Abstract
Co-transcriptional pre-mRNA processing relies on reversible phosphorylation of the carboxyl-terminal domain (CTD) of Rpb1, the largest subunit of RNA polymerase II (RNAP II). In this study, we replaced in live cells the endogenous Rpb1 by S2A Rpb1, where the second serines (Ser2) in the CTD heptapeptide repeats were switched to alanines, to prevent phosphorylation. Although slower, S2A RNAP II was able to transcribe. However, it failed to recruit splicing components such as U2AF65 and U2 snRNA to transcription sites, although the recruitment of U1 snRNA was not affected. As a consequence, co-transcriptional splicing was impaired. Interestingly, the magnitude of the S2A RNAP II splicing defect was promoter dependent. In addition, S2A RNAP II showed an impaired recruitment of the cleavage factor PCF11 to pre-mRNA and a defect in 3′-end RNA cleavage. These results suggest that CTD Ser2 plays critical roles in co-transcriptional pre-mRNA maturation in vivo: It likely recruits U2AF65 to ensure an efficient co-transcriptional splicing and facilitates the recruitment of pre-mRNA 3′-end processing factors to enhance 3′-end cleavage.
INTRODUCTION
In eukaryotic cells, pre-mRNA maturation involves complex sequential steps, including splicing and 3′-end processing (1). Splicing is catalyzed by the spliceosome, a large ribonucleoprotein (RNP) complex. The spliceosome comprises five small nuclear RNP (snRNP) particles and many auxillary factors. Spliceosome assembly starts with the recruitment of U1 snRNP to the 5′ splice site, followed by the U2 snRNP auxiliary factor (U2AF). The U2AF65 and U2AF35 subunits of U2AF, respectively, bind to the polypyrimidine tract and the 3′ splice site. U2AF next recruits U2 snRNP to the branch point to form the pre-spliceosome complex A.
The 3′-end processing of pre-mRNAs involves two tightly coupled steps: 3′-end cleavage and polyadenylation. The mammalian 3′-end processing complex comprises cleavage and polyadenylation specificity factor, cleavage stimulation factor, cleavage factors I and II and poly(A) polymerase (2). Splicing and 3′-end cleavage stimulate each other (3): An intron upstream of the polyadenylation (pA) signal stimulates cleavage (4), and mutation of the pA signals reduces splicing of the last intron in vitro (5).
It is broadly accepted that the carboxyl-terminal domain (CTD) of Rpb1, the largest subunit of RNA polymerase II (RNAP II), is needed to couple pre-mRNA processing to transcription (6,7). The CTD comprises tandem repeats of a heptapeptide with Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 consensus sequence (8,9). Truncation of the CTD impairs both splicing and 3′-end cleavage (10,11). Phosphorylation is the most documented post-translational modification of the CTD. It is involved in co-transcriptional processing (12–15). Phosphorylation of Ser2 increases along genes and peaks toward the end of the gene (16–19), consistent with an involvement in pre-mRNA processing. Indeed, two studies suggested that Ser2 phosphorylation is required for pre-mRNA 3′-end in vivo processing of pre-mRNAs (20,21). One used flavopiridol, a CDK9 inhibitor, and the other used yeast strains where the Ctk1 gene is deleted. Another CDK9 inhibitor, 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), blocks co-transcriptional splicing and 3′-end cleavage (22). However, the effects of either knocking-out or inhibition of Ser2 kinase might be hard to interpret: First, several kinases such as CDK9, Ctk1, CDK12 and CDK13 preferentially phosphorylate Ser2 in vitro (23,24); therefore, blocking CDK9 or deleting Ctk1 might not totally inhibit Ser2 phosphorylation in vivo. Second, inhibitors might affect several targets and have indirect effects (25–28). For example, flavopiridol inhibits phosphorylation of both Ser2 and Thr4 (29), although Thr4 is phosphorylated by the polo-like kinase (19). Furthermore, 3′-end processing of snRNA is impaired by DRB, another CDK9 inhibitor, but Ser7 phosphorylation is involved in this process (30).
An alternative strategy is to replace phosphorylable residues in the CTD heptapeptide repeats by alanines to prevent phosphorylation (31). Therefore, we replaced the endogenous Rpb1 in RNAP II by S2A Rpb1, where the CTD Ser2 had been switched to alanines. Reporter transcripts generated by S2A RNAP II were found defective for both pre-mRNA splicing and 3′-end cleavage. Transcription by S2A RNAP II failed to recruit U2AF65 to transcription sites, and the recruitment efficiency of PCF11 was reduced. Thus, we propose that Ser2 plays a central role in coupling of splicing and 3′-end cleavage to transcription, by enabling the CTD to recruit splicing and 3′-end processing machineries to the transcription machinery.
MATERIALS AND METHODS
Plasmids and cells
Plasmids expressing HA-tagged amanitin-resistant Rpb1, YFP-MS2 and pSVEDA have been described (31–33). pBGwtSV40 and pBGmutSV40 were generated from pBGwt and pBGmut (previously named pTet-on-CFP-MS2wt and pTet-on-CFP-MS2mut, respectively) (29,34) by ApaI–PaeI digestion and ligation. U1-70 K and U2AF35 cDNAs were cloned into pEGFP-C1 (Clontech) to generate pEGFP-U1-70 K and pEGFP-U2AF35. For pU2AF65-Flag, EGFP was replaced by Flag epitope in pEGFP-N1. U2OS Tet-on cells were from Clontech. SJ (previously named 2-6-3) (35), BGwt and BGmut cells were derived from U2OS (34). Concentrations of α-amanitin and doxycycline used were 8 µg/ml and 6 µg/ml, respectively.
