Abstract
Accurate models of the cross-talk between signaling pathways and transcriptional regulatory networks within cells are essential to understand complex response programs. We present a new computational method that combines condition-specific time-series expression data with general protein interaction data to reconstruct dynamic and causal stress response networks. These networks characterize the pathways involved in the response, their time of activation, and the affected genes. The signaling and regulatory components of our networks are linked via a set of common transcription factors that serve as targets in the signaling network and as regulators of the transcriptional response network. Detailed case studies of stress responses in budding yeast demonstrate the predictive power of our method. Our method correctly identifies the core signaling proteins and transcription factors of the response programs. It further predicts the involvement of additional transcription factors and other proteins not previously implicated in the response pathways. We experimentally verify several of these predictions for the osmotic stress response network. Our approach requires little condition-specific data: only a partial set of upstream initiators and time-series gene expression data, which are readily available for many conditions and species. Consequently, our method is widely applicable and can be used to derive accurate, dynamic response models in several species.
Perturbation of the cellular environment typically incites a vast and complex reaction that involves a multitude of proteins operating together to respond to the new condition. Although the widespread availability and falling costs of microarray technologies along with the rise of RNA-seq have made it easier to quantify the transcriptional aspects of a cellular response, measurements of gene expression alone represent only a limited glimpse of the processes employed by a cell in order to adapt to an external or environmental change. To fully explain and ultimately control the response to environmental stress, it is necessary to construct end-to-end models of both the signaling and regulatory mechanisms that are activated.
A popular approach when using high-throughput data to model the interplay between the signaling and regulatory networks is to rely on knockout (KO) experiments. Such experiments provide both a starting point (the knocked-out or knocked-down gene) and a set of endpoints (differentially expressed genes). Computational methods have been developed to search for paths linking the starting points and endpoints and to integrate paths identified in the different KO experiments.
The physical network models (PNM) technique (Yeang et al. 2004) is a pioneering method for using the above strategy to reconstruct signaling and regulatory networks. PNM constructs a skeleton network of physical protein–protein and protein–DNA interactions and searches for directed pathways that connect deleted genes and their targets. SPINE (Ourfali et al. 2007) adopts a similar strategy by relying on KOs as starting points but concentrates on the positive or negative regulatory effects of edges or proteins as opposed to the orientation of protein–protein interactions (PPIs). Peleg et al. (2010) proposed a “network-free” approach to the problem of explaining KO cause–effect pairs, which operates on a functional network instead of explicitly enumerating pathways.
Other methods have used additional types of perturbations as starting points, including data from genetic screens. Motivated by the vast discrepancy between hits in genetic screens and genes that are differentially expressed in response to a stimulus, ResponseNet (Yeger-Lotem et al. 2009) combines these two types of data to generate integrated signaling and regulatory networks for a condition of interest. A related approach combines the genetic hits and differentially expressed genes as the relevant terminal nodes in the network (Huang and Fraenkel 2009). However, this algorithm and others derived from the Steiner tree problem (Yosef et al. 2009; Bailly-Bechet et al. 2011) do not model redundant and parallel pathways to the target nodes regardless of the type of input data used.
Although KO-based methods proved successful in some cases, there are several problems inherent in techniques that primarily depend on gene KOs or other genetic perturbations. First, many genes are essential (e.g., ∼20% of yeast genes) (Giaever et al. 2002), which prohibits their use as starting points even if they are known to play an important role in the response. Second, genes must be perturbed individually in the condition of interest. Due to the large associated costs of profiling numerous deletion strains, the resulting measurements and models are almost always static. Finally, backup mechanisms are employed by regulatory networks (Kafri et al. 2005, 2008; Gitter et al. 2009). Thus, the stress response pathways activated in one KO strain may differ from those activated in the wild-type strain and in other KOs. This makes it very hard to recover the wild-type response pathways from a collection of KOs. Vinayagam et al. (2011) avoided the complications of KOs by orienting PPI with respect to shortest paths between all membrane receptors and transcription factors (TFs). However, their approach relies on general topological features and does not reveal the pathways or regulators most relevant to a specific response.
In addition to techniques proposed for integrating signaling and regulatory networks, several approaches have been proposed to reconstruct dynamic regulatory networks (Ernst et al. 2007; Lin et al. 2007; Wei and Li 2008; for a review see, Gitter et al. 2010). Either these focus on the regulatory network exclusively and thus do not explain how the TFs regulating the response are activated, or they only utilize known (database-derived) pathways. Dependence upon known pathways limits their applications to well-studied networks and species and prevents them from providing new predictions regarding the members and interactions in the signaling network portion of the integrated model.
To overcome the obstacles inherent in genetic perturbation data, we developed the Signaling and Dynamic Regulatory Events Miner (SDREM), which utilizes condition-specific time-series gene expression to reconstruct the signaling and regulatory networks that are activated during stress response. Similar to KOs, temporal data provide causal information. However, unlike individual KO experiments, time-series expression experiments often measure wild-type response and provide information about all pathways simultaneously. SDREM assumes that a small, possibly incomplete set of upstream proteins that play a role in initiating the response is known. SDREM uses the expression data to identify TFs that control the differentially expressed genes. These TFs serve as targets for a network orientation algorithm that links the sensory proteins to the active TFs. SDREM provides a complete picture of the stress response, including directed pathways from upstream source proteins to the TFs, the times at which those TFs are actively regulating their bound genes, the primary temporal expression profiles that characterize the response, and the genes that belong to them.
We applied SDREM to study the responses of Saccharomyces cerevisiae to high osmolarity and rapamycin. Our method successfully recovered the core signaling proteins and TFs that are involved in these well-studied responses, and generated accurate models of the known signaling and regulatory pathways. The models also provide novel hypotheses implicating several additional TFs and proteins in the responses. We experimentally validated several of the osmotic stress model's predictions, shedding new light on the regulation of this response. In addition, we studied the immune response of Arabidopsis thaliana to the pathogen Hyaloperonospora arabidopsidis (Hpa) to demonstrate SDREM's generality.
Results
We developed a new method for integrating time-series expression data with static PPIs and protein–DNA interactions to infer dynamic regulatory networks and the signaling pathways that activate these networks. In addition to the general interaction data, SDREM uses condition-specific time-series data and a small set of proteins that are known to sense the environmental stress, interact with the infecting agent, or play some other role in initiating the response as input. In many cases, such proteins are either known (Kanehisa and Goto 2000) or can be experimentally determined using mass spectrometry or yeast two-hybrid experiments (e.g., in the response to viral infection) (Chatr-aryamontri et al. 2009; Fu et al. 2009; Mukhtar et al. 2011).
Reconstructing dynamic networks and orienting interaction networks
SDREM builds upon two previously developed methods, the Dynamic Regulatory Events Miner (DREM) (Ernst et al. 2007) and a network orientation procedure (Gitter et al. 2011). DREM uses an input–output hidden Markov model (IOHMM) to reconstruct dynamic regulatory networks by identifying bifurcation events, places in the time series where a set of genes that were previously coexpressed diverges. DREM annotates these split events with TFs that are predicted to regulate genes in the outgoing upward and/or downward paths, allowing us to associate temporal information (the time of the splits) with the static protein–DNA interaction data. DREM was successfully applied to reconstruct networks in a large number of species, including yeast (Ernst et al. 2007), Escherichia coli (Ernst et al. 2008), the fly (The modENCODE Consortium et al. 2010), and humans (Gu et al. 2010). Although DREM identifies the active TFs, it does not explain what activated these TFs or consider whether the TFs are consistent with the signaling pathways involved in the response.
The second component is a network orientation algorithm. This algorithm is provided with a PPI data set, a small set of source proteins (e.g., receptors or sensory proteins), and a small set of targets (e.g., TFs). The algorithm then uses a search procedure that orients the undirected protein interaction edges such that the targets can be explained by relatively short, high-confidence pathways that originate at the inputs. These objectives are derived from several biological assumptions observed in reference signaling networks. In studies of yeast pathways where the source and target proteins were given, the orientation algorithm successfully recovered known pathways better than previously suggested pathway prediction methods (Gitter et al. 2011).
