Abstract
Objective: To construct a recombinant lentivirus vector driven by the PSMA promoter carrying mTOR-shRNA, and to obtain the effect on the mTOR gene silencing in human prostate cancer xenografts. Methods: The complimentary oligos of small interference RNA (siRNA) with hairpin structures targeting the mTOR gene and a negative control were synthesized, then ligated with pLV-PSMA-promoter vector and sequenced. The recombinant vectors were then transfected with viral packaging mix into 293T cells, viral supernatant was harvested to determine the titer. Prostate cancer cells infected by virus were harvested and the expression of mTOR (LV-PSMA-shmTOR), target proteins and cell growth were detected by reverse transcription-PCR (RT-PCR), Western blot and MTT separately. In established tumors derived from human prostate cancer cells, concentrated LV-PSMA-shmTOR lentivirus was injected intravenously in the tail vein of C4-2b tumor bearing female severe combined immunodeficient (SCID) mice. Tumor volume and immunohistochemistry was assessed. Results: Sequencing data showed that the constructed plasmids contained the correct sequences of mTOR siRNA transcript templates. A vector producing cell line 293T was established, and the titer for transfection was obtained. RT-PCR, Western blot and MTT analyses demonstrated that mTOR shRNA expression construct could suppress the expression of mTOR and inhibit the prostate cancer cell growth, specially. The tumor growth was suppressed in nude mouse. Conclusion: A PSMA driven lentivirus mediated siRNA targeting mTOR gene was successfully constructed, which decreased the expression of mTOR and induced the prostate cancer cell growth in vitro and in vivo. It has set up a research platform for the gene therapy of tumors which take mTOR as the target in the prostate cancer field.
Keywords: mTOR, PSMA, prostatic carcinoma, apoptosis
Introduction
Apart from digestive system tumors, prostate cancer is the most commonly diagnosed malignancy in men [1]. Despite progress in early diagnosis, and prolongation of patient survival, the disease still carries significant morbidity and mortality, with its advanced and metastatic phase claiming over 30,000 deaths per year in the United States alone [2]. Similar to the genetic heterogeneity of most epithelial malignancies, prostate cancer progresses through a stepwise acquisition of multiple molecular changes, of which insensitivity to androgen deprivation, emergence of an ‘osteomimetic’ phenotype responsible for metastatic tropism to the bone, and deregulated cell proliferation and cell survival, are pivotal traits [3,4].
Prostate specific membrane antigen (PSMA) is a homodimeric type II transmembrane ectopeptidase with both folate hydrolase and N-acetylated, α-linked acidic dipeptidase or NAALADase activities [5-7]. In normal tissue, the expression of PSMA is predominantly restricted to prostatic epithelium, with low expression in kidney, salivary gland, duodenum and the central and peripheral nervous systems. As its name suggests, PSMA is highly over-expressed in prostate cancer where its increased expression correlates with advanced stages of prostate cancer and metastasis. Its function in the prostate remains unknown. Interestingly, PSMA has also been shown to be upregulated on the angiogenic vasculature of most solid tumors [8]. Previous investigations in our lab have shown that mice lacking PSMA are incapable of mounting a pathologic angiogenic response in vivo suggesting that PSMA induction at angiogenic sites is critical for endothelial function in angiogenesis [9].
To determine whether aberrantly potentiated mTOR signaling stimulates prostate cancer growth in vitro and in vivo, we have generated an experimental prostate cancer model in nude mice, through lentivirus mediated mTOR shRNA controlled by PSMA promoter. These findings demonstrate that the genetic mTOR shRNA in the treatment of prostatic tumor growth by downregulated mTOR signaling pathway.
Materials and methods
Cells and cell culture conditions
The RWPE1, LNCap, C4-2b and the human embryonic kidney (HEK) 293T cell lines were purchased from American Type Culture Collection (Rockville, MD, USA). Cells were cultured in RPMI 1640 and Dulbecco’s Modified Eagles’ Medium (Mediatech, USA) respectively and supplemented with 10% FBS (SIGMA, USA), 100 units/ml of penicillin and 100 μg/ml of streptomycin (Invitrogen Corp., Carlsbad, CA). Cells were maintained in a humidified atmosphere of 5% CO2 in air, at 37 °C.
