Abstract
Gamma interferon (IFN-γ) is critical in the immune response against Mycobacterium tuberculosis. In an ongoing trial of aerosol IFN-γ in conjunction with standard drug therapy, we have observed activation of IFN signaling in bronchoalveolar lavage (BAL) cells from tuberculosis (TB) patients. We hypothesized that aerosol IFN-γ treatment of pulmonary TB would increase expression of genes important for the control of TB. We investigated the expression of downstream genes by measuring inducible nitric oxide synthase (iNOS) and the chemokine IFN-inducible 10-kDa protein (IP-10) by real-time quantitative reverse transcription-PCR. In vitro, M. tuberculosis induced IP-10, and IFN-γ stimulated this further, with no effect on iNOS expression. We studied 21 patients with pulmonary TB and 7 healthy subjects. Similar to the in vitro model, IP-10 mRNA was increased in BAL cells from TB patients and was augmented after treatment with aerosolized IFN-γ. TB was also associated with elevated iNOS mRNA, but aerosolized IFN-γ did not further enhance expression. Genomic analysis identified 1,300 of 4,058 genes expressed in BAL cells from six TB patients before and after 1 month of therapy, including aerosolized IFN-γ. However, only 15 genes were differentially regulated by IFN-γ. We conclude that iNOS and IP-10 mRNA expression is increased in TB but that aerosol IFN-γ treatment increases expression of few genes in the human lung.
Gamma interferon (IFN-γ) is the key cytokine in the T-helper type 1 immune response required for adequate control of Mycobacterium tuberculosis infection in the lung (25, 53). Murine models of mycobacterial infection confirmed the importance of IFN-γ, its receptor, signaling molecules, and downstream genes, such as that for inducible nitric oxide synthase (iNOS), using transgenic mice (12, 13, 26, 32, 33, 36). In the murine system, IFN-γ strongly induces iNOS gene transcription, mRNA expression, and enzymatic activity (6, 15). Additionally, in vivo administration of iNOS inhibitors to M. tuberculosis-infected mice increases bacterial burden and early mortality (5). With the advent of microarray technology it is possible to document global regulation of mRNA levels after IFN-γ treatment. Ehrt and colleagues analyzed in vitro gene expression in murine macrophages and found that IFN-γ and/or M. tuberculosis altered expression of 25% of the approximately 10,000 genes assayed. M. tuberculosis infection affected many of the same genes as did IFN-γ or synergized with IFN-γ by altering more genes (20).
In humans, an inherited deficiency of the IFN-γ receptor and polymorphisms of the IFN-γ promoter confer susceptibility to mycobacterial infection (17, 51), but further links to the role of IFN-γ induction of iNOS in human tuberculosis (TB) are less clear. There are conflicting studies describing induction of iNOS in TB infection in vitro (1, 14, 16, 19, 39, 42, 50, 54, 59). In vivo, there is a growing consensus that iNOS is up-regulated in TB. Other investigators have described an increase in iNOS protein levels and catalytic activity in alveolar macrophages (AM) and granulomas from TB patients (8, 21, 41). IFN-γ also induces the chemokine inducible 10-kDa protein (IP-10) and has been found in murine and human lungs infected with M. tuberculosis (49, 52). IP-10 recruits T cells to sites of inflammation and may play a role in generation and function of effector T cells (18, 27, 34, 45).
We have observed an increased number of lymphocytes in the bronchoalveolar lavage (BAL) of subjects with minimally active TB disease compared to extensive cavitary disease, and these cells produce larger amounts of IFN-γ (10). BAL cells from immunocompromised human immunodeficiency virus (HIV)-positive patients with TB produce less IFN-γ than those from immunologically competent patients with TB (35). This led us to the hypothesis that delivering IFN-γ by aerosol to pulmonary TB patients would lead to a successful outcome by induction of IFN-γ-inducible genes. We have found that patients with multiple-drug-resistant TB who had failed second-line treatment to whom IFN-γ is delivered via aerosol have improved clinical, radiological, and mycobacteriologic outcomes (11). In addition, we have detected that aerosol IFN-γ induces the transcription factors STAT-1 and IRF-1 in BAL cells from TB patients (9).
To better define the role of IFN-γ in human TB, we investigated the effect of IFN-γ on BAL cells in vivo and on macrophage models in vitro. We observed an increase in IP-10 mRNA expression in TB and the synergistic effect IFN-γ had on IP-10 expression both in vitro and in vivo. There is a different pattern of regulation of iNOS mRNA. TB patients have elevated iNOS expression, but IFN-γ does not further induce message levels. These data confirm the importance of iNOS in human TB but suggest differences in immune regulation between the mouse model of infection and human disease.
MATERIALS AND METHODS
Subjects.
Twenty-eight subjects were recruited: 21 TB patients and 7 healthy controls (Table 1). For all TB patients, M. tuberculosis was confirmed by sputum culture; 3 had isoniazid-resistant M. tuberculosis. The average age (± standard error of the mean [SEM]) was 40 ± 3 years; 2 were Caucasian, 11 were Black, 10 were Asian, 5 were Hispanic, and the majority were male. Three of the TB patients were HIV-1 positive, and two of them received IFN-γ treatment. Three of the 7 healthy controls and 7 of the 21 TB patients were current smokers. All subjects signed informed consent approved by the NYU Institutional Review Board.
TABLE 1.
Demographic and BAL cell differentials for study subjectsa
| Subject group (no.) | Age (yr) | No. male | No. HIV+ | No. of smokers | BAL | BAL cell differential (%)
|
||
|---|---|---|---|---|---|---|---|---|
| AM | L | PMN | ||||||
| Healthy (7) | 30 ± 2 | 5 | 0 | 3 | 95 | 4 | 1 | |
| TB (10) | 44 ± 5 | 9 | 1 | 2 | IN | 55 | 20 | 24 |
| UN | 90 | 8 | 2 | |||||
| TB receiving IFN-γ (10) | 42 ± 4 | 10 | 2 | 5 | Pre IN | 70 | 25 | 5 |
| Pre UN | 86 | 12 | 2 | |||||
| Post IN | 72 | 22 | 6 | |||||
| Post UN | 80 | 15 | 5 | |||||
| TB with second BAL (1) | 45 | 1 | 0 | 0 | Pre | 84 | 15 | 1 |
| Post | 91 | 9 | 0 | |||||
Values shown are means ± SEM. AM, alveolar macrophages; L, lymphocytes; PMN, neutrophils; IN, involved lobe; UN, uninvolved lobe; Pre, BAL samples obtained before treatment; Post, BAL samples obtained 1 month into treatment.
