Abstract
Background
Although tobacco mosaic virus (TMV) coat protein (CP) has been isolated from virus particles and its crystals have grown in ammonium sulfate buffers for many years, to date, no one has reported on the crystallization of recombinant TMV-CP connecting peptides expressed in E. coli.
Methods
In the present papers genetically engineered TMV-CP was expressed, into which hexahistidine (His) tags or glutathione-S-transferase (GST) tags were incorporated. Considering that GST-tags are long peptides and His-tags are short peptides, an attempt was made to grow crystals of TMV-CP cleaved GST-tags (WT-TMV-CP32) and TMV-CP incorporated His-tags (WT-His-TMV-CP12) simultaneously in ammonium sulfate buffers and commercial crystallization reagents. It was found that the 20S disk form of WT-TMV-CP32 and WT-His-TMV-CP12 did not form high resolution crystals by using various crystallization buffers and commercial crystallization reagents. Subsequently, a new experimental method was adopted in which a range of truncated TMV-CP was constructed by removing several amino acids from the N- or the C-terminal, and high resolution crystals were grown in ammonium sulfate buffers and commercial crystallization reagents.
Results
The new crystallization method was developed and 3.0 Å resolution macromolecular crystal was thereby obtained by removing four amino acids at the C-terminal of His-TMV-CP and connecting six His-tags at the N-terminal of His-TMV-CP (TR-His-TMV-CP19). The Four-layer aggregate disk structure of TR-His-TMV-CP19 was solved. This phenomenon showed that peptides at the C-terminus hindered the growth of high resolution crystals and the peptides interactions at the N-terminus were attributed to the quality of TMV-CP crystals.
Conclusion
A 3.0 Å resolution macromolecular crystal of TR-His-TMV-CP19 was obtained and the corresponding structure was solved by removing four amino acids at the C-terminus of TMV-CP and connecting His-tags at the N-terminus of TMV-CP. It indicated that short peptides influenced the resolution of TMV-CP crystals.
Keywords: GST-tags, His-tags, Peptides, Disk form, Protein crystals, TMV-CP, Truncated protein
Background
Tobacco mosaic virus (TMV) has a rod-like appearance and consists of a single, positive strand RNA of 6395 nucleotides encapsulated in a helical virion by approximately 2130 identical coat protein (CP) subunits [1-7]. CP consists of 158 amino acids that were assembled into four main alpha-helices joined by a prominent loop proximal to the axis of the virion [8-16]. TMV-CP played an important role in the self-assembly of TMV through an initial RNA recognition reaction that triggers the assembly, it was believed to be necessary for virus assembly initiation and elongation [8,17-24]. The biological physical properties of TMV-CP were often determined by the structure of TMV. The TMV structure reported in 1986 was studied based on an electron density map at 3.6 Å resolution by X-ray fiber diffraction [9], and then this structure of the complete virus was determined at 2.9 Å resolution by X-ray fiber diffraction methods [25]. TMV-CP assembly systems, consisting of 34-subunit aggregate of TMV-CP crystallized as a dimer of bilayer disks having 17 subunits per layer, were crystallized and solved at 2.8 Å resolution [2]. The crystalline structure of the Four-layer aggregate of TMV-CP was determined at 2.4 Å resolution by using X-ray diffraction from crystals maintained at cryogenic temperatures. This structure emphasized the importance of water in biological macromolecular assemblies [22,23]. The circular permutants of TMV-CP were crystallized and solved by molecular replacement at 3.0 Å resolution by using X-ray diffraction [26]. The structure of TMV was also obtained by using high resolution transmission electron microscopy [27-29].
TMV-CP was usually propagated and isolated from Nicotiana tabacum (N. tabacum) infected by TMV, and TMV-CP existed as a number of aggregates, depending on PH, ionic strength, temperature, protein concentration, and other factors [5,12,14-21]. At 0.1 mol/L ionic strength orthophosphate solution and pH equal to or greater than 8.0, TMV-CP existed as protein A or 4S protein (a dynamic equilibrium between monomers, trimers, and pentamers of TMV-CP). At 0.1 mol/L ionic strength orthophosphate solution and pH near 7.0, TMV-CP was transformed into the 20S aggregate form (disks consisting of 34 monomers, also named as the 20S structure) with an admixture of 4S protein [30-33]. At 0.1 mol/L ionic strength orthophosphate solution and pH equal to or less than 6.0, TMV-CP was completely transformed into macroscopic aggregate. The 34-subunit disk was virtually crystallized at 1.0 mol/L ionic strength solution and PH near 8.0 [2,3,12,17,19-21,30,31]. The crystals of TMV-CP consisting of Four-layer aggregates were usually obtained at 3.5–14 mg/mL protein concentration (10 mmol/L orthophosphate, pH 7.2) and in crystallization pool solutions consist of 0.2–0.3 mol/L ammonium sulfate and 0.1 mol/L Tris at pH 8.0 for 3 wk to 4 wk at room temperature [23]. The crystals were obtained after been equilibrated overnight against a crystallization pool solution consisting of 0.3 mol/L ammonium sulfate and 0.1 mol/L Tris at pH 8.0 by microdialysis method [23,31-36]. The aformentioned method of obtaining TMV-CP from TMV will take investigators a lot of work and time, and the TMV-CP structures obtained from TMV were aggregate forms [32-36], they could not be easily modified. In this sense, by employing the method of genetically engineered, recombination TMV-CP was expressed, purified and crystallized [37-43].
In the present investigation, the genetically engineered structure of TMV-CP was the concern: a series of the recombinant expression vectors contained TMV-CP genes were constructed and transformed into E.coli, and the recombinant protein of TMV-CP were expressed, and 3.0 Å resolution TR-His-TMV-CP19 (incorporated His-tags at the N-terminus of TMV-CP and truncated four amino acids at the C-terminus of TMV-CP) macromolecular crystals were obtained.
Results and Discussions
Identification to Recombinant Vectors
TMV-RNA has been isolated from TMV particles (propagated in N. tabacum K326) and reverse transcribed into cDNA by primercDNA and reverse transcriptase (TaKaRa). The genetic fragment of wild type TMV-CP (WT-GST-TMV-CP32, with the restriction enzymes of BamH I/Xho I; WT-His-TMV-CP12, with the restriction enzymes of Nde I/Xho I; ) were amplified by using cDNA as template. The following series of genetic fragments, four amino acids truncated at the C-terminus of WT-His-TMV-CP12, (TR-His-TMV-CP19), three amino acids truncated at the N-terminus of WT-His-TMV-CP12 and four amino acids truncated at the C-terminus of WT-His-TMV-CP12 (TR-His-TMV-CP62), three amino acids truncated at the N-terminus of WT-His-TMV-CP12, and five amino acids truncated at the C-terminus of WT-His-TMV-CP12 (TR-His-TMV-CP68), were also amplified by PCR. Compared with the DNA marker, all the PCR products migrated as expected at approximately 500 bp, and the PCR products were marked as the WT-GST-TMV-CP32 (Figure1A), WT-His-TMV-CP12 (Figure 1B), TR-His-TMV-CP19 (Figure1C), TR-His-TMV-CP62, and TR-His -TMV-CP68 separately.
Figure 1.