Cloning, virus preparation, and titration
shRNA against mTOR in a pGenSil-1 basic vector [7] was digested and cloned between BamHI and HindIII (New England Biolabs, Beverly, MA, USA) into a human PSMA promoter driven pRRL vector (Irvin S.Y. Chen, UCLA) (Figure 1A). A vector with shRNA against non-sense sequence (shCON) was used as control [12]. Infectious viral supernatants (DMEM media with 1% FBS) were derived by transient co-transfection of 293T (4×106 in 100 mm3 petri-plates) cells using lipofectamine 2000. A total of 20 μg of plasmid in the proportion of 12 μg of lentiviral vector carrying shRNA, 8 μg of packaging plasmid pCMVΔR8.2 DVPR (VPR deleted) and 2 μg of pCMV-VSVG were used, and viral supernatant collected at 24, 48 and 72 h post transfection. Pooled supernatants were concentrated using an Amicon Ultra-15 100K cutoff filter device (Millipore Billerica, MA, USA). The viral titer of the concentrated supernatant was determined by performing a p24 ELISA kit (Cell BIOLABS, INC. San Diego, USA) to detect the HIV-p24 core protein of the vector.
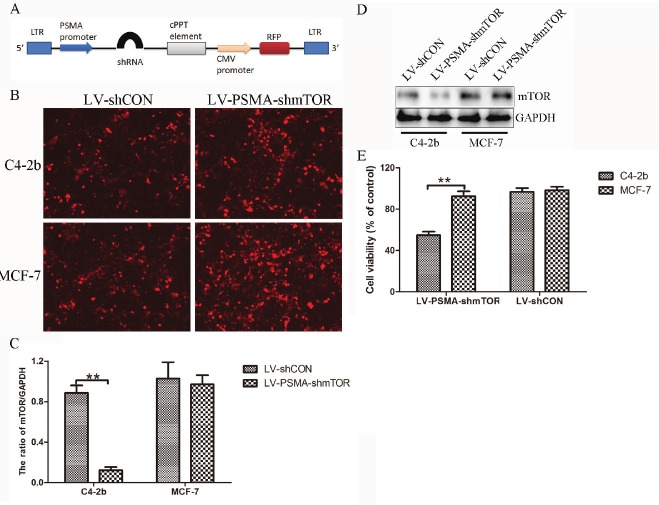
Figure 1.
Molecular characterization of mTOR knockdown in cancer cells following in vitro transduction. A: Schematic map of the lentiviral shuttle vector showing the organization of the different components. B: Representative fluorescent photomicrographs showing the transduction efficiency of both LV-shCON and LV-PSMA-shmTOR as evident by RFP expression in C4-2b and MCF-7 cells. C: qRT-PCR for mTOR mRNA expression in transduced cells (assay performed in triplicates and represented as standard deviation of the mean). Values were obtained relative to the house keeping gene GAPDH. D: Representative immunoblot for mTOR protein. GAPDH immunoblotting was performed as loading control. E: MCF-7 and C4-2b cancer cells were infected with LV-shCON and LV-PSMA-shmTOR at a multiplicity of infection of 5 at 48 h, respectively, post infection, the cell survival was measured by MTT assay. Results were expressed as percentage of untreated control. The data represent the average±s.d. of three independent experiments. (**P<0.01).
Reverse transcription and quantitative RT-PCR (qRT-PCR)
Total tissue RNA was isolated from minced fresh tissue using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Two micrograms of total RNA was digested with DNase I, and reverse transcribed using Superscript First-Strand Synthesis System (Invitrogen). qRT-PCR reactions were done using an Applied Biosystems 7900 sequence detector with 5 ng of cDNA, 200 nM of each primer pair and Power SYBR Green PCR Master Mix or TaqMan Universal PCR Master Mix from Applied Biosystems.
Western blot analysis
Whole-cell lysate (20-40 μg) was resolved by SDS-PAGE and then transferred onto PVDF membranes. PVDF membranes were washed briefly in Tris-buffered saline and 0.1% Tween-20 (TBST) and blocked in a solution of TBST containing 5% nonfat dry milk for 15 min with constant agitation. After blocking, the PVDF membrane was incubated with the following primary antibodies overnight at 4°C: mouse monoclonal mTOR (1:500 dilution in TBST), 4EBP1 (1:800 dilution in TBST), S6K (1:1,000 dilution in TBST), PI3K (1:500 dilution in TBST), AKT (1:1,000 dilution in TBST), (1:500 dilution in TBST) and GAPDH (1:2,000 dilution in TBST) antibody. Membranes were washed in TBST (3 times for 15 min) and were incubated for 1 h with horseradish peroxidase-conjugated secondary antibodies at a 1:10,000 dilution at room temperature with constant agitation before enhanced chemiluminescence (Amersham Biosciences, NJ) and exposure to film.
