Abstract
The number of circulating tumor cells (CTCs) found in blood is known to be a prognostic marker for recurrence of primary tumors, however, most current methods for isolating CTCs rely on cell surface markers that are not universally expressed by CTCs. Dielectrophoresis (DEP) can discriminate and manipulate cancer cells in microfluidic systems and has been proposed as a molecular marker-independent approach for isolating CTCs from blood. To investigate the potential applicability of DEP to different cancer types, the dielectric and density properties of the NCI-60 panel of tumor cell types have been measured by dielectrophoretic field-flow fractionation (DEP-FFF) and compared with like properties of the subpopulations of normal peripheral blood cells. We show that all of the NCI-60 cell types, regardless of tissue of origin, exhibit dielectric properties that facilitate their isolation from blood by DEP. Cell types derived from solid tumors that grew in adherent cultures exhibited dielectric properties that were strikingly different from those of peripheral blood cell subpopulations while leukemia-derived lines that grew in non-adherent cultures exhibited dielectric properties that were closer to those of peripheral blood cell types. Our results suggest that DEP methods have wide applicability for the surface-marker independent isolation of viable CTCs from blood as well as for the concentration of leukemia cells from blood.
INTRODUCTION
The concentration of circulating tumor cells (CTCs) in peripheral blood is prognostic of metastatic disease in breast,1 prostate,2 colon,3 and other tumor types,4, 5, 6 and there is significant interest in developing methods to isolate intact CTCs efficiently from all types of metastatic cancers.7, 8 CTCs carry molecular profiles that reflect their tumors of origin and may be associated with their metastatic potential.9 Furthermore, being obtained by venipuncture, CTCs offer a relatively non-invasive method for studying and monitoring tumor evolution and metastatic potential on an ongoing basis. In addition to providing prognostic and potentially diagnostic information, CTC analysis may allow candidate patients to be identified for targeted therapies based on molecular profiles without invasive biopsies.10
The currently U.S. Food and Drug Administration-approved method for CTC isolation depends on cell surface markers and identifies cells of EpCAM+/cytokeratin+/CD45− phenotype11 more properly termed circulating epithelial cells. While this approach established that a correlation existed between CTC concentration and disease outcome in breast, colorectal, and prostate cancers,12 it cannot isolate EpCAM-negative cells, including tumors of mesenchymal origin13, 14 and cells that have undergone epithelial to mesenchymal transition (EMT). Given that expression of EpCAM and other surface antigens is variable, reliance on these markers for clinical applications is far from ideal. Therefore, antibody-independent methods for isolating unmodified and viable CTCs are being explored.15
Several studies have demonstrated that unmodified, viable tumor cells may be distinguished from blood cells by dielectrophoresis (DEP).16, 17, 18 In early experiments, differential trapping of tumor versus normal cells was achieved on small arrays of microelectrodes in microfluidic embodiments, however this approach had limited cell discrimination and low throughput capacity. Subsequent efforts improved both discrimination and throughput capacity by introducing dielectrophoretic field-flow-fractionation (DEP-FFF),19, 20 a chromatrographic technique in which microscale DEP, sedimentation and hydrodynamic lift forces act to control the position of cells in a hydrodynamic flow profile. In this approach, cells having different properties are carried at different speeds and improved discrimination may be achieved by increasing the length of the isolation chamber. Significantly, only the vertical dimension in which the balance of forces occurs needs to be constrained to the microscale in this approach, allowing the width of the separation channel and microelectrode array to be as large as desired to increase cell capacity. Using the DEP-FFF approach, we demonstrated 92% efficient isolation of 100 cultured breast cancer cells spiked into 105 peripheral blood mononuclear cells (PBMNs) blood21 and successfully removed breast tumor cells from CD34+ hemopoietic stem cells.22 These advances make it feasible for the first time to apply DEP for processing not only small specimens in microfluidic and lab-on-chip devices but also for pre-processing large clinical specimens in a time span that is practical for prognostic and diagnostic determinations of CTCs.
Despite these developments, a systematic study of the DEP properties of a broad range of cancer cell types has been lacking and the biological basis for the observed dielectric differences between tumor and blood cells has not been rationalized, leaving uncertain the extent to which DEP might be applicable to the wide spectrum of human cancers. In this article, we address these questions by showing that the cell types in the widely representative NCI-60 panel of cancers23, 24, 25, 26 are amenable to isolation from blood by DEP. Recently, we modified this DEP-FFF approach to allow continuous-flow sorting of cells to achieve throughputs >106 nucleated cells/min, allowing 10 ml blood specimens to be processed in under an hour,27, 28 a method now being developed commercially.29
MATERIALS AND METHODS
Cell culture
NCI-60 cells were cultured to 50%-70% confluence in RPMI (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Grand Island, NY). Adherent cultures were harvested by rinsing with calcium- and magnesium-free Hank's buffered saline solution, incubation at 37 °C for 5 min with Trypsin/EDTA followed by sharp tapping, and neutralization with RPMI + 10% FBS. Non-adherent cell cultures were spun down from culture and suspended directly in RPMI + 10% FBS. Cells were counted to ensure >98% viability by trypan blue dye exclusion and suspended at ∼106 cell/ml in RPMI + 10% FBS in conical tubes for DEP analysis. Cell line identity was confirmed by short tandem repeat DNA fingerprinting.
