Abstract
Oxidative stress resistance is one of the key properties that enable pathogenic bacteria to survive the toxic reactive oxygen species released by the host. In a previous study characterizing oxidative stress resistance mutants of Helicobacter pylori, a novel potential antioxidant protein (MdaB) was identified by the observation that the expression of this protein was significantly upregulated to compensate for the loss of other major antioxidant components. In this study, we characterized an H. pylori mdaB mutant and the MdaB protein. While the wild-type strain can tolerate 10% oxygen for growth, the growth of the mdaB mutant was significantly inhibited by this oxygen condition. The mdaB mutant is also more sensitive to H2O2, organic hydroperoxides, and the superoxide-generating agent paraquat. Although the wild-type strain can survive more than 10 h of air exposure, exposure of the mutant strain to air for 8 h resulted in recovery of no viable cells. The oxidative stress sensitivity of the mdaB mutant resulted in a deficiency in the ability to colonize mouse stomachs. H. pylori was recovered from 10 of 11 mouse stomachs inoculated with the wild-type strain, with about 5,000 to 45,000 CFU/g of stomach. However, only 3 of 12 mice that were inoculated with the mdaB mutant strain were found to harbor any H. pylori, and these 3 contained less than 2,000 CFU/g of stomach. A His-tagged MdaB protein was purified and characterized. It was shown to be a flavoprotein that catalyzes two-electron transfer from NAD(P)H to quinones. It reduces both ubiquinones and menaquinones with similar efficiencies and preferably uses NADPH as an electron donor. We propose that the physiological function of the H. pylori MdaB protein is that of an NADPH quinone reductase that plays an important role in managing oxidative stress and contributes to successful colonization of the host.
The gastric pathogen Helicobacter pylori is a microaerophilic bacterium that causes peptic ulcers and is a risk factor for adenocarcinomas (11). An important feature of the pathogenesis of H. pylori infection is its persistence in the inflamed gastric mucosa. Infection with H. pylori induces an inflammatory response (gastritis) that leads to an increase in the level of toxic oxygen species in the gastric mucosa and the gastric juice (2, 17, 26). To resist oxidative damage from chronic inflammation, H. pylori relies on a variety of protective enzymatic systems. These include catalase (KatA), superoxide dismutase (SodB), and thioredoxin-dependent peroxiredoxin systems such as alkylhydroperoxide reductase (AhpC) and thioperoxidase (Tpx). These systems in H. pylori have been recently characterized (3, 9, 14, 24, 28). Disruption of the corresponding genes for all of these severely affects the bacterium's ability to colonize the host (15, 25, 28), indicating the significant roles of the oxidative stress resistance factors in H. pylori pathogenesis.
In characterizing the oxidative stress resistance mutants of H. pylori, we observed that H. pylori is able to respond to loss of a major oxidative stress resistance component by up-regulating the expression of other antioxidant proteins (24). For example, disruption of AhpC frequently resulted in increased expression of the iron-binding, neutrophil-activating protein (NapA). Notably, another possible antioxidant protein was identified solely by observing its induction in the ahpC napA double mutant. Two-dimensional gel electrophoresis combined with N-terminal protein sequencing revealed that this induced protein corresponds to HP0630 in the genome sequence of strain 26695.
Sequence analysis of HP0630 revealed that it belongs to a family of NAD(P)H dehydrogenases that, in eukaryotic cells, catalyze the two-electron reduction of quinone and are involved in cellular protection against oxidative stress damage (20). The single bacterial homologue that has been characterized is the MdaB protein of Escherichia coli, which was first identified as a modulator of drug activity (also named mda66 because the gene was mapped at 66 min on the E. coli chromosome) (7). Subsequently, this E. coli protein was shown to be an NADPH-specific quinone reductase that catalyzes the two-electron reduction of quinone to quinols (16). Recent genome sequencing projects revealed that the MdaB homologue is present in the genomes of many pathogenic bacteria, such as Haemophilus influenzae, Pseudomonas putida, Campylobacter jejuni, Salmonella enterica serovar Typhimurium, Neisseria meningitides, Shigella flexneri, and Yersinia pestis. However, the physiological role of MdaB, particularly as an antioxidant factor in pathogenic bacteria, has not been investigated.
The goals of this study were to characterize the H. pylori MdaB protein and investigate the roles of this protein in oxidative stress resistance and in colonization of the host stomach.
MATERIALS AND METHODS
Biochemicals.
Unless otherwise stated, all of the biochemicals and reagents used in this study were from Sigma Chemical Co., St. Louis, Mo.
