Abstract
The foot-and-mouth-disease virus (FMDV) utilizes non-canonical translation initiation for viral protein synthesis, by forming a specific RNA structure called internal ribosome entry site (IRES). Domain 3 in FMDV IRES is phylogenetically conserved and highly structured; it contains four-way junctions where intramolecular RNA–RNA interactions serve as a scaffold for the RNA to fold for efficient IRES activity. Although the 3D structure of domain 3 is crucial to exploring and deciphering the initiation mechanism of translation, little is known. Here, we employ a combination of various modeling approaches to propose candidate tertiary structures for the apical region of domain 3, thought to be crucial for IRES function. We begin by modeling junction topology candidates and build atomic 3D models consistent with available experimental data. We then investigate each of the four candidate 3D structures by molecular dynamics simulations to determine the most energetically favorable configurations and to analyze specific tertiary interactions. Only one model emerges as viable containing not only the specific binding site for the GNRA tetraloop but also helical arrangements which enhance the stability of domain 3. These collective findings, together with available experimental data, suggest a plausible theoretical tertiary structure of the apical region in FMDV IRES domain 3.
INTRODUCTION
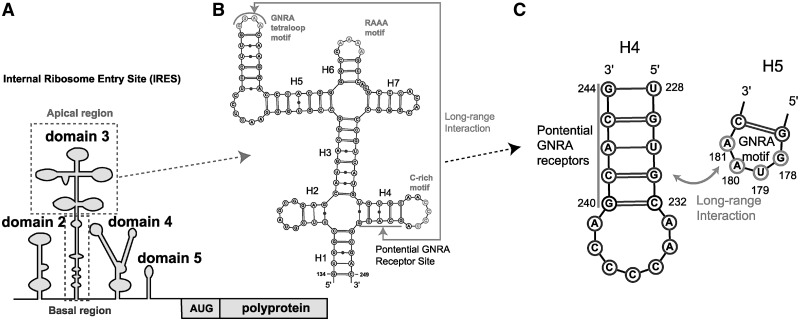
The foot-and-mouth-disease virus (FMDV) belonging to the picornavirus family is the contagious agent of foot-and-mouth-disease, a severe plague for animal farming. The viral replication of FMDV begins with a translation initiation by forming a specific RNA structure called internal ribosome entry site (IRES). FMDV IRES consists of ∼450 nt and can fold in multiple stem–loops organized in five domains (Figure 1A). These domains can host binding proteins such as eukaryotic initiation factors (eIFs) and IRES transacting factors (ITAFs), which play crucial roles in IRES-directed translation (1–7). Among these, the third domain is the largest and contains structural elements critical for IRES activity (8).
Figure 1.
Global organization of FMDV IRES and a secondary structure of truncated domain 3 including conserved RNA motifs for RNA–RNA long-range interactions. (A) Schematic representation of the viral genome organization including four subdomains of FMDV IRES. (B) Secondary structure (deduced from RNA structure probing experiment) of the truncated FMDV IRES domain 3 which consists of a pair of four-way junctions and is a self-folding region containing conserved GNRA and RAAA motif at the apical region for RNA–RNA long-range interactions. (C) Potential long-range interactions between helices H4 and H5. G240CACG244 residue in helix H4 is a potential receptor site of the G178UAA181 tetraloop.
Domain 3 consists of basal and apical regions (Figure 1A). The basal region consists of a long internal loop, and the apical region contains multiple four-way junctions. Recent biochemical data have suggested that it is the apical region that contributes significantly to the structural organization and stability of domain 3, as well as to the critical function of IRES activity (8,9, 10,11).
Specifically, the apical region of domain 3 includes two conserved motifs, GNRA and RAAA motif (8). The GNRA (N is any nucleotides; R is A or G) tetraloop motif is common in folded RNA (12); the loop–helix interactions combine base pairing and stacking to define a tertiary contact that stabilizes the global fold of an RNA molecule. In the IRES domain 3 (Figure 1), the GNRA motif is situated at the apex of a stem–loop motif (13,14). Biochemical studies demonstrate that this motif is critical for IRES function (8,15). RNA probing experimental data further show that the RAAA motif also contributes to enhance IRES activity via RNA–RNA long-range tertiary contacts, but only in the presence of GUAA tetraloop–receptor long-range interactions (9).
Deciphering the contribution of domain 3 to IRES-driven translation has been challenging. Based on the potential capacity for inter and intramolecular RNA–RNA interactions, it has been proposed that this domain stabilizes the entire IRES element (9,16). More recently, structural analysis based on SHAPE probing and microarray data confirmed domain 3’s role in the organization of other domains (10,11).
The GNRA motif in helix H5, along with its potential distal binding region in helix H4 for intramolecular RNA–RNA interactions, is located in the four-way RNA junctions of FMDV IRES domain 3 (Figure 1). RNA junctions in general provide a hub for different double-stranded helical arms to come together (17). Thus, junctions occur in many RNAs, including, for example, the hepatitis C virus IRES for the translation initiation (18). Because the global conformation of a RNA is thought to be largely determined by topological constraints encoded at the secondary structure level (19), an understanding of the 3D structural aspects of RNA junctions in IRES’s domain 3 is essential to decipher the mechanism of IRES-driven translation.
Among recurrent structural elements (or ‘motifs’) common to RNA junctions, coaxial stacking is prominent. Coaxial stacking between two continuous helices stabilized by base stacking in a shared single strand (16) is a major determinant of three or higher-order junctions of RNA. Recent studies of RNA junctions have identified structural patterns in coaxial stacking that define different RNA junction family types (20–22). These classifications also link the RNA junction family type to nucleotide length in a single strand; fewer nucleotides in the single strand between two helices increase the probability of forming coaxial stacking. Although other factors such as protein binding can alter these noted patterns of coaxial stacking arrangements, the above correlation holds in general particularly for self-folding RNA molecules, including transfer RNA (tRNA) (23), P4–P6 domain of the Tetrahymena group I ribozyme (24), hepatitis C virus IRES (18) and FMDV IRES domain 3 (8). The coaxial stacking motif often cooperates with other tertiary motifs including A-minor and loop–helix interaction to enhance the stability of RNAs.
Currently, computational programs cannot predict multiple RNA junction structures well, though there are many useful 3D prediction programs as recently surveyed (25,26). Very recently, our RNA junction structure prediction program Junction-Explorer, based on data mining and bioinformatics, was shown to predict the topology of individual RNA junction domains with ∼70% or more prediction accuracy (27). Here, to construct plausible structures for the two consecutive four-way junctions in the apical region of IRES domain 3, we devise a divide-and-conquer approach that combines various effective computational techniques.
We began with the IRES secondary structure determined by RNA probing (13,28). Considering RNA–RNA long-range interactions involving the GNRA motif, we partitioned RNA into subsystems and then modeled each RNA junction topology on the basis of knowledge from four-way RNA junction classification coupled with Junction-Explorer. Further analysis produced four viable candidates for 3D models constructed using MC-Sym (29).