DEP-FFF
The DEP-FFF device for this study was similar to that described in earlier work and consisted of a thin, flat chamber 300 mm long × 25 mm wide × 0.580 mm high. Its floor was covered by a DEP microelectrode array of plane, parallel, interdigitated gold-plated copper electrodes of 50 μm width and spacing and 1 μm thickness on a kapton substrate.30 The array was energized with AC voltages of 2.8 V p-p in the frequency range 15 kHz to 200 kHz at currents up to 2.5 A root mean square.
The DEP-FFF buffer consisted of an aqueous solution of 9.5% sucrose (S7903, Sigma-Aldrich, St Louis, MO), 0.1 mg/ml dextrose (S73418-1, Fisher, Fair Lawn, NJ), 0.1% pluronic F68 (P1300, Sigma-Aldrich, St Louis, MO), 0.1% bovine serum albumin (A7906, Sigma-Aldrich, St Louis, MO), 1 mM phosphate buffer pH 7.0, 0.1 mM CaAcetate, 0.5 mM MgAcetate, and 100 units/ml catalase (C30, Sigma-Aldrich, St Louis, MO). The buffer was adjusted to a conductivity of 30 mS/m with KCl, which was far below the cell cytoplasmic conductivities (∼1 S/m) in order to facilitate DEP manipulation of the cells (see below). The sucrose compensated for the low ionic concentration by bringing the osmolarity to 315 mOs/kg in the normal physiological range. Dextrose provided a metabolic energy source to maintain cell viability, pluronic acted as a surfactant to protect cells from damage as they flowed near the surfaces of tubing and the DEP chamber, bovine serum albumin maintained a protective coating on chamber and tube surfaces that inhibited cell adhesion. Calcium and magnesium stabilized cell membrane structure and integrity and catalase acted to protect cells from the possibility of low-level H2O2 production by electrochemical processes when the DEP electrodes were energized.
For each DEP-FFF run, a 30 μl aliquot of cell harvest (106 cell/ml) was mixed with 300 μl of DEP buffer, injected into the front of the DEP chamber, and allowed to settle for 8 min. Flow of DEP-FFF buffer was then initiated at 4 ml/min, and one of three DEP signals (see below) was applied to the microelectrode array. As they emerged from the chamber, cells were counted and sized by laser light scatter (PC2400D, ChemTrac Systems, Norcross, GA).
Relationship between cell properties and DEP-FFF behavior
In DEP-FFF, cells are positioned in a parabolic flow velocity profile by the balance of sedimentation, dielectrophoretic, and hydrodynamic lift forces,27, 31, 32
| (1) |
Cells reach different equilibrium heights in the eluate parabolic velocity profile according to their physical properties and are carried through the chamber at corresponding speeds. A heterogeneous mixture of cells, therefore, gives rise to an elution profile that reflects the density, dielectric, and hydrodynamic lift distributions of the cells.27, 31, 32 For a mammalian cell, Eq. 1 may be expressed32 in terms of the height h at which the force balance occurs as
| (2) |
where , , and .
The cell biophysical parameters that determine the three force terms are, respectively, the cell differential density compared with the eluate medium , the cell DEP crossover frequency , and the cell geometrical deformability factor Φ, which govern hydrodymamic lift. Note that each force term depends on the cube of the cell radius (R) so this parameter cancels out and the force balance height is independent of cell size. A, B, and C are constants defined by the chosen experimental conditions and depend on the acceleration due to gravity (g), the DEP microelectrode element width and spacing (d), the electric permittivity of the suspending medium (), the frequency-dependent electrode polarization factor (), the applied DEP voltage (V) of alternating frequency (f), the dynamic viscosity of the suspending medium (η), and the shear rate of fluid flow at the DEP chamber floor ().27, 31, 32 The hydrodynamic lift force term is based on the equation derived by Abkarian et al.33, 34, 35 from exhaustive studies on vesicles. These authors describe the mechanical deformation factor Φ, which accounts for variations in shape that impact hydrodynamic lift at different heights under Poisseiulle flow conditions. We used this approximation for the low wall shear rates of ∼100/s at the constant chamber flow rate of 4 ml/min that was present in all our measurements.
For viable mammalian cells that have an intact membrane barrier function and maintain physiological cytoplasmic ionic conductivity in the DEP-FFF buffer, the dielectric properties are described by the bracketed Claussius-Mossotti factor, which depends on the difference between the so-called cell crossover frequency and the DEP signal frequency f. The direction of the DEP force, towards or away from the microelectrode plane during DEP-FFF, is determined by whether the field frequency is greater or less than the cell crossover frequency.36, 37, 38
At DEP frequencies well below 1 MHz, a mammalian cell that has relaxed to a spherical conformation suspended in DEP buffer has a crossover frequency that may be approximated as , where is the medium conductivity.36, 39, 40, the capacitance per unit area of the cell plasma membrane, varies substantially between different cell types41, 42 and between cells in different states.40, 43, 44, 45, 46, 47, 48 Differences in correlate with plasma membrane area associated with variations in surface features including ruffles, folds, and microvilli.43, 45 Smooth plasma membrane has a capacitance49 of mF m−2 but these cell surface features effectively increase the area by a folding factor , where ϕ is the ratio of the actual cell membrane surface area to that of a smooth sphere of the same radius and is a measure of cell surface “wrinkling.” The effective capacitance per unit area of the cell may be written43 and the crossover frequency as .