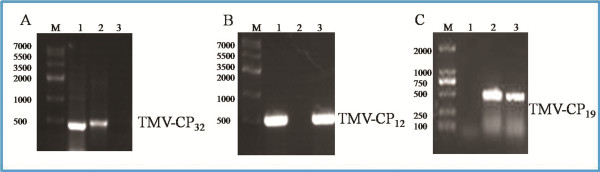
TMV-CP DNA fragments (1% agarose gel). (A) The whole TMV-CP fragments with BamH I and Xho I restriction enzyme cutting sites which have been cloned in PGEX-6P-1. As shown in lane 1, amplified PCR product ran at approximately 500 bp compared with the DNA marker (lane M). Lane 2 is a positive control with DNA template. Lane 3 is a negative control without DNA template. (B) The whole TMV-CP fragments with Nde I and Xho I restriction enzyme cutting sites that have been cloned in pET28a. As shown in lane 1, amplified PCR product ran at approximately 500 bp compared with the DNA marker (lane M). Lane 2 is a negative control without DNA template. Lane 3 is a positive control with DNA template. (C) The truncation of four amino acids from the C-terminus of TMV-CP fragments with Nde I and Xho I restriction enzyme cutting sites that have been cloned in pET28a. Lane 1 is a negative control without DNA template, whereas lane 2 is a positive control with DNA template. Lane 3 is the amplified PCR product that ran at approximately 500 bp compared with the DNA marker (lane M).
The corresponding clones were sequenced by ABI Automatic DNA Sequence Machine, and the correct sequences were obtained and aligned (Figure 2). The DNA sequences of WT-GST-TMV-CP32, WT-His-TMV-CP12, TR-His-TMV-CP19, TR-His-TMV-CP62, and TR-His-TMV-CP68 were similar that of WT-TMV-CP (isolated from TMV), except for the presence of short peptides incorporated at the N-terminal of TMV-CP in their DNA sequences. These correct proteins were successfully cloned to the expression host, E.coli BL21(DE3)-RIL (TakaRa), for protein expression.
Figure 2.

Alignment of the TMV-CP Sequences, the identical residues were marked below by an asterisk.
Confirmation of the Proteins Expressed and Purified by Gel Filtration
Expressed proteins of WT-GST-TMV-CP32, WT-His-TMV-CP12, TR-His-TMV-CP19, TR-His-TMV-CP62, and TR-His-TMV-CP68 were initially assayed by Coomassie brilliant blue method in a small scale experiment in which the final volume was 10 mL(Figure 3A).The protein products including the whole cell lysates and the target proteins were confirmed by 12% sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE). The molecular mass of WT-GST-TMV-CP32 was test by migration at approximately 43.5 kDa (Figure 3B); the molecular mass of WT-His-TMV-CP12 (Figure 3C), TR-His-TMV-CP19 (Figure 3D), TR-His-TMV-CP62, and TR-His-TMV-CP68 were test by in the same migration approximately at 18.5 kDa. The molecular mass of WT-TMV-CP32 (WT-GST-TMV-CP32 cleaved GST-tags) was test by migration at approximately 17.5 kDa (Figure 3E), and the molecular mass of TR-His-TMV-CP12 (Figure 3F) and TR-His-TMV-CP19 (Figure 3G) were test by in the same migration at approximately 18.5 kDa.
Figure 3.
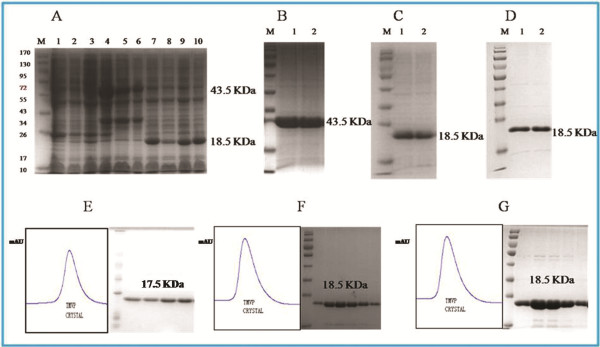
The generation of expression and purification of target protein that was analyzed using 12% SDS-PAGE. (A) Protein molecular weight standards are shown in lane 1. Numbers on the left are the MW of the standards in kDa. Lanes 1, 2, and 3 show the controls without induction by IPTG that were 20 μL aliquots of whole cell lysates from 10 mL PGEX-6P-1-WT-GST-TMV-CP32-BL21(DE3)-RIL, pET28a-His-TMV-CP12-BL21(DE3)-RIL, and pET28a-TR-His-TMV-CP19-BL21(DE3)-RIL cultures, respectively. Lanes 4, 5, and 6 correspond to the cultures after expression that were from 20 μL aliquots of whole cell lysates from 1 L PGEX-6P-1-WT-GST-TMV-CP32-BL21(DE3)-RIL cultures with IPTG. A new protein band at 43.5 kDa corresponds to the target protein GST-TMV-CP. Lanes 7 and 8 correspond to the cultures after expression that were from 20 μL aliquots of whole cell lysates from 1 L pET28a-TR-His-TMV-CP12-BL21(DE3)-RIL and pET28a-TR-His-TMV-CP68-BL21(DE3)-RIL culture with IPTG, respectively. A new protein band at position 18.5 kDa corresponds to the target protein. Lanes 9 and 10 correspond to the cultures after expression that were from 20 μL aliquots of whole cell lysates from 1 L pET28a-TR-His-TMV-CP62-BL21(DE3)-RIL and pET28a-TR-His-TMV-CP19-BL21(DE3)-RIL cultures with IPTG, respectively. A new protein band at position 18.5 kDa corresponds to the target protein. (B) WT-GST-TMV-CP32 protein is shown in lanes 1 and 2 purified using nickel-nitrilotriacetic acid (Ni-NTA) column. (C) WT-His-TMV-CP12 protein is shown in Lanes 1 and 2 purified using Ni-NTA column. (D) TR-His-TMV-CP19 protein is shown in Lanes 1 and 2 purified using Ni-NTA column. (E) WT-TMV-CP32 protein cleaved GST-tags is shown in 12% SDS-PAGE gel filtration purified using HiLoad 16/60 Superdex 200 pg column. (F) WT-His-MV-CP12 protein is shown in 12% SDS-PAGE gel filtration purified using HiLoad 16/60 Superdex 200 pg column. (G) TR-His-MV-CP19 protein is shown in 12% SDS-PAGE gel filtration purified using HiLoad 16/60 Superdex 200 pg column.
Disk state of TR-His-TMV-CP19 in Solution
In the size exclusion chromatography, the retention volume of N-His-TMV-CP19 protein under 20 mM Sodium Phosphate buffer and 100 mM Sodium Chloride solution (PH 8.0) was the oligomeric state (dimmers and monomers), and the retention volume of TR-His-TMV-CP19 was transformed to disks mostly after dialyzing against 0.2–0.3 mol/L ammonium sulfate and 0.1 mol/L Tris ( PH 8.0 ) solution at room temperature for more than 10 hr. The disk state of TR-His-TMV-CP19 was confirmed by the map of SEC (Figure 6A) and Native-PAGE simultaneously (Figure 6B and Figure 6C).
Figure 6.