Antibodies
The following primary antibodies were used: HA (Abcam ab 9110), EGFP (JL-8, Clontech), Flag (F7425, Sigma-Aldrich), α-tubulin (T9026, clone DM1A, Sigma-Aldrich), U2AF65 (ab37530, Abcam), SC35 (sc53518, Santa Cruz), PCF11 (sc161998, Santa Cruz) and SF2/ASF (32-4600, Invitrogen). Primary antibodies were detected with horseradish peroxidase–labeled secondary antibodies (Promega) for western blot or fluorophore-conjugated secondary anti-mouse, anti-rabbit (Invitrogen) and anti-goat (Sigma-Aldrich) antibodies.
Transfection
FuGene 6 (Roche) transfection was used for microscopy; otherwise, LyoVec (Invivogen) was used. An empty plasmid vector was used as carrier DNA to maintain a constant quantity of nucleic acid. For RNAi knockdown, Lipofectamine 2000 (Invitrogen) was used twice, with 24 h as an interval, and U2OS cells were harvested 48 h after the second transfection. Sense sequences of siRNAs (Eurofins MWG Operon) are as follows: U2AF65, 5′-GCACGGUGGACUGAUUCGUdTdT-3′ (36); luciferase, 5′-CUUACGCUGAGUACUUCGAdTdT-3′.
Reverse transcription
Total RNA was extracted using RNeasy kit (Qiagen), and reverse transcribed by Superscript III reverse transcriptase (Invitrogen) after RNase-free DNase I treatment (Fermentas) using random hexamers, Oligo dT or gene-specific primers. Sequences of gene-specific primers are given in the online supplementary material.
PCR and qPCR
PCR without reverse transcription was performed to check the absence of genomic DNA contamination. GSYBR Green qPCR Master kit (Fermentas) was used. Primer sequences and conditions are provided in the online supplementary material.
RNA immunoprecipitation assay
The procedure was adapted from Zhao et al. (37). Cells were transfected with HA-tagged amanitin-resistant Rpb1. Cell nuclei were pelleted and resuspended in 500 µl of RIPA buffer [50 mM Tris–Cl (pH 7.4), 150 mM NaCl, 0.25% Na-deoxycholate, 1% NP-40, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, RNase inhibitor (Fermentas), 1 mM phenylmethylsulfonyl fluoride (PMSF), protease inhibitor cocktail (Sigma)]. Lysates were passed 10 times through a 20-gauge needle and then digested by Turbo DNase (Ambion) on ice for 30 min. After pre-clearance with Dynabeads protein A (Invitrogen) slurry, the lysate was diluted in RIPA buffer to 1 ml and incubated for 2 h at 4°C with 20 µl of Dynabeads protein A (Invitrogen) slurry and 2 µg of anti-HA antibody. After extensive washes, RNA was extracted by RNeasy kit (Qiagen). For plasmid and C-FOS transcripts analysis, 5 × 106 and 2 × 107 cell nuclei were used, respectively.
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) was performed in SJ cells transfected with YFP-MS2 and induced by doxycycline as described by Darzacq et al. (38). Briefly, the phenol red–free Leibovitz’s L15 medium replaced low-glucose DMEM in FRAP. Experiments were carried out at 37°C using a temperature-controlled Delta T4 culture dish system with a heated lid and an objective heater (Bioptechs). An Olympus IX 81 epifluorescence microscope with a PlanApo 100X, 1.4 NA objective (Olympus) and a CoolSNAP HQ camera (Roper Scientific) was used for wide-field imaging. Transcription sites were bleached using a single 488-nm laser pulse in a circle area of 2.5-µm diameter (bleaching time, 5 ms). Stacks of nine images 400-nm apart were acquired every 3–30 s for 15 min. Fluorescence intensities were normalized by setting a pre-bleach value as 1 and the first post-bleach value as 0.
Fluorescence image analysis
Fluorescence in situ hybridization (FISH), immunofluorescence and radial analysis of images have been described previously (34). In brief, rings were centered on the transcription site defined by MS2 hybridization signal. The density of fluorescence in each annulus at a given distance was obtained by dividing the intensity of fluorescence by the area of the same annulus, and then normalized by the average density at 1.6 µm. Transcription sites from >50 cells were used for each analysis. Control sites were selected randomly in the corresponding nuclei. A detailed protocol and probe sequences are provided in the online supplementary material.
RNase protection assay
A radioactive RNA probe was in vitro transcribed using a MAXIscript Kit (Ambion). RNase protection assay (RPA) was carried out using the RPA III kit (Ambion) following the manufacturer’s instructions, except that hybridization of the probe with total RNA was performed overnight at 60°C. Quantification was carried out using a Fuji FLA 3000 phosphorimager. Automated band quantification was performed using ImageQuant TL software.