It is evident from Eq. 2 that the DEP force depends on the square of the applied DEP voltage V, suggesting it might be advantageous to use a high DEP voltage to increase the DEP force. However, cells may be electroporated by intense electric fields and an applied voltage of 2.8 V p-p was chosen for our DEP-FFF experiments because it provided adequate DEP forces without causing evidence of cell damage. Following DEP-FFF profiling at this voltage, cells were viable as judged by trypan blue dye exclusion, and they could be returned to culture and grown (data not shown).
Derivation of cell parameters from DEP-FFF data
To characterize the biophysical properties of each cell type in the NCI-60 panel, elution characteristics were measured under three conditions to allow , , and to be disentangled from one another. This was achieved by applying the following DEP signals:
-
(i)
A constant frequency of 15 kHz throughout the run. Cells are highly levitated and hydrodynamic lift is minimal under this condition causing the elution behavior to be dominated by the cell density and crossover frequency parameters;
-
(ii)
No DEP field. The DEP force is zero, and the elution behavior corresponds to sedimentation DEP-FFF and is controlled by the cell density and mechanical deformability parameters;
-
(iii)
A swept DEP frequency starting far above the DEP crossover frequency of the cells and falling far below it. During the sweep, the DEP force changes from strongly positive, which pulls the cells towards the microelectrode array on the chamber floor and almost immobilizes them, to strongly negative, which levitates the cells high into the eluate flow stream where they are carried rapidly through the chamber. All three cell biophysical parameters control the cell elution behavior in the swept frequency case. Frequency sweeps from100 kHz to 15 kHz over 600 s were used for the NCI-60 cell types and from 300 kHz to 15 kHz over 600 s for blood cell types, reflecting the fact that blood cells generally had much higher crossover frequencies than NCI-60 cells.
To derive cell biophysical parameters from the three resulting cell elution profiles, we applied an approach modified from earlier work27, 31, 32 employing matlab scripts. Starting with seed values of the crossover frequency kHz and mechanical deformability factor ,
-
(a)
Cell elution times for the 15 kHz fixed frequency run were mapped to cell densities based on and Φ;
-
(b)
Using the value derived from (a), cell elution times for the “no DEP” run were mapped to determine revised cell mechanical deformability factors Φ;
-
(c)
Using the and Φ values derived from (a) and (b), cell elution times for the “swept-frequency” run were mapped to determine revised cell crossover frequencies .
-
(d)
Steps (a), (b), and (c) were iterated until , Φ, and converged to solutions that were consistent with the cell elution profiles observed under all three DEP-FFF conditions.
Separation of tumor cells from PBMNs
To demonstrate the separation of tumor cells from PBMNs by batch mode DEP-FFF, a suspension of PBMNs was prepared by standard density gradient isolation of blood from normal donors from the blood bank over Histopaque 1077. PBMNs were counted to ensure >98% viability (by trypan blue), and these were co-suspended with previously harvested cultured cancer cells (either MDA-MB-231 breast cells or HL-60 promyeleocytic leukemia cells) to give a total cell concentration of ∼106 cell/ml in RPMI + 10% FBS in conical tubes. DEP elution profiles illustrating cell separation were then conducted using the same conditions as the other DEP experiments except that fixed operating frequencies of 65 kHz for MDA-MB-231 cells and 80 kHz for HL-60 cells were applied (see later).
RESULTS
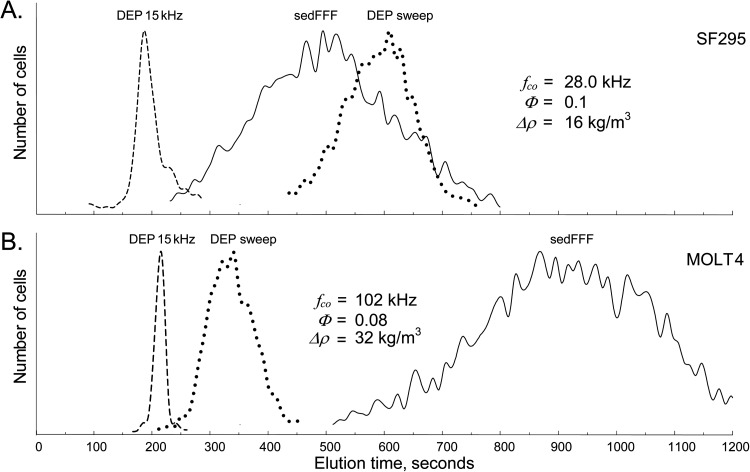
Elution profiles for SF295 glioblastoma and MOLT4 leukemia cell lines under the three DEP-FFF conditions used to characterize the cells are shown in Figure 1 to compare the extremes of variations seen for different cell types in the NCI-60 panel. Cell elution at 15 kHz was rapid because mammalian cells experience DEP repulsion from the microelectrode array in electric fields oscillating below their crossover frequencies, become levitated above the chamber floor and into the hydrodynamic flow stream where they are carried rapidly from the chamber. Denser cells equilibrated nearer to the chamber floor where the eluate flows slowly; that lighter cells that were levitated higher into faster moving regions of the eluate. Therefore, at 15 kHz, the denser MOLT4 leukemia cells are seen to elute more slowly in Figure 1b than the less dense SF295 glioblastoma cells in Figure 1a.
Figure 1.
DEP-FFF elution profiles for (A) SF295 human glioblastoma cells and (B) MOLT4 human acute lymphoblastic leukemia cells demonstrating the behavior of NCI-60 cells having large differences in their density, dielectric, and hydrodynamic lift properties. The elution profiles for 15 kHz DEP reflect predominantly cell density differences; those for no DEP (sedFFF) reflect cell density and hydrodynamic lift effects; and those using a DEP frequency sweep reflect the combination of sedimentation, hydrodynamic lift and DEP crossover frequency effects.