MTT assay
The effect of lV-PSMA-shmTOR was determined based on cycotoxicity to the human prostate cancer cell line using an MTT assay. Briefly, cells were seeded in 96-well tissue culture plates at a density of 5×103 cells/well and then treated with the concentrated virus to the growth medium. The following day, the medium was removed, and 100 μL of fresh medium containing 0.5 mg/mL MTT was added to each well. The cells were incubated at 37 °C in humidified 5% CO2 atmosphere for 4 h, followed by the addition of 150 μL of solubilization solution (0.01 mol/L HCl in 100 g/L sodium dodecyl [SDS]) to each well, and the incubation of cells for a further 10 min at 37°C with gentle shaking. The optical density of the plates was measured using the spectrophotometrical absorbance at 570 nm in the Microplate Reader Model 550 (BIO-RAD; CA, USA).
Establishing prostate cancer xenografts
Approximately 2×106 C4-2b cells in 50 μL of DMEM were inoculated in the upper right thoracic mammary fat pad of age-matched female severe combined immune deficient (SCID) mice. All surgical procedures and animal handling, including viral vector delivery, were performed in accordance with protocols approved by the Xi’an Jiaotong University Institutional Animal Care and Use Committee, and conformed to the Guide for the Care and Use of Laboratory Animals.
Cell and in vivo tumor transduction
For cell studies, 5×106 C4-2b cells were plated in 100 mm3 dishes and 5 ml of viral supernatant with 1 mg/ml of polybrene were added for 4-5 h. This procedure was repeated for three days. The efficiency of transduction was assessed and photomicrographs of RFP expression were recorded using a Nikon TS100 inverted microscope (Nikon, USA) equipped with a charged coupled device (CCD) camera. Images were processed using Image Pro Plus 5.1 (MediaCybernetics, Silver Spring MD) software.
Lentiviral transduction of tumors in vivo was achieved in established C4-2b xenografts. All mice were maintained under specific pathogen-free conditions and given sterile food and water. Briefly, once tumor sizes reached 50 mm3, in vivo transduction of lentivirus was achieved through tail vein injections of 0.2 ml of concentrated viral suspension with a viral titer of 5×1011-lentiviral particles/ml (~5μg of p24/ml) in DMEM, twice a week for up to 30 days. All tumors were collected immediately after death, weighed, and used for fixed in formalin and embedded in paraffin, or frozen in liquid nitrogen. The tumor volumes (in cm3) were calculated using the formula = a × b2/2, where a is the longest diameter, b is the shortest diameter. Each mouse formed one tumor (tumor take rate = 100%). Average tumor volume per mouse was the mean of the tumors formed at the injection site. All tumors from two independent experiments were included for final calculation of tumor volume and weight.
Terminal deoxynucleotidyl transferase (TDT)-mediated dUTP nick-end labeling (TUNEL) assay
C4-2b xenograft tumor sections (5 μm thick) were deparaffinized and digested with proteinase K (20 μg/mL; Roche). Then sections were incubated with TDT (0.3 U/μL) and with biotinylated dUTP (0.2 mM; Roche) in 1× TDT buffer (Invitrogen, Carlsbad, CA) for 1 hour at 37°C. The sections were incubated in 10% normal horse serum to block nonspecific binding, followed by incubation for 1 hour at room temperature with avidin-biotin complex (1:100 dilution) from a Vectastain Elite ABC Kit (Vector Lab, Burlingame, CA). The sections were stained with 0.125% amino-ethyl carbazole (AEC) or AEC buffer (Sigma), counterstained with Mayer’s hematoxylin (DakoCytomation, Carpinteria, CA), and mounted under coverslips in Aqua-mount medium (Thermo Fisher). Sections were examined by microscopy as described above, and digital images at ×100 magnification were obtained using FluoView 1000 software (Olympus Inc, Melville, NY). TUNEL-positive cells were counted in five randomly chosen fields for each tumor section and presented as a percentage of the total number of cells.