H. pylori strains and growth conditions.
H. pylori strains ATCC 43504 and SS1 were used as the wild type. H. pylori was cultured on brucella agar (Difco) plates supplemented with 10% defibrinated sheep blood or 5% fetal bovine serum (called BA plates). Cultures of H. pylori were grown microaerobically at 37°C in a 5% CO2 incubator under different continuously controlled levels of oxygen (between 2 and 12% partial pressure). Chloramphenicol (50 μg/ml) or kanamycin (40 μg/ml) was added to the medium to culture mutants.
DNA techniques.
All DNA manipulations were performed as previously described (21). Chromosomal DNA was extracted from H. pylori with the Aquapure genomic DNA extraction kit (Bio-Rad). Plasmid DNA preparations were carried out with the QiaPrep Spin mini kit (Qiagen). DNA fragments or PCR products were purified from agarose gels with the QIAquick gel extraction kit (Qiagen). PCR was performed with a Perkin-Elmer 2400 thermal cycler with Taq or Pfu DNA polymerase (Fisher). Oligonucleotide primers were synthesized by Integrated DNA Technologies, Coralville, Iowa.
Construction of an H. pylori mdaB mutant.
Primers mdaF (5′-GATTTGCCTAAAGAATACCGC-3′) and mdaR (5′-TCGTAGAACATGACCACTCC-3′) were used to PCR amplify a 1,182-bp fragment containing the H. pylori mdaB gene (HP0630) with genomic DNA from strain ATCC 43504 as the template. The PCR fragment was directly cloned into vector pGEM-T (Promega) in accordance with the manufacturer's instructions to generate pGEM-mdaB (Fig. 1). The host strain used for cloning was E. coli DH5α. Subsequently, a chloramphenicol acetyltransferase cassette was inserted at the unique Eco47III site within the mdaB sequence of pGEM-mdaB (Fig. 1). The recombinant plasmid was then introduced into H. pylori by natural transformation via allelic exchange, and chloramphenicol-resistant colonies were isolated. The disruption of the gene in the genome of the mutant strain was confirmed by PCR showing an increase in the expected size of the PCR product.
FIG. 1.
DNA constructs. The organization of the mdaB gene region in the genome of H. pylori 26695 is shown at the top. The DNA insert fragments that were cloned into corresponding vectors are shown below. pGEM-mdaB::Cm was used for transformation of H. pylori to create H. pylori mdaB::Cm mutants. pET-mdaB-6His was used for overexpression of MdaB protein in E. coli.
Construction of an H. pylori mdaB complementation strain.
The kanamycin resistance cassette (Kan) was inserted behind the mdaB gene in plasmid pGEM-mdaB, yielding pGEM-mdaB-Kan. A fragment containing the mdaB gene and the Kan cassette was then excised from pGEM-mdaB-Kan and ligated into plasmid pEU39, yielding pEU-mdaB-Kan. Plasmid pEU39 (from The Institute for Genomic Research) contains a 2.04-kb fragment of H. pylori genomic DNA covering open reading frame HP0405 (nifS-like gene). Previously, our laboratory showed that disruption of HP0405 in H. pylori had no obvious phenotype. In plasmid pEU-mdaB-Kan, HP0405 was disrupted into two pieces flanking mdaB-Kan. When this plasmid was used to transform H. pylori SS1 mdaB::Cm (selection for Cmr Kanr), the intact mdaB gene (with its promoter) and the Kan cassette were inserted into the genome at the HP0405 locus. This produced a merodiploid strain, SS1 mdaB::Cm-mdaB+-Kan, that contains the original interrupted mdaB gene and an intact copy of mdaB at an unrelated site.
Growth sensitivity to oxygen.
H. pylori wild-type and mutant strains were streaked for isolated colonies onto BA plates and incubated at 37°C in a CO2 incubator with different but continuously controlled oxygen concentrations (2 to 10%). Growth was scored after 48 h as described in Table 1, footnote a.
TABLE 1.
Growth under various O2 concentrations
| Strain | Growtha at O2 concn of:
|
||
|---|---|---|---|
| 2% | 4% | 10% | |
| Wild-type 43504 | +++ | +++ | +++ |
| 43504 mdaB::Cm | +++ | +++ | + |
| 43504 ahpC::Kan | ++ | + | − |
| Wild-type SS1 | +++ | +++ | +++ |
| SS1 mdaB::Cm | +++ | +++ | + |
| SS1 ahpC::Kan | ++ | + | − |
Growth was scored after 48 h as follows: −, no growth; +, growth at site of heaviest inoculum; ++, significant growth and development of isolated, pinpoint colonies (less than 0.5 mm in diameter); +++, healthy growth with individual colonies (>0.5 mm in diameter).
Paper disk assay for peroxide sensitivity.
