Summary: Recruitment of GDP-bound Arf1 from the cytosol to the Golgi apparatus by Arf GAP proteins is essential in maintenance of the Golgi apparatus and protein trafficking through the Golgi apparatus.
Abstract
ADP-ribosylation factor1 (Arf1), a member of the small GTP-binding proteins, plays a pivotal role in protein trafficking to multiple organelles. In its GDP-bound form, Arf1 is recruited from the cytosol to organelle membranes, where it functions in vesicle-mediated protein trafficking. However, the mechanism of Arf1-GDP recruitment remains unknown. Here, we provide evidence that two Glo3p-type Arf GTPase-activating proteins (ArfGAPs), ArfGAP domain8 (AGD8) and AGD9, are involved in the recruitment of Arf1-GDP to the Golgi apparatus in Arabidopsis (Arabidopsis thaliana). RNA interference plants expressing low levels of AGD8 and AGD9 exhibited abnormal Golgi morphology, inhibition of protein trafficking, and arrest of plant growth and development. In RNA interference plants, Arf1 was poorly recruited to the Golgi apparatus. Conversely, high levels of AGD8 and AGD9 induced Arf1 accumulation at the Golgi and suppressed Golgi disruption and inhibition of vacuolar trafficking that was caused by overexpression of AGD7. Based on these results, we propose that the Glo3p-type ArfGAPs AGD8 and AGD9 recruit Arf1-GDP from the cytosol to the Golgi for Arf1-mediated protein trafficking, which is essential for plant development and growth.
ADP-ribosylation factors (Arfs) are small guanine nucleotide-binding proteins that are members of the Ras-like GTPase superfamily and play pivotal roles at various steps in intracellular trafficking pathways (Balch et al., 1992; Lanoix et al., 1999). The mechanism of Arf action is known at the molecular and cellular levels. The cytosolic form of Arf bound to GDP is recruited to the membranes of target organelles, where a guanine nucleotide-exchange factor (GEF) replaces GDP with GTP to produce the active GTP-bound form of Arf (Gillingham and Munro, 2007). Active GTP-bound Arf then interacts with downstream effector proteins, which results in the modulation of various cellular processes. Subsequently, the GTP bound to Arf is hydrolyzed to GDP and Arf-GDP is released from the membranes into the cytosol. For this step, ADP-ribosylation factor GTPase-activating proteins (ArfGAPs) are responsible for the stimulation of the latent intrinsic GTPase activity of Arfs (Cukierman et al., 1995; Goldberg, 1999; Donaldson and Jackson, 2000; Inoue and Randazzo, 2007). Thus, ArfGAPs play a critical role in regulating Arf1 activity and terminate the action of Arfs involved in various biological processes. In addition, the regulation of ArfGAPs themselves is integral to understanding the mechanism of Arf action.
Although ArfGAPs were originally identified as ArfGAPs, they are increasingly implicated in GAP-independent functions. ArfGAP1 plays a role in increasing the affinity of coatomer for cargo, and the Glo3p-type ArfGAPs (Glo3p, ArfGAP2, and ArfGAP3) function in the biogenesis of COATOMER PROTEIN I (COPI) vesicles in animal cells and yeast (Lewis et al., 2004; Lee et al., 2005; Kliouchnikov et al., 2009; Saitoh et al., 2009; Schindler et al., 2009; Spang et al., 2010). A large number of ArfGAPs in both animal and plant cells have been identified and are classified into several subfamilies (Inoue and Randazzo, 2007). All ArfGAPs contain a common GAP domain that is responsible for activation of the Arf GTPase and additional domains that are involved in protein-protein or protein-lipid interactions (Cukierman et al., 1995; Donaldson and Jackson, 2000; Inoue and Randazzo, 2007). Thus, they appear to be regulated by various mechanisms.
In plants, the physiological role of ArfGAP domain (AGD) proteins has been investigated by mutant and overexpression approaches (Jensen et al., 2000; Song et al., 2006; Min et al., 2007; Hwang and Robinson, 2009). The Arabidopsis (Arabidopsis thaliana) genome encodes 15 AGD proteins (Jensen et al., 2000; Song et al., 2006; Min et al., 2007). The physiological roles of some of these AGD isoforms have been demonstrated through their mutant phenotypes: agd3/van3/sfc, nev/agd5, and rpa/agd10 displayed defects in vein formation, floral organ shedding, and the growth of root hairs and pollen tubes, respectively (Koizumi et al., 2005; Sieburth et al., 2006; Song et al., 2006; Min et al., 2007; Yoo et al., 2008; Hwang and Robinson, 2009). In rice (Oryza sativa), OsAGAP is involved in vein patterning and root development (Zhuang et al., 2005). However, the functional mechanisms of these AGDs that underlie the observed developmental processes are largely unknown. A recent study demonstrated that the overexpression of the Golgi-localized AGD7, a close homolog of ArfGAP1 in animal cells (Wesley et al., 2001), caused disassembly of the Golgi apparatus and thereby inhibited vacuolar trafficking (Min et al., 2007).
To expand our understanding of the biological roles of ArfGAPs in plants, we investigated the physiological roles of several AGD isoforms. In particular, we focused on Glo3p-type ArfGAPs in Arabidopsis. Here, we present evidence that two closely related homologs, AGD8 and AGD9, localized to the Golgi apparatus, are required for the maintenance of Golgi morphology. They are involved in the recruitment of Arf1 to the Golgi apparatus in a functionally redundant manner. Thus, AGD8 and AGD9 play essential roles in plant growth and development.
RESULTS
RNA Interference Transgenic Plants Expressing Low Levels of the Glo3p-Type ArfGAPs AGD8, AGD9, and AGD10 Display Defects in Plant Development and Growth
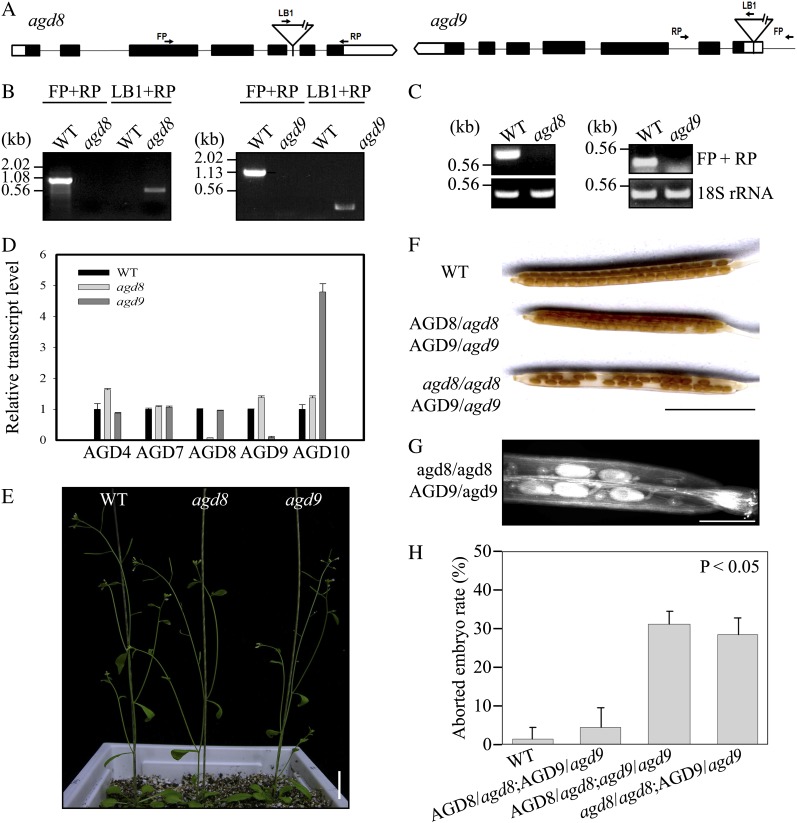
The large number of AGDs in Arabidopsis can be classified into six subfamilies according to their sequence homology (Supplemental Fig. S1). To obtain insight into the physiological roles of AGDs, we focused on one subfamily that contains three AGD isoforms, AGD8, AGD9, and AGD10. They exhibit the closest relationship to the Glo3p-type ArfGAPs that are involved in COPI vesicle biogenesis at the Golgi apparatus (Schindler et al., 2009; Spang et al., 2010). The public microarray data revealed that AGD8 and AGD9 are expressed at high levels in most tissues, whereas AGD10 is strongly expressed in stamens, moderately in root xylem cells, but virtually undetectable in other tissues (Supplemental Fig. S2; www.genevestigator.com). We screened Arabidopsis mutants that had transfer DNA (T-DNA) insertions in AGD8 and AGD9 (Fig. 1, A and B). The lack of AGD8 and AGD9 transcripts in the mutants was confirmed by reverse transcription (RT)-PCR and quantitative reverse transcription (qRT)-PCR (Fig. 1, C and D). The expression of AGD10 was greatly induced in agd9 mutants, suggesting that the lack of AGD9 expression somehow resulted in the induction of AGD10 expression in a compensatory manner. The mutants agd8 and agd9 did not exhibit any noticeable phenotypes (Fig. 1E), which may result from a functional redundancy of AGD8, AGD9, and AGD10, similar to what has been observed for ArfGAP1, ArfGAP2, and ArfGAP3 in animal cells (Saitoh et al., 2009).
Figure 1.
AGD8 and AGD9 display functional redundancy but lethality in double mutation. A, Schematic presentation of T-DNA insertion sites in the AGD8 and AGD9 genes. FP, Forward primer; LB1, left border; RP, reverse primer. B, Screening of agd8 and agd9 mutants. The T-DNA insertions in AGD8 or AGD9 in the putative agd8 and agd9 mutant plants were confirmed via PCR on genomic DNA using gene-specific and left border primers. FP, RP, and LB are forward, reverse, and left border primers, respectively. WT, Wild-type plants. C and D, Lack of AGD8 and AGD9 transcripts in agd8 and agd9 plants. Total RNA from agd8 and agd9 plants and wild-type plants was subjected to RT-PCR (C) or qRT-PCR (D) analyses. As a control for RT-PCR, 18S rRNA was included. As a control for qRT-PCR, AGD4, AGD7, and AGD10 were included. 18S rRNA was used as an internal control for qRT-PCR. E, Phenotype of agd8 or agd9 single mutants. The phenotype of agd8 and agd9 mutant plants was observed 5 weeks after planting in soil. Bar = 2 cm. F to H, Defect in seed production of F2 plants from crosses between adg8 and agd9 single mutants. To obtain the agd8/agd9 double mutant, adg8 (male) and agd9 (female) were crossed. Individual F2 plants that had been confirmed for their genotype by the PCR approach were allowed to self-pollinate, and their siliques were examined for seed production (F). To get better images of aborted seeds, siliques were decolored and images were taken by a scanner (G). To quantify the percentage of abortion, the number of aborted seeds was counted from at least seven siliques (H). Error bars indicate sd. Bars = 5 mm (F) and 1 mm (G).
Accordingly, agd8/agd9 double mutant plants were screened from F2 progeny from the cross of the adg8 (male) and agd9 (female) single homozygote mutants. However, after screening greater than 200 F2 plants, no double homozygotes were obtained. This raised the possibility that the homozygotes of the agd8/agd9 double mutant are lethal. To test this hypothesis, individual F2 plants from the cross of the agd8 and agd9 single mutants were examined for their genotype by the PCR approach and allowed to self-pollinate. Their siliques, representing various genotypes, were examined for seed production. In support of this hypothesis, 31% of the ovules were aborted in the siliques of self-pollinated F2 plants with the genotype AGD8/agd8;agd9/agd9 (Fig. 1, F–H). Similar to that of the heterozygous/homozygous double mutant for AGD8/AGD9, the agd8/agd8;AGD9/agd9 mutant produced 28% aborted seeds in siliques of self-pollinated F2 plants. In contrast, the abortion rate in wild-type plants or heterozygote plants for both loci (AGD8/agd8;AGD9/agd9) was almost negligible. This result indicates that AGD8 and AGD9 are functionally redundant and play essential roles in plant development.