Statistical analysis
Assays were set up in triplicates and the results were presented as mean±S.D. Variance between the experimental groups were determined by two-tailed t-test. P<0.05 was considered statistically significant.
Results
Molecular and functional characterization of transduced prostate cancer cells
To determine the efficacy of viral vectors, viral supernatants prepared from either LV-shCON or LV-PSMA-shmTOR were added to C4-2b and MCF-7 cancer cells. RFP expression in photomicrographs of LV-shCON or LV-PSMA-shmTOR transduced cells shown in Figure 1B confirmed ~ 90% transduction efficiency. To evaluate the silencing efficiency, transduced cells were characterized for mTOR mRNA by qRT-PCR using specific primers against mTOR, and for protein expression in immunoblots obtained using a rabbit polyclonal antibody against mTOR (Figure 1C and 1D). C4-2b cells transduced with mTOR shRNA showed approximately 80% reduction in mTOR mRNA and protein relative to control cells. LV-shCON transduced control cells and MCF-7 cells had comparable expression levels of mTOR mRNA and protein. Moreover, the inhibitory effect on tumor cell proliferation of LV-PSMA-shmTOR is significantly stronger compared with that of LV-shCON (P<0.05) in C4-2b cells at 48 h post infection (Figure 1E).
Antitumor efficacy of LV-PSMA-shmTOR in vivo
The in vivo antitumor efficacy of LV-PSMA-shmTOR was examined by tail vein injections of lentivirus in C4-2b xenograft in nude mice. The tumor growth curves of each treatment group are shown in Figure 2. The tumors grew rapidly and reached 1,600 mm3 within 32 days in the PBS group. However, in LV-PSMA-shmTOR group, tumors grew much slower with 158±43 mm3 at day 32 (Figure 2).
Figure 2.
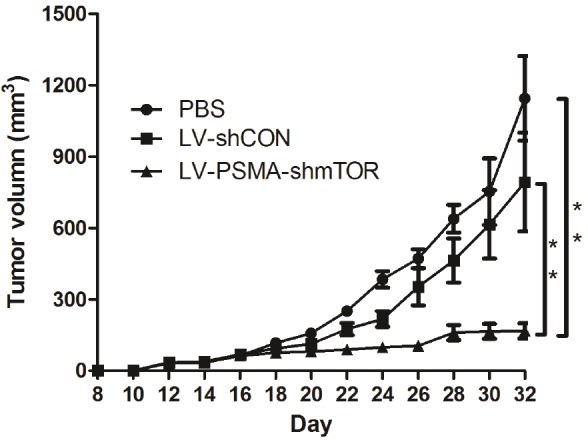
Growth inhibitory effects on prostate cancer cells of lentiviruses in vivo. Tumor model were established by injecting C4-2b tumor cells into female nude mice. At day 5 post-inoculation, the mice were received recombinant lentivirus via tail vein injection. Accurately, the time for the injection of virus was at day 5, 7, 9 and 11 post-inoculation tumor cells, respectively. The tumor size was measured at 2-day intervals and tumor volume error bars represent the SD. (**P<0.01).
PSMA driven mTOR shRNAs are able to inhibit angiogenesis and induce apoptosis. We then examined apoptosis of tumors treated with different lentiviruses. TUNEL staining followed by quantification (Figure 3) showed that the group of LV-PSMA-shmTOR has the highest apoptotic index (23.27±6.91)%. The apoptotic index, expressed as the average percentages of TUNEL-positive cells from 10 random visual fields, were (2.39±0.45)% and (4.34±0.76)% in PBS and LV-shCON, respectively.
Figure 3.