Sensitivity to different oxidative agents was evaluated by disk assay. Sterile filter paper disks (7.5 mm in diameter) were applied to BA plates that had been streaked for confluent growth of H. pylori strains. Ten-microliters each of the agents indicated in Table 2 was applied to each disk. After the plates had been incubated under a 2% O2 condition for 48 h, the clear zones surrounding the disks were measured. The data given represent the distance from the edge of the disk to the end of the clear zone, where growth begins.
TABLE 2.
Disk sensitivity assaya
| Strain | Mean inhibition zone diam (mm) ± SD
|
|||
|---|---|---|---|---|
| 1 M H2O2 | 0.2 M cumene hydroperoxide | 0.2 M t-butyl hydroperoxide | 20 mM paraquat | |
| SS1 | 1.5 ± 0.2 | 8.0 ± 0.4 | 19.0 ± 0.6 | 0.7 ± 0.2 |
| SS1 mdaB::Cm | 3.1 ± 0.4 | 11.8 ± 0.7 | 21.7 ± 1.1 | 2.8 ± 0.4 |
| SS1 ahpC::Kan | 3.5 ± 0.3 | 12.9 ± 0.9 | 27.9 ± 1.5 | 17.5 ± 1.7 |
Zones of inhibition were measured around filter paper disks saturated with 10 μl of the indicated compounds. Water as a control did not yield any zones of growth inhibition. Results are the means ± standard deviations for eight replicate experiments. According to Students t- distribution test, all of the aphC results and three of the four mdaB results are significantly different (greater inhibition zone) from those of the parent strain at the 99% level of confidence (α′ = 0.01). The inhibition zones for mdaB strain with t-butyl hydroperoxide are significantly greater than those for the parent strain at the 95% level of confidence (α′ = 0.05).
Air survival assay.
Survival of nongrowing H. pylori cells under atmospheric oxygen was assayed as follows. Wild-type or mutant H. pylori cells grown under 2% oxygen were suspended in phosphate-buffered saline (PBS) and incubated at 37°C under normal atmospheric conditions. Samples were removed at the times indicated in Fig. 2, serially diluted, and spread onto BA plates. After 3 days of incubation in a 2% oxygen environment, the colony counts were recorded.
FIG. 2.
Survival of nongrowing H. pylori cells under atmospheric oxygen. Cells of wild-type strain SS1 (diamonds), the isogenic mdaB::Cm mutant (triangles), or the mdaB complementation strain (squares) grown under 2% oxygen were suspended in PBS and incubated at 37°C under normal atmospheric conditions. Samples were removed at the times indicated on the x axis and were used for plate counts in a 2% oxygen environment. The data are the means of three experiments with standard deviations.
Mouse colonization.
Mouse colonization assays were performed essentially as described earlier (25, 28). Briefly, wild-type or mdaB::Cm mutant SS1 cells were harvested after 48 h of growth (37°C, 2% oxygen) on BA plates and suspended in PBS to an optical density at 600 nm of 1.7. The headspace in the tube was sparged with Ar gas to minimize oxygen exposure. These suspensions were administered to C57BL/6J mice (1.5 × 108 CFU/mouse; inocula were kept constant for each experiment) via oral gavage. After 3 weeks, the mice were sacrificed and the stomachs were removed, weighed, and homogenized in Ar-sparged PBS. Homogenate was plated on BA plates supplemented with bacitracin (200 μg/ml) and nalidixic acid (10 μg/ml) and was incubated for 5 to 7 days before examination for the presence of H. pylori colonies.
Construction of plasmid for overproduction of H. pylori MdaB-6His.
With genomic DNA from strain ATCC 43504 as the template and primers MDA66F (5′-CGCGCGGCATATGAAAAAAGTACTCATC-3′) and MDA66R2 (5′-ATATAGGCTCGAGGCACTTGCCAAAAGC-3′), a DNA fragment containing the complete mdaB gene (∼600-bp) was amplified by PCR. The PCR product was digested with NdeI and XhoI and cloned into vector pET-21a (Novagen) previously digested with the same restriction enzymes to generate pET-MdaB-6His (Fig. 1). The recombinant plasmid was transformed into E. coli BL21 Origami (Novagen).
Overexpression and purification of H. pylori MdaB-6His.