SDREM: Signaling and Dynamic Regulatory Events Miner
The network orientation algorithm discussed above can complement DREM by linking the identified TFs to the source proteins in order to explain their activation. However, to accurately combine the two, we need to address several computational challenges. First, DREM is a probabilistic model, whereas the network orientation method solves a combinatorial optimization problem. Thus, values computed in one model cannot be directly transferred to the other. In addition, DREM is unable to account for the network connectivity of the TFs (i.e., prefer TFs that are well connected to the upstream sources) because it considers all TFs to be equally likely to be active in the response. Similarly, some active TFs in the DREM model may be implicated more strongly than others in the oriented network model, but the TF enrichment scores DREM calculates cannot incorporate this prior information and are not compatible with the network orientation objective function.
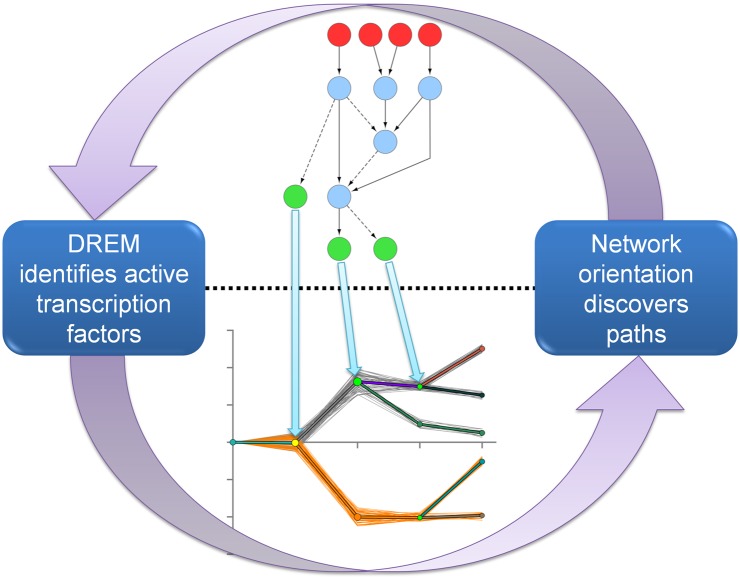
To address these issues, we developed SDREM, which iteratively combines the two methods (Fig. 1). SDREM uses a modified version of DREM to infer the TFs that regulate genes as part of the response, as well as the times at which this regulation takes place. The identified TFs become targets for the network orientation algorithm. The oriented network is then used to determine which of the target TFs are supported by the discovered pathways. There is reason to believe that the secondary TFs involved in stress response or recovery are more likely to be transcriptionally regulated (Farkas et al. 2006; Ernst et al. 2007) so we include both oriented PPI and protein–DNA interactions in the “signaling” pathways. TFs that cannot be explained by the signaling network are penalized so that they are less likely to be selected in the subsequent DREM analysis. This process repeats for a fixed number of iterations, which leads to the final pathways and regulatory network.
Figure 1.
Iterative model for integrating signaling and dynamic regulatory networks. The two components of SDREM iteratively refine an end-to-end model of stress response. DREM identifies active transcription factors (TFs) by analyzing divergence points in dynamic gene expression profiles. Protein–protein interaction (PPI) network orientation is used to connect those TFs to proteins that initiate the response by sensing or interacting with the environment.
In practice, unifying DREM and the network orientation algorithm requires overcoming the aforementioned challenges. We implemented a strategy that allows SDREM to incorporate prior (continuous-valued) information about the TFs during the analysis of the gene expression data. In order to compute these TF activity priors, we developed a method to assign a posterior score for each target TF based on its dominance in the oriented network with respect to random targets. To allow information to flow in the other direction (from DREM to the network orientation), we extended DREM so that it outputs an activity score for each TF at each regulatory path split. We further modified the orientation algorithm to use these scores to prioritize the targets. These new DREM scores provide a set of TFs that are believed to be active as well as a quantitative measure of their activity level.
High osmolarity stress response
To test SDREM, we first applied it to study the response of S. cerevisiae cells to high osmolarity. This response is primarily mediated by the high osmolarity glycerol (HOG) pathway, whose core component is the mitogen-activated protein kinase (MAPK) Hog1. Its main physiological function is to counteract the effects of increased osmolarity, such as water loss and cell shrinking (Hohmann 2009). We collected general (noncondition-specific) PPI data (Stark et al. 2006) and protein–DNA binding data for 117 TFs (MacIsaac et al. 2006). The protein–DNA binding data set was complemented with condition-specific TF–gene interactions for Hot1 and Sko1 (Capaldi et al. 2008). Both PPIs and protein–DNA interactions were allowed in the source–target pathways that explain how TFs are activated via chains of transcriptional and/or post-translational events. In addition, we used condition-specific source proteins (Cdc42, Msb2, Sho1, Sln1, and Ste50) from the Science Signaling Database of Cell Signaling and two complementary time-series gene expression data sets. The first expression data set (Romero-Santacreu et al. 2009) measures gene expression up to 15 min, leading to our short model. The second (Gasch et al. 2000) is used to construct the long model (up to 90 min) because it includes the recovery phase of the response.
Short osmotic stress model
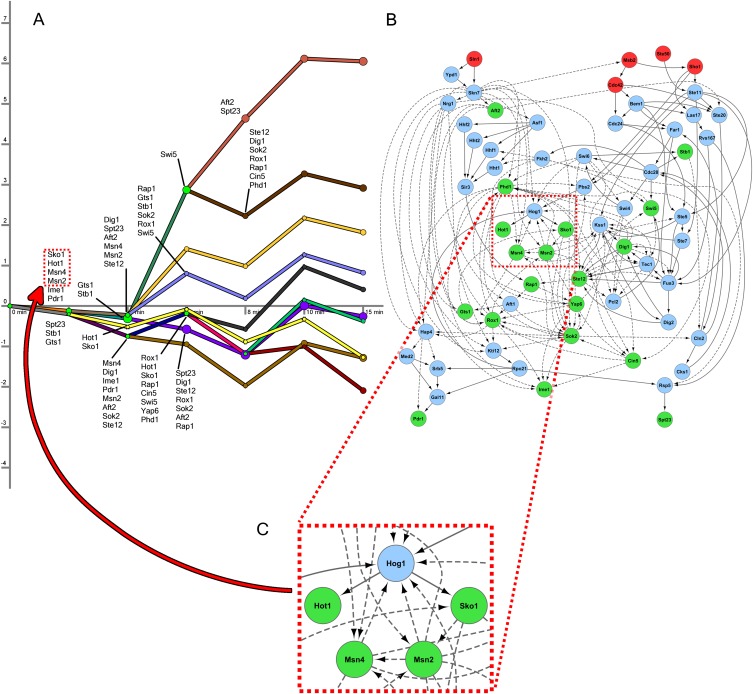
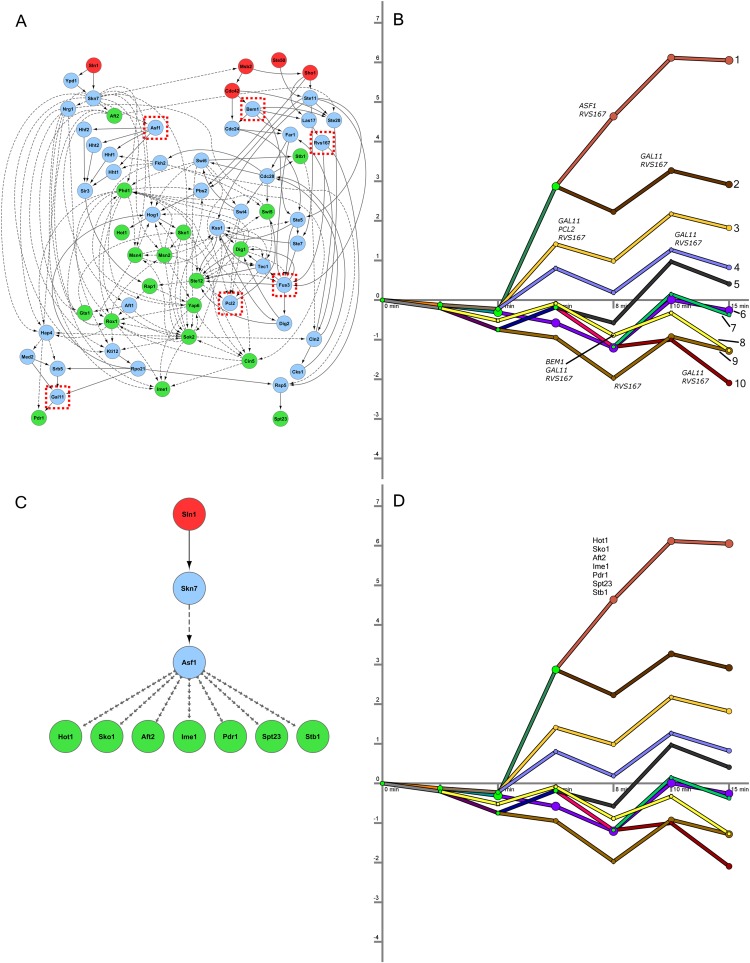
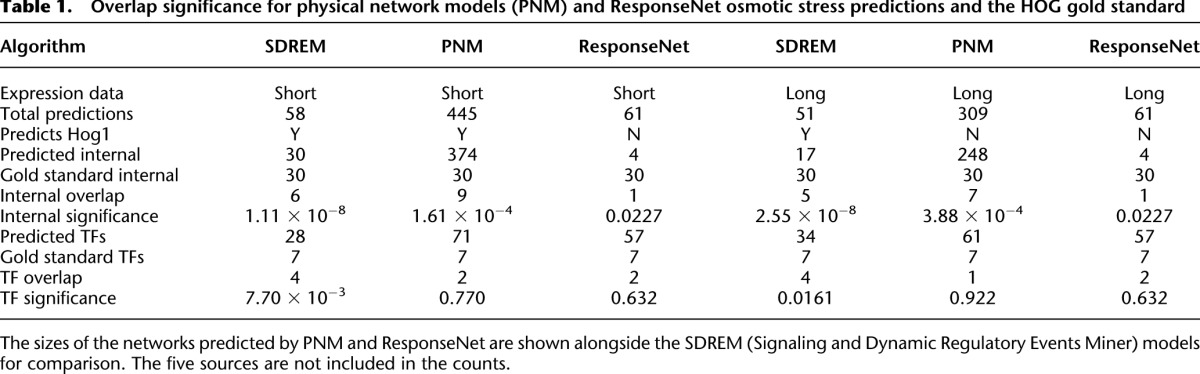