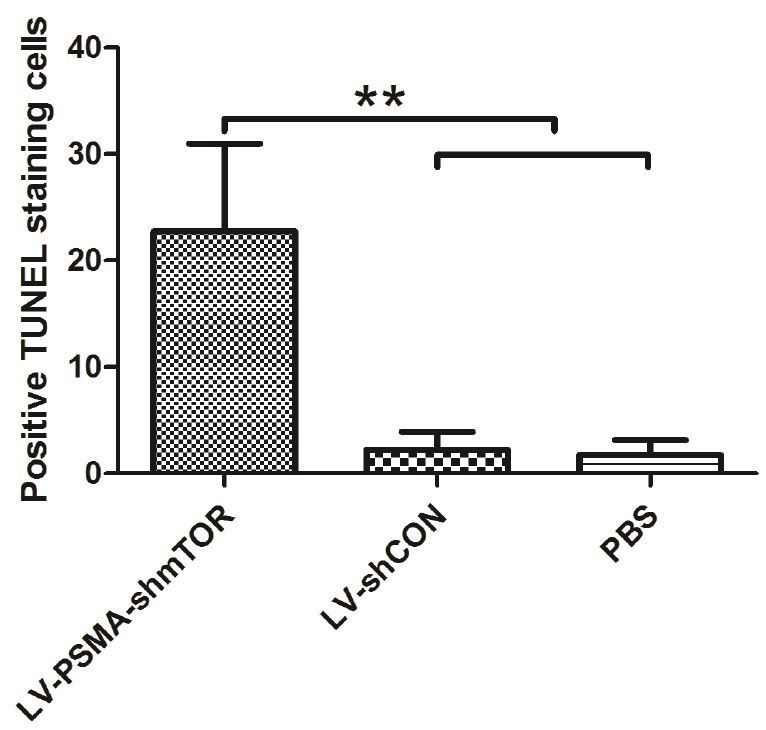
The pathological analysis of xenograft tumors treated by different viruses in vivo. Apoptosis index of C4-2b xenograft tumors in nude mice treated with different viruses. The apoptosis index was represented by the average percentage of TUNEL-positive cells from 10 randomly selected areas. Each bar represents the mean±SD. **P<0.01.
Discussion
Early gene therapy clinical trials for cancer involved lentivirus, adenovirus, AAV and so on, due to safety concerns of nonspecific viral replication in immunocompromised patients; however, these studies were limited by poor viral transduction efficiencies and short express period. To overcome this, lentiviral vectors were developed to kill cancer cells directly and further propagate the vector.
RNAi is the process by which double-stranded RNA induces potent and specific inhibition of eukaryotic gene expression through the degradation of complementary messenger RNA, and is functionally similar to the processes of post-transcriptional gene silencing [10,11]. In the past few years, RNAi has been widely used by researchers to silence the expression of many target genes because of their high specificity and apparent non-toxicity [12,13]. Furthermore, systems based on lentiviral vectors have provided new solutions to achieving stable shRNA-mediated knockdown [14]. In this research, we chose a lentivirus vector as our shRNA delivery vehicle because they can infect both dividing and nondividing cells at a high efficiency and sustain long-term gene expression by integrating into the host genome. shRNA was proved to provide long-lasting silencing and maximal inhibition of gene expression at low concentration [15].
Mammalian target of rapamycin (mTOR) plays a critical role in cell cycle regulation. Rapamycin, a known inhibitor for mTOR [16], can inactivate mTOR specifically. Because mTOR regulates cell proliferation, it has been extensively investigated as a potent target for both anti-cancer [17] and anti-restenotic [18] therapies. Inhibition of mTOR in fibroblasts influences not only proliferation but also collagen production. Rapamycin and its analogues are reported to effectively prevent cardiac and pulmonary fibrosis in vivo [19,20].
In the present study, we observed a strong antitumor effect of mTOR shRNA driven by PSMA promoter on C4-2b prostate cancer cells in vitro and in vivo. Tumor growth was suppressed and tumor apoptosis was increased in nude mice when mTOR mRNA and protein were silenced by PSMA-shmTOR. Therefore, RNAi-directed targeting of mTOR can be used as a potent and specific therapeutic tool for the treatment of prostate cancer.
Conclusion
mTOR plays a critical role in prostate cancer cell proliferation, invasion, and apoptosis. Downregulation of mTOR using the prostate cancer specific promoter PSMA successfully reduces prostate cancer C4-2b cell progression in vitro and in vivo. In conclusion, this study lays the foundation for treatment of prostate cancer through manipulation of mTOR expression.