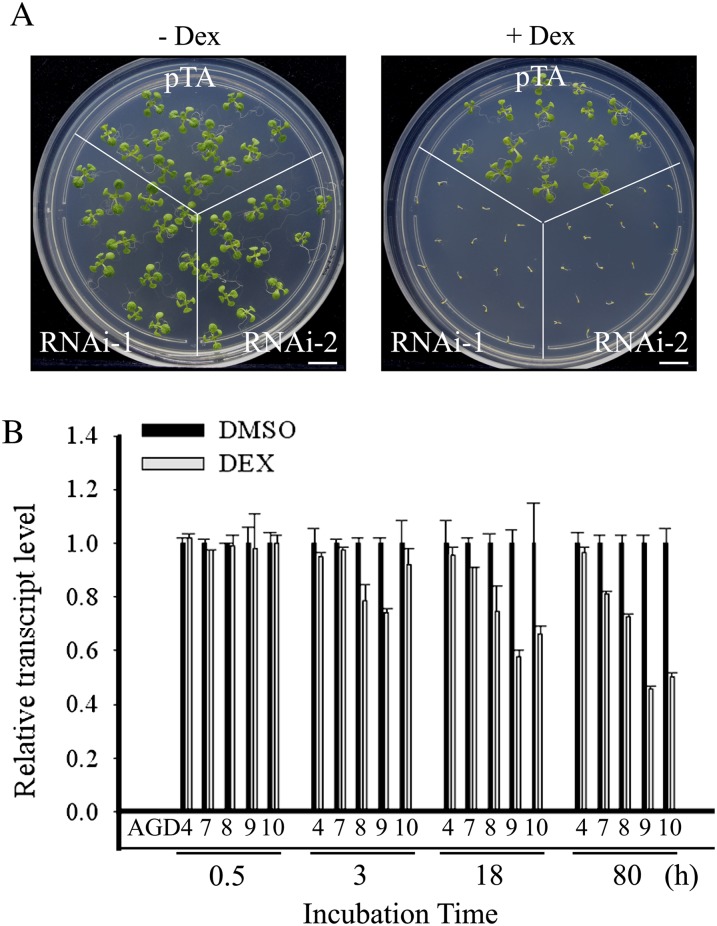
As a result to investigate the physiological role of Glo3p-type ArfGAPs in Arabidopsis, transgenic RNA interference (RNAi) plants were generated that have low expression levels of AGD8 and AGD9, the two abundantly expressed Glo3p-type ArfGAPs (Wesley et al., 2001). To simultaneously knock down both AGD8 and AGD9 transcripts, a 421-bp fragment of AGD9 that contained a high degree of nucleotide sequence homology to AGD8 was used (Supplemental Fig. S3). This fragment was placed under the control of the dexamethasone (dex)-inducible promoter (Aoyama and Chua, 1997). The resulting RNAi construct was cloned into a binary vector and used to generate transgenic plants (RNAi plants). Multiple independent lines were obtained, and their phenotypes were examined in the presence and absence of dex. The RNAi plants did not display any noticeable phenotypes in the absence of dex (Fig. 2A). However, when placed on plates containing 30 μm dex, multiple lines of RNAi plants germinated but failed to develop into mature plants (Fig. 2A). In contrast, wild-type plants germinated and developed normally regardless of dex treatment. qRT-PCR was used to analyze the RNA levels of various AGD isoform genes in the RNAi plants. The transcript levels of AGD9 gradually decreased in dex-treated RNAi plants with increasing time. Similarly, the AGD8 transcript levels were significantly reduced with time, albeit to a slightly lesser degree. The transcript levels of AGD10 also were greatly reduced in dex-treated RNAi plants. In contrast, the transcript levels of AGD4 and AGD7 that belong to two different subfamilies of the AGD family were barely or slightly reduced in the dex-treated RNAi plants, respectively, suggesting that the AGD9 RNAi construct is specific to the members of the Glo3p-type ArfGAP subfamily among AGD isoforms (Fig. 2B). These results are consistent with the inability to generate a double homozygous agd8/agd9 mutant. In comparison with other adg mutants (Koizumi et al., 2005; Sieburth et al., 2006; Song et al., 2006; Yoo et al., 2008), the RNAi plants displayed a much more severe phenotype, raising the possibility that AGD8/AGD9 are involved in more general physiological processes that are necessary for plant growth and development.
Figure 2.
RNAi transgenic plants with low transcript levels of AGD8, AGD9, and AGD10 display severe growth defects. A, Phenotype of RNAi plants. Two independent lines of RNAi plants (RNAi-1 and RNAi-2) and control plants (pTA7002) were planted on MS plates with or without dex (30 μm). The phenotype was observed 10 d after planting. Bars = 1 cm. B, qRT-PCR analysis of transcript levels in RNAi plants. Total RNA was prepared from leaf tissues of RNAi-1 plants that had been treated with dex for the indicated period of time and used for qRT-PCR analysis of transcript levels of five AGD isoform genes, AGD4, AGD7, AGD8, AGD9, and AGD10, using gene-specific primers. 18S rRNA was used as an internal control.
AGD8 and AGD9 Levels Are Crucial for the Maintenance of Golgi Morphology and Protein Trafficking through the Golgi Apparatus
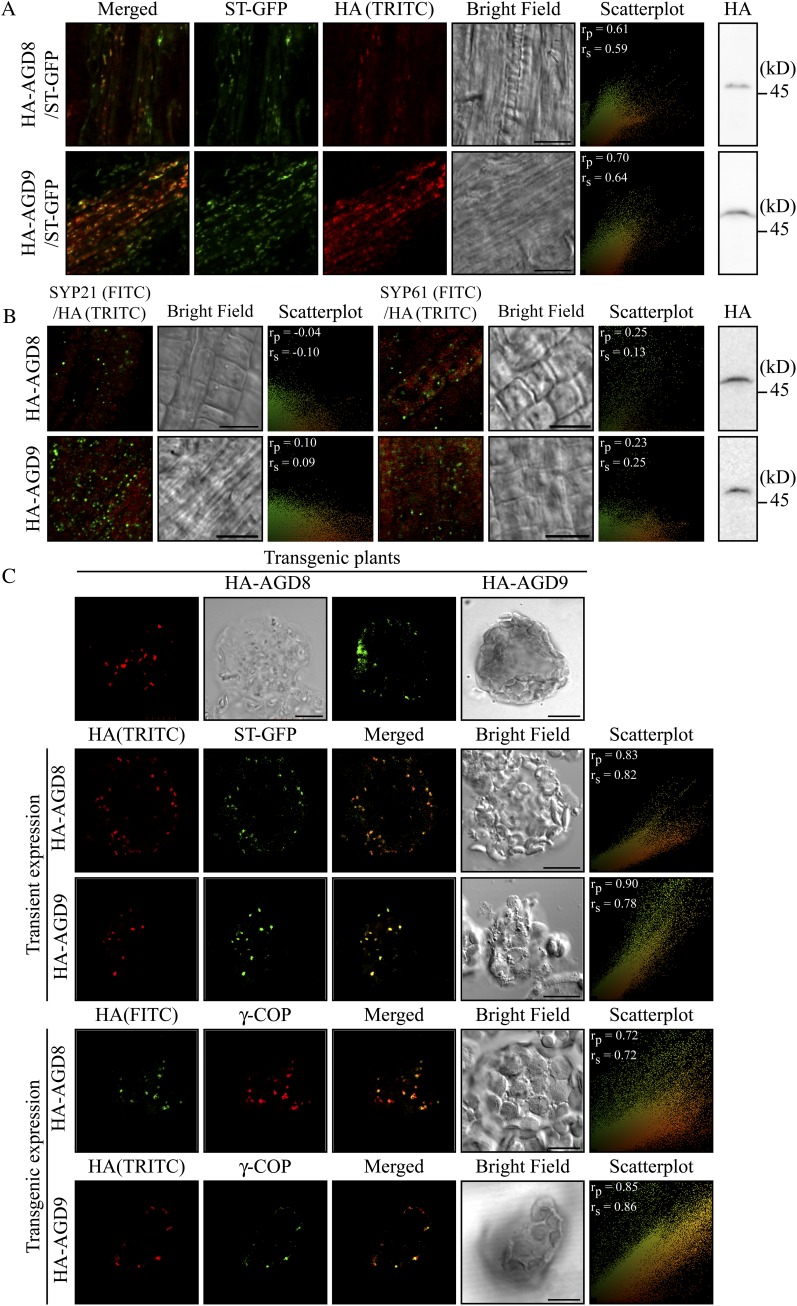
To gain an insight into the physiological role of these proteins, we initially determined the subcellular localization of AGD8/AGD9. Previous studies in Arabidopsis show that two other isoforms (AGD5 and AGD7) localize to the trans-Golgi network (TGN) and Golgi apparatus, respectively (Min et al., 2007; Stefano et al., 2010). AGD8 and AGD9 were fused to the human influenza hemagglutinin (HA) epitope (YPYDVPDYA). HA-AGD8 and HA-AGD9 under the control of their native promoters were transformed into wild-type plants or transgenic plants expressing ST-GFP, a chimeric fusion construct between rat sialyltransferase (ST) and GFP that localizes to the Golgi apparatus (Kim et al., 2001a). In root tissues of transgenic plants, both HA-AGD8 and HA-AGD9 produced punctate staining patterns that closely overlapped with those of stably expressed ST-GFP when detected by using an HA antibody (Fig. 3A). To quantify the degree of overlap between HA-AGD8/AGD9 and ST-GFP, the images from a microscope were subject to Pearson-Spearman correlation (PSC) colocalization analysis (French et al., 2008), and scatterplots of fluorescence pixels across the two channels were obtained. The scatterplots together with the PSC coefficient values of 0.61/0.59 and 0.70/0.64 for colocalization between ST-GFP and HA-AGD8 or AGD9, respectively, confirmed that HA-AGD8 and HA-AGD9 colocalize with ST-GFP (Fig. 3A, scatterplots). The PSC coefficient values range from −1 to +1 for strong negative and positive correlation, respectively. Next, we compared the localization of HA-AGD8 and HA-AGD9 with the prevacuolar compartment (PVC)-localized SYP21 or TGN-localized SYP61 detected with SYP21 and SYP61 antibodies (Sanderfoot et al., 1999, 2001). Neither HA-AGD8 nor HA-AGD9 colocalized with SYP21 or SYP61 (Fig. 3B), consistent with the notion that they do localize to the Golgi apparatus. Again, the degree of overlap was tested by the PSC colocalization analysis. The scatterplots for the colocalization of HA-AGD8 or HA-AGD9 with PVC-localized SYP21 or TGN-localized SYP61 show a separated distribution pattern of two fluorescence pixels and gave the PSC coefficient value close to 0 (Fig. 3B, scatterplots), indicating that AGD8 and AGD9 do not localize to the TGN or PVC. Expression of HA-AGD8 and HA-AGD9 in these transgenic plants was confirmed by western-blot analysis using the HA antibody (Fig. 3B).
Figure 3.