RESULTS
Switching CTD Ser2 into alanines impairs splicing in vivo
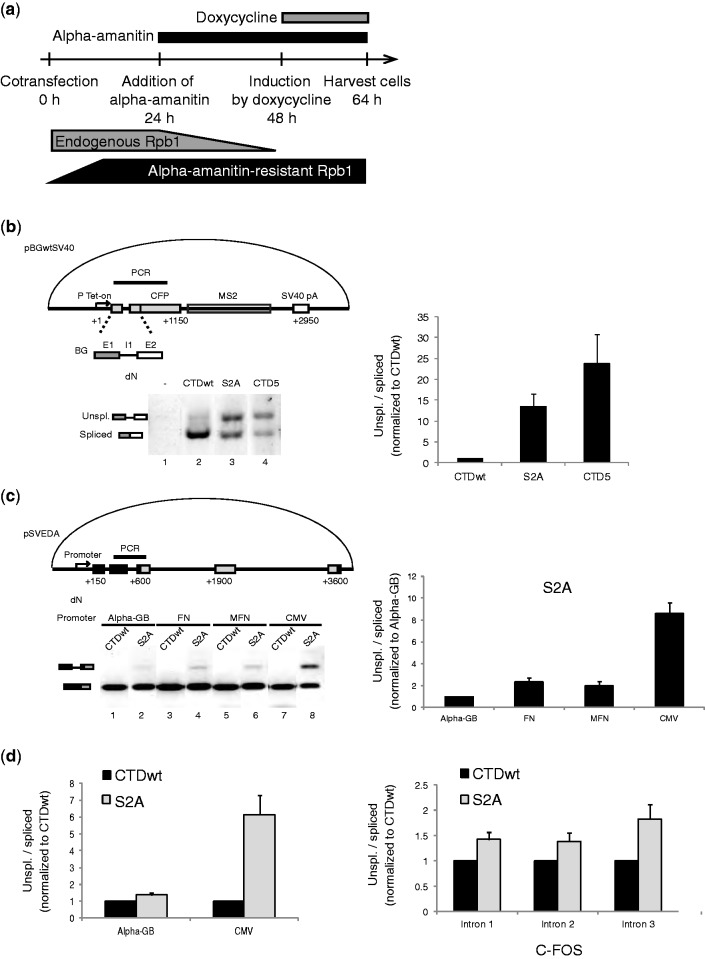
A mutation in Rpb1, the largest subunit of RNAP II confers resistance to α-amanitin (39). This mutation offers an opportunity to functionally replace endogenous Rpb1 in the presence of amanitin by Rpb1 with a modified CTD. To determine whether the Rpb1 derivatives affected splicing in vivo, it was co-transfected with the pBGwtSV40 reporter plasmid. pBGwtSV40 carries the β-globin intron 1 (I1) followed by MS2 repeats and the SV40 polyadenylation signal (Figure 1b, upper panel). After 24 h in amanitin to inactivate endogenous Rpb1, transcription of the reporter gene was induced by doxycycline (Figure 1a). An RT-PCR of RNAs isolated from transfected cells generated two products corresponding to spliced and unspliced transcripts from the reporter gene (Figure 1b, lanes 2–4). No product was amplified from amanitin-treated cell RNAs without transfection of an amanitin-resistant Rpb1 plasmid (Figure 1b, lane 1), indicating that endogenous Rpb1 had indeed been inactivated and that the transfected Rpb1 was functional. Co-transfection of amanitin-resistant Rpb1 having a wild-type CTD (CTDwt) essentially generated spliced reporter transcripts (Figure 1b, lane 2). However, the proportion of unspliced product increased strongly (20-fold) on co-expression of a truncated Rpb1 (CTD5), where only five heptapeptide repeats were left (Figure 1b, lanes 4). Truncation of the CTD impaired reporter expression efficiency and splicing as previously reported (10). The cells were co-transfected next with S2A Rpb1 to mimic a permanently unphosphorylated state (31). The proportion of unspliced product increased 13-fold when generated by S2A RNAP II compared with wild type (Figure 1b, lane 3). This finding suggests that CTD Ser2 is required for an efficient splicing in vivo.
Figure 1.
Switching CTD Ser2 into alanines impairs splicing in vivo. (a) Experimental scheme. U2OS Tet-on cells were co-transfected with α-amanitin–resistant Rpb1 and pBGwtSV40 reporter plasmid. The Tet-on promoter drives the expression of β-globin (BG) exon 1(E1), intron 1 (I1) and exon 2 (E2) fused to CFP-coding sequence followed by MS2 sequences. (b) RT-PCR detection of splicing. CTDwt stands for wild-type Rpb1. S2A designate the mutated Ser2Ala Rpb1. CTD5 carries a deletion of 48 heptad repeats (31). Splicing was analyzed by RT-PCR amplification of a region spanning from β-globin exon 1 to CFP to avoid interference with endogenous β-globin RNAs. Twenty-five amplification cycles were used for lanes 2 and 3, and 28 cycles were used for lanes 1 and 4. Quantification is an average of three experiments with standard error of mean (SEM). Unspliced/spliced ratios were normalized to CTDwt. (c) Splicing defects of S2A are promoter dependent. U2OS cells were co-transfected with pSVEDA plasmids where α-globin (black boxes) and fibronectin (grey boxes) exons were driven by α-globin (Alpha-GB), fibronectin (FN), mutated fibronectin (MFN) or cytomegalovirus (CMV) promoters (33). The PCR-amplified region spans α-globin intron 2. (d) Transcripts co-immunoprecipitated with HA-tagged RNAP II were reverse transcribed with a gene-specific primer downstream α-globin intron 2 (left, for transfected pSVEDA plasmids) or random primers (right, for endogenous C-FOS). cDNAs were amplified by qPCR using intron-specific primers for unspliced or spliced RNAs. Quantification is an average of three experiments with SEM. Unspliced/spliced ratios were normalized to CTDwt.
S2A RNAP II splicing impairment is promoter dependent
To determine whether the splicing deficiency of S2A RNAP II was restricted to the β-globin intron 1 and the Tet-on promoter, we investigated next the splicing of another model (pSVEDA) with different introns (33) (Figure 1c, upper panel). We focused on the α-globin intron 2 that did not experience alternative splicing. In cells expressing CTDwt Rpb1, the spliced product was the sole to be detected (Figure 1c, lower panel). In contrast, unspliced products were seen in transcripts from S2A Rpb1–expressing cells. In the pSVEDA plasmid series, different promoters driving the same reporter gene were available. Interestingly, the magnitude of splicing deficiency in the presence of S2A polymerase was promoter dependent: Transcripts driven by the cytomegalovirus (CMV) promoter showed the most severe splicing deficiency, those driven by the α-globin promoter were the least affected and those driven by fibronectin promoters were intermediate (Figure 1c, lanes 2, 4, 6 and 8). Thus, the magnitude of splicing impairment of transcripts generated by S2A RNAP II might be influenced by factors that define promoter architecture. Importantly, splicing impairment of transcripts generated by S2A RNAP II is not restricted to a particular intron.