The form of TR-His-TMV-CP19 proteins were analyzed by SEC and 17% Native-PAGE and the diffraction analysis of crystals on proteins of TR-His-TMV-CP19 and WT-His-TMV-CP12 (A) The assembly of TR-His-TMV-CP19 was measured by Superdex 200 10/300 GL Column, Blue line represented TR-His-TMV-CP19 dialyzed against 0.2 mol/L ammonium sulfate and 0.1 mol/L Tris solution ( PH 8.0 ) at room temperature for 12 hr. Protein peaks were monitored by UV absorbance at wavelength of 280 nm and retention volumes corresponding to molecular mass were recorded in 20 mM PB and 100 mM sodium chloride, the molecular mass standards were indicated on top of the figure;Red line represents TMV-CP control which did not dialyze against, (B) the 17% Native-PAGE of WT-His-TMV-CP19. As shown in lane M. Numbers on the left were the MW of the standards in kDa, Lanes 1, 2, correspond to the WT-His-TMV-CP19 dialyzed against 0.2 mol/L ammonium sulfate and 0.1 mol/L Tris solution ( PH 8.0 ) at room temperature for 12 hr, (C) the 17% Native-PAGE of WT-His-TMV-CP19. As shown in lane M. Numbers on the left were the MW of the standards in kDa, Lanes 1, 2, correspond to the WT-His-TMV-CP19 did not dialyze against the corresponding solution, (D) X-ray crystal diffraction of WT-His-TMV-CP12 marked in Figure 5B, (E) X-ray crystal diffraction of TR-His-TMV-CP19 marked in Figure 6A, (F) X-ray crystal diffraction of TR-His-TMV-CP19 marked in Figure 6G, (G) X-ray crystal diffraction of TR-His-TMV-CP19 marked in Figure 6J, (H) X-ray crystal diffraction of TR-His-TMV-CP19 obtained from the conditions of Figure 6J, (I) The Four-layer aggregate structure of TR-His-TMV-CP19 incorporated His-tags.
Identification and Diffraction of Crystals
Macromolecular crystals were grown in crystallization buffers with high levels of supersaturation, often reaching several hundred percent. WT-TMV-CP32, WT-His-TMV-CP12, TR-His-TMV-CP62, TR-His-TMV-CP68 crystals (Figure 4) were cultured by using Index Screen (Hampton research) and ammonium sulfate buffers. These crystals were optimized by seeding method, but high resolution crystals were not obtained.
Figure 4.
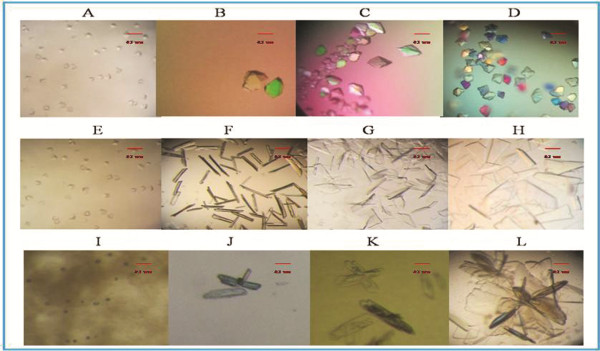
Crystallization of WT-TMV-CP32 (Cleaved GST-tags) and WT-His-TMV-CP12, the scale bar represents 0.1 mm. (A) and (E) Typical octahedral WT-His-TMV-CP12 crystals grown in the crystallization room at 295 K. The crystals did not grow bigger regardless of the time of exposure in the crystallization reagent. (B), (C), and (D) Screening and optimization of WT-His-TMV-CP12 crystallization. Although the size of the crystals improved, the quality of the crystals did not. Conversely, most of the improved crystals have no diffraction. (F), (G), and (H) Optimization of crystallization reagents facilitated growth of WT-His-TMV-CP12 crystals. Results showed that increased salt and ionic strength increased by the crystallization reagent changed the crystallization from octahedral crystals to bar and lamellar crystals. (I) WT-TMV-CP32 microcrystal that was cloned using the vector of PGEX-6P-1, purified using His-tags, cleaved GST-tags by PreScission Protease. In addition, seeding tools were used and crystallization reagents were changed, including the crystallization reagents of Hampton research, in an attempt to improve the quality and size of the crystals or to produce a different crystal form. Only twin crystals or polycrystalline were obtained, as shown in (J), (K), and (L).
On the contrary, TR-His-TMV-CP19 crystals (Figure 5)(protein concentration: 14 mg/mL) with 3.0 Å resolution were obtained in 0.25 mol/L ammonium sulfate and 0.1 mol/L Tris solution at 295 K, PH 7.7 (Figure 6), and the Four-layer aggregate disk structure of TR-His-TMV-CP19 was solved.
Figure 5.
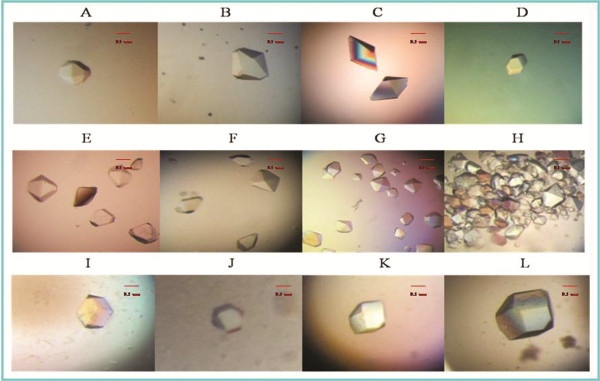
Examples of TR-His-TMV-CP19 crystals with fused short peptides and truncated four amino acids from the C-terminus (Showed in Table4), the scale bar represents 0.1 mm.
TMV-CP has been available as a recombinant protein expressed in E.coli for more than 20 years [37,38]. The incorporation of His-tags at the C-terminal of TMV-CP has been reported recently by introducing His-tags into TMV-CP to facilitate their purification [39]. To date, however, no one has reported on the crystallization of recombinant TMV-CP connecting peptides expressed in E. coli, except on the residues of chemical modification of TMV-CP [26,40-43]. These recombinant proteins connecting with peptides often did not affect the biological activity of the engineered proteins, located at the exterior of the TMV-CP disks. To obtain high resolution crystals, the expression vectors containing TMV-CP fragments were first constructed and expressed. An attempt was made to harvest the WT-His-TMV-CP12 fragments by using thrombin cleavage to cleave His-tags. His-tags were not cleaved when the proportion of thrombin cleavage and recombinant proteins was increased from 1:1 to 8:1. Subsequently, another genetically engineered WT-GST-TMV-CP32 was constructed and expressed. Compared with His-tags, the GST-tags were easily cleaved by PreScission Protease. The crystals of WT-TMV-CP32 and WT-His-TMV-CP12 were grown in ammonium sulfate buffers and commercial crystallization reagents simultaneously. No high resolution crystals were formed in the hanging drops when the proportion of WT-TMV-CP32 to crystallization solution was 1:1. Only tiny octahedral WT-His-TMV-CP12 crystals were grown in the crystallization room at 295 K. No matter how long the growth time was, the crystals did not grow bigger in the crystallization buffers and commercial crystallization reagents. To maintain the physical properties of WT-His-TMV-CP12 according to the crystalline structure of WT-TMV-CP (the amino acids at the terminal of WT-TMV-CP were flexibly) (PDB codes 1EI7 and 1VTM), three kinds of WT-His-TMV-CP12 truncated amino acids at the C-terminal (TR-His-TMV-CP19, TR-His-TMV-CP62, TR-His-TMV-CP68) were constructed, expressed, purified and formed 20S disks. And then the crystals of TR-His-TMV-CP19 appeared when the protein concentration (20 mmol/L orthophosphate and 100 mmol/L sodium chloride, pH 8.0, at 297 K for 12 hr) is 8.0–14 mg/mL and crystallization pool solution (pH 7.7) consisting of 0.1 mol/L ammonium sulfate and 0.1 mol/L Tris at 295 K for 24 hr. The increasing of the ionic strength (1.0) and pH of the crystallization pool buffers resulted in the appearance of TR-His-TMV-CP19 crystals in the circumstance of higher protein concentration. Compared with WT-TMV-CP, the purified protein of TR-His-TMV-CP19 was in the form of protein A (Figure 6A).