The high frequency DEP signal used at the start of the swept-frequency runs attracted cells by positive DEP to the microelectrode array where they barely moved in the slowest-moving region of the eluate. As the frequency swept lower with time, the attractive DEP force diminished, reached zero when the applied frequency coincided with the cell crossover frequency, and then turned repulsive with further decreases in frequency, levitating the cells into fast moving eluate. As a result, each cell exhibited a total elution time under swept frequency DEP-FFF conditions that reflected its crossover frequency. The faster elution of MOLT4 cells compared with SF295 under swept frequency DEP-FFF in Figure 1 shows that MOLT4 cells had a higher crossover frequency and were levitated sooner during the frequency sweep than SF295 cells.
With no DEP signal applied, elution profiles corresponded to sedimentation field-flow fractionation50, 51 and were controlled by the balance of sedimentation and hydrodynamic lift forces. Elution of MOLT4 occurred much more slowly than SF295 under these conditions, reflecting both the lower density and greater mechanical flexibility (higher Φ) of SF295 cells compared with MOLT4.
The density ρ, crossover frequency fco, and cell mechanical deformability Φ parameters were derived by analyzing elution profiles, like those exhibited in Figure 1, for each of the NCI-60 cell types. The mean densities of NCI-60 cell types are shown in ascending order in Figure 2. To verify the validity of these density measurements, elution profiles were also measured for peripheral blood populations for which density values are well known. All cells except BT-549 (one of five breast cancers) and HOP-62 (one of nine non-small cell lung cancers) in the NCI-60 panel had densities in the range 1048 to 1068 kg/m3.
Figure 2.
Densities of the NCI-60 cell types deduced from DEP-FFF analysis of the cells and shown in ascending order. The thickness of each scale bar represents the number of cells at a given density relative to the mode, which has maximum thickness, based on skew-normal distributions. The blue line shows the density of Histopaque 1077 and LSM at 1077 kg/m3.
The DEP crossover frequencies derived for the NCI-60 cell types together with those for peripheral blood cell subpopulations are shown in Figure 3 at an eluate conductivity of 30 mS/m. The values obtained for cell types for which we previously undertook cell dielectric measurements, the DEP-FFF agreed well.41 The shapes of the plotting symbols represent the crossover frequency profile observed for each cell type fitted to a skew-normal distribution, allowing the crossover frequency distributions for different cell types to be visualized and compared. Peripheral blood cell subpopulations exhibited much higher crossover frequencies than all of the NCI-60 cell types that grew in adherent cultures. Only three of the NCI-60 cell lines, namely the leukemias SR, MOLT-4, and CCRF-CEM, had crossover frequencies that approach those of any of the blood cell types (monocytes and CD34+ hemopoietic cells).
Figure 3.
DEP crossover frequencies of the NCI-60 cell types and normal peripheral blood cells deduced from DEP-FFF analysis shown in ascending order at a suspension conductivity of 30 mS/m. The thickness of each scale bar represents the number of cells at a given crossover frequency relative to the mode, which has maximum thickness, based on skew-normal distributions. The blue line at 65 kHz shows a possible choice for a DEP-FFF operating frequency that should be able to isolate NCI-60 cell types, except leukemia lines, from normal blood cells.
Figure 3 shows the DEP crossover frequencies of the cell types expressed in ascending order but it is also helpful, from the perspective of understanding potential differences between tumors associated with different organs, to group the cells according to site of origin as shown in Figure 4.
Figure 4.
Range of DEP crossover frequencies ±1 standard deviation of the NCI-60 cell types and normal peripheral blood cells deduced from DEP-FFF analysis at a suspension conductivity of 30 mS/m by tissue origin. The blue line at 65 kHz shows a possible choice for a DEP-FFF operating frequency that would isolate essentially all NCI-60 cell types except leukemia lines from blood.
An example of the separation of tumor cells from PBMNs using the batch-mode DEP-FFF approach is shown in Figure 5 in which a contrast is drawn between a solid and a leukemic cancer type. Figure 5a shows the DEP-FFF elution profile of a mixture of MDA-MB-231 cells and PBMNs at an applied frequency of 65 kHz. MDA-MB-231 is a triple-negative breast cancer (negative for estrogen, progesterone, and HER-2 receptors) that also does not express EpCAM. The DEP forces levitated the blood cell subpopulations to between 15 and 30 μm above the chamber floor, and they emerged as a single peak from 180 to 400 s after eluate flow was started. MDA-MB-231 cells experienced DEP forces towards the microelectrode array and emerged in a second peak from 520 to 1000 s flowing at between 6 and 11 μm above the chamber floor (approximately the cell radius).
Figure 5.
Batch-mode DEP-FFF elution profiles for two mixtures of cancer cells with normal peripheral blood mononuclear cells: (A) Normal PBMNs + MDA-MB-231 breast tumor cells run at a DEP operating frequency of 65 kHz; (B) Mormal PBMNs + HL-60 promyelocytic leukemia cells run at 80 kHz. Different colors denote cells of differing size ranges. Note that the elution times for the cancer cells have a weak dependency on cell size because their DEP crossover frequencies dominate their elution behaviors. The scale at top shows the height at which the cells travelled through the DEP chamber as deduced from their elution times in the Poiseiulle flow profile. The cancer cells were tracked by prelabelling them with green-fluorescent calcein-AM viable cell stain (L-3224, Molecular Probes, Life Technologies).