BAL.
BAL was performed using a flexible bronchoscope with xylocaine anesthesia, as described previously (48). BAL was performed on TB patients during the first week of antituberculous treatment. Samples were obtained from the radiographically involved and uninvolved (if available) segments of the lung. Ten TB patients were treated with aerosolized IFN-γ at a dose of 500 μg of recombinant IFN-γ (InterMune, Burlingame, Calif.) mixed with 3 ml of normal saline via nebulizer three times a week for 4 weeks in addition to their antituberculous medications. For these subjects, another BAL was then performed within 2 hours of the last aerosol treatment. One subject with active TB had one bronchoscopy before the institution of TB medications, and again after 4 weeks of conventional treatment only, as a control for the IFN-γ-treated patients. The BAL fluid was processed as previously described (48), and cell RNA extraction was begun immediately after acquisition.
Bacterial strains and cell culture.
M. tuberculosis strain TN913, a clinical isolate obtained from the Public Health Research Institute TB Center, was grown as previously described (47). THP-1 cells were treated with 20 nM 12-O-tetradecanoylphorbol 13-acetate (PMA) (Sigma, St. Louis, Mo.) for 24 h, prior to infection with mycobacteria at a multiplicity of infection (MOI) of approximately 3 as previously described (47). After 3 days of infection the cells were harvested or treated with 1 ng of IFN-γ/ml for 2 to 24 h and then harvested for RNA.
Real-time quantitative RT-PCR.
Total RNA from BAL cells was reverse transcribed using oligo-d(T), and first-strand PCR was performed following the manufacturer's instructions for the SuperScript preamplification system (Life Technologies, Carlsbad, Calif.). PCR was carried out with 20% of the cDNA using oligonucleotide primers based on published cDNA sequences (GenBank, National Center for Biotechnology Information [NCBI]). Gene expression was normalized against expression of the human housekeeping gene for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). We used a real-time quantitative reverse transcription (RT)-PCR assay with molecular beacons (57). Sequences for primer and molecular beacons used to detect gene expression are as follows: iNOS forward primer, 5′ TGGCAGCATCAGAGGGGACC 3′; reverse primer, 5′ GCAGGACA GGGGACCACATCGAA 3′; molecular beacon, 5′-(FAM) CCGACGCGTGGAAGCCCAAGT ACGCGTCG (DABCYL)-3′; IP-10 forward primer, 5′ GAGCCTCAGCAGAGGAACC 3′; reverse primer, 5′ GAGTCAGAA AGATAAGGCAGC 3′; molecular beacon, 5′-(FAM) CCGACGGTCTCAGCACCATGAATCA AACGTCGG (DABCYL)-3′; and GAPDH forward primer, 5′ GACCCTCACTGCTGGGGAGT 3′; reverse primer, 5′ACTGTGA GGAGGGGAGATTC 3′; molecular beacon, 5′-(TET) GGACGCGGTGGGGGACTGAGTGTG GCGT CC (DABCYL)-3′.
Nuclear run-on transcription rate assays.
Nuclear run-on experiments were also done as previously described (46). Probes for iNOS, IP-10, the plasmid pGEM as a negative control, and the housekeeping gene for GAPDH were included.
RNA labeling and hybridization for cDNA filters.
Six patients, five of whom received IFN-γ, had serial bronchoscopy and had RNA prepared for hybridization to high-density cDNA arrays according to the manufacturer's instructions (GF211; Research Genetics, Carlsbad, Calif.). These nylon arrays contain 4,058 named human genes. The gel images resulting from the phosphorimager were directly imported into the image analysis software Pathways 3 (Research Genetics). The software locates, calculates, and stores each of the cDNA spot intensities from each gel file. The data were normalized by Cluster to a median intensity of 1 for each filter. TreeView was then used to illustrate the relatedness of the patterns of gene expression determined by the clustering algorithms. We empirically defined the threshold value by comparing the results of four independent experiments on THP-1 macrophages infected with a clinical strain of M. tuberculosis for 3 days. For all genes that produced intensity values between 0.4 and 10, the standard deviation of these replicates was below 30% of the mean, indicating excellent reproducibility within these levels of gene expression. In BAL cells, 1,300 genes had mRNA expression within this range.
Data normalization.
The purpose of normalization was to remove variation due to differences in sample preparation, array production, or processing of the array. Raw data from each array scan were divided by the median expression level of this scan and log (base 2) transformed. In the microarray experiments in which both the involved and uninvolved lung tissue samples were available, the samples from involved lung tissue were additionally normalized against the control samples (from uninvolved lung tissue) for each patient by dividing the expression for each gene in the involved sample by the expression of the same gene in the uninvolved sample from the lung of the same patient. Prior to normalization, the data were explored numerically and graphically using the statistical software packages S-plus and MATLAB, and such preprocessing methods as thresholding and filtering were applied, when necessary, if the data contained negative values, zero, or unusually large values (outliers).
Statistical methods.
The microarray data were examined using various clustering and discrimination methods to identify genes that were differentially expressed with each condition. Analyses of differences in gene expression by the GeneFilter assay between pre-IFN-γ BAL and post-IFN-γ BAL were performed, using the significance analysis of microarrays (SAM) procedure proposed by Tusher et al. (56). SAM assigns a score to each gene on the basis of its change in gene expression relative to the standard deviation of repeated measurements for that gene. Genes whose absolute score is larger than some threshold are called significant. A permutation procedure is used to estimate the false discovery rate (FDR) of the resulting rule for each threshold value, and the FDR can help determine the best choice of threshold. The FDR is defined as the expected proportion of false discoveries among the tests (i.e., genes) that are declared significant (2, 55). The FDR approach incorporated into the SAM procedure is a method of controlling for multiple comparison which offers a sensible balance between the number of true findings and the number of false positives while avoiding a flood of false-positive results. It also provides a measure of statistical significance for each individual gene, called the q value, which, unlike the P value, takes into account the fact that thousands of genes are simultaneously being tested.
Descriptive statistics, including means, SEM, and percentages, were used to summarize the demographic variables of the study subjects, BAL cell differentials, and gene expression data.
RESULTS
IP-10 and iNOS mRNA expression and transcription in vitro following M. tuberculosis infection or IFN-γ.