AGD8 and AGD9 localize to the Golgi apparatus. A and B, Localization of AGD8 and AGD9 in transgenic plants. Native promoter-driven HA-AGD8 and HA-AGD9 constructs were transformed into transgenic plants expressing ST-GFP (A) or wild-type plants (B), and the localization of these proteins was determined by immunostaining using HA antibody followed by FITC-labeled anti-rat IgG (A) or SYP21 or SYP61 antibody followed by TRITC-labeled anti-rabbit IgG (B). ST-GFP was observed directly. Expression of HA-AGD8 and HA-AGD9 was detected by western-blot analysis using an HA antibody. To quantify the degree of overlap between two proteins, PSC colocalization analysis was performed using at least seven individual plants that contain a minimum of 400 punctate stains. The values of fluorescence pixels across the two channels are depicted in an intensity scatterplot. rp, Linear Pearson correlation coefficient; rs, nonlinear Spearman’s rank correlation coefficient. Bars = 10 μm. C, Localization of AGD8/AGD9 in protoplasts. Protoplasts from transgenic plants expressing HA-AGD8 or HA-AGD9 were immunostained with a γ-COP antibody. In addition, HA-AGD8 or HA-AGD9 was transformed into protoplasts from wild-type plants together with ST-GFP, and the localization of HA-AGD8, HA-AGD9, and ST-GFP was examined by immunostaining using HA antibody. The scatterplots were obtained using at least 10 individual protoplasts that contain a minimum of 200 punctate stains. Bars = 10 μm.
To further confirm their localization at the single cell level, protoplasts from transgenic plants expressing HA-AGD8 or HA-AGD9 were immunostained with an HA antibody. Both proteins produced punctate staining patterns in protoplasts, as observed in intact tissues (Fig. 3C). Next, we compared their localization with other Golgi proteins in protoplasts. As Golgi proteins, we used transiently expressed ST-GFP (Kim et al., 2001b), KAM1ΔC-mRFP (a chimeric protein consisting of KATAMARI1 with a C-terminal deletion and monomeric red fluorescent protein; Tamura et al., 2005), and endogenous γ-COP, a component of COPI, detected by γ-COP antibody (Pimpl et al., 2000). In protoplasts, both HA-AGD8 and HA-AGD9 that had been expressed in transgenic plants closely overlapped with transiently expressed ST-GFP (Fig. 3C). Furthermore, HA-AGD9 colocalized with endogenous γ-COP (Fig. 3C). The colocalization of HA-AGD8/AGD9 with Golgi markers was confirmed by the PSC colocalization analysis. Together, these results confirmed that both HA-AGD8 and HA-AGD9 localize to the Golgi apparatus. Next, we examined the localization of transiently expressed HA-AGD8 and HA-AGD9 in protoplasts, since we wanted to examine their behavior after transient expression in protoplasts (Figs. 4 and 5). It has been thought that the transient expression of proteins in protoplasts could lead to mislocalization due to possible overexpression (Denecke et al., 2012). First, HA-AGD8 or HA-AGD9 was transformed into wild-type protoplasts together with ST-GFP, and their localization was examined. Both transiently expressed HA-AGD8 and HA-AGD9 produced punctate staining patterns that closely overlapped with those of ST-GFP (Fig. 3C). Moreover, transiently expressed HA-AGD8 closely overlapped with endogenous γ-COP at the Golgi apparatus (Supplemental Fig. S4). Together, these results confirmed that transiently expressed HA-AGD8 and HA-AGD9 also localize to the Golgi apparatus. Their localization to the Golgi is similar to that of ArfGAP2/ArfGAP3 in animal cells (Frigerio et al., 2007).
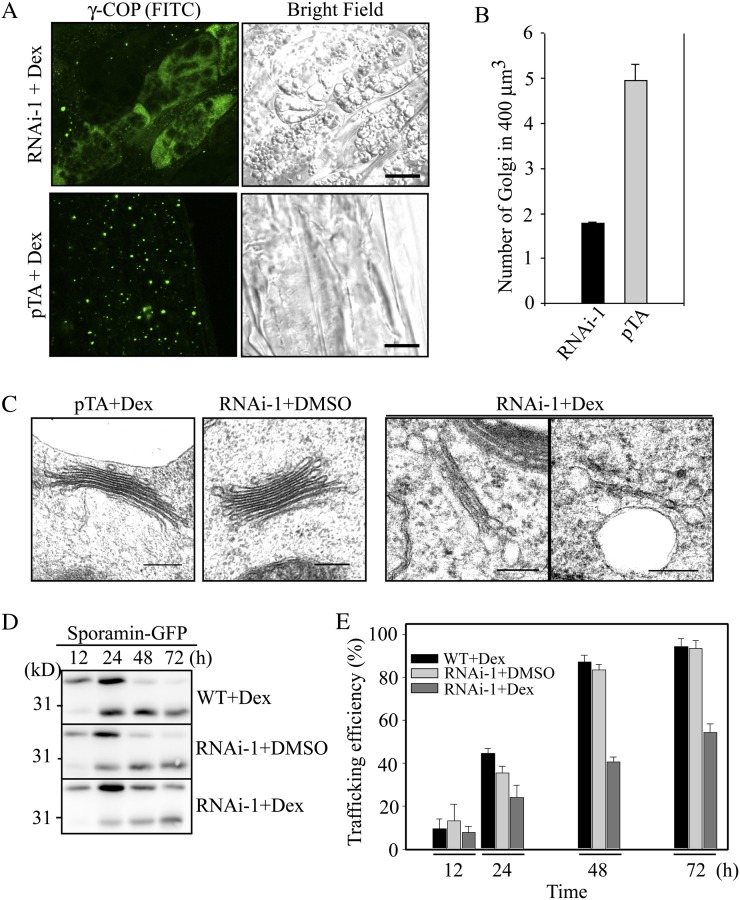
Figure 4.
RNAi plants display severe defects in Golgi morphology and protein trafficking to the vacuole. A, Effect of RNAi on the Golgi apparatus in RNAi plants. RNAi and pTA control plants were grown on MS plates for 1 week and then transferred onto MS plates supplemented with dex (30 μm). Two days after transplanting, the plant root tissues were fixed and immunostained with γ-COP antibody (γ-COP) followed by FITC-labeled secondary anti-rabbit IgG. Bars = 10 μm. B, Quantification of the Golgi apparatus in RNAi plants. To quantify the differences in the Golgi population, serial optical z sections (10 images) obtained at 0.5-μm intervals were used for three-dimensional reconstruction using laser scanning confocal microscopy z-projection software, and the number of punctae in 400 μm3 of 50 cells was counted. Error bars indicate sd (n = 3). C, Morphological alteration of the Golgi apparatus in RNAi plants. RNAi and pTA plants treated with or without dex (30 μm) for 4 d were fixed, and ultrathin sections of root tissues were examined by electron microscopy. Bars = 200 nm. D and E, Inhibition of vacuolar trafficking of sporamin-GFP in RNAi plants. Protoplasts from RNAi that had been treated with or without dex were transformed with sporamin-GFP. D Vacuolar trafficking of sporamin-GFP was examined at various time points by western-blot analysis using a GFP antibody. As a control, protoplasts from the wild type (WT) were included. E, Trafficking efficiency was quantified using the ratio of the processed form over the total amount of expressed protein. Error bars indicate sd (n = 3).
Figure 5.
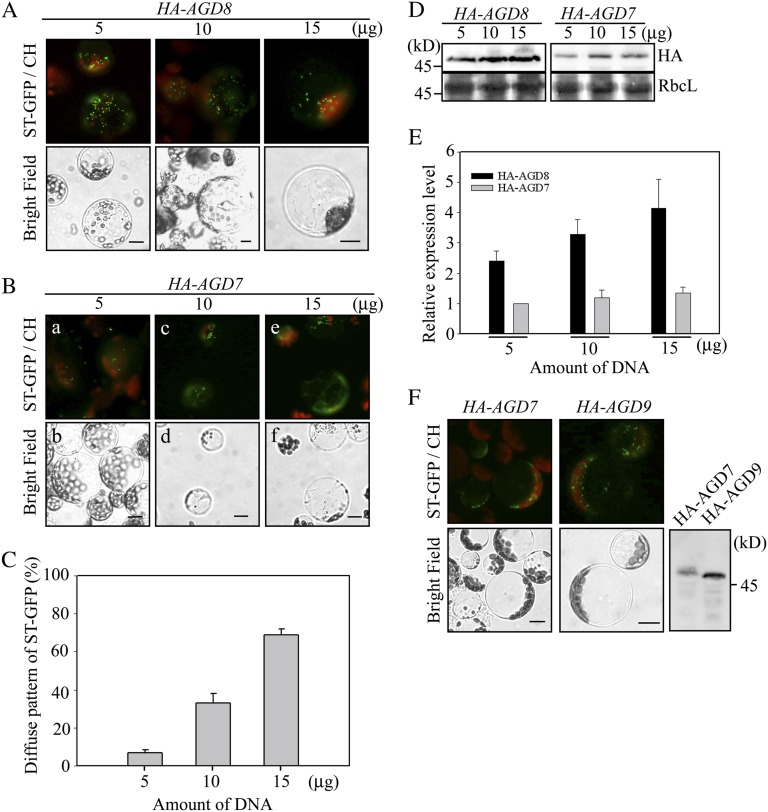
Overexpression of AGD8 and AGD9 does not cause Golgi disassembly. A, Lack of Golgi apparatus disruption by the overexpression of HA-AGD8. ST-GFP was cotransformed into protoplasts with increasing amounts of HA-AGD8, and the localization pattern of ST-GFP was examined. CH, Red autofluorescence of chlorophyll. Bars = 10 μm. B and C, Disruption of the Golgi apparatus by AGD7. B, ST-GFP was introduced into protoplasts together with increasing amounts of HA-AGD7, and the localization of ST-GFP was examined. C, To quantify the degree of Golgi disruption, the number of protoplasts showing the diffuse ST-GFP pattern was counted and expressed as a percentage of the total number of protoplasts counted. When counting the pattern, individual protoplasts were determined whether they had a diffuse pattern or not. As long as there was a diffuse cytosolic signal even in the presence of punctate stains, we counted them as a diffuse pattern. Representative images of punctate staining and diffuse pattern are shown in panels a and e of B, respectively. Three independent transformation experiments were performed, and 40 cells were analyzed each time. Error bars indicate sd (n = 3). D, Expression of AGD7/AGD8. Protein extracts prepared from transformed protoplasts were analyzed by western blotting using an HA antibody. As a loading control, Rubisco complex large subunit (RbcL) was stained with Coomassie blue. E, Quantification of AGD7/AGD8 expression levels in transformed protoplasts. Protein extracts prepared from transformed protoplasts were analyzed by western blotting using an HA antibody. The intensity of the AGD proteins was quantified using software on the LAS3000 and expressed relative to the intensity level of 5 μg of HA-AGD7. F, Lack of Golgi disruption by overexpression of AGD9. ST-GFP was cotransformed into protoplasts with HA-AGD9 or HA-AGD7, and the localization pattern of ST-GFP was examined. Expression of HA-AGD7 and HA-AGD9 was examined by western-blot analysis using an HA antibody. Bars = 10 μm.