Subjecting these four candidate models to molecular dynamics (MD) simulations allowed identification of the most energetically favorable and stable conformational states in the presence of the GNRA tetraloop–receptor long-range interactions. The dynamics data also suggested specific tertiary interactions and helical rearrangements. Only one model emerged as viable, revealing not only the specific binding site for the GNRA tetraloop but also helical junction arrangements that enhance the stability of domain 3 further. We propose this structure, compatible with available experimental data, as a feasible tertiary structure for the apical region in FMDV IRES domain 3.
MATERIALS AND METHODS
RNA target structure
Domain 3 of FMDV IRES is a self-folding RNA that is 214 nt long. We consider the sequence of the FMDV C-S8 IRES and model the apical and basal region separately; the apical region contains two consecutive four-way junction structures, which consists of 116 nt (G134 to C249) and the basal region is a long internal loop containing 98 nt (G86 to U133 and C249 to C299).
RNA sequence alignments and conservation analysis
To assess the significance of the structural key elements involved in long-range RNA–RNA interactions in domain 3, we perform sequence alignments of many IRES sequences and analyze the four-way junctions, focusing on the sequence conservation of the GNRA loop and its binding receptors. Three hundred eighteen FMDV IRES sequences are collected from the GenBank database (30) using the standard Nucleotide Blast webserver with a query sequence of the FMDV C-S8 IRES. Incomplete and identical sequences we removed, and the remaining 318 sequences were aligned using the ClustalW program (31). See Supplementary Tables S2 and S3 for the list of FMDV IRES sequences and aligned sequences.
We use sequence logos to analyze patterns in aligned RNA sequences. The RNA sequence logos consist of stack of four letters (measured in 2 bits)—A, U, G and C—at each position in a sequence. While the overall height of the stack indicates a degree of sequence conservation, the height of each letter within the stack shows a relative frequency at each position. The logos are generated using the RNALogo webserver (32). See Supplementary Figure S1 for the RNA sequence logos of the four-way junctions.
Multiple RNA junction topology modeling for the apical region
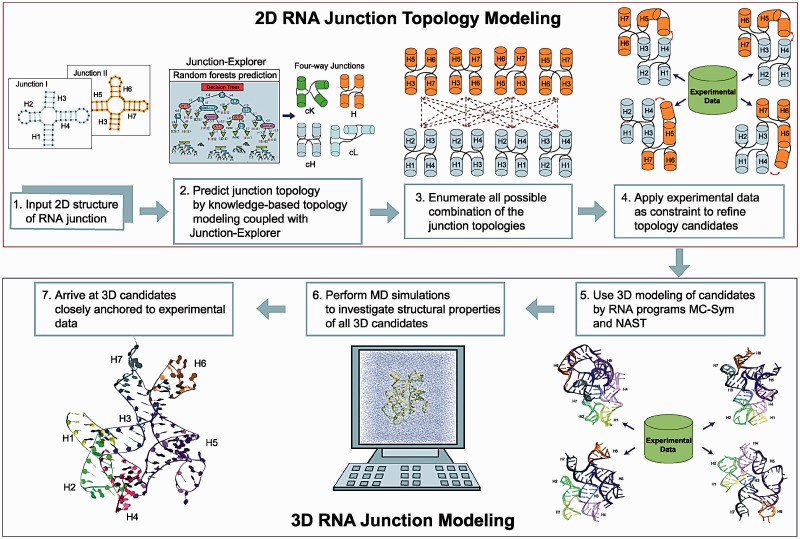
To tackle multiple consecutive four-way RNA junctions, we use a divide-and-conquer approach by partitioning the large complex. Each four-way junction is analyzed with regards to the loop size of single strands between helices; this analysis is coupled to the Junction-Explorer program to help determine coaxial stacking patterns and helical arrangements. Junction-Explorer is based on the random forests data mining algorithm (33) and uses various geometric and energetic parameters as ‘feature vectors’ (which contain information on free energies, loop sizes between junctions and adenine content) for training. Using the predicted topology for each four-way junction, we search for all possible combinations of the multiple four-way junctions to produce combined structures. These potential topologies for the secondary structures are then refined further by incorporating experimental data as constraints (Figure 2).
Figure 2.
Computational procedure for modeling multiple four-way RNA junction structures. (1) Multiple four-way RNA junction structures are separated into individual RNA junction as input. (2) Each junction is analyzed for 2D helical arrangements based on coaxial stacking and junction family type in conjunction with Junction-Explorer. (3) These topologies are processed to enumerate all possible junction combinations. (4) Available experimental data are applied as constraints to refine the topology candidates. (5) 3D models based on the topology candidates are developed using computational programs. (6) MD simulations are performed to arrive at (7) potential 3D structure.
3D modeling of multiple RNA junction structures in the apical region
Using state-of-the-art 3D modeling programs, we build RNA 3D models of FMDV IRES domain 3 combined with experimental data. We primarily use MC-Sym, which utilizes a fragment-based library to obtain all possible structures of RNA junctions (29). To complement the modeling results, we explore conformational space using NAST, a knowledge-based coarse-grained simulation tool (34); these two programs have been shown to perform well in predicting native RNA structures (26). We hypothesize that fewer nucleotides between helices should naturally restrict the orientational flexibility at some degree yielding coaxially stacked helices. Thus, we first model Junctions I and II following the 5′- to 3′-direction without constraints for coaxial stacking arrangement. This yields thousands of structures for each junction. Because helical elements in RNA junctions tend to form coplanar arrangements (18), we ranked the predicted structures for coplanarity and collected the best 1000 structures. These junctions are then assembled by imposing a distance constraint for potential long-range interactions from experimental data (see ‘Modeling atomic junction structures for the apical region’ in ‘Results’ section for more details).
Using NAST, the Nucleic Acid Simulation Tool, we performed a coarse-grained MD simulation for 40 ns (10 × 106 time step) with one tertiary contact between A180 and C232/G240. For the 10 000 coarse-grained templates generated, we filter the templates by a potential energy with the cutoff energy of 1000 kJ.
Molecular dynamics simulations for the apical region
Each system was solvated with the explicit TIP3P water model in a water box of dimension 10 Å on each side. Simulations were performed using the Amber Parmbsc0 force field (35,36) with sodium ions to neutralize the system charge.
We minimize the system in two steps, first over the water and ion molecules holding domain 3 fixed and, second, with all constraints removed. The minimization was performed using the Powell conjugate gradient algorithm. The initial equilibration was achieved over 60 ps at constant temperature (300 K) and pressure (1 atm). Pressure was maintained at 1 atm using the Langevin piston method, with a piston period of 100 fs, damping constant of 50 fs and piston temperature of 300 K. Temperature coupling was enforced by velocity reassignment every 2 ps. Both minimization and equilibration are performed using the NAMD program (37).