We display the resulting networks in two parts corresponding to the signaling and regulatory components of the reconstructed networks (Supplemental Tables S1, S2). TFs serve as the interface between these two models, and some of the connections between the two components are highlighted in Figure 2. In the regulatory network part of the short model, there are 10 distinct paths controlled by a multitude of TFs (Fig. 2A). Figure 2B presents the high-confidence paths leading from the sources to the targets in the protein interaction network, including the inferred PPI orientation. The model predicts that proteins along these paths play an important role in the osmotic stress response. We emphasize that all targets in this network are the same active TFs that can be found along the short model regulatory paths (Fig. 2A; Supplemental Fig. S1).
Figure 2.
Short osmotic stress model. (A) The regulatory part of the model contains 10 paths in the short time-series data, where each path represents a collection of gene expression profiles. The x-axis displays the time points at which gene expression is measured. The y-axis shows log2 fold change in expression. The nodes following a bifurcation event are annotated with the TFs that are predicted to control the split, providing temporal resolution to the static protein–DNA interaction data. TFs are only shown the first time they are active along a regulatory path. (B) This subset of the oriented interaction network contains three types of nodes: upstream proteins given as sources (red), predicted signaling proteins (blue), and active TFs from DREM (green). The blue nodes consist of all proteins that appear in at least 1% of high-scoring paths and are not sources or targets. Dashed edges are protein–DNA interactions, and solid edges are oriented PPIs. (C) An enlarged view of a subsection of the interaction network identified shows that the core transcriptional unit of the HOG pathway was recovered. These TFs were inferred in the regulatory component of the model, and the network displays SDREM's explanation of how they are activated.
SDREM successfully recovered Hog1's control of Hot1, Msn2, Msn4, and Sko1 (Fig. 2C) as part of the core component of the hyperosmotic response (Capaldi et al. 2008). The nodes and edges immediately upstream of Hog1 (Fig. 2B) are consistent with HOG pathway literature as well. The recovered edges Ste50→Ste11, Sho1→Ste11, Sho1→Pbs2, Ste11→Pbs2, and Pbs2→Hog1 compose the majority of the Sho1 input branch of the HOG pathway (Krantz et al. 2009).
To assess the accuracy of the other predicted target TFs and signaling proteins, it is necessary to consider known HOG pathway models as well as other relevant osmotic and general stress proteins that lie outside the HOG pathway. We compiled a gold standard of established HOG pathway members (Supplemental Table S3; Supplemental Fig. S2) derived from KEGG (Kanehisa and Goto 2000), the Science Signaling Database of Cell Signaling, and recent HOG literature and reviews (Hohmann et al. 2007; Hohmann 2009; Krantz et al. 2009; de Nadal and Posas 2010; Rodríguez-Peña et al. 2010). Four of the seven TFs and six of the 30 other signaling proteins in the gold standard were correctly identified by SDREM (P-values 7.70 × 10−3 and 1.11 × 10−8, respectively), indicating that the HOG pathway composes a significant portion of the short model. To account for other proteins involved in the response, we constructed a set of osmotic stress–related genes by incorporating genetic screens (Hillenmeyer et al. 2008). Many of our predictions that are not present in canonical HOG models are indeed supported by these additional data sources (21 predictions are HOG pathway members or screen hits; P-value 5.88 × 10−3). Searching the literature confirmed additional predictions, and in total, 12 of the 19 TFs (63%) and 27 of the 39 predicted signaling proteins (69%) were found to be associated with osmotic stress (Supplemental Tables S1, S2). SDREM's predictions still significantly overlap with prior osmotic stress models when protein–DNA interactions are excluded from the signaling network (Supplemental Fig. S3).
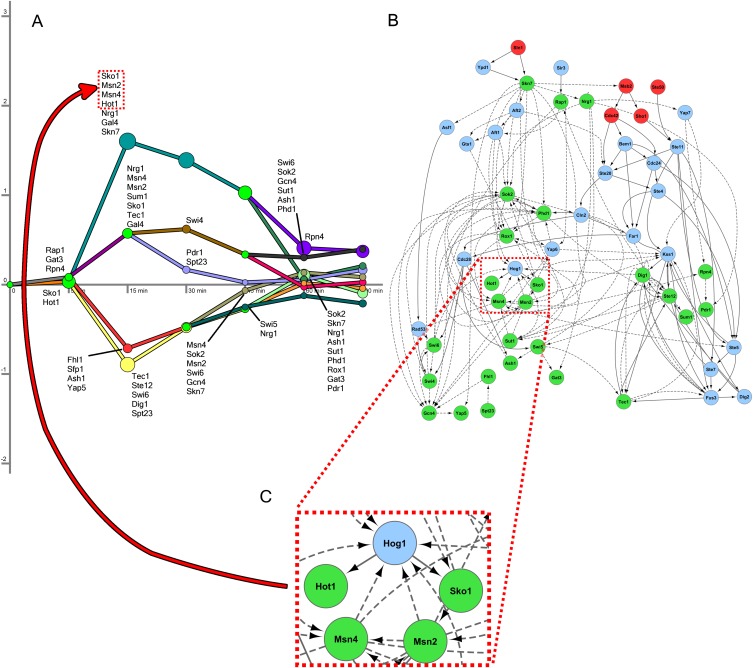
Long osmotic stress model
The model reconstructed from the longer time-series data set is presented in Figure 3. As expected from the fact that it captures the recovery phase and more transcriptional events (Fig. 3A; Supplemental Fig. S4), the long model identified 28 active TFs compared with 19 active TFs in the short model. Many of these additional TFs were determined to be active at the 30- and 45-min time points, indicating their role in restoring gene expression levels to steady-state. In addition, SDREM predicts that Gcn4, Pdr1, Phd1, Sok2, and Swi5—TFs that are only active at the late time points—are activated transcriptionally instead of by signaling cascades (Supplemental Results).
