Abstract
Escherichia coli contains 30 two-component systems (TCSs), each consisting of a histidine kinase and a response regulator. Whereas most TCSs are well characterized in this model organism, little is known about the YpdA/YpdB system. To identify YpdB-regulated genes, we compared the transcriptomes of E. coli cells overproducing either YpdB or a control protein. Expression levels of 15 genes differed by more than 1.9-fold between the two strains. A comprehensive evaluation of these genes identified yhjX as the sole target of YpdB. Electrophoretic mobility shift assays with purified YpdB confirmed its interaction with the yhjX promoter. Specifically, YpdB binds to two direct repeats of the motif GGCATTTCAT separated by an 11-bp spacer in the yhjX promoter. yhjX encodes a cytoplasmic membrane protein of unknown function that belongs to the major facilitator superfamily of transporters. Finally, we characterized the pattern of yhjX expression and identified extracellular pyruvate as a stimulus for the YpdA/YpdB system. It is suggested that YpdA/YpdB contributes to nutrient scavenging before entry into stationary phase.
INTRODUCTION
Two-component systems (TCSs) are the major players in signal transduction in prokaryotes, enabling cells to react adaptively to fluctuating environmental conditions. Each TCS consists of a membrane-bound histidine kinase (HK) and a response regulator (RR). The HK senses specific stimuli and transduces them into a cellular signal via autophosphorylation. Transfer of the phosphoryl group to a conserved aspartate in the cognate RR mediates the intracellular response, generally an alteration in gene expression (1). In Escherichia coli, 30 HKs and 32 RRs have been annotated, and most of these have been studied extensively (2).
However, in spite of its wide distribution among the Gammaproteobacteria, almost nothing is known about the YpdA/YpdB system (Fig. 1). Systematic studies have so far failed to identify either the stimulus sensed by YpdA or any YpdB-regulated target in E. coli (3). Autophosphorylation of YpdA and subsequent phosphotransfer to YpdB have yet to be demonstrated (4), and no detectable differences between a ypdAB deletion mutant and wild-type E. coli were uncovered in a previous phenotypic microarray analysis (5).
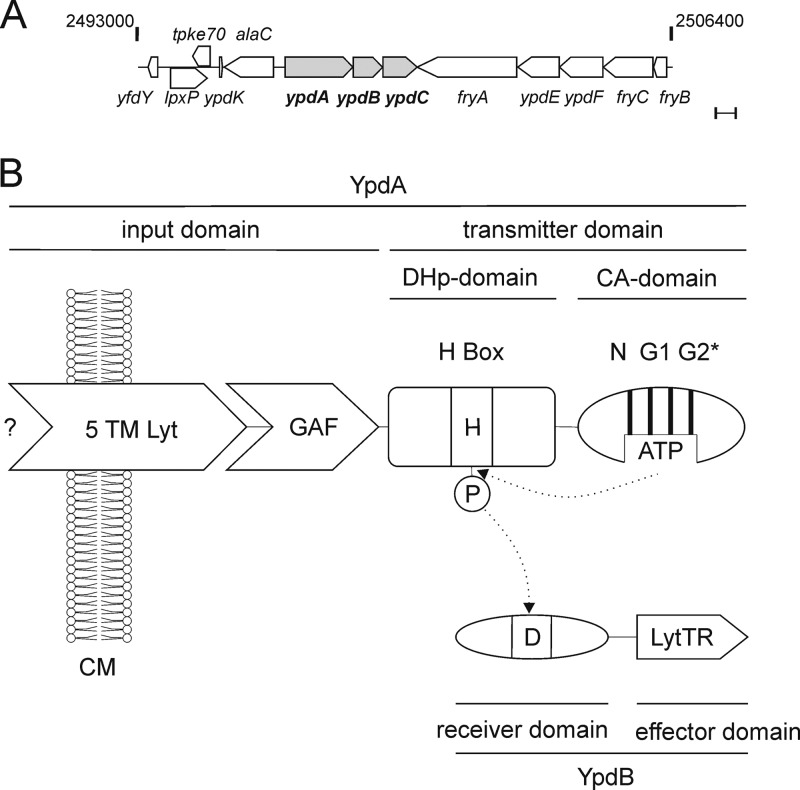
Fig 1.
The YpdA/YpdB system of E. coli. (A) The region between 53.73 and 54.01 centisomes (bp 2493000 to 2506400) around the ypdABC locus of the E. coli MG1655 chromosome (http://www.ecocyc.org/) (23). Bar, 500 bp. (B) Domain structure of YpdA/YpdB. YpdA belongs to the class I HKs, in which DHp and CA domains are connected. The input domain of YpdA consists of a 5TM Lyt (LytS-YhcK) domain and a GAF domain. YpdB consists of a CheY-like receiver domain and a LytTR-type DNA-binding domain. The phosphorylation sites are denoted H (histidine) and D (aspartate). N, G1, and G2 mark the conserved boxes in the CA domain. The G2 box of YpdA is incomplete (G2*). CM, cytoplasmic membrane.
Together with ypdC, ypdA and ypdB form an operon, which is located at 53.56 centisomes in the E. coli MG1655 genome (Fig. 1A). ypdC encodes a protein with a C-terminal AraC-type DNA-binding domain and therefore probably functions as a transcriptional regulator. The neighboring genes are alaC (376 bp upstream of ypdA) and the fryABC-ypdEF operon (3 bp downstream of ypdC).
YpdA and YpdB belong to the family of LytS-like HKs and LytTR-like RRs (6, 7), respectively. In bacterial pathogens of humans and plants, these TCSs are often crucial for host-specific interactions (8). Bioinformatics analyses (using the programs TMHMM, MEMSAT3, and OCTOPUS [9–11]) indicated the presence of at least six transmembrane helices in the YpdA input domain, which belongs to the 5TM Lyt (LytS-YhcK) type (Fig. 1A) (7). In addition, YpdA harbors a GAF domain, also found in cyclic GMP-specific phosphodiesterases, adenylyl cyclases, and FhlA (hence GAF).
YpdB is composed of an N-terminal CheY-like receiver domain and a C-terminal LytTR-like effector domain with DNA-binding affinity (Fig. 1A) (12). Since the elucidation of the crystal structure of Staphylococcus aureus AgrA (a LytTR RR) and its unusual mode of binding to DNA (13), a high degree of variability in the recognition motifs of LytTR-like RRs is predicted (14).
We have recently characterized the YehU/YehT system of E. coli (15). Not only does this LytS/LytTR-like TCS share the same domain structure with the YpdA/YpdB system, their respective components also show over 30% amino acid sequence identity.
This study focuses on the elucidation of the stimulus recognized and the response mediated by the YpdA/YpdB system. We identified yhjX, a gene that codes for an otherwise uncharacterized member of the major facilitator superfamily (MFS), as a target of the system. Moreover, we found that YpdA/YpdB reacts predominantly to the presence of exogenous pyruvate. These results suggest that YpdA/YpdB participates in the carbon control network in E. coli.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
E. coli strains and their genotypes are listed in Table 1. Mutants were constructed with the E. coli Quick and Easy gene deletion kit (Gene Bridges) and the Bac modification kit (Gene Bridges), as reported previously (17). Both kits rely on the Red/ET recombination technique. Plasmids and oligonucleotides used in this work are listed in Tables S1 and S2 in the supplemental material. DNA fragments required for strain constructions were amplified by PCR from genomic DNA using appropriate primers.
Table 1.