The successful localization of AGD8/AGD9 to the Golgi apparatus encouraged us to examine whether lower levels of AGD8/AGD9 in RNAi plants had any effect on the Golgi apparatus. RNAi plants and control plants (pTA plants) harboring the empty vector pTA7002 were grown on Murashige and Skoog (MS) medium and then transplanted onto MS plates supplemented with dex (Aoyama and Chua, 1997). Two days after transplanting, the root tissues were immunostained with γ-COP antibody. In pTA plants, γ-COP antibody produced numerous punctae (Fig. 4A). In contrast, γ-COP antibody in the RNAi plants displayed a diffuse staining pattern and fewer punctae (Fig. 4A). The number of punctae in the RNAi plants was reduced to 30% of the level in pTA plants (Fig. 4B), raising an intriguing possibility that the Golgi apparatus is disassembled in RNAi plants. This is in contrast to previous studies on other ArfGAPs, where overexpression of ArfGAP1 and AGD7 in animal and Arabidopsis cells, respectively, caused disassembly of the Golgi apparatus (Aoe et al., 1997; Min et al., 2007). To confirm this observation, ultrathin sections of root tissue were cut from RNAi and pTA plants that had been treated with or without dex for 4 d and examined for the presence and morphology of the Golgi apparatus by electron microscopy. In control plants (Fig. 4C, pTA+Dex and RNAi-1+DMSO [for dimethyl sulfoxide]), the Golgi apparatus exhibited a normal morphology with five to six stacks. However, in dex-treated RNAi plants, the Golgi apparatus contained only one or two stacks (Fig. 4C, RNAi-1+Dex), indicating that the Golgi apparatus is partially disassembled in dex-treated RNAi plants. These results suggest that normal AGD8 and AGD9 levels are crucial for the maintenance of the Golgi morphology. A previous study in animal cells demonstrated that knockdown of both ArfGAP2 and ArfGAP3 in animal cells caused similar alterations in Golgi morphology (Kartberg et al., 2010). Thus, it is possible that AGD8 and AGD9 are functional orthologs of the Glo3p-type ArfGAPs in animal cells, ArfGAP2/ArfGAP3.
To functionally confirm the Golgi defect in the RNAi plants, protein trafficking in the RNAi plants was examined. If AGD8/AGD9 are Arabidopsis orthologs of Glo3p-type ArfGAPs, they are likely to be involved in a COPI-related process (i.e. in retrograde Golgi-to-endoplasmic reticulum trafficking), as observed for Glo3p ArfGAPs (Frigerio et al., 2007; Kliouchnikov et al., 2009; Kartberg et al., 2010). However, it is technically very difficult to monitor retrograde trafficking in plant cells. Instead, the functionality of the Golgi apparatus was tested by vacuolar trafficking because vacuolar proteins are transported through the Golgi apparatus on their way from the endoplasmic reticulum to the vacuole. For vacuolar cargo, we used sporamin-GFP, a fusion protein between the sweet potato (Ipomoea batatas) protein sporamin and GFP that is targeted to the vacuole and processed to produce the GFP fragment (Sohn et al., 2003). Protoplasts from the RNAi plants were transformed with sporamin-GFP and incubated with or without dex. Sporamin-GFP trafficking to the vacuole was examined by western-blot analysis using a GFP antibody (Jin et al., 2001; Kim et al., 2001a). Wild-type protoplasts were included in the analysis. In dex-treated RNAi protoplasts, the efficiency of protein trafficking was 40%, compared with 85% in dex-untreated RNAi or wild-type protoplasts at 48 h after transformation (Fig. 4, D and E), supporting the notion that the dex-treated RNAi plants contain a functionally defective Golgi apparatus.
To gain an insight into the underlying mechanism of how the lower levels of AGD8/AGD9 cause a defect in Golgi morphology and a perturbation of vacuolar trafficking, in a reverse approach to the RNAi plants, we examined the effect of AGD8/AGD9 overexpression on the Golgi apparatus. Previous studies demonstrated that overexpression of ArfGAPs such as ArfGAP1 and AGD7 caused disassembly of the Golgi apparatus (Aoe et al., 1997; Min et al., 2007). For this experiment, we took advantage of the protoplast system in which protein levels can be easily increased by increasing the amount of plasmid DNA introduced into protoplasts (Lee et al., 2002; Denecke et al., 2012). ST-GFP was cotransformed into protoplasts together with increasing amounts of HA-AGD8, and the localization of ST-GFP was examined in protoplasts. Regardless of the expression levels, HA-AGD8 overexpression did not alter the punctate staining pattern of ST-GFP (Fig. 5A). In contrast, as reported previously (Min et al., 2007), ST-GFP produced a diffuse pattern with a gradual decrease in the number of punctae with increasing amounts of HA-AGD7 (Fig. 5, B and C). The expression levels of HA-AGD8 and HA-AGD7 were determined by western-blot analysis using an HA antibody (Fig. 5, D and E). In fact, the protein levels of HA-AGD8 were 2- to 3-fold higher than those of HA-AGD7 at the same amounts of plasmid DNA in protoplasts. Similar to HA-AGD8, HA-AGD9 did not affect the localization pattern of ST-GFP (Fig. 5F). This result raises the possibility that the physiological role of AGD8/AGD9 is different from that of AGD7.
AGD8 Binds to Both the Wild Type and the GDP-Bound Form of Arf1 at the Golgi Apparatus
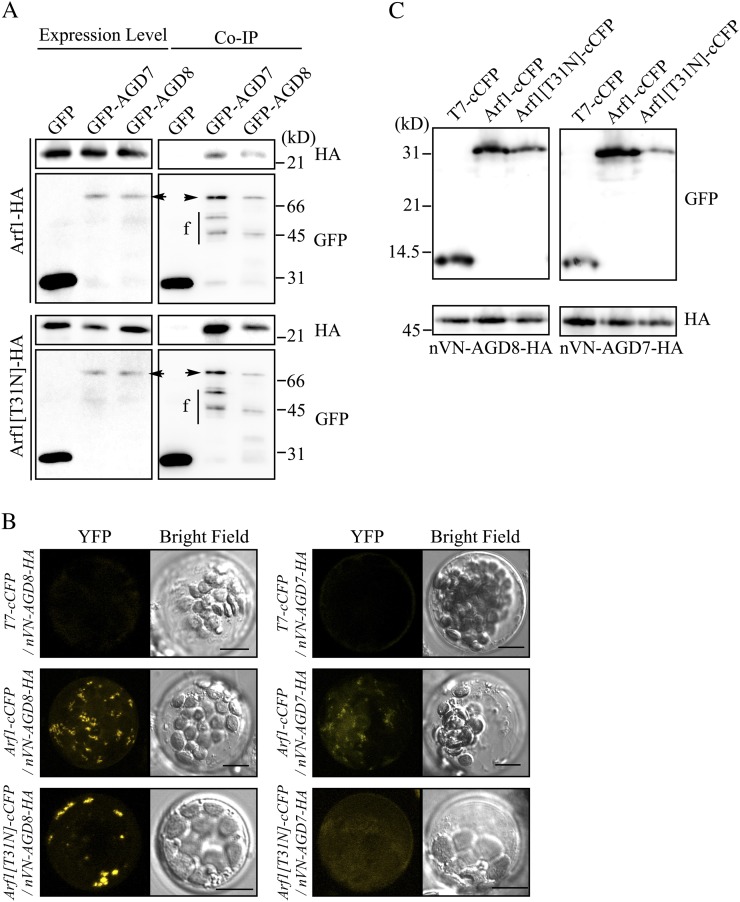
Arf1 plays a crucial role in the maintenance of the Golgi apparatus (Lee et al., 2002; Takeuchi et al., 2002). To gain insight into the molecular mechanisms of AGD8/AGD9 that underlie their roles in the maintenance of the Golgi apparatus at the biochemical level, we examined whether these proteins interact with Arf1 by coimmunoprecipitation experiments. Arf1-HA or Arf1[T31N]-HA was cotransformed into protoplasts together with GFP-AGD7 or GFP-AGD8, and protein extracts from transformed protoplasts were used for immunoprecipitation with a GFP antibody. The immunoprecipitates were analyzed by western blotting using HA and GFP antibodies. GFP-AGD7 was included as a positive control. As reported previously (Min et al., 2007), Arf1-HA was detected in the immunoprecipitates obtained with GFP-AGD7 (Fig. 6A). Arf1-HA was detected in the immunoprecipitates obtained with GFP-AGD8, indicating that both AGD7 and AGD8 interact with Arf1. HA-Arf1[T31N] coprecipitated with both GFP-AGD7 and GFP-AGD8, indicating that both AGD7 and AGD8 interact with Arf1[T31N].
Figure 6.
AGD8 interacts with Arf1 at the Golgi apparatus. A, The interaction between AGD8 and Arf1 was determined by coimmunoprecipitation. Protoplasts were transformed with the indicated constructs, and protein extracts from the transformed protoplasts were subjected to immunoprecipitation using a GFP antibody. The immunoprecipitates (Co-IP) and 5% of total protein extracts (expression control) were analyzed by western blotting using HA and GFP antibodies. Arrowheads indicate full-length proteins of GFP-AGD7 and GFP-AGD8. f, Degradation products of GFP-tagged AGD7 and AGD8; GFP, GFP antibody; HA, HA antibody. B and C, Interaction between AGD8 and Arf1 determined by BiFC. B, Protoplasts were transformed with the indicated constructs, and yellow fluorescent protein (YFP) signals were observed from the live protoplasts 18 h after transformation. The images are projections generated by using 10 serial z section images obtained at 1-μm intervals. Bars = 10 μm. C, To confirm the expression of transformed constructs, protein extracts from transformed protoplasts were analyzed by western blotting using HA and GFP antibodies.
To further confirm and characterize this interaction at the cellular level in vivo, we examined the interaction of AGD7 and AGD8 with Arf1 via a bimolecular fluorescence complementation (BiFC) approach (Kim et al., 2007; Lee et al., 2008). To reduce the nonspecific background signals in the BiFC experiments, the C-terminal half of cyan fluorescent protein (cCFP) and the N-terminal half of the GFP variant Venus (nVN) were used (Lee et al., 2008). AGD8-HA was fused to the C terminus of nVN. AGD7-HA was included as a control. Arf1 and Arf1[T31N] were fused to the N terminus of cCFP. Arf1[T31N], the GDP-bound form of Arf1 (Lee et al., 2002), was used as a control for the interaction between Arf1 and AGD8. As a negative control, cCFP was tagged with a small epitope T7 at the N terminus. Again, we relied on the transient expression of these proteins in protoplasts because transiently expressed HA-AGD8 and HA-AGD9 properly localized to the Golgi apparatus in protoplasts. nVN-AGD8-HA or nVN-AGD7-HA was introduced into protoplasts together with Arf1-cCFP, Arf1[T31N]-cCFP, or T7-cCFP. The amount of nVN-AGD7-HA and nVN-AGD8-HA introduced into protoplasts was carefully controlled because AGD7 overexpression induces disassembly of the Golgi apparatus (Min et al., 2007). The control protoplasts (T7-cCFP + nVN-AGD8-HA or T7-cCFP + nVN-AGD7-HA) did not display any signals (Fig. 6B), confirming that nonspecific interactions between cCFP and nVN were undetectable. When Arf1-cCFP was introduced into protoplasts together with nVN-AGD7-HA or nVN-AGD8-HA, strong BiFC signals were observed as punctate staining patterns in the protoplasts together with weak cytosolic signals (Fig. 6B). Next, Arf1[T31N]-cCFP, the GDP-bound form, was cotransformed into protoplasts together with nVN-AGD7-HA or nVN-AGD8-HA, and BiFC signals were examined. The protoplasts cotransformed with Arf1[T31N]-cCFP and nVN-AGD7-HA produced diffuse cytosolic patterns but did not show any punctate BiFC signals, indicating that Arf1[T31N] and AGD7 may interact in the cytosol but not at the Golgi apparatus. Unexpectedly, protoplasts cotransformed with Arf1[T31N]-cCFP and nVN-AGD8-HA produced strong punctate staining patterns, raising the possibility that AGD8 binds to GDP-bound Arf1 at the Golgi apparatus. However, this interaction between AGD8 and Arf1[T31N] at the Golgi apparatus is not consistent with current ideas about the role of ArfGAPs, which can be demonstrated by the fact that ectopic expression of Arf1[T31N] causes disassembly of the Golgi apparatus (Cukierman et al., 1995; Lee et al., 2002; Inoue and Randazzo, 2007). To confirm the expression of these constructs in protoplasts, protein extracts from transformed protoplasts were analyzed by western blotting using HA and GFP antibodies. As expected, all these proteins were expressed at comparable levels (Fig. 6C).
