Abstract
The etiology of Alzheimer’s disease depends on the relative abundance of different amyloid-β (Aβ) peptide species. These peptides are produced by sequential proteolytic cleavage within the transmembrane helix of the 99 residue C-terminal fragment of the amyloid precursor protein (C99) by the intramembrane protease γ-secretase. Intramembrane proteolysis is thought to require local unfolding of the substrate helix, which has been proposed to be cleaved as a homodimer. Here, we investigated the backbone dynamics of the substrate helix. Amide exchange experiments of monomeric recombinant C99 and of synthetic transmembrane domain peptides reveal that the N-terminal Gly-rich homodimerization domain exchanges much faster than the C-terminal cleavage region. MD simulations corroborate the differential backbone dynamics, indicate a bending motion at a di-glycine motif connecting dimerization and cleavage regions, and detect significantly different H-bond stabilities at the initial cleavage sites. Our results are consistent with the following hypotheses about cleavage of the substrate. First, the GlyGly hinge may precisely position the substrate within γ-secretase such that its catalytic center must start proteolysis at the known initial cleavage sites. Second, the ratio of cleavage products formed by subsequent sequential proteolysis could be influenced by differential extents of solvation and by the stabilities of H-bonds at alternate initial sites. Third, the flexibility of the Gly-rich domain may facilitate substrate movement within the enzyme during sequential proteolysis. Fourth, dimerization may affect substrate processing by decreasing the dynamics of the dimerization region and by increasing that of the C-terminal part of the cleavage region.
Introduction
The amyloid precursor protein (APP) is a single-span membrane protein whose proteolysis by β-secretase produces the C-terminal fragment C99. C99 includes the transmembrane domain (TMD) which is subject to further proteolysis by γ-secretase. Cleavage at multiple sites in the TMD produces the amyloid beta (Aβ) peptides that form toxic oligomers and amyloid plaques, one of the hallmarks of Alzheimer’s disease. Thus, proteolysis of APP is believed to play a fundamental role in the etiology of the disease1–3. The C99 TMD forms an α–helix. Cleavage by γ-secretase is believed to be initiated at either the ε48 (linking T48 and L49) or ε49 (L49/V50) sites and continued by release of tri- and tetra-peptides after proteolysis at alternate ζ- (ζ45 = I45/V46; ζ46 = V46/I47) and γ-sites (γ37 = G37/G38; γ38 = G38/V39; γ40 = V40/I41; γ42 = A42/T43), resulting in two distinct product lines. Successive cleavage leads to release of the most abundant Aβ40 and the minor Aβ42 and Aβ38 peptides plus minor species, both longer and shorter4–7. This 3- to 4-residue stepwise cleavage pattern suggests that the substrate TMD remains essentially helical during processing. Proteolysis is thought to take place within the lumen of the aspartate protease presenilin, the catalytic subunit of the γ-secretase complex8. Proteolysis by aspartate proteases requires proton transfer from one catalytic Asp to the carbonyl oxygen of the scissile bond as well as nucleophilic attack of a water molecule which is H-bonded to another catalytic Asp, at the C1 atom of the same carbonyl. Formation of this tetrahedral intermediate is followed by cleavage of the amide bond9.
It is generally thought that helices cannot be cleaved without first being locally unraveled10. Therefore, it has been proposed that the APP TMD helix has to unwind locally at the cleavage sites to expose the carbonyl. Recent NMR structures were determined for both monomeric C99 and for a homodimeric TMD-containing fragment of C99, both in micelles. The relevance of the monomeric structure to membrane bilayer conditions was directly confirmed by EPR studies of the protein in bilayers11. In both structures, the TMD is fully helical except for a flexible bend centered around G37G3811,12. The helical segment leading up to this bend (sites 29–37, which we refer to in this paper as the “TM-N helix”) is believed to include sites that drive both homodimerization13–17 and cholesterol binding11, perhaps competitively. The helical TM segment starting at G38 and terminating at L52 is here referred to as the “TM-C helix” and contains the various sites for cleavage. A bioinformatic analysis suggested that the cleavage sites of different known γ-secretase substrate TMDs exhibit low helix-forming propensities resulting from an accumulation of potentially helix-destabilizing amino acid types like Ile, Val, and Thr18. Indeed, helix-destabilizing amino acids have been reported to facilitate cleavage of the substrate TMDs of other intramembrane protease including site-2 protease19, rhomboids20,21, and signal peptide peptidase22,23. Thus, the TM-C helix may be subject to transient unraveling that simply was not detected in the NMR structural studies11,12. In agreement with the idea of local unfolding, NMR and FTIR studies of a synthetic fragment (E22 – V64, Aβ numbering) in DMPC/DMPG vesicles by Smith and co-workers showed that the APP TMD unravels downstream of the ε-site15. A more recent ssNMR study by Tycko and co-workers of an APP TMD peptide (K19–K55) was performed in various lipids. In DOPG, the region around the γ-sites was fully helical while mixtures of helical and non-helical conformations of this region were found in neuronal lipid mixtures24.
The APP TMD-C helix contains two consecutive GxxxG motifs starting at G29. A GxxxG motif frequently forms the interface of homodimerizing TMDs25. Indeed, mutating G29 and/or G33 within the APP TMD decreased self-interaction of the helix which was correlated to decreased Aβ40 and Aβ42 production and enhanced Aβ38 formation26,27. It was therefore proposed that monomerization of the dimer by mutating Gly facilitates read-through of γ-secretase; an interpretation which implies that the substrate forms a homodimeric structure within presenilin26. However, it is not yet known with certainty whether C99 binds to the active site of γ-secretase as a monomer, a dimer, or both. Modeling the interface of the wild-type sequence suggested that the G29xxxG33xxxG37 motif (the Gly zipper) forms the dimer interface15,17,26,28, which has been supported by NMR experiments14,15. Other models suggest it is the G38xxxA42 motif is at the interface17,29. Taken together, these experimental studies support a consensus view where the interface of the APP TMD helix homodimer includes the Gly zipper and is characterized by a right-handed crossing angle of the helices. We note that the helices interact in a left-handed crossing angle in the most recent NMR structure of the dimer, where the interface is formed by a heptad-repeat motif of residues involving G38 and A4212. It is possible, therefore, that C99 populates both monomeric and at least two different dimeric states, depending on conditions.
Since a systematic analysis of TMD helix backbone dynamics and its potential dependence on dimerization has not been available, we here examined the APP TMD helix by recording amide exchange kinetics of monomeric recombinant C99 and of synthetic TMD peptides. Surprisingly, we find that the backbone dynamics of the dimerization region TM-N is higher than that of the TM-C helix, even though the sites for γ-secretase cleavage reside in the latter segment. Molecular dynamics (MD) simulations support this finding and suggest that dimerization decreases the dynamics of the dimerization region and enhances the dynamics of the cleavage domain.
Materials and Methods
Amide H/D Exchange Rate Measurements of C99 with NMR Spectoscopy
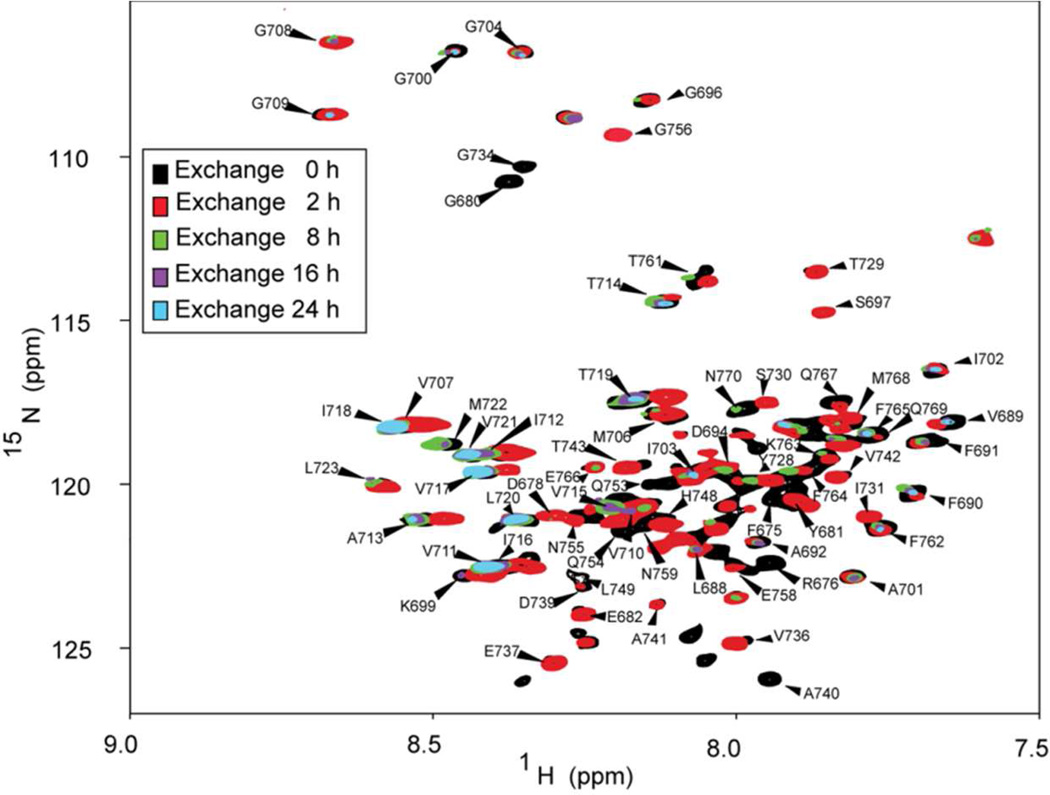
C99 with a C-terminal tag containing His6 was recombinantly expressed in uniformly 15N-labeled form and purified into lyso-myristoylphosphatidylglycerol (LMPG) micelles as described previously14, followed by adjustment of the pH to 6.5. To initiate amide hydrogen-to-deuterium (H/D) exchange a 500 µL micellarU-15N-C99 sample in H2O was mixed with 5 mL of 100% D2O in 100 mM Imidazole, pH 6.5. The sample was then concentrated back to 500 µL using a 30 kDa cutoff filter. Final C99 samples contained 0.2 mM U-15N-C99 in 10% LMPG (w/v), 100 mM Imidazole, pH 6.5 in ca. 90% D2O. 900 MHz 1H,15N-TROSY NMR spectra were recorded serially at times points 0 h, 2 h, 8 h, 16 h, and 24 h after mixing with the D2O, with the goal being to monitor the disappearance of peaks due to H/D exchange as a function of time. The 0 hour time point was recorded on a matched sample in H2O.