For the production run, we simulated a conventional MD trajectory for 100 ns with the Parmbsc0 force field using the NAMD package. The system was simulated at constant temperature (300 K) and volume using weakly coupled Langevin dynamics of non-hydrogen atoms, with a damping coefficient of c = 10 ps−1 with a 2 fs time step maintaining bonds to all hydrogen atoms rigid. Non-bonded interactions are truncated at 12 and 14 Å for van der Waals and electrostatic forces, respectively. Periodic boundary conditions are applied, and the particle mesh Ewald method is used to calculate electrostatic interactions.
All simulations using the NAMD package were run on IBM Blue Gene/L supercomputer at the Computational Center for Nanotechnology Innovations based in Rensselaer Polytechnic Institute, NY.
Stability of the simulated structures for the apical region
Our simulated structures contain 116 nt, of which 82 involve base pairs. To justify the stability of simulations maintaining secondary structure in trajectories, we computed average distance of base-paired residues in helices (5 bp in H1, 3 bp in H2, 8 bp in H3, 5 bp in H4, 13 bp in H5, 3 bp in H6 and 4 bp in H7) and measured RMS deviations (RMSDs) considering all residues and only base pairs with reference to the starting structure as we hypothesize unpaired residues contribute to increase RMSD.
The average distances of all helices in our simulations are <3 Å and most of them are <2.5 Å (Supplementary Figure S2A). In addition, RMSD values (considering all except hydrogen atoms) are for the entire system (116 residues) including the paired 82 residues indicating overall stability (Supplementary Figure S2B).
Very recently, problems in χ torsion angles of MD simulations for RNA systems have been reported (namely, ladder-like structures that loose the helical twist of A-form RNA conformation, especially in long RNA MD simulations) (38,39), and improved force fields have been introduced (40,41). We have carefully monitored these potential problems but have not observed in our dynamics data.
Entire sequence modeling including the basal region
We model the basal region containing 98 nt (G86 to U133, C249 to C299) based on 2D information of FMDV C-S8 IRES domain 3 using MC-Sym. Evaluating all the 717 structures based on RMSD and clustering analysis yields four candidate models that were chosen from the first four large clusters containing at least 10 structures (Supplementary Figures S3–S4). Since overall shapes of these four candidates were similar, we chose a representative model from the largest cluster (Supplementary Figure S4A) to build a complete 3D model of domain 3. Structures of the apical and basal region were merged using a python library of modeRNA (42). Both minimization and equilibration were performed on the entire domain 3 following the protocol in the ‘Molecular dynamics simulation for the apical region’ section above.
RESULTS
Sequence conservation analysis of the apical region
Sequence similarity provides evidence for structural conservation and hence essential biological function. Sequence logos of the aligned 318 sequences of FMDV IRES systems suggest that the apical region is largely conserved (Supplementary Figure S1), implying that its 2D structure is constrained under an evolutionary pressure to carry an important biological function for non-canonical IRES-mediated translation initiation. In particular, conservation of the potential binding receptors (G229 to C232 and G240 to C243) of the GNRA loop is near perfect (316 out of 318 sequences); the sequence logos of H4 marked in the red box (Supplementary Figure S1) indicate that the entire hairpin including the binding nucleotides are conserved almost fully. We also observe that the GUAA sequence appears most frequently with 233 instances (73.2%) followed by GUGA (17%), GCAA (7%), GCGA (2.5%) and GAGA (0.3%) (Supplementary Table S1).
Multiple four-way junction topology prediction at secondary structure level for the apical region
Initial guess of each four-way junction topology
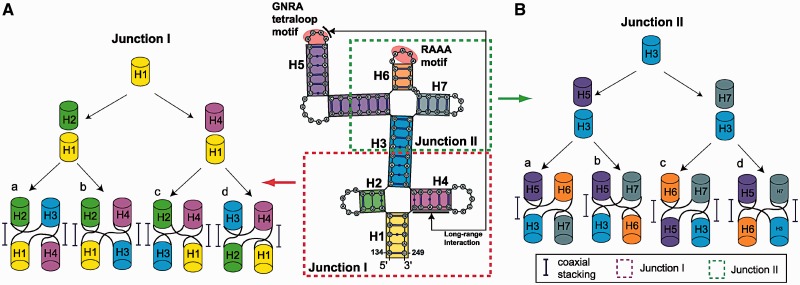
We partition domain 3’s 2D structure into two four-way junctions and list all possible junction topologies. We denote the two four-way junctions as Junctions I and II following the 5′- to 3′-direction (Figure 3). As loop size dictates orientation and flexibility of helices in RNA junctions (21,22), we build candidate topological models accordingly. Because very few nucleotides are present between helices in both junctions, we consider two coaxial stacking patterns, parallel to each other with a possible crossing at the point of single-strand exchange.
Figure 3.
Piecing the possible helical arrangements for the two four-way junctions in domain 3 of IRES. (A) Possible junction topologies of junction I with two coaxial stacking following the 5′- to 3′-direction: (a) H1H4 and H2H3 without crossing in the single-stranded region, (b) H1H4 and H2H3 with crossing, (c) H1H2 and H3H4 with crossing and (d) H1H2 and H3H4 without crossing. (B) Predicted junction topologies of junction II with one or two coaxial stacking following the 5′- to 3′-direction: (a) H3H5 and H6H7 without crossing, (b) H3H5 and H7H6 with crossing, (c) H5H6 and H7H3 without crossing and (d) H6H5 and H7H3 with crossing.
For Junction I, two types of pairwise coaxial stacking patterns are likely (because no nucleotides are present between helices). Helix H1 can coaxially stack with either H2 or H4 (Figure 3A). This results in two coaxial stacking: H1H2 with H3H4 or H1H4 with H2H3, as shown in the figure.
Similarly, for Junction II we consider H3H5 with H6H7 or H3H7 with H5H6 (Figure 3B). However, we speculate that the latter pattern is more likely due to the presence of 2 nt in a single-strand loop between H6 and H7, whereas no nucleotides are in other single strands between coaxially stacked helices H3H7, H5H6 and H3H5 (see enlarged view in the middle of Figure 3); a strong preference for coaxial stacking has been observed with a smaller loop size (21,22,43). The Junction-Explorer program also predicts a pair of coaxial stacking formation for both four-way junctions, parallel to each other. On the basis of these combined models, we arrive at four candidate helical arrangements for each junction (Figure 3) that correspond to H and cH family types containing two coaxially stacked helices (see Supplementary Figure S5 for the nine major junction family types) based on our four-way junction classification study (21). Note that only three four-way junction families H, cH and cL contain two coaxially stacked helices and are distinguished by the angle between the two stacked helices with roughly 0°, 180° and 90°, respectively. To achieve a particular configuration for each family, different lengths of single strands between stacked helices are required. While the shape of family H and cH can be achieved with a relatively short single strand, family cL requires a long single strand. Because the junctions in domain 3 contain 2 nt in single-stranded regions at most, we do not consider the cL family as a candidate. Next, we consider these combinations of configurations compatible with experiment.