Bacterial strains used in this study
| E. coli strain | Relevant genotype and/or description | Reference or source |
|---|---|---|
| MG1655 | F− λ− ilvG rfb-50 rph-1 | 16 |
| MG1655 rspL150 | F− λ− ilvG rfb-50 rph-1 rpsL150; Strr | 17 |
| MG20 | MG1655 ΔypdABC | This work |
| MG21 | MG1655 rpsL150 ΔypdBC::rpsL-neo; Kanr Strs | This work |
| MG22 | MG1655 rpsL150 ΔypdB::rpsL-neo; Kanr Strs | This work |
| MG23 | MG1655 rpsL150 ypdA-H371Q; Kans Strr | This work |
| MG24 | MG1655 rpsL150 ypdB-D53E; Kans Strr | This work |
| MG25 | MG1655 rpsL150 ypdB-D53N; Kans Strr | This work |
| MG26 | MG1655 ΔyhjX | This work |
| MG1655-ΔlacZ | MG1655 ΔlacZ::Tetr | Gift from K. Jahreis |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | 18 |
| DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | 19 |
Molecular biological techniques.
Plasmid DNA and genomic DNA were isolated using the Hi-Yield plasmid minikit (Suedlabor) and the DNeasy blood and tissue kit (Qiagen), respectively. DNA fragments were purified from agarose gels using the Hi-Yield PCR cleanup and gel extraction kit (Suedlabor). Phusion high-fidelity DNA polymerase or Phire hot-start DNA polymerase (Finnzymes) was used according to the supplier's instructions. Restriction enzymes and other DNA-modifying enzymes were purchased from New England BioLabs and used according to the manufacturer's directions.
Growth conditions and RNA isolation.
E. coli MG1655 and mutant strains MG21 and MG22 were transformed with plasmid pBAD24-ypdB, pBAD24-yehS, or pBAD24. Strains were grown overnight in lysogeny broth (LB) and subsequently used for inoculation of 100 ml of fresh LB medium. Cells were grown aerobically at 37°C to the early exponential growth phase (optical density at 600 nm [OD600] of 0.5). The overexpression of ypdB and yehS was then induced by the addition of 0.2% (wt/vol) l-arabinose to the medium, and growth was allowed to continue for 45 min. Cells were harvested, and total RNA was isolated essentially as described previously (20) and treated with DNase I for 30 min to remove residual chromosomal DNA. Subsequently, RNA was purified using the RNA Pure kit (Suedlabor).
Preparation of fluorescence-labeled cDNA, hybridization, and microarray analysis.
Preparation of fluorescence-labeled cDNA, hybridization, and microarray analysis were performed by the Kompetenzzentrum für Fluoreszente Bioanalytik (KFB, Regensburg, Germany), using Affymetrix E. coli 2.0 chips with Affymetrix chemistry. RNA derived from three biological replicates was prepared and hybridized. Statistical analysis was also performed by the KFB (Regensburg, Germany), using MAS5 software (Affymetrix).
Northern blot analysis.
mRNA transcripts of potential target genes were quantitatively analyzed by Northern blotting. This technique reveals only relative induction levels by using a radioactively labeled probe that is specific for the mRNA of interest. The protocol used for Northern analysis was described previously (21). Briefly, 20-μg samples of RNA were fractionated by electrophoresis on 1.2% (wt/vol) agarose gels containing 1.1% (vol/vol) formaldehyde in MOPS (4-morpholinepropanesulfonic acid) buffer. RNA was transferred onto a Hybond nylon membrane (GE Healthcare) by capillary blotting. Hybridization was performed according to a standard protocol (22), using an [α-32P]dCTP-labeled PCR fragment specific for the first 99 to 1,000 bp of the target mRNA. The radioactive probes were synthesized by using the Rediprime II DNA labeling system (GE Healthcare) and purified with a PCR cleanup and gel extraction kit. Radioactive labeling was quantified after exposure to a phosphorscreen using the Typhoon Trio variable-mode imager (GE Healthcare). As a control, the expression of rpoD, a housekeeping gene encoding the sigma 70 subunit of the E. coli RNA polymerase (23), was analyzed.
Purification and phosphorylation of 6His-YpdB, 6His–YpdB-D53E, and 6His–YpdB-D53N.
Proteins were purified according to a protocol described previously for 6His-YehT (15). Proteins were about 95% pure, as judged by SDS-PAGE (24) and Western blotting using the anti-His tag antibody.
[32P]acetyl phosphate was synthesized by using a protocol modified from a method described previously (25) and resuspended in buffer (50 mM Tris-HCl [pH 7.5], 5% [vol/vol] glycerol, 0.1 mM EDTA, 1 mM dithiothreitol [DTT]). The concentration was determined as described previously (26). Phosphorylation of 6His-YpdB (1 mg ml−1) was performed at 37°C with 20 mM MgCl2 and 20 mM [32P]acetyl phosphate for a maximum of 60 min. The reaction was stopped by the addition of SDS sample buffer to the mixture (24). Samples were then loaded onto an SDS gel. Phosphorylated YpdB was detected by Western blotting and visualized after exposure to a phosphorscreen with the Typhoon Trio variable-mode imager (GE Healthcare).
Electrophoretic mobility shift assay (EMSA).
The upstream regulatory regions of all genes identified in the transcriptome analysis (Table 2) were amplified by using a primer labeled with the 6-isomer of carboxyfluorescein (6-FAM) (6-FAM uni24) and an unlabeled primer (rev24) (see Table S2 in the supplemental material) with the corresponding pUC19 plasmids as the template. Similarly, short DNA fragments comprising parts of the yhjX upstream regulatory sequence were cloned into vector pUC19 and amplified by PCR as described above. After PCR amplification, the DNA fragments were purified by electrophoresis on 7% (wt/vol) polyacrylamide gels according to the protocol provided with the GenElute gel extraction kit (Sigma).
Table 2.
Genes most affected by the overexpression of ypdB
| Genea | b no.a | rFb (arbitrary units) |
Log2 ratioc | Pd | Function | Transcriptional regulatione | |
|---|---|---|---|---|---|---|---|
| YpdB | YehS | ||||||
| ypdB | b2381 | 12,890 | 150 | 7.5 | 0.01 | Predicted response regulator | YpdB ↑ (positive control) |
| yhjX | b3547 | 1,040 | 30 | 4.9 | ≤10−3 | Uncharacterized member of the MFS of transporters | YpdB ↑ |
| yjiY | b4354 | 2,080 | 110 | 4.3 | ≤10−3 | Predicted inner membrane protein | YpdB ↑ |
| fhuF | b4367 | 1,670 | 500 | 1.7 | ≤10−3 | Ferric iron reductase | ↑↓ |
| guaC | b0104 | 2,100 | 670 | 1.6 | ≤10−3 | GMP reductase | ↑↓ |
| entC | b0593 | 250 | 90 | 1.6 | 0.01 | Isochorismate synthase 1 | ↑↓ |
| fhuA | b0150 | 1,260 | 570 | 1.1 | 0.01 | Outer membrane porin | ↑↓ |
| cpxP | b3913 | 1,000 | 490 | 1.0 | 0.01 | Regulator of the Cpx response | ↑↓ |
| fecB | b4290 | 1,230 | 650 | 0.9 | ≤10−3 | Periplasmic substrate-binding component of the iron dicitrate ABC transporter | ↑↓ |
| entE | b0594 | 110 | 60 | 0.9 | 0.02 | 2,3-Dihydroxybenzoate-AMP ligase | ↑↓ |
| ynjH | b1760 | 740 | 1,860 | −1.3 | ≤10−3 | Hypothetical protein | ↑↓ |
| ygbL | b2738 | 130 | 340 | −1.4 | ≤10−3 | Hypothetical protein | ↑↓ |
| yahM | b0327 | 210 | 580 | −1.5 | ≤10−3 | Hypothetical protein | ↑↓ |
| iraP | b0382 | 330 | 1,030 | −1.6 | ≤10−3 | Antiadaptor protein for σS stabilization | ↑↓ |
| ygbK | b2737 | 220 | 690 | −1.7 | 0.01 | Hypothetical protein | ↑↓ |
| yehS | b2124 | 1,680 | 16,130 | −3.3 | ≤10−3 | Hypothetical protein | ↑↓ (control) |
Gene names/b numbers and gene product functions were taken from the EcoCyc database (http://www.ecocyc.org/) (23) and the Affymetrix Expression Analysis Sequence Information Database (27).
rF, relative fluorescence (arbitrary units).