AGD8 and AGD9 Enhance the Recruitment of Arf1-GFP to the Golgi Apparatus
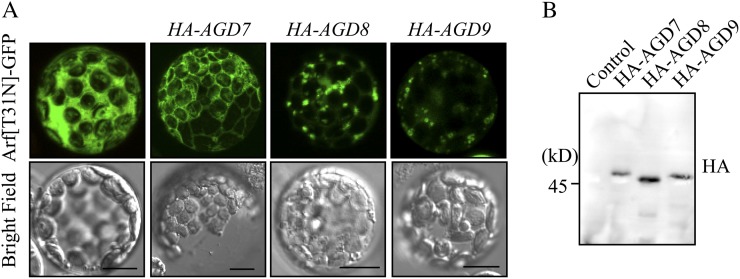
During cycling between the GDP- and GTP-bound forms, Arf1-GDP transiently localizes to the Golgi membrane, where the bound GDP is replaced with GTP by a GEF (Donaldson and Jackson, 2000; D’Souza-Schorey and Chavrier, 2006; Gillingham and Munro, 2007). Thus, it is possible that AGD8/AGD9 are involved in the recruitment of GDP-bound Arf1 to the Golgi apparatus. To test for this possibility, the GDP-bound form of Arf1, Arf1[T31N], was fused to superfolder GFP (sGFP), and the resulting construct, Arf1[T31N]-sGFP, was introduced into protoplasts alone or together with HA-AGD7, HA-AGD8, or HA-AGD9, and then the localization of Arf1[T31N]-sGFP was examined. A previous study demonstrated that Arf1[T31N], a dominant negative mutant form of Arf1, caused disassembly of the Golgi apparatus in plant cells (Lee et al., 2002; Takeuchi et al., 2002). Moreover, protoplasts expressing Arf1[T31N]-sGFP alone produced a diffuse pattern (Fig. 7A). Coexpression of HA-AGD7 did not change the diffuse pattern of Arf1[T31N]-sGFP in protoplasts. However, in the presence of HA-AGD8 or HA-AGD9, Arf1[T31N]-sGFP primarily produced the punctate staining patterns and weak cytosolic patterns, indicating that the majority of Arf1[T31N]-sGFP localizes to the Golgi apparatus. This confirms that high levels of AGD8/AGD9 cause the accumulation of Arf1[T31N] at the Golgi apparatus. Expression of HA-AGD7, HA-AGD8, and HA-AGD9 was confirmed by western-blot analysis using an HA antibody (Fig. 7B). The localization of Arf1[T31N] to the Golgi apparatus contradicted the previous results showing that overexpression of Arf1[T31N] causes disassembly of the Golgi apparatus in plant cells (Lee et al., 2002; Takeuchi et al., 2002). However, it should be noted that wild-type protoplasts should contain endogenous Arf1, and when Arf1[T31N] is ectopically expressed in protoplasts, the effect observed should be the result of competition between endogenous Arf1 and ectopically expressed Arf1[T31N]. Therefore, the disassembly of the Golgi apparatus should be highly dependent on the concentration of Arf1[T31N]. One possible explanation for the presence of the Golgi apparatus in protoplasts expressing Arf1[T31N]-sGFP is that the effective concentration of Arf1[T31N]-sGFP in the protoplasts coexpressing AGD8 or AGD9 should be lower that of Arf1[T31N]-sGFP in the absence of AGD8/AGD9, because these AGDs bind to Arf1[T31N] (Fig. 7), which in turn results in prevention of the Golgi disruption to a certain extent. In addition, there could be other mechanisms for the protection of the Golgi. In yeast cells, Glo3p-type Arf1 GAP was identified as a high-copy suppressor of temperature-sensitive arf1-3, a GDP-bound dominant negative mutant (Zhang et al., 1998). This is very much reminiscent of the effect we observed with AGD8/AGD9 in protoplasts. Furthermore, Glo3p-type ArfGAPs interact with coat proteins through the C-terminal domain. Therefore, overexpression of Glo3p-type ArfGAPs may bring coat proteins to the Golgi apparatus, which in turn results in protection of the Golgi even in the presence of Arf1[T31N].
Figure 7.
AGD8/AGD9 cause localization of Arf1[T31N]-sGFP to the Golgi apparatus. A, Arf1[T31N]-sGFP was introduced into protoplasts alone or together with HA-AGD7, HA-AGD8, or HA-AGD9, and the localization of Arf1[T31N]-sGFP was examined. Bars = 10 μm. B, Expression of HA-AGD7, HA-AGD8, and HA-AGD9 was examined by western-blot analysis using an HA antibody (HA). Control indicates protoplasts transformed with Arf1[T31N]-sGFP alone.
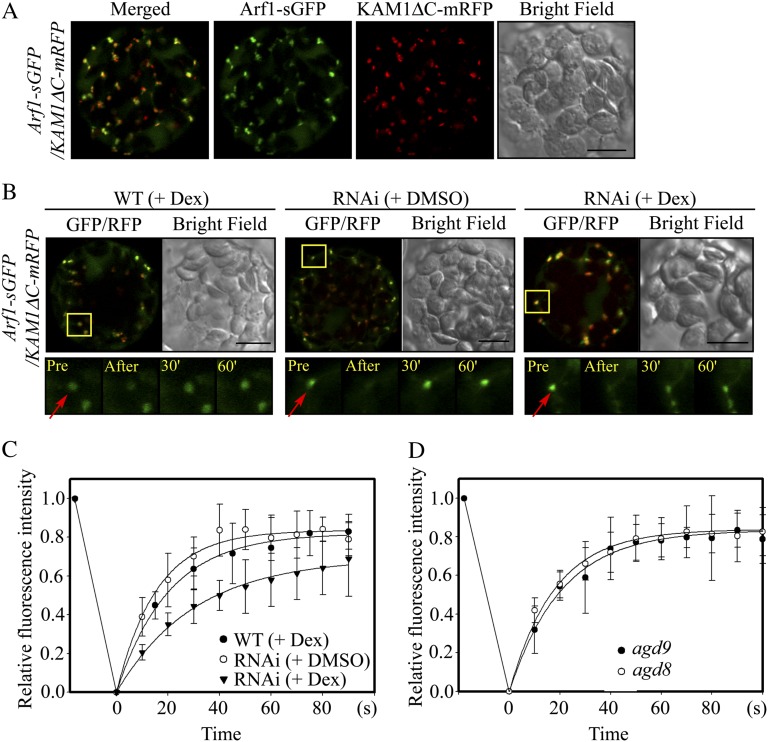
To obtain more direct evidence that AGD8/AGD9 are involved in recruiting Arf1-GDP to the Golgi apparatus, the targeting rate of Arf1 to the Golgi apparatus was examined in wild-type, agd8, agd9, and RNAi plants using fluorescence recovery after photobleaching (FRAP) with Arf1-sGFP (Presley et al., 2002). First, localization of Arf1-sGFP to the Golgi apparatus was examined in protoplasts. Protoplasts from wild-type plants were cotransformed with Arf1-sGFP and KAM1ΔC-mRFP, and localization of these proteins was examined. Both Arf1-sGFP and KAM1ΔC-mRFP produced punctate staining patterns that closely overlapped (Fig. 8A), confirming that Arf1-sGFP primarily localizes to the Golgi apparatus in protoplasts from leaf tissues. A minor portion of Arf1-sGFP did not colocalize with KAM1ΔC-mRFP, suggesting that a minor pool of Arf1 may localize in the cytosol or to other organelles. Based on this finding, FRAP experiments were performed using protoplasts from wild-type or RNAi plants that were cotransformed with Arf1-sGFP and KAM1ΔC-mRFP and incubated with dex or DMSO for 20 h. The punctae where both Arf1-sGFP and KAM1ΔC-mRFP colocalized were then photobleached using a laser. Subsequently, the time course of recovery of Arf1-sGFP localization to the Golgi apparatus was examined (Fig. 8B, boxed areas). The half-recovery times were 39, 15.6, and 18 s in dex-treated RNAi plants, DMSO-treated RNAi plants, and dex-treated wild-type plants, respectively. Thus, the recovery time in dex-treated RNAi plants was delayed by more than 2-fold with respect to the recovery time of dex-treated wild-type or DMSO-treated RNAi plants (Fig. 8C). These results confirm the idea that AGD8/AGD9 are involved in the targeting of Arf1 to the Golgi apparatus. As controls, agd8 or agd9 mutant plants without dex or DMSO treatment were included in the FRAP analysis. The half-recovery times in adg8 and agd9 plants were 16.2 and 19 s, respectively (Fig. 8D), indicating that the recovery time in these mutants is not significantly different from that of wild-type plants.
Figure 8.
RNAi plants display defects in targeting Arf1-sGFP to the Golgi apparatus. A, Localization of Arf1-sGFP to the Golgi apparatus. Protoplasts from wild-type plants were cotransformed with Arf1-sGFP and KAM1ΔC-mRFP, and localization of these proteins was examined. Bar = 20 μm. B, Images of Arf1-sGFP recovery to the Golgi apparatus after photobleaching. Arf1-sGFP and KAM1ΔC-mRFP were introduced into protoplasts from leaf tissues of wild-type (WT) or RNAi plants and incubated in medium supplemented with dex (30 μm) or DMSO for 20 h. The signals of Arf1-sGFP and KAM1ΔC-mRFP were examined using a laser scanning confocal microscope. For FRAP experiments, punctate stains in boxes were photobleached using a laser attached to a laser scanning confocal microscope, and recovered GFP fluorescence in the bleached areas was measured at various time points. Representative images of FRAP experiments in wild-type and RNAi plants with dex or DMSO are shown in the bottom panels. Pre and After indicate before and after photobleaching, respectively; arrows indicate punctae that were photobleached. Bars = 10 μm. C and D, Signal recovery curves after photobleaching. Signal intensities obtained in wild-type or RNAi plants treated with dex or DMSO (C) or adg8 and agd9 plants (D) were plotted with time. The curves were fitted to a single exponential rise to maximum by using a SigmaPlot algorithm. Half-time for recovery was 18, 15.6, 39, 16.2, and 19 s for WT + DEX, RNAi + DMSO, RNAi + DEX, agd8, and agd9 plants, respectively. Error bars indicate sd (n = 6).