E. coli BL21 Origami cells harboring pET-MdaB-6His were grown at 37°C to an optical density at 600 nm of 0.5 in 500 ml of Luria-Bertani medium with ampicillin (100 μg/ml) and kanamycin (40 μg/ml). Expression of the MdaB protein was induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the medium, followed by further incubation for 3 h, and the cells were harvested by centrifugation (5,000 × g, 15 min, 4°C). All subsequent steps were performed at 4°C. Cells were washed with 200 ml of 20 mM Na2HPO4 (pH 7.4)-500 mM NaCl-5 mM imidazole (buffer A) and resuspended in 5 ml of the same buffer. Cells were lysed by two passages through a cold French pressure cell at 18,000 lb/in2. Cell debris was removed by centrifugation at 20,000 × g. The supernatant was applied to a nickel-nitrilotriacetic acid affinity column (Qiagen), and buffer A was used to wash the resin until the A280 reached the baseline. Proteins were washed with buffer B (buffer A with 30 mM imidazole) until the A280 reached the baseline and finally eluted with buffer C (buffer A with 250 mM imidazole). Extracts of E. coli BL21 Origami containing the vector only did not result in retrievable proteins from this purification (Ni affinity) procedure. Fractions were analyzed by gel electrophoresis, and the samples of interest were pooled. Protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce).
Enzyme activity assays.
The enzymatic activity of MdaB protein was mainly determined by measuring NAD(P)H oxidation with Quinone as the electron acceptor (16). The reaction mixture contained 20 mM Tris-HCl (pH 7.5), 0.2 mM NADPH or NADH, 0.1 mM quinone (or other electron donors, listed in Table 3), and 10 μl of the appropriately diluted MdaB protein in a total volume of 1.0 ml. The reaction was started by addition of the enzyme and carried out at room temperature. Enzyme activity was calculated from the decrease in absorbance at 340 nm with an absorbance coefficient of 6.22 mM−1 cm−1. One unit of activity was defined as the amount of enzyme catalyzing the oxidation of 1 μmol of NADPH or NADH per min per mg of protein.
TABLE 3.
NADPH-quinone reductase activities of purified Hp MdaB
| Electron acceptor | Enzyme activity with following electron donor:
|
|
|---|---|---|
| NADPH | NADH | |
| Coenzyme Q0 | 74.4 ± 6.8 | 6.4 ± 0.5 |
| Coenzyme Q1 | 74.4 ± 7.3 | 6.2 ± 0.7 |
| Menadione | 56.0 ± 4.7 | 4.6 ± 1.2 |
| 1,4-Naphthoquinone | 53.6 ± 8.1 | 8.6 ± 0.8 |
| Ferricyanide | 58.4 ± 4.5 | 3.8 ± 0.6 |
| Dichlorophenolindophenol | 1.4 ± 0.2 | 0.2 ± 0.1 |
Enzyme activity was determined spectrometrically by measuring the oxidation of NADPH or NADH (decrease at 340 nm). One unit of activity is defined as the amount of enzyme catalyzing the oxidation of 1 μmol of NADPH or NADH per minute per milligram of protein. Each value is the mean of three sets of experiments with the standard deviation.
The enzymatic activity of MdaB protein was also determined by measuring the reduction of certain quinone-like compounds. Menadione reduction was monitored in a coupled assay under aerobic conditions (27). Reduced menadione, menadiol, in turn reduces cytochrome c. Measurements were made in mixture of 20 mM Tris-HCl (pH 7.5), 0.2 mM NADPH, 0.1 mM menadione, and 0.1 mM cytochrome c. Reduction of cytochrome c was monitored by the increase in absorbance at 550 nm (ɛ550 = 29.5 mM−1 cm−1). For the assay of dichlorophenolindophenol (DCIP) reductase activity, the reaction mixture contained 20 mM Tris-HCl (pH 7.5), 0.2 mM NADPH, and 0.15 mM DCIP. Reduction of DCIP was monitored by the decrease in absorbance at 600 nm (ɛ600 = 21.5 mM−1 cm−1).
RESULTS
H. pylori mdaB mutant is more sensitive to oxidative stress.
To investigate the physiological roles of MdaB in H. pylori, we created an mdaB mutant. The mdaB gene in type strain ATCC 43504 and mouse-adapted strain SS1 was disrupted by insertion of a chloramphenicol acetyltransferase cassette at the unique Eco47III site (Fig. 1). The mutant strains were obtained by screening transformants under 2 or 4% O2 partial pressure conditions.
To characterize the phenotype of the mdaB mutant in oxidative stress resistance, we first determined its ability to tolerate oxygen, compared to that of the wild-type strain (Table 1). In the previous study, the ahpC mutant was shown to be hypersensitive to oxygen, so that the mutant can only be obtained under a 2% O2 atmosphere condition (24). The ahpC mutant (type II) strains (both ATCC 43504 and SS1) were included here as a reference. Under 2 or 4% O2 conditions, the mdaB mutant cells grew well, similar to the wild type. While the wild type grew in a healthy manner at 10% O2, the mdaB mutants grew poorly, indicating that MdaB is involved in oxidative stress resistance. Compared to the ahpC mutant, the mdaB mutant displayed a moderate sensitivity to oxygen.