NMR acquisition parameters were the same for the all spectra collected, with 128 scans and 256 increments for each. To quantify the extent of exchange for each backbone amide resonance, peak intensity was monitored at each time point and compared to the intensity for the reference sample prior to D2O addition (time = 0 h). For each NMR experiment the total acquisition time was approximately 6 h, such that the temporal resolution of observed H/D exchange process is low. For this reason, we limited our analysis to comparison of the exchange states represented by the TROSY spectra collected over the 2–8 h and 16–22 h time ranges (see Fig. 1). Previously completed NMR resonance assignments14 allowed the data to be interpreted in a site-specific manner.
Figure 1. Use of NMR to monitor exchange of C99 backbone amide protons for deuterons as a function of time.
This figure depicts the overlay of five 900 MHz 1H,15N-TROSY NMR spectra of C99, collected at different points of time following dilution of C99 into D2O. The black spectrum shows the reference spectrum collected in an H2O solution. Red, green, purple, and cyan spectra reveal the state of changes after 2, 8, 16, and 24 h, respectively. All NMR resonances that could be accurately assigned are labeled. It should be noted that because each spectrum required 6 h of acquisition time, the actual time reflected by the data is a window spanning the designated time plus 6 h, with the observed spectra being weighted towards the early part of the 6 hour window because of the way the 2-D NMR data is collected (most of the signal is detected in the early increments of the 2D experiment).
From the experimental ratio r of populations at t = 16 h with respect to t0 = 0 h, an estimate of the exchange rate is: kHX = (1-r(t))/(t-t0). To compare these values with the MD-derived rates in Fig. 5 C, we scaled their logarithmic values linearly between −3 and +2.
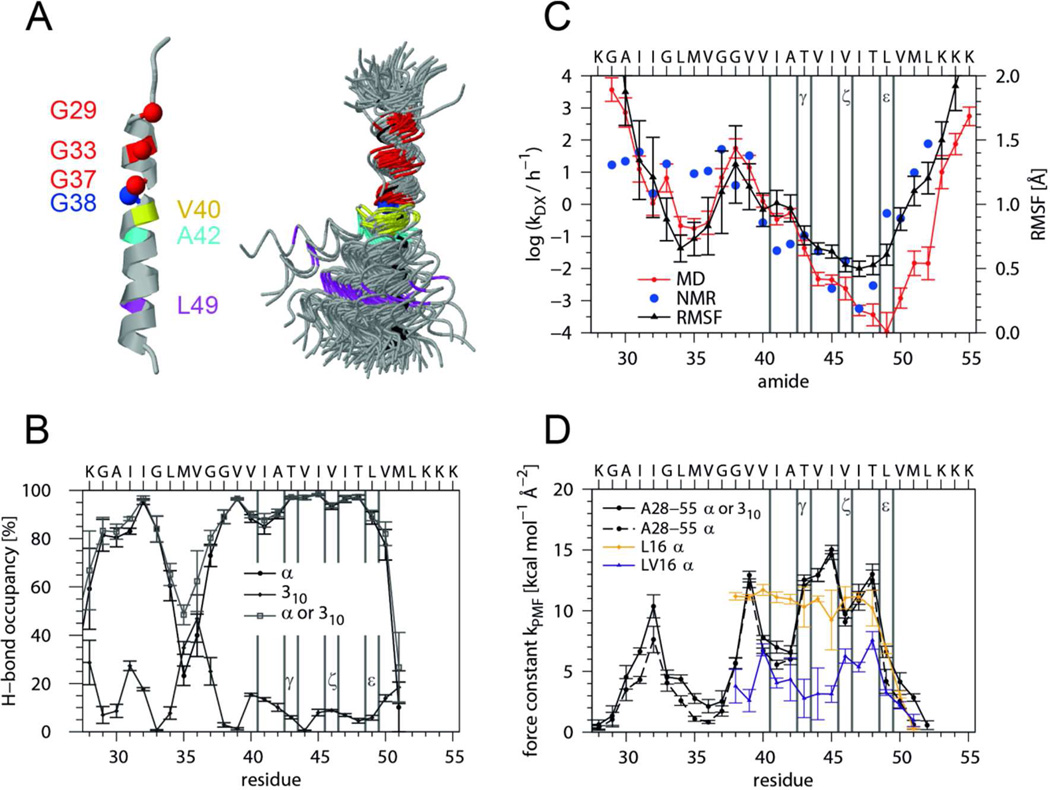
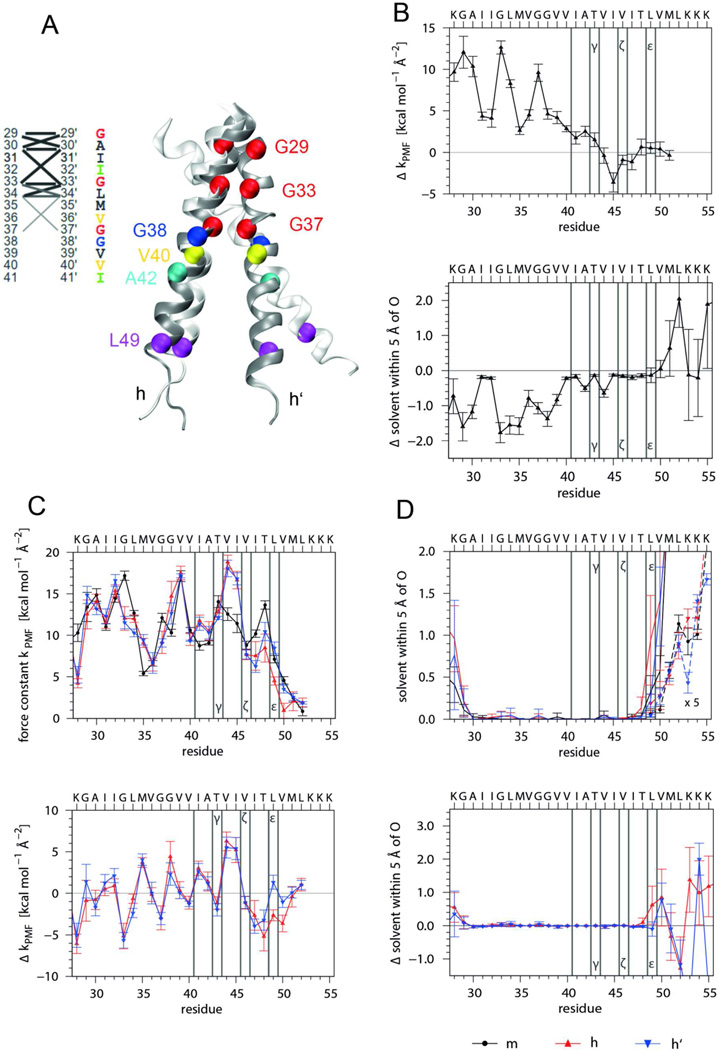
Figure 5. The backbone dynamics of the APP TMD helix probed by MD simulation.
A) Overall helix backbone conformation. The superposition (left graph) was obtained by overlaying backbone traces from consecutive frames taken every 1.5 ns, each of which was oriented with a rigid body fit of residues 29–37 to the Cα atoms of an ideal α–helix (black trace). The Gly residues and amino acids at major cleavage sites are colored. Note the anisotropic helix bending with preferential bending over the hinge, i.e. towards the Cα atom of G38. The average structure (right) identifies the region around G37G38 as hinge while the helical regions are represented by a ribbon. B) Population of H-bonds that extend from the carbonyl oxygen of residue i to the amide hydrogen of residue i+4 (α-helix) or i+3 (310-helix). The probability that at least one of these H-bonds is formed is referred to as “α or 310”. C) Local DHX rate constants of amide hydrogens at positions i and RMSF of C1 carbons at residues i-1 around their average positions as calculated from the MD simulation. For comparison, values roughly characterizing HDX rates in detergent micelles after 16 h are given. D) Force constants characterizing the potential of mean force of H-bonds extending from the carbonyl oxygen at residue i to the closest amide of residue i+3 (α) or i+4 (310). For comparison, force constants were calculated for residues 5 through 23 of L16 (K3WL16K3) and LV16 (K3W(LV)8K3) helices whose C-terminal Lys-tripletts are overlaid onto the Lys-triplett of A28-55. Error bars indicate standard deviations calculated from 30 ns block averages. Vertical lines denote cleavage sites.
Peptide Synthesis
Peptides were synthesized by Fmoc chemistry (A28-55, A28-44 and A37-55 were from PSL, Heidelberg, Germany, AL-peptides were from Dr. Sven Rothemund, IZKF, Leipzig, Germany) and were >90 % pure as judged by mass spectrometry.
Membrane Reconstitution of AL-Peptides
Deuterated AL-peptides were incorporated at a molar peptide/lipid ratio of ~0.02 into liposomal membranes composed of dilauryl-phosphatidylcholine (DLPC), dilauryl-phosphatidylethanolamine (DLPE), and dilauryl-phosphatidylserine (DLPS) at a 3:1:1 molar ratio in 50 mM ND4Ac, pD 7.5. The lipids were dissolved in 800 µl cyclohexane and combined with peptide solutions in deuterated hexafluoroisopropanol at a 20:1 (v/v) ratio, mixed, and lyophilized. Hydrating the lyophilsate with 400 µl 50 mM ND4Ac, pD 7.5 and subsequent sonication gave the liposomes. The peptide/lipid ratios were determined as described30.
CD Spectroscopy
For CD spectroscopy in solution, peptides were dissolved in 80% (v/v) 2,2,2-trifluoroethanol (TFE) and 10 mM NH4Ac, pH 5 at 30 µM. For each sample, 10 accumulated CD spectra from 185–260 nm were obtained using a Jasco J-710 CD spectrometer with a 0.1 data pitch, 1 sec response, 100 nm/min scan velocity, 100 mdeg/cm sensitivity, and a path length of either 0.5 mm or 1 mm. Mean molar residue ellipticities were calculated based on peptide concentrations as determined by UV spectroscopy using extinction coefficients of 5,600 M−1cm−1 for Trp. Secondary structure contents were estimated by deconvoluting the CD spectra using the program CDNN/PEPFIT that is based on peptide-derived reference spectra31. CD experiments of peptides incorporated into liposomes were recorded from 190–260 nm with 20 accumulated CD spectra at 70°C and a path length of 0.5 mm. Acquisition parameters were the same as for peptide solutions.