Figure 3.
Long osmotic stress model. (A) The regulatory model for the long osmotic stress expression data contains nine paths. The initial splits overlap with those in the short model in terms of the TFs predicted to control them. (B) The sources, signaling proteins, and active TFs in the long model are shown. Again, there is a large overlap with the signaling model from the short time-series data set. (C) The primary TFs of the osmotic stress response are recovered in the long model as well. Hog1 and Sko1 are shown a second time along the uppermost regulatory path to emphasize the connection between the signaling and regulatory components.
Although the two expression data sets were collected in diverse experimental settings and each contains many unique differentially expressed genes (Supplemental Methods), there was very good agreement between the networks reconstructed by SDREM. Specifically, 16 of the 19 (84%) TFs identified in the short model were also identified in the long model. Four of the long model TFs (P-value 0.0161) and five signaling proteins (P-value 2.55 × 10−8) are HOG pathway members, and 17 predictions are supported by the HOG models or osmotic stress screens (P-value 0.0292). Including the literature, the osmotic stress evidence supports 13 TFs (46%) and 17 signaling proteins (74%) identified in this model (Supplemental Tables S4, S5). In the long model, the network orientation procedure again correctly orients the PPI Hog1-Hot1 and Hog1-Sko1 (Fig. 3B,C). Thus, both models point to SDREM's ability to correctly identify HOG pathway members and osmotic stress responders while at the same time reconstructing the networks by which they are activated.
Validating predicted osmotic stress TFs
Although many proteins in SDREM's reconstructed networks were supported by the HOG gold standard, they also included novel predictions. To validate these predictions, we first focused on TFs that were predicted to regulate either the response (in both models) or the recovery (in the long model). We thus selected four TFs from the short and long models—Cin5, Gcn4, Rox1, and Spt23—that are absent from the HOG gold standard, as well as Hog1 as a control.
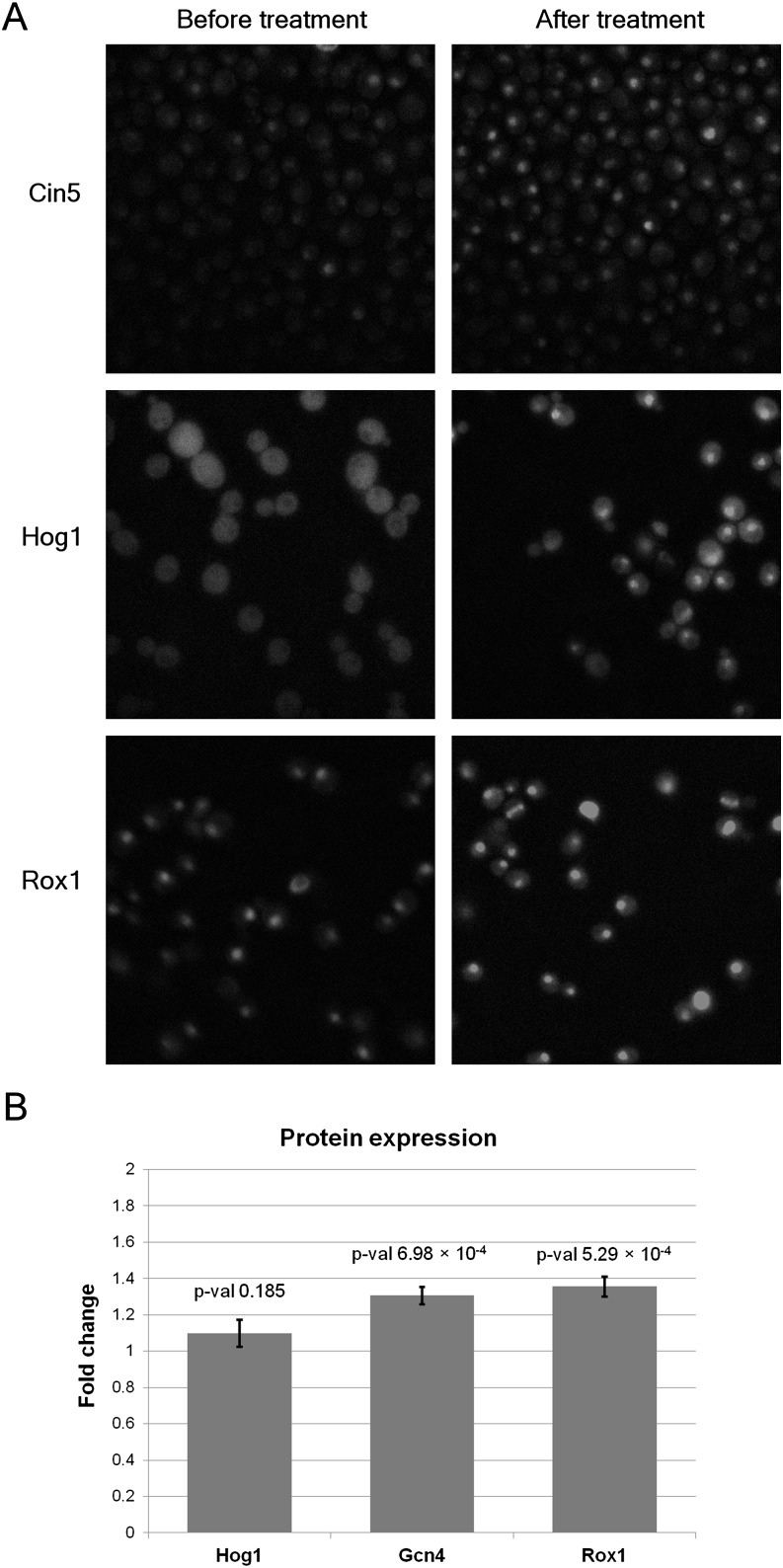
We used fluorescence microscopy to determine whether these proteins were differentially localized following sorbitol treatment at the times predicted by our models. Cin5, Hog1, and Rox1 displayed significant nuclear localization patterns following treatment with sorbitol (P-values of 1.87 × 10−11, 2.67 × 10−7, and 1.02 × 10−15, respectively, using a one-tailed t-test) as predicted by SDREM (Fig. 4A; Supplemental Fig. S5; Supplemental Methods) and in accordance with Hog1's known rapid import into the nucleus in osmotic stress (Hohmann 2009). In contrast, we did not observe a significant change in localization for Gcn4 or Spt23.
Figure 4.
Differential nuclear localization and protein expression after treatment with sorbitol. (A) Each row corresponds to localization of the predicted osmotic stress responder before and after sorbitol treatment. The images were taken 50 min after treatment for Cin5, 21 min for Hog1, and 26 min for Rox1. (B) FACS reveals increased protein levels for Gcn4 and Rox1. The y-axis is the protein level ratio relative to the level before sorbitol treatment. (Error bars) SD of the protein level ratios over all replicates.
In addition to microscopy, we also performed fluorescence-activated cell sorting (FACS) to determine whether protein levels of the four TFs and Hog1 increased following sorbitol treatment. The levels of Gcn4 and Rox1 were found to increase significantly (P-values 6.98 × 10−4 and 5.29 × 10−4, respectively, using a one-tailed t-test) at times consistent with SDREM's predictions (Fig. 4B; Supplemental Results). Hog1, whose protein expression is stable after sorbitol treatment (Westfall et al. 2008), served as a negative control and was not significantly affected (P-value 0.185). In summary, we validated that four of our five predicted osmotic stress–activated regulators (including the control Hog1) are indeed activated following treatment with sorbitol.