Full junction topology prediction
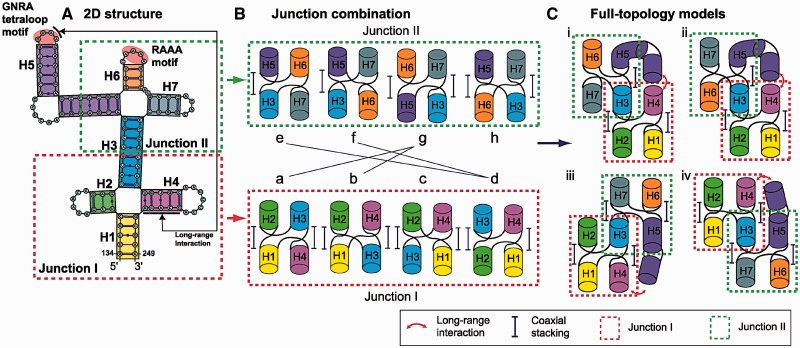
Considering the nine major junction family types in four-way RNA junctions (21), the number of ways to pair two four-way junctions is (9 family types × 2 different helical arrangements)2 = 324 when no other information is considered. Using the two possible family types for each junction (Figure 3A and B), the number of likely conformations becomes (2 family types × 2 different helical arrangements)2 = 16 (Figure 4B). We further consider the potential RNA–RNA long-range interactions between the GNRA motif and its distal receptor region from experiment (9) to eliminate some of these 16.
Figure 4.
Four candidate models derived from combinations of two four-way Junctions I and II of Figure 3. (A) Secondary structure of domain 3 in FMDV IRES. (B) Combinations of the two four-way junctions considered. To accommodate the long-range interactions between helices H4 and H5, helix H3 must be either parallel or perpendicular to the helices H4 and H5 in space. Two arrangements, c and h, of junctions I and II do not satisfy the long-range interactions and are thus eliminated. (C) Four complete junction topology models where Junctions I (dotted red box) and II (dotted green box) are stitched via helix H3 considering the GNRA tetraloop long-range interaction between H4 and H5.
Given that the GNRA tetraloop and its potential receptors are located in helices H4 and H5 (Figure 4A), we can eliminate some helical arrangements. To make the long-range interactions possible, both H4 and H5 are required, in positions either parallel or perpendicular with respect to H3. Thus, the two configurations c and h in Figure 4B can be eliminated because the bridging helix H3 between Junction I and II is diagonal to H4 and H5 [see the helical arrangements in third row (for c) and fourth column (for h) in Supplementary Figure S6]; these two models are not eligible to make tertiary contacts between H4 and H5 due to either the orientation of these two helices that are opposite one another [(c, e), (c, f), (c, h), (a, h) and (b, h) in Supplementary Figure S6] or some steric clashes [(c, g) and (d, h) in Supplementary Figure S6]. In addition, the five pairs of helical arrangements (a, e), (a, f), (b, e), (b, f) and (d, g) in Figure 4B can also be excluded because helices H3, H4 and H5 are aligned in the same direction (either to parallel or perpendicular). Four viable models remain (Figure 4C); the junctions in the final topology models correspond to either H or cH family types, and thus H/H or H/cH combinations are possible overall. See Supplementary Figure S6 for all 16 combinations elaborated from Figure 4B.
Modeling atomic junction structures for the apical region
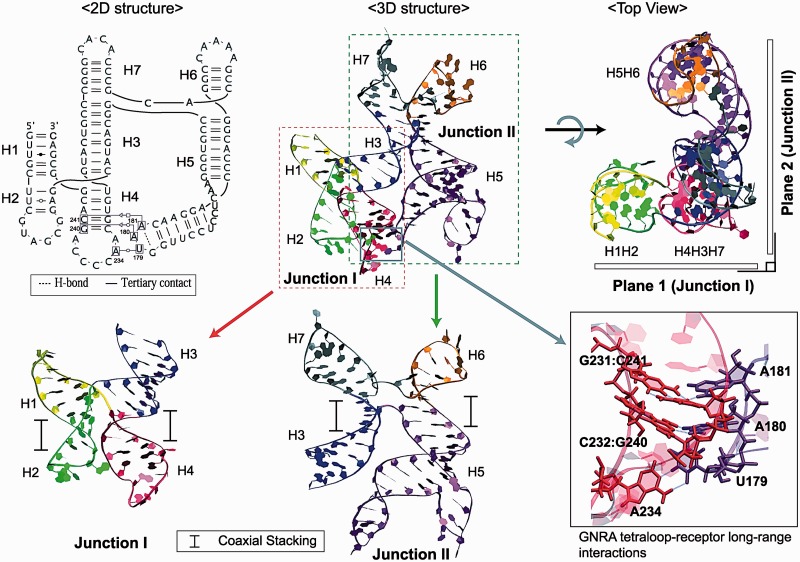
Mutational analysis proposed a non-specific receptor site, G240CACG244 in H4 of Junction I, for the G178UAA181 tetraloop of H5 in Junction II (9). The two adenosines in the GUAA tetraloop prefer to interact with a pair of C/G base pairs or alternatively a combination of C/G and G/C base pairs. In the potential receptor site, we identify a combination of C232/G240 and G231/C241 base pairs that was reported as receptors of GUAA loop by an in vitro selection experiment (44). Note that the C232/G240 pair is highly conserved in 130 FMDV sequences while the G231/C241 pair is invariant (45) and thus probably significant for correct RNA folding.
Because the energetics of tertiary interactions have not yet been considered, at this stage we model the RNA–RNA long-range interactions by imposing a loose distance constraint of 10 Å between helices H4 and H5, specifically between A180 and C232G240 using C1′ atoms.
Sampling these constrained models using MC-Sym reduces the number of models to 267: in Junction I, 160 of these contain stacked helices of H1H2 with H3H4 while the remaining 107 structures contain stacking of H1H4 with H2H3. These numbers may reflect the preference of RNA’s helical arrangements in Junction I. Until now, the coaxial stacking pattern—H1H2 and H3H4—appears to dominate the possibilities when long-range interactions are considered. After evaluating all the 267 structures by structural similarity based on RMSD, clustering analysis and visual inspection (Supplementary Figures S7 and S8), we arrive at the consensus with initial junction topology models (Figure 5). Some variations in H6 and H7 may occur due to flexibility introduced by two unpaired nucleotides in the single-stranded region.
Figure 5.
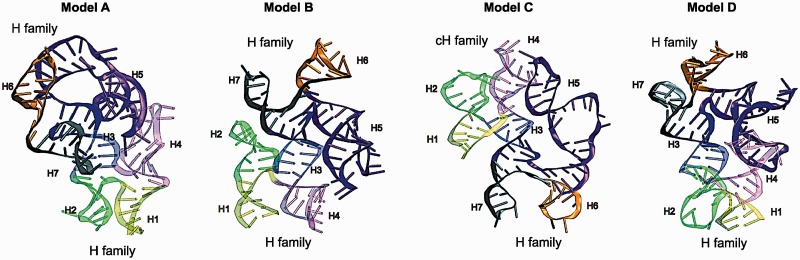
Four candidate 3D models of the FMDV IRES domain 3. Models A, B and C correspond to our junction topology models (Figure 4C), whereas Model D is new. All structures have two coaxial stacking both in Junctions I and II. A combination of H/H or H/cH, but not cH/cH family types is observed in the junctions.