ESI-MS-Monitored DHX Experiments for Synthetic Peptides and Related Data Analysis
Solutions of deuterated peptide (100 µM in 80% (v/v) dTFE in 10 mM ND4Ac) were diluted 1:20 with protonated solvent (80% (v/v) TFE in 10 mM NH4Ac, pH 5.0 unless specified otherwise) to a 5 µM final peptide concentration at 20°C. Aliquots were removed after 0, 10 s, 20 s, and 30 s and quenched by chilling on ice and by adding 0.5% (v/v) formic acid to lower the pH to ~2.6. Time points > 40 s were recorded in continuous mode by injecting reaction mixtures into the ion source of the mass spectrometer for 4 h32. The DHX kinetics of AL-peptides in liposomes was recorded at 70°C in discontinuous mode. Mass spectra were acquired in positive-ion mode using a Waters Q-ToF Ultima with one scan/second and evaluated as described33. For continuous measurements, 5 scans were accumulated and smoothed, and centered mass/charge ratios were obtained for intervals of ten seconds. The triply charged isotope patterns were smoothed with the Savitzky-Golay algorithm with 25 measuring points and a data pitch of 5. The numbers of remaining deuterons was calculated as described33. The distributions of DHX rate constants were calculated from the DHX kinetics using a maximum entropy method (kindly provided by Dr. A. Marshall)32 or by fitting with a 3-exponential function.
Molecular Dynamics Simulations and Trajectory Analysis
Solvent simulations were performed in 80% (v/v) TFE/water as described34 using the CHARMM force field35. The monomer with initial ideal α-helical backbone conformation was solvated in a rectangular solvent box (10.3 nm × 6.0 nm × 6.0 nm) containing 2314 water and 2314 TFE molecules and 6 neutralizing chloride ions. Peptide termini were charged. Peptide and solvent were equilibrated for 12 ns with gradual release of constraints on backbone atoms (1 ns with a force constant k = 5 kcal/(mol Å2), 1 ns with k = 2 kcal/(mol Å2), 10 ns without restraints) followed by 150 nsec of free dynamics at constant temperature and pressure (T= 293 K, p = 0.1 MPa). The dimer was constructed using the program CHI36. One cluster with a right-handed crossing angle and G29, G33 and G37 of both helices in close contact was averaged and used as the starting structure. This dimer was solvated in a cubic solvent box of 10.3 nm side length (7165 water and 7165 TFE molecules, 12 chloride ions) and subjected to 15 ns equilibration. For the first 3 ns positional constraints were imposed on all backbone atoms (1 ns with k = 5 kcal/(mol Å2), 2 ns with k = 2 kcal/(mol Å2) followed by 2 ns where only the Gly residues in the interface were constrained with k = 1 kcal/(mol Å2). Finally all constraints were cleared and the equilibration monitored for 10 ns before recording free dynamics for 95 ns.
For membrane simulations, monomer or dimer were inserted into a rectangular patch of POPC molecules (10 × 10 nm, 260 lipids) with water boxes extending for 2 nm to either side of the bilayer (~36 waters/lipid) and containing 6 or 12 neutralizing Cl− ions for monomer and dimer, respectively. For the first 7.2 ns, positional constraints were imposed on all backbone atoms (0.2 ns with k = 5 kcal/(mol Å2) on backbone atoms, k = 2 kcal/(mol Å2) on lipids, water, ions; 5 ns with k = 5 kcal/(mol Å2) on backbone atoms, k = 2 kcal/(mol Å2) on water and ions; 2 ns with k = 2 kcal/(mol Å2) on backbone atoms, k = 2 kcal/(mol Å2) on water and ions). Finally, all constraints were cleared and the equilibration monitored for ~40 ns before recording free dynamics for 110 ns (dimer) or 100 ns (monomer). Free dynamics was calculated with the use of the SHAKE algorithm, periodic boundary conditions, and particle mesh Ewald electrostatics (short-range real-space interactions were cut off at 1.2 nm using a switching function between 0.8 and 1.2 nm). The temperature was kept constant (310 K) using a Langevin thermostat and a Nose-Hoover piston was employed for pressure control (NPT, 0.1 MPa). The CHARMM protein35 and lipid37 force field was used. An integration step of 1 ps was used for the first ~4 ns after which the reversible multiple time-step algorithm38 was employed to integrate the equations of motion with time steps of 1 fs for bonded forces, 2 fs for short-range non-bonded forces and 4 fs for long-range electrostatic forces. All simulations were performed with the program NAMD39. Structures were saved every ps for analysis.
To assess the convergence of the simulations, the mean distance between all pairs of average backbone structures taken from nonoverlapping time windows were analyzed as function of sequence position and window size34,40. For the hydrophobic core of the TMDs mean distances <0.03 nm (monomer) or <0.06 nm (dimer) were reached within 30 ns. Larger deviations up to 0.1 nm indicate deficient sampling of an enlarged conformational space in the C- and N-terminal parts resulting from helix fraying (residues 28–30 and 52–55). In the case of the membrane simulations, the membrane-thickness, measured as the distance between two least-square planes through the phosphate atoms in upper and lower leaflets, was constant after the equilibration phase (4.14 nm ± 0.04 nm). The area per lipid was 0.66 ± 0.03 nm2,which matches experimental data41. The cell dimensions did not fluctuate more than 3% in the bilayer plane and 1% normal to the bilayer.
Analysis of H-bond populations, H-bond length distributions, secondary structure, helix bending, rotation and crossing angles, and root mean-square fluctuations was carried out with routines provided with the CHARMM software42. For analysis of helix bending, TM-N and TM-C helices were defined using the Cα atoms of residues 31–36 and 41–48, respectively.
An intrahelical H-bond is considered as closed if the H…O distance is below 0.26 nm and the N-H … O angle is in the range of 180°±60°. The free energy profiles W(di) (potential of mean force, PMF) of the intrahelical H-bonds were calculated from the distribution of the closest distances di between backbone carbonyl oxygens at position i and amide hydrogens at positions i+4 (α-H-bond) or i+3 (310 H-bond). The PMF W(di) follows from the probability P(di) to observe a given distance di: W(di) = - RT ln [P(di) / P(di,0)] (RT = 0.6 kcal/mol, di,0 = most probable distance). For a Gaussian distribution, the PMF resembles a harmonic potential characterized by a force constant kPMF(i) which is related to the variance σi2 of the length distribution: kPMF(i) = 1/σi2. Since the length distributions differ only slightly from Gaussians (correlation > 95% in most of the cases) we calculated the corresponding force constants directly from the variances σi2 without fit of the PMF to a parabolic function.
Statistical uncertainties were calculated by dividing the trajectories into non-overlapping windows of 30 ns length and evaluating the standard deviations (SD) of the mean values (66% confidence interval).
Calculation of DHX rates from MD simulations
The exchange rate kDX for an amide deuteron is determined by (1) the probability fop by which an amide H-bond opens, and (2) the concentration of exchange catalyst [OH-]: kDX (i) = fop (i) kB (i) [OH-](i). kB is the rate constant for base-catalyzed DHX in poly-D,L-alanine at the experimental temperature T=293 K, corrected for effects of neighboring side chains others than Ala43. The factor fop (i) is derived from the MD simulations and denotes the probability by which an amide H to has a distance dHO > 0.3 nm to the closest intrahelical (α or 310) H-bond accepting carbonyl oxygens34,44,45. In bulk water [OH-] is directly related to the pH-value and the autoprotolysis constant KW of H2O: [OH-]bulk /cø = KW /10−pH (cø = 1 mol/l). Several factors contribute to the deviation of local [OH-](i) around each amide hydrogen from the bulk value: (1) the 80% TFE (v/v) solution contains water only as a cosolvent with 1/5 of the molarity of bulk water; (2) hydrophobic side chains promote local dehydration46,47, and (3) preferential solvation by TFE48,49 increases the local dehydration in a side-chain specific manner. Under the assumption that the water dissociation equilibrium as well as the pH will not change in 80% TFE, the ratio of local and bulk hydroxide concentration is given by the ratio of the concentration of water molecules: [OH-] (i) / [OH-]bulk = ([HOH] (i) / [HOH]bulk)2. From the MD simulations we calculated the number of water molecules within a distance < 0.7 nm of amide hydrogens46. To obtain the local water concentration we take the volume excluded by the peptide into account. If the numbers of molecules counted are nHOH and nTFE, respectively, the volume Vsolv occupied by solvent molecules (water and TFE) around the amide H is Vsolv = nHOH Vm,HOH + nTFE Vm,TFE with the molecular volumes Vm,HOH = 31.56 Å3 and Vm,TFE = 116.28 Å3 48). We observed that the local hydroxide concentration at the hydrophobic peptide cores is only 1/10 to 1/100 of the bulk concentration while it could exceed the bulk value by a factor of ~10 near the charged termini. The presence of TFE can exert additional effects: (1) The rates for H- or D- transfer reactions (e.g. kB and water dissociation rate) might be influenced by the changed electrostatics. (2) Local concentrations might be different from local activities. These TFE effects are contained in a correction factor δ common for all peptides. A value δ = 0.01 was found to be optimal as it minimizes the reduced chi-square value χ2red50 between experimental and calculated DHX kinetics for previously investigated LV-peptides34 as well as for A28-55 and derivatives to values in the order of 10−1 to 10−2. Fig. S2 shows the results for L16, LV16, and A28-55.
Results
The rationale of the present study was to determine the backbone dynamics of the APP TMD helix by recording amide exchange kinetics, which is a powerful way to analyze the conformational equilibria along a protein sequence. The exchange kinetics of amides that are potentially involved in intramolecular H-bonding report local and transient unfolding of secondary structure30,33,51,52. MD simulations provide insights into the H-bond dynamics along the backbone as well as into solvation34,44. Both determine the local exchange rates which can be calculated from the simulations to complement the experimental analysis.
Monitoring Backbone Amide Hydrogen/Deuterium-Exchange for C99 in Detergent Micelles
C99 was recombinantly expressed in uniformly 15N-labeled form and purified into LMPG micelles. LMPG is a close analog of natural phospholipids and is generally regarded as a mild detergent. C99 dimerizes with only very modest affinity in LMPG micelles14,53 and hydrogen/deuterium exchange (HDX) studies were carried out using a high (1000:1) LMPG:C99 mole ratio in which only the monomeric form of the protein was present11. An aliquot of concentrated U-15N-C99 stock solution was diluted into D2O and 1H,15N-TROSY spectra were recorded at time intervals, allowing monitoring of the disappearance of the backbone amide 1H,15N cross peaks due to replacement of the amide protons with deuterons (Fig. 1). Because each TROSY spectrum required 6 h of acquisition time, exact quantification of amide exchange rates from these data is not possible. However, comparison of data from the 2 and 16 h time points provides a very clear assessment of the relative rates of amide exchange in C99 in the micelles (Fig. 2).