Log2 ratio of transcript levels for the ypdB and yehS overexpression strains. Log2 was calculated from the ratio of the mean fluorescence intensity of the respective transcript in the ypdB overexpression strain to that measured in the yehS overexpression strain. A negative (or positive) value denotes a decrease (or increase) in transcription level upon overproduction of YpdB in comparison to that seen in the YehS-overproducing strain.
P value significance (t test) of single rF values.
Effect of YpdB overproduction on the transcription levels of the respective genes in E. coli MG21 cells compared to control cells (E. coli MG21/pBAD24), as determined by Northern blot analysis. YpdB-dependent induction (YpdB ↑), YpdB-independent induction/repression (↑↓), or YpdB-dependent repression (YpdB ↓) of the gene is indicated (Fig. 2A).
YpdB-DNA binding assays were carried out with a total volume of 27.5 μl containing 50 mM NaCl, 10 mM Tris-HCl (pH 7.5), 6% (vol/vol) glycerol, 5.45 ng μl−1 salmon sperm DNA (as a nonspecific competitor), 1.1 nM 6-FAM-labeled DNA, 135 μg ml−1 bovine serum albumin (BSA), and increasing concentrations of 6His-YpdB. DNA-protein binding assay mixtures were incubated at room temperature for 15 min. Complexes were resolved by electrophoresis on native Tris-acetate-EDTA (TAE) polyacrylamide gels (5% [wt/vol]), which had been prerun at 70 V for 1 h. After 3 h of electrophoresis at a constant voltage (10 V cm−1) in 0.5× TAE buffer, gels were scanned with a Typhoon Trio variable-mode imager equipped with 488-nm (excitation) and 526-nm (emission) filters. Quantification of free DNA and protein-bound DNA was performed using ImageQuant 5.0 analysis software (Molecular Dynamics).
DNase I footprinting.
The DNase I footprinting protocol using 6-FAM-labeled DNA and an ABI3730 DNA sequencer was adapted from the work of Zianni et al. (50). Briefly, a DNA fragment including the upstream region of yhjX (bases −264 to +36) was amplified from the corresponding pUC19 derivative and purified as described above. Aliquots (550 fmol) of this 6-FAM-labeled DNA fragment were incubated with purified 6His-YpdB (or BSA as a control), at concentrations of 300 nM, 600 nM, 1,200 nM, or 2,400 nM, in a total volume of 27.5 μl (the other components were 50 mM NaCl, 10 mM Tris-HCl [pH 7.5], 6% [vol/vol] glycerol, 135 μg ml−1 BSA, 0.05 mM CaCl2, and 0.25 mM MgCl2) at 25°C for 15 min.
Upon the addition of DNase I (0.01 U), the reaction mixture was incubated at 25°C for 2 min. The reaction was stopped by the addition of 137.5 μl of DF buffer (Hi-Yield PCR cleanup and gel extraction kit; Suedlabor) to the mixture. After purification, the DNA fragments were analyzed with an ABI3730 DNA sequencer. The sequence ladders were analyzed with PeakScanner software 1.0 (Applied Biosystems). Protected positions were mapped using the GeneScan 500 LIZ size standard (Applied Biosystems), which was co-run with each sample as a molecular ruler.
β-Galactosidase activity assay.
Cells grown overnight from cultures of E. coli MG1655 ΔlacZ transformed with pBAD33-ypdB and pRS415 encoding various yhjX promoter::lacZ fusions were inoculated into LB medium (OD600 of 0.05). Cultures were grown aerobically in Erlenmeyer flasks at 37°C to the mid-exponential growth phase. YpdB overproduction was induced by the addition of 0.2% (wt/vol) l-arabinose for 45 min. Cells were harvested, and β-galactosidase activities were measured, as described previously (28). Values were calculated according to methods described previously by Miller (29) and normalized to values for wild-type promoter activity.
Production and detection of YhjX-6His in the membrane fraction.
E. coli BL21(DE3) cells transformed with pBAD24-yhjX were grown to an OD600 of 0.5 in LB medium, and gene expression was induced by the addition of 0.2% (wt/vol) l-arabinose. After growth for a further 3 h, cells were harvested by centrifugation, disrupted, and fractionated, as described above. At each step, pellets were resuspended in equal volumes of TG buffer (50 mM Tris-HCl [pH 7.5], 10% [vol/vol] glycerol). An equal volume of each fraction was subjected to SDS-PAGE (24) and analyzed by Western blotting using a primary anti-His antibody.
In vivo yhjX expression studies.
In vivo yhjX expression was probed with a luciferase-based reporter gene assay using plasmid pBBR yhjX-lux in E. coli MG1655 and the indicated mutants. For complementation studies, the ypdAB operon was expressed under the control of the PBAD promoter in the absence of l-arabinose as an inducer. Cells from a culture grown overnight in M9 minimal medium with 0.5% (wt/vol) glucose as the C source were inoculated into M9 minimal medium (supplemented with different C sources and additives [for concentrations, see Table S3 in the supplemental material]) or LB medium to an OD600 of 0.05. Cells were then incubated under aerobic growth conditions at 37°C, and OD600 and luminescence were measured continuously. Optical densities of cultures were determined with a microplate reader (Tecan Sunrise) at 600 nm. Luminescence levels were determined by using a Centro LB960 instrument (Berthold Technology) for 0.1 s and are reported as relative light units (RLU) (counts s−1). Solutions of peptidoglycan fragments from Bacillus subtilis, Lactobacillus casei, and E. coli were isolated as described previously (30).
Determination of extracellular pyruvate concentrations.
We used the pyruvate oxidase-based Pyruvate Assay kit (Biovision), according to the manufacturer's directions, to determine the pyruvate concentration in cell-free culture supernatants. The experimental values were calculated from a standard curve.
Microarray data accession number.
The complete microarray data set is listed in the ArrayExpress database (31) under accession no. E-MTAB-1347.
RESULTS AND DISCUSSION
Identification of YpdA/YpdB target genes.