Similarly, when we consider another invariant—G229/C243 base pair—near the junction core for tertiary interactions, we obtain 52 viable structures; about one-fifth of the structures targeting the C232/G240 base pair. We speculate that the relatively small number of sampled structures targeting near the junction core may explain unfavorable potential binding receptor. In fact, the representative 3D structures show that helical arrangements in Junction II are rather distorted than structured (Supplementary Figures S9–S11).
We also arrive at a new candidate model, a modified combination of the junction topologies in Figure 4B(d) and (g) predicted by MC-Sym [see Supplementary Figure S6(d, g)*]. Although excluded while modeling junction topologies, this model may be possible due to a versatile nature of RNA molecules.
The four resulting candidate 3D models are shown in Figure 5. A, B and C correspond to the junction topology models in Figure 4C(i), (iii) and (iv), respectively, while D is a new 3D model. Note that we select four 3D models from six clusters (Supplementary Figure S9) considering similarity of overall helical arrangement. All structures have two coaxial stacking in Junctions I and II. Interestingly, the 3D model corresponding to the topology model in Figure 4C (ii) is not predicted by MC-Sym; this model, different from the three other topology models, has a crossing at the point of single-strand exchange in Junction II. We speculate that this particular helical arrangement in Junction II makes it difficult to satisfy the distance constraint criteria in 3D space.
We also explored different RNA conformations by simulating models over 40 ns by one-bead coarse-grained MD simulations using NAST. These simulations yield three representative conformations where helical arrangements are identical in Junction I, but some variations in Junction II (data not shown). Overall, these simulations lead further support to the models in Figure 5B and C.
Assessment of structural properties of the apical region using MD simulations
We use MD to supplement the structural studies above and to further explore the feasibility of our structural candidates. Despite algorithmic approximations as well as force field imperfections, MD is widely used to provide further insights into atomic-level interactions and energetic aspects that are not readily revealed from other techniques (46). Hence, we perform 100 ns MD simulations for all four candidate structures (Models A–D) in Figure 5; specifically to investigate structure stability and potential long-range interactions suggested by experimental data.
Long-range interactions including a novel tertiary contact revealed by MD simulation
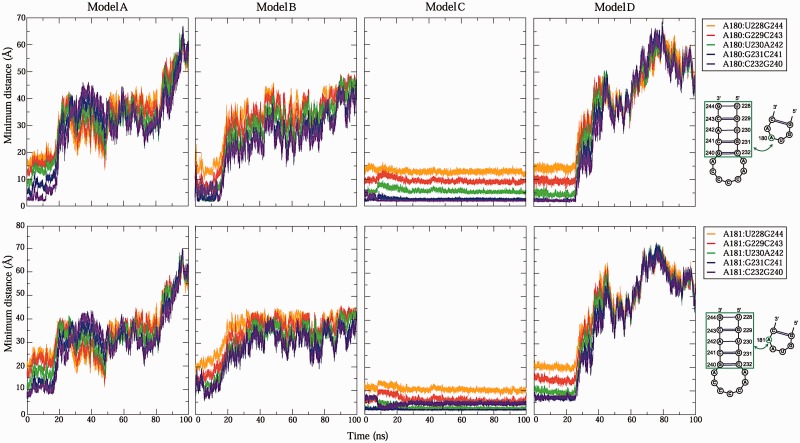
Experimental data have proposed intramolecular long-range interactions in domain 3 of FMDV IRES (9,13). Although this tertiary contact is required for efficient IRES activity, the specific binding receptor of GNRA tetraloop is yet unknown. To explore this, we track for each trajectory the distances between the GUAA hairpin in H5 and each of potential target receptors in H4, specifically between A180A181 and G240CACG244 (including their complementary residues). Only for Model C we detected two receptor candidates—G231/C241 and C232/G240 base pairs—interacting with A180A181 residues in GUAA tetraloop (Figure 6). The trajectory for Model C shows that the two adenosines retain a distance <3 Å. In contrast, only the first adenosine A180 of Models A, B and D retain a distance <4 Å during the initial 12, 15 and 26 ns, respectively. In Model C, the average distance between C232/G240 pair and A180 is 2.1 ± 0.59 Å while C231/G241 pair and A181 is 2.0 ± 0.20 Å. These findings suggest that the C232/G240 and C231/G241 pairs may be the target receptors of A180 and A181 residues, respectively.
Figure 6.
RNA–RNA long-range interactions identified by distance measures of atoms between two adenosine—A180 and A181—in GUAA tetraloop and its potential receptors during the MD trajectories.
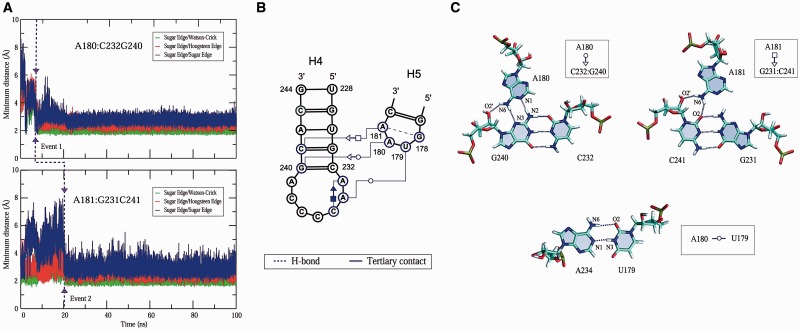
To further explore the tertiary interaction of Model C, we consider the Leontis/Westhof nomenclature (47) and analyze the three edges—Watson–Crick, Hoogsteen and Sugar edge—for potential hydrogen bonding interactions. The measured minimum distances between the Sugar edge of each C232/G240 and C231/G241 base pair with three edges of each A180 and A181 over the 100 ns time course in Figure 7 show tightly formed hydrogen bonding interactions for the Sugar edge/Watson–Crick between the C232/G240 pair and A180 and Sugar edge/Hoogsteen edge tertiary interactions between G231/C241 pair and A181. In addition, we observe tertiary contacts between U179 and A234 residues via trans Watson–Crick/Watson–Crick edge interactions at ∼22 ns. These long-range interactions occur sequentially: at ∼7, 20 and 22 ns, involving A180, A181 and U179, respectively (Figure 7A). These cooperative long-range interactions help stabilize the IRES domain 3.
Figure 7.
Intramolecular RNA–RNA long-range interactions involving GUAA hairpin loop during the Model C MD simulation. (A) A minimum distance of atoms for the GUAA tetraloop–receptor long-range interactions. (B) Long-range interactions in a time-averaged secondary structure obtained from the dynamics data. (C) Atomic details of these three tertiary contacts involving U179, A180 and A181 residues in GUAA loop and their binding receptors.