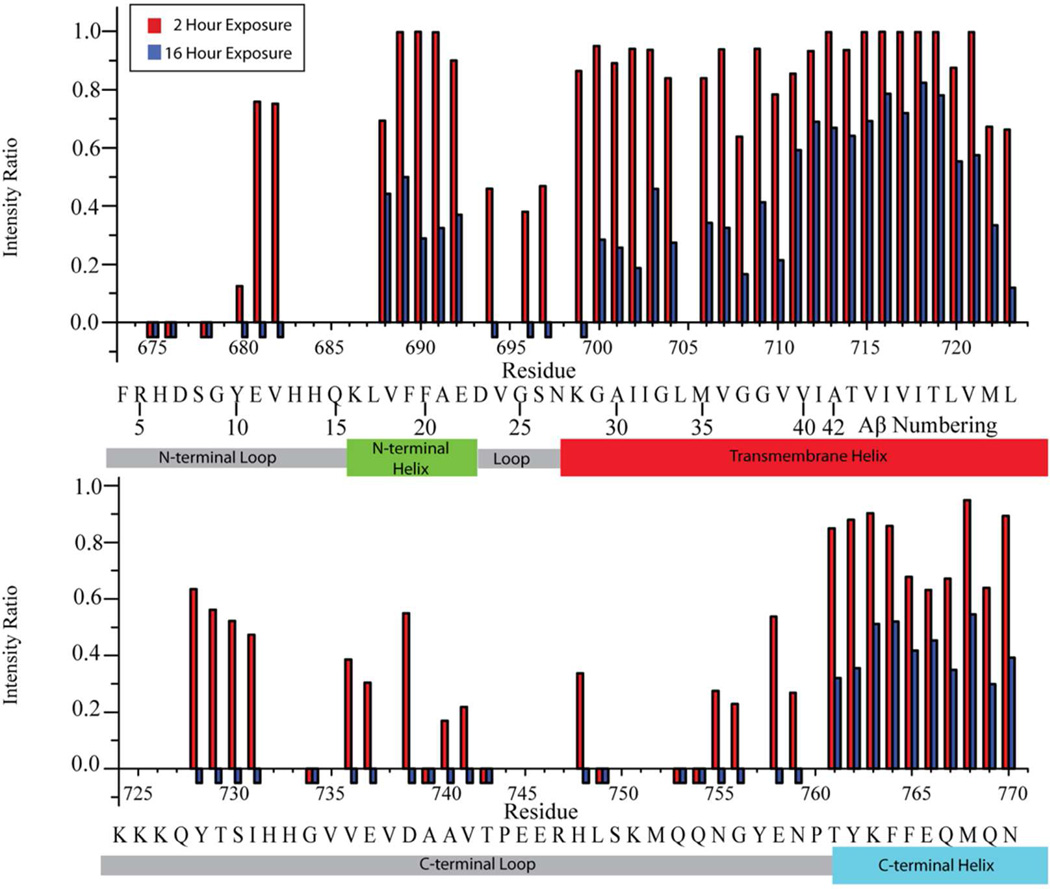
Figure 2. Time dependent replacement of amide protons with deuterons upon dilution of C99 into D2O solution.
The intensity ratio is for the peak intensity observed after exchange with D2O for 2 (red bars) or 16 (blue bars) h relative to a non-exchanged “time = 0” reference sample. Negative bars indicate that the peaks had disappeared completely, while the absence of either a positive or negative bar indicates that data was not measured for that site because of difficulties due to peak overlap or assignment. The known topology of C99 is shown below each graph, with helical regions indicated by large, colored rectangles and dynamic loops indicated by grey, small rectangles. The numbering given in the upper line refers to full-length APP numbering while the lower line gives Aβ numbering. Data was collected for 0.2 mM U-15N-C99 in 10% LMPG micelles at pH 6.5 and 45°C.
C99 contains three domains in which DHX rates are relatively rapid, reaching completion at 16 h. These are (i) the N-terminus (through Q15, Aβ numbering), (ii) the loop that connects a short surface-associated helix with the N-terminus of the TMD (G25–K28), and (iii) the long connector between the end of the TMD and the surface-associated helix at the extreme C-terminus. Within these segments are sites that are completely exchanged even at 2 h and other sites for which significant retention of protons are seen at 2 h. These results are consistent with the results of previous NMR studies of micellar C99 and of the isolated intracellular domain in solution having suggested that these segments do not adopt stable secondary tertiary structure, although there some local transient structure is evident14,54,55. Further, based on significant protection from exchange even after 16 h at 318 K the exchange data are consistent with the presence of both a surface-associated α-helix just prior to the TMD (K16 to D23) and one formed by the 11 residues at the extreme C-terminus11. We note that the C-terminal purification tag of C99 used in this work14 might attenuate exchange at the extreme C-terminus. However, recent EPR studies of C99 in lipid vesicles confirm that the C-terminus is surface-associated even when a C-terminal tag is not present11.
The exchange data for the TMD (G29 through L52) is complex. While exchange is at no position complete at 16 h, the N-terminal portion of the TMD (TM-N) extending from G29 to V40 shows much lower protection (most evident for the 16 h data of Fig. 2) than the I41 to I47 segment of TM-C that includes γ- and ζ-cleavage-sites. Some evidence for “fraying” of TM-C is evident in the transition from I47 (at which protection is high even after 16 h) to the end of this domain at L52. These results indicate significantly greater helix backbone dynamics at, and/or greater access of water to, sites located in TM-N compared to TM-C, except near the frayed C-terminus of the TMD.
Monitoring Backbone Amide Deuterium/Hydrogen-Exchange for Synthetic APP TMD Peptides in Isotropic Solution
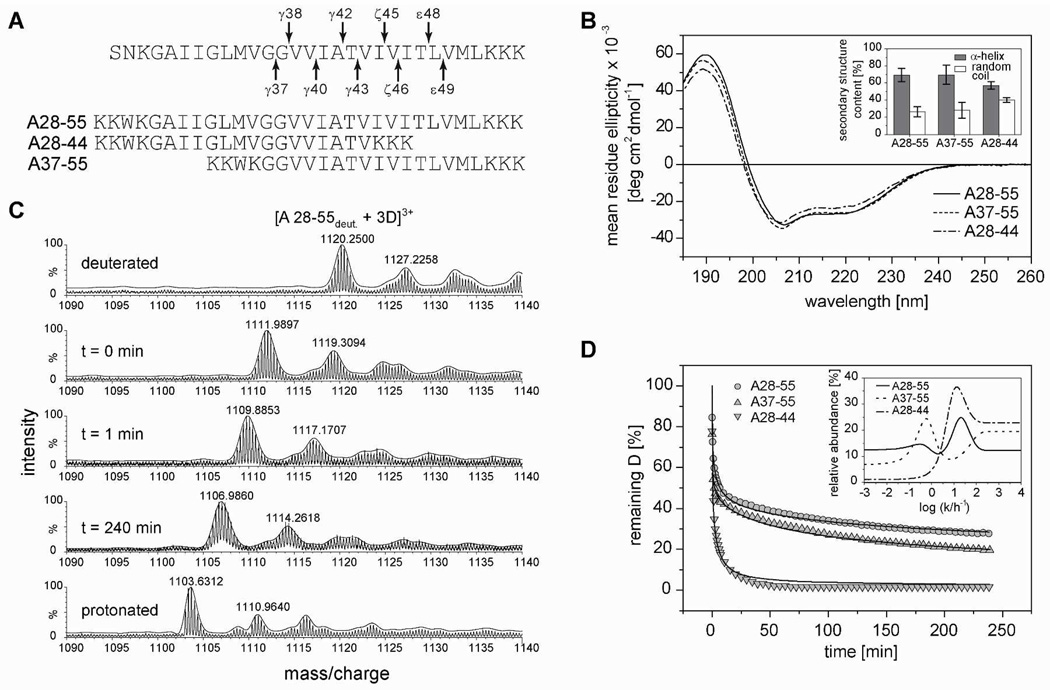
Here, we analyzed the secondary structures and DHX kinetics of synthetic peptides that either represent the full APP TMD or contain mainly the homodimerization (A28-44) or cleavage (A37-55) domains, respectively (Fig. 3 A, A28-55). The helical APP TMD is thought to be located within a water-filled cavity at the active site of presenilin56,57. Since hydrophobic peptides precipitate in water, we dissolved the APP TMDs in 80% (v/v) trifluoroethanol (TFE) in aqueous buffer as exercised previously with other TMD peptides30,33,58. The TFE water mixture mimics the aqueous environment while maintaining helicity and preventing aggregation. Circular dichroism (CD) spectroscopy revealed that A28-55 and A37-55 form ~70% helix while the helicity of A28-44 is decreased to ~55% in favor of random coil (Fig. 3 B). DHX kinetics were recorded in the same solvent at a concentration of 5 YM where the TMD remains monomeric as shown by fluorescence resonance transfer experiments of labeled peptides (Fig. S1). Deuterium-to-hydrogen exchange (DHX) of exhaustively (>95%) deuterated peptides were continuously monitored by determining the molecular masses of the triply charged peptide ions using electrospray ionization mass spectrometry (ESIMS). As exemplified by A28-55 spectra, the isotope envelopes gradually shift with incubation time towards lower mass/charge values (Fig. 3 C). A gradual mass shift is diagnostic of uncorrelated exchange which suggests fast local, rather than slower global, helix unfolding59. Kinetics were normalized to the respective numbers of potentially H-bonded amides and follow the rank order A28-44 >> A37-55 ≥ A28-55 (Fig. 3 D). For quantitative evaluation, the kinetics were deconvoluted by employing a maximum entropy (MEM) method. This procedure returns a distribution of exchange rate constants for each peptide60. The distribution of A28-55 rate constants shows two major populations of deuterons exchanging within the incubation time (Fig. 3 D, inset). The mean rate constants of the more slowly exchanging deuterons peak at log (k/h−1) = 0.36 (corresponding to a mean half-time of DHX of t1/2 = 96 min) while the faster deuterons peak at log (k/h−1) = 1.27 (t1/2 = 2.2 min). While the slow peak of the complete A28-55 TMD closely matches the main peak seen with the A37-55 fragment, the fast A28-55 peak corresponds to the major peak of A28-44. Flat regions represent either deuterons that do not exchange within 240 min (t1/2 > 12 h, log (k/h−1) < −1.2) or very fast deuterons (t1/2 < 5 s, log (k/h−1) > 2.7), respectively. While a quantitative comparison of the full A28-55 TMD and its fragments is difficult due to the different lengths of the peptides, it is evident that the fragment harboring the dimerization domain (A28-44) exchanges considerably faster than the one containing the cleavage domain (A37-55), as seen with the full length protein in LMPG micelles. The good agreement obtained with C99 in micelles and isolated TMD peptides in solution shows that the experimental design yields self-consistent results. The recombinant protein would not be stable in our TFE/water mixture, nor in water alone.