Impairment of C-FOS co-transcriptional splicing
A defective co-transcriptional splicing might be responsible for the promoter effect on S2A-generated splicing impairment. To test this hypothesis, we investigated the splicing status of nascent transcripts co-immunoprecipitated with HA-tagged RNAP II wild type or S2A mutant (40). There was small increase (×1.3) in the unspliced/spliced ratio of nascent transcripts generated by the S2A RNAP II with α-globin promoter (Figure 1d, left). In contrast, a 6-fold increase in unspliced/spliced ratio was observed for nascent transcripts driven by the CMV promoter as described earlier in the text for the bulk transcripts (Figure 1c).
We next investigated splicing of an endogenous gene transcribed by S2A RNAP II. C-FOS was selected because its mRNA has a very short half-life. When C-FOS nascent transcripts were generated by S2A RNAP II, the unspliced/spliced ratio increased (Figure 1d, right). Such an increase was observed with all three C-FOS introns, intron 3 being the most affected. To conclude, the splicing of both endogenous and the reporter transcripts investigated here is co-transcriptional. Both the S2A-driven splicing impairment and its promoter dependence are detected at the co-transcriptional level.
Transcription by S2A RNAP II impairs the recruitment of U2AF65 and U2 snRNA, but not U1 snRNA onto sites of transcription
We previously reported that recruitment of essential splicing factors, such as U2AF65 and U2 snRNA, to transcription sites is impaired when splicing is blocked by mutation of the donor and acceptor splice sites (34). To determine the mechanism of splicing inhibition by S2A mutant, we examined whether S2A mutant would affect recruitment of splicing factors in BGwt cells expressing S2A or wild-type Rpb1. In BGwt cells, the pBGwt plasmid has been stably integrated as a gene array into a unique locus (34). It contains the β-globin intron 1 fused to MS2 repeats. Transcripts were detected by FISH with an MS2-specific probe (Supplementary Figure S1). Transcription sites were not observed in amanitin-treated cells that did not express an amanitin-resistant Rpb1 (data not shown), but were seen in cells expressing amanitin-resistant Rpb1 (Supplementary Figure S1). Splicing components were detected by FISH or immunofluorescence in the same cells (Supplementary Figure S1). Co-localizations between transcription sites and splicing factors were quantified by image processing analysis of >50 nuclei (34). Splicing components involved in the earliest steps of spliceosome assembly such as U1 snRNA and SF2/ASF (alias SRSF1) co-localized with transcription sites in BGwt cells when either CTDwt or S2A Rpb1 were expressed (Figure 2 and Supplementary Figure S1a and b). In contrast, recruitment of components involved in subsequent steps, such as U2AF65 (alias U2AF2), U2 snRNA and SC35 (alias SRSF2), was impaired in S2A Rpb1– but not in CTDwt Rpb1–expressing cells (Figure 2 and Supplementary Figure S1c to e). These findings suggested that CTD Ser2 is required for the assembly of the pre-spliceosome complex at transcription sites.
Figure 2.
S2A RNAP II transcription impairs recruitment of U2AF65 and U2 snRNA, but not of U1 snRNA, to transcription sites. Radial analysis of U1 snRNA (a and b), U2AF65 (c and d) or U2 snRNA (e and f) recruitment to transcription sites in BGwt cells. Typical fluorescence microscopy images are shown in Supplementary Figure S1. For radial analysis, the mean relative enrichments are plotted against the distance from the center of transcription sites (closed symbols) defined by the MS2 hybridization signal or from randomly selected control sites (open symbols). SEM of data for U1 snRNA, U2AF65 and U2 snRNA using 64, 59 and 64 cells for CTDwt and 77, 63 and 77 cells for S2A, respectively.
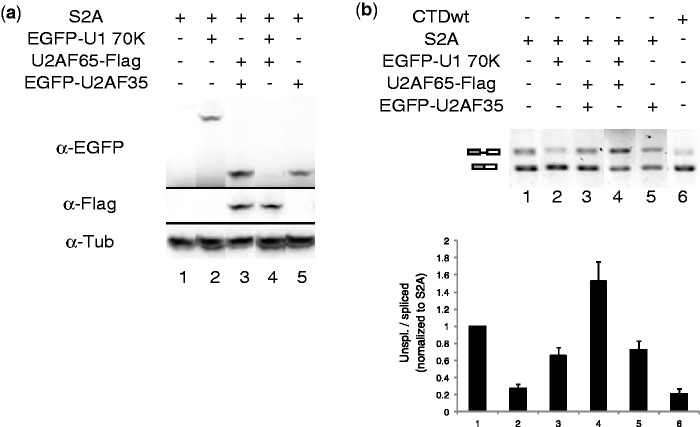
To determine whether the lack of U2AF recruitment could be compensated by increased levels of this factor, we overexpressed U2AF subunits in cells co-transfected with S2A mutant and pBGwtSV40. Plasmids expressing U2AF65-flag or EGFP-U2AF35 were co-transfected with S2A mutant, and their expression was confirmed by western blot (Figure 3a, lanes 3–5). Splicing of reporter gene RNA extracted from the same sample was tested using RT-PCR (Figure 3b). Overexpression of U2AF subunits alone or their combination did not significantly improve splicing in S2A Rpb1–expressing cells (Figure 3b, lanes 3–5). However, overexpression of U1-70 K, a protein interacting with U1 snRNA, which is enriched on transcription sites of S2A Rpb1, partially rescued splicing of transcripts generated by S2A (Figure 3b, lane 2) to the level observed with CTDwt (lane 6).