AGD8 Overexpression Suppresses the Inhibition of Vacuolar Trafficking Caused by AGD7 Overexpression
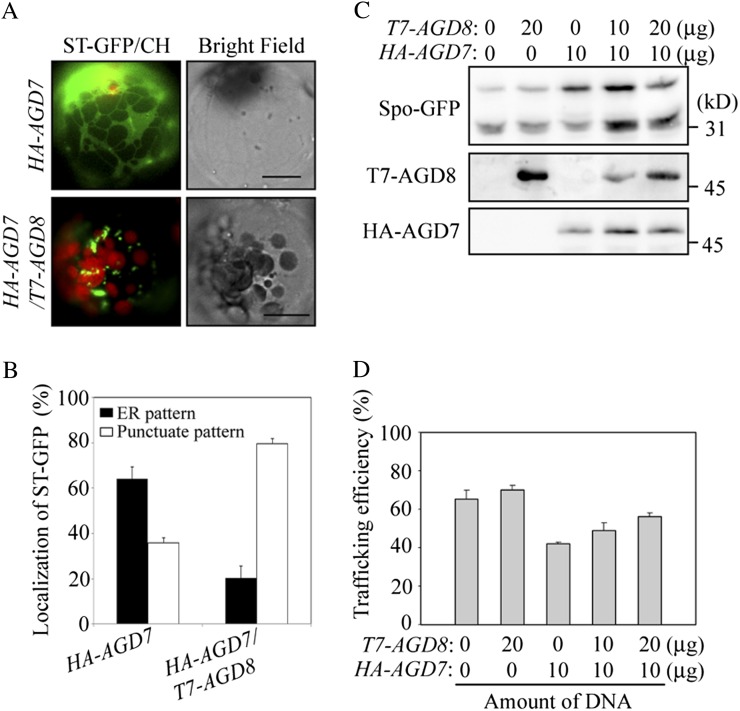
Overexpression of AGD7 causes disassembly of the Golgi apparatus through premature hydrolysis of GTP bound to Arf1 and results in an inhibition of vacuolar trafficking (Min et al., 2007). Thus, we tested whether the overexpression of AGD8 prevents the Golgi disassembly caused by AGD7 overexpression. ST-GFP and HA-AGD7 were cotransformed into protoplasts with or without T7-AGD8, and the localization of ST-GFP was examined. In protoplasts expressing only HA-AGD7, ST-GFP produced a network pattern, an indication of its localization to the endoplasmic reticulum, that resulted from disassembly of the Golgi apparatus as demonstrated previously (Min et al., 2007). In contrast, in protoplasts expressing both T7-AGD8 and HA-AGD7, ST-GFP exhibited primarily the punctate staining pattern (Fig. 9, A and B), indicating that the overexpression of AGD8 suppressed the AGD7-induced Golgi disassembly. To confirm this at the functional level, we determined the trafficking efficiency of sporamin-GFP to the vacuole in the presence of AGD7 alone or together with AGD8. Increasing amounts of T7-AGD8 were introduced into protoplasts together with HA-AGD7 and sporamin-GFP, and the vacuolar trafficking efficiency of sporamin-GFP was examined by western-blot analysis using a GFP antibody (Kim et al., 2001b). As observed previously (Min et al., 2007), expression of HA-AGD7 caused a reduction in the vacuolar trafficking efficiency of sporamin-GFP (from 70% down to 40%) compared with control protoplasts without HA-AGD7 at 24 h after transformation (Fig. 9, C and D). However, in protoplasts coexpressing both HA-AGD7 and T7-AGD8, the trafficking efficiency of sporamin-GFP to the vacuole increased gradually with increasing amounts of T7-AGD8 (Fig. 9, C and D). These results support the notion that overexpression of T7-AGD8 suppressed HA-AGD7-induced Golgi disassembly, thereby resulting in an increase in the trafficking efficiency of sporamin-GFP to the vacuole. Of course, we cannot completely exclude the possibility that overexpression of AGD8 activates a Golgi-independent route for the trafficking of sporamin-GFP to the vacuole.
Figure 9.
AGD8 overexpression suppresses the disruption of the Golgi apparatus and inhibition of vacuolar trafficking caused by AGD7 overexpression. A, Localization of ST-GFP to the Golgi apparatus. ST-GFP and HA-AGD7 were introduced into protoplasts with or without T7-AGD8, and localization of ST-GFP was examined. Bars = 10 μm. B, Quantification of the localization pattern of ST-GFP in the presence of HA-AGD7 alone or both HA-AGD7 and T7-AGD8. To quantify the localization pattern of ST-GFP, two plasmids, ST-GFP (10 μg) and HA-AGD7 (15 μg), were introduced into protoplasts together with or without T7-AGD8 (15 μg), and the localization pattern of ST-GFP was determined at 24 h after transformation from 50 protoplasts each time. Three independent transformation experiments were performed. Error bars indicate sd (n = 3). C, Trafficking of sporamin-GFP. Protoplasts were transformed with the indicated types and amounts of constructs. The trafficking efficiency of sporamin-GFP (Spo-GFP) was analyzed 36 h after transformation by western-blot analysis using various antibodies. D, Quantification of trafficking efficiency. To quantify trafficking efficiency, the percentage of the 30-kD processed form relative to the total amount of expressed sporamin-GFP was determined using band intensities. Error bars indicate sd (n = 3).
DISCUSSION
Arf1 and AGD isoforms locate to multiple organelles such as the Golgi apparatus, TGN, endosomes, and plasma membrane (Min et al., 2007; Matheson et al., 2008; Hwang and Robinson, 2009; Stefano et al., 2010). The localization of AGD isoforms may provide clues to the cellular processes or trafficking pathways in which the AGD isoforms are involved. AGD8 and AGD9 localized to the Golgi apparatus in plant cells. Another AGD isoform, AGD7, also localized to the Golgi apparatus (Min et al., 2007). However, the physiological roles of AGD8 and AGD9 differ from that of AGD7. This conclusion is based on differences in the effects of the AGD8/AGD9 protein levels on the Golgi apparatus. Lower levels of AGD8/AGD9 in AGD9 RNAi plants cause disassembly of the Golgi apparatus, whereas overexpression of AGD8 and AGD9 in protoplasts does not. In fact, high levels of AGD8 and AGD9 suppress AGD7-induced Golgi disassembly. It is known that the overexpression of ArfGAPs causes disassembly of the Golgi apparatus through premature hydrolysis of GTP bound to Arf1 in both animal and plant cells (Cukierman et al., 1995; Aoe et al., 1997; Liu et al., 2005; Min et al., 2007). Thus, the inability to induce Golgi disassembly suggests that AGD8 and AGD9 may not function as typical GAPs for Arf1, as exemplified by AGD7 and animal ArfGAP1.
The large number of AGD isoforms in Arabidopsis can be classified into six subfamilies. AGD8, AGD9, and AGD10 constitute a subfamily that is different from the subfamily of AGD7. AGD7 constitutes another subfamily together with AGD6. AGD8 and AGD9 are most closely related to animal ArfGAP2 and ArfGAP3, which are known as Glo3p-type ArfGAPs, whereas AGD7 is more closely related to animal ArfGAP1, a Gcs1p-type ArfGAP (Saitoh et al., 2009). In animal cells, the physiological roles and biochemical properties of ArfGAP2/ArfGAP3 are thought to be different from those of ArfGAP1 (Frigerio et al., 2007; Weimer et al., 2008; Kliouchnikov et al., 2009; Kartberg et al., 2010). ArfGAP1 contains the ALPS domain that is involved in membrane binding (Bigay et al., 2003). In contrast, ArfGAP2/ArfGAP3 contain the BoCCS domain that is involved in the interaction with coatomer, cargo, and SNARE; thus, they are thought to be involved in cargo-dependent COPI coat assembly (Kartberg et al., 2010). However, despite this difference in the physiological roles and biochemical properties between ArfGAP1 and ArfGAP2/ArfGAP3, there is a certain degree of functional redundancy; knockdown of the two Glo3p-type ArfGAPs (ArfGAP2 and ArfGAP3) resulted in only minor defects (Saitoh et al., 2009), but knockdown of two Glo3p-type ArfGAPs together with the Gcs1p-type ArfGAP1 resulted in a lethal phenotype. This is in contrast with the result showing that knockdown of the three Glo3p-type ArfGAPs, AGD8, AGD9, and AGD10, in RNAi plants was sufficient to cause a severe growth arrest.
A clue to the physiological role of these Glo3p-type ArfGAPs in plants was obtained by studying the binding properties of AGD8 with Arf1 and the GDP-bound form of Arf1 (Arf1[T31N]) and the localization patterns of Arf1 and Arf1[T31N] in the presence of AGD8. AGD8 interacts with both Arf1 and Arf1[T31N]. Moreover, this binding occurs at the Golgi apparatus. By contrast, at the Golgi apparatus, AGD7 interacts only with wild-type Arf1 but not with Arf1[T31N], although AGD7 also interacts with Arf1[T31N] in the cytosol. Another intriguing property of AGD8/AGD9 is that they caused the accumulation of both Arf1 and Arf1[T31N] at the Golgi apparatus. These results led to the hypothesis that the Glo3p-type ArfGAPs, AGD8/AGD9, play roles in the targeting of Arf1-GDP to the Golgi apparatus. These proposed new roles of AGD8/AGD9 differ from that of AGD7, which is mainly involved in the hydrolysis of Arf1-bound GTP to GDP (Min et al., 2007). Previous studies show that Arf1-GDP binds to Golgi membrane-localized proteins such as p23 and membrin (Gommel et al., 2001; Honda et al., 2005). Thus, these proteins are thought to function as receptors of Arf1-GDP at the Golgi membrane. However, it is not clear how Arf1-GDP proteins are recruited from the cytosol to the receptors localized at the Golgi membranes.
Targeting of Arf1-GDP to the Golgi apparatus can be explained by two possibilities. One possibility is that the diffusion of Arf1-GDP to receptors localized at the Golgi membranes constitutes the in vivo targeting mechanism. The second possibility is that a specific targeting mechanism operates to target Arf1-GDP to specific organelles among multiple potential target compartments, such as the Golgi apparatus, TGN, endosomes, and plasma membrane (D’Souza-Schorey and Chavrier, 2006; Gillingham and Munro, 2007; Matheson et al., 2008). In a previous study, ArfGAP1 was shown to facilitate membrane recruitment of Arf1-ΔN17 (Rein et al., 2002). Moreover, in AGD9 RNAi plants that have low levels of all three Glo3p-type ArfGAPs (AGD8, AGD9, and AGD10), recruitment of Arf1-GFP was significantly reduced compared with the wild-type plants. These results are consistent with the second possibility, that Glo3p-type ArfGAPs may function as the factor involved in delivering Arf1-GDP to the Golgi membranes, where Arf1-GDP may bind to Golgi membrane-localized Arf1-GDP receptors such as p23 and membrin that are found in animal cells (Gommel et al., 2001; Honda et al., 2005). Further support of this hypothesis is that AGD8 overexpression counteracted the effect of excess AGD7, which induces Golgi disassembly and inhibits vacuolar trafficking. One possible way to counteract the effect of excess AGD7 is to rapidly supply Arf1-GDP to the Golgi apparatus in order to maintain appropriate levels of Arf1-GTP at the Golgi apparatus, as long as GDP-GTP exchange by GEFs is not the rate-limiting step.
Correct targeting of Arf1-GDP to Golgi membranes represents a previously unidentified novel component of Arf1 action in protein trafficking. In animal cells and yeast, Glo3p-type ArfGAPs played roles in the assembly of COPI vesicles and also copurified with COPI vesicles (Lewis et al., 2004; Frigerio et al., 2007; Kliouchnikov et al., 2009; Saitoh et al., 2009; Schindler et al., 2009; Kartberg et al., 2010). Furthermore, in yeast cells, Glo3p at high levels can suppress the temperature-sensitive arf1-3 mutant, a GDP-bound dominant negative form (Zhang et al., 1998). Thus, their role in animal and yeast cells appears to be different from the proposed role of the Glo3p-type ArfGAPs AGD8/AGD9 in plants. However, the difference in the proposed roles of the Glo3p-type ArfGAPs in plant and animal cells may not necessarily be irreconcilable. It is possible that the ability of Glo3p-type ArfGAPs to recruit GDP-bound Arf1 is the underlying mechanism by which high levels of Glo3p suppress the arf1-3 mutant in yeast cells. In addition, in assisting COPI vesicle assembly, the C-terminal BoCCS domain of ArfGAP2/ArfGAP3 is crucial because the BoCCS domain directly interacts with coatomer and cargo. In this study, we have not addressed the question of whether AGD8/AGD9 interact with any of these ArfGAP2/ArfGAP3-interacting proteins. Nevertheless, sequence analysis strongly suggests that the C-terminal domains of AGD8/AGD9 contain a region with the important amino acid residues found in the BoCCS domain. Thus, the results in this study in plants may expand the role of the Glo3p-type ArfGAPs in the biogenesis of COPI vesicles already explored in animal cells and yeast. Furthermore, these results demonstrate that the AGD8/AGD9-mediated recruitment of Arf1-GDP to the Golgi apparatus is essential for the maintenance of the Golgi apparatus and protein trafficking, processes that are essential for plant growth and development.