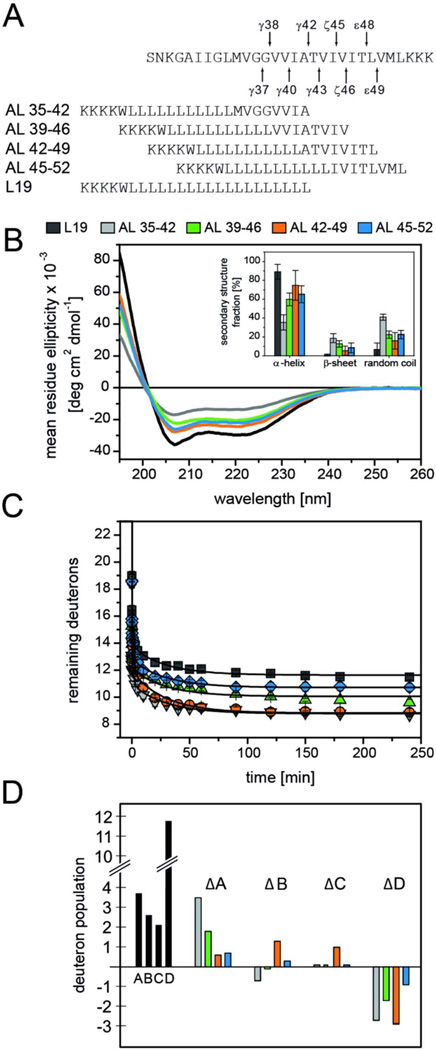
Figure 3. Secondary structure and backbone dynamics of the APP TMD helix.
A) Sequences of the predicted APP TMD (arrows indicate cleavage sites leading to the Aβ40 and Aβ42 product lines, respectively) and the model peptides used here. All peptides contain an additional N-terminal KKW(K) sequence for better solubility and quantification; A28-44 contains a non-natural C-terminal KKK sequence. B) Averaged CD spectra and calculated secondary structure contents (inset). n = 3–6, means±SD. C) Representative mass spectra of the triply charged A28-55 ion from different time points of a DHX experiment. D) DHX kinetics where the absolute numbers of remaining deuterons were normalized to the maximum numbers of H-bonded amide deuterons in an ideal α-helix (A28-44 and A37-55, 19 D; A28-55, 27 D). Data points at t=0 min correspond to the numbers of amide deuterons seen after exchange under quench conditions. Data were fit with a MEM assuming Dt = 0 min equaling the maximum numbers of potentially H-bonded amide deuterons (continuous lines). n = 3, SD ≤ 0.3 D (error bars not shown). Inset, distribution of exchange rate constants determined by fitting the data with MEM.
Next, the dynamics of the cleavage region was mapped using a set of hybrid peptides. These hybrids are based on an invariant oligo-Leu host sequence that forms a rather rigid α–helix30,34. Eight C-terminal Leu of the parental L19 peptide were replaced by different octa-residue fragments that cover the APP cleavage region and comprise γ-, ζ-, or ε-sites, respectively (Fig. 4 A). Thus, the DHX kinetics of these hybrid peptides are thought to reflect the backbone dynamics around the respective cleavage sites. The free C-termini of the TMD fragments mimic the free C-termini of fragments that form after proteolysis of C99 at γ-, ε-, or ζ-sites. Since CD spectroscopy revealed that some of the hybrid peptides did not form stable secondary structures in 80% TFE, we reconstituted them into liposomal membranes. CD spectroscopy determined the helicity of the hybrid peptides in the membranes to follow the rank order L19 > AL39-46 ≈ AL42-49 ≈ AL45-52 > AL35-42 (Fig. 4 B). DHX kinetics were recorded over 240 min where ~9 to ~11 potentially H-bonded amide deuterons exchanged. Since three non-H-bonded amide-deuterons at the helix N-termini exchange within the dead-time of the experiment, another ~9 to ~11 remaining deuterons were protected by the bilayer (AL-peptides contain 23 amides) (Fig. 4 C). Assuming that exchange can occur with similar efficiency at both termini of a membrane-spanning helix and that the N-terminal Trp residue is located at the headgroup / acyl chain boundary leads us to the conclusion that ~3 to ~4 H-bonded deuterons are likely to exchange at the N-termini and ~6 to ~7 deuterons at the C-termini. Accordingly, the APP octa-peptide amides seem to exchange almost completely within 240 min. For quantitative evaluation, the kinetics of exchangeable deuterons were fit with a triple exponential function, which describes some of them better than the MEM (Table S1). This fitting procedure subdivides each peptide’s amide deuterons into three classes (A, B, C) that exchange with different rate constants (kA, kB, kC) plus a class D that does not exchange within 240 min (Table S1). For a better comparison of the numbers of deuterons within classes A, B, and C, we averaged the DHX rate constants of each class over all peptides and recalculated the respective numbers of deuterons. Now, the numbers of the fastest deuterons (~4 to ~7 in class A) follow the rank order AL35-42 > AL39-42 > AL42-49 ≈ AL45-52 > L19 (Table S1). Fig. 4 D visualizes the differences between the numbers of class A, B, C, and D deuterons of the hybrid peptides and L19. These differences show that i) AL39-46, AL42-49, and AL45-52 contain more class B deuterons than AL35-42 and that ii) AL42-49 contains most class C deuterons. Since AL39-46, AL42-49, and AL45-52 contain one or two Thr, the shift of fast class A deuterons into intermediate class B and slow class C deuteron populations may relate to Thr backbonding.
Figure 4. Secondary structure and backbone dynamics of hybrid peptides.
A) Peptide sequences. B) CD spectra and calculated secondary structure contents (inset) of AL-peptides in DLPC/DLPS/DLPE (3/1/1) membranes at P/L ~0.03 in 50 mM ND4Ac, pD 7.5 at 70°C. C) DHX kinetics of AL-peptides recorded in liposomal DLPC/DLPS/DLPE (3/1/1) membranes at P/L ~0.03 in 50 mM NH4Ac, pH 7.5 at 70°C. Data points at t=0 min correspond to the numbers of amide deuterons seen after exchange under quench conditions. Lines connecting the data points were obtained by fitting the kinetics with a triple exponential function that was characterized by a reduced chi-square value <0.1 for all peptides. n = 3, means±SD. D) A representation of the numbers of deuterons within the kinetically distinct classes A, B, C, and class D, which represents deuterons that do not exchange within 4 h. To make them comparable, the numbers were calculated after averaging the respective DHX rate constants over all peptides (Table S1). Black bars represent the numbers found for the L19 reference. To facilitate the comparison between AL peptides, we plot the differences between the numbers seen with AL-peptides and L19.
Modeling Backbone Dynamics and Homodimerization of the APP TMD
MD simulations of the A28-55 helix were performed in 80% (v/v) TFE in water, i.e. in a solvent matching the experimental conditions of peptide DHX. The objective was to obtain detailed insights into helix dynamics at the carbonyl carbon (C1), which is attacked by acidified water during proteolysis, and into H-bonding at the carbonyl oxygen, which is the target of one catalytic presenilin Asp.
In order to validate our MD simulations, we first calculated the global exchange kinetics and compared them to the experimental observation. Previously, calculation of exchange rates from MD using H-bond occupancies were moderately successful34,44. Here, a significant improvement was made by using the local water concentration around the amide to calculate the local hydroxide concentration rather than using the hydroxide concentration of bulk water. (see Materials & Methods). The close agreement between predicted and experimental exchange kinetics of A28-55 is documented by a chi-square value of 0.06 (Fig. S2); this demonstrates that the results obtained by modeling agree very well with the experimental data.
First, we characterized the global helix conformation (Fig. 5 A). The helix bends at a hinge near G37G38 where its TM-N and TM-C parts move relative to each other within a ns timescale. The average bending angle is 20° and ~40% of the conformations show a kink >20°. The rotation of TM-N relative to TM-C is largely anisotropic and mostly “over the hinge” so that G37 and G38 are at the concave side of the bend.
Second, we determined the occupancy of intrahelical H-bonds, which indicates ≥ 80% α-helical structure (O(i) to HN(i+4) bonding) from G29 to G33 and from G38 to V50. A significant drop in α-helicity from L34 to G37 is partially compensated for by pure 310-helix (O(i) to HN(i+3) bonding) (Fig. 5 B). Helicity tends to be lower within TM-N compared to TM-C. Of note, the stretch from V40 to A42 harboring γ-sites is somewhat less α-helical than ζ- and ε-sites. The amide hydrogens and side-chain hydroxyls of T43 and T48 form simultaneous H-bonds to the carbonyl oxygens of their respective i-4 residues in >97% of all frames. This suggests helix stabilization by side chain / main chain backbonding.
Third, we determined the site-specific backbone dynamics of the TMD i) by calculating block-averaged root mean square fluctuations (RMSF) of the C1 atoms around their positions in the average structure and ii) by computing local exchange rates. Both sets of values correlate better with each other for pairs of residues at positions i and i-1 than for other pairs of residues. Together, both values describe the dynamics of the amide bond between neighboring residues. The results corroborate that the backbone dynamics of TM-N is generally higher than that of TM-C and that both are connected by the highly dynamic G37G38 hinge. The helix termini exhibit the highest dynamics while the region from residue 44–50 is likely to correspond to the seven deuterons that do not exchange on the time-scale of the peptide DHX experiment (Fig. 5 C). To compare MD- and NMR-derived exchange kinetics, we estimated NMR exchange rates from the intensity ratios at 16 h (see: Fig. 2). Both sets of position-specific exchange rates show similar trends although they differ at some points which could be due to different access of the catalytic hydroxide ions to amides within LMPG micelles and isotropic solvent, respectively. Differences within TM-N could also result from interaction of TM-N with the short preceding interfacial helix in C9911. In any case, the data suggest a gradient of decreasing amide bond dynamics from the hinge region toward the ε-sites.
Fourth, the dynamics of intra-helical H-bonds was mapped by calculating the free energy profiles W(di) (that correspond to the potential of mean force, PMF) from the distribution of the closest distance di between carbonyl O(i) and the potential amide H-bond donors at positions i+3 and i+4. The force constants thus obtained describe the full dynamics of H-bonds while DHX rates are computed from the abundance of amides that only exchange when the O … H–N distances and angles exceed given limits (see Materials & Methods). Lower force constants, indicating more strongly fluctuating H-bonds, are seen within TM-N compared to TM-C (Fig. 5 C). The force constants were compared to values from LV model TMDs that were previously designed for different backbone dynamics30,34. In general, the APP TMD exhibits a more uneven distribution of force constants compared to L16 (K3WL16K3) and LV16 (K3W(LV)8K3). Specifically, the H-bonds extending from the carbonyl oxygens of G38, V40 to A42, and L49 are characterized by values close to the more dynamic LV16 while H-bonds of T43 to T48 are similar to the ones of the more rigid L16 or more stable than those.