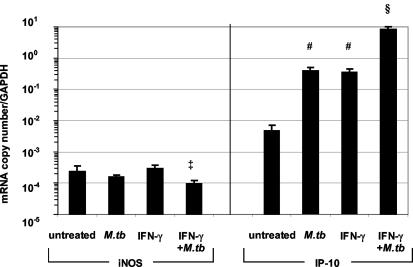
To characterize the regulation of IP-10 and iNOS expression in human macrophages, we used the THP-1 cell line, differentiated to macrophages. Previous investigations have demonstrated that THP-1 macrophages are an accurate model of the interferon response in AM (28, 29, 58). iNOS mRNA levels in THP-1 macrophages were unaffected by either IFN-γ or infection with M. tuberculosis (Fig. 1, left panel), while iNOS levels were reduced significantly in cells treated with both IFN-γ and M. tuberculosis (P ≤ 0.05). Similar results were observed from ex vivo stimulations of human AM (data not shown). In contrast, IP-10 expression was increased 80-fold by M. tuberculosis infection (P ≤ 0.03) or IFN-γ treatment compared to that for untreated cells (P ≤ 0.03) (Fig. 1, right panel). The combination of both M. tuberculosis and IFN-γ led to synergistic 1,000-fold induction of IP-10 mRNA levels compared to those for untreated cells (P ≤ 0.01).
FIG. 1.
iNOS and IP-10 levels measured in vitro by real-time quantitative PCR. PMA-differentiated THP-1 cells were infected with M. tuberculosis (MOI of 3:1) for 3 days (M.tb), treated with 1 ng of IFN-γ/ml for 2 h (IFN-γ), or both (IFN-γ+M.tb). All experiments were repeated three to five times. Levels of IP-10 were significantly increased in THP-1 cells after M. tuberculosis or IFN-γ treatment (#, P ≤ 0.03), and the combination resulted in a further increase (§, P ≤ 0.005 compared to results with no treatment, TB treatment, or IFN-γ treatment). Values were corrected for GAPDH expression and are expressed as means ± SEM. ‡, P = 0.03 compared to results with IFN-γ.
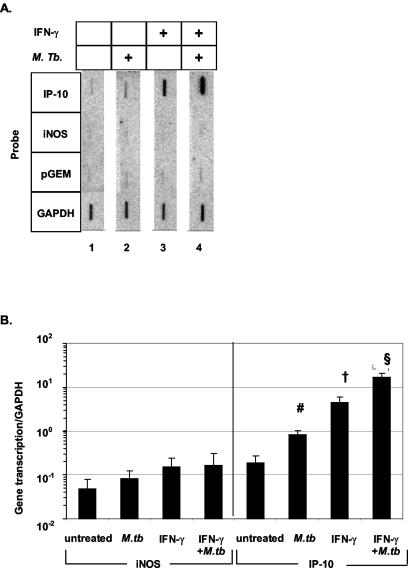
To determine if changes in RNA expression were due to transcriptional regulation, transcription rates of iNOS and IP-10 genes were measured by nuclear run-on in THP-1 macrophages stimulated with either IFN-γ, M. tuberculosis, or both. Figure 2A depicts a representative nuclear run-on assay. iNOS expression was at the background levels of the assay with all of the conditions. However, IP-10 gene transcription was increased with M. tuberculosis infection (lane 2) or IFN-γ treatment (lane 3) alone. The combination of both IFN-γ and M. tuberculosis synergistically induced IP-10 expression (lane 4). Figure 2B summarizes the data from six separate nuclear run-on experiments. Again, iNOS gene transcription was not induced by any of the conditions, yet IP-10 gene transcription was induced in THP-1 macrophages with M. tuberculosis infection (P = 0.01), IFN-γ stimulation (P ≤ 0.04), and the combination of both (P ≤ 0.02) compared to that in untreated cells. Therefore, these data suggest that alterations in mRNA are due to changes in gene transcription.
FIG. 2.
Nuclear run-on assay of iNOS and IP-10 in THP-1 macrophages. (A) Representative nuclear run-on assay of PMA-differentiated THP-1 cells. Probes for the IP-10, iNOS, the plasmid pGEM as a negative control, and the GAPDH housekeeping gene are shown. Lane 1, untreated; lane 2, infected with a clinical strain of M. tuberculosis (MOI of 3:1) for 3 days; lane 3, treated with 1 ng of IFN-γ/ml for 2 h; lane 4, both infected with M. tuberculosis and treated with IFN-γ. (B) Cumulative data from six separate experiments with THP-1 macrophages for iNOS (left) and IP-10 (right) nuclear transcription. IP-10 transcription was significantly induced by M. tuberculosis (#, P = 0.01) and IFN-γ (†, P ≤ 0.04 compared to results with no treatment and TB treatment), and results with the combination were significantly increased compared to results with either one alone and with untreated cells §, P < 0.02). Values are expressed as means ± SEM.
M. tuberculosis but not IFN-γ increased iNOS mRNA expression in vivo. We next investigated the mechanism underlying the beneficial effect of aerosolized IFN-γ seen in the treatment of pulmonary TB (11). For 21 TB patients, BAL was performed in the radiographically involved lung at the time of diagnosis, and for 10 patients, BAL was performed again after 1 month of treatment with conventional antituberculous medications and 500 μg of IFN-γ. In addition, BAL was performed for seven healthy control subjects. The BAL from the involved lobe of all TB patients contained fewer macrophages (61% ± 6% versus 89% ± 4%; P = 0.01) and more lymphocytes (21% ± 5% versus 4% ± 1%; P = 0.03) than BAL from healthy subjects (Table 1). There were no significant changes in BAL cell differentials after 1 month of aerosolized IFN-γ and antituberculous treatment.
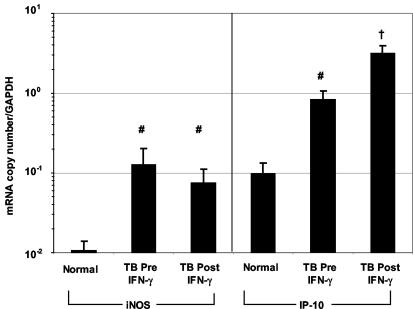
To determine iNOS gene expression in BAL cells during TB, iNOS mRNA was measured by real-time quantitative RT-PCR (Fig. 3, left panel). iNOS expression was significantly higher in the BAL cells of TB patients than in those of healthy subjects (P = 0.01) and did not change after 1 month of treatment with aerosolized IFN-γ plus anti-TB medications. Correction for cell populations did not alter the results. Measurements of BAL fluid nitrite levels by Griess reagent were below the limits of detection (3 μM) for all subjects (data not shown).