Figure 5b shows the DEP elution profile for promyelocytic leukemia line HL-60 cells mixed with PBMNs at an 80 kH operating frequency. The DEP crossover frequency for HL-60 is 70 kHz and the 80 kHz operating frequency was needed to bring these cells to the microelectrode array. This higher frequency still levitated lymphocytes to between 12 and 25 μm above the chamber floor where they emerged at from 250 to 500 s after eluate flow started. However, other mononuclear blood cells were levitated between only 9 and 20 μm above the floor and eluted in a much broader blood peak, from between 300 and 750 s, than that seen at 60 kHz. Even so, Figure 4b shows that the HL-60 cells eluted later than the PBMN peak, flowing through the chamber at between 5.5 and 9 μm above the chamber floor and emerging between 730 and 1100 s after eluate flow began.
DISCUSSION
The first step for achieving efficient isolation of CTCs from blood by DEP is debulking the specimen by removing erythrocytes (which constitute approximately 40% of the volume of whole blood) from the very much smaller concentration of PBMNs and cancer cells. This is necessary because, even though we will see later that DEP can isolate cancer cells from all types of blood cells, electric dipole interactions between cells result in poor DEP discrimination when cells in the suspension are less than 5 diameters apart.21 Debulking can be accomplished by erythrocyte lysis or by centrifugation of the specimen over histopaque or lymphocyte separation medium (LSM, Ficoll-Hypaque) having a density of 1077 kg/m3. Each method has disadvantages. The viability of tumor cells may be reduced during erythrocyte lysis if their dynamic volume response mechanism to osmotic stress is impaired and this is problematical if the goal is to recover viable tumor cells for proteinomic or gene expression analysis. On the other hand, for the density gradient method to enable efficient isolation of CTCs, the cancer cells must have a low enough density to be collected in the PBMN fraction and not in the pellet. Our DEP-FFF analysis is the first to profile the cell density distributions of a large number of different cancer cell types. Significantly, the cell density values found for all NCI-60 cell lines (Figure 2) lie below 1077 kg/m3, showing that all these cell types will indeed be collected with PBMNs during density gradient separation of whole blood specimens. This supports the use of density gradient isolation for debulking blood specimens for CTC analysis of different types of cancer without substantial loss of target cells assuming that the range of cell densities found within the NCI-60 panel is similar to that of CTCs in clinical specimens. Indeed, CTCs are successfully recovered from blood using density gradient centrifugation alone.52 Nevertheless, Figure 2 also suggests that protocols designed to reduce the number of blood cells in CTC isolates by centrifugation over medium of density significantly below 1077 kg/m3 might lose CTCs because many of the NCI-60 cell lines have densities greater than monocytes (1063 kg/m3) and very close to lymphocytes (1071 kg/m3).
The DEP force imposed on a cell depends on the frequency f of the DEP energizing voltage and the dielectric properties of the cell. Under conditions applicable for mammalian cell sorting, the cell dielectric properties may be described in terms of the cell crossover frequency . If cancer cells are to be separated efficiently by differential DEP forces from blood cells, the cancer and blood cells must have significantly different crossover frequencies. Figure 3 shows cell crossover frequency distributions measured for the NCI-60 cell types by DEP-FFF in an isotonic sucrose eluate having a conductivity of 30 mS/m. With the notable exception of cell lines of leukemic origin, all of the NCI-60 cell types have crossover frequencies that lie far below those of normal peripheral blood cell subpopulations. This is further emphasized by Figure (4), which shows the means and standard deviations of the crossover frequencies for the cell lines of each tissue type represented in the NCI-60 panel and demonstrates the clear distinction between the DEP crossover frequencies of cells derived from organs and those of blood with leukemia cell types lying in between. Under our conditions, a DEP operating frequency of 65 kHz (vertical lines in Figures 34) lay above the crossover frequencies of the non-leukemic NCI-60 cell lines and below the crossover frequencies for all peripheral blood cell subpopulations, showing that this frequency would provide DEP forces that attract cancer cells towards the microelectrodes while repelling blood cells. Therefore, this frequency is a suitable choice for driving the isolation of non-leukemic cancer cell types from blood by DEP-FFF under our conditions.
This result shows not only that all non-leukemia NCI-60 cell types are amenable to isolation from blood by DEP-FFF but also, remarkably, that a single set of DEP-FFF operating conditions may be used for tumor cell isolation regardless of the cancer type. If CTCs in clinical specimens have properties that are broadly similar to the cell types in the NCI-60 panel, the implication is that DEP-FFF is applicable to the isolation of all non-leukemic tumor types using a single set of operating parameters without needing prior knowledge of the cancer. Successful experiments to isolate CTCs from clinical specimens from a number of different cancers in our laboratory so far support this conclusion (data not shown). The standard deviations of the crossover frequencies for the blood cell types (see the skew normal distributions represented by the symbols in Figure 3) were small and well-separated from the crossover frequency distributions of the cancer cells. This shows that, even at the large concentration excesses of PBMNs compared to CTCs in clinical specimens, it should be possible to isolate CTCs from blood at high purity.