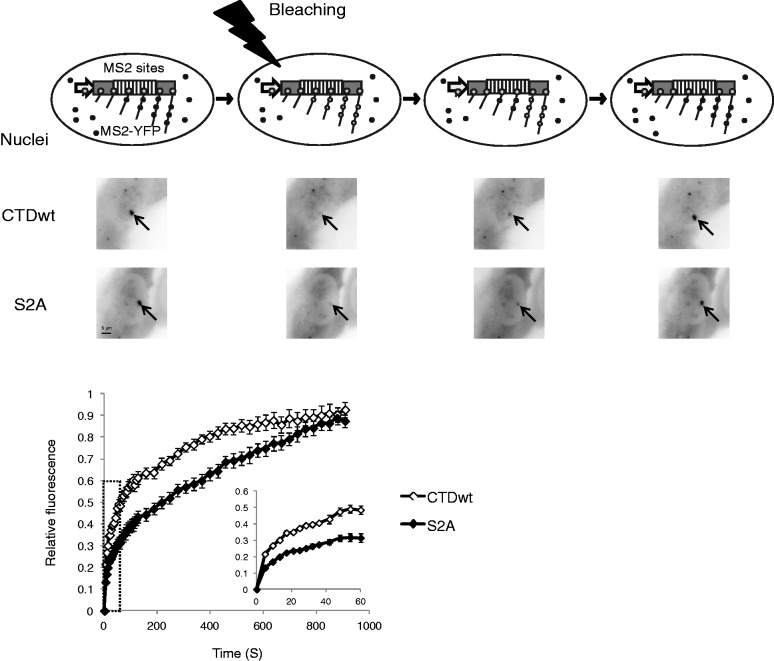
Figure 4.
FRAP evaluation of CTDwt and S2A RNAP II transcription rates. Upper scheme represents the principle of FRAP. A typical image set is shown underneath. SJ cells were co-transfected with α-amanitin–resistant Rpb1, YFP-MS2 and the reverse tetracycline chimeric activator (rTetR). Half-recovery time is 111 s and 268 s for CTDwt and S2A, respectively. The inlet shows the recovery curves of the first 60 s. Curves show the mean with SEM using data from 10 and 16 cells for CTDwt and S2A RNAP II, respectively.
Figure 3.
Overexpression of U1-70 K rescues splicing defects of S2A RNAP II. (a) Western blot of overexpressed U1-70 K, U2AF65 and U2AF35. U2OS Tet-on cells were co-transfected with pBGwtSV40, CTDwt or S2A Rpb1, and with expression vectors for U2AF35, U2AF65 and U1-70 K. (b) Splicing patterns on U2AF65, U2AF35 or U1-70 K overexpression. Error bars represent SEM of four experiments.
Ser2Ala RNAP II transcription rate in living cells
It has been reported that a deletion of β-globin intron 2 limits the release of unprocessed RNA from transcription sites (41). Hence, unspliced transcripts generated by the S2A RNAP II might accumulate on the transcription sites. To address this possibility, we followed FRAP of MS2-YFP labeled nascent RNA on transcription sites (38). To visualize transcription sites, we used U2OS-derived SJ cells that had 200 copies of a reporter fused with MS2 sequence and a β-globin exon/intron cassette inserted into a single locus (35). SJ cells were co-transfected with plasmids expressing MS2-YFP protein and either S2A mutant or wild-type Rpb1. Transcription sites were only observed in cells transfected with amanitin-resistant Rpb1. During the first 32 s after photobleaching, which corresponds to the estimated elongation time in SJ cells (38), MS2-YFP fluorescence recovery was one-third weaker with S2A RNAP II than with wild type (CTDwt) (0.26 versus 0.39 at 32 s) (Figure 4), suggesting the elongation rate of S2A RNAP II is slower than that of wild-type RNAP II. YFP fluorescence on S2A transcription sites recovered close to pre-bleach values after 15 min. Therefore, the S2A mutant transcribes fairly well, and unspliced RNA generated by S2A RNAP II are released from transcription sites. Yet, elongation rate is slower for S2A than for CTDwt.
Transcription by S2A RNAP II impairs 3′-end cleavage
Previous studies suggested that Ser2 phosphorylation is essential for pre-mRNA 3′-end processing (20). To test whether S2A RNAP II transcripts showed a 3′-end processing defect of pre-mRNA, we used RT-qPCR. All transcripts from cells transfected with CTDwt or S2A were amplified from cDNA libraries using the RT1 gene–specific primer upstream the pA signal. Uncleaved read-through transcripts were amplified from cDNAs using gene-specific primers (GSPs) (RT2, RT3 and RT4) downstream the pA signal (Figure 5a, upper panel). A 2-fold increase of the read-through/total RNA ratio was observed with S2A RNAP II transcripts compared with CTDwt (Figure 5a, lower panel). In contrast, a small but reproducible decrease in polyadenylated/total RNA ratio (dT primer) was observed.
Figure 5.
S2A RNAP II transcription impairs 3′-end cleavage. (a) Transcripts of S2A in transfected U2OS cells show more read-through than CTDwt, as quantified by RT-qPCR. Positions of the amplicon and RT primers are shown in the upper diagram. cDNAs were reverse transcribed using various GSPs. A primer upstream the SV40 pA (RT1) reverse transcribed all transcripts (processed and read-through). GSPs downstream the pA signal (RT2, RT3 and RT4) only transcribed uncleaved read-through transcripts. Oligo-dT only transcribed polyadenylated transcripts. Real-time PCR product values for RT2, RT3, RT4 and dT cDNAs were divided by the value obtained with RT1. The ratio of S2A was normalized to CTDwt. SEM of four experiments is shown. (b) RNase protection assay for 3′-end cleavage. Cell treated as in Figure 1a. Probe and protected products are positioned in the reporter diagram. Read-through transcripts protect a longer fragment than transcripts of the 5′-end (Total). RNAs from pBGwtSV40 (Wt. S.S.) and pBGmutSV40 (Mut. S.S.), with wild-type or mutated splice sites, respectively, were analyzed. SEM of three experiments.