FIG. 3.
iNOS and IP-10 mRNA levels in vivo measured by real-time quantitative PCR. iNOS (left) and IP-10 (right) expression was measured by quantitative PCR using molecular beacons in mRNA from BAL cells. Data are from 17 of 21 TB patients and 8 of 10 patients who received IFN-γ. Values were corrected for GAPDH gene expression and are expressed as means ± SEM. iNOS and IP-10 mRNA levels were higher in TB patients than in normal volunteers (#, P ≤ 0.03), and IP-10 was increased further post-IFN-γ treatment (Post IFN-γ) (†, P ≤ 0.01 compared to normal and TB pre-IFN-γ treatment [Pre IFN-γ] results).
IP-10 mRNA expression was evaluated to establish a biological effect of aerosolized IFN-γ. IP-10 expression, normalized to GAPDH, was increased threefold in BAL cells from TB patients compared to that in normal BAL cells (P = 0.03) (Fig. 3, right panel). Treatment with aerosolized IFN-γ enhanced IP-10 expression another fourfold in TB patients (P = 0.01). As a control, one pulmonary TB patient who received only anti-TB medications and had a second BAL had no change in either iNOS or IP-10 expression after 1 month of conventional therapy (data not shown).
Genomic assessment of genes regulated by IFN-γ in vivo.
In order to identify other genes regulated by aerosolized IFN-γ treatment, mRNA from the BAL cells of five TB patients obtained from the radiographically involved and uninvolved segments of the lung before and after 1 month of treatment with aerosolized IFN-γ and antituberculous medications was hybridized to high-density cDNA arrays.
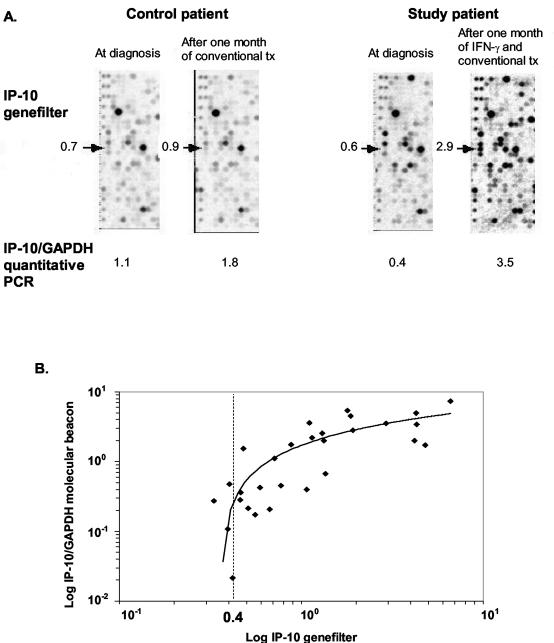
We compared genomics to quantitative PCR for IP-10 mRNA expression. Representative GeneFilter panels with the IP-10 spot indicated are shown in Fig. 4A. The two panels on the left of the figure are from a TB patient before and after 1 month of conventional antituberculous treatment and show no change in IP-10 expression. The two panels on the right are from a patient before and after 1 month of aerosolized IFN-γ in addition to conventional treatment and show a fivefold induction of IP-10. The respective IP-10/GAPDH ratios assayed by quantitative RT-PCR on the same RNA samples are shown below each panel. The values for the two assays correlate well, and IP-10 expression increased after aerosolized IFN-γ administration. In Fig. 4B, IP-10 expression in 29 BAL samples from 16 healthy subjects and TB patients measured by quantitative PCR (y axis) and cDNA array (x axis) exhibited a strong association (r2 = 0.53; P < 0.001), although the sensitivity of the cDNA decreased at a relative expression of 0.4, supporting our threshold value calculated for gene detection. Using this cutoff, 1,300 out of 4,058 genes (32%) were expressed in BAL cells. Microarray data were then analyzed by SAM to calculate the FDR, which is the expected proportion of false discoveries among the genes that are declared significant. We chose an FDR value of less than or equal to 70% because of the greater number of IFN-inducible genes differentially expressed at this FDR value when comparing BAL gene expression patterns before and after IFN-γ treatment. Yet a high FDR means the gene has a greater likelihood of being false positive. To further censor our results, we decided to look at those genes that changed by more than twofold after IFN-γ treatment.
FIG. 4.
Validation of GeneFilter assays. (A) Representative panels from a GeneFilter membrane for two TB patients before and after 1 month of conventional TB treatment (tx) or conventional treatment and aerosolized IFN-γ. The IP-10 gene probe is marked by the arrow. Numeric values are given for relative expression of cDNA hybridized to the spot. Below the panels are the respective IP-10/GAPDH ratios calculated by quantitative RT-PCR. (B) IP-10/GAPDH measured by quantitative PCR (y axis) and IP-10 expression measured by GeneFilter (x axis) (n = 29) were significantly correlated (r2 = 0.53; P < 0.001). The value 0.4 shows the cutoff for sensitivity of the GeneFilter assay and corresponds to the threshold value selected for gene detection.
Only six genes in BAL cells were significantly up-regulated by IFN-γ, with an FDR of ≤70% and a post-IFN-γ treatment/pre-IFN-γ treatment ratio of >2 (Table 2). As expected, the IP-10 gene was included in this group in addition to the gene for the inflammatory chemokine MCP-1. As shown earlier in Fig. 4A, the BAL cells from the lobes of a patient who received only conventional TB treatment failed to show an increase in IP-10 expression after 1 month of therapy. Nine genes were down-regulated, with an FDR of ≤70% and a ratio of less than 0.5 (Table 3).
TABLE 2.
Genes significantly up-regulated in BAL cells after aerosol IFN-γ treatment
| Gene product | Fold change in gene expression | FDR (%) |
|---|---|---|
| RAD51-like 3 | 2.2 | 70 |
| CREB 3 | 2.5 | 70 |
| Monocyte chemotactic protein 1 | 4.2 | 70 |
| Growth arrest-specific 6 | 2.5 | 70 |
| Fibronectin 1 | 2.4 | 70 |
| IP-10 | 3.7 | 70 |
TABLE 3.