Since the ypdAB deletion mutant displayed no obvious phenotype and the stimulus recognized by YpdA/YpdB was unknown, we used transcriptome analysis in combination with overproduction of the RR YpdB to identify target genes of this system. Overproduction of RRs often leads to target gene expression irrespective of the phosphorylation state of the RR and, hence, in the absence of the stimulus that activates the HK (15). Such an artificial microarray-based strategy was successfully utilized for the identification of target genes of Rap proteins in Bacillus subtilis (32) and of the YehU/YehT system in E. coli (15), among others. To avoid side effects due to protein overproduction per se, cells with the same genetic background that overproduced YehS instead of YpdB were used as a control. YehS, an 18-kDa protein of unknown function, was chosen as the control because its overproduction has no effect on the growth rate (data not shown), and it lacks a DNA-binding domain. Overexpression was induced by adding arabinose to cultures of cells containing pBAD24-derived plasmids bearing the ypdB or yehS gene, and total RNA was prepared 45 min later. The RNA was then hybridized to Affymetrix E. coli 2.0 gene chips (see Materials and Methods). Growth rates and levels of protein overproduction were comparable between the two strains (data not shown). In evaluating differential gene expression levels between the two strains, we excluded intergenic regions and considered only genes that were well expressed (fluorescence, ≥100 arbitrary units) and that showed a highly significant (P ≤ 0.02 by t test) and at least a 1.8-fold difference in expression. In all, 15 genes met these criteria, with expression differences ranging from maximally 30-fold induction to 3-fold repression (Table 2). As expected, the expression level of ypdB mRNA increased 180-fold. The complete microarray data set is listed in the ArrayExpress database (31) under accession no. E-MTAB-1347.
Validation of YpdB-dependent target gene expression by Northern blotting.
Because overproduction of RRs is an artificial procedure, the microarray data had to be carefully validated. Therefore, the transcription levels of all 15 genes previously flagged as YpdB dependent on microarrays were measured by Northern analysis in strain MG22 (ΔypdB) cells transformed with pBAD24-ypdB or the empty vector pBAD24 (Fig. 2A and Table 2; see also Fig. S1 in the supplemental material). This revealed that only yhjX and yjiY were directly dependent on YpdB. Expression of yjiY, which codes for a putative carbon starvation protein, was induced 4-fold, while yhjX, encoding a putative MFS transport protein, was induced 24-fold upon overproduction of YpdB (Fig. 2A). Expression levels of the genes fhuF, guaC, entC, fhuA, cpxP, fecB, entE, ynjH, ygbL, yahM, and ygbK differed by 0.8- to 1.2-fold between the two strains (see Fig. S1 in the supplemental material). No iraP transcript was detected (data not shown).
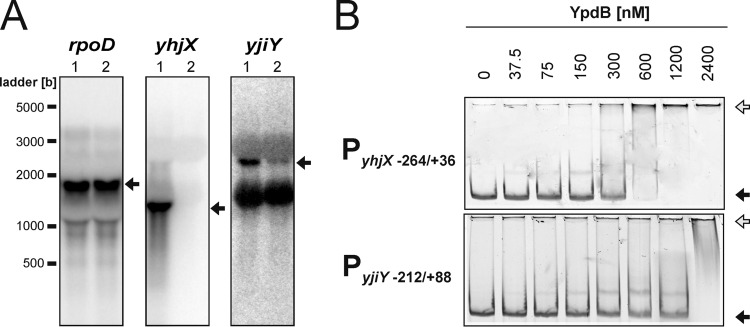
Fig 2.
Evaluation of potential YpdB target genes. (A) Northern blot analysis was used to measure the effect of overproduction of YpdB on expression of the genes yjiY, yhjX, and rpoD (control) in E. coli MG22 (ΔypdB). The expression levels of these genes were assessed upon overproduction of YpdB (lanes 1) or in the absence of YpdB (MG22 transformed with the empty pBAD24 vector) (lanes 2). Twenty micrograms of total RNA was loaded per lane, and the transcripts were detected with the corresponding gene-specific DNA probes. The transcripts of each of the genes are marked by an arrow. (B) EMSA. Fluorescence-labeled DNA fragments comprising the indicated regions of the 5′-regulatory sequences were incubated with increasing concentrations of purified 6His-YpdB and then fractionated by gel electrophoresis. The numbers indicate nucleotide positions relative to the transcriptional start site at position +1. The positions of free DNA (black arrows) and YpdB-DNA complexes (white arrows) are marked.
Thus, Northern blot analysis indicated that the overproduction of YpdB has a marked effect on the transcription of yhjX and a weaker effect on yjiY. Expression of the other identified genes seemed not to be directly dependent on YpdB.
Quantitative YpdB-promoter DNA interaction studies.
We used electrophoretic mobility shift assays (EMSAs) as an independent approach to validate the targets putatively recognized by YpdB. Fluorescently labeled DNA fragments encompassing the transcriptional start point, the putative −35 and −10 boxes, and sufficiently large upstream and downstream regions of all potential target promoters identified by transcriptome analysis were incubated with increasing concentrations of purified 6His-YpdB. Since the purified protein could be phosphorylated with [32P]acetyl phosphate, we assume it to be in a native state (see Fig. S2 in the supplemental material). Nevertheless, it should be mentioned that purified 6His-YpdB has a high tendency to aggregate (data not shown), which makes it difficult to estimate precisely the percentage of active protein in the preparations used, and KD (equilibrium dissociation constant) values might be too high. 6His-YpdB bound tightly to the yhjX promoter region (KD, 175 nM) (Fig. 2B), whereas the KD values for all other potential target gene promoters, including yjiY (Fig. 2B), were determined to be >800 nM, essentially equivalent to the measured affinity of 6His-YpdB for the negative-control fragment (data not shown), a 276-bp fragment of the lysP promoter (33).
The affinity of YpdB for the yhjX promoter region is comparable to those of other LytTR RRs for their targets, e.g., the monomeric DNA-binding domain of AgrA (KD, 80 nM) (13) or full-length YehT (KD, 75 nM) (15). However, phosphorylation of 6His-YpdB did not notably increase its affinity for the yhjX promoter (data not shown), which contrasts with the behavior of other RRs of the LytTR type (34).
Taken together, these results indicate that 6His-YpdB binds specifically and with high affinity to the yhjX promoter, which supports the idea that yhjX expression is dependent on YpdB. We therefore did subsequent, more detailed analyses on the yhjX promoter.
Characterization of the yhjX promoter and localization of the YpdB-binding site.
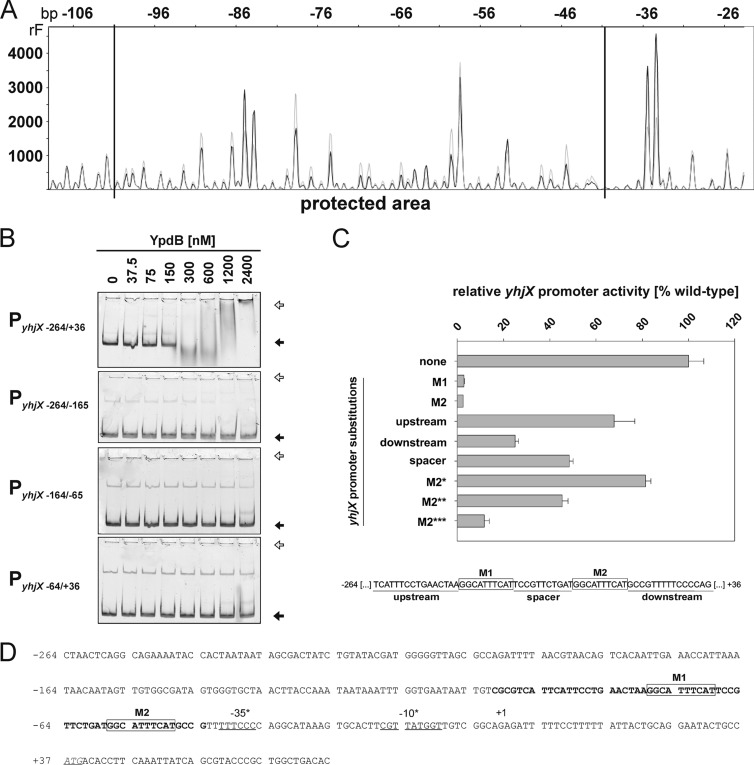
To identify the YpdB-binding site within the yhjX promoter, we used DNase I footprinting. A DNA fragment comprising the segment at positions −264 to +36 of the yhjX promoter/control region was incubated with increasing concentrations of 6His-YpdB. YpdB specifically protected a 59-bp stretch between positions −101 and −43 (Fig. 3A and D) from the action of DNase I. The degree of protection increased with the concentration of 6His-YpdB (data not shown). Within the protected stretch, two copies of the 10-bp motif GGCATTTCAT, designated M1 and M2, were identified, separated by an intervening spacer of 11 bp. The disposition of these sequence elements is typical for protein-binding sites in general (35).