The crystals of TR-His-TMV-CP19 were obtained at 2.9–7.0 Å resolution (Figure 6E-6H) and the crystal of WT-His-TMV-CP12 was obtained without diffraction. A Four-layer aggregate crystal structure of TR-His-TMV-CP19 was obtained by removing four amino acids at the C-terminal of His-TMV-CP and connecting short peptides at the N-terminal of His-TMV-CP (TR-His-TMV-CP19). A diagram of the growth curve of TR-His-TMV-CP19 crystallization was drawn (Figure 7).
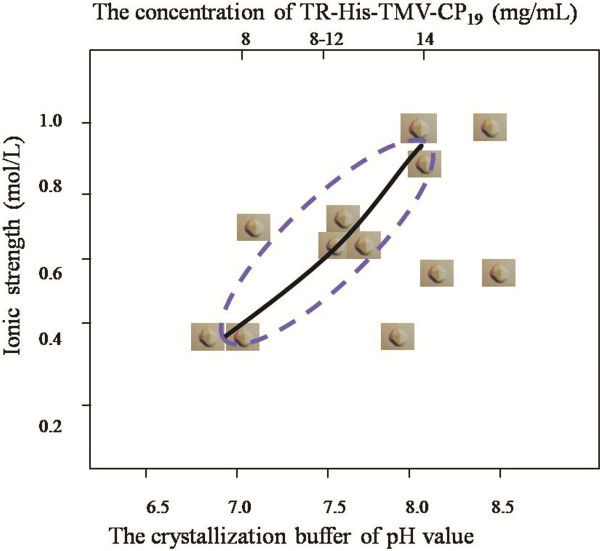
Figure 7.
Diagram showing the growth curve of the crystallization of TR-His-TMV-CP19. The predominant species at protein concentration conditions of 8.0–14 mg/mL at pH 6.8–7.8 with ionic strength of 0.4–1.0 are the perfect crystals, identified as higher resolution crystals. As the pH falls below pH 7.0, TR-His-TMV-CP19 did not grow in the form of crystals. On the contrary, the TR-His-TMV-CP19 crystals grew with increasing protein concentration, pH (pH 7.8–8.0), and ionic strength (0.7–1.0) of the crystallization pool buffers. The crystals can also be harvested when the protein concentration is 14 mg/mL because high resolution crystals are formed at this concentration. When the crystallization pool buffers conditions are at pH 8.5 with ionic strength of 0.7–1.0, no high resolution crystals can be harvested.
Compared with the crystals of WT-TMV-CP (isolated from TMV particles), the Propagation, purification and acquisition the macromolecular crystals of TR-His-TMV-CP19 from E.coli were very facile process. By investigating, it was found that the crystals of WT-His-TMV-CP12 had close relationships with protein concentration, ionic strength and PH of the solution. After truncating four amino acids at the C-terminal of WT-His-TMV-CP12, a Four-layer aggregate structure of TR-His-TMV-CP19 was determined at 3.0 Å resolution by using the technique of Hanging-drop vapor diffusion and seeding methods, but the high resolution crystals of WT-TMV-CP32, WT-His-TMV-CP12, TR-His-TMV-CP62, and TR-His-TMV-CP68 did not obtain.
Conclusions
The crystallization and stability of recombinant WT-TMV-CP were influenced by various factors such as the charge, isoelectric point, hydrophilicity of hybrid CP, and especially the length of the amino sequence. Based on the crystallization methods of WT-TMV-CP (isolated from TMV particles), the crystals of TR-His-TMV-CP (TR-His-TMV-CP19, TR-His-TMV-CP62, TR-His-TMV-CP68) were grown in the crystallization buffers as mentioned above. The crystals of TR-His-TMV-CP19 were grown by the technique of Hanging-drop vapor diffusion and seeding methods with crystallization reagents including several kinds of ammonium sulfate and Tris solutions. A map of the growth curve of the crystallization of TR-His-TMV-CP19 was drawn. In this map, the proteins of 20s disk form of TR-His-TMV-CP19 (8.0–14 mg/mL) were firstly prepared in solutions (pH 8.0) containing 20 mmol/L orthophosphate and 100 mmol/L sodium chloride and then dialyzed against 0.2–0.3 mol/L ammonium sulfate and 0.1 mol/L Tris solutions ( PH 8.0 ) at 277 K for 12 hr. The corresponding crystals were grown in crystallization pool solutions containing 0.1–0.3 mol/L ammonium sulfate and 0.1 mol/L Tris at pH 6.5–8.5 by the technique of Hanging-drop vapor at 277 and 295 K. Seeding methods were performed at 295 K after 1–7 d, the good macromolecular crystals appeared under different conditions. 2.9–7.0 Å resolution of TR-His-TMV-CP19 (concentration 14 mg/mL) macromolecular crystals were obtained in crystallization pool solution consisting of 0.25 mol/L ammonium sulfate and 0.1 mol/L Tris at pH 7.7. Then, using the same method, the crystals of WT-His-TMV-CP12 without resolution and some tiny WT-TMV-CP32 crystals, were also obtained by series of seeding experiments. It was showed from the experiment that: the genetically engineered proteins of TR-His-TMV-CP19 could grow high resolution crystals. Hence, the present investigations suggest that the C-terminal of TMV-CP was unstable for crystallization buffer, and the amino acids at the C-terminus were hypothesized to be very flexible. Additionally, the inserted sites of short peptides of TMV-CP could access the grown crystals. Short peptides have a positive influence on the stability of the biophysical properties of TMV-CP. Compared with WT-TMV-CP isolated from TMV particles, this recombinant protein of TR-His-TMV-CP19 is easy to purify and grow crystals. Thus, the latter can be applied to structural biology and structure-based drug design.
Methods
TMV (common strain) was isolated from N. tabacum K326 leaves infected by TMV, which were cultivated in the Greenhouse of Center for Research and Development of Fine Chemicals of Guizhou University, and purified by the method described by Gooding [44], and modified by Shire [16,45]. TMV-CP was prepared by Scheel [18]. TMV-RNA was extracted from purified virus by treating with phenol and SDS [46-48]. In order to obtain the generation of full-length viral cDNA sequence, TMV-RNA was reverse transcribed using primercDNA (Table 1) in 50 mmol/L Tris at pH 8.0, 8.0 mmol/L magnesium chloride, 75 mmol/L potassium chloride, 10 mmol/L DL-Dithiothreitol, 1.0 mmol/L dNTPs, 0.5 unit/μL AMV reverse transcriptase (TaKaRa), and 1.0 unit/μL RNase inhibitor (TaKaRa) for 1.5 hr at 315 K.