Genes significantly down-regulated in BAL cells after aerosol IFN-γ treatment
| Gene product | Fold change in gene expression | FDR (%) |
|---|---|---|
| Cytochrome P450, subfamily XXIV (vitamin D 24-hydroxylase) | 0.49 | 62 |
| Acyl-coenzyme A dehydrogenase, C-2 to C-3 short chain | 0.49 | 62 |
| Beta-galactosidase 1 | 0.45 | 62 |
| Fructose-bisphosphatase 1 | 0.49 | 62 |
| Dihydropyrimidinase-like 2 | 0.47 | 62 |
| Macrophage receptor with collagenous structure | 0.43 | 62 |
| RAN binding protein 2-like 1 | 0.41 | 66 |
| Pyridoxal (pyridoxine, vitamin B6) kinase | 0.44 | 70 |
| Homeo box D3 | 0.49 | 70 |
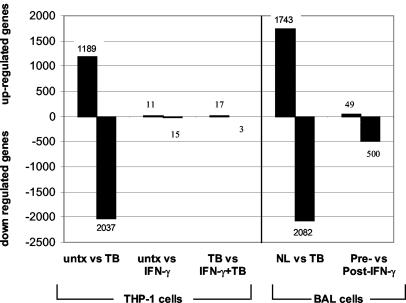
To test if this small number of regulated genes was due to insensitivity of the cDNA arrays, we assayed the effect of IFN-γ and M. tuberculosis infection on THP-1 macrophages. The number of genes significantly regulated (FDR of ≤70%) in THP-1 cells and in patients is shown in Fig. 5. Only a few genes were regulated by IFN-γ alone. In contrast, TB infection regulated many more genes in THP-1 cells. We also examined the gene expression patterns from the BAL of three of the healthy volunteers and compared their values to those of the TB patients at diagnosis. Again, TB infection affected the regulation of many genes, while treatment with aerosolized IFN-γ did not affect as many, just as in THP-1 cells. These data confirm that the high-density cDNA arrays are able to detect changes of a large number of genes after stimulation, as evidenced by M. tuberculosis infection, and also suggest that IFN-γ treatment has little influence on gene expression.
FIG. 5.
IFN-γ treatment regulated few genes. The numbers of genes significantly regulated in THP-1 cells and in BAL from patients are shown in the graph. Many more genes were regulated by M. tuberculosis infection (TB) in THP-1 cells and in BAL cells, but few genes were regulated by IFN-γ. untx, untreated.
DISCUSSION
In this study, we investigated the effects of aerosolized IFN-γ in patients with pulmonary TB. In support of the importance of iNOS in human TB, we found that iNOS mRNA was increased during M. tuberculosis infection in vivo. This result fits well with pathological investigations that demonstrate an increase in iNOS protein and nitrosilation products in human TB granulomas (8, 21). Unlike the case with the mouse model (15, 24, 38), IFN-γ treatment did not increase iNOS mRNA levels in cell culture or in the lungs of patients. Furthermore, we found that M. tuberculosis infection did not increase iNOS gene transcription or mRNA expression in vitro. The discrepancies between the in vivo observation, the in vitro model, and the mouse model suggest interspecies differences in iNOS regulation.
The regulation of iNOS in human cells has been controversial. Multiple investigations have shown that iNOS was not induced in human macrophages in vitro by IFN-γ, M. tuberculosis infection, or stimulation with bacterial products (1, 39, 54). The human mononuclear cell line U937 transfected with iNOS cDNA could produce high levels of iNOS RNA, protein, and enzyme activity but required supplemental tetrahydrobiopterin, an essential cofactor (3). However, two studies using human peritoneal macrophages were unable to induce NO production despite supplemental tetrahydrobiopterin (54, 59). We used the human mononuclear cell line THP-1 for our in vitro studies. Primed THP-1 cells are known to produce NO upon stimulation with silica or lipopolysaccharide (7). Rich et al. showed that human AM are capable of producing NO, but this did not affect the intracellular growth of M. tuberculosis (50). A potential problem with these studies is that many of them measure NO production with the colorimetric Griess assay, which has been shown to be insensitive (31). This could explain why we were unable to detect NO in BAL fluid. It has been shown that the addition of activated lymphocytes and IFN-γ to M. tuberculosis-infected human macrophages can increase iNOS expression and mycobacterial killing and induce a T-helper type 1 immune response, suggesting that models of cellular immunity can up-regulate iNOS (4). In addition, the inflammatory zone of tuberculous granulomas and the nongranulomatous pneumonitis zone are areas in the human lung where macrophages, lymphocytes, and multinucleated giant cells reside, and iNOS and nitrotyrosine can be detected by immunohistochemistry (8). These cells also stain positive for the eNOS isoform, indicating that other isoforms of NOS can be transcriptionally regulated, while our molecular beacon assay was specific for iNOS; eNOS expression was not increased by IFN-γ using the genomics assay. Finally, TB granulomas that stained positive for iNOS also expressed IFN-γ but not interleukin 4 (21). The results from these in vivo studies implied to us that cellular immunity was essential for iNOS activity and supported the finding of elevated iNOS expression in TB BAL in our study as well as others (41). Our observation that IFN-γ treatment did not further induce iNOS in the setting of TB may be explained by an already maximized cellular immune response late in the disease process.
Expression of IFN-regulated genes, such as that for IP-10, is increased in TB and are further augmented by aerosolized IFN-γ. This revealed that IFN-γ reached the alveolus and had a significant effect on resident cells. Previous reports demonstrated that aerosolized IFN-γ increases IP-10 expression in BAL cells of normal volunteers (30). An increase in IP-10 was not observed in the samples from a TB patient who received only antituberculous medications, suggesting that the increase in IP-10 in the experimental patients was due to IFN-γ. Unlike iNOS, IP-10 regulation is similar in murine and human systems (22, 37, 40, 43). In vitro induction of IP-10 mRNA and transcription after infection by M. tuberculosis suggested that it was part of the innate immune response. IP-10 expression is seen early following infection with a variety of pathogens (Taxoplasma gondii, M. tuberculosis, Mycobacterium leprae, viral hepatitis, and HIV) and lipopolysaccharide, recruits activated lymphocytes to sites of inflammation, and may play a role in generation and function of effector T cells by promoting antigen-specific proliferation and IFN-γ secretion (18, 27, 34, 40). Taken together, IP-10 plays an important role in the innate immune response and bridges to the adaptive cellular response.