To test whether the mdaB mutant has increased sensitivity to other oxidative reagents (such as peroxides), a series of disk inhibition assays were performed. Inhibition zones were measured around agent-saturated paper disks (Table 2). Both the mdaB mutant and the ahpC mutant showed moderate sensitivity to H2O2, cumene hydroperoxide, and t-butyl hydroperoxide. The ahpC mutant is highly sensitive to the superoxide-generating agent paraquat, whereas the mdaB mutant is moderately sensitive.
To determine the oxygen-dependent cell-killing effect on the mdaB mutant, we measured the ability of nongrowing cells to survive periods of air exposure (Fig. 2). The wild type is O2 labile but still shows viable cells upon 10 h of air exposure, with a loss of viability of about 2 orders of magnitude. This agrees with our previous studies of the wild-type strains (25, 28). The viability of the mdaB mutant cells upon air exposure dropped much more rapidly, about 4 orders of magnitude after 6 h, and no viable cells were detected after 8 h of exposure to air. These results combined with disk assays clearly showed that MdaB in H. pylori has a physiological role in mediating oxidative stress.
To ensure that the observed sensitivity to oxidative stress is completely attributed to inactivation of mdaB, we introduced a functional copy of the mdaB gene back into the mdaB mutant strain for complementation. Strain SS1 mdaB::Cm-mdaB+-Kan contains a mutated mdaB gene at the original locus and an intact mdaB gene at an unrelated site (see Materials and Methods). We determined the oxygen-dependent cell-killing effect on this strain. Upon exposure to air, this strain exhibited the same phenotype as the wild-type strain (Fig. 2). This indicated that introduction of a functional mdaB gene at another locus restored the cell's ability to resist oxidative stress.
H. pylori mdaB mutant is defective in host colonization.
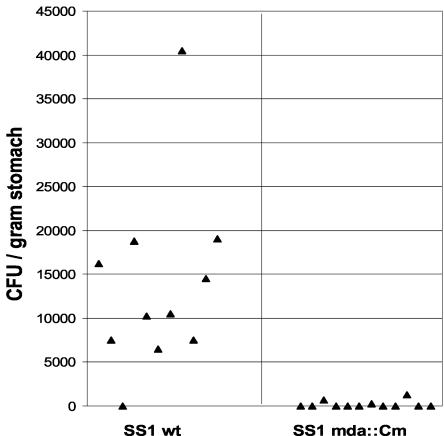
Previously, our laboratory showed that H. pylori sodB and ahpC mutant strains are both defective in host colonization (25, 28). To determine whether loss of MdaB activity is important for H. pylori colonization in the host, the relative abilities of the wild-type and mdaB-deficient mutant strains to colonize the mouse stomach were evaluated (Fig. 3). Eleven C57BL/6J mice were inoculated with wild-type strain SS1, and 12 mice were inoculated with mutant strain SS1 mdaB::Cm; the inoculants were incubated in low O2 for administration to the animals. H. pylori cells were recovered from the mouse stomachs 3 weeks after oral administration. To avoid O2 exposure of the stomach isolates, the stomachs were homogenized under low-O2 conditions and the cells plated from the stomach homogenate dilutions were rapidly transported to an incubator containing 2% O2. H. pylori was recovered from the stomachs of 10 of the 11 mice inoculated with the wild-type strain, with numbers ranging from 5,000 to 45,000 CFU/g of stomach. However, only 3 of the 12 mice that were inoculated with the mdaB mutant strain were found to harbor H. pylori. In addition, the colonization titers of these H. pylori-positive stomachs were all below 2,000 CFU/g of stomach, much lower than those for the wild-type strain. Colonies isolated from the three mouse stomachs inoculated with the mutant strain were confirmed to be the mdaB mutant strain on the basis of their chloramphenicol resistance. These results indicated that MdaB plays an important role in H. pylori colonization of the host.
FIG. 3.
Mouse colonization assay of wild-type (wt) H. pylori SS1 and its isogenic mdaB::Cm mutant. The mice were inoculated with H. pylori two times with a dose of 5 × 108 cells. Data are presented as a scatter plot of CFU per gram of stomach as determined by plate counts.
H. pylori MdaB protein is an NADPH quinone reductase.
To examine the biochemical activities of the MdaB protein, the H. pylori mdaB gene was cloned into pET21a, expressed in E. coli strain BL21 Origami, and purified to near homogeneity with the His tag and nickel-nitrilotriacetic acid resin (Fig. 4). The six-His-tagged MdaB protein migrated at approximately 24 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (the predicted molecular mass of MdaB is 22 kDa). Most of the expressed protein present was soluble, although some was contained in the membrane fraction (data not shown).
FIG. 4.