Fig 3.
Characterization of the YpdB-binding site within the yhjX promoter. (A) DNase I digestion patterns were determined for a yhjX promoter fragment comprising bp −264 to +36 in the presence of 2.4 μM 6His-YpdB (black) or BSA (gray). The protected region lies within the horizontal lines (bp −101 to −43). rF, relative fluorescence. (B) EMSA of fluorescence-labeled DNA fragments comprising different yhjX promoter fragments after incubation in the presence of increasing concentrations of purified 6His-YpdB. The positions of free DNA (black arrows) and YpdB-DNA complexes (white arrows) are marked. (C) The in vivo effects of nucleotide substitutions (purines for pyrimidines and vice versa) in motifs 1 and 2 and the upstream, spacer, and downstream regions within the yhjX promoter were assayed using a yhjX promoter::lacZ fusion. The locations of motifs M1 and M2 and the other regions are depicted in the scheme below the graph. Pairwise substitutions, beginning at each end, were also introduced into motif 2, resulting in M2* (TGCATTTCAG), M2** (TTCATTTCCG), and M2*** (TTAATTTACG) (letters in boldface type mark the substituted nucleotides). Bacteria were cotransformed with pBAD33-ypdB and cultivated under aerobic growth conditions in LB medium at 37°C until the logarithmic growth phase was reached. ypdB overexpression was then induced by the addition of 0.2% (wt/vol) l-arabinose to the culture, and cells were harvested after 45 min. The β-galactosidase activity was determined (three independent experiments) and normalized to the wild-type yhjX promoter activity (set to 100%). (D) Nucleotide sequence of the yhjX upstream region (positions −264 to +77). The region protected from digestion by DNase I is marked in boldface type (59 bp). The start codon is shown in italics and underlined. +1 indicates the start of transcription identified previously by Kraxenberger et al. (15) via 5′ rapid amplification of cDNA ends, and binding motifs M1 and M2 are boxed and comprise a repeat of the sequence GGCATTTCAT.
In parallel, the same 300-bp yhjX promoter/control sequence was split into three 100-bp fragments, and each was tested in EMSAs. As before, 6His-YpdB bound tightly to the PyhjX−264/+36 fragment (KD, 175 nM) (Fig. 3B), but it showed little affinity for any of the individual segments (KD, >1,200 nM). These data suggested that subdivision of the fragment in this way destroys the YpdB-binding site, which is in accord with the arrangement of the binding motifs identified by footprinting analysis (Fig. 3D).
To characterize the putative YpdB-binding sites in more detail, the effects of alterations within the yhjX promoter were tested in vivo. Reporter strains carrying various mutated yhjX promoter::lacZ fusions were tested for β-galactosidase activity (Fig. 3C). A reporter with the wild-type YpdB-binding motif (PyhjX) displayed maximal yhjX promoter activity, which was set to 100%. Substitution of purines for pyrimidines (and vice versa) in motif M1 or M2 eliminated promoter activity. Reporter strains carrying yhjX::promoter variants with replacements of the spacer or replacements of 15 bp upstream of M1 or downstream of M2 were still able to induce the yhjX promoter albeit with reduced activity. Finally, we introduced pairwise substitutions (shown in boldface type) in motif M2, beginning at each end, to give M2* (TGCATTTCAG), M2** (TTCATTTCCG), and M2*** (TTAATTTACG). The more substitutions present in the variant, the greater the decrease in promoter activity (Fig. 3C).
Many of the LytTR-type family members described so far bind to pairs of direct repeats of 9 to 10 bp separated by 12- or 13-bp spacers (13, 15, 34, 36, 37). In its architecture and overall length, the newly identified YpdB-binding site conforms to these criteria. It is, however, conceivable that structural properties of the surrounding DNA also influence YpdB-dependent yhjX expression, as we also observed reductions in promoter activity when the upstream or downstream region or the spacer between the M1 and M2 motifs was mutated (Fig. 3C). We therefore suggest that binding of YpdB to the yhjX promoter is modulated by the sequence-dependent structure of the surrounding DNA. Similar effects have been observed for other LytTR-type RRs (14, 15).
Strikingly, a search of the E. coli genome revealed that the YpdB-binding motif occurs only in the yhjX promoter. In addition, when we checked all differentially expressed genes identified by our microarray analysis (Table 2), no related motif was found in the regulatory regions of any of these genes. The same was true for the microarray data set of E. coli BW25113 versus the isogenic ypdAB deletion strain (3). Remarkably, a YpdB-binding site was also not found within the yjiY promoter. Since we found an increase in the level of yjiY expression (which is under the control of the YehU/YehT system [15]) upon overproduction of YpdB (Table 2 and Fig. 2A), there might be interconnectivity between the two similar YpdA/YpdB and YehU/YehT systems. A pattern search for two repetitive motifs gapped by 10 to 12 nucleotides with maximally three mismatches per motif within the first 600 nucleotides of 5′ untranslated regions (5′UTRs) identified the promoters of only two genes, ypdE (which is located in close proximity to the ypdABC operon) and yahL. However, we found no evidence that expression of these genes is dependent on YpdB, and no YpdB binding to either promoter was detectable (data not shown). Therefore, yhjX seems to be the sole target gene regulated by the YpdA/YpdB system.
Characterization of YhjX.