By using cDNA filter arrays, we confirmed that this assay was able to measure IFN-γ-induced IP-10 mRNA expression. Surprisingly, very few other genes were altered in the BAL of TB patients following aerosolized IFN-γ and antituberculous medications. One reason for the few significantly altered genes is the statistical analysis that we performed on data from a small number of patients. Using a simple paired t test for each gene would yield a large number of “significant” genes (i.e., P value of <0.05), but many of these genes would be false positives. On the other hand, controlling for multiple comparisons by increasing the stringency (e.g., Bonferroni correction) would decrease the number of false-positive values, but at the same time many true positives could be lost. The FDR approach incorporated into the SAM procedure offered a sensible balance between the number of true findings and the number of false positives that was automatically calibrated and easily interpreted. A larger number of genes were found to be significant if we chose to include genes with a high FDR. However, because of the high FDR, they have a greater chance of being false positive. In the case of IP-10 expression, which was significantly elevated in BAL cells after aerosol IFN-γ treatment with an FDR of 70%, the greater than twofold increase in expression and the confirmed elevated gene expression after IFN-γ in vivo and in vitro by real-time quantitative RT-PCR together verify the true-positive conclusion. Many more genes were regulated in THP-1 cells and in BAL cells with M. tuberculosis infection. However, the number of regulated genes dropped significantly after treatment with IFN-γ (Fig. 5). This suggests that IFN-γ treatment has little influence on gene expression and does not account for the success of aerosol IFN-γ treatment. We have shown that IFN-γ signaling pathways are up-regulated in BAL after aerosol treatment (9). Both IP-10 and MCP-1 were elevated. This may be sufficient to account for the clinical response.
An increase in IP-10 also has been demonstrated in mice, in addition to other chemokines and iNOS. However, a considerably higher number of genes were regulated by IFN-γ in mice than we observed in humans (20). This is not surprising, since both inbred mice and cell lines do not harbor the genetic diversity found in a heterogeneous population. Other possible explanations for the smaller number of IFN-regulated genes observed in this clinical study include the following: the IFN-γ system is highly activated in TB and therefore difficult to further stimulate; differences in IFN-γ signaling between humans and mice, as has been observed with other interferons (23, 44); differences in IFN-γ concentration, routes of administration, and time after last dose; and human genetic diversity obscuring all but the most pronounced effects of IFN-γ.
Another possibility for the small number of genes affected by IFN-γ is that the cDNA arrays yielded variable results. We directly tested reproducibility in this system and found that only genes with very low levels of expression were not precisely measured. In addition, we compared the expression profile from the same lung segment before and after therapy, and most patients demonstrated remarkable stability (data not shown). Finally, comparison of quantitative PCR with GeneFilter assays produced excellent correlation between these two assays. It was therefore unlikely that the small number of regulated genes was due to variability or instability in the genomic assay used in these experiments.
In summary, IP-10 was induced significantly after aerosol IFN-γ treatment. IP-10 is one of the few genes that is consistently up-regulated more than twofold by IFN-γ in vivo. IP-10 may recruit more immune effector cells to the site of infection and could be the reason for the favorable clinical effect seen with aerosol IFN-γ. There are a number of other genes up-regulated and down-regulated that may account for the beneficial effect, but the magnitude of the increase is small and the biological effect is unclear. The data presented, however, clearly demonstrate significant differences between humans and mice in the response of iNOS to IFN-γ both in vitro and in vivo. In addition, we observed far fewer genes regulated by IFN-γ in human TB than would be expected from experiments with mice. These results emphasize the need to validate the conclusions derived from mouse models with humans with disease.
Acknowledgments
This work was supported by NIH/NCRR grants M01 RR00096 and HL-59832, HL-57879, and HL-68517; the ALA Career Investigator Award to M.D.W.; the NYC Speakers Award to B.R.; the Uehara Memorial Foundation Award to Y.H.; the NIH/NYU AIDS Institutional Training Grant and Parker B. Francis Award to J.A.G.; and the Doris Duke Clinical Scientist Award to R.C.
We thank Andrew Lubin, Eleni Michailidis, and Sharmila Basu for their assistance.
Editor: V. J. DiRita
REFERENCES
- 1.Aston, C., W. N. Rom, A. T. Talbot, and J. Reibman. 1998. Early inhibition of mycobacterial growth by human alveolar macrophages is not due to nitric oxide. Am. J. Respir. Crit. Care Med. 157:1943-1950. [DOI] [PubMed] [Google Scholar]
- 2.Benyamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a powerful and practical approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 3.Bertholet, S., E. Tzeng, E. Felley-Bosco, and J. Mauel. 1999. Expression of the inducible NO synthase in human monocytic U937 cells allows high output nitric oxide production. J. Leukoc. Biol. 65:50-58. [DOI] [PubMed] [Google Scholar]
- 4.Bonecini-Almeida, M. G., S. Chitale, I. Boutsikakis, J. Geng, H. Doo, S. He, and J. L. Ho. 1998. Induction of in vitro human macrophage anti-Mycobacterium tuberculosis activity: requirement for IFN-gamma and primed lymphocytes. J. Immunol. 160:4490-4499. [PubMed] [Google Scholar]
- 5.Chan, J., K. Tanaka, D. Carroll, J. Flynn, and B. R. Bloom. 1995. Effects of nitric oxide synthase inhibitors on murine infection with Mycobacterium tuberculosis. Infect. Immun. 63:736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, J., Y. Xing, R. S. Magliozzo, and B. R. Bloom. 1992. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J. Exp. Med. 175:1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, F., D. C. Kuhn, L. J. Gaydos, and L. M. Demers. 1996. Induction of nitric oxide and nitric oxide synthase mRNA by silica and lipopolysaccharide in PMA-primed THP-1 cells. APMIS 104:176-182. [DOI] [PubMed] [Google Scholar]