Expression and purification of the H. pylori MdaB protein. A 12% polyacrylamide gel containing 1% sodium dodecyl sulfate stained with Coomassie brilliant blue included the following samples: crude extract of noninduced (lane 1) and induced (lane 2) E. coli BL21 Origami (pET-mdaB-6His) and 6 μg of purified, six-His-tagged H. pylori MdaB protein (lane 3). Lane M, molecular mass markers (Bio-Rad).
A conserved-domain search (4) for the H. pylori MdaB protein sequence revealed that it consists of a domain with a flavodoxin-like fold (pfam02525), and the E. coli MdaB protein was shown to contain flavin adenine dinucleotide cofactor (16). We examined the absorption spectrum of the purified H. pylori MdaB protein (Fig. 5). The MdaB protein showed a characteristic flavin absorption spectrum with a peak at 376 nm and a major peak at 456 nm with shoulders at 429 and 484 nm. This indicated that the H. pylori MdaB protein contains flavin adenine dinucleotide as a cofactor.
FIG. 5.
Absorbance spectrum of the purified, six-His-tagged H. pylori MdaB protein. The spectrum was recorded at a concentration of 1.3 mg of protein/ml.
The menadione reductase activity of the H. pylori MdaB protein was determined in a coupled assay in which menadione is reduced (with NADPH as the electron donor) to menadiol and, in turn, menadiol reduces cytochrome c. Reduction of cytochrome c was monitored by an increase in absorbance at 550 nm (ɛ550 = 29.5 mM−1 cm−1). On the basis of this assay, the H. pylori MdaB protein showed an activity of 83.5 ± 6.7 μmol of cytochrome c reduced per min per mg of protein. Apparently, the H. pylori MdaB protein reduces menadione by a two-electron transfer pathway, similar to the E. coli MdaB protein and the DT-diaphorase from animal cells.
The substrate specificity of the H. pylori MdaB protein was determined with different electron acceptors and donors (Table 3). The enzyme activities were measured by oxidation of NADPH or NADH (decrease in absorbance at 340 nm). With NADPH as the electron donor, the H. pylori MdaB protein can reduce coenzyme Q0, coenzyme Q1, menadione, 1,4-naphthoquinone, or ferricyanide, with a similar specific activity ranging from 53 to 75 U/mg of protein, and it has a much lower activity (1.4 U/mg of protein) with dichlorophenol-indophenol. NADH can also be used as an electron donor, but it is much less efficient, with the activities about 10% of those with NADPH. No activity was detected when the electron acceptor (quinone-like compound) was omitted from the reaction mixture or when no enzyme was used in the reaction mixture. The results indicated that the H. pylori MdaB protein is a quinone reductase that preferably uses NADPH (over NADH) as an electron donor.
DISCUSSION
In this study, we characterized MdaB, a potential antioxidant protein in H. pylori. The data showed that loss of MdaB function results in the oxygen-sensitive phenotype in vitro and deficiency in the bacterium's ability to colonize the mouse stomach. Biochemical studies on the purified MdaB protein demonstrated that, like its homologue in E. coli, it is able to reduce quinone to quinol, preferably with NADPH as the electron donor.
A body of evidence indicates that NADPH, rather than NADH, is the physiological electron donor to the respiratory chain in H. pylori (6, 8, 18, 29), unlike in many other bacteria. Although a cluster of genes encoding a potential NADH-quinone oxidoreductase was found in the genome sequence of H. pylori (1, 33), the examination of the deduced proteins led Finel (12) to conclude that this complex has no function of NADH oxidation. One reason is the lack of the NADH-binding subunits (NuoF and NuoE), a flavin mononucleotide prosthetic group and an FeS cluster for electron transfer. Summarizing the available data, Chen et al. (8) proposed a model for the electron transfer chain in H. pylori in which a potential NADPH dehydrogenase transfers electrons to a quinone pool. Our data reported here indicate that MdaB could play this role in the electron transfer chain. The E. coli MdaB protein was shown to be NADPH specific, having no activity at all with NADH as the electron donor (16). Our results showed that the H. pylori MdaB protein has a 10-fold preference for NADPH over NADH as the electron donor. On the basis of this specificity and the notion that NADPH is the physiological electron donor in H. pylori, we conclude that the physiological function of the H. pylori MdaB protein is as an NADPH quinone reductase.
The quinone composition, quinone content, and quinone biosynthetic pathways in H. pylori are not very clear (19). Bacteria contain two main types of quinones, ubiquinone (UQ) and menaquinone (MK), that mediate electron transfer between dehydrogenases and reductase or oxidase components of respiratory chains (30). Some evidence supported a notion that MK is the major quinone in H. pylori (22, 23); however, genes for the biosynthesis of MK have not been identified in the genome sequence (19). Perhaps H. pylori obtains MK or the precursors for its synthesis from the host. Although genes for the biosynthesis of UQ were annotated in the H. pylori genomes (1, 33), no UQ was detected in H. pylori (19). Nevertheless, our data demonstrated that the H. pylori MdaB protein reduces both UQ and MK with similar proficiencies.