Acknowledgements
Funding: This work was supported by the following: National Science Foundation of China (grant number: 30901500/H1619; URL: http://www.nsfc.gov.cn); Science and Technology Program of Shaan-Xi Province (grant number: 2009JQ4002; URL: http://www.sninfo.gov.cn); The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
Conflict of interest statement
The authors have declared that no competing interests exist.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Carter HB, Ferrucci L, Kettermann A, Landis P, Wright EJ, Epstein JI, Trock BJ, Metter EJ. Detection of life-threatening prostate cancer with prostate-specific antigen velocity during a window of curability. J Natl Cancer Inst. 2006;98:1521–1527. doi: 10.1093/jnci/djj410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer. 2002;2:584–593. doi: 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- 4.Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, Golub TR, Loda M, Lane HA, Sellers WR. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 5.Heston WD. Characterization and glutamyl preferring carboxypeptidase function of prostate specific membrane antigen: a novel folate hydrolase. Urology. 1997;49:104–112. doi: 10.1016/s0090-4295(97)00177-5. [DOI] [PubMed] [Google Scholar]
- 6.Lundwall A, Malm J, Clauss A, Valtonen-Andre C, Olsson AY. Molecular cloning of complementary DNA encoding mouse seminal vesicle-secreted protein SVS I and demonstration of homology with copper amine oxidases. Biol Reprod. 2003;69:1923–1930. doi: 10.1095/biolreprod.103.019984. [DOI] [PubMed] [Google Scholar]
- 7.Pinto JT, Suffoletto BP, Berzin TM, Qiao CH, Lin S, Tong WP, May F, Mukherjee B, Heston WD. Prostate-specific membrane antigen: a novel folate hydrolase in human prostatic carcinoma cells. Clin Cancer Res. 1996;2:1445–1451. [PubMed] [Google Scholar]
- 8.Murphy GP, Elgamal AA, Su SL, Bostwick DG, Holmes EH. Current evaluation of the tissue localization and diagnostic utility of prostate specific membrane antigen. Cancer. 1998;83:2259–2269. [PubMed] [Google Scholar]
- 9.Conway RE, Petrovic N, Li Z, Heston W, Wu D, Shapiro LH. Prostate-specific membrane antigen regulates angiogenesis by modulating integrin signal transduction. Mol Cell Biol. 2006;26:5310–5324. doi: 10.1128/MCB.00084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, Siomi MC, Carninci P, Gilmour DS, Corona DF, Orlando V. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen T, Heller E, Beronja S, Oshimori N, Stokes N, Fuchs E. An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration. Nature. 2012;485:104–108. doi: 10.1038/nature10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Lai YK, Zhang J, Wang H, Lin MC, He ML, Kung HF. Targeting S100P inhibits colon cancer growth and metastasis by Lentivirus-mediated RNA interference and proteomic analysis. Mol Med. 2011;17:709–716. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrocca F, Lieberman J. Promise and challenge of RNA interference-based therapy for cancer. J. Clin. Oncol. 2011;29:747–754. doi: 10.1200/JCO.2009.27.6287. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Rossi JJ. Lentiviral vector delivery of siRNA and shRNA encoding genes into cultured and primary hematopoietic cells. Methods Mol Biol. 2008;433:287–299. doi: 10.1007/978-1-59745-237-3_18. [DOI] [PubMed] [Google Scholar]
- 15.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 16.Dumont FJ, Su Q. Mechanism of action of the immunosuppressant rapamycin. Life Sci. 1996;58:373–395. doi: 10.1016/0024-3205(95)02233-3. [DOI] [PubMed] [Google Scholar]
- 17.Law BK. Rapamycin: an anti-cancer immunosuppressant? Crit Rev Oncol Hematol. 2005;56:47–60. doi: 10.1016/j.critrevonc.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Windecker S, Roffi M, Meier B. Sirolimus eluting stent: a new era in interventional cardiology? Curr Pharm Des. 2003;9:1077–1094. doi: 10.2174/1381612033455107. [DOI] [PubMed] [Google Scholar]
- 19.Gao XM, Wong G, Wang B, Kiriazis H, Moore XL, Su YD, Dart A, Du XJ. Inhibition of mTOR reduces chronic pressure-overload cardiac hypertrophy and fibrosis. J Hypertens. 2006;24:1663–1670. doi: 10.1097/01.hjh.0000239304.01496.83. [DOI] [PubMed] [Google Scholar]
- 20.Simler NR, Howell DC, Marshall RP, Goldsack NR, Hasleton PS, Laurent GJ, Chambers RC, Egan JJ. The rapamycin analogue SDZ RAD attenuates bleomycin-induced pulmonary fibrosis in rats. Eur Respir J. 2002;19:1124–1127. doi: 10.1183/09031936.02.00281602. [DOI] [PubMed] [Google Scholar]