integrin-binding-protein-1 (CIB1) [76]. In the case of CaBP1 and CIB1, these proteins are myristoylated, providing an additional potential mechanism for localizing Myo1c in a calcium-dependent manner. It is also worth noting that the heavy chain of vertebrate Myo1c can be phosphorylated at a residue near the calmodulin binding site, providing another possible mechanism for regulating light chain binding [4]. Whether the different light chains affect the mechanics and/or calcium-binding properties of the LCBD is still an outstanding question.
Conclusions
Our understanding of myosin-I regulation has grown dramatically in the past few years. Despite structural and kinetic similarities between myosin-I isoforms, there is great diversity of function imparted by the motor and LCBDs and further characterization of myosin-I family members may reveal additional regulatory mechanisms (Box 2). How this diversity is manifested in vivo is not clear. Experiments in both lower [77] and higher eukaryotes [78] have shown that deletion of one myosin-I isoform can lead to redistribution of other myosin-Is to compensate for the loss. While this redistribution does not completely rescue the wild type phenotype, it strongly suggests that certain myosin-Is are able to compensate for other isoforms. Investigation of the mechanical and kinetic properties of other myosin-I family members will likely shed light on these results. Future studies will surely help to illuminate our understanding of myosin-I, possibly illuminating the evolutionary imperatives that resulted in the retention of 8 distinct myosin-I family members in humans.
Box 2. Outstanding Questions.
New experiments are providing a clearer understanding of the cellular roles of myosin-I isoforms, but what are the molecular roles of myosin-I in these cellular processes?
Why do vertebrates require eight myosin-I genes? What are the cellular consequences of alternative splicing? What are the structural elements responsible for the biochemical and mechanical diversity of myosin-I isoforms?
What is the relationship of myosin-I to the regulation of actin assembly dynamics and filament organization? What are the regulatory and/or structural roles of membrane binding in these processes?
What is the relationship between myosin-I localization and the differential sorting of tropomyosin and actin isoforms? Do these proteins sort myosins, or do myosins have a role in directing localization of these proteins? Also, how are the different myosin-I isoforms sorted in the cell?
What are the myosin-I binding partners and how do these proteins, lipids, and nucleic acids affect myosin-I function and vice versa?
How are myosin-I mechanics, localization, and function affected by membrane binding and the physical state of the membrane?
How do myosin-I isoforms interact with other members of the myosin family and the microtubule motors dynein and kinesin?
Acknowledgments
The authors would like to thank Betsy Buechler, Elizabeth Feeser, Serapion Pyrpassopoulos, Abbey Weith, and Allison Zajac for helpful comments. This work was supported by NIGMS grants PO1 GM087253 (E.M.O.) and F32 GM097889 (M.J.G.).
Footnotes
Conflicts of Interest
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berg JS, et al. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McConnell RE, et al. The enterocyte microvillus is a vesicle-generating organelle. J Cell Biol. 2009;185:1285–1298. doi: 10.1083/jcb.200902147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng J, et al. Myosin 1E coordinates actin assembly and cargo trafficking during clathrin-mediated endocytosis. Molecular biology of the cell. 2012;23:2891–2904. doi: 10.1091/mbc.E11-04-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yip MF, et al. CaMKII-mediated phosphorylation of the myosin motor Myo1c is required for insulin-stimulated GLUT4 translocation in adipocytes. Cell metabolism. 2008;8:384–398. doi: 10.1016/j.cmet.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Brandstaetter H, et al. Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. J Cell Sci. 2012 doi: 10.1242/jcs.097212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyrpassopoulos S, et al. Membrane-Bound Myo1c Powers Asymmetric Motility of Actin Filaments. Current biology : CB. 2012 doi: 10.1016/j.cub.2012.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almeida CG, et al. Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat Cell Biol. 2011;13:779–789. doi: 10.1038/ncb2262. [DOI] [PubMed] [Google Scholar]