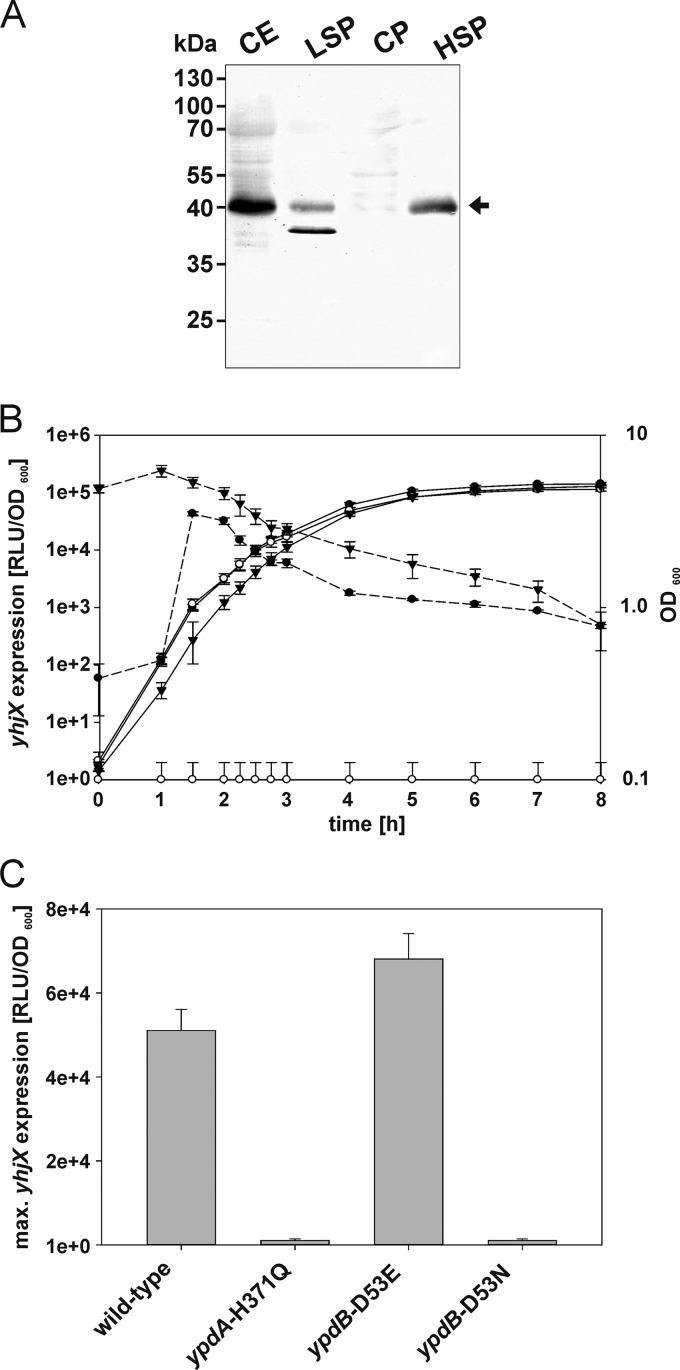
At 79.94 centisomes on the E. coli chromosome, yhjX is located quite far away from the ypdABC operon. Strikingly, however, in other bacteria, e.g., Yersinia enterocolitica, Aeromonas hydrophila, and Pectinobacterium species, the ypdABC operon and yhjX or a gene encoding a similar sequence are colocalized on the chromosome (38), providing further support for a functional relationship between YpdA/YpdB and YhjX. yhjX codes for an inner membrane protein with 12 predicted transmembrane helices belonging to the MFS transporters (39). Cell fractionation experiments confirmed that YhjX-6His is an integral membrane protein (Fig. 4A). Based on sequence similarities to the oxalate:formate antiporter OxlT in Oxalobacter formigenes, a function of YhjX as an exchanger for carboxylic acids has been postulated but never proven (23, 39). Future studies will concentrate on characterizing the function of YhjX, its mode of transport (importer or exporter), its specific substrate, and its mode of energization.
Fig 4.
Localization of YhjX-6His and characterization of yhjX expression in vivo. (A) yhjX-6His was overexpressed in E. coli BL21(DE3) cells. The cells were then disrupted and fractionated, and the subcellular localization of YhjX was analyzed by SDS-PAGE and Western blotting. An equal volume of each fraction was loaded into each lane, and a monoclonal mouse antibody against the 6His tag was used for detection. CE, crude cell extract; LSP, low-speed pellet; HSP, high-speed pellet; CP, cytoplasmic fraction. (B and C) A luciferase-based reporter assay was used to monitor yhjX expression in growing E. coli strains. All strains contained plasmid pBBR yhjX-lux. In addition, E. coli MG1655 (●) and E. coli MG21 (ΔypdABC) (○) were transformed with plasmid pBAD24 or pBAD24-ypdAB, and the latter was used for complementation of E. coli strain MG21 (▼) (B). Furthermore, E. coli strain MG1655 rpsL150 (wild type), E. coli strain MG23 (ypdA-H371Q), E. coli strain MG24 (ypdB-D53E), and E. coli strain MG25 (ypdB-D53N) were tested (C). Bacteria were cultivated in LB medium under aerobic conditions, and growth (solid lines) and the activity of the reporter enzyme luciferase (dashed lines) were continuously monitored. The maximal luciferase activity normalized to an optical density of 1 (RLU/OD600) served as the measure for yhjX expression. All experiments were performed at least three times, and the error bars indicate the standard deviations of the means.
Stimulus response analysis of the YpdA/YpdB system.
To gain insight into the in vivo expression pattern of yhjX, a transcriptional fusion was constructed, in which the yhjX promoter (PyhjX−264/+36) is coupled to the luciferase luxCDABE operon (plasmid pBBR yhjX-lux). E. coli MG1655 was transformed with this plasmid, and growth and luminescence (as a measure of yhjX expression) in 118 different media (see Table S3 in the supplemental material) were monitored under aerobic conditions over time. The maximal luciferase activity/OD600 was used as an indicator of the degree of induction of yhjX (Fig. 4B).
In wild-type cells, induction of yhjX in tryptone-yeast extract (LB) medium was observed, whereas yhjX induction was completely prevented in the isogenic ypdABC deletion mutant strain MG20 (Fig. 4B). Complementation of mutant strain MG20 with ypdAB in trans restored yhjX expression (Fig. 4B). Moreover, inactivation of the conserved histidine (H371) in the HK YpdA by replacement with glutamine, as well as substitution of the conserved aspartate in the RR YpdB (YpdB-D53N), prevented yhjX induction (Fig. 4C). In contrast, the putatively phosphorylation-independent YpdB-D53E variant induced yhjX (Fig. 4C). This yhjX induction pattern clearly indicates the importance of the phosphorylation sites in the YpdA/YpdB system.
Our comprehensive carbon source evaluation (see Table S3 in the supplemental material) revealed that yhjX was strongly induced when the reporter strain was grown in medium containing pyruvate, yeast extract, tryptone-yeast extract (LB), gluconate, or glucuronate as the carbon source (Fig. 5A; see also Table S3 in the supplemental material). Other sugars or amino acids, acetate and lactate, or toxic compounds, e.g., benzoate (40), peptidoglycan fragments (41) (Fig. 5A; see also Table S3 in the supplemental material), or the toxic peptides ShoB, LdrD, or IbsC (42), did not induce yhjX expression (data not shown).