To confirm 3′-end cleavage impairment, an RNase protection assay of 3′-end processing was set up next. Read-through transcripts that failed to be cleaved after the pA signal would be extended further, and because the plasmid is circular, some read-through transcripts would be extended to sequences from upstream to downstream of the transcription start site. Hence, an antisense RNA probe that overlaps the transcription start site on pBGwtSV40 should give longer protected fragments by read-through transcripts than those initiated at the transcription start site (Figure 5b, upper panel). The ratio of read-through to total transcripts was higher for S2A Rpb1 than for CTDwt Rpb1-expressing cells (Figure 5b, lanes 1 and 2). It is worth to note that the 3′-end processing impairment of S2A was weaker than that of CTD5 (Supplementary Figure S2b).
Because splicing stimulates pre-mRNA 3′-end processing in vitro (42), it was questioned whether S2A RNAP II transcription affects 3′-end processing directly or because of the splicing deficiency. Thus, the RNase protection assay was repeated with pBGmutSV40 plasmids where both 5′ and 3′ splice sites have been mutated to completely abolish splicing (34). Again, S2A transcription resulted in 3′-end processing defect of pre-mRNA from the pBGmutSV40 reporter as well (Figure 5b, lanes 3 and 4). To determine whether this 3′-end processing defect was restricted to SV40 polyadenylation (pA) signals, we tested the 3′-end processing of pre-mRNA transcribed by S2A mutant from a pBGwt plasmid where the SV40 pA sequence had been replaced with the bovine growth hormone one. Similar observations were made (Supplementary Figure S2a), showing that S2A RNAP II transcription decreases 3′-end processing efficiency with different pA signals and independently of splicing.
Decreased 3′-end cleavage efficiency in the absence of U2AF65
It has been shown that introns stimulate 3′-end processing in vitro (42). Indeed 3′-end cleavage of non-spliced pBGmutSV40 transcripts was less efficient than that of spliced pBGwtSV40 transcripts in CTDwt-expressing cells (Figure 5b, compare lanes 1 and 3). We then hypothesized that a failure of U2AF recruitment may lead to a deficient 3′-end processing of S2A-generated transcripts. This, because U2AF65 has been shown to bind the 3′-end processing machinery in vitro (43) and because it is not recruited to S2A transcription sites. Therefore, U2AF65 expression was knocked down by siRNA in cells expressing endogenous Rpb1 (Figure 6a). It resulted in a 3-fold decreased splicing efficiency compared with a control siRNA (Figure 6b). Furthermore, the U2AF65 knockdown increased the ratio of read-through pre-mRNA to total transcripts (Figure 6c, left and middle). In contrast, it resulted in a small but reproducible decrease in polyadenylated mRNA (right). A U2AF65 knockdown leads to a decreased 3′-end processing efficiency.
Figure 6.
U2AF65 knockdown impairs 3′-end cleavage. (a) Western blot of U2AF65 siRNA knockdown. U2OS cells were transfected twice, with 24 h as an interval, and harvested 48 h after the last transfection. (b) U2AF65 knockdown impaired splicing. U2OS cells were transfected with pBGwtSV40 plasmid and induced by doxycycline 24 h before harvest. (c) Left—RNase protection assay for 3′-end cleavage of pBGwtSV40 transcripts after knockdown of U2AF65. Middle—Quantification of the RNase protection assay. Right—Ratio qPCR using oligo dT and RT1 as RT primers like in Figure 5. SEM of three experiments is shown.
Both Ser2Ala switch and the absence of splicing decrease PCF11 recruitment to transcription sites
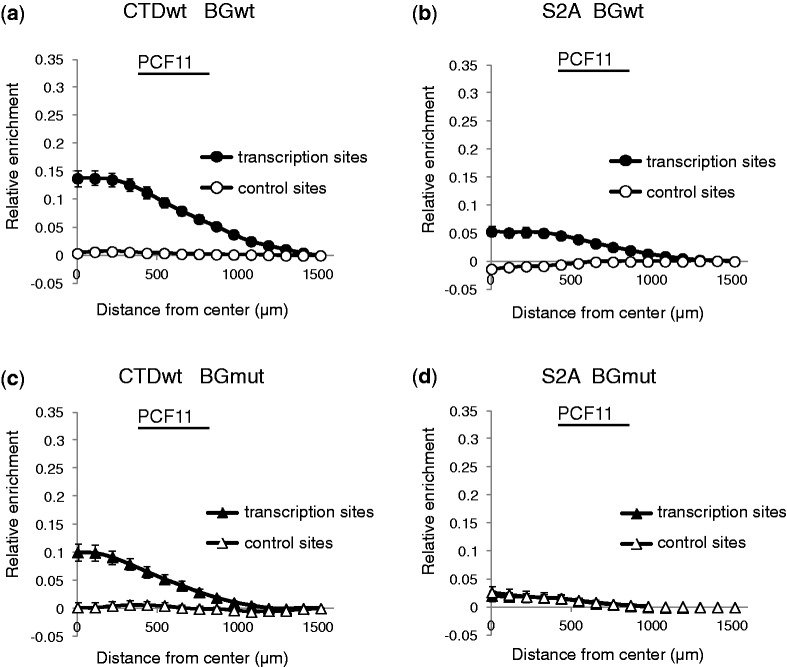
The 3′-end cleavage of non-spliced pBGmutSV40 transcripts was less efficient than that of spliced pBGwtSV40 transcripts. We then hypothesized that it might relate to a deficient 3′-end processing factor recruitment on transcription sites. PCF11, an essential component of the 3′-end processing machinery, was investigated in cells expressing wild-type Rpb1. Indeed, enrichment of PCF11 on transcription sites was greater in BGwt cells (splicing competent) than in BGmut cells (splicing incompetent) (Figure 7 compare a and c). A similar pattern was observed on sites transcribed by endogenous RNAP II (Supplementary Figure S2a and b). Thus, splicing appears to enhance PCF11 recruitment on transcription sites.