- 8.Shifrin DA, Jr, et al. Enterocyte microvillus-derived vesicles detoxify bacterial products and regulate epithelial-microbial interactions. Current biology : CB. 2012;22:627–631. doi: 10.1016/j.cub.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McConnell RE, Tyska MJ. Myosin-1a powers the sliding of apical membrane along microvillar actin bundles. J Cell Biol. 2007;177:671–681. doi: 10.1083/jcb.200701144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batters C, et al. A model of stereocilia adaptation based on single molecule mechanical studies of myosin I. Philos Trans R Soc Lond B Biol Sci. 2004;359:1895–1905. doi: 10.1098/rstb.2004.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olety B, et al. Myosin 1G (Myo1G) is a haematopoietic specific myosin that localises to the plasma membrane and regulates cell elasticity. FEBS letters. 2010;584:493–499. doi: 10.1016/j.febslet.2009.11.096. [DOI] [PubMed] [Google Scholar]
- 12.Sirotkin V, et al. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. The Journal of cell biology. 2005;170:637–648. doi: 10.1083/jcb.200502053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan Y, et al. Myo1c facilitates G-actin transport to the leading edge of migrating endothelial cells. The Journal of cell biology. 2012;198:47–55. doi: 10.1083/jcb.201111088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokac AM, et al. Myosin-1c couples assembling actin to membranes to drive compensatory endocytosis. Dev Cell. 2006;11:629–640. doi: 10.1016/j.devcel.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagan GN, et al. A Rictor-Myo1c complex participates in dynamic cortical actin events in 3T3-L1 adipocytes. Mol Cell Biol. 2008;28:4215–4226. doi: 10.1128/MCB.00867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pestic-Dragovich L, et al. A myosin I isoform in the nucleus. Science. 2000;290:337–341. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 17.Tyska MJ, et al. Myosin-1a is critical for normal brush border structure and composition. Mol Biol Cell. 2005;16:2443–2457. doi: 10.1091/mbc.E04-12-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bose A, et al. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature. 2002;420:821–824. doi: 10.1038/nature01246. [DOI] [PubMed] [Google Scholar]
- 19.Chen XW, et al. Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev Cell. 2007;13:391–404. doi: 10.1016/j.devcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Krendel M, et al. Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett. 2007;581:644–650. doi: 10.1016/j.febslet.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korn ED. Coevolution of head, neck, and tail domains of myosin heavy chains. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12559–12564. doi: 10.1073/pnas.230441597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hokanson DE, et al. Myo1c Binds Phosphoinositides through a Putative Pleckstrin Homology Domain. Mol Biol Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hokanson DE, Ostap EM. Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc Natl Acad Sci U S A. 2006;103:3118–3123. doi: 10.1073/pnas.0505685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Mezgueldi M, et al. The kinetic mechanism of Myo1e (human myosin-IC) J Biol Chem. 2002;277:21514–21521. doi: 10.1074/jbc.M200713200. [DOI] [PubMed] [Google Scholar]
- 25.Jontes JD, et al. Kinetic characterization of brush border myosin-I ATPase. Proc Natl Acad Sci U S A. 1997;94:14332–14337. doi: 10.1073/pnas.94.26.14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis JH, et al. Temperature dependence of nucleotide association and kinetic characterization of myo1b. Biochemistry. 2006;45:11589–11597. doi: 10.1021/bi0611917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin T, et al. Biochemical and motile properties of Myo1b splice isoforms. J Biol Chem. 2005;280:41562–41567. doi: 10.1074/jbc.M508653200. [DOI] [PubMed] [Google Scholar]
- 28.Ostap EM, et al. Mechanism of regulation of Acanthamoeba myosin-IC by heavy-chain phosphorylation. Biochemistry. 2002;41:12450–12456. doi: 10.1021/bi0262193. [DOI] [PubMed] [Google Scholar]