The corresponding time-averaged secondary structure from the 100 ns dynamics data underscores these three long-range interactions involving the GUAA tetraloop (Figure 7B). The A180A181 residues in the GUAA tetraloop form hydrogen bonds via non-canonical base pairing interactions with the C232/G240 and G231/C241 base pairs, respectively; specifically, trans Sugar edge/Watson–Crick edge where N1 and N6 atoms of A180 interact with N2, N3 and O2′ atoms of G240 and Sugar edge/Hoogsteen edge interaction where N6 atom of A181 forms hydrogen bonds with O2 and O2′ atoms of C241 (Figure 7C). The U179 and A234 residues interact via trans Watson–Crick/Watson–Crick edge interactions involving N1 and N6 atoms of A234 with N3 and O2 atoms of U179. As demonstrated by in vitro selection experiment (44), this U179:A234 tertiary contact promotes the loop–helix long-range interactions.
How might these long-range interactions affect the structural organization of domain 3?
Because the single-stranded region is more dynamic and flexible than double-stranded helices, we speculate that the five hairpins and one long internal loop present in the system may contribute to the overall exhibit of the structure. Thus, the tertiary contacts in domain 3 of IRES may restrict these fluctuations and therefore help recruit ribosomes for viral protein synthesis. Below we further analyze dynamics data for all four models to discern the contributions of the long-range interactions to structural stability as well as organization based on root-mean-square (RMS) fluctuation and radius of gyration (Rg).
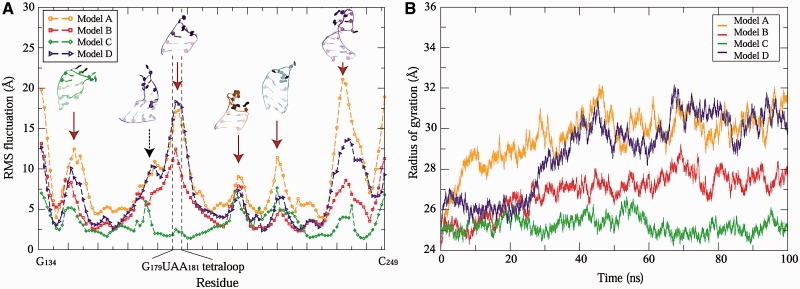
In the RMS fluctuation plot (Figure 8A), we observe six peaks that correspond to hairpins and an internal loop. Among them, two highest peaks are from hairpins located in the helices H4 and H5. Interestingly, these helices involve in the long-range interactions and have been emphasized for its important role in IRES activity.
Figure 8.
RMS fluctuations (A) and Rg (B) measures for four candidate 3D models. In the RMS fluctuations, high peaks (dotted black arrow for internal loop and solid brown arrows for hairpins) correspond to unpaired regions shown as solid color in the structures.
Overall, the RMS fluctuations of all four models follow a similar trend, albeit at different scales. Overall, Models A, B and D fluctuate widely with the values from ∼2.5 to ∼21 Å, whereas Model C ranges between ∼2 and ∼7.5 Å which is about a four-fold decrease. Notably, the GUAA tetraloop in H4 fluctuates between 12 and 18 Å for Models A, B and D, whereas Model C experiences only ∼2.5 Å deviation; this underscores the potential stabilizing role of the tertiary contacts. The long-range interactions appear to stabilize not only adjacent stem–loops but also the entire structure of IRES domain 3.
From the combined data above, involving bioinformatic, experimental and MD modeling data, we propose a theoretically feasible tertiary structure for the apical region in FMDV IRES domain 3 (Figure 9). Here the non-canonical long-range interactions occur between the GUAA tetraloop in helix H5 and the distal region in helix H4. The overall configuration is highly structured; each four-way junction contains two coaxial stacks parallel to each other and Junction I has a crossing at the point of strand exchange. Junctions I and II are classified as family type cH and H, respectively, according to the nomenclature in (21). The three helices H3, H4 and H7 are coaxially stacked together. Each of these two four-way junctions is nearly planar and these two planes are perpendicular to each other.
Figure 9.
Time-averaged tertiary structure of domain 3 taken from the 100 ns dynamics data (top middle), where the long-range interactions occur between helices H4 and H5 (details shown at bottom right). Both Junctions I and II contain two coaxial stacking, parallel to each other and Junction I with a crossing in the single-stranded region (bottom-left and -middle for Junctions I and II, respectively). Both junctions are planar locally and are arranged in a perpendicular orientation to each other globally (top right); note that the three helices H4, H3 and H7 are coaxially stacked all together.
Modeling of domain 3 including the basal region
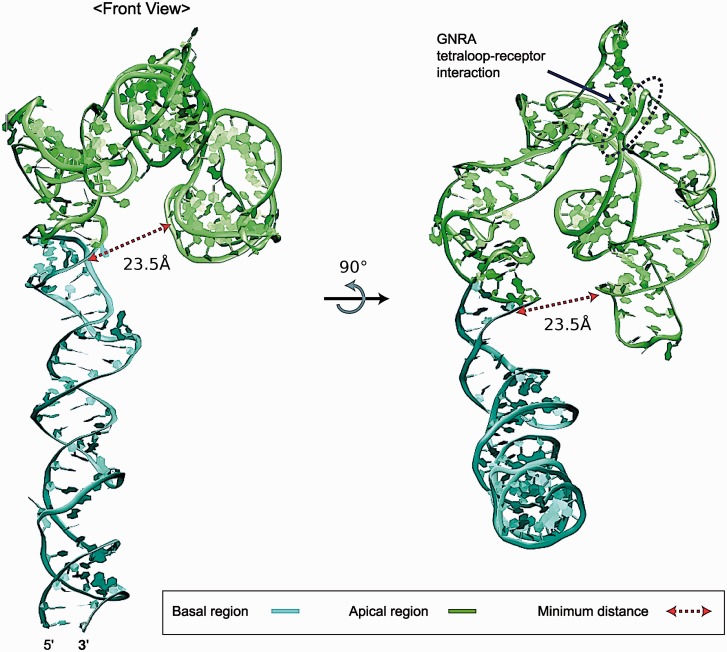
The basal region contains a long internal loop formed by 98 nt (G86 to U133 and C249 to C299). Sampling this region using MC-Sym produces 717 models from which four representative structures were selected from the four largest clusters (Supplementary Figures S3–S4). The overall shape and orientation of the helical axis in these four structures are relatively similar, with differences explained by the flexibility of bending in unpaired bases. Thus, our candidate model for the basal region is chosen from the largest cluster (Supplementary Figure S4A), and then the apical region model (Figure 9) was combined to it to complete a 3D model of the entire domain 3; the minimum distance between the basal (C250) and apical (G194) regions is 23.5 Å; the orientation of the basal region (turned away from the apical region) suggests that the former region is unlikely to be involved in RNA folding of the junctions of the latter region (Figure 10).
Figure 10.
3D model of the entire sequence in domain 3. Domain 3 consists of basal and apical regions; the corresponding structural elements are an internal loop and four-way junctions, respectively. The minimum distance between the two regions is 23.5 Å that the basal region is not likely involved in RNA folding of the junctions.