- 8.Choi, H. S., P. R. Rai, H. W. Chu, C. Cool, and E. D. Chan. 2002. Analysis of nitric oxide synthase and nitrotyrosine expression in human pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 166:178-186. [DOI] [PubMed] [Google Scholar]
- 9.Condos, R., B. Raju, A. Canova, B. Y. Zhao, M. Weiden, W. N. Rom, and R. Pine. 2003. Recombinant gamma interferon stimulates signal transduction and gene expression in alveolar macrophages in vitro and in tuberculosis patients. Infect. Immun. 71:2058-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condos, R., W. N. Rom, Y. M. Liu, and N. W. Schluger. 1998. Local immune responses correlate with presentation and outcome in tuberculosis. Am. J. Respir. Crit. Care Med. 157:729-735. [DOI] [PubMed] [Google Scholar]
- 11.Condos, R., W. N. Rom, and N. W. Schluger. 1997. Treatment of multidrug-resistant pulmonary tuberculosis with interferon-gamma via aerosol. Lancet 349:1513-1515. [DOI] [PubMed] [Google Scholar]
- 12.Cooper, A. M., D. K. Dalton, T. A. Stewart, J. P. Griffin, D. G. Russell, and I. M. Orme. 1993. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 178:2243-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalton, D. K., S. Pitts-Meek, S. Keshav, I. S. Figari, A. Bradley, and T. A. Stewart. 1993. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science 259:1739-1742. [DOI] [PubMed] [Google Scholar]
- 14.Denis, M. 1994. Human monocytes/macrophages: NO or no NO? J. Leukoc. Biol. 55:682-684. [DOI] [PubMed] [Google Scholar]
- 15.Denis, M. 1991. Interferon-gamma-treated murine macrophages inhibit growth of tubercle bacilli via the generation of reactive nitrogen intermediates. Cell Immunol. 132:150-157. [DOI] [PubMed] [Google Scholar]
- 16.Denis, M. 1991. Tumor necrosis factor and granulocyte macrophage-colony stimulating factor stimulate human macrophages to restrict growth of virulent Mycobacterium avium and to kill avirulent M. avium: killing effector mechanism depends on the generation of reactive nitrogen intermediates. J. Leukoc. Biol. 49:380-387. [DOI] [PubMed] [Google Scholar]
- 17.Dorman, S. E., and S. M. Holland. 1998. Mutation in the signal-transducing chain of the interferon-gamma receptor and susceptibility to mycobacterial infection. J. Clin. Investig. 101:2364-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dufour, J. H., M. Dziejman, M. T. Liu, J. H. Leung, T. E. Lane, and A. D. Luster. 2002. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 168:3195-3204. [DOI] [PubMed] [Google Scholar]
- 19.Dumarey, C. H., V. Labrousse, N. Rastogi, B. B. Vargaftig, and M. Bachelet. 1994. Selective Mycobacterium avium-induced production of nitric oxide by human monocyte-derived macrophages. J. Leukoc. Biol. 56:36-40. [DOI] [PubMed] [Google Scholar]
- 20.Ehrt, S., D. Schnappinger, S. Bekiranov, J. Drenkow, S. Shi, T. R. Gingeras, T. Gaasterland, G. Schoolnik, and C. Nathan. 2001. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194:1123-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Facchetti, F., W. Vermi, S. Fiorentini, M. Chilosi, A. Caruso, M. Duse, L. D. Notarangelo, and R. Badolato. 1999. Expression of inducible nitric oxide synthase in human granulomas and histiocytic reactions. Am. J. Pathol. 154:145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farber, J. M. 1997. Mig and IP-10: CXC chemokines that target lymphocytes. J. Leukoc. Biol. 61:246-257. [PubMed] [Google Scholar]
- 23.Farrar, J. D., J. D. Smith, T. L. Murphy, S. Leung, G. R. Stark, and K. M. Murphy. 2000. Selective loss of type I interferon-induced STAT4 activation caused by a minisatellite insertion in mouse Stat2. Nat. Immunol. 1:65-69. [DOI] [PubMed] [Google Scholar]
- 24.Flesch, I. E., and S. H. Kaufmann. 1991. Mechanisms involved in mycobacterial growth inhibition by gamma interferon-activated bone marrow macrophages: role of reactive nitrogen intermediates. Infect. Immun. 59:3213-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93-129. [DOI] [PubMed] [Google Scholar]
- 26.Flynn, J. L., J. Chan, K. J. Triebold, D. K. Dalton, T. A. Stewart, and B. R. Bloom. 1993. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 178:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gangur, V., F. E. Simons, and K. T. Hayglass. 1998. Human IP-10 selectively promotes dominance of polyclonally activated and environmental antigen-driven IFN-gamma over IL-4 responses. FASEB J. 12:705-713. [DOI] [PubMed] [Google Scholar]
- 28.Honda, Y., L. Rogers, K. Nakata, B. Y. Zhao, R. Pine, Y. Nakai, K. Kurosu, W. N. Rom, and M. Weiden. 1998. Type I interferon induces inhibitory 16-kD CCAAT/enhancer binding protein (C/EBP)beta, repressing the HIV-1 long terminal repeat in macrophages: pulmonary tuberculosis alters C/EBP expression, enhancing HIV-1 replication. J. Exp. Med. 188:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshino, Y., K. Nakata, S. Hoshino, Y. Honda, D. B. Tse, T. Shioda, W. N. Rom, and M. Weiden. 2002. Maximal HIV-1 replication in alveolar macrophages during tuberculosis requires both lymphocyte contact and cytokines. J. Exp. Med. 195:495-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaffe, H. A., R. Buhl, A. Mastrangeli, K. J. Holroyd, C. Saltini, D. Czerski, H. S. Jaffe, S. Kramer, S. Sherwin, and R. G. Crystal. 1991. Organ specific cytokine therapy. Local activation of mononuclear phagocytes by delivery of an aerosol of recombinant interferon-gamma to the human lung. J. Clin. Investig. 88:297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jagannath, C., J. K. Actor, and R. L. Hunter, Jr. 1998. Induction of nitric oxide in human monocytes and monocyte cell lines by Mycobacterium tuberculosis. Nitric Oxide 2:174-186. [DOI] [PubMed] [Google Scholar]
- 32.Kamijo, R., H. Harada, T. Matsuyama, M. Bosland, J. Gerecitano, D. Shapiro, J. Le, S. I. Koh, T. Kimura, S. J. Green, et al. 1994. Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science 263:1612-1615. [DOI] [PubMed] [Google Scholar]