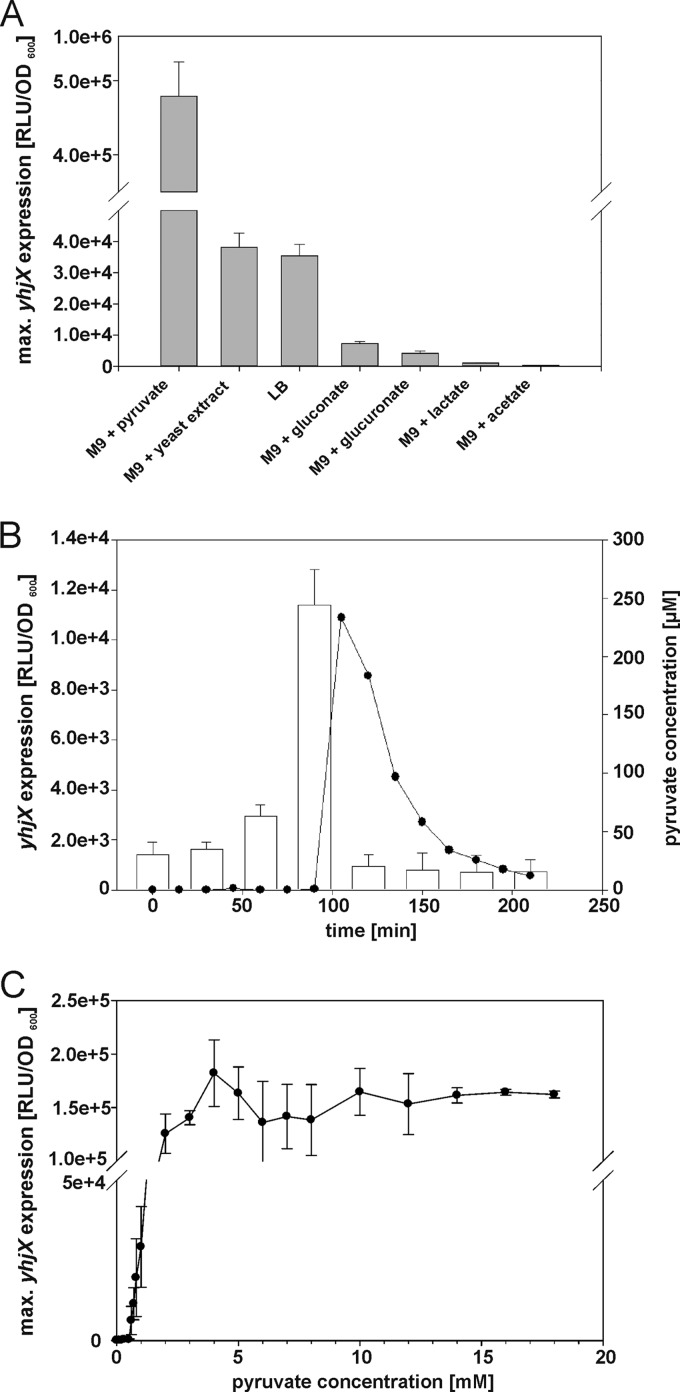
Fig 5.
Influence of extracellular pyruvate on yhjX expression. E. coli MG1655/pBBR yhjX-lux cells were grown as described in the legend of Fig. 4B. (A) E. coli MG1655/pBBR yhjX-lux cells were cultivated in LB medium or M9 minimal medium with the indicated carbon sources (0.4% [wt/vol]). The histogram shows the maximal levels of yhjX expression recorded in each case. (B) At the indicated times, samples were taken to determine the pyruvate concentration in cell-free culture supernatants (white bars) and to measure yhjX expression in LB medium (●). (C) A luciferase-based reporter assay was used to determine the dependence of yhjX expression on the concentration of pyruvate in the medium (M9 minimal medium with pyruvate and glucose as the carbon source [together 20 mM]). All experiments were performed at least three times, and the error bars indicate the standard deviations of the means.
Furthermore, we tested whether YhjX, the putative transporter, had an effect on yhjX induction. Induction of the PyhjX-luxCDABE reporter was found to be almost identical in the yhjX mutant and the parental strain (see Fig. S3 in the supplemental material). Therefore, it seems unlikely that YhjX exerts feedback regulation on its own expression.
In summary, expression of yhjX is induced by the YpdA/YpdB system in response to pyruvate (see below) and under conditions (gluconate or glucuronate as the sole C source) in which the Entner-Doudoroff pathway is required for carbon metabolism. Although the in vitro phosphorylation of YpdA was very weak and phosphotransfer to YpdB was undetectable (data not shown), our in vivo data indicate the functional importance of the phosphorylation sites.
Pyruvate-dependent induction of yhjX in E. coli MG1655.
When E. coli cells were grown in LB medium, yhjX expression was induced when the culture was in the mid-exponential growth phase (Fig. 4B), whereas in pyruvate-containing medium, no such delay was observed (data not shown). Therefore, we measured the extracellular pyruvate concentration in a culture of E. coli MG1655/pBBR yhjX-lux growing in LB medium. We detected a basal concentration of about 25 μM pyruvate, which increased sharply to 250 μM after 90 min. Shortly afterwards, yhjX expression was detected (Fig. 5B), and strikingly, the extracellular pyruvate concentration subsequently returned to the basal level (Fig. 5B). These data revealed that induction of yhjX is correlated with a relatively abrupt increase in the concentration of extracellular pyruvate and is followed by a gradual fall in pyruvate levels in the medium. We then set out to determine the threshold concentration of pyruvate necessary to induce yhjX expression. Cells were cultivated on glucose as the primary C source and supplemented with increasing concentrations of pyruvate (total C source, 20 mM). The threshold concentration of pyruvate required for induction was determined to be 600 μM. Below this concentration, no yhjX expression was detectable, but the expression level increased further at higher concentrations (the concentration that resulted in half-maximal expression was determined to be 1.6 ± 0.4 mM) (Fig. 5C). The threshold concentration for pyruvate was found to be in the same range as the highest concentration of extracellular pyruvate measured in E. coli cultures growing in LB medium. The 2.4-fold difference might be explained by (i) an underestimation of the concentration of extracellular pyruvate by the coupled enzymatic assay used and (ii) differences in growth media (LB versus minimal medium). Consequently, we determined the extracellular pyruvate concentration in E. coli cells grown on other C sources (Table 3). When glucose was present as the C source, the extracellular pyruvate level did not increase, and yhjX induction was undetectable. In contrast, gluconate and glucuronate as C sources resulted in increased extracellular pyruvate levels and induction of yhjX. These findings suggest that extracellular pyruvate functions as a stimulus for the YpdA/YdpB system.
Table 3.
Extracellular pyruvate levels in growing E. coli cultures correlate with yhjX induction
| Medium or C source | Pyruvate concn (μM) |
||
|---|---|---|---|
| Before yhjX induction | After 30 min of yhjX induction | After 60 min of yhjX induction | |
| LB (tryptone/yeast extract) | 244 | 20 | 17 |
| M9 + gluconate | 214 | 105 | 53 |
| M9 + glucuronate | 185 | 52 | 18 |
| M9 + glucosea | 20 | 18 | 17 |
As no yhjX induction was detectable, samples were analyzed at the equivalent growth phases. Data were obtained from at least three independent experiments, and average values are presented; the standard deviation was less than 20%.
An increase in the extracellular pyruvate level occurs when the rate of production of pyruvate temporarily exceeds the bacterium's capacity to metabolize the compound (43). Under conditions of overflow metabolism, pyruvate, like acetate, is a major component of the exometabolome in a broad range of bacteria (44). Sugar acids, e.g., gluconate or glucuronate, are predominantly metabolized via the Entner-Doudoroff pathway in E. coli, resulting in the production of 2 pyruvates per sugar acid (45). Pyruvate is a precursor of several compounds and a central metabolite; therefore, its cellular concentration has to be tightly controlled (46). Nevertheless, exponentially growing bacteria secrete pyruvate into the medium under conditions of overflow metabolism. When nutrients become limiting, bacteria have to adapt accordingly. It was suggested previously that a switch to carbon starvation follows a two-stage protocol (47). The first response is scavenging, a process in which the level of production of proteins that forage for the limiting nutrient is increased (47). The carbon-scavenging regulon includes cyclic AMP (cAMP)/cAMP receptor protein (CRP), which mediates the use of alternative carbon sources (47). Furthermore, previously excreted compounds (e.g., pyruvate) are rapidly taken up to drive continuing growth (44). The second response includes global reprogramming for starvation and the transition into the stationary phase (48). To prevent premature initiation of the second response, the levels of extracellular nutrients have to be precisely monitored, a process in which the YpdA/YdpB system participates by sensing the presence of a specific exometabolite and inducing the expression of an appropriate transporter. To our knowledge, this is the first description of a pyruvate-sensing HK, although a correlation between pyruvate utilization and LytS/LytTR-like signaling was reported previously for Staphylococcus epidermidis (49). Future experiments will determine whether pyruvate binds directly to YpdA.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Exc114/1).