MATERIALS AND METHODS
Plant Growth and Generation of Transgenic Plants
Arabidopsis (Arabidopsis thaliana ecotype Columbia) was grown on MS plates at 22°C in a culture room or in a greenhouse under conditions of 70% relative humidity and a 16/8-h light/dark cycle. The generation of transgenic plants expressing ST-GFP was described previously (Lee et al., 2002).
Construction of Plasmids
The construction of Arf1[T31N] has been described previously (Lee et al., 2002). The AGD8 (GenBank accession no. AF360177) complementary DNA was amplified via PCR using the primers AGD8-5 and AGD8-3. AGD9 (GenBank accession no. AY099693) was amplified via PCR using the primers AGD9-5 and AGD9-3 To tag AGD8 with a small epitope T7 (with amino acid sequence MASMTGGQQMG) at the N terminus, AGD8 was inserted into the pET21a vector (Novagen) using EcoRI and SalI sites, and subsequently, T7-AGD8 was placed under the control of the cauliflower mosaic virus 35S promoter in a pUC vector using XbaI and SalI sites (Jin et al., 2001; Kim et al., 2001a). To generate HA-AGD8 and HA-AGD9, AGD8 and AGD9 were amplified by PCR using primer sets Sma-AGD8-5/Xho-AGD8-3 and Sma-AGD9-5/Xho-AGD9-3, respectively. The resulting PCR products were ligated to a plasmid that contains the cauliflower mosaic virus 35S promoter and the epitope HA after digestion with SmaI and XhoI. The Arf1-sGFP and Arf1[T31N]-sGFP constructs were generated by incorporating Arf1 without the termination codon into the N terminus of sGFP. The HA tag was added to Arf1 and Arf1[T31N] by two sequential PCRs, first with primers HA-3-1 and 35S-p and then with primers HA-3-2 and 35S-p. To generate GFP-AGD8, the AGD8 fragment amplified by PCR using primers Sma-AGD8-5 and Xho-AGD8-3 was ligated to the C terminus of GFP without the termination codon.
The constructs used for BiFC experiments were generated using Venus, a variant of yellow fluorescent protein, and cyan fluorescent protein, a variant of GFP (Lee et al., 2008). Briefly, the N-terminal region (VN) of Venus was amplified using specific primers Xba-VN-5 and Sma-VN-3. The resulting PCR product was fused to the N termini of AGD7 or AGD8 using XbaI and SmaI restriction sites. Subsequently, VN-AGD7 and VN-AGD8 were digested with XbaI and HpaI and ligated to an identically digested C-terminal HA-tagging vector. cCFP was amplified using the specific primers Bam-cCFP-5 and Xho-CFP-3. The resulting PCR product was ligated to the C termini of Arf1 or Arf1[T31N] after digesting with BamHI and ScaI. T7-cCFP was generated by replacing AGD8 of T7-AGD8 with cCFP of Arf1-cCFP. The sequences of primers are shown in Supplemental Table S1. All PCR products were sequenced.
Generation of RNAi Plants and Transcript-Level Analysis
To generate RNAi plants suppressing both AGD8 and AGD9, an RNAi construct was generated using a 421-bp fragment of AGD9 that shows a high degree of sequence homology to AGD8. The 421-bp AGD9 fragment was amplified by PCR using two sets of primers, Bam-RNAi-5/Cla-RNAi-3 and Xho-RNAi-5/Kpn-RNAi-3. The two amplified products were introduced sequentially into the BamHI/ClaI sites and XhoI/KpnI sites of RNAi vector pHannibal (Wesley et al., 2001). The AGD9 RNAi construct in the pHannibal vector was then transferred to pTA7002, a dex-inducible binary vector. RNAi transgenic plants were generated by the floral dip method (Clough and Bent, 1998). To induce expression of the AGD9 fragment in RNAi plants, RNAi transgenic plants grown on MS plates were transferred onto dex (30 μm)-containing plates. Total RNAs from the RNAi transgenic plants were prepared using the RNeasy Plant Mini Kit (Qiagen). Complementary DNA was prepared with the SuperScript II reverse transcriptase kit (Invitrogen) according to the manufacturer’s instructions.
To quantify gene expression levels in RNAi, wild-type, and mutant plants, the StepOne and StepOnePlus real-time PCR systems (Applied Biosystems) were used with Power SYBR Green PCR master mix (Applied Biosystems). The data were analyzed with StepOne software version 2.1 using the 2-ΔΔCT method (Livak and Schmittgen, 2001). Amplified 18S ribosomal RNA (rRNA) fragments (primers 18S-f-2 and 18S-r-2) were used as endogenous controls to normalize the expression of the tested genes. AGD4, AGD7, AGD8, AGD9, and AGD10 transcript levels in wild-type, agd8, and agd9 plants were determined using specific primers: AGD4-f and AGD4-r for AGD4; AGD7-f and AGD7-r for AGD7; AGD8-f and AGD8-r for AGD8; AGD9-f and AGD9-r for AGD9; and AGD10-f and AGD10-r for AGD10 (Supplemental Table S1).
Isolation of Single Mutants
Arabidopsis mutants agd8 (SALK-053104) and agd9 (SAIL-660_G01) containing T-DNA insertions in AGD8 and AGD9, respectively, were obtained from the Arabidopsis Biological Resource Center (Ohio State University). Homozygotes of the agd8 and agd9 mutants were screened by PCR-based genotyping using left border primers (LB for the SALK line and LB1 for the SAIL line) and gene-specific primers (forward primer of AGD8, FP-AGD8; reverse primer of AGD8, RP-AGD8; forward primer of AGD9, FP-AGD9; reverse primer of AGD9, RP-AGD9).
Transient Expression in Protoplasts and Western-Blot Analysis
For transient expression experiments in protoplasts (subcellular localization and BiFC), plasmid DNA was purified using the Qiagen column and was then transformed into protoplasts derived from leaf tissues (Hyunjong et al., 2006). Protein extracts were prepared from transformed protoplasts and used for western-blot analysis using anti-GFP (Bio-Application) and anti-HA (Roche) antibodies as described previously (Kim et al., 2005).
Coimmunoprecipitation
Protein extracts in immunoprecipitation buffer (150 mm KCl, 20 mm HEPES, pH 7.4, 1 mm MgCl2, 0.2% Triton X-100, 0.2% Nonidet P-40, 1 mm dithiothreitol, 1 mm EGTA, and 1× protease inhibitor cocktail) were incubated at 4°C for 4 h on a rotating platform with GFP antibody (Invitrogen) and protein A-Sepharose beads. Immune complexes were isolated using protein A-Sepharose beads. Beads were washed five times with IP buffer. After washing, proteins were released by 10 min of incubation in SDS sample buffer at 98°C and analyzed by western blotting using rat HA (Roche) or GFP (Clontech) antibodies.
FRAP Analysis
To prevent movement of the Golgi apparatus, transformed protoplasts were incubated with 25 μm latrunculin B in W5 solution for 1 h prior to photobleaching experiments (Brandizzi et al., 2002). Cells were visualized using a laser scanning confocal microscope (LSM 510 Meta System; Carl Zeiss). Bundle software (LSM 510 version 3.2) was used to record prebleach and postbleach signals and to modulate laser beam intensity.
Generation of Transgenic Plants Expressing AGD8p::HA-AGD8 or AGD9p::HA-AGD9
A DNA fragment containing the AGD8 promoter region (AGD8p; 1.25 kb) was amplified by PCR using primers Pst-up-AGD8-5 and up-AGD8-HA-3, and a DNA fragment containing HA-AGD8 was amplified by PCR using primers HA-5 and Stu-nos-t-3. To generate AGD8p::HA-AGD8, these two DNA fragments were fused by the second round of PCR using primers Pst-up-AGD8-5 and Stu-nos-t-3. AGD9p::HA-AGD9 was constructed in a similar way; the primers were Pst-up-AGD9-5 and up-AGD9-HA-3 for a DNA fragment containing the AGD9 promoter region (2.05 kb), HA-5 and Stu-nos-t-3 for HA-AGD9, and Pst-up-AGD9-5 and Stu-nos-t-3 for the second round of PCR to generate AGD9p::HA-AGD9. The amplified fragments were digested with PstI and StuI and ligated to PstI- and StuI-digested pCR-ccD gateway donor vector (Invitrogen). Inserts of these constructs were introduced into the pCAMVIA1300 binary Gateway vector by LR recombination (Invitrogen). HA-AGD8 and HA-AGD9 under the control of the native promoter were introduced into wild-type or ST-GFP transgenic plants by the floral dip method (Clough and Bent, 1998).
Immunohistochemistry
Protoplasts were allowed to adhere to poly-l-Lys-coated slides (Sigma) for 30 min. Protoplasts were then fixed for 30 min in W6 buffer (10 mm HEPES, pH 7.2, 0.9% NaCl, 125 mm CaCl2, 5 mm KCl, and 5 mm Glc) containing 4% paraformaldehyde, washed three times with W6 buffer, and permeabilized in TSW buffer (10 mm Tris-HCl, pH 7.4, 0.9% NaCl, 0.25% gelatin, 0.02% SDS, and 0.1% Triton X-100) for 10 min. Fixed protoplasts were incubated overnight at 4°C in the presence of primary antibodies (T7 antibody, Novagen; rat HA antibody, Roche; γ-COP antibody, Agrisera) in TSW buffer. After four washes in TSW buffer, cells were incubated with secondary antibodies (i.e. fluorescein isothiocyanate [FITC]-conjugated anti-mouse or anti-rat IgG or tetramethyl rhodamine isothiocyanate [TRITC]-conjugated anti-rabbit IgG; Zymed Laboratories) for 1 h at room temperature. After four washes in TSW buffer, cells were mounted in Mowiol (Hoechst) containing 2.5% 1,4-diazobicyclo-[2.2.2]-octane (Sigma). Stained cells were visualized using a Zeiss fluorescence microscope and a laser scanning confocal microscope (Meta System; Zeiss).
For immunostaining intact root tissues, transgenic plants (7 d old) were transferred to MS plates supplemented with or without dex (30 μm). Two days after transplanting, plant root tissues were fixed with 4% paraformaldehyde in PEX buffer (5 mm MgSO4, 5 mm EGTA, and 100 mm PIPES, pH 6.8) for 2 h at 4°C, washed three times with PEX buffer, and treated with enzyme solution (2% macerozyme R-10 [Yakult Honsha] and 1% cellulase R-10 [Yakult Honsha]) in PEX buffer for 30 min at 23°C. The partially digested plant root tissues were attached to poly-l-Lys-coated glass slides. For blocking, the slides with plant root tissues were submerged in TSW buffer overnight at 4°C followed by incubation with the primary antibodies (HA antibody, Roche; SYP21 antibody [Sanderfoot et al., 1999], SYP61 antibody [Sanderfoot et al., 2001], and γ-COP antibody, Agrisera) overnight at 4°C. After three washes with TSW buffer, the root tissues were incubated with secondary antibodies (FITC- or TRITC-conjugated anti-rat or anti-rabbit IgG, respectively; Zymed Laboratories). Images were captured using a Zeiss LSM 510 META laser scanning confocal microscope. For observations of mesophyll protoplasts and root tissues, the combinations of excitation wavelength/emission filter were 488 nm (argon/ion laser)/505 to 530 bandpass for GFP and FITC and 543 nm/560 to 615 bandpass for mRFP and TRITC. Images of protoplasts were obtained using a cooled CCD camera and an Axioplan fluorescence microscope (Carl Zeiss). The filter sets were XF116 (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23) and XF33/E (exciter, 535DF35; dichroic, 570DRLP; emitter, 605DF50 [Omega]) for GFP/FITC and TRITC, respectively. Red autofluorescence of chlorophyll was observed using filter set Lumar 15 (exciter, 546/12; emitter, LP 590). The data were processed using Adobe Photoshop software, and the images are presented in pseudocolor.