Table 1.
DNA sequences of the primers
| serial number | Name | Primers |
|---|---|---|
| PrimercDNA |
TMV-cDNA |
5’-TCGACATAGGGACATCTTC-3’ |
| Primer1a,c |
WT-TMV-CP-FOR |
5’-GGAATTCCATATGTCTTACAGTATCACTACTCC-3’ |
| Primer2b,d |
WT-TMV-CP-REV |
5’- CCGCTCGAGTCAAGTTGCAGGACCAGAGG-3’ |
| Primer3b |
TR-TMV-CP-FOR |
5’-CGGGATCCATGTCTTACAGTATCACTACTCC-3’ |
| Primer6a,d,e |
TR-His-TMV-CP-FOR |
5’-GGAATTCCATATGATCACTACTCCATCACAGTTCG-3’ |
| Primer8e |
TR-His-TMV-CP-REV |
5’-CCGCTCGAGTCAACCAGAGGTCCAAACCAA-3’ |
| Primer9c | TR-His-TMV-CP-REV | 5’-CCGCTCGAGTCAAGTTGCAGGACCAGAGG-3’ |
aThe WT-His-TMV-CP12 fragment was amplified by using primer1 and primer2.
bThe WT-GST-TMV-CP32 fragment was amplified by using primer3 and primer2.
cThe TR-His-TMV-CP19 fragment was amplified by using primer1and primer9.
dThe TR-His-TMV-CP62 fragment was amplified by using primer6 and primer2.
eThe TR-His-TMV-CP68 fragment was amplified by using primer6 and primer8.
Using the generation of full-length viral cDNA sequence and the corresponding primers by the same method, TR-His-TMV-CP19, WT-GST-TMV-CP32, WT-His-TMV-CP12, TR-His-TMV-CP62, and TR-His-TMV-CP68 sequences were amplified by PCR technology (Table 2).
Table 2.
Protein building blocks used or referenced herein
| Building block | Origin | Abbreviation |
|---|---|---|
| Wild type TMV coat protein |
N. tabacum K326 |
WT-TMV-CPa |
| Wild type TMV coat protein with GST tags |
E. coliBL21(DE3)-RIL |
WT-GST-TMV-CP32b |
| Wild type TMV coat protein with His tags |
E. coliBL21(DE3)-RIL |
WT-His-TMV-CP12c |
| Truncated 19 TMV coat protein with His tags |
E. coliBL21(DE3)-RIL |
TR-His-TMV-CP19d |
| Truncated 62 TMV coat protein with His tags |
E. coliBL21(DE3)-RIL |
TR-His-TMV-CP62e |
| Truncated 68 TMV coat protein with His tags | E. coliBL21(DE3)-RIL | TR-His-TMV-CP68f |
a The wild type TMV-CP was purified from N. tabacum K326.
bThe wild type TMV-CP expressed and purified from reconstructed prokaryotic expression vector PGEX-6P-1-TMV-CP32 that was expressed in competent cell BL21(DE3)-RIL.
cThe wild type TMV-CP expressed and purified from reconstructed prokaryotic expression vector pET28a-TMV-CP12 that was expressed in competent cell BL21(DE3)-RIL.
dThe truncated TMV-CP expressed and purified from reconstructed prokaryotic expression vector pET28a-TMV-CP19 that was expressed in competent cell BL21(DE3)-RIL.
eThe truncated TMV-CP expressed and purified from reconstructed prokaryotic expression vector pET28a-TMV-CP62 that was expressed in competent cell BL21(DE3)-RIL.
fThe truncated TMV-CP expressed and purified from reconstructed prokaryotic expression vector pET28a-TMV-CP68 that was expressed in competent cell BL21(DE3)-RIL.
The dsDNA of correct length was purified and identified by 1% agarose gel electrophoresis. Both plasmid pET28a (Novagen) and CP were digested with Nde I (NEB, 10 units/μL)/Xho I (NEB, 10 units/μL) and cloned into the same sites in pET28a (pET28a-WT-His-TMV-CP12, pET28a-TR-His-TMV-CP19, pET28a-TR-His-TMV-CP62, and pET28a-TR-His-TMV-CP68). Both plasmid PGEX-6P-1 (Novagen) and CP were digested with BamH I (NEB, 10 units/μL)/Xho I (NEB, 10 units/μL) and cloned into the same sites in PGEX-6P-1 (PGEX-6P-1-WT-GST-TMV-CP32). Transcription reactions were performed by using the corresponding transcription system. E. coli BL21(DE3)-RIL (TaKaRa) cultures were transformed into vectors involving aforementioned recombinant plasmid. Expression plasmids were grown in Luria-Bertani (LB) medium containing 30 μg/mL kanamycin at 310 K until the OD600 reached 0.65–1.0. After cooling the cultures to 289 K, the expression product was induced by the addition of 1.0 mmol/L IPTG, and the culture was incubated for 16 hr. The cells were harvested by centrifugation and resuspended in 35 mL lysis buffer (100 mmol/L sodium chloride, 50 mmol/L phosphate buffer, pH 8.0, 10 mmol/L ß-mercaptoethanol). Then, the cells were thawed, lysed by supersonic device, and then centrifuged at 15000 rpm for 30 min at 277 K. The supernate was then passed through 0.22 mm syringe filters (Millipore) and loaded onto a Ni Sepharose High performance column (GE Healthcare, 5mL), washed with five column volumes of 40 mmol/L imidazole, and eluted with 400 mmol/L imidazole. The N-terminal His-tags failed to cleaved with thrombin (1.0 unit/mg) and N-terminal GST-tags was cleaved successfully with PreScission Protease (1.0 unit/mg) by incubating overnight at 277 K. The cleaved GST-tags and uncleaved His-tags were removed by the same chelating column, and the flow-through was concentrated in an Amicon Ultra centrifugal filter device (Millipore) with a 10 kDa filter and then loaded onto a HiLoad 16/60 Superdex 200 pg column equilibrated in the dialysis solution (20 mmol/L orthophosphate and 100 mmol/L sodium chloride, pH 8.0). The protein was concentrated to 5.0–25 mg/mLfor the crystallization trials by using Amicon Ultra centrifugal filter units (Millipore) with a 10 kDa molecular weight cutoff. The target proteins were briefly stored at 277 K.