Figures 34 show that the NCI-60 leukemia lines had crossover frequencies that lay closer to those of peripheral blood cell types than the other cancers in the panel. Nevertheless, by using DEP operating frequencies above 65 kHz, concentration (rather than efficient isolation) of the leukemia cells is possible, as we demonstrated in earlier work on HL-60 leukemia cells.53 Figure 5 shows the separation of tumor cells from PBMNs using the batch-mode DEP-FFF approach and draws a contrast between a solid and a leukemic cancer type. MDA-MB-231, shown in Figure 5a, is a triple-negative breast cancer (negative for estrogen, progesterone, and HER-2 receptors) that also is very low in EpCAM expression. Cell isolation efficiency is high and essentially no blood cells eluted with MDA-MB-231 cells. This serves to emphasize that the DEP-FFF method is independent of cell surface markers, such as EpCAM, as expected.
The HL-60 leukemia cell line exhibited a DEP crossover frequency of 70 kHz and an operating frequency of 80 kHz was needed to achieve DEP-FFF separation. This frequency is approaching the DEP crossover frequency of granulocytes and monocytes, and Figure 5b shows that while the HL-60 cells eluted later than the PBMN peak, a small overlap between the PBMN and cancer cell elution peaks occurred and that a small proportion of granulocytes and monocytes co-eluted with the HL-60 leukemia cells. In general, then, our results show that DEP-FFF is suitable for enriching, rather than isolating, leukemia cells from patient blood specimens.
The crossover frequencies of cells in suspension arise from the two variables that determine cell membrane capacitance, namely the cell radius, r, and the membrane folding factor, ϕ (see earlier). We showed earlier that the membrane folding factor of cells recently released into suspension reflects their external morphology prior to release.41 Cells such as CTCs originate from growth sites in solid tumors and tend to have far more complex surface morphologies in association with neighboring cells during growth that give rise to much larger values of ϕ than is the case for cells native to the peripheral blood. Leukemia cells in various stages of the division cycle tend to have larger sizes than resting circulating blood cells but their site of origin leads to small membrane folding factors. These general principles seem to underlie the crossover frequency properties of the NCI-60 cells shown in Figure 3.
While this study uses batch mode DEP-FFF to characterize the cell properties, Figure 3 also reveals the conditions under which continuous flow DEP-FFF can be achieved.27 In that method, the PBMN specimen containing cancer cells is fed continuously into the bottom of the chamber and forms a thin laminar flow over the microelectrode array. After cells reach equilibrium flow conditions, the flow stream is split so that cancer cells are skimmed off through a slot in the chamber bottom while blood cells flow to a waste port. We recently introduced a method for continuous flow DEP-FFF to isolate CTCs from clinical specimens in a phase I clinical trial at a processing rate of more than 10 ml blood per hour.28, 29 As with batch-mode DEP-FFF, the discrimination of the continuous flow DEP-FFF method depends upon differences in the cell crossover frequencies, which translate to differences in cell transport heights above the DEP microelectrodes as shown in Figure 5. Provided the continuous flow DEP-FFF chamber is not overloaded with cells, isolation will proceed regardless of the number of blood or cancer cells present, allowing very small numbers of CTCs to be isolated from the 40 million or so PBMNs present in 10 ml clinical blood specimens. The continuous flow approach is described in the companion article to this one.28
CONCLUSIONS
Our results show that the dielectric properties of the non-leukemic cell types of the NCI-60 panel are sufficiently different from those of peripheral blood cells for them to be isolated from blood by DEP methods with high efficiency. Furthermore, similar operating conditions are applicable for isolating different types of non-leukemic cancers, implying that prior knowledge of the cancer characteristics should not be a requirement for the application of DEP-based isolation. Leukemic cells exhibit dielectric properties that permit them to be enriched from blood but not isolated at high purity. In all cases, DEP is independent of cell surface marker expression such as EpCAM and, therefore, DEP methods have the potential to be universally applicable for tumor cell analysis. Given the relative ease with which DEP methods can be electronically controlled and readily integrated into microfluidic devices, our results emphasize the significant potential for DEP methods in micrototal analysis systems for cancer applications and for sample preprocessing for these systems.
ACKNOWLEDGMENTS
We thank Tom Anderson for design, fabrication, and engineering in creating and maintaining the DEP-FFF instrumentation used in this study. NCI-60 cells are under MTA with Dr. Gordon B. Mills. This work was supported by Grant RP100934 from the Cancer Prevention and Research Institute of Texas (CPRIT) and funding from the Kleberg Center for Molecular Markers. K.S.H. is also supported by a Stand Up to Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0209). STR DNA fingerprinting was done by the Cancer Center Support Grant-funded Characterized Cell Line core, NCI # CA016672. P.G. and F.F.B. qualify to receive royalty distributions from patents assigned to the Regents of the University of Texas and licensed for commercial development of DEP-FFF technology.