DISCUSSION
Picornavirus IRES elements are considered as efficient regulatory RNAs which make possible initiation of translation for viral RNAs. FMDV requires RNA-binding proteins such as translation initiation factors (eIFs) and ITAFs that can affect IRES activity; for instance, domains 2, 4 and 5 provide binding sites for cellular proteins including PTB, eIF4G, eIF3 and eIF4B (48).
The FMDV IRES domain 3, often denoted as a central domain, consists of two structural elements—a long internal loop in the ‘basal’ region and four-way junctions in the ‘apical’ region; each of which is ∼50% of the entire sequence. Sequence logos of the 318 aligned FMDV IRES sequences show that the apical region of domain 3 is highly conserved (Supplementary Figure S1); in particular, hairpin loop H4 that contains the potential binding receptors of the GNRA tetraloop is nearly perfectly conserved. The GUAA loop in H5 is also strongly preferred in FMDV IRES systems. This analysis suggests that these structural elements provide an important role in maintaining the functional 3D structure of FMDV IRES domain 3.
It was determined by biochemical experiments that the apical region is a self-folding structural element due to the intramolecular RNA–RNA interactions involving the crucial GNRA motif (13,49,50). This region has thus been suggested to contribute significantly to the structural organization and stability of domain 3, and to the critical function of IRES activity (8–11). IRES-mediated translation initiation is closely linked to structural organization in domain 3, specifically the apical region formed by two four-way junctions enabling the RNA–RNA intramolecular interactions. Thus, we focused on the apical region of domain 3 to decipher the spatial arrangement of the RNA fold that is a prerequisite essential step to understand the initiation mechanism of translation.
Grounded in our recent RNA four-way junction classification study and the Junction-Explorer program (21,27), we have constructed possible junction topologies for the apical region in domain 3 where a pair of coaxial stacks are arranged parallel to each other in the presence and absence of a crossing at the point of strand exchange (Figure 9). Utilizing only the information for the helical arrangements—H and cH family types, we built 16 candidate topologies (Figure 4) and these were reduced to four after applying constraints from experimental data regarding the GNRA tetraloop–receptor long-range interactions. Our next step in modeling was employing MC-Sym to explore conformational space using experimental data. The combined data from junction topology and 3D modeling produced four representative structures where three of the four confirmed the junction topology models. We speculate that the excluded model does not satisfy geometric criteria due to steric clashes. Using MD simulations, we attempted to identify geometrically accessible binding receptors to the GNRA tetraloop for long-range interactions. Among the five residues in the potential receptor site, the bases near the hairpin loop emerged viable over one near the junction core in helix H4; they also have great potential to form long-range interactions with the GNRA tetraloop in H5.
Specifically, the MD simulations revealed a GUAA tetraloop-binding site in addition to a novel tertiary interaction in Model C (Figure 5). The two adenosines A180 and A181 form hydrogen bonds with the receptors C230/G242 and G231/C241 base pair, respectively. The dynamics data also suggest a U:A tertiary contact which enhances the structural stability (U179 in GUAA tetraloop interacts with A234 in a hairpin loop of H4), an interaction also observed by an in vitro selection experiment (44). Interestingly, these tertiary interactions form sequentially.
A previous study suggested that RNA–RNA long-range interactions involving an RAAA motif occur in the presence of GNRA tetraloop long-range interactions as well as Mg2+ ions (9). We have not observed this RAAA motif, but speculate that the distant contacts associated with the RAAA motif might occur when the current RNA system (G134 … C249 residues) is extended to include 25 more residues (U121 … G133, U250 … A261). Such an extended system has greater potential for the long-range interactions (Supplementary Supplementary Figure S12). Due to current limitations of the force field for treating divalent ions (51–54), we have not attempted to include magnesium ions in our system.
In picornavirus, types 1 and 2 of IRES species exist. FMDV IRES belongs to type 2, whereas poliovirus IRES belongs to type 1. Although the overall contents of sequence and 2D structure are different, these two IRES systems share the GNRA motif. NMR data of stem–loop in domain IV revealed the L-shaped conformations are required in order to provide a protein-binding site. However, little is known how the L shape is achieved and maintained. In FMDV IRES domain 3, helix H5 contains similar 2D structure of loop B which includes the GNRA motif. In our dynamics simulations, we observe that H5 forms an L-shape configuration in the presence of long-range RNA–RNA interactions. The overall shape of H5 agrees well with the NMR data (Supplementary Figure S13). However, the shape of H5 in the absence of long-range interactions is variable, with potential diverse phases (S-shaped or U-shaped). Thus, we speculate that long-range interactions involving the GNRA motif have a role in stabilizing the L-shaped loop B in poliovirus IRES.
Based on the above modeling of the apical, self-folding region of IRES domain 3 containing four-way junctions and the experimental data discussed above (13,49,50) combined with our extended modeling of the entire sequence of FMDV IRES domain 3, we hypothesize that the influence of the basal region on structural stability and organization of the junctions is not primary. This is because the basal region is set apart from the junction domains in the apical region with a minimum distance of 23.5 Å (Figure 10).
Although our combined modeling strategy involves many proven approaches and is closely anchored to available experimental data, it is not possible to rule out other plausible overall 3D structures. Further studies using the candidate models for long-time MD studies or with advanced sampling techniques and investigation of potential receptors for RAAA motif may be useful. Yet, the overall 3D configuration reached in Figure 9 and the suggested long-range interactions in the central domain of FMDV IRES provide insights into the potential role of the long-range interactions for structural stability and organization of IRES domain 3 and thus may help in further analysis of the structure, mechanism, and function of viral RNAs. Ultimately, structures may lead to the development of antiviral drugs that inhibit IRES activity and thus virus multiplication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–14 and Supplementary PDB file.
FUNDING
The National Science Foundation [EMT award CCF-0727001 to T.S.]; National Institutes of Health [grants GM081410 and GM100469 to T.S.]; MacCracken Fellowship (to S.J.). Funding for open access charge: National Science Foundation, National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Christian Laing for discussions related to the virus IRES structure, Dr Marc Parisien for help with MC-Sym and Mr Elmetwaly for technical assistance. The computations were made possible by support of the IBM Blue Gene/L supercomputer at the Computational Center for Nanotechnology Innovations based in Rensselaer Polytechnic Institute, NY. Molecular images were generated using the VMD program (55) and Gromacs tool packages (56) were used to analyze the dynamics data.