Figure 7.
PCF11 recruitment is diminished in the absence splicing or on S2A RNAP II transcription. Radial analysis of PCF11 recruitment. Typical images are shown in Supplementary Figure S3. SEM of data from 67 BGwt (a) and 61 BGmut (c) cells for CTDwt and from 61 BGwt (b) and 60 BGmut (d) cells for S2A.
Because S2A RNAP II transcription resulted in impaired splicing and 3′-end processing, we hypothesized that it might also lead to a deficient 3′-end processing factor recruitment on transcription sites. Indeed, recruitment of PCF11 in BGwt cells (splicing competent, Figure 7, compare a and b) was weaker in the presence of S2A compared with wild-type Rpb1 (CTDwt). The effect of Ser2 phosphorylation and splicing on PCF11 recruitment is synergistic. PCF11 was not detectable at the transcription sites in BGmut cells (splicing incompetent) expressing S2A Rpb1 (Figure 7d). These observations suggested that both Ser2 and splicing contribute to recruit PCF11 for an efficient pre-mRNA 3′-end cleavage.
DISCUSSION
Replacing Rpb1, the largest subunit of RNAP II, with a Ser2Ala CTD mutant, we provide evidence that Ser2 is important for splicing and 3′-end cleavage in vivo. We took advantage of an Rpb1 mutation that confers resistance to α-amanitin. Although α-amanitin has been reported to affect the level of short-lived proteins, and therefore indirectly impair pre-mRNA splicing (44), our results suggest that the splicing machinery is intact in our experimental conditions. Indeed, no splicing defect was observed when pre-mRNAs were synthesized by the α-amanitin–resistant wild-type Rpb1. Phosphorylation of the CTD has been proposed to favor splicing (20,21), although it is unclear which phosphorylated residue in the CTD repeat is required. Ser2 phosphorylation is a good candidate. It occurs after capping (45–47) and increases along the genes (16,17,19). The splicing impairment we observe with the S2A RNAP II mutant might be caused by loss of CTD Ser2 phosphorylation, although one may not exclude the involvement of factors interacting with the Ser2 hydroxyl group.
Splicing can occur co-transcriptionally (7). Indeed, an intron probe only detects RNA molecules at the transcription sites of cells that synthesize a pre-mRNA with a functional intron (34). Here we find that a large amount of nascent transcripts co-immunoprecipitated with RNAP II is spliced. Consistently, the spliceosome snRNAs (U2, U4, U5, U6) and auxiliary factors such U2AF are recruited to a transcription site that has a functional intron. As U2AF65 is not enriched on transcription sites when Ser2 is replaced by alanine, we propose that binding of U2AF65 to Ser2-phosphorylated CTD is important. First, U2AF65 plays a role in early steps of spliceosome assembly, just after 5′ splice site recognition by U1 snRNP (1). U2AF65 stabilizes U2 snRNP association with the pre-mRNA (48). Second, U2AF65 binds preferentially to the phosphorylated Ser2 CTD in vitro (49). However, Ser2 phosphorylation is necessary but not sufficient. Functional splice sites in pre-mRNA are also required for U2AF65 enrichment on gene arrays transcribed by wild-type RNAP II (32,34).
In contrast to U2 snRNA, U1 snRNA is enriched on an intron-less gene array transcribed by wild-type RNAP II (32,34) or on an intron-containing gene array transcribed by S2A RNAP II (this work). Recruitment of U1 snRNA might relate to the constitutive association of U1 snRNP with RNAP II (34,50). Our data suggest that Ser2 phosphorylation is not involved in this association. Interestingly, overexpression of U1-70 K, a U1 snRNP–specific protein, partially compensates splicing defects of S2A mutant transcripts. However, the mechanism of this observation is unclear. Experiments involving a model gene driven by various promoters indicate that the magnitude of the splicing defect of transcripts generated by S2A RNAP II is promoter dependent. The first intron of transcripts driven by the α-globin promoter remains spliced correctly, whereas splicing of the same transcripts driven by the CMV promoter is impaired. Interestingly, the constructs used in this study show a promoter-dependent alternative splicing of their other introns (33). There are several possible explanations. First, a multiplicity of interactions is involved in U snRNP and SR protein recruitment to the nascent transcript. For instance, our data show that both Ser2 and a functional splice site are required to recruit U2AF and U2 snRNP. Other interactions with activators, co-activators and transcription factors that are promoter specific might be involved. For instance, U snRNP have been shown to interact directly or indirectly with modified histones (51,52). Histone modifier loading as well as chromatin modifications on genes is dependent on promoter characteristics, and chromatin modifications influence the efficiency of co-transcriptional splice site recognition (53). Along this line of thoughts, the rescuing effect of U1-70 K overexpression suggests that some promoter-dependent transcription factor combinations might enhance the loading or the activity of U1 snRNP and thereby increase the splicing of S2A transcripts.
We observed a deficient 3′-end processing of S2A transcripts. This is in agreement with a requirement in Ser2 phosphorylation that might be attributed to the specific binding of PCF11 to Ser2-phosphorylated CTD in vitro (42,54,55).