Quinone metabolism within a cell has a direct effect on the cell's ability to deal with oxidative stress (30). Quinones can be reduced by certain oxidoreductases via a one-electron transfer reaction to semiquinone radicals, which are able to react with molecular oxygen to form superoxide radicals. On the other hand, NAD(P)H quinone oxidoreductase, known as DT-diaphorase in eukaryotes, catalyze the two-electron reduction of quinones to quinols, thus competing with the potentially toxic one-electron reduction pathway and protecting against the cytotoxic and carcinogenic effects of semiquinones (5, 10). It is also well known in eukaryotes that the reduced UQ (ubiquinol) functions as a scavenger for lipid peroxyl radicals and thereby prevents lipid peroxidation (13). Reduced UQ also participates as a direct antioxidant within E. coli cells (31). Our results demonstrated that the H. pylori MdaB protein, like its homologue in E. coli and DT-diaphorase in eukaryotes, catalyzes two-electron transfer from NADPH to quinone, whose reduced status is undoubtedly important for managing oxidative stress. The enzymatic activity of the MdaB protein is consistent with the observed phenotype of the H. pylori mdaB mutant, which showed increased sensitivity to oxidative stress. This also explains why the expression of MdaB was upregulated in ahpC napA mutant cells, in which the bacterium's ability to remove lipid peroxidation was severely impaired by the mutation (24).
Oxidative stress resistance is one of the key properties that enable pathogenic bacteria to survive the effects of the production of reactive oxygen by the host (32). Therefore, the proteins (enzymes) that are involved in oxidative stress resistance are usually important factors in bacterial colonization and pathogenesis. Microaerophilic organisms, like H. pylori, are particularly vulnerable to the detrimental effects of oxygen and oxidative stress. Nevertheless, they can maintain persistent infection by using some enzymatic machineries that eliminate or minimize toxic oxygen-derived products. Recent studies on H. pylori catalase (KatA), superoxide dismutase (SodB), alkylhydroperoxide reductase (AhpC), and thioperoxidase (Tpx) demonstrated that they are important factors conferring resistance to oxidative stress, and loss of any of these functions leads to a significant defect in colonization ability (15, 25, 28). Here we showed that NADPH quinone reductase (MdaB) is another important factor that confers oxidative stress resistance and contributes to colonization of the host. As MdaB homologues are present in the genomes of many pathogenic bacteria, investigating the function and mechanism of this protein in oxidative stress resistance will likely have broad implications for our understanding of bacterial infection and pathogenesis.
Acknowledgments
This work was supported by NIH grant 1-RO1-DK60061-01.
We thank Nalini Metha, James P. Walton, Adriana Olczak and Sue Maier for expertise and assistance.
Editor: B. B. Finlay
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Bagchi, D., G. Bhattachatya, and S. J. Stohs. 1996. Production of reactive oxygen species by gastric cells in association with Helicobacter pylori. Free Radic. Res. 24:439-450. [DOI] [PubMed] [Google Scholar]
- 3.Baker, L. M., A. Raudonikiene, P. S. Hoffman, and L. B. Poole. 2001. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 183:1961-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyer, R. E., J. Segura-Aguilar, S. Di Bernard, M. Cavazzoni, R. Fato, D. Fiorentini, M. C. Galli, M. Setti, L. Landi, and G. Lenaz. 1996. The role of DT-diaphorase in the maintenance of the reduced antioxidant form of coenzyme Q in membrane systems. Proc. Natl. Acad. Sci. USA 93:2528-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, H. T., S. W. Marcelli, A. A. Davison, P. A. Chalk, R. K. Poole, and R. J. Miles. 1995. Kinetics of substrate oxidation by whole cells and cell membranes of Helicobacter pylori. FEMS Microbiol. Lett. 129:33-38. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, P. K., and N. L. Sternberg. 1995. A general genetic approach in Escherichia coli for determining the mechanism(s) of action of tumoricidal agents: application to DMP 840, a tumoricidal agent. Proc. Natl. Acad. Sci. USA 92:8950-8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, M., L. P. Andersen, L. Zhai, and A. Kharazmi. 1999. Characterization of the respiratory chain of Helicobacter pylori. FEMS Immunol. Med. Microbiol. 24:169-174. [DOI] [PubMed] [Google Scholar]
- 9.Comtois, S. L., M. D. Gidley, and D. J. Kelly. 2003. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology 149:121-129. [DOI] [PubMed] [Google Scholar]