KOs support signaling protein predictions
To validate predicted proteins that are not target TFs (which we term signaling proteins regardless of their specific mechanistic function), we used KO expression experiments. Because SDREM produces an oriented network, each signaling protein has a well-defined set of TFs that are downstream from it in the signaling cascades. By comparing the genes predicted to be regulated by these downstream TFs with those affected by the deletion, we can determine whether the KO effects agree with the proposed SDREM models.
We selected six genes that SDREM determined to be involved in separate high-confidence paths: the nucleosome assembly factor ASF1, the cell polarity-related BEM1, the MAPK FUS3, Mediator complex member GAL11, the cyclin PCL2, and the actin-associated RVS167 (Fig. 5A). All six are absent from the HOG gold standard. Microarrays were used to profile wild-type and KO strains treated with sorbitol.
Figure 5.
Knockouts affect downstream expression of genes on the recovered regulatory paths. (A) The six proteins from different regions of the signaling network were selected for deletion. The short network model, reproduced from Figure 2B, is shown here, and the positions of knocked-out genes are highlighted with red boxes. (B) Five knockouts significantly affected the genes assigned to the regulatory paths in the short model. Numbered paths are annotated with the knockouts where we found significant overlap between path members and knockout-affected genes. (C) The subnetwork affected by the ASF1 deletion. Only the relevant subset of the downstream TFs is shown, and the edges connecting Asf1 to the TFs are omitted for clarity. (D) The seven TFs predicted to control path 1's split from path 2 are displayed above path 1. All seven are downstream from Asf1 in the oriented network.
We compared the differentially expressed genes with the short and long models to determine whether the KO-affected genes significantly overlapped the genes assigned to the regulatory paths in the SDREM models. In order to ensure that any observed overlaps could be attributed to the osmotic stress response and recovery as opposed to the general stress response, we analyzed only osmotic stress–specific genes (Supplemental Table S21). For the short model, we found that there was significant overlap (P-value <0.05 using Fisher's exact test with correction for multiple hypothesis testing) for five of the six deletions: ASF1, BEM1, GAL11, PCL2, and RVS167 (Fig. 5B; Supplemental Fig. S6). Seven of the 10 paths in the regulatory network were significantly associated with at least one KO experiment (P-value <10−5 compared with enrichment of random paths) (Supplemental Table S6). Similar results were obtained for the long model, where BEM1, FUS3, GAL11, and RVS167 KOs significantly overlapped one or more regulatory paths (Supplemental Figs. S7, S8). Together, we found significant overlap for all six genes in at least one of the two models, although the support for FUS3 and PCL2 was weaker than the others (Supplemental Table S21).
We highlight the ASF1 KO to explicitly demonstrate how the overlap with the SDREM regulatory paths confirms Asf1's osmotic stress involvement and the inferred network orientation. Asf1 is downstream from the source Sln1 and upstream of numerous TFs in the oriented network, including the crucial HOG pathway TFs Hot1 and Sko1 (Fig. 5C). Our model predicts that ASF1 deletion is likely to partially affect many of these TFs and consequently perturb the genes (and regulatory paths) they control in the osmotic stress response. Indeed, we find that differentially expressed genes in the asf1Δ mutant significantly overlap with regulatory path 1 in the short model (Fig. 5B), and all seven TFs predicted to control this path's split from path 2 (Fig. 5D) are downstream from Asf1 (Fig. 5C), supporting the SDREM model. Similar explanations for the BEM1, GAL11, and RVS167 overlaps are presented in the Supplemental Results. However, in general it is difficult to make any broad claims about the specificity of the KO effects because there are a large number of TFs downstream in the interaction network for some of the deletions and many TFs that are active on each regulatory path (Supplemental Table S21).
Rapamycin response
Although we have primarily focused on the osmotic stress response, we also used SDREM to study the target of rapamycin (TOR) response pathway in yeast using temporal expression data (Urban et al. 2007), condition-specific TF binding interaction for 14 TFs (Methods), and the general PPI and protein–DNA interaction network to demonstrate SDREM's flexibility and generality. Similar to its success in reconstructing the osmotic stress response, SDREM recovers a detailed model of TOR signaling that agrees well with previous experimental and literature evidence (Supplemental Figs. S9, S10). We found support for rapamycin response involvement for 74% of the predicted target TFs and 56% of the internal nodes, and the overlap between SDREM's model and a collection of existing TOR evidence is significant (P-value 2.55 × 10−3). For a detailed discussion of the rapamycin response model, including the full list of predicted proteins and the degree of support for each prediction, see the Supplemental Results and Supplemental Tables S8 and S9.
A. thaliana immune response
Because SDREM requires relatively little condition-specific data, it is readily applicable to higher-order species as well. We modeled the A. thaliana immune response to Hpa infection by combining PPI between Hpa effectors and Arabidopsis proteins (Mukhtar et al. 2011) with the Arabidopsis PPI network (Arabidopsis Interactome Mapping Consortium 2011) and protein–DNA binding interactions (Yilmaz et al. 2011). The Hpa effectors are used as the sources because the downstream transcriptional changes (Wang et al. 2011) are caused by the host's detection and response to these pathogen proteins. Despite the sparse plant interaction network and the lack of a comprehensive gold standard, SDREM was still able to correctly identify several important proteins in the reconstructed networks (Supplemental Figs. S11, S12; Supplemental Table S10). Of the 83 predicted signaling proteins and TF targets, six have already been functionally validated as being relevant to the specific Hpa infection response (P-value 7.29 × 10−8) (Mukhtar et al. 2011) and another seven have been annotated with the Gene Ontology term “defense response” (Supplemental Methods; Ashburner et al. 2000).
Discussion
Approaches for the combined reconstruction of signaling and regulatory networks that depend on KO data are susceptible to the effects of redundancy, which may lead to models that do not accurately represent wild-type behavior. In order to avoid the complications of KOs, we employ condition-specific dynamic gene expression data to infer causal relationships between TFs and differentially expressed genes. Since most active TFs are influenced by the upstream signaling mechanisms that initiate stress response, we link the proteins that sense the environment with the downstream expression outcomes of these stresses. With very limited condition-specific data as input, we were able to accurately reconstruct models for stress responses in yeast and A. thaliana. The models identified both the core regulatory component and many of the signaling cascades that are activated as part of these responses. By use of gold standard data and our own follow-up experiments, we validated several of SDREM's predictions, shedding new light on the proteins and pathways involved in osmotic stress response.
SDREM improves upon previously suggested methods
Neither component of SDREM on its own can accurately recover the osmotic stress response network. By modeling the upstream pathways, the set of TFs that DREM identifies improves substantially from the initial application (when the signaling network is not yet utilized) to the final iteration. In the short model, there were 17 TFs selected by DREM in the first iteration that were dropped in subsequent iterations due to lack of support in the oriented PPI network. Of these only one, Mcm1, is present in the HOG gold standard, and even Mcm1 is considered a HOG pathway member in only one of the seven gold standard sources (Supplemental Table S3). On the other hand, there were eight active TFs in the final model that were missed in the first iteration. These eight TFs include Cin5 and Rox1, TFs for which our experimental results strongly support their role as active regulators in the osmotic stress response. Thus the signaling network context leads to more accurate regulatory models, which in turn provide new targets allowing for better reconstruction of the signaling pathways.
Likewise, although the network orientation algorithm performs very well when given a set of sources and targets, its applicability and utility are greatly reduced if it is limited to conditions in which the target TFs are completely known. In the osmotic stress response, DREM detected active TFs such as Cin5, Gcn4, Nrg1, Rox1, and Yap6 that play a role in the response and recovery but are not included in canonical HOG pathway representations and would not be included in the target set for the network orientation algorithm. In the Arabidopsis immune response, there is even less prior knowledge of which TFs should serve as targets.