REFERENCES
- 1.Kolupaeva VG, Hellen CU, Shatsky IN. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez de Quinto S, Martinez-Salas E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6:1380–1392. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez de Quinto S, Lafuente E, Martinez-Salas E. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7:1213–1226. doi: 10.1017/s1355838201010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stassinopoulos IA, Belsham GJ. A novel protein-RNA binding assay: functional interactions of the foot-and-mouth disease virus internal ribosome entry site with cellular proteins. RNA. 2001;7:114–122. doi: 10.1017/s1355838201001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreev DE, Fernandez-Miragall O, Ramajo J, Dmitriev SE, Terenin IM, Martinez-Salas E, Shatsky IN. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pacheco A, Lopez de Quinto S, Ramajo J, Fernandez N, Martinez-Salas E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res. 2009;37:582–590. doi: 10.1093/nar/gkn979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez de Quinto S, Martinez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Miragall O, Ramos R, Ramajo J, Martinez-Salas E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez N, Fernandez-Miragall O, Ramajo J, Garcia-Sacristan A, Bellora N, Eyras E, Briones C, Martinez-Salas E. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res. 2011;39:8572–8585. doi: 10.1093/nar/gkr560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez N, Garcia-Sacristan A, Ramajo J, Briones C, Martinez-Salas E. Structural analysis provides insights into the modular organization of picornavirus IRES. Virology. 2011;409:251–261. doi: 10.1016/j.virol.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Correll CC, Swinger K. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 A resolution. RNA. 2003;9:355–363. doi: 10.1261/rna.2147803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Miragall O, Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Z, Ulyanov NB, Yu J, Andino R, James TL. NMR structures of loop B RNAs from the stem-loop IV domain of the enterovirus internal ribosome entry site: a single C to U substitution drastically changes the shape and flexibility of RNA. Biochemistry. 2004;43:5757–5771. doi: 10.1021/bi0363228. [DOI] [PubMed] [Google Scholar]
- 15.Robertson ME, Seamons RA, Belsham GJ. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 17.Lilley DM, Clegg RM, Diekmann S, Seeman NC, Von Kitzing E, Hagerman PJ. A nomenclature of junctions and branchpoints in nucleic acids. Nucleic Acids Res. 1995;23:3363–3364. doi: 10.1093/nar/23.17.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 19.Bailor MH, Sun X, Al-Hashimi HM. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- 20.Laing C, Jung S, Iqbal A, Schlick T. Tertiary motifs revealed in analyses of higher-order RNA junctions. J. Mol. Biol. 2009;393:67–82. doi: 10.1016/j.jmb.2009.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laing C, Schlick T. Analysis of four-way junctions in RNA structures. J. Mol. Biol. 2009;390:547–559. doi: 10.1016/j.jmb.2009.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SH, Sussman JL, Suddath FL, Quigley GJ, McPherson A, Wang AH, Seeman NC, Rich A. The general structure of transfer RNA molecules. Proc. Natl Acad. Sci. USA. 1974;71:4970–4974. doi: 10.1073/pnas.71.12.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 25.Laing C, Schlick T. Computational approaches to RNA structure prediction, analysis, and design. Curr. Opin. Struct. Biol. 2011;21:306–318. doi: 10.1016/j.sbi.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laing C, Schlick T. Computational approaches to 3D modeling of RNA. J. Phys. Condens. Matter. 2010;22:283101. doi: 10.1088/0953-8984/22/28/283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laing C, Wen D, Wang JT, Schlick T. Predicting coaxial helical stacking in RNA junctions. Nucleic Acids Res. 2011;40:487–498. doi: 10.1093/nar/gkr629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilipenko EV, Blinov VM, Chernov BK, Dmitrieva TM, Agol VI. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 1989;17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 30.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Chang TH, Horng JT, Huang HD. RNALogo: a new approach to display structural RNA alignment. Nucleic Acids Res. 2008;36:W91–W96. doi: 10.1093/nar/gkn258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 34.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA. 2009;15:189–199. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, 3rd, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995:5179–5197. [Google Scholar]
- 37.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mlynsky V, Banas P, Hollas D, Reblova K, Walter NG, Sponer J, Otyepka M. Extensive molecular dynamics simulations showing that canonical G8 and protonated A38H+ forms are most consistent with crystal structures of hairpin ribozyme. J. Phys. Chem. B. 2010;114:6642–6652. doi: 10.1021/jp1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reblova K, Sponer J, Lankas F. Structure and mechanical properties of the ribosomal L1 stalk three-way junction. Nucleic Acids Res. 2012;40:6290–6303. doi: 10.1093/nar/gks258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zgarbova M, Otyepka M, Sponer J, Mladek A, Banas P, Cheatham TE, 3rd, Jurecka P. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yildirim I, Kennedy SD, Stern HA, Hart JM, Kierzek R, Turner DH. Revision of AMBER torsional parameters for RNA improves free energy predictions for tetramer duplexes with GC and iGiC base pairs. J. Chem. Theory Comput. 2012;8:172–181. doi: 10.1021/ct200557r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rother M, Rother K, Puton T, Bujnicki JM. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39:4007–4022. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SH, Cech TR. Three-dimensional model of the active site of the self-splicing rRNA precursor of Tetrahymena. Proc. Natl Acad. Sci. USA. 1987;84:8788–8792. doi: 10.1073/pnas.84.24.8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burks JM, Zwieb C, Muller F, Wower IK, Wower J. In silico analysis of IRES RNAs of foot-and-mouth disease virus and related picornaviruses. Arch. Virol. 2011;156:1737–1747. doi: 10.1007/s00705-011-1043-7. [DOI] [PubMed] [Google Scholar]
- 46.Schlick T, Collepardo-Guevara R, Halvorsen LA, Jung S, Xiao X. Biomolecularmodeling and simulation: a field coming of age. Q. Rev. Biophys. 2011;44:191–228. doi: 10.1017/S0033583510000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol. 2008;16:230–237. doi: 10.1016/j.tim.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramos R, Martinez-Salas E. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES) RNA. 1999;5:1374–1383. doi: 10.1017/s1355838299991240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrano P, Gomez J, Martinez-Salas E. Characterization of a cyanobacterial RNase P ribozyme recognition motif in the IRES of foot-and-mouth disease virus reveals a unique structural element. RNA. 2007;13:849–859. doi: 10.1261/rna.506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDowell SE, Spackova N, Sponer J, Walter NG. Molecular dynamics simulations of RNA: an in silico single molecule approach. Biopolymers. 2007;85:169–184. doi: 10.1002/bip.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sponer J, Burda JMS, Leszczynski J, Hobza P. Interaction between the guanine-cytosine Watson-Crick DNA base pair and hydrated group IIa (Mg2+, Ca2+, Sr2+, Ba2+) and group IIb (Zn2+, Cd2+, Hg2+) metal cations. J. Phys. Chem. A. 1998;102:5951–5957. [Google Scholar]
- 53.Reblova K, Lankas F, Razga F, Krasovska MV, Koca J, Sponer J. Structure, dynamics, and elasticity of free 16 s rRNA helix 44 studied by molecular dynamics simulations. Biopolymers. 2006;82:504–520. doi: 10.1002/bip.20503. [DOI] [PubMed] [Google Scholar]
- 54.Razga F, Koca J, Sponer J, Leontis NB. Hinge-like motions in RNA kink-turns: the role of the second a-minor motif and nominally unpaired bases. Biophys. J. 2005;88:3466–3485. doi: 10.1529/biophysj.104.054916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 56.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.