- 29.Ostap EM, Pollard TD. Biochemical kinetic characterization of the Acanthamoeba myosin-I ATPase. J Cell Biol. 1996;132:1053–1060. doi: 10.1083/jcb.132.6.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batters C, et al. Myo1c is designed for the adaptation response in the inner ear. Embo J. 2004;23:1433–1440. doi: 10.1038/sj.emboj.7600169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coluccio LM, Geeves MA. Transient kinetic analysis of the 130-kDa myosin I (MYR-1 gene product) from rat liver. A myosin I designed for maintenance of tension? J Biol Chem. 1999;274:21575–21580. doi: 10.1074/jbc.274.31.21575. [DOI] [PubMed] [Google Scholar]
- 32.Durrwang U, et al. Dictyostelium myosin-IE is a fast molecular motor involved in phagocytosis. J Cell Sci. 2006;119:550–558. doi: 10.1242/jcs.02774. [DOI] [PubMed] [Google Scholar]
- 33.Tsiavaliaris G, et al. Mechanism, regulation, and functional properties of Dictyostelium myosin-1B. The Journal of biological chemistry. 2008;283:4520–4527. doi: 10.1074/jbc.M708113200. [DOI] [PubMed] [Google Scholar]
- 34.Greenberg MJ, et al. Myosin IC generates power over a range of loads via a new tension-sensing mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2433–2440. doi: 10.1073/pnas.1207811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mooseker MS, Coleman TR. The 110-kD protein-calmodulin complex of the intestinal microvillus (brush border myosin I) is a mechanoenzyme. J Cell Biol. 1989;108:2395–2400. doi: 10.1083/jcb.108.6.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 37.Bloemink MJ, Geeves MA. Shaking the myosin family tree: biochemical kinetics defines four types of myosin motor. Semin Cell Dev Biol. 2011;22:961–967. doi: 10.1016/j.semcdb.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bement WM, Mooseker MS. TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil Cytoskeleton. 1995;31:87–92. doi: 10.1002/cm.970310202. [DOI] [PubMed] [Google Scholar]
- 39.Attanapola SL, et al. Ste20-kinase-dependent TEDS-site phosphorylation modulates the dynamic localisation and endocytic function of the fission yeast class I myosin, Myo1. Journal of cell science. 2009;122:3856–3861. doi: 10.1242/jcs.053959. [DOI] [PubMed] [Google Scholar]
- 40.Laakso JM, et al. Myosin I can act as a molecular force sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laakso JM, et al. Control of myosin-I force sensing by alternative splicing. Proc Natl Acad Sci U S A. 2010;107:698–702. doi: 10.1073/pnas.0911426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis JH, et al. Calcium regulation of myosin-I tension sensing. Biophysical journal. 2012;102 doi: 10.1016/j.bpj.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruppert C, et al. Identification, characterization and cloning of myr 1, a mammalian myosin-I. J Cell Biol. 1993;120:1393–1403. doi: 10.1083/jcb.120.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takagi Y, et al. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys J. 2006;90:1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 46.Veigel C, et al. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 47.Veigel C, et al. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 48.Lin T, et al. A hearing loss-associated myo1c mutation (R156W) decreases the myosin duty ratio and force sensitivity. Biochemistry. 2011;50:1831–1838. doi: 10.1021/bi1016777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruppert C, et al. Localization of the rat myosin I molecules myr 1 and myr 2 and in vivo targeting of their tail domains. J Cell Sci. 1995;108 ( Pt 12):3775–3786. doi: 10.1242/jcs.108.12.3775. [DOI] [PubMed] [Google Scholar]
- 50.Adamek N, et al. Calcium sensitivity of the cross-bridge cycle of Myo1c, the adaptation motor in the inner ear. Proc Natl Acad Sci U S A. 2008;105:5710–5715. doi: 10.1073/pnas.0710520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holt JR, et al. A chemical-genetic strategy implicates myosin-1c in adaptation by hair cells. Cell. 2002;108:371–381. doi: 10.1016/s0092-8674(02)00629-3. [DOI] [PubMed] [Google Scholar]
- 52.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 53.Tyska MJ, Warshaw DM. The myosin power stroke. Cell Motil Cytoskeleton. 2002;51:1–15. doi: 10.1002/cm.10014. [DOI] [PubMed] [Google Scholar]
- 54.Veigel C, et al. The motor protein myosin-I produces its working stroke in two steps. Nature. 1999;398:530–533. doi: 10.1038/19104. [DOI] [PubMed] [Google Scholar]