We compared SDREM to two other methods for reconstructing pathways; however, neither is intended to directly connect upstream proteins in a signaling network to temporal gene expression changes. PNM (Yeang et al. 2004) infers directed, signed pathways from KOs to affected genes, and ResponseNet (Yeger-Lotem et al. 2009) links genetic screen hits to differentially expressed genes. Thus, comparing them to SDREM requires preprocessing and transforming the input data (Methods). PNM and ResponseNet were only able to identify one or two TFs involved in the HOG pathway, which yielded insignificant overlaps (Table 1). Only SDREM correctly recovered all four core HOG TFs, demonstrating that modeling the dynamic transcriptional response enhances identification of active TFs. PNM recovered Hog1 when run on the short expression data, but ResponseNet omitted Hog1 in both models. Although ResponseNet predicted Hog1 when nondefault settings were used (Supplemental Results), it was still unable to recover a significant portion of the HOG TFs under a variety of settings (Supplemental Table S11).
Table 1.
Overlap significance for physical network models (PNM) and ResponseNet osmotic stress predictions and the HOG gold standard
A popular approach for identifying the important TFs in a dynamic process is to search for time-lagged dependencies in the expression data of regulator-target gene pairs (Schmitt et al. 2004; Balasubramaniyan et al. 2005; Huang et al. 2010; Zoppoli et al. 2010). To emphasize the benefits of incorporating known physical interactions, we compared SDREM's TF predictions to those of GeneReg (Huang et al. 2010), an algorithm that analyzes only the temporal expression data. GeneReg identified TF–gene interactions for 46 differentially expressed TFs in the long osmotic stress data, but at each of three thresholds—top 10 TFs, top 28 (the number of targets SDREM predicts), and all 46—the overlaps between the GeneReg TFs and the HOG gold standard are insignificant (P-values 0.302, 0.249, and 0.194, respectively) (see Supplemental Table S12). GeneReg and all other algorithms that rely on expression alone are inherently limited because they cannot identify TFs that are post-translationally activated and do not exhibit changes in their own expression levels (e.g., Sko1 in this particular response).
Limitations of the learned models
Although SDREM identified the majority of the gold standard proteins, it missed two important HOG pathway proteins, Ssk1 and Ssk2, that are present in all seven gold standard sources. The most likely explanation for their absence is that both proteins have a low degree in the protein interaction network. Consequently, it is unlikely that these proteins will have a large number of source–target paths through them in the directed network, which means that they will have low connectivity scores and will not be recognized as important HOG members. This suggests a possible bias in our technique against low-degree proteins. The gold standard members Ctt1, Glo1, Gpd1, and Msn1 were found to be left out of SDREM's models because they were not in the input interaction networks.
Source–target paths that include a protein–DNA interaction imply that the bound gene is transcribed and translated in response to the environmental perturbation and subsequently affects the next protein on the path. To improve the biological interpretability of SDREM models, future applications could filter the protein–DNA edges in the network to retain only interactions where the bound gene is differentially expressed in the time-series expression data. SDREM could also be extended to automatically predict which TFs are transcriptionally activated. In addition, a coarse level of transcriptional feedback could be modeled by integrating the predicted TF activation times, the times at which genes are differentially expressed, and the protein–DNA edges in the network. However, many nodes in the signaling network are not transcriptionally activated, and learning the full network dynamics would likely require including additional types of data such as condition-specific measurements of phosphorylation dynamics (Olsen et al. 2006).
Extending the algorithm for other species
SDREM's success in reconstructing dynamic response networks in yeast and A. thaliana opens the door to applications in other species, including mammals. Recent studies provide information about inputs to the signaling networks for a number of human infections (Chatr-aryamontri et al. 2009; Fu et al. 2009), and corresponding temporal expression data are also available. Although mammalian PPI networks are larger than the yeast network (making it harder to search for high-scoring pathways), we can incorporate additional data sources, including genome-wide RNA interference screens (Karlas et al. 2010; König et al. 2010), to determine which nodes in the network are relevant to the response.
The ability to reconstruct accurate, dynamic models of cellular response networks helps reveal the components and mechanisms involved in such responses. SDREM is a step in this direction, allowing us to correctly reconstruct regulatory and signaling networks involved in stress response. The method is general and can be used for any species for which PPI and protein–DNA interaction data are available. An open source software implementation of SDREM is available from http://sb.cs.cmu.edu/sdrem.
Methods
DREM: Dynamic Regulatory Events Miner
DREM uses protein–DNA binding interactions and time-series gene expression data to learn which TFs control the differential expression of their bound genes and the time(s) at which they do so. DREM utilizes an IOHMM (Bengio and Frasconi 1995), which unlike traditional HMMs also includes additional observed input data that can influence transition probabilities. In DREM, protein–DNA interactions serve as the static input data that influence transitions between hidden states. An L1-regularized logistic regression classifier is trained at all expression profile bifurcations to assign transition probabilities to genes based on the set of TFs that bind them. DREM searches the state space of possible splits in gene expression profiles to predict a compact set of diverging regulatory paths and the TFs that control them.
Network orientation
The network orientation algorithm directs all undirected edges in a physical interaction network so as to optimally connect sources to target proteins (in this case, TFs from DREM). All reasonably short source–target paths (five or fewer edges here) are assigned a weight 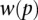 , which is based on the confidence in each PPI along the path. The objective is to maximize the function
, which is based on the confidence in each PPI along the path. The objective is to maximize the function
where P is the set of all short paths between sources and targets, and 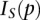 is an indicator function that has the value 1 if path p is satisfied. A path is satisfied if all of its edges are oriented toward the target. The orientation procedure itself involves random restarts of a local search technique, which has been shown to accurately determine edge direction (Gitter et al. 2011).
is an indicator function that has the value 1 if path p is satisfied. A path is satisfied if all of its edges are oriented toward the target. The orientation procedure itself involves random restarts of a local search technique, which has been shown to accurately determine edge direction (Gitter et al. 2011).
Reconstructing signaling and dynamic regulatory networks using SDREM
To link the two methods, we first extended DREM in order to make it suitable for our iterative approach. Originally DREM only accepted either binary input for TF–gene binding interactions or ternary input (−1, 0, 1) if the TFs are known to be activators or repressors. We modified this so that continuous TF activity priors can be accepted. Initially all priors are set to 0.5, but in subsequent iterations, modified priors are derived from the oriented network as described below.
The activity priors influence the transition probabilities in the IOHMM as well as the new activity scores that DREM now calculates. The activity score is calculated for each TF at each bifurcation point in the gene expression profiles. The score at a particular split is the likelihood ratio
where a reflects whether the TF t is active at this split, and Gt is the set of genes bound by the TF that pass through the split. These probabilities are calculated using the activity prior (which is derived from the signaling network component) and the paths followed by Gt out of the split, which tell how well the TF explains the behavior at the split. The final activity score for each TF is the maximum score for the TF across all bifurcation points. We use a randomization procedure to determine the significance of a specific activity score (Supplemental Methods).
We also extended the network orientation algorithm so that in addition to the edge weights, it incorporates the target (TF) weights from DREM. Specifically, the modified path weight used in the objective function for orientation is
where p is a source–target path, t is the target on that path, v is a vertex on the path, and e is an edge on the path. In our current analyses, 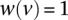 for all nodes in the network. However, the SDREM software supports vertex-specific weights for studies in which the user has prior knowledge that some proteins are more or less likely to be involved in the stress response.
for all nodes in the network. However, the SDREM software supports vertex-specific weights for studies in which the user has prior knowledge that some proteins are more or less likely to be involved in the stress response. 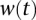 is a normalized version of the activity score from DREM that ranges from 0 to k, the number of edges allowed in a source–target path. For the yeast models presented here, we included protein–DNA interactions in the interaction network, which have a known directionality (TF→gene) and do not need to be oriented algorithmically.
is a normalized version of the activity score from DREM that ranges from 0 to k, the number of edges allowed in a source–target path. For the yeast models presented here, we included protein–DNA interactions in the interaction network, which have a known directionality (TF→gene) and do not need to be oriented algorithmically.