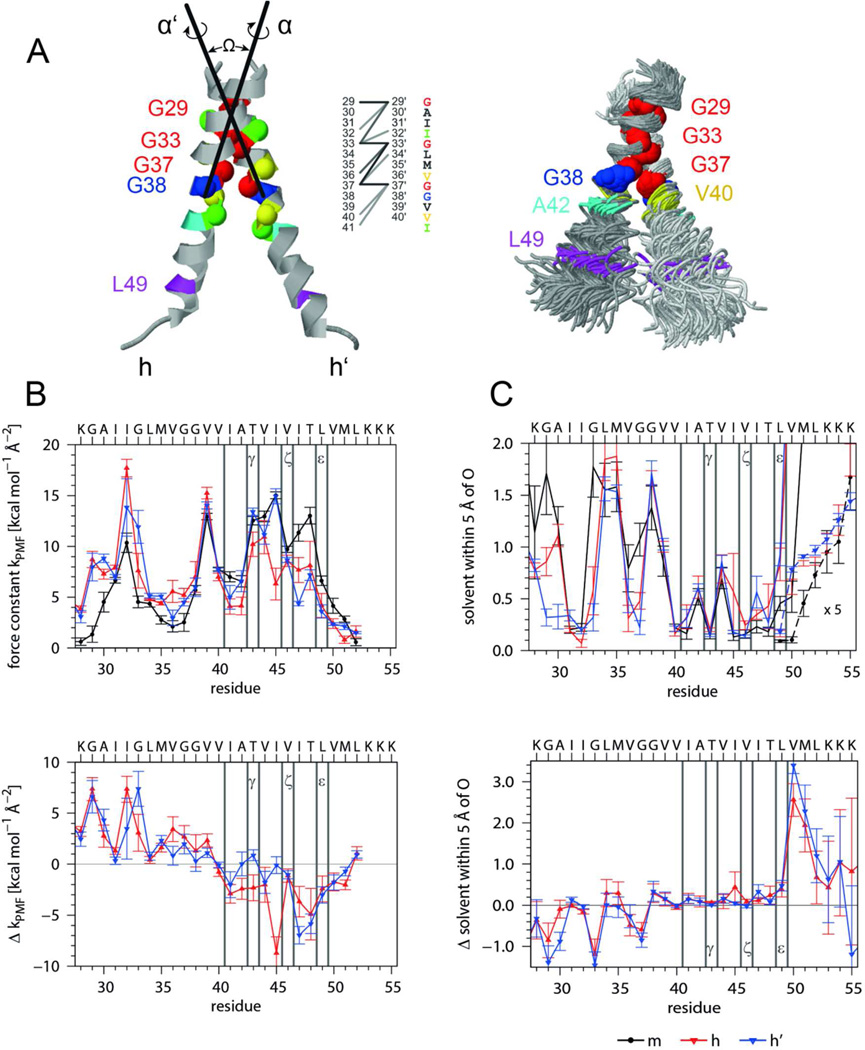
Fifth, we simulated an APP TMD homodimer to assess the potential impact of helix-helix interaction on helix dynamics. To this end, we constructed initial dimer models from the A28-55 helix by short global-searching MD simulations in vacuo36. By averaging the similar structures of one cluster we obtained a dimer where G29, G33, and G37 are close to an interface between a right-handed pair of helices (Ω = −19°). This model is close to most dimer structures proposed earlier15,17,26,28 and was subjected to MD simulation in 80% (v/v) TFE in water. The simulation produced two slightly different dimer structures (see legend of Fig. 6 for details). Average Cα − Cα' distances of Gly residues of the G29xxxG33xxxG37 motif are <0.55 nm (at 50% occupancy throughout the trajectory) while a few extra residues contact each other at distances from 0.55 to 0.60 nm, including V40 and I41 (Fig. 6 A). The side chains of T43 and T48 are not part of the interface and backbond to the carbonyl oxygens of their respective i-4 residues in >97% of all frames. The average helix-bending is reduced to 16°, only 25% of the structures bend >20°, and “over-the-hinge bending” with G37G38 on the concave side is preserved. The helices in the dimer do not deviate significantly from monomeric helices in terms of amide H-bond occupancies and connectivities (data not shown). However, the H-bond length fluctuations, that represent a more sensitive measure of backbone dynamics, reveal that most H-bonds of dimer subunits fluctuate less within TM-N and more within TM-C compared to the monomer (Fig. 6 B). To uncover the potential cause underlying these differences, we assessed solvation of the helix backbones by counting the average numbers of solvent H-bond donors within 0.5 nm of main-chain carbonyl oxygens (Fig. 6 C). The Gly residues of the G29xxxG33xxxG37 motif are less solvated than equivalent sites of the monomeric helix while the dimer exhibits more efficient solvation downstream of L49, in particular at V50M51, which might weaken H-bonds within TMC.
Figure 6. Interhelical interactions, backbone dynamics, and solvation of the A28-55 dimer characterized by MD simulation in isotropic solvation.
A) Average structure and helix-helix contacts (left graph). On average there are 15 contacts formed at any frame of the trajectory as defined by Cα-Cα' distances < 0.6 nm with >50% occupancy (dark, d < 0.5 nm; medium, 0.5 < d/nm < 0.55; light, 0.55 < d/nm < 0.6). The superposition (right graph) shows 100 snapshots taken every ns oriented by a rigid body fit to the Cα atoms of residues 29–37 of the average structure. As compared to the monomer (Fig. 5 A) the bending anisotropy of the helices is reduced. The average interface is slightly asymmetric since interfacial residues of one subunit also contact residues closer to the N-terminus of the partner subunit in addition to their equivalent counterparts. The simulation produced two slightly different dimer populations. The main population (65%) has a crossing angle −50° < Ω < −30° (most probable: −45°) and a center of mass (COM-COM) distance of 0.71 ± 0.03 nm. The helices are oriented (rotation angles α/α’ = −15±15°/ −130±15°) such that the G29xxxG33xxxG37 motifs point toward each other and optimize contact. The minor population with −80° < Ω < −50° (most probable: −60°) has a larger COM-COM distance up to 0.95 nm and the second helix is rotated by 270 ° (α/α’ = −30±10°/ +150±10°) diminishing the contact at G29. B) Helix dynamics characterized by force constants of intra-helical H-bonds extending from carbonyl oxygens at residue i to the amide of residue i+4 or i+3 (upper panel, m = monomeric helix, h, h’ = helices in the dimer) as well as by the differences between mean dimer and monomer values (lower panel). C) Solvation of helices as determined by the numbers of solvent (water or TFE) oxygens within 0.5 nm of the backbone carbonyl oxygens (upper panel, values at V50 to K55 are multiplied by a factor of 5) and differences between mean dimer and monomer values (lower panel). Note that the variation of H-bond strengths of the individual helices is related to the extent of solvation. Error bars indicate standard deviations calculated from 30 ns block averages. Vertical lines denote cleavage sites.
Sixth, monomer and homodimer were subjected to MD simulation in a solvated membrane patch consisting of phosphatidylcholine (POPC). In the membrane, the monomer helix adopts a mean tilt angle of 13 ± 4° and induces moderate positive mismatch as membrane thickness increases by 0.4 nm for lipids within 1.0 nm around the TMD. Compared to the peptides in solvent, helix-bending in the membrane is drastically reduced (Fig. 7 A). Monomeric and dimeric helices have a major population with a bending angle of ~10°. Minor populations (monomer, 4%; dimer, 8%) show bending >20°. Like in the solvent system, larger bending is asymmetric and occurs mainly in direction of the helical face including the G37G38 hinge. The dimer interface (Fig. 7 A) is more symmetric and shorter compared to the one in solvent. Only G29, A30, G33, and L34 are connected to residues of the partner helix over Cα − Cα' distances from 0.45 to 0.55 nm while G37 contacts L34 at a distance from 0.55 to 0.60 nm. The membrane strongly rigidifies the monomeric TM-N helix as indicated by the H-bond strength measured by the force constants that change by ~10 kcal/(Å2 mole) at the positions of Gly residues (Fig. 7 B, upper panel); on the other hand, the membrane has only a minor impact on TM-C dynamics (Fig. 7 B, lower panel). In line with this, membrane-embedding decreases monomer backbone solvation within TM-N. Solvation changes very little for most of TM-C, which is also scarcely solvated in aqueous solvent; however, the membrane enhances solvation at the C-terminus (Fig. 7 B, lower panel). Dimerization stabilizes TM-N to a lesser extent in the membrane compared to solvent and stabilizes TM-C at V44 and I45. A feature that is conserved between solvent and membrane is destabilization of helices in the dimer from I47 to V50 (Fig. 7 C, upper and lower panels). Also, helix destabilization close to the C-terminus is paralleled by increased solvation of terminal regions at, or close to, Lys residues (Fig. 7 D, upper and lower panels). A novel feature not seen in solvent is destabilization at K28 along with increased solvation.
Figure 7. MD simulation of monomeric and dimeric A28-55 in a POPC membrane – comparison to solvent.
A) Comparison of dimer structures in the membrane (dark grey) and in 80% TFE (light grey). The average structure obtained in the membrane was superimposed by a rigid body fit to the Cα atoms of residues 29–37 of helix h of the average structure obtained in TFE. Note that the membrane environment keeps the C-terminal parts of both helices in the dimer closer together. Helix-helix contacts are shown for the membrane (see the legend Fig. 6 A for coding of average Cα-C'α distances). As compared to the helix contacts in TFE (Fig. 6 A) the membrane environment favors a highly symmetric interaction pattern with a unique crossing angle of −40.9 ± 4.2° and a reduced center of mass (COM-COM) distance of 0.68 ± 0.03 nm. The interface is shifted and orients the helices in membrane such that the G29xxxG33 motifs point toward each other (major population: rotation angles α/α’ = −39±19°/ +125±25°; <10% shift to symmetry-related rotation angles α/α’ = +39±18°/ −129±20°). B) Influence of the membrane on helix dynamics and solvation of the monomer as characterized by force constants of intra-helical H-bonds (upper panel, compare Fig. 6 B) and the number of solvent oxygens within 0.5 nm of backbone carbonyl oxygens (lower panel, compare Fig. 6 C). Shown are the differences between mean values in membrane and solvent. Negative values indicate destabilization or desolvation of the TMD in the membrane. Despite the pronounced influence on dynamics, the average structures in both solvents are very similar (the backbone RMSD of core residues equals 0.03 nm). C) Impact of dimerization on helix dynamics in the membrane as characterized by force constants of intra-helical H-bonds (upper panel, m = monomeric helix, h, h’ = helices in the dimer) as well as by the differences between mean dimer and monomer values (lower panel). D) Solvation of helices of monomer and dimer in the membrane as determined by the numbers of solvent (water or TFE) oxygens within 0.5 nm of the backbone carbonyl oxygens (upper panel, values at V50 to K55 are multiplied by a factor of 5) and differences between mean dimer and monomer values (lower panel). Error bars indicate standard deviations calculated from 25 ns block averages. Vertical lines denote cleavage sites.