U2AF65 interacts with the cleavage machinery through poly(A) polymerase, and this interaction has been proposed to mediate the coupling of splicing and 3′-end processing (56). Indeed, U2AF65 knockdown impaired 3′-end cleavage in vivo. When splice sites of a reporter gene are mutated, U2AF65 is not enriched on transcription sites and 3′-end processing of these transcripts is less efficient. This further supports that U2AF65 mediates the stimulation of 3′-end processing by splicing. We now show that splicing also enhances PCF11 recruitment on transcription sites. PCF11 might be recruited to the transcription machinery through two distinct but cooperating mechanisms: a direct interaction with the Ser2-phosphorylated CTD (55) and an indirect mechanism, which requires the presence of a functional intron. In contrast to U2AF65 (which requires both), either Ser2 phosphorylation or a functional intron is sufficient to recruit PCF11 to transcription sites.
In summary, we propose that CTD Ser2 phosphorylation is essential for co-transcriptional processing in vivo (Figure 8): It is required to recruit U2AF65 and support splicing; U2AF65, in turn, facilitates the recruitment of pre-mRNA 3′-end processing factors such as PCF11 to enhance 3′-end cleavage. Meanwhile, splicing factors, such as U2AF65, stimulate 3′-end processing and couple it to splicing.
Figure 8.
Ser2 phosphorylation couples splicing and 3′-end processing to transcription. A model. This scheme is restricted to factors related to this study. Py, polypyrimidine track; SS, splice site. The arrow in the upper diagram means functional enhancement. Ser2 phosphorylation is necessary to recruit U2AF65 to support splicing, and it facilitates the recruitment of pre-mRNA 3′-end processing factor such as PCF11 to enhance 3′-end cleavage. Meanwhile, splicing factors, such as U2AF65, stimulate 3′-end processing and couple it to splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3 and Supplementary Methods.
FUNDING
Ligue Nationale contre le Cancer [RS10/75-15 to O.B.]; Association pour la Recherche sur le Cancer [ARC 1010 to O.B.]; Deutsche Forschungsgemeinschaft, SFB-TR5 (to D.E.); People’s Republic of China Scholarship Council [2008623021 to B.G.]. Funding for open access charge: CNRS—French research agency.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Drs Didier Auboeuf, Xavier Darzacq, Francesca Fiorini, Alberto Kornblihtt, Sui Huang, Hervé Le Hir, John Lis, Yaron Shav-Tal and Zhen Wang and their colleagues for materials and fruitful discussions.
REFERENCES
- 1.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinson HG. An active role for splicing in 3′-end formation. Wiley Interdiscip. Rev. RNA. 2011;2:459–470. doi: 10.1002/wrna.68. [DOI] [PubMed] [Google Scholar]
- 4.Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 5.Niwa M, Berget SM. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 6.de Almeida SF, Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–1976. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 7.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 10.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 11.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 12.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 13.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:630–642. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, et al. Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 2010;17:1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 19.Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31:2784–2797. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 21.Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 22.Bird G, Zorio DA, Bentley DL. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartkowiak B, Mackellar AL, Greenleaf AL. Updating the CTD story: from tail to epic. Genet. Res. Int. 2011;2011:623718. doi: 10.4061/2011/623718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25:2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Ann. Rev. Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 32.Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc. Natl Acad. Sci. USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiluttini B, Gu B, Belagal P, Smirnova AS, Nguyen VT, Hébert C, Schmidt U, Bertrand E, Darzacq X, Bensaude O. Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J. Cell. Sci. 2010;123:2085–2093. doi: 10.1242/jcs.061358. [DOI] [PubMed] [Google Scholar]
- 35.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pacheco TR, Coelho MB, Desterro JM, Mollet I, Carmo-Fonseca M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol. Cell. Biol. 2006;26:8183–8190. doi: 10.1128/MCB.00350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartolomei MS, Corden JL. Localization of an a-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol. 1987;7:586–594. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bittencourt D, Auboeuf D. Analysis of co-transcriptional RNA processing by RNA-ChIP assay. Methods Mol. Biol. 2012;809:563–577. doi: 10.1007/978-1-61779-376-9_36. [DOI] [PubMed] [Google Scholar]
- 41.Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat. Struct. Mol. Biol. 2011;18:1115–1123. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
- 42.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 43.Millhouse S, Manley JL. The C-terminal domain of RNA polymerase II functions as a phosphorylation-dependent splicing activator in a heterologous protein. Mol. Cell. Biol. 2005;25:533–544. doi: 10.1128/MCB.25.2.533-544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsao DC, Park NJ, Nag A, Martinson HG. Prolonged alpha-amanitin treatment of cells for studying mutated polymerases causes degradation of DSIF160 and other proteins. RNA. 2012;18:222–229. doi: 10.1261/rna.030452.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pei Y, Schwer B, Shuman S. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J. Biol. Chem. 2003;278:7180–7188. doi: 10.1074/jbc.M211713200. [DOI] [PubMed] [Google Scholar]
- 46.Guiguen A, Soutourina J, Dewez M, Tafforeau L, Dieu M, Raes M, Vandenhaute J, Werner M, Hermand D. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 2007;21:1552–1559. doi: 10.1038/sj.emboj.7601627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenasi T, Peterlin BM, Barboric M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb) J. Biol. Chem. 2011;286:22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valcarcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected] Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 49.David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011;25:972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 51.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gunderson FQ, Merkhofer EC, Johnson TL. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc. Natl Acad. Sci. USA. 2011;108:2004–2009. doi: 10.1073/pnas.1011982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allemand E, Batsche E, Muchardt C. Splicing, transcription, and chromatin: a menage a trois. Curr. Opin. Genet. Dev. 2008;18:145–151. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 55.Lunde BM, Reichow SL, Kim M, Suh H, Leeper TC, Yang F, Mutschler H, Buratowski S, Meinhart A, Varani G. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2010;17:1195–1201. doi: 10.1038/nsmb.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.