We defined two types of connectivity scores: target scores and node scores. Target connectivity scores are calculated by adding random targets. Each random target has 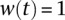 , and the number of random targets is equal to the number of real targets (active TFs from DREM). Satisfied paths are ranked by path weight, and the top 5T paths, where T is the number of real and random targets, are considered high-confidence paths. A target's score is the sum of all path weights of satisfied top-ranked paths that end at that target. These target connectivity scores are averaged over 10 runs for the real targets, and the scores of the random targets in all 10 runs are used to create a target connectivity score distribution. Node connectivity scores are obtained using a separate set of 10 orientations using the real targets. The node connectivity score calculation sums over all satisfied paths that include the node. The percentage of high-confidence paths that contain a particular node is the node's connectivity score.
, and the number of random targets is equal to the number of real targets (active TFs from DREM). Satisfied paths are ranked by path weight, and the top 5T paths, where T is the number of real and random targets, are considered high-confidence paths. A target's score is the sum of all path weights of satisfied top-ranked paths that end at that target. These target connectivity scores are averaged over 10 runs for the real targets, and the scores of the random targets in all 10 runs are used to create a target connectivity score distribution. Node connectivity scores are obtained using a separate set of 10 orientations using the real targets. The node connectivity score calculation sums over all satisfied paths that include the node. The percentage of high-confidence paths that contain a particular node is the node's connectivity score.
Activity priors for the next iteration of DREM are then increased or decreased according to both the target and node connectivity scores. A TF's activity prior is increased if its target score is high (compared with the random distribution) or its node score is high (i.e., it is involved in many high-scoring paths). Any target that does not meet these criteria has its prior halved. The binding priors of all other TFs that were not identified as active targets are not changed. Sensitivity analysis indicates that SDREM is robust to variations in the values of these and other parameters (Supplemental Tables S13–S16; Supplemental Fig. S13).
SDREM settings
For all analyses here, we considered a maximum path length of 5, and SDREM was run for 10 iterations. In practice, the TFs and signaling proteins predicted in each iteration tend to converge upon a stable, well-supported set as the iterations proceed (Supplemental Tables S17, S18). Only binary splits were allowed in the regulatory paths. Genes were filtered if they were missing data for more than one time point or if they were not differentially expressed after exposure to the external stimulus. The default values were used for all other parameters (Supplemental Table S13).
Yeast interaction networks
All protein–protein interaction data were taken from BioGRID (Stark et al. 2006) and weighted as previously described (Gitter et al. 2011). We complemented a general genome-wide ChIP-chip binding data set (MacIsaac et al. 2006) with condition-specific binding data for osmotic stress (Capaldi et al. 2008) and rapamycin (Harbison et al. 2004). For the weighting schemes and additional details, see Supplemental Table S19 and Supplemental Methods.
Osmotic and rapamycin stress data
The osmotic stress response analysis source proteins were Cdc42, Msb2, Sho1, Sln1, and Ste50 (Science Signaling Database of Cell Signaling, http://stke.sciencemag.org/cgi/cm/stkecm;CMP_14620). The osmotic stress validation data sets included the gold standard (Supplemental Table S3), osmotic stress screens (Supplemental Methods), and literature-based evidence (see Supplemental Tables S1, S2, S4, and S5; Supplemental Methods). TORC1 complex members were used as sources in our rapamycin response modeling—Kog1, Lst8, Tco89, Tor1, and Tor2 (Zaman et al. 2008)—and the SDREM predictions were again evaluated using literature models (Supplemental Tables S8, S9) and screens (Supplemental Results; Supplemental Methods). ChIP-chip data from rapamycin-treated cells for 14 TFs (Harbison et al. 2004)—Dal80, Dal81, Dal82, Fhl1, Gat1, Gcn4, Gln3, Gzf3, Hap2, Msn2, Msn4, Rtg1, Rtg3, and Uga3—were merged with the noncondition-specific protein–DNA interactions.
Strains
The S. cerevisiae single KO strains used in this study were taken from the Yeast GFP Library and Yeast Deletion Library. These strains were constructed from a BY4741 background (MATa his3-D1 leu2-D0 met15-D0 ura3-D0).
Microscopy
Cells were grown to logarithmic phase in synthetic complete (SC) medium, washed, and resuspended in SC medium with 1 M sorbitol. Pictures were taken before and after suspension in sorbitol. Images were taken using DeltaVision system package (Applied Precision). ImageJ (http://rsbweb.nih.gov/ij/) was used for all image post-processing and analysis (see Supplemental Methods; Supplemental Fig. S5).
Flow cytometry
FACS analysis was done by BD LSRII system (BD Biosciences). Flow cytometry was conducted with excitation at 488 nm and emission at 525 ± 25 nm for GFP samples. For each protein, we calculated mean protein expression before (pControl) and after sorbitol treatment (pSor) as well as the background before (bgControl) and after treatment (bgSor). Fold change (fc) was then calculated as
P-values were calculated using a paired one-tailed t-test on the log2 protein levels. For additional details, see Supplemental Methods and Supplemental Table S20.
KOs and microarray analysis
The six genes we deleted were selected because they are nonessential, are members of many high-confidence pathways, and are predicted to belong to different levels of the signaling network hierarchy (Supplemental Methods). Cells were grown to logarithmic phase in SC medium (OD = 0.5). The cells were harvested, pelleted, and frozen for further analysis. Sorbitol experiments were similarly performed by growing to logarithmic phase in SC medium and then washing and resuspending in SC medium with 1 M sorbitol for 30 min. Total RNA was extracted using MasterPure yeast RNA purification Kit (Epicentre). The samples were amplified, labeled, hybridized to yeast microarrays (Gene Expression Omnibus platform GPL13340), and scanned using standard Agilent protocols, reagents, and instruments.
To focus specifically on the osmotic stress response, we only analyzed genes that were differentially expressed in the short and long time-series expression data sets, and removed environmental stress response genes (Gasch et al. 2000). Significance analysis of microarrays (Tusher et al. 2001) was used to identify significantly differentially expressed genes. We used the Bonferroni correction to correct for multiple hypothesis testing when calculating regulatory pathway overlaps. For the overlaps between the regulatory paths and KO-affected genes and the lists of osmotic stress–specific genes, see the Supplemental Methods and Supplemental Table S21.
Comparison to benchmark algorithms
PNM was designed for gene KO data; thus, we transformed our data to create a “pseudo-knockout” data set using the time-series expression data. The five HOG pathway sources were given as the deleted genes, and each one was assigned identical transcriptional effects in the form of P-values from EDGE (Leek et al. 2006). For ResponseNet, we created weighted targets where the weight of a gene was its maximum magnitude log2 fold change across all time points. The default parameters were used (gamma = 10 and capping = 0.7). GeneReg identified TF–gene interactions for 46 of the 47 TFs that were differentially expressed. To evaluate the highest-confidence predictions, we ranked these TFs by the strength of their predicted involvement (i.e., the number of genes each TF was predicted to control). For additional details, see the Supplemental Methods.
Network images
All signaling network images were generated using Cytoscape (Shannon et al. 2003). Supplemental Data S1 contains Cytoscape-formatted files that can be used to load and manipulate these networks.
Data access
Gene expression data have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE28213.
Acknowledgments
We thank Jason Ernst for his assistance with DREM as well as Itay Tirosh and Yoav Voichek for their helpful microarray normalization discussions. This work was supported by National Institutes of Health (1RO1 GM085022 to Z.B.-J.) and the National Science Foundation (DBI-0965316 award to Z.B.-J.). In addition, this material is based upon work supported under a National Science Foundation Graduate Research Fellowship to A.G.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138628.112.