Taken together, modelling confirms that the APP TMD contains a dynamic homodimerization/cholesterol-binding domain that is connected by a highly flexible linker to a less dynamic cleavage domain. Homodimerization tends to dampen H-bond fluctuations within the former while increasing H-bond fluctuations within the latter.
Discussion
Our results reveal that a highly dynamic dimerization, TM-N, and a less dynamic cleavage, TM-C, domain of the APP TMD connect at G37G38 which is the most flexible site in the TMD with the exception of the frayed helix termini. The hinge has previously been detected in monomeric C9911 and in a dimeric APP fragment (G12-K55)12. It has been proposed that the hinge might precisely position the TMD within a curved lumen of presenilin11. Thereby, the ε-sites may be positioned such that they are close to the active site for initial cleavage. What could be the functional significance of the pronounced TM-N dynamics? We envision four, not mutually exclusive, scenarios. First, if the G37G38 hinge is initially positioned at a curved site within presenilin, sequential proteolysis would require sliding of the TMD past this curved site, a process that would be facilitated by a flexible TM-N domain. Second, a flexible helix could improve cholesterol and drug binding to this region11,61. Third, a higher backbone dynamics could enhance the rate of dimerization by increasing the probability by which randomly colliding helices enter a stable association; a similar mechanism has been suggested for interactions between partially unstructured proteins62. Finally, high backbone dynamics could facilitate the release of peptides from presenilin following cleavage at γ-sites since a flexible helix may readily convert to extended water-soluble structures. Given that substrate helix unraveling is considered to promote proteolysis, one surprising finding of this study was the relatively low and unevenly distributed helix dynamics of the cleavage region. What are the implications of these data for cleavage? Our C99 HDX experiments and TMD simulations reveal that backbone dynamics strongly increases downstream of the ε-sites which agrees with earlier data obtained by NMR spectroscopy15. This local unraveling at the helix C-terminus results from absent H-bonds and side-chain / side-chain interactions between the respective i and i+3,4 residues as well as from increased solvation. It is plausible, that this fraying of the substrate helix promotes the initial endoproteolytic cuts at ε-sites. The newly formed C-termini of the resulting C48 and C49 fragments would then be frayed themselves which would facilitate ζ-cleavages; this, in turn, would produce frayed C45 and C46 fragments etc. The order of DHX kinetics seen with the octa-peptide fragments (AL-peptides, Fig. 4 C) suggests that, once initiated, sequential cleavage becomes more facile as it proceeds from the C-terminus towards the γ-sites. One may wonder why the efficiency of sequential cleavage drops strongly upstream of the γ40 site4–7. Previously, this has been ascribed to steric hindrance as a result of helix-helix interaction. The substrate was proposed to be cleaved as a dimer since mutating the Gly residues within the G29xxxG33xxxG37 motif led to shorter cleavage products concurrent with decreasing self-interaction of the TMD13,27. Also, movement of the substrate helix could be prohibited after cleavage at γ-sites due to interaction of K28 with presenilin63 or Aβ fragments containing the relatively hydrophilic TM-N could readily detach from the enzyme. In addition, our results suggest that cleavage may be reduced by rigidification of the TM-N helix after dimerization.
Do our results reveal why different Aβ peptide variants are produced with different abundance? Experimental studies have shown that Aβ40 is more abundant than Aβ42 and Aβ38 and that shorter sequences are rare4–7. One factor defining the Aβ40/Aβ42 ratio could be differential cleavability at γ40 and γ42 sites. Our data indicate similar dynamics at both sites in terms of the C1 dynamics (Fig. 5 B), H-bond occupancy (Fig. 5 B), H-bond length fluctuations (Fig. 5 D and 6 D), and local exchange rates at the respective i+4 residues V44 and V46 (Fig. 5 C). It is presently unclear how the efficiency of proteolysis depends on these properties and our data do not indicate that the Aβ40/Aβ42 ratio depends on the local TMD dynamics. However, the situation may be different at ε-sites where cleavage is initiated. A close inspection of our data reveals that i) the L49 amide exchanges faster, ii) the L49 C1 carbon fluctuates more strongly (Fig. 5 B), iii) the H-bond extending from the L49 carbonyl oxygen is more dynamic (Fig. 5 D and 6 B), and that iv) the L49 carbonyl oxygen is better solvated (Fig. 6 C) compared to the values seen at T48. These differences may favor cleavage at ε49 over ε48 and thus facilitate entry into the product line generating Aβ40. Differences in H-bond dynamics are also seen at respective positions of the L16 and LV16 controls. Therefore, they may not only arise from sequence-specific local changes in helix stability but also from the more sequence-independent C-terminal helix fraying. In any case, the differences between T48 and L49 may facilitate the initial cut at ε49, thereby enhancing Aβ40 production.
What could be the impact of TMD dimerization on TMD processing? Apart from steric hindrance, as discussed above, dimerization could affect proteolysis more directly. This is suggested by our simulations of a dimer that conforms to most experimentally supported models13–17 although alternative models exist12. Dimerization increases the level of solvation at V50M51L52 carbonyl oxygens (Fig. 6 C). Increased solvation is suited to destabilize upstream regions64, as revealed by stronger amide H-bond fluctuations within TM-C (Fig. 6 B), which could generally promote cleavage. This destabilization is also evident from longer H-bonds in the dimer compared to the monomer in the recent NMR structure12. For comparison, we also simulated monomer and dimer in a POPC membrane patch. The membrane has a strong stabilizing effect within TM-N but little impact on TM-C. TM-N stabilization is strongest at Gly residues, this coincides which the observation that desolvation by the membrane is most significant at Gly sites. This suggests that helix flexibility around Gly is not only related to missing intrahelical side-chain – side-chain interactions; rather, Gly can destabilize a helix in aqueous solvent by enhancing backbone solvation. The effect of dimerization on solvation at the C-terminus and on dynamics of the cleavage region is qualitatively similar in the membrane and in solvent. Since solvation by dimerization is somewhat less pronounced in the membrane, however, the strongest impact of dimerization on the dynamics of the cleavage region is probably felt in the aqueous lumen of presenilin.
It is clear that embedding of the substrate helix within presenilin8 could also affect its dynamics and hydration. Thus, our results obtained with free substrate should be interpreted with caution. Indeed, the Aβ40/Aβ42 ratio is influenced by hereditary presenilin mutations that lead to early-onset AD65–67 and by drugs that may intercalate between enzyme and substrate68,69. On the other hand, other disease-causing point mutations that change the Aβ42/Aβ40 ratio are located within the APP TMD27,70,71. Thus, the efficiency by which the different sites are cleaved results from a complex interplay of substrate and enzyme.
Conclusion
In sum, processing of the APP TMD could depend on its primary structure at several stages. The hinge between the APP TMD dimerization and cleavage domains may precisely position the substrate within presenilin such that its catalytic center can initially only access the ε-sites. The ratio of cleavage products could be influenced by different extents of solvation and of H-bond stabilities at alternate ε-sites. Substrate movement during sequential proteolysis may facilitated by the flexibility of the TM-N helix. Finally, dimerization may affect substrate processing by decreasing the dynamics of the dimerization region while increasing that of the C-terminal part of the cleavage region.
Supplementary Material
Acknowledgements
We thank Dr. Steven Verhelst for critical reading of the manuscript and his valuable comments and Matthias Möch and Walter Stelzer for help with graphics. This work was supported by grant LA699/14-1 of the Deutsche Forschungsgemeinschaft, by grant 01GI0724 of the Bundesministerium fø Forschung und Technologie, the State of Bavaria and the Center of Integrative Protein Science Munich (CIPSM) (DL) and by US NIH grants PO1 GM080513 (CRS) and F31 NS077681 (to PJB). We also thank the Leibniz Rechenzentrum, Garching, for computing resources.