- 55.Warzecha CC, et al. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA biology. 2009;6:546–562. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perreault-Micale C, et al. Truncation of a mammalian myosin I results in loss of Ca2+-sensitive motility. J Biol Chem. 2000;275:21618–21623. doi: 10.1074/jbc.M000363200. [DOI] [PubMed] [Google Scholar]
- 57.Collins K, et al. Calmodulin dissociation regulates brush border myosin I (110-kD-calmodulin) mechanochemical activity in vitro. J Cell Biol. 1990;110:1137–1147. doi: 10.1083/jcb.110.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stoffler HE, Bahler M. The ATPase activity of Myr3, a rat myosin I, is allosterically inhibited by its own tail domain and by Ca2+ binding to its light chain calmodulin. J Biol Chem. 1998;273:14605–14611. doi: 10.1074/jbc.273.23.14605. [DOI] [PubMed] [Google Scholar]
- 59.Black DJ, et al. Monitoring the total available calmodulin concentration in intact cells over the physiological range in free Ca2+ Cell Calcium. 2004;35:415–425. doi: 10.1016/j.ceca.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Tang N, Ostap EM. Motor domain-dependent localization of myo1b (myr-1) Curr Biol. 2001;11:1131–1135. doi: 10.1016/s0960-9822(01)00320-7. [DOI] [PubMed] [Google Scholar]
- 61.Stark BC, et al. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Molecular biology of the cell. 2010;21:989–1000. doi: 10.1091/mbc.E09-10-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hodges AR, et al. Tropomyosin Is Essential for Processive Movement of a Class V Myosin from Budding Yeast. Current biology : CB. 2012 doi: 10.1016/j.cub.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pelham RJ, Jr, et al. A high molecular mass non-muscle tropomyosin isoform stimulates retrograde organelle transport. J Cell Sci. 1996;109 ( Pt 5):981–989. doi: 10.1242/jcs.109.5.981. [DOI] [PubMed] [Google Scholar]
- 64.Fanning AS, et al. Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil Cytoskeleton. 1994;29:29–45. doi: 10.1002/cm.970290104. [DOI] [PubMed] [Google Scholar]
- 65.Kollmar M, et al. Crystal structure of the motor domain of a class-I myosin. EMBO J. 2002;21:2517–2525. doi: 10.1093/emboj/21.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lieto-Trivedi A, et al. Myosin surface loop 4 modulates inhibition of actomyosin 1b ATPase activity by tropomyosin. Biochemistry. 2007;46:2779–2786. doi: 10.1021/bi602439f. [DOI] [PubMed] [Google Scholar]
- 67.Ostap EM. Tropomyosins as discriminators of myosin function. Adv Exp Med Biol. 2008;644:273–282. doi: 10.1007/978-0-387-85766-4_20. [DOI] [PubMed] [Google Scholar]
- 68.McKillop DF, Geeves MA. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baines IC, et al. Quantification and localization of phosphorylated myosin I isoforms in Acanthamoeba castellanii. The Journal of cell biology. 1995;130:591–603. doi: 10.1083/jcb.130.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oberholzer U, et al. Myosin I is required for hypha formation in Candida albicans. Eukaryotic cell. 2002;1:213–228. doi: 10.1128/EC.1.2.213-228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Novak KD, Titus MA. The myosin I SH3 domain and TEDS rule phosphorylation site are required for in vivo function. Mol Biol Cell. 1998;9:75–88. doi: 10.1091/mbc.9.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grosshans BL, et al. TEDS site phosphorylation of the yeast myosins I is required for ligand-induced but not for constitutive endocytosis of the G protein-coupled receptor Ste2p. The Journal of biological chemistry. 2006;281:11104–11114. doi: 10.1074/jbc.M508933200. [DOI] [PubMed] [Google Scholar]
- 73.Cyr JL, et al. Myosin-1c interacts with hair-cell receptors through its calmodulin-binding IQ domains. J Neurosci. 2002;22:2487–2495. doi: 10.1523/JNEUROSCI.22-07-02487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sammons MR, et al. A calmodulin-related light chain from fission yeast that functions with myosin-I and PI 4-kinase. Journal of cell science. 2011;124:2466–2477. doi: 10.1242/jcs.067850. [DOI] [PubMed] [Google Scholar]
- 75.Crawley SW, et al. Identification and characterization of an 8-kDa light chain associated with Dictyostelium discoideum MyoB, a class I myosin. The Journal of biological chemistry. 2006;281:6307–6315. doi: 10.1074/jbc.M508670200. [DOI] [PubMed] [Google Scholar]