To quantify the degree of colocalization of two different proteins on an image, the ImageJ analysis program (Abramoff et al., 2004) and the PSC colocalization plug-in software (French et al., 2008) were used. Using these two software programs, we calculated the linear Pearson correlation coefficient and nonlinear Spearman’s rank correlation coefficient of the signal intensity from the green and red channels and obtained a scatterplot with the values of fluorescence pixels across the two channels distributed between PSC coefficient values −1 to +1 for strong negative and positive correlations, respectively. The masking of areas was performed prior to analysis using the selection brush tool (Abramoff et al., 2004). The threshold level was set to 10, below which pixel values were considered noise and excluded in the statistical analysis.
Electron Microscopy
Five-day-old RNAi or wild-type plants were transferred onto MS plates supplemented with or without dex (30 μm). Four days after transplanting, root tissues were fixed with 2% paraformaldehyde and 2% glutaraldehyde-containing sodium cacodylate buffer (50 mm sodium cacodylate, pH 7.2) at 4°C for 4 h. Samples were washed three times with sodium cacodylate buffer followed by postfixation with 1% osmium tetroxide in sodium cacodylate buffer at 4°C for 2 h. After washing briefly with distilled water, the samples were treated with 0.5% uranyl acetate at 4°C for 30 min, followed by dehydration with a graded series of ethanol. Dehydrated samples were infiltrated with a graded series of ethanol and Spurr’s resin mixtures before final embedding in 100% Spurr’s resin. The infiltrated samples were polymerized at 70°C for 24 h. After ultrathin sectioning with an ultramicrotome (MT-X; RMC), samples were stained with 2% uranyl acetate and Reynolds’ lead citrate and then examined with a transmission electron microscope (JEM-1010; JEOL) operating at 80 kV.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AGD7, NP181291; AGD8, AF36177; AGD9, AY099693; AGD10, AEC09080; and Arf1, AAA32729.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of Arabidopsis GAP domain proteins.
Supplemental Figure S2. AGD transcript levels in wild-type plants.
Supplemental Figure S3. Nucleotide sequence alignment of AGD8, AGD9, and AGD10.
Supplemental Figure S4. Transiently expressed HA-AGD8 and HA-AGD9 localize to the Golgi apparatus.
Supplemental Table S1. PCR primers for cloning, generation of various mutants, and confirming of mutant plants.
Glossary
- RT
reverse transcription
- qRT
quantitative reverse transcription
- RNAi
RNA interference
- dex
dexamethasone
- TGN
trans-Golgi network
- HA
hemagglutinin
- ST
sialyltransferase
- PSC
Pearson-Spearman correlation
- PVC
prevacuolar compartment
- MS
Murashige and Skoog
- DMSO
dimethyl sulfoxide
- BiFC
bimolecular fluorescence complementation
- cCFP
C-terminal half of cyan fluorescent protein
- nVN
N-terminal half of the GFP variant Venus
- FRAP
fluorescence recovery after photobleaching
- FITC
fluorescein isothiocyanate
- mRFP
monomeric red fluorescent protein
- T-DNA
transfer DNA
- rRNA
ribosomal RNA
- sGFP
superfolder GFP
References
- Abramoff MD, Magelhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Aoe T, Cukierman E, Lee A, Cassel D, Peters PJ, Hsu VW. (1997) The KDEL receptor, ERD2, regulates intracellular traffic by recruiting a GTPase-activating protein for ARF1. EMBO J 16: 7305–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Balch WE, Kahn RA, Schwaninger R. (1992) ADP-ribosylation factor is required for vesicular trafficking between the endoplasmic reticulum and the cis-Golgi compartment. J Biol Chem 267: 13053–13061 [PubMed] [Google Scholar]
- Bigay J, Gounon P, Robineau S, Antonny B. (2003) Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature 426: 563–566 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N. (2002) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cukierman E, Huber I, Rotman M, Cassel D. (1995) The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science 270: 1999–2002 [DOI] [PubMed] [Google Scholar]
- Denecke J, Aniento F, Frigerio L, Hawes C, Hwang I, Mathur J, Neuhaus JM, Robinson DG. (2012) Secretory pathway research: the more experimental systems the better. Plant Cell 24: 1316–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. (2000) Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol 12: 475–482 [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. (2006) ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 7: 347–358 [DOI] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. (2008) Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Frigerio G, Grimsey N, Dale M, Majoul I, Duden R. (2007) Two human ARFGAPs associated with COP-I-coated vesicles. Traffic 8: 1644–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. (2007) The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol 23: 579–611 [DOI] [PubMed] [Google Scholar]
- Goldberg J. (1999) Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell 96: 893–902 [DOI] [PubMed] [Google Scholar]
- Gommel DU, Memon AR, Heiss A, Lottspeich F, Pfannstiel J, Lechner J, Reinhard C, Helms JB, Nickel W, Wieland FT. (2001) Recruitment to Golgi membranes of ADP-ribosylation factor 1 is mediated by the cytoplasmic domain of p23. EMBO J 20: 6751–6760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Al-Awar OS, Hay JC, Donaldson JG. (2005) Targeting of Arf-1 to the early Golgi by membrin, an ER-Golgi SNARE. J Cell Biol 168: 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Robinson DG. (2009) Transport vesicle formation in plant cells. Curr Opin Plant Biol 12: 660–669 [DOI] [PubMed] [Google Scholar]
- Hyunjong B, Lee DS, Hwang I. (2006) Dual targeting of xylanase to chloroplasts and peroxisomes as a means to increase protein accumulation in plant cells. J Exp Bot 57: 161–169 [DOI] [PubMed] [Google Scholar]
- Inoue H, Randazzo PA. (2007) Arf GAPs and their interacting proteins. Traffic 8: 1465–1475 [DOI] [PubMed] [Google Scholar]
- Jensen RB, Lykke-Andersen K, Frandsen GI, Nielsen HB, Haseloff J, Jespersen HM, Mundy J, Skriver K. (2000) Promiscuous and specific phospholipid binding by domains in ZAC, a membrane-associated Arabidopsis protein with an ARF GAP zinc finger and a C2 domain. Plant Mol Biol 44: 799–814 [DOI] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartberg F, Asp L, Dejgaard SY, Smedh M, Fernandez-Rodriguez J, Nilsson T, Presley JF. (2010) ARFGAP2 and ARFGAP3 are essential for COPI coat assembly on the Golgi membrane of living cells. J Biol Chem 285: 36709–36720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I. (2001a) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Park M, Kim SJ, Hwang I. (2005) Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell 17: 888–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Roh HE, Lee MN, Hur MW. (2007). New fast BiFC plasmid assay system for in vivo protein-protein interactions. Cell Physiol Biochem 20: 703–714 [DOI] [PubMed] [Google Scholar]
- Kim YW, Park DS, Park SC, Kim SH, Cheong GW, Hwang I. (2001b) Arabidopsis dynamin-like 2 that binds specifically to phosphatidylinositol 4-phosphate assembles into a high-molecular weight complex in vivo and in vitro. Plant Physiol 127: 1243–1255 [PMC free article] [PubMed] [Google Scholar]
- Kliouchnikov L, Bigay J, Mesmin B, Parnis A, Rawet M, Goldfeder N, Antonny B, Cassel D. (2009) Discrete determinants in ArfGAP2/3 conferring Golgi localization and regulation by the COPI coat. Mol Biol Cell 20: 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K, Naramoto S, Sawa S, Yahara N, Ueda T, Nakano A, Sugiyama M, Fukuda H. (2005) VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development 132: 1699–1711 [DOI] [PubMed] [Google Scholar]
- Lanoix J, Ouwendijk J, Lin CC, Stark A, Love HD, Ostermann J, Nilsson T. (1999) GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COP I vesicles. EMBO J 18: 4935–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB. (2008) Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4: 24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Min MK, Lee YJ, Jin JB, Shin DH, Kim DH, Lee KH, Hwang I. (2002) ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol 129: 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Yang JS, Hong W, Premont RT, Hsu VW. (2005) ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J Cell Biol 168: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM, Poon PP, Singer RA, Johnston GC, Spang A. (2004) The ArfGAP Glo3 is required for the generation of COPI vesicles. Mol Biol Cell 15: 4064–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Duden R, Phair RD, Lippincott-Schwartz J. (2005) ArfGAP1 dynamics and its role in COPI coat assembly on Golgi membranes of living cells. J Cell Biol 168: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matheson LA, Suri SS, Hanton SL, Chatre L, Brandizzi F. (2008) Correct targeting of plant ARF GTPases relies on distinct protein domains. Traffic 9: 103–120 [DOI] [PubMed] [Google Scholar]
- Min MK, Kim SJ, Miao Y, Shin J, Jiang L, Hwang I. (2007) Overexpression of Arabidopsis AGD7 causes relocation of Golgi-localized proteins to the endoplasmic reticulum and inhibits protein trafficking in plant cells. Plant Physiol 143: 1601–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl P, Movafeghi A, Coughlan S, Denecke J, Hillmer S, Robinson DG. (2000) In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Ward TH, Pfeifer AC, Siggia ED, Phair RD, Lippincott-Schwartz J. (2002) Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature 417: 187–193 [DOI] [PubMed] [Google Scholar]
- Rein U, Andag U, Duden R, Schmitt HD, Spang A. (2002) ARF-GAP-mediated interaction between the ER-Golgi v-SNAREs and the COPI coat. J Cell Biol 157: 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh A, Shin HW, Yamada A, Waguri S, Nakayama K. (2009) Three homologous ArfGAPs participate in coat protein I-mediated transport. J Biol Chem 284: 13948–13957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. (2001) Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell 12: 3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV. (1999) The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol 121: 929–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Rodriguez F, Poon PP, Singer RA, Johnston GC, Spang A. (2009) The GAP domain and the SNARE, coatomer and cargo interaction region of the ArfGAP2/3 Glo3 are sufficient for Glo3 function. Traffic 10: 1362–1375 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Muday GK, King EJ, Benton G, Kim S, Metcalf KE, Meyers L, Seamen E, Van Norman JM. (2006) SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18: 1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, et al. (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XF, Yang CY, Liu J, Yang WC. (2006) RPA, a class II ARFGAP protein, activates ARF1 and U5 and plays a role in root hair development in Arabidopsis. Plant Physiol 141: 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Shiba Y, Randazzo PA. (2010) Arf GAPs: gatekeepers of vesicle generation. FEBS Lett 584: 2646–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano G, Renna L, Rossi M, Azzarello E, Pollastri S, Brandizzi F, Baluska F, Mancuso S. (2010) AGD5 is a GTPase-activating protein at the trans-Golgi network. Plant J 64: 790–799 [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Yahara N, Nakano A. (2002) Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J 31: 499–515 [DOI] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I. (2005) KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer C, Beck R, Eckert P, Reckmann I, Moelleken J, Brügger B, Wieland F. (2008) Differential roles of ArfGAP1, ArfGAP2, and ArfGAP3 in COPI trafficking. J Cell Biol 183: 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Yoo CM, Wen J, Motes CM, Sparks JA, Blancaflor EB. (2008) A class I ADP-ribosylation factor GTPase-activating protein is critical for maintaining directional root hair growth in Arabidopsis. Plant Physiol 147: 1659–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CJ, Cavenagh MM, Kahn RA. (1998) A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J Biol Chem 273: 19792–19796 [DOI] [PubMed] [Google Scholar]
- Zhuang X, Xu Y, Chong K, Lan L, Xue Y, Xu Z. (2005) OsAGAP, an ARF-GAP from rice, regulates root development mediated by auxin in Arabidopsis. Plant Cell Environ 28: 147–156 [DOI] [PubMed] [Google Scholar]