The purification proteins were dialyzed against the appropriate high-salt solution at room temperature to obtain the Four-layer aggregate (20S disk) [35,36,49-52]. The 20S disk form proteins were confirmed by Size Exclusion Chromatography (SEC) and Native-polyacrylamide gel Electrophoresis (Native-PAGE) method. SEC was performed at room temperature by using a calibrated Superdex 200 10/300 GL column (GE Healthcare) attached to an AKTApurifier fast protein liquid chromatography system (GE Healthcare). The column was equilibrated with a solution containing 20 mM orthophosphate (pH 8.0), 100 mM NaCl solution. Molecular mass standards (Bio-Rad) used are: Thyroglobulin (669 kDa), Ferritin (440 kDa), BSA (67 kDa), β-lactoglobulin (35 kDa), Ribonuclease A (13.7 kDa), Cytochrome (13.6 kDa), Aprotinin (6.51 kDa) and Vitamin B12 (1.36 kDa). Protein was monitored by absorbance at the wavelength of 280 nm. The crystals of purified proteins were obtained by the technique of Hanging-drop vapor diffusion. The protein concentration was 5.0–25 mg/mL, and the crystallization solutions contained 0.1–0.3 mol/L ammonium sulfate and 0.1 mol/L Tris at PH 6.5–8.5 (Table 3) for 1–7 d at 293–298 K. The crystals (Table 4) were first soaked with cryoprotection (the reservoir solution containing an extra 30% glycerol), and then mounted and flash-frozen in liquid nitrogen [33,34]. Diffraction data were collected at Shanghai Synchrotron Radiation Facility beamline 17U. All the X-ray data were processed by using HKL2000 program suite and converted to structure factors within the CCP4 program (Table 5).
Table 3.
The crystallization solutions of ammonium sulfate
| Sample |
Protein concentration and buffera |
Crystallization bufferb |
Temperature (K) |
|---|---|---|---|
| WT-TMV-CP32 (cleaved GST) |
8 –14 mg/mL, 20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0 |
0.10–0.35 mol/L ammonium sulfate, 0.1 mol/L Tris buffer, pH 6.5–8.5 |
277/295 |
| WT-His-TMV-CP12 |
8–14 mg/mL, 20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0 |
0.10–0.35 mol/L ammonium sulfate, 0.1 mol/L Tris buffer, pH 6.5–8.5 |
277/295 |
| TR-His-TMV-CP19 |
8–14 mg/mL, 20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0 |
0.10–0.35 mol/L ammonium sulfate, 0.1 mol/L Tris buffer, pH 6.5–8.5 |
277/295 |
| TR-His-TMV-CP68 |
8–14 mg/mL, 20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0 |
0.10–0.35 mol/L ammonium sulfate, 0.1 mol/L Tris buffer, pH 6.5–8.5 |
277/295 |
| TR-His-TMV-CP62 | 8–14 mg/mL, 20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0 | 0.10–0.35 mol/L ammonium sulfate, 0.1 mol/L Tris buffer, pH 6.5–8.5 | 277/295 |
a The buffers were provided with filter membrane.
b The buffers were provided with filter membrane.
Table 4.
Examples of TR-His-TMV-CP19 crystals in Figure5
| Examples | Protein Concentration | Crystal Conditions | Crystal appearance | Resolutions |
|---|---|---|---|---|
| A |
8.0 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.1 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 6.8 |
The typical dodecahedral crystals were rapid growth (approximately 24 hr) at 295 K |
>7.0 Å |
| B |
8.0 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.1 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 7.5. |
The irregular octahedral crystals were rapid growth (approximately 24 hr) at 295 K |
-- |
| C |
8.0 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.1 mol/L ammonium sulfate and 0.2 mol/L Tris/HCl, pH 7.0. |
The irregular octahedral crystals were rapid growth (approximately 24 hr) at 295 K |
-- |
| D |
8.0 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.3 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 7.7. |
The typical octahedral crystals were rapid growth (approximately 24 hr) at 295 K |
-- |
| E |
8.0 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.2 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 6.8. |
The octahedral crystals were rapid growth (approximately 24 hr) at 295 K |
-- |
| F |
8.0 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.2 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 7.7. |
The octahedral crystals were rapid growth (approximately 24 hr) at 295 K |
-- |
| G |
8.0 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.2 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 6.8. |
The Octahedral crystals were rapid growth (approximately 24 hr) at 295 K |
>5.0 Å |
| H |
14 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.3 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 8.0. |
The splintered off and crowded together crystals were rapid growth (approximately 24 hr) at 295 K |
-- |
| I |
22 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.3 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 7.7. |
The hexahedral octahedral crystals were Slow growth (approximately 7 d) at 295 K |
-- |
| J |
14 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.25 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 7.7. |
The typical octahedral crystals were Slow growth (approximately 1-2 d) at 295 K |
2.9–3.0 Å |
| K |
22 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) |
0.2 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 7.7. |
The dodecahedral crystals were slow growth (approximately 24 hr) at 295 K |
-- |
| L | 22 mg/mL (20 mmol/L sodium phosphate buffer, 100 mmol/L sodium chloride, pH 8.0) | 0.2 mol/L ammonium sulfate and 0.1 mol/L Tris/HCl, pH 7.7. | The dodecahedral crystals were Slow growth (approximately 9d) at 295 K | -- |
- stand for the crystals which didn't been diffraction by X-ray methods.
Table 5.
Data collection and refinement statistics of TR-His-TMV-CP19 crystals in Figure6I
| TR-His-TMV CP19 | |
|---|---|
|
Data collection | |
| Space group |
P21212 |
| Cell dimensions |
|
|
a, b, c (Å) |
171, 311, 314 |
| α, β, γ (°) |
90, 90, 90 |
| Resolution (Å) |
50.0-3.06 (3.17-3.06) |
|
Rsym |
0.11 (0.196) |
|
I/σ(I) |
10.1 (6.4) |
| Completeness (%) |
99.7 (99.2) |
| Redundancy |
4.4 (3.84) |
|
Refinement statistics | |
| Resolution (Å) |
20.0-3.06 (3.14-3.06 ) |
| No. reflections |
155016 |
|
Rwork/Rfree |
20.4/25.5 |
| No. atoms |
36918 |
| B-factors |
47.9 |
| R.m.s deviations | |
| Bond lengths (Å) |
0.013 |
| Bond angles (º) | 1.499 |
*Highest resolution shell is shown in parenthesis.
Abbreviations
TMV: Tobacco mosaic virus; CP: Coat protein; His: Hexahistidine; GST: Glutathione-S-transferase; Ni-NTA: Nickel-nitrilotriacetic acid; dsDNA: Double-stranded DNA; cDNA: Complementary DNA; IPTG: Isopropyl ß-D-thiogalactopyranose; SDS: Dodecylsulfate; PAGE: Polyacrylamide gel electrophoresis; Native-PAGE: Native-polyacrylamide gel Electrophoresis; SEC: Size Exclusion Chromatography; WT: Wild type; TR: Truncation; Tris: Tris(hydroxymethyl)aminomethane; DTT: DL-dithiothreitol; LB: Luria-Bertani; E.coli: Escherichia coli; N. tabacum: Nicotiana tabacum.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BAS and SY designed this study. XYL carried out the clone and protein expression studies, participated in the sequence alignment and drafted the manuscript. ZC,ZCW,MJZ and DDY performed Crystallization tests. LHJ collected and analyzed the data of genetically engineered TMV-CP crystals. BAS, SY, DYH, and ZC critically revised the manuscript. All of the authors read and approved the final version of the manuscript.
Contributor Information
Xiangyang Li, Email: michael.lee1983@yahoo.com.cn.
Baoan Song, Email: songbaoan22@yahoo.com.