- 10.Dinkova-Kostova, A. T., and P. Talalay. 2000. Persuasive evidence that quinone reductase type 1 (DT diaphorase) protects cells against the toxicity of electrophiles and reactive forms of oxygen. Free Radic. Biol. Med. 29:231-240. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finel, M. 1998. Does NADH play a central role in energy metabolism in Helicobacter pylori? Trends Biochem. Sci. 23:412-4133. [DOI] [PubMed] [Google Scholar]
- 13.Forsmark-Andrée, P., G. Dallner, and L. Ernster. 1995. Endogenous ubiquinol prevents protein modification accompanying lipid peroxidation in beef heart submitochondrial particles. Free Radic. Biol. Med. 19:749-757. [DOI] [PubMed] [Google Scholar]
- 14.Harris, A. G., F. E. Hinds, A. G. Beckhouse, T. Kolesniow, and S. L. Hazell. 2002. Resistance to hydrogen peroxide in Helicobacter pylori: role of catalase (KatA) and Fur, and functional analysis of a novel gene product designated ′KatA-associated protein,' KapA (HP0874). Microbiology 148:3813-3825. [DOI] [PubMed] [Google Scholar]
- 15.Harris, A. G., J. E. Wilson, S. J. Danon, M. F. Dixon, K. Donegan, and S. L. Hazell. 2003. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology 149:665-672. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, M., H. Ohzeki, H. Shimada, and T. Unemoto. 1996. NADPH-specific quinone reductase is induced by 2-methylene-4-butyrolactone in Escherichia coli. Biochim. Biophys. Acta 1273:165-170. [DOI] [PubMed] [Google Scholar]
- 17.Hazell, S. L., A. G. Harris, and M. A. Trend. 2001. Evasion of the toxic effects of oxygen, p. 167-175. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C.
- 18.Hughes, N. J., C. L. Clayton, P. A. Chalk, and D. J. Kelly. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180:1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly, D. J., N. J. Hughes, and R. K. Poole. 2001. Microaerobic physiology: aerobic respiration, anaerobic respiration, and carbon dioxide metabolism, p. 113-124. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 20.Li, R., M. A. Bianchet, P. Talalay, and L. M. Amzel. 1995. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction. Proc. Natl. Acad. Sci. USA 92:8846-8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Marcelli, S. W., H. T. Chang, T. Chapman, P. A. Chalk, R. J. Miles, and R. K. Poole. 1996. The respiratory chain of Helicobacter pylori: identification of cytochromes and the effects of oxygen on cytochrome and menaquinone levels. FEMS Microbiol. Lett. 138:59-64. [DOI] [PubMed] [Google Scholar]
- 23.Moss, C. W., M. A. Lambert-Fair, M. A. Nicholson, and G. O. Guerrant. 1990. Isoprenoid quinones of Campylobacter cryaerophila, C. cinaedi, C. fennelliae, C. hyointestinalis, C. pylori, and “C. upsaliensis.” J. Clin. Microbiol. 28:395-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olczak, A. A., J. W. Olson, and R. J. Maier. 2002. Oxidative-stress resistance mutants of Helicobacter pylori. J. Bacteriol. 184:3186-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olczak, A. A., R. W. Seyler, J. W. Olson, and R. J. Maier. 2003. Association of Helicobacter pylori antioxidant activities with host colonization proficiency. Infect. Immun. 71:580-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramarao, N., S. D. Gray-Owen, and T. F. Meyer. 2000. Helicobacter pylori induces but survives the extracellular release of oxygen radicals from professional phagocytes using its catalase activity. Mol. Microbiol. 38:103-113. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez, L. B., H. Elmendorf, T. E. Nash, and M. Muller. 2001. NAD(P)H:menadione oxidoreductase of the amitochondriate eukaryote Giardia lamblia: a simpler homologue of the vertebrate enzyme. Microbiology 147:561-570. [DOI] [PubMed] [Google Scholar]
- 28.Seyler, R. W., Jr., J. W. Olson, and R. J. Maier. 2001. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect. Immun. 69:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, M. A., and D. I. Edwards. 1997. Oxygen scavenging, NADH oxidase and metronidazole resistance in Helicobacter pylori. J. Antimicrob. Chemother. 39:347-353. [DOI] [PubMed] [Google Scholar]
- 30.Soballe, B., and R. K. Poole. 1999. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145:1817-1830. [DOI] [PubMed] [Google Scholar]
- 31.Soballe, B., and R. K. Poole. 2000. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology 146:787-796. [DOI] [PubMed] [Google Scholar]
- 32.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-60. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 33.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]