- 76.Tang N, et al. CIB1 and CaBP1 bind to the myo1c regulatory domain. J Muscle Res Cell Motil. 2007;28:285–291. doi: 10.1007/s10974-007-9124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ostap EM, Pollard TD. Overlapping functions of myosin-I isoforms? J Cell Biol. 1996;133:221–224. doi: 10.1083/jcb.133.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benesh AE, et al. Differential localization and dynamics of class I myosins in the enterocyte microvillus. Molecular biology of the cell. 2010;21:970–978. doi: 10.1091/mbc.E09-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McConnell RE, Tyska MJ. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends Cell Biol. 2010;20:418–426. doi: 10.1016/j.tcb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doberstein SK, Pollard TD. Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoeba myosin IC. J Cell Biol. 1992;117:1241–1249. doi: 10.1083/jcb.117.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feeser EA, et al. Myo1e binds anionic phospholipids with high affinity. Biochemistry. 2010;49:9353–9360. doi: 10.1021/bi1012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mazerik JN, Tyska MJ. Myosin-1A Targets to Microvilli Using Multiple Membrane Binding Motifs in the Tail Homology 1 (TH1) Domain. J Biol Chem. 2012;287:13104–13115. doi: 10.1074/jbc.M111.336313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McKenna JM, Ostap EM. Kinetics of the interaction of myo1c with phosphoinositides. J Biol Chem. 2009;284:28650–28659. doi: 10.1074/jbc.M109.049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Komaba S, Coluccio LM. Localization of myosin 1b to actin protrusions requires phosphoinositide binding. The Journal of biological chemistry. 2010;285:27686–27693. doi: 10.1074/jbc.M109.087270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Patino-Lopez G, et al. Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent pleckstrin homology (PH) domain (Myo1PH) J Biol Chem. 2010;285:8675–8686. doi: 10.1074/jbc.M109.086959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brzeska H, et al. Acanthamoeba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge. J Biol Chem. 2008;283:32014–32023. doi: 10.1074/jbc.M804828200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tyska MJ, Mooseker MS. A role for myosin-1A in the localization of a brush border disaccharidase. J Cell Biol. 2004;165:395–405. doi: 10.1083/jcb.200310031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakamori Y, et al. Myosin motor Myo1c and its receptor NEMO/IKK-gamma promote TNF-alpha-induced serine307 phosphorylation of IRS-1. J Cell Biol. 2006;173:665–671. doi: 10.1083/jcb.200601065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arif E, et al. Motor protein Myo1c is a podocyte protein that facilitates the transport of slit diaphragm protein Neph1 to the podocyte membrane. Molecular and cellular biology. 2011;31:2134–2150. doi: 10.1128/MCB.05051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Etournay R, et al. PHR1, an integral membrane protein of the inner ear sensory cells, directly interacts with myosin 1c and myosin VIIa. J Cell Sci. 2005;118:2891–2899. doi: 10.1242/jcs.02424. [DOI] [PubMed] [Google Scholar]
- 91.Rump A, et al. Myosin-1C associates with microtubules and stabilizes the mitotic spindle during cell division. Journal of cell science. 2011;124:2521–2528. doi: 10.1242/jcs.084335. [DOI] [PubMed] [Google Scholar]
- 92.Xu P, et al. The myosin-I-binding protein Acan125 binds the SH3 domain and belongs to the superfamily of leucine-rich repeat proteins. Proc Natl Acad Sci U S A. 1997;94:3685–3690. doi: 10.1073/pnas.94.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jung G, et al. The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;153:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jontes JD, Milligan RA. Brush border myosin-I structure and ADP-dependent conformational changes revealed by cryoelectron microscopy and image analysis. J Cell Biol. 1997;139:683–693. doi: 10.1083/jcb.139.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jontes JD, et al. Three-dimensional structure of Acanthamoeba castellanii myosin-IB (MIB) determined by cryoelectron microscopy of decorated actin filaments. J Cell Biol. 1998;141:155–162. doi: 10.1083/jcb.141.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]