Deyu Hu, Email: fcc.dyhu@gzu.edu.cn.
Zhenchao Wang, Email: wzc.4884@163.com.
Mengjiao Zeng, Email: 1094185612@qq.com.
Dandan Yu, Email: caoyuanyexia@163.com.
Zhuo Chen, Email: gychenzhuo@yahoo.com.cn.
Linhong Jin, Email: fcc.jinlh@gzu.edu.cn.
Song Yang, Email: fcc.syang@gzu.edu.cn.
Acknowledgements
We thank Professor Chuan He from the University of Chicago and Professor Caiguang Yang from the Shanghai Institute of Materia Medica (CAS) for the crystallographic discussion and assistance. We also acknowledge user support for the beam line BL17U at the Shanghai Synchrotron Radiation Facility as well as In House X-Ray at Shanghai Institute of Materia Medica, China. The authors also wish to thank the National Key Program for Basic Research (No. 2010CB 126105) and Special Fund for Agro-scientific Research in the Public Interest (No.201203022) and the National Natural Science Foundation of China (No.21132003) and the Key Technologies R&D Program (No. 2011BAE06B05-6) for the financial support.
References
- Caspar DLD. Assembly and stability of the tobacco mosaic virus particle. Adv Protein Chem. 1963;18:37–121. doi: 10.1016/s0065-3233(08)60268-5. [DOI] [PubMed] [Google Scholar]
- Bloomer AC, Champness JN, Bricogne G, Stoden R, Klug A. Protein disk of tobacco mosaic virus at 2.8 Å resolution showing the interaction within and between subunits. Nature. 1978;276:362–368. doi: 10.1038/276362a0. [DOI] [PubMed] [Google Scholar]
- Culver JN. Tobacco mosaic virus assembly and disassembly: Determinants in pathogenicity and resistance. Annu Rev Phytopathol. 2002;40:287–308. doi: 10.1146/annurev.phyto.40.120301.102400. [DOI] [PubMed] [Google Scholar]
- Harrison BD, Wilson TM. Milestones in the research on tobacco mosaic virus. Philos Trans R Soc B. 1999;354:521–529. doi: 10.1098/rstb.1999.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele RB, Schuster TM. Kinetics of protein subunit interaction: simulation of a polymerized overshoot. Biopolymers. 1974;13:275–288. doi: 10.1002/bip.1974.360130204. [DOI] [PubMed] [Google Scholar]
- Pattanayek R, Stubbs G. Structure of the U2 strain of tobacco mosaic virus refined at 3.5 Å resolution using x-ray fiber diffraction. J Mol Biol. 1992;228:516–528. doi: 10.1016/0022-2836(92)90839-C. [DOI] [PubMed] [Google Scholar]
- Stubbs G, Warren S, Holmes K. Structure of RNA and RNA binding site in tobacco mosaic virus from a 4 Å map calculated from X-ray fiber diagrams. Nature. 1997;267:216–221. doi: 10.1038/267216a0. [DOI] [PubMed] [Google Scholar]
- Culver JN, Stubbs G, Dawson WO. Structure-function relationship between tobacco mosaic virus coat protein and hypersensitivity in Nicotiana sylvestris. J Mol Biol. 1994;242:130–138. doi: 10.1006/jmbi.1994.1564. [DOI] [PubMed] [Google Scholar]
- Namba K, Stubbs G. Structure of tobacco mosaic virus at 3.6 Å resolution:implications for assembly. Science. 1986;231:1401–1406. doi: 10.1126/science.3952490. [DOI] [PubMed] [Google Scholar]
- Klug A, Caspar DLD. The structure of simple viruses. Adv Virus Res. 1960;13:1–63. doi: 10.1016/s0065-3527(08)60012-3. [DOI] [PubMed] [Google Scholar]
- Mandelkow E, Holmes KC, Gallawitz U. A new helical aggregate of tobacco mosaic virus protein. J Mol Biol. 1976;102:265–285. doi: 10.1016/S0022-2836(76)80053-8. [DOI] [PubMed] [Google Scholar]
- Butler PJG, Durham ACH. Tobacco mosaic virus protein aggregation and the virus assembly. Adv Protein Chem. 1977;31:188–251. doi: 10.1016/s0065-3233(08)60219-3. [DOI] [PubMed] [Google Scholar]
- Mandelkow E, Stubbs G, Warren S. Structures of the helical aggregates of tobacco mosaic virus protein. J Mol Biol. 1981;152:375–386. doi: 10.1016/0022-2836(81)90248-5. [DOI] [PubMed] [Google Scholar]
- Butler PJG. The current picture of the structure and assembly of tobacco mosaic virus. J Gen Virol. 1984;65:253–279. doi: 10.1099/0022-1317-65-2-253. [DOI] [PubMed] [Google Scholar]
- Bloomer AC, Butler PJG. In: The plant viruses. 2. Regenmortel MHV, Fraenkel-Conrat H, editor. New York: Chapman and Hall; 1986. Tobacco mosaic virus structure and self-assembly; pp. 19–57. [Google Scholar]
- Shire SJ, Steckert JJ, Adams ML, Schuster TM. Kinetics and mechanism of tobacco mosaic virus assembly: direct measurement of relative rates of incorporation of 4S and 20S protein. Proc Natl Acad Sci USA. 1979;76(6):2745–2749. doi: 10.1073/pnas.76.6.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DLD, Namba K. Switching in the self-assembly of tobacco mosaic virus. Adv Biophys. 1990;26:157–185. doi: 10.1016/0065-227x(90)90011-h. [DOI] [PubMed] [Google Scholar]
- Scheele RB, Lauffer MA. Acid-base titrations of tobacco mosaic virus and tobacco mosaic virus protein. Biochemistry. 1967;6:3076–3081. doi: 10.1021/bi00862a014. [DOI] [PubMed] [Google Scholar]
- Durham ACH, Finch JT, Klug A. States of aggregation of tobacco mosaic virus protein. Nature New Biol. 1971;229:37–42. doi: 10.1038/newbio229037a0. [DOI] [PubMed] [Google Scholar]
- Durham ACH, Klug A. Polymerisation of tobacco mosaic virus protein and its control. Nature New Biol. 1971;229:42–46. doi: 10.1038/newbio229042a0. [DOI] [PubMed] [Google Scholar]
- Durham ACH. The cause of irreversible polymerization of tobacco mosaic virus protein. FEBS Lett. 1972;25:147–152. doi: 10.1016/0014-5793(72)80473-3. [DOI] [PubMed] [Google Scholar]
- Stubbs G. Molecular structures of viruses from the tobacco mosaic virus group. Semin Virol. 1990;1:405–412. [Google Scholar]
- Bhyravbhatla B, Stanley JW, Caspar DLD. Refined atomic model of the four-layer aggregate of the tobacco mosaic virus coat protein at 2.4 Å resolution. Biophys J. 1998;74:604–615. doi: 10.1016/S0006-3495(98)77819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba K, Caspar DLD, Stubbs G. Enhancement and simplification of macromolecular images. Biophy J. 1998;53:469–475. doi: 10.1016/S0006-3495(88)83125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba K, Pattanegek R, Stubbs G. Visualization of protein RNA interaction in a virus: Refined structure of TMV at 2.9 Å resolution by X-ray fiber diffraction. J Mol Biol. 1989;208:307–325. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- Dedeo MT, Duderstadt KE, Berger JM, Francis MB. Nanoscale protein assemblies from a circular permutant of the tobacco mosaic virus. Nano Lett. 2010;10:181–186. doi: 10.1021/nl9032395. [DOI] [PubMed] [Google Scholar]