References
- Dawood S., Broglio K., Valero V., Reuben J., Handy B., Islam R., Jackson S., Hortobagyi G. N., Fritsche H., and Cristofanilli M., Cancer 113, 2422–2430 (2008). 10.1002/cncr.23852 [DOI] [PubMed] [Google Scholar]
- de Bono J. S., Scher H. I., Montgomery R. B., Parker C., Miller M. C., Tissing H., Doyle G. V., Terstappen L. W., Pienta K. J., and Raghavan D., Clin. Cancer Res. 14, 6302–6309 (2008). 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- Meropol N. J., Clin. Adv. Hematol. Oncol. 7, 247–248 (2009). [PubMed] [Google Scholar]
- Xu W., Cao L., Chen L., Li J., Zhang X. F., Qian H. H., Kang X. Y., Zhang Y., Liao J., Shi L. H., Yang Y. F., Wu M. C., and Yin Z. F., Clin. Cancer Res. 17, 3783–3793 (2011). [DOI] [PubMed] [Google Scholar]
- Koyanagi K., O'Day S. J., Boasberg P., Atkins M. B., Wang H. J., Gonzalez R., Lewis K., Thompson J. A., Anderson C. M., Lutzky J., Amatruda T. T., Hersh E., Richards J., Weber J. S., and Hoon D. S., Clin. Cancer Res. 16, 2402–2408 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara T., Itoi T., Sofuni A., Itokawa F., Tsuchiya T., Tsuji S., Ishii K., Ikeuchi N., Tsuchida A., Kasuya K., Kawai T., Sakai Y., and Moriyasu F., J. Hepatobiliary Pancreat Surg. 15, 189–195 (2008). 10.1007/s00534-007-1250-5 [DOI] [PubMed] [Google Scholar]
- Bednarz-Knoll N., Alix-Panabieres C., and Pantel K., Breast Cancer Res. 13, 228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterlini-Brechot P. and Benali N. L., Cancer Lett. 253, 180–204 (2007). 10.1016/j.canlet.2006.12.014 [DOI] [PubMed] [Google Scholar]
- Leversha M. A., Han J., Asgari Z., Danila D. C., Lin O., Gonzalez-Espinoza R., Anand A., Lilja H., Heller G., Fleisher M., and Scher H. I., Clin. Cancer Res. 15, 2091–2097 (2009). 10.1158/1078-0432.CCR-08-2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachmann K., Camara O., Kroll T., Gajda M., Gellner A. K., Wotschadlo J., and Runnebaum I. B., J. Cancer Res. Clin. Oncol. 137, 1317–1327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. C., Doyle G. V., and Terstappen L. W., J. Oncol. 2010, 617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. J., Punt C. J., Iannotti N., Saidman B. H., Sabbath K. D., Gabrail N. Y., Picus J., Morse M., Mitchell E., Miller M. C., Doyle G. V., Tissing H., Terstappen L. W., and Meropol N. J., J. Clin. Oncol. 26, 3213–3221 (2008). 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- Raimondi C., Gradilone A., Naso G., Vincenzi B., Petracca A., Nicolazzo C., Palazzo A., Saltarelli R., Spremberg F., Cortesi E., and Gazzaniga P., Breast Cancer Res. Treat 130, 449–455 (2011). [DOI] [PubMed] [Google Scholar]
- Bonnomet A., Brysse A., Tachsidis A., Waltham M., Thompson E. W., Polette M., and Gilles C., J. Mammary Gland Biol. Neoplasia 15, 261–273 (2010). [DOI] [PubMed] [Google Scholar]
- Riethdorf S. and Pantel K., Pathobiology 75, 140–148 (2008). 10.1159/000123852 [DOI] [PubMed] [Google Scholar]
- Becker F. F., Wang X. B., Huang Y., Pethig R., Vykoukal J., and Gascoyne P. R., Proc. Natl. Acad. Sci. USA 92, 860–864 (1995). 10.1073/pnas.92.3.860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H. S., Kwon K., Kim S. I., Han H., Sohn J., Lee S., and Jung H. I., Lab Chip 11, 1118–1125 (2011). 10.1039/c0lc00345j [DOI] [PubMed] [Google Scholar]
- Wang L., Lu J., Marchenko S. A., Monuki E. S., Flanagan L. A., and Lee A. P., Electrophoresis 30, 782–791 (2009). 10.1002/elps.200800637 [DOI] [PubMed] [Google Scholar]
- Wang X. B., Vykoukal J., Becker F. F., and Gascoyne P. R., Biophys. J. 74, 2689–2701 (1998). 10.1016/S0006-3495(98)77975-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. B., Yang J., Huang Y., Vykoukal J., Becker F. F., and Gascoyne P. R., Anal. Chem. 72, 832–839 (2000). 10.1021/ac990922o [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne P. R. C., Noshari J., Anderson T. J., and Becker F. F., Electrophoresis 30, 1388–1398 (2009). 10.1002/elps.200800373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang J., Wang X. B., Becker F. F., and Gascoyne P. R., J. Hematother. Stem Cell Res. 8, 481–490 (1999). 10.1089/152581699319939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokilde R., Kaczkowski B., Podolska A., Cirera S., Gorodkin J., Moller S., and Litman T., Mol. Cancer Ther. 10, 375–384 (2011). 10.1158/1535-7163.MCT-10-0605 [DOI] [PubMed] [Google Scholar]
- Lorenzi P. L., Reinhold W. C., Varma S., Hutchinson A. A., Pommier Y., Chanock S. J., and Weinstein J. N., Mol. Cancer Ther. 8, 713–724 (2009). 10.1158/1535-7163.MCT-08-0921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankavaram U. T., Reinhold W. C., Nishizuka S., Major S., Morita D., Chary K. K., Reimers M. A., Scherf U., Kahn A., Dolginow D., Cossman J., Kaldjian E. P., Scudiero D. A., Petricoin E., Liotta L., Lee J. K., and Weinstein J. N., Mol. Cancer Ther. 6, 820–832 (2007). 10.1158/1535-7163.MCT-06-0650 [DOI] [PubMed] [Google Scholar]