We thank Ingrid Weitl for excellent technical assistance, Nicola Lorenz for her support with strain constructions, Tobias Bauer for his help with the expression studies, and Björn Schwalb, Thomas Engleitner, and Achim Tresch for bioinformatics advice.
Footnotes
Published ahead of print 7 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02051-12.
REFERENCES
- 1. Jung K, Fried L, Behr S, Heermann R. 2012. Histidine kinases and response regulators in networks. Curr. Opin. Microbiol. 15: 118–124 [DOI] [PubMed] [Google Scholar]
- 2. Heermann R, Jung K. 2010. Stimulus perception and signaling in histidine kinases, p 135–161 In Krämer R, Jung K. (ed), Bacterial signaling. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 3. Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46: 281–291 [DOI] [PubMed] [Google Scholar]
- 4. Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280: 1448–1456 [DOI] [PubMed] [Google Scholar]
- 5. Zhou L, Lei X-H, Bochner BR, Wanner BL. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185: 4956–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Riley M, Abe T, Arnaud MB, Berlyn MKB, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, Mori H, Perna NT, Plunkett G, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart D, Wanner BL. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot. Nucleic Acids Res. 34: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anantharaman V, Aravind L. 2003. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics 4: 34 doi:10.1186/1471-2164-4-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galperin MY. 2008. Telling bacteria: do not LytTR. Structure 16: 657–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krogh A, Larsson B, von Heijne G, Sonnhammer E. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Evol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- 10. Jones DT. 2007. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23: 538–544 [DOI] [PubMed] [Google Scholar]
- 11. Viklund H, Elofsson A. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24: 1662–1668 [DOI] [PubMed] [Google Scholar]
- 12. Nikolskaya AN, Galperin MY. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30: 2453–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sidote DJ, Barbieri CM, Wu T, Stock AM. 2008. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure 16: 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Del Papa MF, Perego M. 2011. Enterococcus faecalis virulence regulator FsrA binding to target promoters. J. Bacteriol. 193: 1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kraxenberger T, Fried L, Behr S, Jung K. 2012. First insights into the unexplored two-component system YehU/YehT in Escherichia coli. J. Bacteriol. 194: 4272–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blattner F, Plunkett G, Bloch C, Perna N, Burland V, Riley M, Collado-Vides J, Glasner J, Rode C, Mayhew G, Gregor J, Davis N, Kirkpatrick H, Goeden M, Rose D, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1474 [DOI] [PubMed] [Google Scholar]
- 17. Heermann R, Zeppenfeld T, Jung K. 2008. Simple generation of site-directed point mutations in the Escherichia coli chromosome using Red/ET recombination. Microb. Cell Fact. 7: 14 doi:10.1186/1475-2859-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189: 113–130 [DOI] [PubMed] [Google Scholar]
- 19. Meselson M, Yuan R. 1968. DNA restriction enzyme from E. coli. Nature 217: 1110–1114 [DOI] [PubMed] [Google Scholar]
- 20. Aiba H, Adhya S, de Crombrugghe B. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256: 11905–11910 [PubMed] [Google Scholar]
- 21. Jung K, Krabusch M, Altendorf K. 2001. Cs+ induces the kdp operon of Escherichia coli by lowering the intracellular K+ concentration. J. Bacteriol. 183: 3800–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23. Keseler IM, Bonavides-Martínez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, Peralta-Gil M, Santos-Zavaleta A, Shearer AG, Karp PD. 2009. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 37: D464–D470 doi:10.1093/nar/gkn751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 25. Stadtman ER. 1957. Preparation and assay of acetyl phosphate. Methods Enzymol. 3: 228–231 [Google Scholar]
- 26. Lipmann F, Tuttle LC. 1945. A specific micromethod for the determination of acyl phosphates. J. Biol. Chem. 159: 21–28 [Google Scholar]
- 27. Cheng J, Sun S, Tracy A, Hubbell E, Morris J, Valmeekam V, Kimbrough A, Cline MS, Liu G, Shigeta R. 2004. NetAffx Gene Ontology Mining Tool: a visual approach for microarray data analysis. Bioinformatics 20: 1462–1463 [DOI] [PubMed] [Google Scholar]
- 28. Tetsch L, Koller C, Haneburger I, Jung K. 2008. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol. Microbiol. 67: 570–583 [DOI] [PubMed] [Google Scholar]
- 29. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria, 4th ed. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 30. Shah IM, Laaberki MH, Popham DL, Dworkin J. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135: 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, Kurbatova N, Lukk M, Malone J, Mani R, Pilicheva E, Rustici G, Sharma A, Williams E, Adamusiak T, Brandizi M, Sklyar N, Brazma A. 2011. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 39: D1002–D1004 doi:10.1093/nar/gkq1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Auchtung JM, Lee CA, Grossman AD. 2006. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J. Bacteriol. 188: 5273–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruiz J, Haneburger I, Jung K. 2011. Identification of ArgP and Lrp as transcriptional regulators of lysP, the gene encoding the specific lysine permease of Escherichia coli. J. Bacteriol. 193: 2536–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. 2004. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 186: 7549–7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, Jimenez-Jacinto V, Salgado H, Juárez K, Contreras-Moreira B, Huerta AM, Collado-Vides J, Morett E. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4: e7526 doi:10.1371/journal.pone.0007526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheung JK, Rood JI. 2000. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 182: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Diep DB, Havarstein LS, Nes IF. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178: 4472–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39: D561–D568 doi:10.1093/nar/gkq973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pao SS, Paulsen IT, Saier MH., Jr 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62: 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kannan G, Wilks JC, Fitzgerald DM, Jones BD, Bondurant SS, Slonczewski JL. 2008. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 8: 37 doi:10.1186/1471-2180-8-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brunskill E, Bayles K. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. 2008. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 70: 1076–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Holms H. 1996. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol. Rev. 19: 85–116 [DOI] [PubMed] [Google Scholar]
- 44. Paczia N, Nilgen A, Lehmann T, Gatgens J, Wiechert W, Noack S. 2012. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Fact. 11: 122 doi:10.1186/1475-2859-11-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murray EL, Conway T. 2005. Multiple regulators control expression of the Entner-Doudoroff aldolase (Eda) of Escherichia coli. J. Bacteriol. 187: 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA. 2006. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl. Environ. Microbiol. 72: 3653–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peterson CN, Mandel MJ, Silhavy TJ. 2005. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J. Bacteriol. 187: 7549–7553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66: 373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu T, Lou Q, Wu Y, Hu J, Yu F, Qu D. 2010. Impact of the Staphylococcus epidermidis LytSR two-component regulatory system on murein hydrolase activity, pyruvate utilization and global transcriptional profile. BMC Microbiol. 10: 287 doi:10.1186/1471-2180-10-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17: 103–113 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.