- Sachse C, Chen JZ, Coureux PD, Stroupe ME, Fändrich M, Grigorieff N. High-resolution electron microscopy of helical specimens: A fresh look at tobacco mosaic virus. J Mol Biol. 2007;371:812–835. doi: 10.1016/j.jmb.2007.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare DK, Orlova EV. 4.6 Å cryo-EM reconstruction of tobacco mosaic virus from images recorded at 300 keV on a 4k × 4k CCD camera. J Struct Biol. 2010;171:303–308. doi: 10.1016/j.jsb.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc Natl Acad Sci. 2011;1008:9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y. Molecular assembly of tobacco mosaic virus in vitro. Adv Biophys. 1986;22:95–145. doi: 10.1016/0065-227x(86)90004-3. [DOI] [PubMed] [Google Scholar]
- Raghavendra K, Adams ML, Schuster TM. Tobacco mosaic virus protein aggregates in solution: Structural comparison of 20S aggregates with those near conditions for disk crystallization. Biochemistry. 1985;24:3298–3304. doi: 10.1021/bi00334a034. [DOI] [PubMed] [Google Scholar]
- Sperling R, Klug A. States of aggregation of the Dahlemense strain of tobacco mosaic virus protein and their relation to crystal formation. J Mol Biol. 1975;96:425–430. doi: 10.1016/0022-2836(75)90169-2. [DOI] [PubMed] [Google Scholar]
- Raghavendra KDM, Salunke DLD, Caspar STM. Disk aggregates of tobacco mosaic virus coat protein in solution: electron microscopy observations. Biochemistry. 1986;25:6276–6279. doi: 10.1021/bi00368a066. [DOI] [PubMed] [Google Scholar]
- Raghavendra K, Kelly JA, Khairallah L, Schuster TM. Structure and function of disk aggregates of the coat protein of tobacco mosaic virus. Biochemistry. 1988;27:7583–7588. doi: 10.1021/bi00420a002. [DOI] [PubMed] [Google Scholar]
- Correia JJ, Shire S, Yphantis DA, Schuster TM. Sedimentation equilibrium measurements of the intermediate-size tobacco mosaic virus protein polymers. Biochemistry. 1985;24:3292–3297. doi: 10.1021/bi00334a033. [DOI] [PubMed] [Google Scholar]
- Durham ACH. Structures and roles of the polymorphic forms of tobacco mosaic virus protein. I. Sedimentation studies. J Mol Biol. 1972;67:289–305. doi: 10.1016/0022-2836(72)90242-2. [DOI] [PubMed] [Google Scholar]
- Shire SJ, McKay P, Leung DW, Cachianes GJ, Jackson E, Woods WI, Raghavendra K, Khairallah L, Schuster TM. Preparation and properties of recombinant DNA rerived Tobacco Mosaic Virus Coat Protein. Biochemistry. 1990;29:5119–5126. doi: 10.1021/bi00473a017. [DOI] [PubMed] [Google Scholar]
- Hwang DJ, Roberts IM, Wilson TMA. Expression of tobacco mosaic virus coat protein and assembly of pseudo virus particles in Escherichia Coli. Proc Natl Acad Sci USA. 1994;91:9067–9071. doi: 10.1073/pnas.91.19.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckman MA, Soto CM, McDowell H, Liu JL, Ratna BR, Korpany KV, Zahr OK, Blum AS. Role of hexahistidine in directed nanoassemblies of tobacco mosaic virus coat protein. Nano Lett. 2011;5(3):1606–1616. doi: 10.1021/nn1025719. [DOI] [PubMed] [Google Scholar]
- Endo M, Wang H, Fujitsuka M, Majima T. Pyrene-stacked nanostructures constructed in the recombinant tobacco mosaic virus rod scaffold. Chem Eur J. 2006;12:3735–3740. doi: 10.1002/chem.200501309. [DOI] [PubMed] [Google Scholar]
- Lee SY, Choi J, Royston E, Janes DB, Culver JN, Harris MT. Deposition of platinum clusters on surface-modified tobacco mosaic virus. J Nanosci Nanotechnol. 2006;6:974–981. doi: 10.1166/jnn.2006.146. [DOI] [PubMed] [Google Scholar]
- Miler RA, Presley AD, Francis MB. Self-assembling light-harvesting systems from synthetically modified tobacco mosaic virus coat proteins. J Am Chem Soc. 2007;129:3104–3109. doi: 10.1021/ja063887t. [DOI] [PubMed] [Google Scholar]
- Schlick TL, Ding Z, Kovacs EW, Francis MB. Dual-Surface modification of the tobacco mosaic virus. J Am Chem Soc. 2005;127:3718–3723. doi: 10.1021/ja046239n. [DOI] [PubMed] [Google Scholar]
- Gooding GV, Hebert TT. A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathologigal notes. 1967. pp. 1285–1286. [PubMed]
- Shire SJ, Steckert JJ, Schuster TM. Mechanism of self assembly of tobacco mosaic virus protein. II. Characterization of the metastable polymerization nucleus and the initial stages of helix formation. J Mol Biol. 1979;127:487–506. doi: 10.1016/0022-2836(79)90233-X. [DOI] [PubMed] [Google Scholar]
- Fraenkel-Conrat HB, Singer, Tsugita A. Purification of viral RNA by means of bentonite. Virology. 1961;14:54–58. doi: 10.1016/0042-6822(61)90131-3. [DOI] [PubMed] [Google Scholar]
- Hebert TT. Precipitation of plant viruses by polyethylene glycol. Phytopathology. 1963;53:36. [Google Scholar]
- Wilcockson JRH. The rapid isolation of plant virus RNAs using sodium perchlorate. J Gen Virol. 1974;23:107–111. doi: 10.1099/0022-1317-23-1-107. [DOI] [PubMed] [Google Scholar]
- Schuster TM, Scheele RB, Adams ML, Shire SJ, Steckert JJ, Potschka M. Studies on the mechanism of assembly of tobacco mosaic virus. Biophys J. 1980;32:313–329. doi: 10.1016/S0006-3495(80)84959-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster TM, Scheele RB, Khairallah LH. Mechanism of self-assembly of tobacco mosaic virus protein. I. Nucleation-controlled kinetics of polymerization. J Mol Biol. 1979;127:461–468. doi: 10.1016/0022-2836(79)90232-8. [DOI] [PubMed] [Google Scholar]
- Schuster TM, Scheele RB, Adams ML, Shire SJ, Steckert JJ, Potschka M. Studies on the mechanism of assembly of tobacco mosaic virus. Biophys J. 1980;10:313–317. doi: 10.1016/S0006-3495(80)84959-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel WK, Schoot P. Physical regulation of the self-assembly of tobacco mosaic virus coat protein. Biophys J. 2006;91:1501–1512. doi: 10.1529/biophysj.105.072603. [DOI] [PMC free article] [PubMed] [Google Scholar]