- Roschke A. V., Tonon G., Gehlhaus K. S., McTyre N., Bussey K. J., Lababidi S., Scudiero D. A., Weinstein J. N., and Kirsch I. R., Cancer Res. 63, 8634–8647 (2003). [PubMed] [Google Scholar]
- Gascoyne P. R. C., “Isolation and characterization of cells by dielectrophoretic field-flow fractionation,” in Field-Flow Fractionation in Biopolymer Analysis (Springer-Verlag, 2012). [Google Scholar]
- Shim S., Stemke-Hale K., Tsimberidou A. M., Noshari J., Anderson T. E., and Gascoyne P. R. C., Biomicrofluidics 7, 011807 (2013). 10.1063/1.4774304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Jafferji I., Garza M., Melnikova V., Hasegawa D., Pethig R., and Davis D., Biomicrofluidics 6, 024133 (2012). 10.1063/1.4731647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vykoukal J., Vykoukal D. M., Freyberg S., Alt E. U., and Gascoyne P. R., Lab Chip 8, 1386–1393 (2008). 10.1039/b717043b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne P. R., Anal. Chem. 81, 8878–8885 (2009). 10.1021/ac901470z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S., Gascoyne P., Noshari J., and Hale K. S., Integr. Biol. 3, 850–862 (2011). 10.1039/c1ib00032b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader M. A., Vitkova V., Abkarian M., Viallat A., and Podgorski T., Eur. Phys. J. E 19, 389–397 (2006). 10.1140/epje/i2005-10058-x [DOI] [PubMed] [Google Scholar]
- Abkarian M. and Viallat A., Biophys. J. 89, 1055–1066 (2005). 10.1529/biophysj.104.056036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abkarian M., Lartigue C., and Viallat A., Phys. Rev. Lett. 88, 068103 (2002). 10.1103/PhysRevLett.88.068103 [DOI] [PubMed] [Google Scholar]
- Jones T. B. and Kallio G. A., J. Electrostat. 6, 18 (1979). 10.1016/S0005-2760(97)00092-1 [DOI] [Google Scholar]
- Chan K. L., Gascoyne P. R., Becker F. F., and Pethig R., Biochim. Biophys. Acta 1349, 182–196 (1997); available at http://www.sciencedirect.com/science/article/pii/S0005276097000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethig R., Biomicrofluidics 4, 022811 (2010). 10.1063/1.3456626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek P., Zielinsky J. J., Fikus M., and Tsong T. Y., Biophys. J. 59, 982–987 (1991). 10.1016/S0006-3495(91)82312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wang X. B., Becker F. F., and Gascoyne P. R., Biochim. Biophys. Acta 1282, 76–84 (1996); available at http://www.sciencedirect.com/science/article/pii/0005273696000478. 10.1016/0005-2736(96)00047-8 [DOI] [PubMed] [Google Scholar]
- Gascoyne P. R. C., Shim S., Noshari J., Becker F. F., Huang H., Pethig R., Vykoukal J., Gascoyne P. R. C., and Stemke-Hale K., “Correlations between the dielectric properties and exterior morphology of cells revealed by dielectrophoretic field-flow fractionation,” Electrophoresis (in press). [DOI] [PMC free article] [PubMed]
- Vykoukal D. M., Gascoyne P. R. C., and Vykoukal J., Integr. Biol. 1, 477–484 (2009). 10.1039/b906137a [DOI] [PubMed] [Google Scholar]
- Wang X. B., Huang Y., Gascoyne P. R., Becker F. F., Holzel R., and Pethig R., Biochim. Biophys. Acta 1193, 330–344 (1994). 10.1016/0005-2736(94)90170-8 [DOI] [PubMed] [Google Scholar]
- Cristofanilli M., De Gasperis G., Zhang L., Hung M.-C., Gascoyne P. R. C., and Hortobagyi G. N., Clin. Cancer Res. 8, 615–619 (2002). [PubMed] [Google Scholar]
- Huang Y., Wang X. B., Gascoyne P. R., and Becker F. F., Biochim. Biophys. Acta 1417, 51–62 (1999). 10.1016/S0005-2736(98)00253-3 [DOI] [PubMed] [Google Scholar]
- Kim U., Shu C.-W., Dane K. Y., Daugherty P. S., Wang J. Y. J., and Soh H. T., Proc. Natl. Acad. Sci. USA 104, 20708–20712 (2007). 10.1073/pnas.0708760104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Becker F. F., and Gascoyne P. R. C., Biochim. Biophys. Acta 1564, 412–420 (2002). 10.1016/S0005-2736(02)00495-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui-ock S., Ruchirawat M., and Gascoyne P., Anal. Chem. 80, 7727–7734 (2008). 10.1021/ac801095p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethig R. and Kell D. B., Phys. Med. Biol. 32, 933–970 (1987). 10.1088/0031-9155/32/8/001 [DOI] [PubMed] [Google Scholar]
- Tong X. and Caldwell K. D., J. Chromatogr. B: Biomed. Appl. 674, 39–47 (1995). 10.1016/0378-4347(95)00297-0 [DOI] [PubMed] [Google Scholar]
- Chianea T., Assidjo N. E., and Cardot P. J., Talanta 51, 835–847 (2000). 10.1016/S0039-9140(99)00335-5 [DOI] [PubMed] [Google Scholar]
- Rosenberg R., Gertler R., Friederichs J., Fuehrer K., Dahm M., Phelps R., Thorban S., Nekarda H., and Siewert J. R., Cytometry 49, 150–158 (2002). 10.1002/cyto.10161 [DOI] [PubMed] [Google Scholar]
- Becker F. F., Huang H., Pethig R., Vykoukal J., Gascoyne P.R.C., J. Phys. D: Appl. Phys. 27, 2659–2662 (1994). 10.1088/0022-3727/27/12/030 [DOI] [Google Scholar]