References
- Arabidopsis Interactome Mapping Consortium 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: Tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M, Borgs C, Braunstein A, Chayes J, Dagkessamanskaia A, François J-M, Zecchina R 2011. Finding undetected protein associations in cell signaling by belief propagation. Proc Natl Acad Sci 108: 882–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniyan R, Hullermeier E, Weskamp N, Kamper J 2005. Clustering of gene expression data using a local shape-based similarity measure. Bioinformatics 21: 1069–1077 [DOI] [PubMed] [Google Scholar]
- Bengio Y, Frasconi P 1995. An input output HMM architecture. Adv Neural Inf Process Syst 7: 427–434 [Google Scholar]
- Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O'Shea EK 2008. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet 40: 1300–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, Sacco F, Tinti M, Smolyar A, Castagnoli L, Vidal M, et al. 2009. VirusMINT: A viral protein interaction database. Nucleic Acids Res 37: D669–D673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Posas F 2010. Multilayered control of gene expression by stress-activated protein kinases. EMBO J 29: 4–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Vainas O, Harbison CT, Simon I, Bar-Joseph Z 2007. Reconstructing dynamic regulatory maps. Mol Syst Biol 3: 74 doi: 10.1038/msb4100115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Beg QK, Kay KA, Balázsi G, Oltvai ZN, Bar-Joseph Z 2008. A semi-supervised method for predicting transcription factor-gene interactions in Escherichia coli. PLoS Comput Biol 4: e1000044 doi: 10.1371/journal.pcbi.1000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas IJ, Wu C, Chennubhotla C, Bahar I, Oltvai ZN 2006. Topological basis of signal integration in the transcriptional-regulatory network of the yeast, Saccharomyces cerevisiae. BMC Bioinformatics 7: 478 doi: 10.1186/1471-2105-7-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Sanders-Beer BE, Katz KS, Maglott DR, Pruitt KD, Ptak RG 2009. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res 37: D417–D422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Gitter A, Siegfried Z, Klutstein M, Fornes O, Oliva B, Simon I, Bar-Joseph Z 2009. Backup in gene regulatory networks explains differences between binding and knockout results. Mol Syst Biol 5: 276 doi: 10.1038/msb.2009.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitter A, Lu Y, Bar-Joseph Z 2010. Computational methods for analyzing dynamic regulatory networks. Methods Mol Biol 674: 419–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitter A, Klein-Seetharaman J, Gupta A, Bar-Joseph Z 2011. Discovering pathways by orienting edges in protein interaction networks. Nucleic Acids Res 39: e22 doi: 10.1093/nar/gkq1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Hsu H-K, Hsu P-Y, Wu J, Ma Y, Parvin J, Huang Tim, Jin V 2010. Inference of hierarchical regulatory network of estrogen-dependent breast cancer through ChIP-based data. BMC Syst Biol 4: 170 doi: 10.1186/1752-0509-4-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne J-B, Reynolds DB, Yoo J, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St. Onge RP, Tyers M, Koller D, et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S 2009. Control of high osmolarity signalling in the yeast Saccharomyces cerevisiae. FEBS Lett 583: 4025–4029 [DOI] [PubMed] [Google Scholar]
- Hohmann S, Krantz M, Nordlander B 2007. Yeast osmoregulation. Methods Enzymol 428: 29–45 [DOI] [PubMed] [Google Scholar]
- Huang SC, Fraenkel E 2009. Integration of proteomic, transcriptional, and interactome data reveals hidden signaling components. Sci Signal 2: ra40 doi: 10.1126/scisignal.2000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Tao, Liu L, Qian Z, Tu K, Li Y, Xie L 2010. Using GeneReg to construct time delay gene regulatory networks. BMC Res Notes 3: 142 doi: 10.1186/1756-0500-3-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Bar-Even A, Pilpel Y 2005. Transcription control reprogramming in genetic backup circuits. Nat Genet 37: 295–299 [DOI] [PubMed] [Google Scholar]
- Kafri R, Dahan O, Levy J, Pilpel Y 2008. Preferential protection of protein interaction network hubs in yeast: Evolved functionality of genetic redundancy. Proc Natl Acad Sci 105: 1243–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner K-P, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, et al. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463: 818–822 [DOI] [PubMed] [Google Scholar]
- König R, Stertz S, Zhou Y, Inoue A, Hoffmann H-H, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, et al. 2010. Human host factors required for influenza virus replication. Nature 463: 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz M, Ahmadpour D, Ottosson L-G, Warringer J, Waltermann C, Nordlander B, Klipp E, Blomberg A, Hohmann S, Kitano H 2009. Robustness and fragility in the yeast high osmolarity glycerol (HOG) signal-transduction pathway. Mol Syst Biol 5: 281 doi: 10.1038/msb.2009.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Monsen E, Dabney AR, Storey JD 2006. EDGE: extraction and analysis of differential gene expression. Bioinformatics 22: 507–508 [DOI] [PubMed] [Google Scholar]
- Lin L-H, Lee H-C, Li W-H, Chen B-S 2007. A systematic approach to detecting transcription factors in response to environmental stresses. BMC Bioinformatics 8: 473 doi: 10.1186/1471-2105-8-473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac K, Wang T, Gordon DB, Gifford D, Stormo GD, Fraenkel E 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113 doi: 10.1186/1471-2105-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis A-R, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. 2011. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Ourfali O, Shlomi T, Ideker T, Ruppin E, Sharan R 2007. SPINE: a framework for signaling-regulatory pathway inference from cause-effect experiments. Bioinformatics 23: i359–i366 [DOI] [PubMed] [Google Scholar]
- Peleg T, Yosef N, Ruppin E, Sharan R 2010. Network-free inference of knockout effects in yeast. PLoS Comput Biol 6: e1000635 doi: 10.1371/journal.pcbi.1000635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña JM, García R, Nombela C, Arroyo J 2010. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes. Yeast 27: 495–502 [DOI] [PubMed] [Google Scholar]
- Romero-Santacreu L, Moreno J, Pérez-Ortín JE, Alepuz P 2009. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15: 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt WA, Raab RM, Stephanopoulos G 2004. Elucidation of gene interaction networks through time-lagged correlation analysis of transcriptional data. Genome Res 14: 1654–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M 2006. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34: D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26: 663–674 [DOI] [PubMed] [Google Scholar]
- Vinayagam A, Stelzl U, Foulle R, Plassmann S, Zenkner M, Timm J, Assmus HE, Andrade-Navarro MA, Wanker EE 2011. A directed protein interaction network for investigating intracellular signal transduction. Sci Signal 4: rs8. [DOI] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li Hairi, Tör M, Caldelari D, Lee D, Fu X-D, Dong X 2011. Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Li Hongzhe 2008. A hidden spatial-temporal Markov random field model for network-based analysis of time course gene expression data. Ann Appl Stat 2: 408–429 [Google Scholar]
- Westfall PJ, Patterson JC, Chen RE, Thorner J 2008. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci 105: 12212–12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeang C-H, Ideker T, Jaakkola T 2004. Physical network models. J Comput Biol 11: 243–262 [DOI] [PubMed] [Google Scholar]
- Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, et al. 2009. Bridging high-throughput genetic and transcriptional data reveals cellular responses to α-synuclein toxicity. Nat Genet 41: 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E 2011. AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Ungar L, Zalckvar E, Kimchi A, Kupiec M, Ruppin E, Sharan R 2009. Toward accurate reconstruction of functional protein networks. Mol Syst Biol 5: 248 doi: 10.1038/msb.2009.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR 2008. How Saccharomyces responds to nutrients. Annu Rev Genet 42: 27–81 [DOI] [PubMed] [Google Scholar]
- Zoppoli P, Morganella S, Ceccarelli M 2010. TimeDelay-ARACNE: Reverse engineering of gene networks from time-course data by an information theoretic approach. BMC Bioinformatics 11: 154 doi: 10.1186/1471-2105-11-154 [DOI] [PMC free article] [PubMed] [Google Scholar]