- 33.Kamijo, R., J. Le, D. Shapiro, E. A. Havell, S. Huang, M. Aguet, M. Bosland, and J. Vilcek. 1993. Mice that lack the interferon-gamma receptor have profoundly altered responses to infection with Bacillus Calmette-Guerin and subsequent challenge with lipopolysaccharide. J. Exp. Med. 178:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan, I. A., J. A. MacLean, F. S. Lee, L. Casciotti, E. DeHaan, J. D. Schwartzman, and A. D. Luster. 2000. IP-10 is critical for effector T cell trafficking and host survival in Toxoplasma gondii infection. Immunity 12:483-494. [DOI] [PubMed] [Google Scholar]
- 35.Law, K. F., J. Jagirdar, M. D. Weiden, M. Bodkin, and W. N. Rom. 1996. Tuberculosis in HIV-positive patients: cellular response and immune activation in the lung. Am. J. Respir. Crit. Care Med. 153:1377-1384. [DOI] [PubMed] [Google Scholar]
- 36.MacMicking, J. D., R. J. North, R. LaCourse, J. S. Mudgett, S. K. Shah, and C. F. Nathan. 1997. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc. Natl. Acad. Sci. USA 94:5243-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majumder, S., L. Z. Zhou, P. Chaturvedi, G. Babcock, S. Aras, and R. M. Ransohoff. 1998. p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein, 10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J. Immunol. 161:4736-4744. [PubMed] [Google Scholar]
- 38.Moreira, A. L., L. Tsenova, P. J. Murray, S. Freeman, A. Bergtold, L. Chiriboga, and G. Kaplan. 2000. Aerosol infection of mice with recombinant BCG secreting murine IFN-gamma partially reconstitutes local protective immunity. Microb. Pathog. 29:175-185. [DOI] [PubMed] [Google Scholar]
- 39.Murray, H. W., and R. F. Teitelbaum. 1992. L-arginine-dependent reactive nitrogen intermediates and the antimicrobial effect of activated human mononuclear phagocytes. J. Infect. Dis. 165:513-517. [DOI] [PubMed] [Google Scholar]
- 40.Neville, L. F., G. Mathiak, and O. Bagasra. 1997. The immunobiology of interferon-gamma inducible protein 10 kD (IP-10): a novel, pleiotropic member of the C-X-C chemokine superfamily. Cytokine Growth Factor Rev. 8:207-219. [DOI] [PubMed] [Google Scholar]
- 41.Nicholson, S., G. Bonecini-Almeida Mda, J. R. Lapa e Silva, C. Nathan, Q. W. Xie, R. Mumford, J. R. Weidner, J. Calaycay, J. Geng, N. Boechat, et al. 1996. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J. Exp. Med. 183:2293-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nozaki, Y., Y. Hasegawa, S. Ichiyama, I. Nakashima, and K. Shimokata. 1997. Mechanism of nitric oxide-dependent killing of Mycobacterium bovis BCG in human alveolar macrophages. Infect. Immun. 65:3644-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohmori, Y., and T. A. Hamilton. 1995. The interferon-stimulated response element and a kappa B site mediate synergistic induction of murine IP-10 gene transcription by IFN-gamma and TNF-alpha. J. Immunol. 154:5235-5244. [PubMed] [Google Scholar]
- 44.O'Shea, J. J., and R. Visconti. 2000. Type 1 IFNs and regulation of TH1 responses: enigmas both resolved and emerge. Nat. Immunol. 1:17-19. [DOI] [PubMed] [Google Scholar]
- 45.Piali, L., C. Weber, G. LaRosa, C. R. Mackay, T. A. Springer, I. Clark-Lewis, and B. Moser. 1998. The chemokine receptor CXCR3 mediates rapid and shear-resistant adhesion-induction of effector T lymphocytes by the chemokines IP10 and Mig. Eur. J. Immunol. 28:961-972. [DOI] [PubMed] [Google Scholar]
- 46.Pine, R., T. Decker, D. S. Kessler, D. E. Levy, and J. E. Darnell, Jr. 1990. Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell. Biol. 10:2448-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiao, Y., S. Prabhakar, E. M. Coccia, M. Weiden, A. Canova, E. Giacomini, and R. Pine. 2002. Host defense responses to infection by Mycobacterium tuberculosis. Induction of IRF-1 and a serine protease inhibitor. J. Biol. Chem. 277:22377-22385. [DOI] [PubMed] [Google Scholar]
- 48.Raju, B., C. F. Tung, D. Cheng, N. Yousefzadeh, R. Condos, W. N. Rom, and D. B. Tse. 2001. In situ activation of helper T cells in the lung. Infect. Immun. 69:4790-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhoades, E. R., A. M. Cooper, and I. M. Orme. 1995. Chemokine response in mice infected with Mycobacterium tuberculosis. Infect. Immun. 63:3871-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rich, E. A., M. Torres, E. Sada, C. K. Finegan, B. D. Hamilton, and Z. Toossi. 1997. Mycobacterium tuberculosis (MTB)-stimulated production of nitric oxide by human alveolar macrophages and relationship of nitric oxide production to growth inhibition of MTB. Tuber. Lung Dis. 78:247-255. [DOI] [PubMed] [Google Scholar]
- 51.Rossouw, M., H. J. Nel, G. S. Cooke, P. D. van Helden, and E. G. Hoal. 2003. Association between tuberculosis and a polymorphic NFkappaB binding site in the interferon gamma gene. Lancet 361:1871-1872. [DOI] [PubMed] [Google Scholar]
- 52.Sauty, A., M. Dziejman, R. A. Taha, A. S. Iarossi, K. Neote, E. A. Garcia-Zepeda, Q. Hamid, and A. D. Luster. 1999. The T cell-specific CXC chemokines IP-10, Mig, and I-TAC are expressed by activated human bronchial epithelial cells. J. Immunol. 162:3549-3558. [PubMed] [Google Scholar]
- 53.Schluger, N. W., and W. N. Rom. 1998. The host immune response to tuberculosis. Am. J. Respir. Crit. Care Med. 157:679-691. [DOI] [PubMed] [Google Scholar]
- 54.Schneemann, M., G. Schoedon, S. Hofer, N. Blau, L. Guerrero, and A. Schaffner. 1993. Nitric oxide synthase is not a constituent of the antimicrobial armature of human mononuclear phagocytes. J. Infect. Dis. 167:1358-1363. [DOI] [PubMed] [Google Scholar]
- 55.Storey, J. 2002. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B 64:479-498. [Google Scholar]
- 56.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 58.Weiden, M., N. Tanaka, Y. Qiao, B. Y. Zhao, Y. Honda, K. Nakata, A. Canova, D. E. Levy, W. N. Rom, and R. Pine. 2000. Differentiation of monocytes to macrophages switches the Mycobacterium tuberculosis effect on HIV-1 replication from stimulation to inhibition: modulation of interferon response and CCAAT/enhancer binding protein beta expression. J. Immunol. 165:2028-2039. [DOI] [PubMed] [Google Scholar]
- 59.Weinberg, J. B., M. A. Misukonis, P. J. Shami, S. N. Mason, D. L. Sauls, W. A. Dittman, E. R. Wood, G. K. Smith, B. McDonald, K. E. Bachus, et al. 1995. Human mononuclear phagocyte inducible nitric oxide synthase (iNOS): analysis of iNOS mRNA, iNOS protein, biopterin, and nitric oxide production by blood monocytes and peritoneal macrophages. Blood 86:1184-1195. [PubMed] [Google Scholar]