Abbreviations
- AD
Alzheimer’s disease
- APP
Amyloid Precursor Protein
- CD
circular dichroism
- COM
center of mass
- (d)TFE
(mono-deuterated) 2,2,2-trifluoroethanol
- DHX
deuterium/hydrogen-exchange
- ESI-MS
electrospray ionization mass spectrometry
- HDX
hydrogen/deuterium-exchange
- LMPG
lyso-myristoylphosphatidylglycerol
- MD
molecular dynamics
- MEM
maximum entropy method
- P/L
peptide/lipid
- DLPC
di-lauryl-phosphatidylcholine
- DLPE
di-lauryl-phosphatidylethanolamine
- DLPS
di-lauryl-phosphatidylserine
- POPC
phosphatidylcholine
- PMF
potential of mean force
- RMSF
root mean square fluctuation
- TMD
transmembrane domain
- SD
standard deviation.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional data are shown in Figs. S1 and S2 and Table S1. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Haass C, Selkoe DJ. Nat Rev Mol Cell Biol. 2007;8:101. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenthaler SF, Haass C, Steiner H. J Neurochem. 2011;117:779. doi: 10.1111/j.1471-4159.2011.07248.x. [DOI] [PubMed] [Google Scholar]
- 3.Kaden D, Munter LM, Reif B, Multhaup G. Eur J Cell Biol. 2011;91:234. doi: 10.1016/j.ejcb.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Funamoto S, Morishima-Kawashima M, Tanimura Y, Hirotani N, Saido TC, Ihara Y. Biochemistry. 2004;43:13532. doi: 10.1021/bi049399k. [DOI] [PubMed] [Google Scholar]
- 5.Qi-Takahara Y, Morishima-Kawashima M, Tanimura Y, Dolios G, Hirotani N, Horikoshi Y, Kametani F, Maeda M, Saido TC, Wang R, Ihara Y. J Neurosci. 2005;25:436. doi: 10.1523/JNEUROSCI.1575-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takami M, Nagashima Y, Sano Y, Ishihara S, Morishima-Kawashima M, Funamoto S, Ihara Y. J. Neurosci. 2009;29:13042. doi: 10.1523/JNEUROSCI.2362-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukumori A, Fluhrer R, Steiner H, Haass C. J Neurosci. 2010;30:7853. doi: 10.1523/JNEUROSCI.1443-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H. Proc Natl Acad Sci U S A. 2006;103:6889. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh R, Barman A, Prabhakar R. J Phys Chem B. 2009;113:2990. doi: 10.1021/jp811154w. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Phylip LH, Lees WE, Winther JR, Dunn BM, Wlodawer A, Kay J, Gustchina A. Nat Struct Biol. 2000;7:113. doi: 10.1038/72378. [DOI] [PubMed] [Google Scholar]
- 11.Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. Science. 2012;336:1168. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadezhdin KD, Bocharova OV, Bocharov EV, Arseniev AS. FEBS Lett. 2012;586:1687. doi: 10.1016/j.febslet.2012.04.062. [DOI] [PubMed] [Google Scholar]
- 13.Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G. EMBO J. 2007;26:1702. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beel AJ, Mobley CK, Kim HJ, Tian F, Hadziselimovic A, Jap B, Prestegard JH, Sanders CR. Biochemistry. 2008;47:9428. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato T, Tang TC, Reubins G, Fei JZ, Fujimoto T, Kienlen-Campard P, Constantinescu SN, Octave JN, Aimoto S, Smith SO. Proc Natl Acad Sci U S A. 2009;106:1421. doi: 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyashita N, Straub JE, Thirumalai D, Sugita Y. J Am Chem Soc. 2009;131:3438. doi: 10.1021/ja809227c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Barreyro L, Provasi D, Djemil I, Torres-Arancivia C, Filizola M, Ubarretxena-Belandia I. J Mol Biol. 2011;408:879. doi: 10.1016/j.jmb.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beel AJ, Sanders CR. Cell. Mol. Life Sci. 2008;65:1311. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS. Proc. Natl. Acad. Sci. U S A. 2000;97:5123. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urban S, Freeman M. Molecular Cell. 2003;11:1425. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 21.Akiyama Y, Maegawa S. Mol Microbiol. 2007;64:1028. doi: 10.1111/j.1365-2958.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 22.Lemberg MK, Martoglio B. FEBS Letters. 2004;564:213. doi: 10.1016/S0014-5793(04)00192-9. [DOI] [PubMed] [Google Scholar]
- 23.Fluhrer R, Martin L, Klier B, Haug-Kroeper M, Grammer G, Nuscher B, Haass C. J. Biol. Chem. 2012;287:5156. doi: 10.1074/jbc.M111.328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu JX, Yau WM, Tycko R. Biophys J. 2011;100:711. doi: 10.1016/j.bpj.2010.12.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langosch D, Arkin IT. Protein Sci. 2009;18:1343. doi: 10.1002/pro.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G. EMBO J. 2007;26:1702. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page RM, Gutsmiedl A, Fukumori A, Winkler E, Haass C, Steiner H. J Biol Chem. 2010;285:17798. doi: 10.1074/jbc.M110.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kienlen-Campard P, Tasiaux B, Hees Jv, Li M, Huysseune S, Sato T, Fei JZ, Aimoto S, Courtoy PJ, Smith SO, Constantinescu SN, Octave J-N. J. Biol. Chem. 2008;283:7733. doi: 10.1074/jbc.M707142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorman PM, Kim S, Guo M, Melnyk RA, McLaurin J, Fraser PE, Bowie JU, Chakrabartty A. BMC Neurosci. 2008;9:17. doi: 10.1186/1471-2202-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poschner BC, Quint S, Hofmann M, Langosch D. J. Mol. Biol. 2009;386:733. doi: 10.1016/j.jmb.2008.12.077. [DOI] [PubMed] [Google Scholar]
- 31.Poschner B, Reed J, Langosch D, Hofmann MW. Analyt. Biochemistry. 2007;363:306. doi: 10.1016/j.ab.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 32.Hofmann MW, Poschner BC, Hauser S, Langosch D. Biochemistry. 2007;46:4204. doi: 10.1021/bi602539n. [DOI] [PubMed] [Google Scholar]
- 33.Stelzer W, Poschner BC, Stalz H, Heck AJ, Langosch D. Biophys. J. 2008;95:1326. doi: 10.1529/biophysj.108.132928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quint S, Widmaier S, Minde D, Langosch D, Scharnagl C. Biophys. J. 2010;99:2541. doi: 10.1016/j.bpj.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiókiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, Engelman DM, Brüger AT. Proteins. 1996;26:257. doi: 10.1002/(SICI)1097-0134(199611)26:3<257::AID-PROT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 37.Feller SE, MacKerell AD. J. Phys.Chem.B. 2000;104:7510. [Google Scholar]
- 38.Tuckerman M, Berne BJ. J. Chem. Phys. 1992;97:1990. [Google Scholar]
- 39.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. J. Comp. Chem. 2005:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faraldo-Gomez JD, Forrest LR, Baaden M, Bond PJ, Domene C, Patargias G, Cuthbertson J, Sansom MS. Proteins. 2004;57:783. doi: 10.1002/prot.20257. [DOI] [PubMed] [Google Scholar]
- 41.Kucerka N, Nieh MP, Katsaras J. Biochim Biophys Acta. 20111808:2761. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 42.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J Comput Chem. 2009;30:1545. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teilum K, Kragelund BB, Poulsen FM. Protein Folding Handbook. Weinheim, Germany: Wiley-VCH Verlag GmbH; 2008. Application of Hydrogen Exchange Kinetics to Studies of Protein Folding; p. 634. [Google Scholar]
- 44.Sessions RB, Gibbs N, Dempsey CE. Biophys J. 1998;74:138. doi: 10.1016/S0006-3495(98)77775-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dempsey CE. Progr. Nucl. Magn. Res. Spectr. 2001;39:135. [Google Scholar]
- 46.Fernandez A, Berry RS. Biophys J. 2002;83:2475. doi: 10.1016/S0006-3495(02)75258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez A, Scheraga HA. Proc Natl Acad Sci U S A. 2003;100:113. doi: 10.1073/pnas.0136888100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehrnejad F, Naderi-Manesh H, Ranjbar B. Proteins. 2007;67:931. doi: 10.1002/prot.21293. [DOI] [PubMed] [Google Scholar]
- 49.Roccatano D, Colombo G, Fioroni M, Mark AE. Proc Natl Acad Sci U S A. 2002;99:12179. doi: 10.1073/pnas.182199699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bevington PR, Robinson DK. Data reduction and error analysis for the physical science. New York, USA.: McGraw-Hill Higher Education; 1969. [Google Scholar]
- 51.Bahar I, Wallqvist A, Covell DG, Jernigan RL. Biochemistry. 1998;37:1067. doi: 10.1021/bi9720641. [DOI] [PubMed] [Google Scholar]
- 52.Demmers JA, Haverkamp J, Heck AJ, Koeppe RE, 2nd, Killian JA. Proc. Natl. Acad. Sci. U S A. 2000;97:3189. doi: 10.1073/pnas.050444797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhuang T, Jap BK, Sanders CR. J Am Chem Soc. 2011;133:20571. doi: 10.1021/ja208972h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kroenke CD, Ziemnicka-Kotula D, Xu J, Kotula L, Palmer AG., 3rd Biochemistry. 1997;36:8145. doi: 10.1021/bi9705669. [DOI] [PubMed] [Google Scholar]
- 55.Ramelot TA, Gentile LN, Nicholson LK. Biochemistry. 2000;39:2714. doi: 10.1021/bi992580m. [DOI] [PubMed] [Google Scholar]
- 56.Tolia A, Chavez-Gutierrez L, De Strooper B. J Biol Chem. 2006;281:27633. doi: 10.1074/jbc.M604997200. [DOI] [PubMed] [Google Scholar]
- 57.Sato C, Morohashi Y, Tomita T, Iwatsubo T. J Neurosci. 2006;26:12081. doi: 10.1523/JNEUROSCI.3614-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stelzer W, Langosch D. Protein Sci. 2012;21:1097. doi: 10.1002/pro.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao H, Hoerner JK, Eyles SJ, Dobo A, Voigtman E, Mel'Cuk AI, Kaltashov IA. Prot. Sci. 2005;14:543. doi: 10.1110/ps.041001705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z, Li W, Logan TM, Li M, Marshall AG. Protein Sci. 1997;6:2203. doi: 10.1002/pro.5560061015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richter L, Munter LM, Ness J, Hildebrand PW, Dasari M, Unterreitmeier S, Bulic B, Beyermann M, Gust R, Reif B, Weggen S, Langosch D, Multhaup G. Proc Natl Acad Sci U S A. 2010;107:14597. doi: 10.1073/pnas.1003026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou HX. Trends Biochem Sci. 2012;37:43. doi: 10.1016/j.tibs.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kukar TL, Ladd TB, Robertson P, Pintchovski SA, Moore B, Bann MA, Ren Z, Jansen-West K, Malphrus K, Eggert S, Maruyama H, Cottrell BA, Das P, Basi GS, Koo EH, Golde TE. J. Biol. Chem. 2011;286:39804. doi: 10.1074/jbc.M111.274696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanbonmatsu KY, Garcia AE. Proteins. 2002;46:225. doi: 10.1002/prot.1167. [DOI] [PubMed] [Google Scholar]
- 65.Selkoe DJ. Physiol Rev. 2001;81:741. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 66.Tanzi RE, Bertram L. Cell. 2005;120:545. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 67.Goedert M, Spillantini MG. Science. 2006;314:777. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 68.Weggen S, Eriksen JL, Das P, Sagi SA, Wang R, Pietrzik CU, Findlay KA, Smith TE, Murphy MP, Bulter T, Kang DE, Marquez-Sterling N, Golde TE, Koo EH. Nature. 2001;414:212. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- 69.Heneka MT, Kummer MP, Weggen S, Bulic B, Multhaup G, Munter L, Hull M, Pflanzner T, Pietrzik CU. Curr Alzheimer Res. 2011;8:115. doi: 10.2174/156720511795256099. [DOI] [PubMed] [Google Scholar]
- 70.Kumar-Singh S, De Jonghe C, Cruts M, Kleinert R, Wang R, Mercken M, De Strooper B, Vanderstichele H, Lofgren A, Vanderhoeven I, Backhovens H, Vanmechelen E, Kroisel PM, Van Broeckhoven C. Hum Mol Genet. 2000;9:2589. doi: 10.1093/hmg/9.18.2589. [DOI] [PubMed] [Google Scholar]
- 71.Munter LM, Botev A, Richter L, Hildebrand PW, Althoff V, Weise C, Kaden D, Multhaup G. J Biol Chem. 2010;285:21636. doi: 10.1074/jbc.M109.088005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.